Abstract
Meiotic recombination plays a central role in the evolution of sexually reproducing organisms. The two recombination outcomes, crossover (CO) and noncrossover (NCO), increase genetic diversity, but have the potential to homogenize alleles by gene conversion. While CO rates are known to vary considerably across the genome, NCOs and gene conversions have only been identified in a handful of loci. To examine recombination genome-wide and at high spatial resolution, we generated maps of COs, CO-associated gene conversion and NCO gene conversion using dense genetic marker data collected from all four products of 56 yeast meioses. Our maps reveal differences in the distributions of COs and NCOs, showing more regions where either COs or NCOs are favoured than expected by chance. Furthermore, we detect evidence for interference between COs and NCOs, a phenomenon previously only known to occur between COs. Up to 1% of the genome of each meiotic product is subject to gene conversion in a single meiosis, with detectable bias towards GC nucleotides. The maps represent the first high-resolution, genome-wide characterization of the multiple outcomes of recombination in any organism. In addition, because NCO hot spots create holes of reduced linkage within haplotype blocks, our results stress the need to incorporate NCOs into genetic linkage analysis.
In most eukaryotes, homologous chromosomes exchange genetic information through recombination during meiosis. This process increases genetic diversity by breaking haplotypes, but it may also homogenize alleles through gene conversion1,2. Furthermore, recombination is fundamental to sexual reproduction because it provides physical connections between homologs during the first meiotic division, contributing to correct chromosome segregation3. In the current model, meiotic recombination starts with the formation of a double-strand break (DSB)4,5. The break is then repaired through a series of steps, involving resection, synthesis and ligation, using the homologous chromosome as a template. Repair results in either a crossover (CO) — reciprocal exchange accompanied by a tract subject to gene conversion — or a noncrossover (NCO) — a tract subject to conversion but not associated with reciprocal exchange4,6. At least two pathways form COs: the Msh4/Msh5-dependent pathway, which proceeds through a double Holliday junction, and the Mus81/Mms4-dependent pathway7,8. In contrast, NCOs are thought to be the result of synthesis-dependent strand annealing (SDSA)9. It is known that DSB10–14 and CO rates15 vary along chromosomes. NCOs and CO-associated gene conversions have not been characterized genome-wide, however, because this requires monitoring recombination between closely spaced markers along the genomes of all four meiotic products2.
High-resolution mapping of recombination
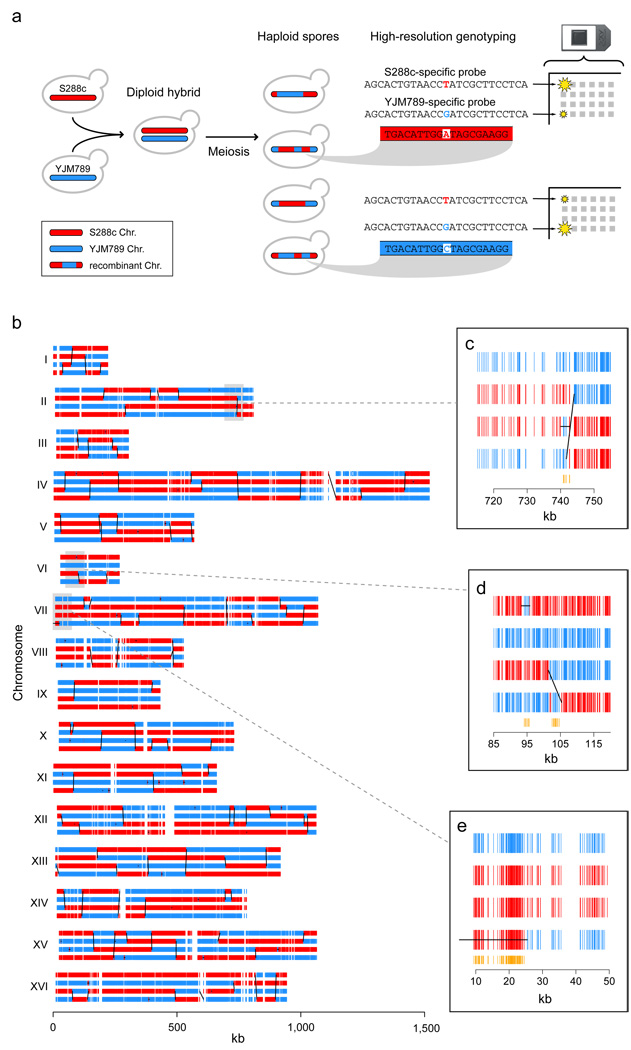
In Saccharomyces cerevisiae, we achieved a detailed characterization of recombination outcomes by genotyping ∼52,000 markers in all four viable spores derived from 51 meioses of an S288c/YJM789 hybrid strain16,17 (Figure 1). Genomic DNA from parental strains and each of the 204 spores was hybridized to high-density microarrays that tile the genomes of both S288c and YJM789 with a median probe offset of 4 bp. To infer genotypes from the hybridization intensities of the probes covering each marker (8 probes per marker on average), we developed a new algorithm, ssGenotyping, based on semi-supervised clustering (see Methods). The high density of polymorphism and probes resulted in spore genotypes with a median distance of 78 bp between consecutive markers (Supplementary Figure 1). This resolution is over 20 times higher than in the current yeast genetic map15 and more than 360 times higher than in the most recent human CO map18.
Figure 1.
High-resolution mapping of meiotic recombination along the yeast genome. (a) Schematic description of recombination mapping approach. Meiotic products from a hybrid derived from highly polymorphic strains were individually genotyped using microarrays. (b) Genotype calls at ∼52,000 markers for all four spores resulting from a single meiosis. Diagonal/vertical black lines are inferred COs, and horizontal lines, NCOs. Close-ups of (c) a CO overlapped by an independent NCO in a third spore; (d) a CO with complex gene conversion tract, and a nearby, independent NCO; and (e) a long NCO at the end of the chromosome. (See Methods for full annotation procedure.) In close-ups, orange vertical segments denote markers with non-Mendelian ratios (1:3 or 0:4).
Due to their high resolution, our maps invert the traditional relationship between markers and recombination events: there are multiple markers within each recombination event rather than vice versa. This allows characterization of both CO-associated and NCO gene conversion tracts, which are typically thought to be only 1–2 kb long2. Genotype calls from all four spores in each wildtype tetrad were used to infer a total of 4,163 COs and 2,126 NCOs (see Methods). We expect to have detected nearly all COs but, because NCOs have no effect on flanking markers, to have missed NCOs that completely fell between two markers, or NCOs in which mismatch repair restored the original genotype. We observed an average of 90.5 COs and 46.2 NCOs per meiosis. 30.1% of observed COs occurred between two consecutive markers, and therefore had no detectable conversion tract. Taking this percentage as an estimate of the fraction of unobserved NCOs, we obtained a corrected total (90.5 COs plus 66.1 NCOs) which is remarkably similar to a recent estimate of 140–170 DSBs per meiosis13.
All chromosomes but one had at least one CO, in agreement with the essential role that COs play in chromosome segregation3. The average number of COs was linearly related to chromosome length, with an intercept of 1.0, corresponding to one obligate crossover, plus an additional 6.1 COs per megabase (Supplementary Figure 5). Notably, NCOs behaved similarly (3.4 NCOs per Mb), but with a lower intercept (0.3).
The median size of conversion tracts was 2.0 kb for those associated with COs, and 1.8 kb for NCO conversion tracts (see Methods). The difference in medians is statistically significant (Wilcoxon rank-sum p < 0.0001). These sizes are consistent with previous estimates made at a single yeast hot spot19, but are considerably larger than single-locus estimates in human20. Our finding that CO tracts tend to be larger than NCO tracts also corroborates previous, single-locus observations in yeast and human20,21.
We observed 57 NCO conversion tracts larger than 5 kb in size, the largest being 40.8 kb (minimal length). Three of these were found at the end of chromosomes, suggesting that they could be the result of meiotic break-induced replication, as has been proposed for long NCO tracts at the HIS4 locus22 (Figure 1e). Three also showed complete loss of allelic variation across all four meiotic products (4:0 segregation), consistent with either mitotic or complex meiotic events.
We also observed that 11.5% of the conversion tracts accompanying COs exhibited complex patterns of genotype change (Figure 1d). 11.1% had more than one genotype change on just one of the involved chromatids, and 0.4%, on both chromatids. Such tracts are predicted to result from the resolution of a double Holliday junction due to multiple distinct patches of heteroduplex in a single CO event6, but they could also possibly result from mismatch repair alternating between conversion and restoration. 3.4% of single-chromatid NCO events were also detected to have complex conversion tracts.
Recombination hot spots
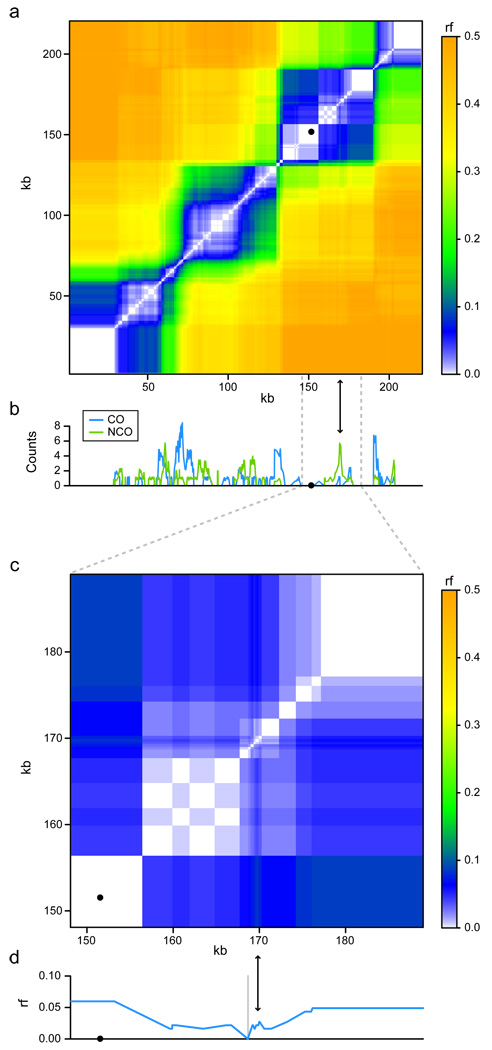
To estimate the local recombination rate along the genome, we counted the events overlapping each intermarker interval and adjusted for the size of the interval (see Methods, Figure 2b, Supplementary Figure 8, Supplementary Figure 9). This novel approach was necessary because recombination events typically overlapped multiple markers, making existing rate estimation methods designed for low-resolution data inappropriate. Recombination hot spots were defined as runs of contiguous intermarker intervals involved in more recombination events than expected under a homogeneous genomic rate (p < 0.001, see Methods and Supplementary Information). We identified hot spots for CO, NCO, and overall recombination activity separately. At the hottest of the 179 resulting overall recombination hot spots, 27.7% of spores showed observable evidence of involvement in a CO or NCO event (58.7% of meioses). At the hottest CO and NCO hot spots, 21.7% and 8.7% of spores showed observable evidence of a CO or a NCO, respectively. This corresponds to 21.7% and 17.4% of spores being involved in a CO or NCO event, because a single CO produces two spores with observable evidence while a NCO, only one. Given that some NCOs may have been missed, we therefore observed similar rates for both outcomes at their hottest locations in the genome.
Figure 2.
CO and NCO rates along chromosome I and their effect on recombination fraction (rf). (a) rf was calculated for every pair of markers as the portion of segregants in which the markers have opposite genotype. Black dots denote centromeres. (b) CO (blue) and NCO (green) counts, adjusted for varying intermarker interval size (see Methods). (c) close-up of a NCO-biased region (indicated by arrows) on chromosome I. (d) rf relative to a single reference marker (grey vertical line) upstream of the NCO-biased region, showing the non-monotonic relationship between genetic and physical maps caused by such a region.
It is known that most DSBs occur in promoter regions10,11, and indeed, 84% of hot spots overlap a promoter. Nonetheless, hot spot intervals primarily overlap coding sequence: only 25% of the bases in hot spot intervals overlap promoters, while 68% overlap coding sequences.
Centromere-proximal regions showed low recombination rates, and no recombination event overlapped a centromere on any chromosome (Supplementary Figure 8, Supplementary Figure 9; Supplementary Table 1). However, many chromosomes did have at least one event less than 4 kb away, including a CO only 341 bp from CEN5 (Supplementary Table 1). Telomeres could not be directly interrogated due to repetitive sequence. We did, however, observe some chromosomes with a complete lack of recombination activity well before the telomeres; others showed strong activity near a telomere (Supplementary Figure 8, Supplementary Figure 9).
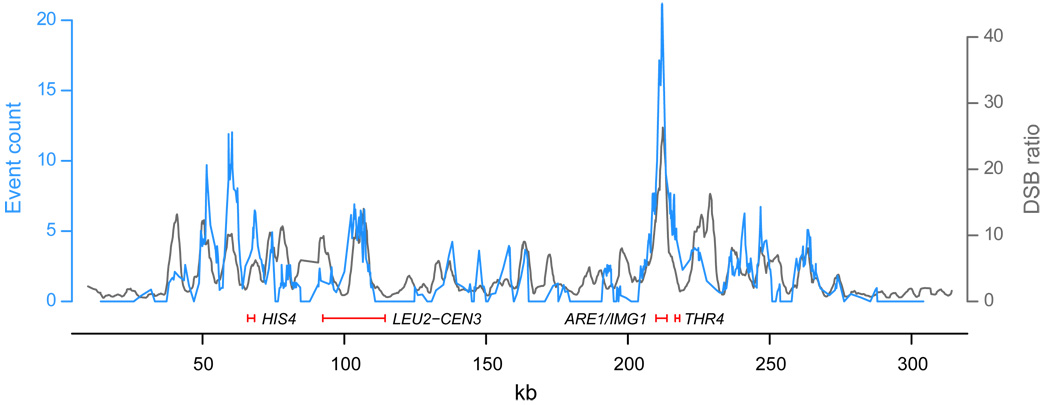
Validating our approach, all previously known yeast recombination hot spots except for HIS2 are within or adjacent to one of our hot spots (HIS4, ARG4, CYS3, DED81, ARE1/IMG1, CDC19, THR4, LEU2-CEN3)23. Furthermore, despite differences in strain background and the numerous heterozygosities in our hybrid strain, our recombination rates are in close agreement with a recently generated genome-wide DSB rate map in a homozygous SK1 strain13 (Figure 3). In addition to showing correspondence between the initiation of recombination and its resolution, this agreement suggests that the distribution of meiotic recombination is largely persistent within a species. Some fine-scale differences, however, do exist, possibly reflecting within-species variation in recombination rate18.
Figure 3.
Comparison of DSB and recombination rates along chromosome III. DSB smoothed fluorescence ratios in a SK1 strain (dmc1Δ, grey)13 are compared with our recombination event counts (blue), after adjusting the latter for varying intermarker interval size (see Methods). Peak locations largely agree despite distinct strain backgrounds, although some fine-scale differences exist. Previously known hot spots are indicated by red segments.
Distinct CO and NCO distributions
It is expected that the distribution of meiotic recombination is determined by the location of initiating DSBs as well as by how the DSBs are repaired4. It has not been clear, however, whether COs and NCOs always occur in similar proportions or whether there are CO- or NCO-specific hot spots. While a recent study reported mild CO/NCO differences for two hot spots24, our maps allow investigation of such differences genome-wide (Figure 2b). Using an approach which accounts for unobserved NCOs which fall completely between two markers, we identified regions with biased CO/NCO ratios, and found more intermarker intervals with extreme ratios than expected by chance (p < 0.0005, see Methods). We observed an average excess of ∼60 intervals favoring COs, and ∼170 intervals favoring NCOs, spanning ∼100 kb of genomic sequence in total (see Methods). Strikingly, we estimated that such differences affect at least 1.4% of the genomic regions exhibiting one or more recombination events. The CO/NCO event ratios at the regions showing the strongest evidence of bias, after accounting for the effect of marker spacing, were 14:0 and 0:7. Our findings therefore suggest that a significant fraction of the genome exhibits differences in CO/NCO ratio.
The observed dissimilarity in CO/NCO distribution has implications for linkage analysis. In contrast to CO hot spots, regions with high NCO frequency can be expected to have reduced linkage to their surroundings, but to maintain linkage between loci to either side. By estimating the recombination fraction between all pairs of markers on each chromosome, we show that CO hot spots are associated with linkage block boundaries, while NCO-biased regions correspond to regions with reduced linkage within blocks (Figure 2). NCO-biased regions result in a nonmonotonic relationship between the genetic and physical distance, and create holes within linkage blocks. Over generations, NCO-biased hot spots would form genomic regions with low linkage disequilibrium (LD) relative to their surroundings, and thus be difficult to track with markers outside the NCO-biased region25,26.
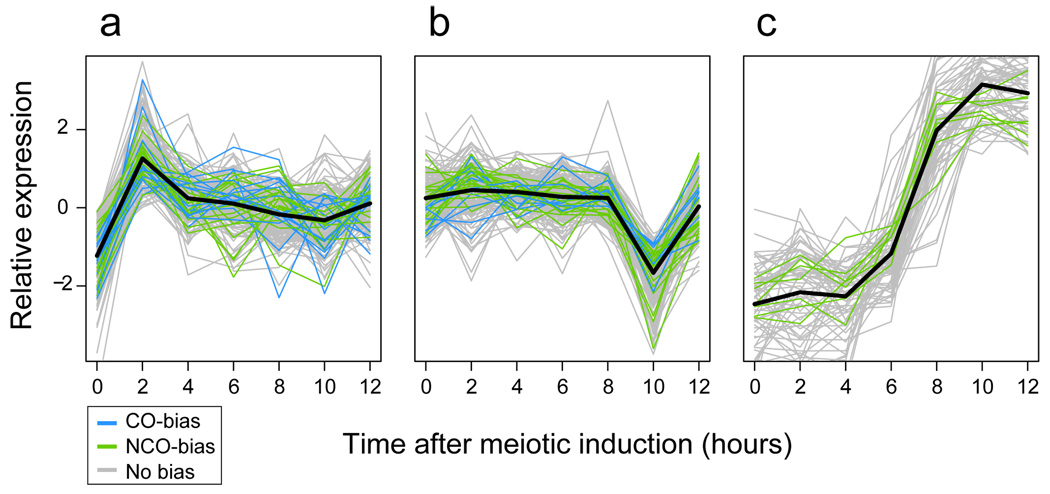
The existence of regions with a CO/NCO bias suggests that the bifurcation between the two outcomes could, in fact, be a controlled process, influenced by local chromosomal properties. Recombination hot spots were found to contain short poly(A) stretches (20–41 bp) more frequently than expected, and to be significantly associated with several gene ontology (GO) terms (see Supplementary Information). Nonetheless, we found no sequence motifs to be specifically associated with CO- or NCO-biased regions, and only one GO term (“cell aging”) to exhibit a significant association with such regions. A comparison of our results with measurement of transcriptional activity during meiosis in W303 and SK1 strains27 showed that hot-spot-proximal genes were significantly enriched in two specific expression profiles: a transcription peak around two hours after meiotic induction (p < 0.0001, see Supplementary Information, Figure 4a), and a transcription decrease between 8 and 10 hours (p = 0.0046, Figure 4b). In addition, a cluster with genes upregulated four hours after meiotic induction contained genes from NCO-biased regions, but no genes from CO-biased regions (Fisher exact test p = 0.015, Figure 4c). This relationship between specific transcriptional behavior and proximity to recombination hot spots supports a role for chromatin accessibility and transcription factor binding in meiotic recombination28.
Figure 4.
Association between gene expression and recombination activity. After centering by their mean over time points, genes were clustered (see Supplementary Information) by their W303 expression profile during meiosis27. (a, b) Two clusters enriched for genes overlapped by overall recombination hot spots. Genes overlapped by regions of CO or NCO bias, however, are present in similar proportion in these clusters. (d) Gene cluster containing genes overlapped by regions with NCO bias, but no genes overlapped by regions with CO bias.
COs and NCOs in recombination mutants
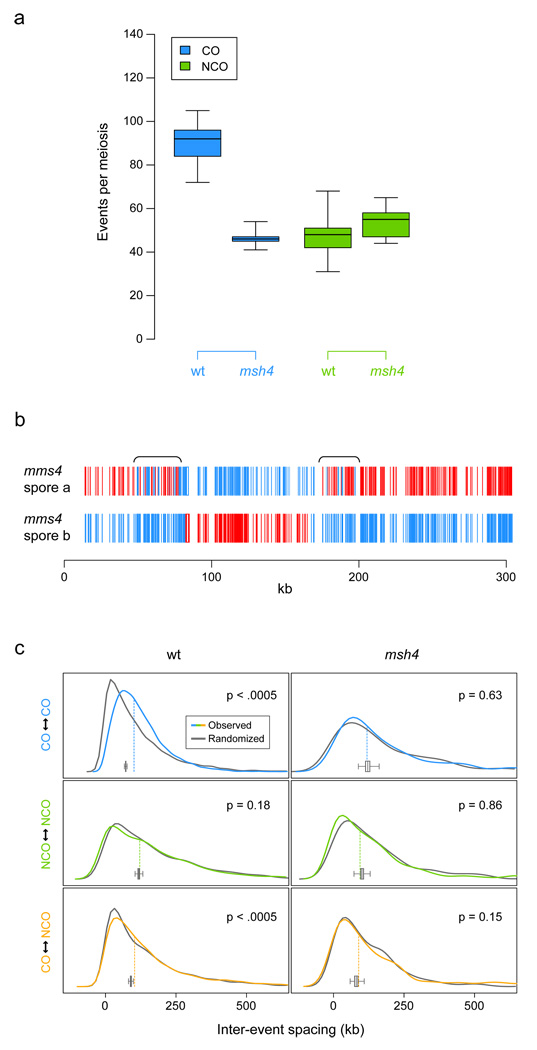
To further assess differences between the generation of COs and NCOs, we mapped recombination events in msh4 and mms4 null mutants, in which either the Msh4/Msh5-dependent or the Mus81/Mms4-dependent CO pathway is disturbed7. Five full tetrads of the msh4 mutant were genotyped. Given the role of MSH4 in CO generation, its deletion is expected to reduce the number of COs but maintain the number of NCOs29. Consistent with this expectation, we observed that the NCO frequency showed no statistically significant change (t-test p = 0.12), while the average number of COs per meiosis was drastically reduced from 90.5 in the wildtype to 46.6 in msh4 (t-test p < 0.0001, Figure 5a). Furthermore, in contrast to the wildtype, all msh4 tetrads except one had one or more chromosomes with no COs at all (6.3% of all chromosomes). Unexpectedly, the median size of msh4 CO conversion tracts was 479 bp larger than for wildtype (Wilcoxon rank-sum p = 0.0003). The median size of msh4 NCO recombination tracts, however, was 338 bp shorter than for wildtype (Wilcoxon rank-sum p = 0.0008). Therefore, deletion of MSH4 reduced genome-wide frequency of COs, as expected given its role in the Msh4/Msh5-dependent pathway, but affected tract size of both COs and NCOs (Supplementary Figure 6).
Figure 5.
Meiotic recombination in msh4 and mms4 strains. (a) Genomic frequencies of COs and NCOs, per meiosis (wildtype n = 46, msh4 n = 5; boxplots show minimum, first quantile, median, third quantile, and maximum). (b) Genotype calls (S288c/YJM789, red/blue) along chromosome III in an mms4 dyad showing large regions of frequent genotype change (calls in brackets validated by sequencing). (c) Density estimates for the distance between adjacent recombination events, to measure interference. Colored lines show real data distribution, while grey lines denote one corresponding tetrad-randomized distribution. Dashed vertical lines show real data median, and the boxplot, the distribution of medians for 2000 randomizations. P-values for difference in median distance are given within each panel (see Supplementary Information).
The observation that, in the msh4 mutant, the frequency of one event type was altered with respect to wildtype while the other was not has two important implications. First, since Msh4 is thought to function downstream of DSB formation30, we expect the msh4 null mutant to have the same number of DSBs as the wildtype. (This is known to be the case for MSH5, the functional partner of MSH431.). Our data therefore suggest that a fraction of DSBs are not resolved towards COs or NCOs, but may instead be repaired by alternative mechanisms such as sister chromatid exchange32 or non-homologous end joining4. Second, we have perturbed a DSB-resolution pathway and seen strong but distinct effects on the global CO/NCO balance. If this pathway has regional preferences, this may contribute to observed CO/NCO bias.
The mms4 mutant exhibited low sporulation efficiency and spore viability, which impeded recovery of complete tetrads, so we only genotyped 6 dyads (12 spores) and 8 single mms4 spores. Surprisingly, the mms4 spores showed several regions (∼7 per spore) exhibiting unusually frequent genotype changes (Figure 5b) — up to ∼70 kb in size and typically associated with apparent COs. For example, one such 63 kb region contained a total of 31 genotype changes. The mechanism responsible for these genotype changes is not known, but their presence may help elucidate the way in which the Mus81/Mms4 nuclease complex generates COs8. We chose not to pursue recombination event inference for the mms4 spores, due to both the presence of such regions and the inherent difficulty in distinguishing between single NCOs and pairs of nearby COs in a single-spore context.
CO and NCO interference
Interference, where a recombination event reduces the probability that an additional event occurs nearby33, is an important determinant of the distribution of meiotic recombination, and could also contribute to differences in CO/NCO rates. So far, interference has been reported only between COs34. To assess interference, we considered the distances between adjacent, same-tetrad recombination events. These distances were compared with those in tetrad-randomized data sets (see Supplementary Information). Tetrad randomization preserves hot and cold spot structure along the genome, but removes interference effects. The distance between consecutive COs was larger in wildtype meioses than expected by chance: a median inter-CO distance of 101.1 kb in observed data versus 71.8 kb under tetrad-randomization (p < 0.0005, see Supplementary Information, Figure 5c). No such effect was seen for NCOs. Surprisingly, and in contrast to previous reports34, COs and NCOs also exhibited interference: the median observed distance from a CO to the nearest NCO was 4.4 kb larger in real data than under tetrad-randomization (p < 0.0005). In the msh4 null mutant, COs did not show interference (p = 0.63). This is consistent with the hypothesis that only COs generated by the Msh4/Msh5-dependent pathway exhibit interference7. Furthermore, in the msh4 mutant, evidence of interference between COs and NCOs disappears as well (p = 0.15). These results support the existence of at least two types of COs with differences in interference, and yield genome-wide evidence for interference between COs, and among COs and NCOs.
We also observed an overrepresentation of overlapping events within the same meiosis in the wildtype strain, which is surprising given the observed patterns of interference. For example, 2.6% of CO conversion tracts had an overlapping NCO partner on a third spore, and an additional 0.6% had an overlapping CO partner involving the other two spores (Figure 1c, Supplementary Figure 12). Such overlapping events could result from paired DSBs in two different chromatids; but, they could also be the consequence of a single DSB whose resolution involves multiple rounds of strand invasion and extension from different templates35. We also observed 110 pairs of partially or exactly overlapping NCOs with reciprocal genotypes. The existence of such pairs is relevant to current models for NCO formation (see Supplementary Information Section 8 for discussion).
Genomic effect of gene conversion
Having observed differences in CO and NCO distributions as well as interference between events, we next considered the effects of gene conversion tracts. We determined the portion of the yeast genome that is involved in CO-associated and NCO gene conversion. 2.1% of the polymorphic positions were converted to the opposite genotype per meiosis. Furthermore, across the genomes of all four wildtype meiotic products, CO tracts covered between 92 kb and 320 kb per meiosis (minimal and maximal), and the NCO tracts, between 62 kb and 148 kb. Therefore, as much as 1% of a meiotic product’s genome may be subject to conversion in a single meiosis.
Genomic regions active in gene conversion are susceptible to the effect of gene conversion on allelic frequency, and also to mutation-prone processes36. We therefore analyzed GC content and single-nucleotide polymorphism (SNP) density in converted regions and hot spots. For both CO-associated and NCO gene conversions, we detected mismatch repair bias favoring GC nucleotides (Supplementary Information). Relative to the base content at SNP positions in the parental genomes, we observed a 1.4% GC increase in the converted sequences of the spores (χ2 p = 0.0001, Supplementary Table 2). This bias could contribute to the association between recombination hot spots and GC-richness that we observed (χ2 p < 0.0001) — an association that has also been found for DSBs11. While on an evolutionary timescale, GC bias could potentially homogenize alleles, comparison to low-depth genome sequences of 37 S. cerevisiae strains showed that our hot spots were actually associated with greater genetic diversity (see Supplementary Information). Therefore, GC conversion bias may be counteracted by other processes, such as those that increase AT content37,38. In summary, we find no evidence of allelic homogenization at recombination hot spots, despite the presence of GC bias during mismatch repair.
Conclusion
The recombination maps presented here constitute the first survey of NCOs and both CO-associated and NCO gene conversion across an entire genome in any organism. In addition to permitting detection and characterization of gene conversion, the high resolution of our approach reveals phenomena which would otherwise be difficult to observe, such as complex conversion tracts and large regions of frequent genotype changes (Figure 1d and Figure 5b). The data uncover regions of interest for further investigation, and the approach is applicable to other mutants and conditions. It could thus contribute to answering questions about the mechanisms of interference and CO homeostasis24,39, or possible alternative DSB-resolution pathways4–6.
While the degree of polymorphism between the parental strains results in unprecedented marker resolution, polymorphisms may also affect recombination propensity40,41. Nonetheless, several observations suggest that recombination is not drastically perturbed in our hybrid: the agreement between our maps and the DSB map from a homozygous SK1 strain13; consistency between our overall number of COs and the number generated from genetic-map estimates42; and the detection of previously known recombination hot spots23. Furthermore, outside laboratory conditions, most sexually reproducing organisms are heterozygous. Individuals in natural populations may, therefore, resemble our hybrid more than they do a homozygous strain.
Our maps show the existence of locations with distinct preferences for either COs or NCOs, suggesting a role for genomic position in determining DSB resolution outcome. Given that chromatin conformation is known to be important for recombination generally28, it is plausible that local chromosomal properties could influence the CO/NCO bifurcation. Such properties may not, however, be the sole determinants of CO/NCO bias. Through interference, both CO-CO and CO-NCO, the decision could also depend on recombination activity in nearby regions.
Our maps also stress the relevance of NCOs, and gene conversion generally, in genetic analysis. CO is the major determinant of linkage disequilibrium, but both CO-associated and NCO gene conversion weaken LD between nearby loci. Models which incorporate gene conversion will therefore be able to more accurately relate LD and physical distance. Further, CO-associated and NCO conversion tracts have different effects on the fine structure of haplotypes26. As shown in Figure 2, gene conversion at CO hot spots softens the boundaries of linkage blocks, while NCO-biased regions create holes within blocks. Both phenomena have implications for genetic association analyses. While these regions are highly localized and impact only a fraction of meioses, their effect can accumulate over generations, hiding genetic variants with phenotypic relevance (e.g., disease genes). Having a higher density of markers in regions with frequent gene conversion may thus help to uncover genetic factors contributing to phenotypic variation.
METHODS SUMMARY
A S96/YJM789 hybrid strain was sporulated43, and genomic DNA — from 51 wildtype and 5 msh4 tetrads as well as from 20 mms4, 13 S96 parental, and 12 YJM789 parental spores — was extracted from single-colony cultures and hybridized to a custom-designed tiling microarray44. (S96 is isogenic to S288c16,17.) Normalized45 fluorescence intensities corresponding to the set of probes covering each polymorphism were analyzed by applying multivariate semi-supervised clustering to the combined parental and segregant data. Segregant genotypes were assigned using posterior probability of class membership. To reduce genotyping errors, we applied filters to whole arrays, to probe sets and to individual genotype calls. DNA sequencing of ∼60 kb confirmed 100% of filtered genotype calls. After grouping data by tetrad, pairs of genotype change points isolated from all other changes were called NCOs if they involved one spore, or COs if they involved two. Complex groups of genotype changes were annotated as described in Supplementary Figure 3. To calculate event rate along the genome, it was necessary to adjust for varying intermarker interval size. Because individual recombination events typically overlapped multiple intermarker intervals, a novel adjustment procedure was used (Supplementary Information). We defined three types of hot spots — CO, NCO, and overall recombination events — by identifying runs of contiguous intermarker intervals involved in more recombination events than expected under a homogeneous genomic rate. To assess CO/NCO bias, we compared the number and size of intermarker intervals exhibiting more/fewer COs than expected to the corresponding null distribution, generated via simulation. We tested for interference — between consecutive events of the same type and also between COs and NCOs — by comparing the median distance between adjacent, same-tetrad events to medians computed after tetrad label randomization. This randomization strategy preserved hot and cold spot structure but removed interference.
Methods
Strains and media
The hybrid strain S288c/YJM789 was generated by crossing S96, isogenic to S288c17, with YJM78916. To generate the homozygous msh4Δ and mms4Δ hybrid strains, the corresponding gene was replaced by a natMX4 or kanMX4 drug-resistant marker48 in each of the haploid parental strains, which were then crossed. Sporulation was induced by transferring overnight cultures from liquid YEPD to 2% potassium acetate43.
DNA extraction and hybridization
51 complete wildtype and 5 complete msh4 tetrads were dissected for genotyping. 20 mms4 viable spores were also selected, as were 13 S96 and 12 YJM789 parentals. Spores were allowed to grow in YEPD solid medium and then streaked out to obtain single colonies, only one of which was used for genotyping. Note that starting from a single colony prevented analysis of heterozygosities within a single spore arising from post-meiotic segregation. Genomic DNA was extracted from an overnight, 100 ml, YEPD, saturated culture of each spore using a QIAGEN Genomic-tip according to manufacturer’s protocol. 10µg of genomic DNA were fragmented, biotin-labeled and hybridized to a custom Affymetrix microarray, as described previously44. All probes were remapped (exonerate49) to the S288c genome and the aligned portion of the YJM789 genome50. Only probes with one exact match (25 matching bases) and no near matches (22 to 24) were retained, yielding 287,000 S288c–specific probes, 112,000 YJM789-specific probes, and 2.37 M probes interrogating non-polymorphic sequence.
Genotyping
Fluorescence intensities were normalized with vsn45. SNPs, insertions, and deletions were identified using the S288c/YJM789 alignment50, and for each polymorphism, a probe set was formed from probes interrogating the position(s) involved. Nearby polymorphisms producing identical probe sets were treated as a single marker. Genotype labels were available for parental data, so to genotype segregants, semi-supervised clustering was applied to the combined parental and segregant data. For each probe set, a two-component Gaussian mixture model — with fixed mixture proportions (0.5) but distinct covariance matrices — was fit using the EM algorithm. For the small fraction of probe sets with >10 probes (probe sets interrogating large indels), principal components dimension reduction (d = 10) was applied first. Segregant genotypes were assigned using posterior probability of class membership. For mms4, genotypes were assigned in a supervised fashion, using the distributions previously estimated from the wildtype and msh4 data.
Filtering of genotype calls
We deliberately opted for a high no-call rate with fewer errors, to reduce the chance of spurious short NCOs. Five wildtype and two mms4 arrays exhibiting excessive genotype switching and large Mahalanobis residuals were set aside. A small fraction (0.7%) of probe sets exhibiting >2 classes — likely due to cross-hybridization with unlinked loci — were discarded (Supplementary Figure 2c). Misclassification rates were estimated using inferred mixture distributions, and probe sets (4.6%) for which this estimate exceeded 1% were also discarded (Supplementary Figure 2b). For retained probe sets, individual calls (4.9%) were discarded if the posterior probability of assigned class membership was too far from 1, or if the Mahalanobis residual was large (Supplementary Figure 2a). In two sequencing validation data sets covering ∼60 kb — one focused on calls from 16 different wildtype spores, and another, on two regions of an mms4 segregant exhibiting frequent genotype switching — 100% of filtered genotype calls were confirmed.
Recombination event annotation
After collecting genotype data into tetrads, genotype change points were grouped by proximity. Most cases were simple: pairs of changes isolated from all other changes were called NCOs if they involved one spore, or COs if they involved two (Supplementary Figure 4a). A fraction of cases, however, were more complex, admitting several distinct interpretations. To systematically treat such cases, cutoff-based rules reflecting basic assumptions about the recombination process were used. See Supplementary Information Section 1 for details. Importantly, we explored a variety of plausible alternative annotation sets, and found no qualitative change in our main results.
Conversion tract length
Tract size estimates obtained using midpoints of flanking intermarker intervals were used for most calculations (see Supplementary Information Section 3). Where indicated, we also computed lower and upper bounds, using the regions spanned by converted markers (minimal), and delimited by the two nearest unconverted markers (maximal)2. For summary statistics, we combined simple (Supplementary Figure 4a) and complex (Supplementary Figure 4b, c) conversion tracts.
Event rate adjustment
Intermarker interval size affects the probability of involvement in recombination events. To adjust for this, we used a semi-parametric statistical model (Supplementary Information) to relate size to the probabilities of (i) involvement in and (ii) detection of recombination events. The model’s extension length distribution was estimated empirically. Given this estimate, we then counted recombination events overlapping each intermarker interval, and estimated remaining parameters by Poisson regression.
Defining hot spots
Using model parameter estimates, expected CO and NCO counts were computed for each intermarker interval under a null hypothesis of rate homogeneity. We identified three types of hot spots: CO, NCO, and overall recombination events. To identify CO hot spots, we performed a one-tailed test (α = 0.001) using the Poisson distribution and the expected CO counts. Hot intermarker intervals separated by <500 bp were merged. NCO and overall recombination hot spots were identified similarly. Note that the three types of hot spots are statistically related, but CO and NCO hot spot counts need not sum to the overall count.
CO/NCO bias testing
To assess CO/NCO bias, we used expected CO and NCO counts to compute expected CO fractions. Conditioning on the observed number of events overlapping each interval, we then compared observed and expected counts using two one-tailed binomial distribution tests. The resulting p-values correspond to either an excess or deficiency of COs. Despite the large sample size, individual intermarker intervals were rarely involved in >10 events, so we chose to treat CO/NCO bias p-values collectively rather than individually. We simulated data (B = 2000) under the same binomial distributions used for p-value calculations — conditioning on observed counts so that rate inhomogeneity across the genome was preserved — and examined (i) the average number of simulated p-values falling below 0.10, and (ii) the average total size of intermarker intervals associated with such p-values. The former permitted estimation of false discovery rate, and the latter, estimation of the total size of intermarker intervals associated with true CO/NCO bias.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Clauder-Münster, M. Granovskaia, M. Sieber, T. Bähr-Ivacevic, M. Nguyen, V. Benes, Z. Xu, L. Ettwiller, P. McGettigan and the EMBL Genomics Core Facility for technical help; M. Knop for discussions; A. Akhtar, A. Ladurner, A. De Luna and M. Knop for critical comments on the manuscript; E. Louis, R. Durbin and D. Carter for making data from the Saccharomyces Genome Resequencing Project available; and the contributors to the Bioconductor46 and R47 projects for making their software available. This work was supported by grants to L.M.S. from the National Institutes of Health and the Deutsche Forschungsgemeinschaft, and to W.H. from the Human Frontier Science Program; and by a Darwin Trust’s Jeff Shell Scholarship awarded to E.M.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
REFERENCES
- 1.Gordo I, Charlesworth B. Genetic linkage and molecular evolution. Curr Biol. 2001;11(17):R684. doi: 10.1016/s0960-9822(01)00408-0. [DOI] [PubMed] [Google Scholar]
- 2.Chen JM, et al. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8(10):762. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 3.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301(5634):785. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 4.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15(5):565. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 5.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117(1):9. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 6.Whitby MC. Making crossovers during meiosis. Biochem Soc Trans. 2005;33(Pt 6):1451. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 7.Argueso JL, Wanat J, Gemici Z, Alani E. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics. 2004;168(4):1805. doi: 10.1534/genetics.104.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18(2):117. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106(1):47. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 10.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci U S A. 1997;94(10):5213. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerton JL, et al. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97(21):11383. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borde V, et al. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13(3):389. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 13.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5(12):e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blitzblau HG, et al. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol. 2007;17(23):2003. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 15.Cherry JM, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387(6632 Suppl):67. [PMC free article] [PubMed] [Google Scholar]
- 16.McCusker JH, Clemons KV, Stevens DA, Davis RW. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics. 1994;136(4):1261. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortimer RK, Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113(1):35. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coop G, et al. High-Resolution Mapping of Crossovers Reveals Extensive Variation in Fine-Scale Recombination Patterns Among Humans. Science. 2008 doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 19.Borts RH, Haber JE. Length and distribution of meiotic gene conversion tracts and crossovers in Saccharomyces cerevisiae. Genetics. 1989;123(1):69. doi: 10.1093/genetics/123.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36(2):151. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 21.Terasawa M, et al. Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation. Proc Natl Acad Sci U S A. 2007;104(14):5965. doi: 10.1073/pnas.0611490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merker JD, Dominska M, Petes TD. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics. 2003;165(1):47. doi: 10.1093/genetics/165.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichten M, Goldman AS. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:423. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 24.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126(2):285. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ardlie K, et al. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am J Hum Genet. 2001;69(3):582. doi: 10.1086/323251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall JD. Close look at gene conversion hot spots. Nat Genet. 2004;36(2):114. doi: 10.1038/ng0204-114. [DOI] [PubMed] [Google Scholar]
- 27.Primig M, et al. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26(4):415. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 28.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2(5):360. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 29.Ross-Macdonald P, Roeder GS. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79(6):1069. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 30.Kunz C, Schar P. Meiotic recombination: sealing the partnership at the junction. Curr Biol. 2004;14(22):R962. doi: 10.1016/j.cub.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 31.Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117(1):29. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 32.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90(6):1123. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 33.Hillers KJ. Crossover interference. Curr Biol. 2004;14(24):R1036. doi: 10.1016/j.cub.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 34.Malkova A, et al. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics. 2004;168(1):49. doi: 10.1534/genetics.104.027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh SD, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130(2):259. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurles M. How homologous recombination generates a mutable genome. Hum Genomics. 2005;2(3):179. doi: 10.1186/1479-7364-2-3-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19(7):1181. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 38.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 39.Kleckner N, et al. A mechanical basis for chromosome function. Proc Natl Acad Sci U S A. 2004;101(34):12592. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borts RH, Haber JE. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987;237(4821):1459. doi: 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151(4):1299. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77(7):977. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 43.Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9(21):2684. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- 44.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103(14):5320. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber W, et al. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18 Suppl 1:S96. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 46.Gentleman Robert C, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Team R. Austria: Vienna; 2007. Development Core, R: A Language and Environment for Statistical Computing ({R Foundation for Statistical Computing} [Google Scholar]
METHODS REFERENCES
- 48.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15(14):1541. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 49.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei W, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci U S A. 2007;104(31):12825. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.