Abstract
Chemicals and radiation can damage DNA leading to the formation of adducts/lesions, which – if not removed by DNA repair pathways – usually block replicative DNA polymerases (DNAPs). To overcome such potentially lethal blockage, cells have lesion bypass DNAPs, which are often in the Y-Family and include several classes. One class includes human DNAP κ and E. coli DNAP IV, and they insert dCTP in the non-mutagenic pathway opposite [+ta]-B[a]P-N2-dG, which is the major adduct formed by the environmental carcinogen benzo[a]pyrene. Another class includes hDNAP η and ecDNAP V, and they insert dATP opposite [+ta]-B[a]P-N2-dG in the dominant G → T mutagenic pathway. Herein we develop a hypothesis for why the IV/κ-class preferentially does cellular dCTP insertion. On the minor groove side of the active site, Y-Family DNAPs have a cleft/hole that can be analyzed based on an analogy to a “chimney.” Our models of DNAP IV show a large chimney opening from which the pyrene of [+ta]-B[a]P-N2-dG can protrude, which allows canonical adduct-dG:dCTP pairing. In contrast, our models of DNAP V have small chimney openings that forces adduct-dG downward in the active site such that canonical adduct-dG:dCTP pairing is not possible. Based on X-ray structures, sequence alignment and our modeled structures of Y-Family DNAPs, chimney opening size seems primarily controlled by one amino acid (“flue-handle”), which dictates whether nearby amino acids (“flue”) plug the chimney or not. Based on this analysis, a correlation is apparent: the flue is closed in V/η-class DNAPs giving small chimney openings, while the flue is open for the IV/κ-class giving large chimney openings. Secondarily, a hypothesis is developed for why the V/η-class might preferentially do cellular dATP insertion opposite [+ta]-B[a]P-N2-dG: the small chimney forces adduct-dG lower in the active site, possibly leading to catalysis using a non-canonical dNTP shape that permits syn-adenine:adduct-dG base pairing. In summary, a hypothesize is developed that the pyrene moiety of [+ta]-B[a]P-N2-dG protrudes from the large chimney opening of DNAP IV, thus permitting canonical dCTP:adduct-dG pairing, while the small chimney opening of DNAP V forces [+ta]-B[a]P-N2-dG lower down in the active site, in which syn-adenine can pair with adduct-dG via a non-canonical dNTP shape.
Keywords: Cancer, Y-Family DNA polymerases, Mutations, Genotoxins, Adducts, Lesion-bypass
1. Introduction
Cells possess many DNA polymerases (DNAPs); e.g., human cells, yeast (S. cerevisiae) and E. coli have at least fifteen, eight and five, respectively [1–3]. Some DNAPs are present to do translesion synthesis (TLS) past DNA lesions/adducts that result from radiation and chemical insults [1–9]. Most lesion-bypass DNAPs are in the Y-Family [1–9], where humans have three members (hDNAPs η, ι and κ), yeast has one (scDNAP η) and E. coli has two (ecDNAPs IV and V).
Y-Family DNAPs have a conserved ∼350aa core, which includes the polymerase active site (representative Refs.: [10–18]). As with all DNA polymerases, Y-Family members resemble a right-hand with thumb, palm and finger domains, although their “stubby” fingers and thumb result in more solvent accessible surface around the template/dNTP-binding pocket [7]—presumably to accommodate the bypass of bulky and/or deforming DNA adducts/lesions, which protrude into these open spaces. Y-Family DNAPs grip DNA with an additional domain [7,10,11,15], usually called the “little finger”. Steps in the mechanism of Y-Family DNAPs have been proposed for both protein structural changes based on a series of X-ray structures [19,20] and for chemical catalysis based on theoretical studies [21].
Our work has focused on benzo[a]pyrene (B[a]P), which is a well-studied DNA damaging agent that is a potent mutagen/carcinogen and an example of a polycyclic aromatic hydrocarbon (PAH), a class of ubiquitous environmental substances produced by incomplete combustion [22,23]. PAHs in general and B[a]P in particular induce the kinds of mutations thought to be relevant to carcinogenesis and may be important in human cancer (representative Ref.: [24]). B[a]P mutational spectra were established with the major metabolite that reacts with DNA, namely (+)-anti-B[a]PDE, in E. coli [25], yeast [26,27] and mammalian (CHO) cells ([28] and references therein). Mutagenesis has also been studied with [+ta]-B[a]P-N2-dG (+BP, Fig. 1), the major adduct of (+)-anti-B[a]PDE, and G → T mutations predominate in most cases ([29], and references therein).
Fig. 1.
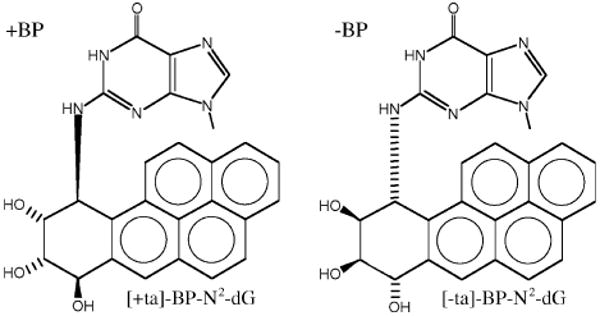
Structures of +BP ([+ta]-B[a]P-N2-dG) and −BP ([-ta]-B[a]P-N2-dG).
DNAPs IV and V of E. coli are both involved in translesion synthesis (TLS) with B[a]P-N2-dG adducts, although evidence suggests they play very different roles. In studies with purified proteins, DNAP IV inserted dCTP (>99%) opposite both +BP and [-ta]-B[a]P-N2-dG (−BP) in a 5′-CGA sequence, while DNAP V inserted dATP (>99%) [30], and this tendency is evident in E. coli where DNAP V inserts dATP opposite +BP in the G → T pathway [31], while in the non-mutagenic pathway DNAP IV does dCTP insertion opposite +BP [32–36], −BP [34] and other N2-dG adducts [35,36]. Regarding the non-mutagenic pathway, only DNAP IV is required for efficient TLS with −BP [34], while DNAP V is required in addition to DNAP IV with +BP [32–36]. Why are two DNAPs required for non-mutagenic TLS with +BP: certain lesions need one DNAP for insertion and a second for extension [37,38]. Thus, if DNAP IV does dCTP insertion [30,32–36], then DNAP V must do extension, which is sensible given kinetic findings with purified proteins, where DNAP V can be up to ∼1500 times better than DNAP IV at the step directly following adduct-G:C formation (i.e., extension) in the case of +BP compared to −BP (discussed in greater detail in Ref. [34]). Finally, random mutagenesis studies with [+anti]-B[a]PDE suggest that most G → T mutations with B[a]P-adducts require SOS-inducible Y-Family DNAPs, although a minor non-SOS-inducible G → T pathway does exist (discussed in Ref. [34]), which has been studied with +BP in a 5′-GGA sequence [32].
The study of E. coli's Y-Family DNAPs may provide insights about Y-Family DNAPs in general. Human DNAP κ was originally discovered because its sequence closely resembles E. coli DNAP IV [39–41], and dNTP insertion opposite a variety of adducts/lesions, including +BP, is remarkably similar for the DNAP IV/κ pair (Table 1), suggesting they are functional orthologs (discussed in Ref. [42]). This notion was substantiated when the identical mutation in a conserved residue (the “steric gate”, which excludes rNTPs) in the active site of DNAP IV and DNAP κ had a similar effect on lesion bypass vs. normal replication both in vitro and in cells [35]. DNAPs IV and κ have been shown to accurately bypass a variety of N2-dG-adducts [32–36], including those formed via various cellular trioses [36], which may be the main cellular rationale for the genesis of the IV/κ-class. E. coli DNAP V and human DNAP η are also functional orthologs, based on their similarity of dNTP insertion opposite a variety of adducts/lesions (Table 1 [42]), and a strong case has been made that the main cellular rationale for the V/η-class is TLS of UV-induced CPDs (discussed in Ref. [43]).
Table 1.
Dominant dNTP insertions opposite various DNA adducts/lesions by E. coli DNAPs IV and V and human DNAPs κ and η.a.
| Lesion | DNAP V | DNAP η | DNAP IV | DNAP κ |
|---|---|---|---|---|
| [+ta]-BP-N2-Dg | A/C | A >G | C | C |
| AAF-C8-dG | C | C | C/T | C/T |
| AF-C8-dG | – | – | C | C |
| TT-CPD | AA | AA | n | n |
| T(6-4)T | AG | nG | n | n |
| AP site | A | A | n | A* |
Dominant dNTP insertion using purified DNAPs, where “n” indicates “no” or low activity, “A*” indicates bypass by an unusual mechanism, and “–” indicates data unavailable. Data as reviewed in Ref. [41].
There must be structural reasons why the insertion preference opposite adducts/lesions is different for the DNAP IV/κ pair vs. the DNAP V/η pair (Table 1 [42]). However, no X-ray structures exist for UmuC (the polymerase subunit of DNAP V), DNAP IV or hDNAP η, which prompted us to build models taking a homology modeling approach [42]. Analysis of X-ray structures, modeled structures and sequence alignment suggests that Y-Family DNAPs lend themselves to accurate homology modeling [42,43]. An X-ray structure does exist for hDNAP κ with DNA [18].
Herein a molecular modeling study is presented that suggests a hypothesis for why the IV/κ-class correctly inserts dCTP opposite B[a]P-N2-dG adducts. To form an adduct-dG:dCTP base pair, the B[a]P moiety must be in the developing minor groove, since the adduction site (N2-dG) is in the minor groove in a Watson–Crick base pair. On the minor groove side, Y-Family DNAPs have an opening (or gap) next to the active site between the fingers and little finger domains. This opening looks like an elliptical hole of varying size in Dpo4 [11–14], Dbh [10], hDNAP ι [17] and in our models of DNAP IV and UmuC(V) (see below), while it looks like a slot in hDNAP κ [18]. The character of this opening can be analyzed based on analogy to a “chimney,” where a cluster of nearby amino acids can be thought of as a “flue” that either plugs the chimney leaving a small opening, or does not plug the chimney leaving a large opening. Next to the flue is a single amino acid, which can be thought of as a “flue-handle” that controls whether the flue is open or closed. Three regions of the protein make up the chimney as shown in Fig. 2 for our model of DNAP IV: an upper lip (turquoise), a lower lip (dark blue) and a left lip (blue) [43]. It is not unreasonable to think that chimney opening size and shape might influence dNTP insertional mechanism given that the bulky B[a]P moiety (red, Fig. 2) must fit in it.
Fig. 2.
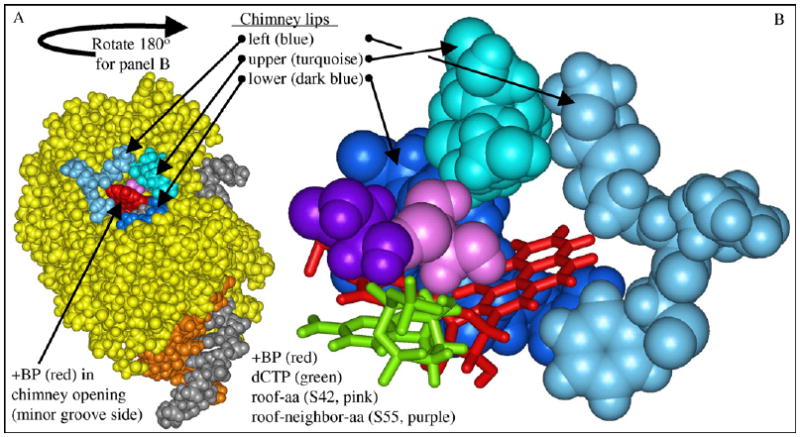
DNAP IV model showing the “chimney” and “roof” regions. Panel A: View from the minor groove side of DNAP IV (yellow), showing the “chimney” opening (cleft or hole), which is encircled by the upper lip (turquoise), left lip (blue) and lower lip (dark blue), along with +BP (red) emerging from the opening. The template (gray) and primer (brown) are also shown. The view in panel A is rotated approximately 180° to give panel B, which shows the chimney (white space) between the three chimney lips. Also shown are the roof-aa (S42, pink), which is a positionally conserved residue that lies above the nucleobase of the dNTP, the “roof-neighbor-aa” (S55, purple), which is next to the roof-aa and also contacts the dNTP nucleobase (dCTP, green), and the +BP (red), whose pyrene moiety must fit in the chimney in order for the dG moiety of +BP to pair with dCTP.
Herein we show that DNAP IV has a large chimney opening, which allows +BP to pair with dCTP, when dCTP adopts the canonical shape observed in all other families of DNAPs. In contrast, our molecular models suggest that UmuC(V) has a small chimney opening, which forces +BP downward in the active site into a position where catalysis is less likely to be facile.
Secondarily, we reflect on the conundrum: if the small chimney opening of DNAP V enforces a structure that seems unlikely to be active, then how might DNAP V insert dATP opposite +BP? Dpo4 is the best studied Y-Family DNAP in terms of structure (representative Refs.: [11–14]). Though the dNTPs in the active site of many Dpo4 structures adopt the canonical “chair-like” dNTP shape (Fig. 3, green), which is similar to the shape of dNTPs in the active sites of other families of DNAPs (Fig. 3, insert), a second non-canonical “goat-tail-like” shape has also been observed (Fig. 3, yellow), as noted by others [13]. (For simplicity we refer to the “chair-like” shape as S1-dNTP and the “goat-tail-like” shape as S2-dNTP.) Based on X-ray structures, modeled structures, sequence alignment and experimental results, a logical framework emerges from the hypothesis that the syn-adenine orientation of S2-dNTP pairs with adduct-dG, since S2-dNTP sits lower down in the active site and permits +BP to fit under the small chimney of UmuC(V). This hypothesis depends on the non-canonical S2-dNTP shape being active, which is considered in Section 4. The goal of the modeling work herein was to probe the insertional differences between the V/η-class and IV/κ-class of Y-family DNAPs and develop hypotheses, which we intend to test experimentally.
Fig. 3.
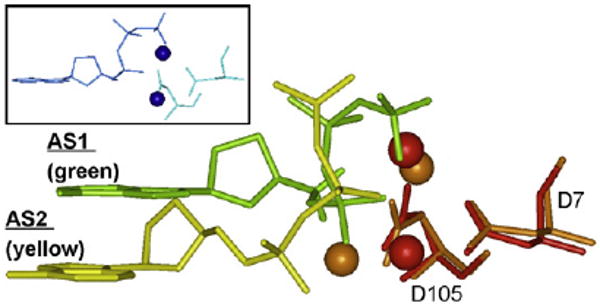
Side view of a dNTP in the “chair-like” shape S1-dNTP (green) vs. the “goat-tail-like” shape S2-dNTP (yellow). Key amino acids (only two are shown) from a Dpo4 structure adopting the S1-dNTP shape were superimposed on the same amino acids in a Dpo4 structure adopting the S2-dNTP. Spheres are divalent cations (S1-dNTP/red and S2-dNTP/brown). The insert shows that dNTP shape in the T7 DNAP active site resembles S1-dNTP (green). X-ray coordinates are from 1SOM-B for Dpo4/S1-dNTP, 1RYS-A for Dpo4/S2-dNTP, and 1T7P for T7 DNAP.
2. Methodology
Methods were identical to those described previously [43], except as noted below. Molecular dynamics (MD) runs were done with CHARMM 30 using Boston University's IBM Power4 p690 or IBM BlueGene. Starting coordinates for DNAP IV and UmuC(V) were derived from refined structures in our previous work [42], from which unadducted DNA was removed and replaced with adducted S1-dNTP or S2-dNTP DNA derived from our modeling work with Dpo4 [43]. Water was added [43]. The MD protocol (outlined here) is slightly modified from our previous work [43]. Water alone was minimized (200 steps of ABNR), after which water and protein were minimized (200 steps of steepest descent), and then, finally, water, protein and the phosphates of the dNTP were minimized (200–500 steps of steepest descent). The structures were heated to 300 K over 40 ps using the velocity Verlet integration method (time step 1 fs) with a harmonic force constraint of 5 kcal/mol per Å2 on all nucleobase atoms. Thereafter, MD was conducted at 300 K for 250 ps, whereupon the constraint was reduced to 0.5 kcal/mol per Å2 for 50 ps. The run was completed with an additional 1000 ps of unconstrained MD (e.g., see Fig. 4). During MD, electrostatic, van der Waals interactions and non-bonded Lennard–Jones interactions were smoothed to zero at 11 Å, and the SHAKE algorithm was used to constrain bonds with hydrogen atoms using a tolerance of 0.0005 Å. Each structure in a figure is a snapshot taken from the last 50 ps of a MD run and is representative, as all structures were more-or-less similar, except as noted in Section 3. Structures are viewed after minimization (200 steps of ABNR) to remove distortions due to the elevated temperatures.
Fig. 4.
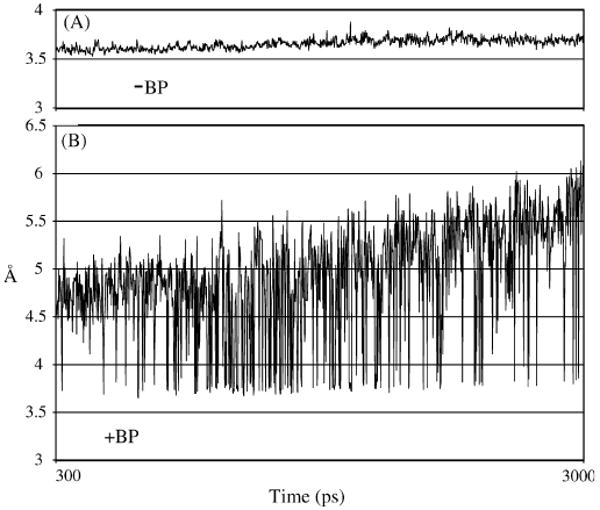
Distance between the O3′-primer and Pα-dCTP during the final 1 ns of the MD runs with −BP (panel A) and +BP (panel B) for DNAP IV/S1-dNTP.
X-ray coordinates in some figures were taken from the RSCB Protein Data Bank [44], and include Dpo4 with an S1-dNTP shape (1SOM-B), Dpo4 with an S2-dNTP shape (1RYS-A), Dpo4 with +BP (2IA1)), T7 DNAP (1T7P), hDNAP κ (2OH2), yDNAP η (1JIH).
3. Results
3.1. The canonical S1-dNTP “Chair-like” shape
In duplex DNA, the pyrene moieties of −BP and +BP (when paired with dC) are oriented toward the 3′- and 5′-base [45]. Herein we assume these same orientations for the pyrene moieties in UmuC(V) and DNAP IV. (Though we have not completed a thorough study, −BP in the BPmi5 conformation looks reasonably similar to +BP in BPmi5, while +BP in a BPmi3 conformation looks reasonably similar to –BP in BPmi3. No observations from the work herein depend on the unique assignment of −BP to BPmi3 or +BP to BPmi5, and, thus, this assumption is not critical to our analysis.)
DNAP IV inserts dCTP opposite −BP (Section 1), and this possibility was modeled using the canonical S1-dNTP shape. During the final 1 ns of the MD run, the structure is stable, appeared reasonable, and showed no significant distortions (Fig. 5C) when compared to the control structure (Fig. 5A) in which unadducted-dG was paired with S1-dCTP. (In each panel of Fig. 5, the left structure shows the region around the roof-aa/roof-neighbor-aa, while a ∼90 °C rotation on the horizontal gives the right structure, which is of the chimney region.) For example, the distance between primer-O3′ and Pα-dNTP was reasonably constant for the −BP structure (Fig. 4A) and averaged 3.66 Å, which was comparable to the dG no-adduct control structure that averaged 3.70 Å. These distances are close to a van der Waals contact (∼3.5 Å) and can be considered “reaction-ready.”
Fig. 5.

(A–L) Regions of DNAP IV and UmuC(V) from our MD runs, in S1-dNTP or S2-dNTP with no adduct, −BP, or +BP. In each panel, the left structure shows the roof-aa (pink), the roof-neighbor-aa (purple), the dCTP (green), the primer (yellow), and the template adduct-dG (red), while the right structure shows the three lips of the chimney opening (turquoise/blue/dark blue) for DNAP IV (aa31-34/aa72-75/aa244-247) or UmuC(V) (aa30-33/aa71-74/aa255-258).
DNAP IV also inserts dCTP opposite +BP (Section 1), which we modeled using the BPmi5 orientation opposite S1-dCTP. In this case, the MD run oscillated between two structures (Fig. 4B), one being reaction-ready (Fig. 5E, primer-O3′/dNTP-Pα distance, 3.66 Å), while the second was not reaction-ready (not shown, 5.12 Å). Since the reaction-ready structure presents a significant fraction of time (∼20%), we only consider it. (With the exception of dCTP insertion opposite +BP by DNAP IV, all other MD runs looked stable as exemplified by Fig. 4A.)
With UmuC(V) and S1-dNTP, both −BP and +BP gave structures (Fig. 5D and F, respectively) with O3′-Pα distances that were not reaction-ready (4.93 Å and 5.12 Å, respectively). Why did UmuC(V)/S1-dNTP not give reaction-ready distances, when DNAP IV/S1-dNTP did? DNAP IV has a large chimney opening (right side structures in Fig. 5A, C and E), while UmuC(V) has a smaller chimney opening (right side structures in Fig. 5B, D and F) into which (e.g.) the pyrene moiety of +BP does not readily fit without distortion/reorientation. During the MD run, this problem is resolved by opposing motions: the chimney opens slightly, but principally +BP is pushed downward and away from the chimney/minor groove and toward the major groove. As the adduct-dG moves so does its paired dCTP, thus forcing Pα away from the primer-O3′. In fact, during the S1-dNTP/UmuC(V) MD run, adduct-dG:dCTP moves about halfway toward S2-dNTP/UmuC(V) positioning. Other distortions result, including the loss of co-planarity between the adduct-dG:dCTP and the base pair below it (not shown). −BP experiences similar problems, though it is the saturated ring of the B[a]P moiety that clashes with the small chimney (Fig. 5D).
3.2. The non-canonical S2-dNTP “Goat-tail-like” shape
A non-canonical S2-dNTP shape (“goat-tail-like”) is also frequently observed in Dpo4 X-ray structures (Introduction), which we also consider. With the S2-dNTP shape in UmuC(V), both −BP and +BP show reaction-ready structures (Fig. 5J and L, respectively) with primer-O3′/dNTP-Pα distances of 3.77 Å and 3.85 Å, respectively. Why did S2-dNTP/UmuC(V) give reaction-ready distances, when S1-dNTP/UmuC(V) did not? The dNTP shape in S2-dNTP is lower down compared to S1-dNTP (Fig. 3), and, thus, +BP is also lower down, allowing the pyrene to fit comfortably below the small chimney opening (Fig. 5F), with the consequence being that Pα-dCTP stays near the primer-O3′. With dCTP being lower down in S2-dNTP, a gap exists above the nucleobase in the initial structure; this gap diminishes during the MD run due to slight rearrangements of the bulky sidechains of the roof-aa I38 and its roof-neighbor-aa M51. The observations are similar for −BP (Fig. 5J).
With the S2-dNTP shape, both −BP and +BP gave distorted structures in DNAP IV (Fig. 5I and K, respectively) and non-reaction-ready primer-O3′/dNTP-Pα distances of 4.52 Å and 4.85 Å, respectively. Why did DNAP IV/S2-dNTP not give reaction-ready distances and reasonable structures, when DNAP V/S2-dNTP and DNAP IV/S1-dNTP did? DNAP IV has a less bulky roof-aa (S42) and a less bulky roof-neighbor-aa (S55), neither of which has a sidechain that is big enough to rearrange and fill the gap above the nucleobase of dCTP in S2-dNTP. During the MD run, the deoxyribose of dCTP rises to try to fill in this gap (though the gap is still large in the final structure, Fig. 5I and K) and the phosphates rearrange into a “straight-chain” orientation, which is non-canonical and has not been observed to the best of our knowledge.
4. Discussion
We first consider a hypothesis for why chimney opening size might be different in DNAP IV vs. UmuC(V), before assessing how chimney size might affect dNTP insertion pattern. Finally, we discuss why in cells DNAP IV might insert dCTP, while DNAP V inserts dATP (Fig. 6), which are hypotheses that we intend to test experimentally, which was the goal of the modeling work described herein.
Fig. 6.
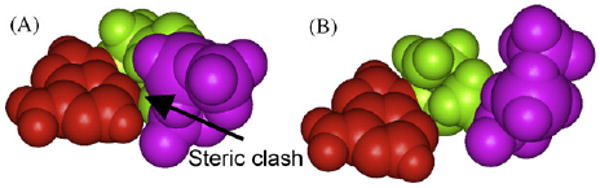
Adenine (red) in the syn-orientation sterically clashes with the deoxyribose (green) and Pα (purple) when dATP is in the canonical “chair-like” shape S1-dNTP (panel A), while no such steric clash exists in the “goat-tail-like” shape S2-dNTP (panel B).
4.1. Structural basis for a large vs. a small chimney opening
What structural difference(s) in DNAP IV vs. UmuC(V) might result in a large vs. a small chimney opening, and is this structural difference(s) conserved in other Y-Family DNAPs in the IV/κ-class vs. the DNAP V/η-class? The chimney has three sides (Figs. 2 and 5): the upper lip (numbered aa32–35 in DNAP IV, turquoise) and left lip (aa73–76, blue) are in the fingers domain, while the lower lip (aa244–247, dark blue) is in the little finger domain. The amino acids associated with the chimney lips can be assigned reliably based on conserved amino acids. For example, the region surrounding the chimney upper lip for the IV/κ-class and V/η-class (as shown in Fig. 7) have 22 and 18 conserved amino acids, respectively, and, of these, seven are identical between the sets (boxed in gray in Fig. 7A and B). The assignment of amino acids to the left lip and lower lip are similarly anchored by the alignment of nearby conserved amino acids.
Fig. 7.
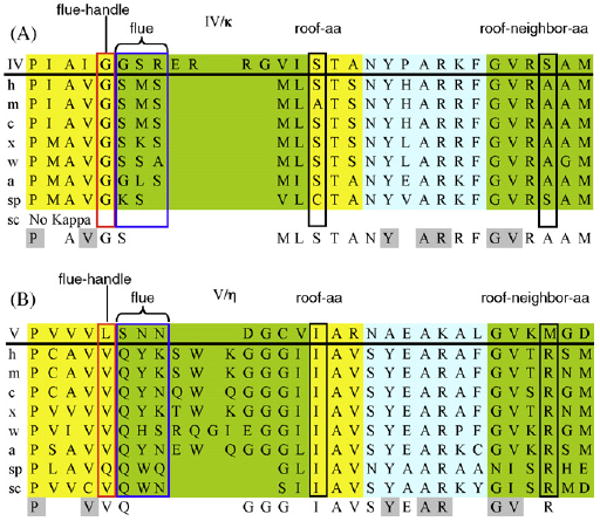
Amino acid alignment of several key regions of representative Y-Family DNAPs in the IV/κ-class (panel A) and the V/η-class (panel B). The R-groups of the final three amino acids (“flue”) of the chimney's upper lip either don't plug (IV/κ-class) or plug (V/η-class) the chimney, depending upon whether the first amino acid in the upper lip (“flue-handle”) is glycine (flue open, IV/κ-class) or is a bulky aa (often valine, flue closed, V/η-class). The lowest line in each panel shows conserved residues, with residues conserved (≥75%) between the IV/κ-class and the V/η-class are highlighted in gray. Abbreviations: ss (Sulfolobus solfataricus) h, human (Homo sapien); m, mouse (Mus musculus); c, chicken (Gallus gallus); x, frog (Xenopus laevis); w, worm (Caenorhabditis elegans); a, plant (Arabidopsis thaliana); sp, fission yeast (Saccharomyces pombe); sc, budding yeast (Saccharomyces cerevisae). Colors: blue, α-helicies; yellow, extended (β); and green, loops.
The chimney upper lip (turquoise, Fig. 5) is closest to the active site, and principally defines whether the chimney can accommodate the bulky B[a]P moiety. The first amino acid in the upper lip of DNAP IV is glycine (G32), and all IV/κ-class members from all of the species listed in Fig. 7A have a glycine at this position, often embedded in a VGS sequence. The one X-ray structure available for this class, hDNAP κ [18], shows that this glycine (G131, turquoise, Fig. 8A) is followed by upward curvature of the chimney upper lip (red arrow, Fig. 8A). This glycine can be thought of as a “flue-handle” whose backbone properties allow this upward curvature (discussed below), with the consequence being that the R-groups on the next several amino acids (“flue”; S132/R133, blue in Fig. 8B) are turned away from the chimney opening, which remains open. Our models of DNAP IV also have this upward curvature (Fig. 5) with an open flue, which depends on an analogous flue-handle glycine (G32).
Fig. 8.
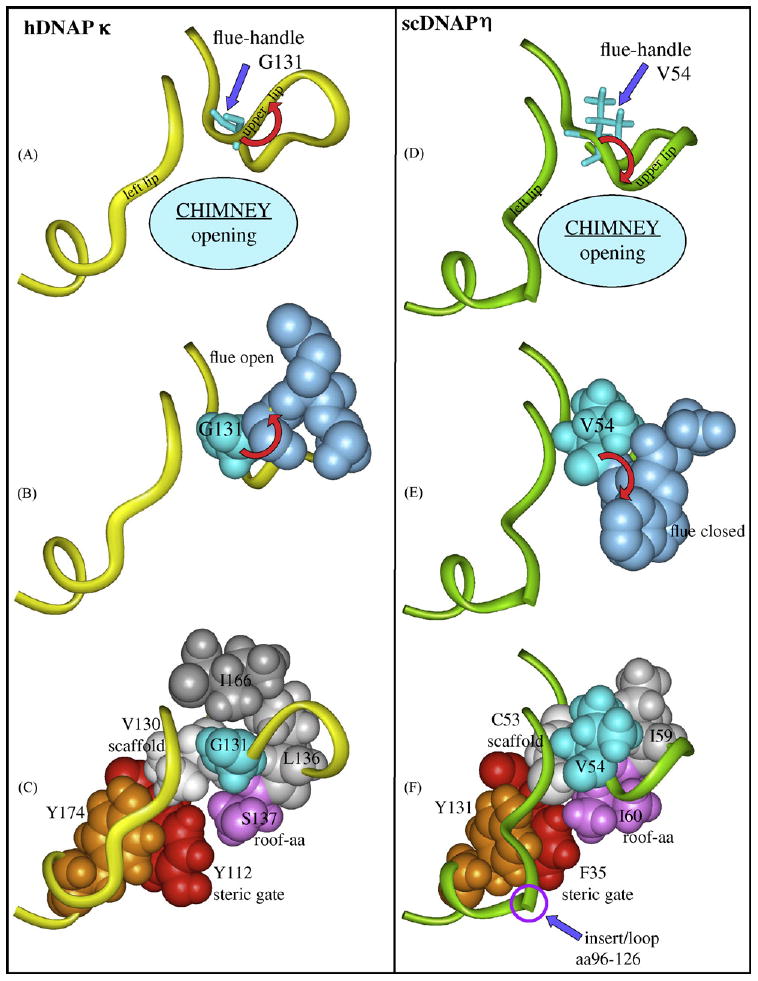
Regions of hDNAP κ (panels A and B), scDNAP η (panels C and D) and hDNAP η (insert) that show why the chimney opening is large or small. Panels A–C: Y-Family DNAPs in the IV/κ-class have a glycine “flue-handle,” such as G131 (turquoise) in hDNAP κ, which leads to upward curvature of the protein backbone in the chimney upper lip (red arrow, panel A) and results in the “flue” amino acids (S132/M133, blue, panel B) to point away from the chimney, giving a large chimney opening. Panel C shows V130 (white) that serves as a scaffold to organize the chimney's upper lip and left lip (yellow ribbons in panel A), along with the roof-aa (S137, purple), the steric gate (Y112, red) and a conserved tyrosine (Y174, brown), which stacks on the backbone of the left lip and orients it. V130 forms a rectangle with the G131 flue-handle, L136 (gray) and the S137 roof (pink) upon which I166 stacks (dark gray). Panels D–F: DNAP V/η-class DNAPs have a bulky “flue-handle,” such as V54 in scDNAP η (turquoise, panel D), which causes downward curvature of the chimney upper lip (red arrow, panel D), and results in the “flue” amino acids (Q33/W34, blue, panel E) to plug the chimney, giving a small opening. Panel E shows scaffold C53 (white) organizing the chimney's upper lip and left lip (green ribbons in panel C), along with the roof-aa (I60, purple), the steric gate (F35, red) and a conserved tyrosine (Y131, brown), which stacks on the backbone of the left lip and orients it. C53 forms a rectangle with the V54 flue-handle (turquoise), I59 (gray) and the I60 roof (pink). scDNAP η has a large insert/loop (aa96–126) in the left lip, which is represented as a discontinuity. X-ray coordinates are from 2OH2 for hDNAP κ [18] and from 1JIH for scDNAP η [16], where hydrogens were added using insightII.
In contrast all DNAP V/η-class members have bulky “flue-handles,” which is usually valine in a VVQ sequence (Fig. 7B). The one X-ray structure in the V/η-class, scDNAP η [16], shows that its bulky V54 flue-handle (turquoise, Fig. 8D) is associated with downward curvature of the chimney upper lip (red arrow), which forces the “flue” (Q55/Y56, blue in Fig. 8E) to plug the chimney. In UmuC(V) the sequence is slightly different (VLSN), though the outcome is the same: the bulky L30 flue-handle causes downward curvature, and an asparagine (N32) plugs the chimney giving a closed flue (Fig. 5B).
Glycine has greater flexibility in its ϕ/φ-angles compared to other amino acids, which appears to be the fundamental conformational reason why a glycine vs. a non-glycine flue-handle gives different curvature of the chimney upper lip. Fig. 9 is a Ramachandran plot for all valines, leucines and cysteines from X-ray structures of Dpo4 [11] hDNAP κ [18] and scDNAP η [16]. Fig. 9 also shows the ϕ/φ-angles for the flue-handles in scDNAP η (V54, blue circle), hDNAP η (V37, red circle), UmuC(V) (L30, yellow circle) and Dpo4 (C31, pink circle), which are all clustered in the region expected for extended (β) secondary structure, with the consequence being that this combination of ϕ/φ-angles leads to downward curvature of the chimney's upper lip, causing the flue to plug the chimney and the chimney opening to be small. In contrast, the angles for the glycine flue-handles are outside this region for hDNAP κ (G131, green square) and DNAP IV (G32, purple square), which results in upward curvature of the chimney's upper lip, thus opening the flue and giving a large chimney opening. (Given the discontinuous nature of Ramachandran plots, the ϕ/φ-angles for DNAPs IV and κ look farther from the valine/leucine/cysteine cluster than they actually are.)
Fig. 9.
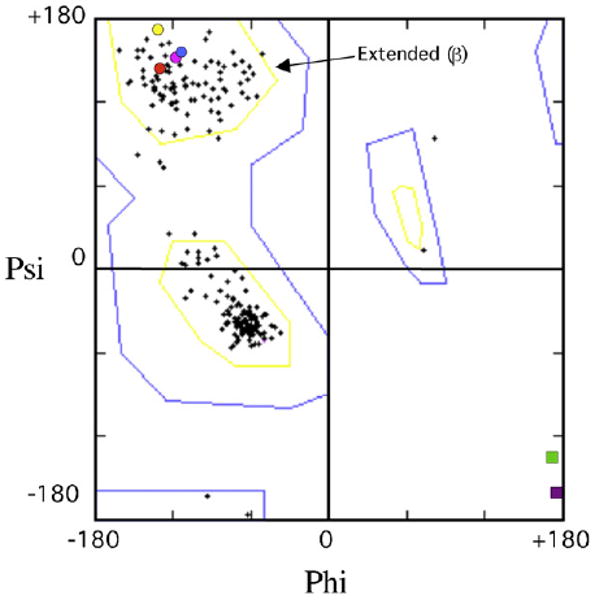
Ramachandran plot showing ϕ/φ-angles for all valines, leucines and cysteines (crosses) from X-ray structures of Dpo4 [11] hDNAP κ [18] and scDNAP κ [15], along with the flue-handles for scDNAP η (blue circle, V54), hDNAP η (red circle, V37), UmuC(V) (yellow circle, L30) Dpo4 (pink circle, C31), which are all clustered in the region expected for extended (β) secondary structure. The glycine flue-handles for hDNAP κ (G131, green square) and DNAP IV (G32, purple square) lie outside this clustered region.
Glycines are not excluded from adopting the ϕ/φ-angles observed in the valine/leucine/cysteine cluster, so in principle IV/κ-class DNAPs could have a closed flue leading to a small chimney opening as in the DNAP V/η-class. Our model for DNAP IV began with the X-ray backbone coordinates of Dpo4, whose non-glycine flue-handle (C31) leads to downward curvature of the upper lip and a closed flue in X-ray structures ([11–14], as well as other Dpo4 structures). In spite of the downward curvature in the starting structure (derived from Dpo4) in our DNAP IV model, its flue-handle (G32) always rearranged during MD runs [42] to give upward curvature (e.g., Fig. 5A), resulting in a final structure with an open flue, which resembles the hDNAP κ X-ray structure (Fig. 8B). This finding implies that glycine flue-handles prefer the “up” orientation, which appears to be stabilized by several interactions—notably by a R34/E25 salt bridge in our model of DNAP IV. In DNAP κ, a M133/I166 hydrophobic interaction is the major contact favoring the up-orientation for the glycine flue-handle.
We could find no other reasonable structural rationale for the difference in chimney opening size. For example, loop-size per se does not correlate, since UmuC(V) and DNAP κ in most cases have relatively small loops, while DNAPs IV and DNAP η have relatively large loops (leftmost green region in Fig. 7).
4.2. Roof-aa and roof-neighbor-aa
The roof-aa (Fig. 2, pink) is a positionally conserved residue that lies above the nucleobase of the dNTP, as seen in the active site of Dpo4 [11–14], yDNAP η [16], hDNAP ι [17] and hDNAP κ [18]. The roof-aa is next to the chimney (see Section 4.3). The “roof-neighbor-aa” (Fig. 2, purple) also contacts the nucleobase of the dNTP. It is not unreasonable to imagine that the roof-aa and roof-neighbor-aa could influence dNTP insertion patterns, and in fact a correlation exists. The roof-aa is isoleucine for all members of the DNAP V/η class listed in Fig. 7B, and the roof-neighbor-aa is always bulky (arginine or methionine). Bulky roof-aa/roof-neighbor-aa can rearrange to fill in the gap above the nucleobase in the S2-dNTP shape (Fig. 5B). The roof-aa is non-bulky (usually serine) for all IV/κ class members (Fig. 7A) and the roof-neighbor-aa is non-bulky (usually alanine). Non-bulky amino acids work well for S1-dNTP, but do not fill in the gap above the nucleobase in the S2-dNTP shape, making it less viable.
4.3. The interconnected architecture of the chimney and roof regions
To understand the architecture of the chimney/roof region of Y-Family DNAPs, it is useful to focus on a bulky aliphatic amino acid, usually a valine (e.g., V29/UmuC(V), I30/DNAP IV, V37/scDNAP η and V130/hDNAP κ, Fig. 7), that plays a scaffolding role as revealed in X-ray structures [10–18] and in our models [42,43]. Using hDNAP κ as an example (Fig. 8C), this scaffolding valine (V130, white) begins a loop that ends with the roof-aa, with which it forms a hydrogen bond (scaffold-C=O:HN-roof). This type of hydrogen bond is also observed in X-ray structures from Dpo4 [11–14], hDNAP κ [18] and scDNAP η [15,16]. The flue-handle (G131) is adjacent to scaffold-V130, which also contacts the bulky amino acid L136 (Fig. 8C, gray) next to roof-S137. Thus, the loop is anchored by a square of four amino acids (V130/G131/L136/S137), above which sits the R-group of I166 (dark gray, Fig. 8C) giving a rectangular pyramid of amino acids that orient the flue-handle/flue/roof-aa region. In our model of DNAP IV, the rectangle includes I31/G32/I41/S42 with L71 sitting above it. This region in scDNAP η (Fig. 8F) looks similar, though only the rectangle C53/V54/I59/I60 exists, because the bulky R-group of flue-handle/V54 serves the role of the fifth amino acid (i.e., the R-group of flue-handle-V54 takes the place of I166 in hDNAP κ). In our model of UmuC(V), the rectangle involves V29/L30/V37/I38.
Scaffold-V130 in hDNAP κ (white, Fig. 8C) also helps organize the steric gate (Y12, red), which face-stacks with Y174, a highly conserved tyrosine whose other aromatic face contacts the backbone of the left lip of the chimney (i.e., aa168-171 in hDNAP κ), thus helping to orient it. The scDNAP η structure is similar (Fig. 8F) except the chimney left lip has an insert/loop (aa96–126, which is indicated by the circle). In spite of this insert/loop, the chimney left lip is similarly positioned in the vicinity of the upper lip. (Of the DNAPs shown in Fig. 7, only DNAPs η from yeast (both sc and sp) have an insert at this position.)
Amino acid structure in the upper lip/left lip/roof region is similar in all Y-Family DNAPs, for which X-ray structures [10–18] or modeled structures [42,43] exist.
4.4. dCTP insertion by DNAP IV using S1-dNTP
DNAP IV can pair dCTP with both −BP and +BP using S1-dNTP (Fig. 5C/E), importantly because DNAP IV's large chimney opening can accommodate B[a]P bulk in the developing minor groove. In contrast, DNAP V does not give a satisfactory structure when either −BP or +BP are paired with S1-dNTP (Fig. 5C/E), because DNAP V's small chimney opening forces the bulky pyrene moiety downward. In fact, UmuC(V) did not retain normal S1-dNTP-positioning in the active site during MD; rather the small chimney forced template BP-dG and its paired dCTP to move about halfway toward S2-dNTP-positioning with one consequence being that primer-O3′/dNTP-Pα distance elongated to a non-reaction-ready distance of ∼5 Å. These observations provide a reasonable rationale for why DNAP IV preferentially does dCTP insertion, since it can do so using the S1-dNTP shape, which is presumably active, given that it is found in other DNAP families (e.g., T7 DNAP, insert Fig. 3), while DNAP V's small chimney enforces the use of S2-dNTP, which is expected to be less active (or inactive).
4.5. How does UmuC(V) do dATP insertion?
−BP and +BP cannot be paired using the S1-dNTP shape in UmuC(V) without significant distortions to the adduct, the dNTP, the DNA and/or the protein, because B[a]P bulk does not fit in UmuC(V)'s small chimney opening. N31 is the main problem, as its sidechain plugs the UmuC(V) chimney leading to clashes with −BP/+BP when paired using the S1-dNTP shape. In the unadducted structure (Fig. 5B), N31 adopts its lowest energy rotational conformer with respect to the Cα–Cβ bond. The presence of +BP causes a rotation about the Cα–Cβ bond (Fig. 5D); however, no other rotations are possible to get N31 farther out of the way.
These observations lead to the question: how does UmuC(V) insert a dATP opposite +BP and −BP? One possibility involves S2-dATP pairing with adduct-dG, which allows the pyrene moiety to lie under UmuC(V)'s small chimney opening (Fig. 5J and L), since S2-dNTP lies lower down in the active site than S1-dNTP (Fig. 3). Adduct-dG can Hoogsteen pair with syn-adenine in S2-dATP. Interestingly, syn-adenine has steric clashes with atoms in the deoxyribose and the α-phosphate in the S1-dATP shape (Fig. 6A), but no such steric clashes exist with the S2-dATP shape (Fig. 6B and discussed in Ref. [43]). MD runs with adduct-dG:syn-dATP pairing in UmuC(V) gave reasonable structures; e.g., the primer-O3′/dNTP-Pα distances averaged 3.64 Å and 3.61 Å, respectively (structures not shown, though virtually identical to Fig. 5J and L, respectively).
S2-dATP Hoogsteen pairing of syn-adenine with adduct-dG is a plausible first step in the G → T pathway. The notion that syn-dATP can be present in S2-dNTP is supported by one Dpo4 X-ray structure (1RYS-A), in which syn-dATP pairs with the 5′-T of a TT-CPD using S2-dNTP [12]. In fact, Y-Family DNAPs in the DNAP V/η-class may have evolved a catalytically active S2-dNTP shape to permit syn-dATP insertion opposite (e.g.) TT-CPDs, since one of the roles of the DNAP V/η-class is to minimize UV-light mutagenesis by just such a mechanism (discussed in Ref. [43]). Perhaps, G → T mutations with BP-adducts are the unfortunate by-product of the occasional use of the S2-dNTP insertion pathway.
We presume that the S1-dNTP shape is optimal for catalytic efficiency given that it is observed in all families of DNAPs. But could S2-dNTP possibly be active? In previous work we analyzed thirty-four Dpo4 X-ray structures [43] and concluded that those adopting the S2-dNTP shape had accompanying protein components that should allow phosphoester bond making and breaking, notably with protein elements needed: (1) to ensure proper geometry between the nucleophilic primer-O3′ and the electrophilic dNTP-Pα; (2) to enhance electrophilicity of Pα of the dNTP during nucleophilic attack; (3) to dampen negative charge on the pyrophosphate leaving group; and (4) to orient the Pα–Pβ–Pγ phosphate tail. In fact, S2-dNTP seemed to have all of the necessary requirements to be catalytically active. For example, in fact four out of five S2-dNTP structures had primer-O3′/dNTP-Pα distances of less than 4.0 Å (average: 4.0 Å, range: 3.66–4.69 Å), while even the best S1-dNTP structure had a longer distance of 4.41 Å with an average of 5.9 Å (range: 4.41–7.95 Å). We note, however, that there is no evidence that S2-dNTP is active, and it is merely a hypothesis that it might be.
Other G:A mispairings are also possible. In principle, anti-dATP can pair with syn-guanine in adduct-dG, which requires the pyrene moiety to be in the major groove. Anti-dATP can also pair with anti-guanine in adduct-dG in an elongated mispair. Thus, there are scenarios other than the one involving our syn-dATP:anti-adduct-dG hypothesis.
Our modeled structures show that, in principle, either dCTP or syn-dATP can pair with adduct-dG using S2-dNTP. In a 5′-GGT sequence [30], purified DNAP V inserts both dCTP and dATP ([dATP]/[dCTP] = 0.75 and 0.18 for −BP and +BP, respectively). In a 5′-CGA sequence, however, purified DNAP V inserts dATP > 100-fold more efficiently than dCTP opposite −BP and +BP [30]. In cells we know that DNAP V does not efficiently insert dCTP in our 5′-TGC sequence, because – if it did – the non-mutagenic pathway would not have declined to ∼1% and ∼3% efficiency for −BP and +BP, respectively, in E. coli defective in DNAP IV [34]. It will be of interest to investigate whether an S2-dNTP structure might explain these differences in dATP vs. dCTP insertion efficiency.
We note, however, that one published finding is hard to rationalize based on the mechanism proposed in this section. Of mismatched insertions, dATP is most often inserted opposite a B[a]P-dA adduct by DNAP V [30]; there is no obvious way to pair dATP with an adduct-dA, not even via Hoogsteen or wobble pairings. It is impossible to know whether this observation implies that a more nuanced variation of the mechanism proposed in this section is required, or whether it implies that the mechanism is wrong.
4.6. Dpo4 compared to DNAPs IV and V
Based on biochemical and X-ray findings [14], Dpo4 insertion opposite +BP was proposed to follow a “dislocation” or “templated” pathway. Dislocation/templated insertion ([46,47] and references therein) involves DNAP stalling at an adduct, slippage to the next 5′-base along the template, and its use to direct incorporation (e.g., dATP insertion opposite the 5′-T in a 5′-TG sequence context), whereupon the newly incorporated dA slips back to form an adduct-G:A mispair, from which extension yields the mispair that ultimately gives a G → T mutation. Dpo4 preferentially inserted dCTP, dTTP, dATP and dGTP opposite +BP in 5′-GG, 5′-AG, 5′-TG and 5′-CG sequences, respectively [14], which is consistent with a dislocation/templated mechanism.
It was suggested that the dislocation/templated mechanism might operate widely with B[a]P-N2-dG adducts [14], which-if true-would mean that the mechanism proposed herein is off-track. Though the dislocation/templated mechanism is attractive for Dpo4, considerable evidence suggests it cannot dominate in E. coli, as we have discussed previously [29,31,34]. (All findings below not specifically attributed were reviewed in Ref. [29].) (1) In five cases using various B[a]P-N2-dG isomers in 5′-TG sequences, G → T mutations dominate, which is consistent with a dislocation/templated mechanism. However, G → T mutations dominate in eleven cases using various B[a]P-N2-dG isomers in various 5′-CG sequences; these findings are not consistent with a dislocation/templated mechanism, which predicts a G → C mutation. G → A mutations dominated with +BP and −BP in one 5′-AG sequence, which is consistent with a dislocation/templated mechanism. However, G → A mutations dominate in one 5′-CG sequence using three different B[a]P-N2-dG isomers, which is inconsistent with a dislocation/templated mechanism. Thus, where B[a]P-N2-dG has been studied site-specifically about twice as many examples are inconsistent than are consistent with the dislocation/templated mechanism. (Of course, even if a finding is “consistent” it does not mean it is the true mechanism.) (2) In mutagenesis studies with (+)-anti-B[a]PDE [25], where a broad range of sequence contexts were sampled, G → T mutations dominated in 5′-TG sequence contexts, consistent with “dislocation/templated” mutagenesis. However, if this were a general mechanism, it should operate in other sequence contexts, which is not the case: G → C and G → A mutations did not dominate in 5′-CG and 5′-AG sequences, respectively, while 5′-GG sequences were not principally mutagenically silent [25]. In fact, G → T mutations dominated in non-5′-TG sequences with (+)-anti-B[a]PDE, implying that it cannot be the only dATP insertion mechanism, even if the dislocation/templated pathway does occur. (3) Purified DNAP V preferentially inserted dATP opposite +BP and −BP in a 5′-CG sequence [30], which is not consistent with a dislocation/templated mechanism. DNAP V inserted dCTP opposite +BP and −BP in a 5′-GG sequence [30], which is consistent with a dislocation/templated mechanism, though dATP insertion was also significant in both cases, which is not consistent. Importantly, these findings demonstrate that DNAP V, which inserts dATP opposite +BP in the G → T pathway in E. coli [31], has a viable means of doing so that does not depend on a dislocation/templated mechanism. (4) Purified DNAP IV correctly inserted dCTP opposite +BP and −BP in 5′-CG and 5′-GG sequences [30], where only the latter is consistent with a dislocation/templated mechanism.
Thus, while a dislocation/templated mechanism is consistent with some findings in E. coli, the fact that it fails to rationalize the majority of the findings implies that it is not a unifying hypothesis. Furthermore, the dislocation/templated mechanism is not compelling given that its invocation forces one to propose that the dominant G → T mutational pathway follows one mechanism (dislocation/templated) in one sequence context (i.e., 5′-TG), but some other mechanism in other sequence contexts, when there is no obvious reason why the mechanism would change. The mechanism proposed herein (i.e., adduct-dG pairing with syn-dATP using S2-dNTP) would give G → T mutations independent of sequence context.
Returning to Dpo4, the biochemical evidence was clear: Dpo4 appears to use a dislocation/templated mechanism with +BP [14], as summarized in the first paragraph of this subsection. Why might Dpo4 be different than DNAPs IV and V of E. coli? Both the roof-aa and roof-neighbor-aa for Dpo4 are non-bulky (A44/A57), while its bulky flue-handle (C31) causes downward curvature of the chimney upper lip, leading to a closed flue (V32) and a small chimney opening (based on structures from Refs. [11–14], and other Dpo4 structures). Thus, Dpo4 is a hybrid with a roof similar to the IV/κ-class and a chimney similar to the V/η-class.
In fact, M76, which is the second amino acid in Dpo4's left lip, also rests in the chimney (Fig. 10), making its chimney even more plugged than those of DNAP IV and UmuC(V), which have non-bulky G74 and S72, respectively, in the equivalent position. In fact one X-ray structure for +BP/Dpo4 reveals that the pyrene (light red, Fig. 10) is so excluded that it also drags the adduct-dG moiety (red, Fig. 10) into the chimney resulting in a gap in the template strand. (This is an “extension structure” with dATP paired with a template-5′-T in the active site, while the +BP moiety is formally in the [n + 1] position, though displaced into the chimney. Extension structures, which we are also modeling (data not shown), are more similar to S2-dNTP structures, since both have the adduct-dG in the [n + 1] position.) Furthermore, the V32 flue of Dpo4 is also inserted deeper into the chimney than (e.g.) the N32 flue of UmuC(V). The excessively plugged chimney of Dpo4 does not permit S2-dNTP insertion structures similar to UmuC(V) (Fig. 5J and L), which may explain why the dG moiety of +BP in the chimney is not available for pairing and why Dpo4 follows a dislocation/templated mechanism. We hasten to add that Dpo4 actually has a large cleft between the fingers and little finger domains, but it begins below the C31/M76 plugs.
Fig. 10.
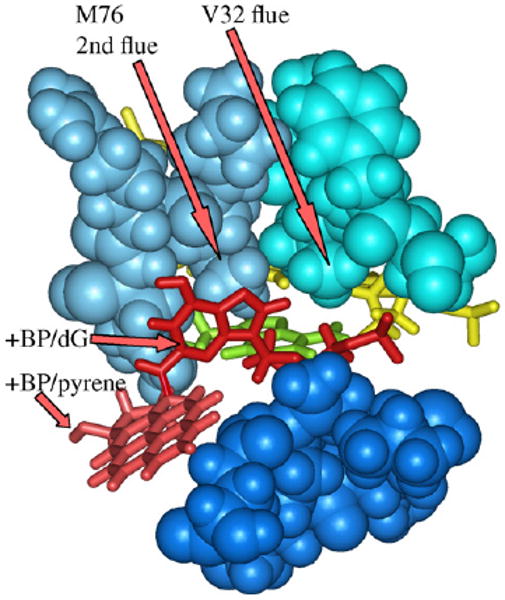
Chimney region of Dpo4 from X-ray coordinates 2IA1 [14] showing that the dG moiety (red) in addition to the pyrene (light red) of +BP are excluded from the active site, probably because Dpo4's chimney is particularly plugged, since it has both a regular flue (V32) in the upper lip (turquoise) and a second flue (M76), the latter being the second amino acid in the left lip (blue). The lower lip includes the positionally conserved four amino acids (dark blue, aa-246–249) along with aa271, the latter because the pyrene moiety stretches so far down. This is an “extension structure” with a 5′-template-dT paired with dATP (yellow) in the active site, while the +BP moiety is in the [n + 1] position. Hydrogens were added using insightII.
This analysis suggests a reason for caution when applying conclusions from Dpo4 to other Y-Family DNAPs, especially those purely in the IV/κ-class or the V/η-class. Perhaps Dpo4 evolved its hybrid roof/chimney structure to bypass a unique set of lesions encountered by a thermophilic bacteria. Alternatively, perhaps the structure of Dpo4 at physiologically relevant elevated temperatures is different than at the temperature at which it was crystallized (r.t.) and assayed (37°C), and this affects its structure and behavior.
4.7. Summary
The IV/κ-class of Y-Family DNAPs have a key glycine “flue-handle” that keeps adjacent bulky amino acids (“flue”) out of an opening on the minor groove side of the active site (“chimney”), which is thus large, can accommodate the pyrene, ands allows B[a]P-dG:dCTP pairing using the canonical “chair-like” S1-dNTP shape. The V/η-class of DNAPs have a bulky flue-handle (usually valine) that force the flue amino acids to plug the chimney, which gives a small opening that forces +BP/−BP downward in the active site, thus preventing proper geometry for primer-O3′ attack on Pα when dCTP is in the S1-dNTP shape. This may explain why IV/κ-class DNAPs do cellular dCTP insertion opposite N2-dG adducts, such as +BP and −BP. The small chimney opening of the V/η-class may force +BP/−BP so far down in the active site that adduct-dG can pair with the non-canonical “goat-tail-like” S2-dNTP shape, which lies lower in the active site than S1-dNTP. The S2-dNTP shape permits Hoogsteen syn-dATP pairing with B[a]P-dG and might be the first step in the G → T mutational pathway.
Acknowledgments
This work was supported by United States Public Health Services Grant R01ES03775.
Abbreviations
- B[a]P
benzo[a]pyrene
- +BP
[+ta]-B[a]P-N2-dG (Fig. 1)
- −BP
[-ta]-B[a]P-N2-dG (Fig. 1)
- TLS
translesion synthesis: the insertion of a base opposite a DNA adduct as well as subsequent elongation
- DNAP
DNA polymerase
- S1-dNTP
dNTP active shape 1
- S2-dNTP
dNTP active shape 2
- MD
molecular dynamics
- TT-CPD
thymine–thymine cyclopyrimidine dimer
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier's archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- 1.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 3.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 4.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-Family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 6.Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- 7.Yang W. Damage repair DNA polymerases. Curr Opin Struct Biol. 2003;13:23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 8.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 9.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-Family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 11.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-Family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 12.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis–syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 13.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;25:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H. A structural gap in Dpo4 supports mutagenic bypass of a major benzo [a]pyrene dG adduct in DNA through template misalignment. Proc Natl Acad Sci USA. 2007;104:14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 16.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cis-platin by DNA polymerase eta. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 17.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 18.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;23:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:25–42. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J Mol Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Yu X, Hu P, Broyde S, Zhang Y. A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J Am Chem Soc. 2007;129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey RG. Polycyclic Aromatic Hydrocarbons: Chemistry and Cancer. Wiley–VCH Inc.; New York: 1997. [Google Scholar]
- 23.Dipple A. Polycyclic aromatic hydrocarbon carcinogens. In: Harvey RG, editor. Polycyclic Aromatic Hydrocarbons and Carcinogenesis. American Chemical Society Press; Washington, DC: 1985. pp. 1–17. [Google Scholar]
- 24.Pfeifer GP, Hainaut P. On the origin of G-to-T transversions in lung cancer. Mutat Res. 2003;526:39–43. doi: 10.1016/s0027-5107(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez H, Loechler EL. Mutagenesis by the (+)-anti-diol epoxide of benzo[a]-pyrene: what controls mutagenic specificity? Biochemistry. 1993;32:373–383. doi: 10.1021/bi00058a009. [DOI] [PubMed] [Google Scholar]
- 26.Xie Z, Braithwaite E, Guo D, Zhao B, Geacintov NE, Wang Z. Mutagenesis of benzo [a]pyrene diol epoxide in yeast: requirement for DNA polymerase zeta and involvement of DNA polymerase eta. Biochemistry. 2003;42:11253–11262. doi: 10.1021/bi0346704. [DOI] [PubMed] [Google Scholar]
- 27.Yoon JH, Lee CS, Pfeifer GP. Simulated sunlight and benzo[a]pyrene diol epoxide induced mutagenesis in the human p53 gene evaluated by the yeast functional assay: lack of correspondence to tumor mutation spectra. Carcinogenesis. 2003;24:113–119. doi: 10.1093/carcin/24.1.113. [DOI] [PubMed] [Google Scholar]
- 28.Schiltz M, Cui XX, Lu YP, Yagi H, Jerina DM, Zdzienicka MZ, Chang RL, Conney AH, Wei SJ. Characterization of the mutational profile of (+)-7R,8S-dihydroxy-9S, 10R-epoxy-7,8,9 10-tetrahydrobenzo[a]pyrene at the hypoxanthine (guanine) phosphoribosyltransferase gene in repair-deficient Chinese hamster V-H1 cells. Carcinogenesis. 1999;20:2279–2286. doi: 10.1093/carcin/20.12.2279. [DOI] [PubMed] [Google Scholar]
- 29.Seo KY, Nagalingam A, Loechler EL. Mutagenesis studies on four stereoisomeric N2-dG benzo[a]pyrene adducts in the identical 5′-CGC sequence used in NMR studies: although adduct conformation differs, mutagenesis outcome does not as G → T mutations dominate in each case. Mutagenesis. 2005;20:441–448. doi: 10.1093/mutage/gei061. [DOI] [PubMed] [Google Scholar]
- 30.Shen X, Sayer JM, Kroth H, Ponten I, O'Donnell M, Woodgate R, Jerina DM, Goodman MF. Efficiency and accuracy of SOS-induced DNA polymerases replicating benzo[a]pyrene-7,8-diol 9 10-epoxide A and G adducts. J Biol Chem. 2002;277:5265–5674. doi: 10.1074/jbc.M109575200. [DOI] [PubMed] [Google Scholar]
- 31.Yin J, Seo KY, Loechler EL. A role for DNA polymerase V in G-to-T mutagenesis from the major benzo[a]pyrene N2-dG adduct when studied in a 5′-TGT sequence in Escherichia coli. DNA Rep. 2004;3:323–334. doi: 10.1016/j.dnarep.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Lenne-Samuel N, Janel-Bintz R, Kolbanovskiy A, Geacintov NE, Fuchs RP. The processing of a benzo(a)pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol Microbiol. 2000;38:299–307. doi: 10.1046/j.1365-2958.2000.02116.x. [DOI] [PubMed] [Google Scholar]
- 33.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo KY, Nagalingam A, Miri Shadi, Yin Jun, Kolbanovskiy Alexander, Shastry Anant, Loechler EL. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Rep. 2006;5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 36.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc Natl Acad Sci USA. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 38.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase eta. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 39.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta. Proc Natl Acad Sci USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee CH, Chandani S, Loechler EL. Homology modeling of four lesion-bypass DNA polymerases: structure and lesion bypass findings suggest that E. coli pol IV and human Pol k are orthologs, and E. coli pol V and human Pol η are orthologs. J Mol Graph Model. 2006;25:87–102. doi: 10.1016/j.jmgm.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Chandani S, Lee CH, Loechler EL. Molecular modeling benzo[a]pyrene N2-dG adducts in two partially overlapping active sites of the Y-Family DNA polymerase Dpo4. J Mol Graph Model. 2007;25:658–670. doi: 10.1016/j.jmgm.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 46.Papanicolaou C, Ripley LS. An in vitro approach to identifying specificity determinants of mutagenesis mediated by DNA misalignments. J Mol Biol. 1991;221:805–821. doi: 10.1016/0022-2836(91)80177-v. [DOI] [PubMed] [Google Scholar]
- 47.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]


