Abstract
Osterix (Osx) is an osteoblast-specific transcription factor required for osteoblast differentiation and bone formation. Osx null mice develop a normal cartilage skeleton but fail to form bone and to express osteoblast-specific marker genes. To better understand the control of transcriptional regulation by Osx, we identified Osx-interacting proteins using proteomics approaches. Here, we report that a Jumonji C (JmjC)-domain containing protein, called NO66, directly interacts with Osx and inhibits Osx-mediated promoter activation. The knockdown of NO66 in preosteoblast cells triggered accelerated osteoblast differentiation and mineralization, and markedly stimulated the expression of Osx target genes. A JmjC-dependent histone demethylase activity was exhibited by NO66, which was specific for both H3K4me and H3K36me in vitro and in vivo, and this activity was needed for the regulation of osteoblast-specific promoters. During BMP-2-induced differentiation of preosteoblasts, decreased NO66 occupancy correlates with increased Osx occupancy at Osx-target promoters. Our results indicate that interactions between NO66 and Osx regulate Osx-target genes in osteoblasts by modulating histone methylation states.
Keywords: demethylase, Jumonji, NO66, osteoblast, Osterix
Introduction
Bone formation occurs through two distinct processes. Many skeletal elements form by endochondral ossification, which involves a cartilage intermediate. The other skeletal elements, which mainly consist of craniofacial bones, are formed by intramembranous ossification, whereby bones form directly from mesenchymal condensations (Fang and Hall, 1997). Despite these differences, osteoblast differentiation is controlled by an identical set of transcription factors: Runx2, Osterix (Osx), and β-catenin that have essential roles in both processes (Ducy et al, 1997; Nakashima et al, 2002; Hill et al, 2005; Komori, 2006; Rodda and McMahon, 2006).
Runx2 is required at an early step and Osx is needed at a later step of osteoblast differentiation. The Osx is specifically expressed in osteoblast lineage cells and at lower levels in prehypertrophic chondrocytes. The Osx contains a proline- and serine-rich transactivation domain located in the N-terminal part of the protein and three DNA-binding C2H2-type zinc fingers at its C-terminus. Osx-null mice, which die at birth, have a normal cartilage skeleton, including the development of normal hypertrophic cartilage, but completely lack bone formation. In all skeletal elements of Osx-null embryos, precursor cells are arrested at their differentiation and consequently fail to express osteoblast-specific marker genes (Nakashima et al, 2002). Several factors, including HDACs, Twist 1 and 2, ATF4, NFATc, Msx, and Dlx also have important roles in the differentiation and function of osteoblasts (Nakashima and de Crombrugghe, 2003; Hassan et al, 2004; Vega et al, 2004; Yang and Karsenty, 2004; Koga et al, 2005; Cheng et al, 2008).
In addition to the various classes of transcription factors, the transcriptional control of gene expression also depends on the state of histone modifications, which provides a dynamic control of chromatin activity. Methylation of H3K9, H3K27, or H4K20 is often associated with inactive chromatin, whereas methylation of H3K4, H3K36, H3K79, and H3R17 is largely associated with active gene transcription (Strahl and Allis, 2000; Lachner and Jenuwein, 2002; Martin and Zhang, 2005).
The identification of histone lysine demethylases, including lysine specific demethylase1 (LSD1) and jumonji (JmjC)-domain containing proteins has provided strong evidence that histone methylation is reversible and dynamically regulated, much like histone acetylation and phosphorylation (Shi et al, 2004; Klose et al, 2006). Several JmjC-domain-containing histone demethylases (JHDMs) reverse methylation states at lysines 4, 9, 27, 36 and 79 of H3 and thus regulate the activation of target genes in chromatin (Fodor et al, 2006; Klose et al, 2006; Tsukada et al, 2006; Whetstine et al, 2006; Yamane et al, 2006; Agger et al, 2007; De Santa et al, 2007; Iwase et al, 2007; Lan et al, 2007; Lee et al, 2007).
In osteoblasts, histone acetylases and deacetylases (HDACs) control the activity of Runx2 (Shen et al, 2002; Schroeder et al, 2004; Kang et al, 2005). In addition, HDAC4 was shown to inhibit the differentiation of hypertrophic chondrocytes by negatively regulating Runx2 activity (Vega et al, 2004). Moreover, the histone lysine methytransferase Wdr5 has been reported to be involved in osteoblast differentiation (Zhu et al, 2008), but no histone demethylases have been shown to regulate osteoblast-specific genes.
In a screen for polypeptides that associate with Osx, we identified the JmjC domain-containing protein NO66. This protein was previously described as a component of nucleoli in Xenopus embryos that co-fractionated biochemically with preribosomal particles (Eilbracht et al, 2004). The NO66 is highly conserved in eukaryotes, and immunofluorescence studies revealed that the protein is present in both the granular part of nucleoli and in the nucleoplasm of tissue culture cells (Eilbracht et al, 2004). However, not much is known about its function. Here we show that NO66, which is present in all bones as observed on the basis of histochemical studies, inhibits transcription of Osx-dependent promoters. We present evidence that NO66 is a histone demethylase for H3K4me and H3K36me, and that this activity is required for the inhibition of the transcriptional activation by Osx. Importantly, our experiments strongly support a physiological role for NO66 during osteoblast differentiation.
Results
JmjC-domain containing protein NO66 is an Osx-interacting protein
To define the regulation of Osx transactivation in osteoblast differentiation, we stably transfected Flag–HA–Osx into the mouse C2Cl2 line, which differentiates into osteoblasts on BMP-2 addition. The induction of Flag–HA–Osx in this cell line (referred to as C2-Osx) occurs after tetracycline withdrawal. Using tandem affinity purification followed by SDS–PAGE fractionation and mass spectrometry, we identified Osx-interacting polypeptides in nuclear extracts of this cell line. One of the proteins co-purifying with Flag–HA–Osx was the Jumonji C (JmjC) domain-containing protein NO66 (Figure 1A).
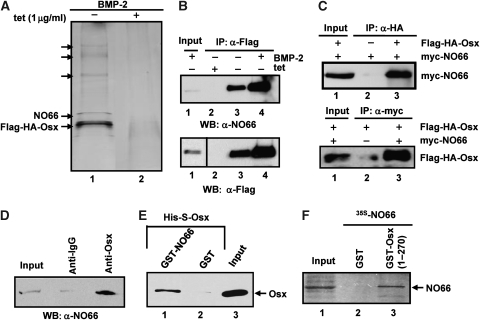
Figure 1.
Identification of NO66 as an Osx-interacting protein. (A) Polypeptides co-purified with Flag–HA–Osx by tandem immunoaffinity from nuclear extracts of BMP-2-treated C2-Osx cells were separated on a 4–20% gradient SDS gel followed by silver staining. (B) Total cell lysates of C2-Osx cells, grown as indicated above, from each lane were immunoprecipiated using α-Flag agarose followed by immunoblot with α-NO66 antibody (top panel). The blot was stripped and then probed with α-Flag antibody to detect Flag–HA–Osx immunoprecipitates (bottom panel). Different exposure time was used for each antibody. The faint band in the input lane was from a diluted lysate sample. A longer exposure time was used to detect input signal in bottom panel. (C) Co-immunoprecipitation of myc-NO66 and HA-Osx in lysates of co-transfected HEK293T cells. Top panel—immunoprecipitation of Flag–HA–Osx with HA-tagged antibody followed by immunoblotting with myc-tagged antibody for myc-NO66, Bottom panel—immunoprecipitation of myc-NO66 using anti-myc antibody followed by immunoblotting with HA-tagged antibody for Flag–HA–Osx. Lane 1 in both cases represents 5% of the cell lysate used in lanes 2 and 3. (D) Interactions of endogenous NO66 and Osx in rat osteosarcoma UMR 106 cells were analysed by immunoprecipitation of cell lysates with rabbit α-Osx followed by immunoblot with α-NO66. Input lane represents 10% of the lysate used in other two lanes. (E) GST pulldown of purified Osx by purified GST-NO66. The complexes retained by glutathione Sepharose beads were separated by SDS–PAGE and analysed by western blot with S-epitope antibody for Osx. Lane 3 represents 10% of the purified Osx used in lane 1 and 2. (F) GST pull down of labelled NO66 by recombinant GST–Osx (1–270). Input lane represents 10% of lysate used in the two lanes.
The co-purification of NO66 with Osx in lysates of C2-Osx cells grown without tetracycline and with or without BMP-2 treatment was confirmed by immunoprecipitation with α-Flag antibody followed by western blot using α-NO66 antibody. No NO66 was detected after immunoprecipitation of tetracycline-treated C2-Osx cell lysates, confirming that NO66 was co-immunoprecipitated with Flag–HA–Osx, and that NO66 and Osx are part of a common complex (Figure 1B). In a similar experiment, other co-purified proteins, such as Yb-1 and Flightless, were also detected in α-Flag immunoprecipitates, thus confirming our mass spectrometry analysis (Supplementary Figure S1). An association of NO66 and Osx was also observed in HEK293T cells co-transfected with Flag–HA–Osx and myc–NO66 expression vectors (Figure 1C). Myc-tagged NO66 was found in the α-HA immunoprecipitates of whole cell lysates (Figure 1C, top panel), and Flag–HA–Osx was present in α-myc immunoprecipitates (bottom panel). In addition, co-immunoprecipitation of endogenous Osx and endogenous NO66 was also observed in rat osteosarcoma UMR 106 cells (Figure 1D), which are highly differentiated osteoblasts, further indicating that NO66 is associated with Osx in differentiated osteoblasts. Purified recombinant GST–NO66 retained purified recombinant Osx in a pull-down assay (Figure 1E) indicating that these two proteins interacted directly with each other. Moreover, another pull-down experiment showed that 35S-labelled NO66 interacted with a segment of Osx (1–270) that contains the transactivation domain but lacks the DNA-binding domain, indicating that this Osx segment was sufficient for NO66 interactions (Figure 1F).
NO66 inhibits the transcriptional activation of Osx
Our previous studies showed that Osx is expressed in all endochondral and membranous skeletal elements during mouse embryonic development beginning at E13.5 and continuing after birth (Nakashima et al, 2002). In situ hybridizations using an anti-sense RNA probe showed that NO66 RNA co-localized with Osx RNA starting at E14.5. The NO66 RNA was present in all developing endochondral and membranous bones, such as E15.5 vertebrae and mandible and E18.5 femur, tibia, and fibula (Figure 2A). Immunohistochemistry of skeletal tissue sections using an NO66-specific antibody confirmed the presence of NO66 in developing bones (Figure 2B). Northern blot analyses using a human NO66 cDNA probe also showed expression of NO66 in the human Saos-2 and rat UMR106 ostesarcoma cell lines, as well as in RNA preparations from human trabecular bone (Supplementary Figure S2). Others have observed expression of NO66 in several mouse tissues and cell lines (Eilbracht et al, 2004). All together these studies indicate that NO66 is widely, but not ubiquitously, expressed.
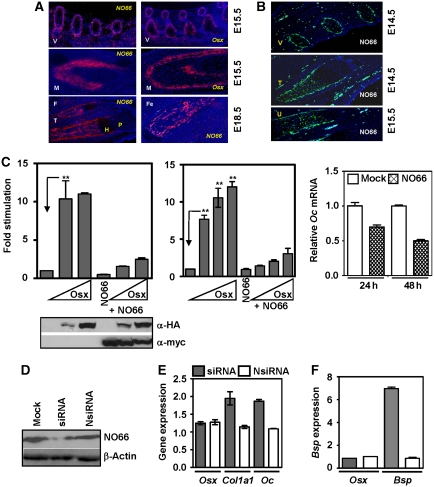
Figure 2.
NO66 is expressed in developing bone and inhibits Osx-target gene activation. (A) NO66 is expressed in all developing mouse skeletal tissues as shown by in situ RNA hybridizations of NO66 in mouse vertebrae (V) and mandible (M) at E15.5, and fibula (F), tibia (T), and femur (Fe) at E18.5. H, hypertrophic cartilage; P, prehypertrophic cartilage. Expression of Osx is also shown in parallel sections of mouse vertebrae (V) and mandible (M) at E15.5. (B) Immunohistochemistry to monitor NO66 protein expression in vertebrae (V) and tibia (T) at E14.5, and ulna (U) at E15.5. (C) NO66 inhibits transcriptional activation by Osx. Stimulation of a 2-kb mouse Bsp promoter (left) and a 1.1-kb mouse Osteocalcin promoter (middle) by increasing amounts of Osx (pTriEX Flag–HA–Osx) in HEK293T cells and inhibition by co-expression of NO66. Levels of Osx and NO66 in transfected cells were analysed by western blotting (below, left panel). The P-value (<0.05) for statistical difference was calculated using one way ANOVA test and is indicated by asterisks. Quantitative PCR of Osteocalcin mRNA in BMP-2-treated C2C12 cells transfected with a NO66 expression plasmid (right panel). The error bars represent s.e.m. values of at least triplicate experiments. (D) Western blot of the knockdown of NO66 in lysates of BMP-2-treated C2C12 cells by a NO66-specific RNA. NsiRNA is non-specific siRNA. β-Actin serves as loading control. (E) Increased expression of Osx target genes by knockdown of NO66. Representative quantitative RT–PCR of Osx, Col1a1, and Osteocalcin (Oc) RNAs in C2C12 cells transfected with siRNAs and treated with BMP-2. (F) Same as (E), but for Bone sialoprotein (Bsp) and Osx RNAs in MC3T3 cells. RNA levels were measured relative to those of mock-transfected cells that were also treated with BMP-2.
In Osx null embryos, expression of osteoblast-specific marker genes such as Osteocalcin (Oc) and Bone sialoprotein (Bsp) is completely lost, indicating that Osx is required for the expression of these genes (Nakashima et al, 2002). To further examine the effects of Osx on transcription of osteoblast-specific promoters, we transfected HEK293T cells with an Osx expression vector. Osx strongly stimulated the 2-kb Bsp promoter (Figure 2C, left panel) as well as the 1.1-kb Osteocalcin promoter in a dose-dependent manner (middle panel). Although the basal activity of these reporters showed little change in the presence of NO66, co-expression of NO66 with Osx inhibited Osx-mediated activation of the reporters by 70–80%. The overexpression of NO66 in these cells did not affect the level of Osx (left, bottom panel). In control experiments, NO66 also did not inhibit the activity of different reporters driven by Sox9, ATF6, or GAL4-DBD-VP16 (Supplementary Figure S3A–C). These results indicate that although NO66 markedly inhibited transcriptional activation by Osx, it is not a general transcriptional inhibitor.
Previously we have shown that a proline-rich activation domain spanning amino-acid residues 27–192 in Osx was required for the activation of the 5XGAL4-Luc reporter (Nakashima et al, 2002). Using GAL4-DBD-Osx (27–192) and 5XGAL4-Luc in reporter assays, addition of either sodium butyrate (inhibitor of class I and II HDACs) or nicotinamide (inhibitor of class III HDACs) did not relieve the NO66-mediated inhibition of Osx-dependent transactivation (Supplementary Figure S3D). These data suggest that the inhibitory effects of NO66 on Osx-dependent transcription were not mediated by HDACs and that NO66 did not recruit HDACs to repress Osx-mediated transactivation.
To verify that NO66 inhibits the expression of endogenous osteoblast-specific genes, we measured expression of the endogenous Osteocalcin (Oc) gene in BMP-2-induced osteoblasts that overexpressed NO66. The transfection of C2C12 cells with NO66 expression vector followed by BMP-2 treatment for 24 and 48 h decreased Oc mRNA by almost 50% compared with mock-transfected cells after 48 h of BMP-2 treatment (Figure 2C, right panel).
To complement the above experiments, we next questioned whether loss of NO66 expression would result in increased expression of Osx-dependent genes in osteoblasts. The C2C12 cells were first either mock transfected or transfected with control or NO66-specific siRNAs, followed by treatment with BMP-2 to induce osteoblast differentiation. Immunoblots of whole-cell lysates using an α-NO66 antibody indicated that the NO66-specific siRNA, but not the control siRNA (Nsi-non-specific), significantly inhibited NO66 expression (Figure 2D). Strikingly, knockdown of NO66 by siRNA increased the levels of Col1a1 and Oc RNAs compared with those in control siRNA-transfected C2C12 cells (Figure 2E). Similar experiments were performed in pre-osteoblast mouse cell line, MC3T3-E1, which can also differentiate into osteoblasts after BMP-2 treatment. The transfection of NO66-specific siRNA followed by BMP-2 addition led to marked stimulation of Bsp expression, when compared with control siRNA-transfected cells, which were also treated with BMP-2 (Figure 2F). However, expression of Osx was not affected by transfection of either NO66-specific or control siRNAs in either cell line (Figure 2E and F). Thus, knockdown of NO66 in osteoblasts caused increased expression of osteoblast-specific marker genes, suggesting an important role for NO66 in the regulation of osteoblast differentiation.
Knockdown of NO66 accelerates osteoblast differentiation and maturation
To further test our hypothesis that NO66 has a physiological role as a negative regulator of osteoblast differentiation and maturation, we generated MC3T3 cell lines expressing stably transfected NO66-specific and non-specific shRNAs. Levels of NO66 were markedly decreased in NO66sh cells compared with control Nsh cells (Figure 3A). Histochemical and quantitative analyses showed that the production of alkaline phosphatase, an early marker of osteoblast differentiation, was accelerated in NO66-shRNA-expressing cells (NO66sh) compared with control Nsh cells, when these cells were cultured in osteogenic medium for the indicated periods (Figure 3B and C). Further, the extracellular matrix mineralization, a late event, as shown by alizarin red S staining was also accelerated in NO66sh-treated cells compared with control Nsh-treated cells (Figure 3D). These results suggest that knockdown of NO66 in preosteoblast MC3T3 cells caused these cells to differentiate earlier than control cells.
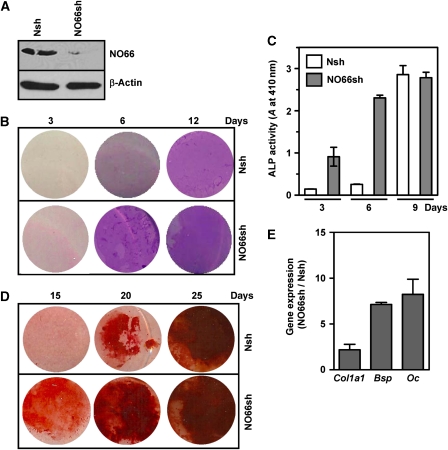
Figure 3.
Stable depletion of NO66 by shRNA accelerates osteoblast differentiation. (A) Immunoblot of NO66 in MC3T3 cell line stably transfected with NO66-specific shRNA (NO66sh) compared with control cells stably transfected with non-specific shRNA (Nsh). β-Actin serves as loading control. (B, C) Left, representative staining of alkaline phosphatase activity (ALP); right, quantification of ALP, and (D) representative staining of extracellular matrix deposition by 1.4% Alizarin S red in NO66sh and Nsh cells after culture for indicated times in osteogenic medium. Post-staining, photograph was taken with computer-assisted scanning and images with equal area in each case are shown. (E) Representative quantitative RT–PCR of Bsp and Oc RNAs after 10 days culture of these cells in osteogenic medium.
In addition, expression levels of the Osx-dependent Bsp and Osteocalcin matrix forming genes were markedly stimulated in NO66sh cells relative to in control cells (Figure 3E), further indicating our interpretation that acceleration of osteoblast differentiation in NO66-deficient cells was due to increased expression of these matrix genes. All together, these data strongly indicate that knockdown of NO66 accelerates the rate of osteoblast differentiation and maturation of MC3T3 cells.
NO66 has histone demethylase activity that is required for inhibition of Osx-dependent activation
Given the existence of the JmjC domain in NO66, and the presence of conserved amino acids that have been implicated as co-factor-binding sites in several other JmjC-containing proteins (Klose et al, 2006 and Figure 4A), we tested NO66 for histone demethylase activity. Using calf thymus histones as substrates, we found that recombinant NO66 very efficiently demethylated H3K4me3 and H3K4me1, and to a lesser extent H3K4me2 (Figure 4B). NO66 also demethylated H3K36me3 and H3K36me2. In contrast, no activity was observed towards H3K9me3 or H3K27me3 (Figure 4C, lanes 1–2 and 3–4, respectively). These results showed that NO66 is a histone demethylase specific for lysine 4 and 36 of histone H3.
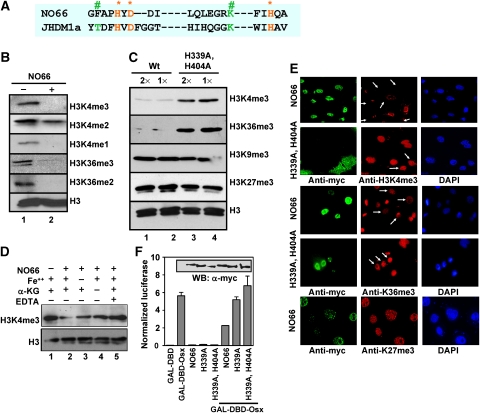
Figure 4.
NO66 has H3K4me and H3K36me demethylase activity. (A) Comparison of conserved catalytic amino acids within JmjC domain of NO66 and JHDM1A (* sites for Fe++ binding and # for a-ketoglutarate binding). (B) NO66 histone demethylase activity was assayed with recombinant NO66 purified from insect cells and calf thymus bulk histones (Sigma) followed by immunoblotting with site-specific methyl antibodies. (C) The demethylase activity of NO66 mutant protein with double substitution mutations (H339A, H404A) was compared with the activity of wild-type NO66 (Wt). The GST-tagged proteins were expressed and purified from Sf9 cells (see details in Supplementary Figure S3). The demethylase activity was assayed by western blotting using specific antibodies as indicated in the panel. All lanes in this panel came from the same gel. (D) Recombinant His-tagged NO66, affinity purified from transfected HEK 293T cells, was used for demethylase reaction assay in the presence or absence of co-factors and EDTA (50 μM) as indicated in the panel. Ascorbate was added to each reaction mixture. The demethylase activity was assayed by western blotting. (E) Overexpression of wild-type NO66 in transfected COS7 cells decreased the intensity of H3K4me3 and H3K36me3 (arrows) immunostaining, but not that of H3K27me3, whereas mutant NO66 (H339A, H404A) did not decrease the intensity of H3K4me3 and H3K36me3. Both wild-type (WT and mutant NO66 were tagged with myc-epitope at their C-terminus and their expression was detected with anti-myc antibody. (F) NO66 demethylase activity mediated the inhibition of the transcriptional activity of Osx. DNA transfection experiments were performed in HEK 293T cells with WT NO66 and NO66 demethylase mutants (H339A, and H339 plus H404A), as well as with GAL4–DBD–Osx (27–192) using the 5xGAL4-Luc reporter. Immunoblot shows expression levels of NO66 and mutant proteins (inset).
We next tested whether, as in other JHDMs, the potential co-factor binding sites in the JmjC domain of NO66 were essential for demethylase activity by comparing the activity of wild-type NO66 with that of mutant NO66 protein (see Supplementary Figure S4 for enzyme preparation). Our results showed that a mutant protein bearing two histidine-to-alanine substitutions in conserved sites critical for ferrous binding (H339A, H404A; Figure 4A) showed no demethylase activity towards H3K4me3 or K36me3 (Figure 4C, lanes 3 and 4). This indicated that the mutations crippled the activity of NO66. The omission of co-factor α-KG from the demethylase reaction mixture inhibited the demethylase activity of NO66 (Figure 4D, compare lane 2 and 4). In the absence of added Fe++ ions the inhibition of NO66 activity was less pronounced, maybe because intrinsic Fe++ ions remained bound to our preparation of NO66 (compare lane 2 and 3). However, addition of EDTA, which chelates Fe++ ions, markedly inhibited the demethylase activity of NO66 for H3K4me3 (compare lane 2 and 5). The level of H3K4me3 in the presence of EDTA was similar to that present in control reaction with no enzyme (lane 1 and 5). These data strongly suggest that NO66 is a co-factor-dependent JmjC histone demethylase and that co-factors Fe++ ions and α-KG are needed for full enzymatic activity.
To determine whether NO66 has demethylase activity in cultured cells, we overexpressed wild-type NO66 or a mutant in COS7 cells and monitored histone methylation levels by immunofluorescence using site-specific antibodies. The overexpression of wild-type NO66 clearly reduced the intensity of H3K4me3 and to lesser extent H3K36me3 staining, but not the intensity of H3K27me3 staining (Figure 4E). Similarly, the intensity of anti-H3K4me2, K4me1 and K36me2 staining was also reduced by overexpression of wild-type NO66 in COS7 cells (Supplementary Figure S5). In contrast, overexpression of the NO66 (H339A/H404A) mutant, which was inactive in the in vitro assay, did not reduce immunostaining of H3K4me3 or H3K36me3 (Figure 4E). Thus, both in vitro assays and in-cell assays indicate that NO66 exhibited H3K4- and H3K36-specific demethylase activity.
To test whether the demethylase activity of NO66 was required for the inhibition of transcriptional activation by Osx, we examined the effect of NO66 demethylase mutants on Osx-dependent reporter activation using 5XGAL4-Luc co-transfected with Osx (27–192) and wild-type or mutant NO66. In contrast to wild-type NO66, which exhibited demethylase activity for H3K4me3 and H3K36me3 (Figure 4) and repressed Osx-mediated activation (Figure 4F and Supplementary Figure S3D), NO66 mutants (H339A or H339A/H404A) did not inhibit Osx-mediated transcriptional activation (Figure 4F).
Domain of NO66 that interacts with the activation domain of Osx
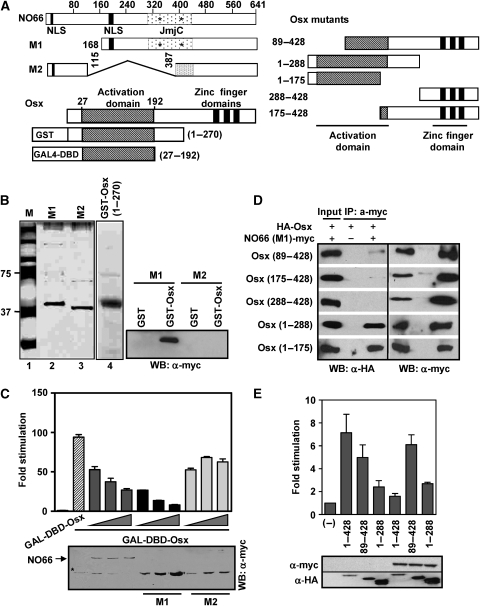
As described above, Figure 1F showed that the transcription activation domain of Osx interacted with NO66. To further identify the domain of NO66 that interacts with the activation domain of Osx, we performed an in vitro interaction assay using recombinant polypeptides. Two NO66 mutants (M1 and M2) were tested for their ability to physically interact with GST–Osx (1–270) in pull-down assay (Figure 5A and B). The NO66 mutant M1, which retained an intact JmjC domain and the C-terminus of NO66, interacted with GST–Osx (1–270), whereas the mutant M2, which had an internal deletion including a part of the JmjC domain, did not interact with GST–Osx (1–270), indicating that the segment between 168 and 386 containing the JmjC domain of NO66 was necessary for strong interactions with the activation domain of Osx.
Figure 5.
JmjC domain of NO66 interacts with the activation domain of Osx. (A) Schematic of NO66 and Osx and mutant forms of these proteins used in this study. Conserved histidines within the catalytic JmjC domain are indicated by asterisk. (B) Left, recombinant His-tagged NO66 mutants were affinity purified from transfected HEK293T cells and separated on 4–20% gradient SDS-gel followed by silver staining. Lane 2 and 3 represent 15% of proteins used in right panel. Lane 4 shows preparation of recombinant GST–Osx (1–270) in Escherichia coli, which was analysed on a separate gel followed by Coomassie staining. Lane 1 refers to protein markers (M). Right, GST pull down of NO66 mutants (M1 and M2) by purified GST–Osx (1–270) followed by western blot with myc-epitope antibody to detect mutant NO66. (C) JmjC domain of NO66 was essential for inhibition of Osx activation, shown by reporter assays using 5xGAL4-Luc co-transfected with increasing amounts of WT or mutant NO66 (50–200 ng), and GAL4–DBD–Osx (27–192). Western blot showing level of expressed proteins in the lysates of transfected cells (bottom). Asterisk indicates non-specific bands co-migrating with mutant M1 and M2. (D) The N-terminus segment of Osx interacts with NO66. HEK 293T cells were transfected with Osx mutants as indicated in the panel together with NO66 mutant M1. Co-immunoprecitations were performed in lysates of transfected cells using myc antibody followed by western blotting with HA antibody (left panel), and also with myc antibody to detect the immunoprecipitates (right panel). (E) NO66 functionally interacts with N-terminus of Osx. Reporter assay was carried out using Bsp-Luc co-transfected with WT and mutant Osx, and NO66 (top). Western blot showing level of expressed proteins in the lysates of transfected cells (bottom).
Next we examined the effect of full-length NO66 or M1 and M2 mutants on the ability of a GAL4–Osx fusion polypeptide (27–192) to activate a 5XGAL4-Luc reporter gene. This domain of Osx was shown previously to be necessary and sufficient for the activation of this reporter (Nakashima et al, 2002). The co-transfection of GAL4–Osx (27–192) with wild-type NO66 or mutant M1 resulted in dose-dependent inhibition of 5XGAL4-Luc reporter activity, thereby strongly suggesting that inhibition by NO66 was mediated through the activation domain of Osx (27–192) (Figure 5C). It should be noted that both polypeptides were able to interact physically with Osx (Figures 1E and F, and 5B). In contrast, the mutant M2, which did not interact with Osx owing to a partial deletion of the JmjC domain, showed weak inhibition of Osx-dependent activation even with the highest levels tested.
To map the NO66-interacting segment present in Osx, we used Osx deletion mutants together with NO66 mutant M1 in co-immunoprecipitation experiments (Figure 5D). These results indicated that Osx C-terminal truncation mutants (1–175 and 1–288) that lacked the zinc finger DNA-binding domains co-immunoprecipitated with the NO66 M1 mutant, suggesting that an Osx segment spanning amino acids 1–175 residues was sufficient to interact with NO66. However, Osx mutants bearing N-terminal deletions (89–428, 175–428, and 288–428) were not co-immunoprecipitated by the NO66 mutant. Two of these mutations (89–428 and 175–428) remove parts of the activation domain in Osx. These results strongly indicate that an N-terminal segment of Osx that includes the activation domain of Osx is required for interactions with NO66.
Furthermore, we examined the effects of NO66 on the ability of the Osx mutants (89–428 and 1–288) to transcriptionally activate a Bsp-Luc reporter (Figure 5E). The Osx (89–428) mutant was still able to stimulate Bsp-Luc reporter activity, even though the activation domain is partially truncated in this mutant, though the activation was less than that observed with wild-type Osx (1–428). The co-transfection of NO66 inhibited the activity of wild-type Osx in the reporter assay, but NO66 was not able to inhibit the activity of the Osx (89–428) mutant, consistent with the inability of this mutant to interact with NO66. These results indicate that NO66 functionally interacts with the N-terminus of Osx; hence it is able to interact with and inhibit the activity of wild-type Osx (1–428) but not mutant Osx (89–428). Another mutant Osx (1–288), which contains an intact activation domain but lacks a DNA-binding domain, was unable to stimulate reporter activity, as expected, and NO66 did not further reduce the activity of this mutant. Taken together, these data suggest that under the conditions of our assay, functional interactions between NO66 and Osx are required to inhibit Osx activation of target genes.
Association of NO66 with the chromatin of Osx target genes is inversely correlated with their activation in osteoblasts
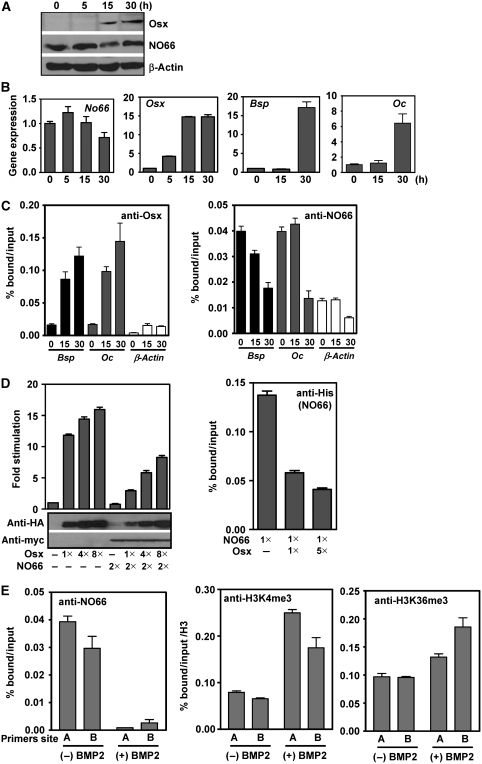
The addition of BMP-2 to MC3T3 preosteoblastic cells triggers the sequential activation of osteoblast-specific genes resulting in the differentiation of these cells into osteoblasts (Katagiri et al, 1994; Nakashima et al, 2002; Figure 6A and B). Before BMP-2 addition, the RNA levels of Osx, Bsp, and Oc were either undetectable or very low. At 15 h after BMP-2 addition, the levels of Osx RNA were already high but levels of Bsp and Oc RNAs were still very low. At 30 h after BMP-2 addition, RNA levels of Osx, Bsp, and Oc were all high. In contrast, cellular levels of NO66 protein levels showed little change during BMP-2-induced differentiation, although some decrease in NO66 RNA level was observed at 30 h after BMP-2 addition (Figure 6A and B).
Figure 6.
Association of NO66 with Osx target genes prevents their activation in osteoblasts. (A) Immunoblot analyses show the levels of Osx and NO66 expression in lysates of MC3T3 cells at different times after BMP-2 addition. β-Actin served as loading control. (B) NO66, Osx, Bsp, and Oc RNA levels in MC3T3 cells at different times after BMP-2 addition. As measured by quantitative RT–PCR. (C) Chromatin immunoprecipitation (ChIP) experiments showing the relative binding of Osx (left) and NO66 (right) at promoters of the Bsp and Oc genes in MC3T3 cells at different times after BMP-2 addition. Primers used correspond to region approximately −1.5 kb upstream of the Bsp and at −0.5 kb upstream of the Oc gene. (D) Left, inhibition of Osx activation by NO66 depends on intracellular levels of Osx. Reporter assays using Bsp-Luc in HEK 293T cells after transfection with NO66 and increasing levels of Osx (1 × =25 ng). Right, ChIP assays were performed using anti-His antibody for NO66 after transfection of HEK 293 cells as described in left panel (1 × =250 ng). (E) Representative ChIP experiments of NO66 (left), H3K4me3 (middle), and H3K36me3 (right) occupancy at the Bsp gene before and after BMP2 treatment of MC3T3 cells for 40 h. A and B refer to two chromatin segments within the promoter (−1.0 kb, A) and the coding region (+12 kb, B) of the Bsp gene. Error bars represent s.e.m. values of triplicate qPCR of representative ChIP assays. The data are presented as percent of input after subtracting control IgG values. Levels of H3K4me3 and H3K36me3 were normalized with bound histone H3 after immunoprecipitation with histone H3 antibody.
To gain further insights into the physiological role of NO66 during osteoblast differentiation, we tested whether changes occurred in the occupancy of Osx and NO66 at the promoters of the Osx target genes Bsp and Oc. Chromatin immunoprecipitations from MC3T3 cells after they were treated with BMP-2 for 0, 15, and 30 h (Figure 6C) indicated that NO66 occupancy at the two promoters was high before treatment, whereas as expected, occupancy of Osx at these promoters was negligible. After 15 h, Osx occupancy had markedly increased, whereas NO66 occupancy started to decrease at the Bsp promoter, but interestingly, not yet at the Oc promoter. At 30 h after BMP-2 treatment, Osx occupancy continued to increase whereas NO66 occupancy was markedly reduced. The expression of Bsp and Oc genes was seen only after 30 h of BMP-2 addition, at a time when significant depletion of NO66 from promoters of these genes was observed. The interactions of Osx or NO66 with the promoter of the β-actin gene appeared to be within background levels, confirming the specificity of our ChIPs. Other experiments showed that occupancy of Osx was much higher at the promoter of the Oc gene than at the 3′UTR region (Supplementary Figure S6), which is consistent with other biochemical experiments, indicating that the binding of Osx occurs at the promoter of Oc gene and that mutations in the Osx-binding site results in a loss of Osx-dependent promoter activity (Sinha et al, unpublished observations). Thus, these results revealed a marked correlation between decreased NO66 occupancy and increased Osx occupancy at two Osx target promoters.
These observations suggest that Osx and NO66 may compete for occupancy of target promoters. To test this idea, we performed a reporter assay using the same osteoblast-specific reporter (Bsp-Luc) used in the experiment described in Figure 2C (left). In these transfection experiments, an amount of NO66 plasmid (2X, Figure 6D) that was sufficient to inhibit Osx activation was used together with increasing amounts of Osx (Figure 6D, also see Supplementary Figure S7 for titration of NO66 dose required to inhibit Osx-dependent activation). Our data showed that inhibition of the Osx-dependent activation by NO66 was overcome, at least in part, by increasing amount of Osx (Figure 6D, left, bars 6–8). We also questioned whether in these transfection experiments Osx affects the occupancy of NO66 at the promoter of the reporter gene. Hence, we performed ChIP experiments in cells co-transfected with the reporter, NO66 and Osx. Our results showed that interactions of NO66 with transfected Bsp promoter (Bsp-luciferase), which occurred in the absence of Osx, were decreased by 60–70% in the presence of Osx (Figure 6D, right panel).
In independent experiments using BMP2-treated MC3T3 preosteoblasts, we also observed that levels of H3K4me3 and H3K36me3 increased in parallel with the decrease in NO66 occupancy of the Bsp gene. Thus, depletion of NO66 at the Bsp gene was associated with enhanced methylation of histone H3 lysine residues that are linked with transcriptionally active chromatin (Figure 6E). Furthermore, levels of H3 acetylation, RNA polymerase II and Wdr5 (a BMP2-inducible histone lysine methyl transferase) increased at the promoter of Bsp gene in BMP-2-treated MC3T3 cells, consistent with active transcription of this gene (Supplementary Figure S8). However, levels of H3K27me3, which is not a substrate for NO66-mediated demethylation in vitro or in transfected cells, were not changed by the addition of BMP-2 (Supplementary Figure S9). In other control experiments, NO66 occupancy was very low at the promoters of Osx (a BMP-inducible gene) and Col2a1 genes, and was not changed by BMP-2 addition. Col2a1 is a chondrocyte-specific marker gene and a target of Sox9 activation; it remains transcriptionally inactive in osteoblast cells (data not shown). Taken together, our data suggest that NO66 interactions specifically regulate two Osx target genes, Bsp and Oc, and that expression of these genes depends on chromatin remodelling activities that mediate histone acetylation and methylation.
Interestingly, both Osx and NO66 seemed to occupy the same region of Osx target promoters at 15 h post BMP-2 addition (Figure 6C). To obtain additional evidence that Osx and NO66 are able to occupy the same chromatin fragment, we performed a re-ChIP experiment in C2-Osx cells. These data confirmed co-localization of Osx and NO66 in the same segment of the Bsp promoter (Supplementary Figure S10), suggesting the possibility that both proteins could be part of the same chromatin bound complex, consistent with the physical interactions we observed between NO66 and Osx (Figure 1).
Discussion
Our experiments provide the first evidence that transcriptional activation by Osx can be regulated by the JmjC-domain containing protein, NO66, which acts as a histone demethylase. Given the essential function of Osx in activating a large repertoire of genes in osteoblasts, our data strongly suggest that NO66 has a major role in osteoblast differentiation and function, adding a new layer of control in the regulation of the genetic programme of osteoblasts. Our results further emphasize the idea that chromatin remodelling is crucial for the control of Osx function.
The negative regulation of Osx activity by NO66 in transient expression experiments required both physical interactions between Osx and NO66, and the demethylase activity of NO66. In these experiments the chromatinization of reporter DNA templates should be less extensive than in the genome. Moreover, although the inhibition of the transcriptional activity of Osx might certainly be due to demethylation of histone H3, it could possibly also be caused by demethylation of proteins that form an active complex with Osx at the Bsp and Oc promoters or even by demethylation of Osx itself. Previous experiments by others have shown that LSD1 demethylates K370me2 in p53 to regulate its activity (Huang et al, 2007). As NO66 does not affect the activation of several other promoters by specific activators, the inhibition of Osx activity by NO66 in transient transfection experiments was highly specific, which we attribute to direct interaction between Osx and NO66.
A physiological role for NO66 during osteoblast differentiation is strongly supported by the acceleration of differentiation and the marked increase in expression of osteoblast marker genes during the spontaneous differentiation of MC3T3 preosteoblasts stably transfected with a NO66-shRNA. One possible reason among others for the premature increase in alkaline phosphatase in MC3T3 cells containing a NO66-specific shRNA would be that NO66 regulates the activity of other factors needed for upregulation of early markers in osteoblasts, a hypothesis that needs to be tested. In this context, it is important to note that ectopically expressed Osx in bone marrow stromal cells (BMSC) increased expression and activity of alkaline phosphatase (Tu et al, 2006).
During osteoblast differentiation both in intact mouse embryos and after BMP-2 addition to preosteoblasts in culture, there is a sequential activation of osteoblast marker genes starting with Runx2 and Osx and continuing later with Osx target genes (Nakashima et al, 2002). Despite the constant cellular levels of NO66 after BMP-2 addition to MC3T3 cells, the occupancy of NO66 at the endogenous Bsp and Oc promoters decreased. In contrast, occupancy of Osx, acetylation of histone H3, Wdr5, and Pol II at these promoters increased, consistent with active transcription of these genes during osteoblast differentiation. Our data further suggest that the hypothesis that depletion of NO66 from these genes could be due to recruitment of Osx and chromatin modifying enzymes involving histone or factor acetylation and methylation, thus altering the chromatin from a repressed to an active state, although, the exact mechanism by which this occurs is not known.
The conserved histidine and lysine residues that are needed for co-factor binding in other JHDMs are also present in NO66. As substitution mutations in these conserved residues abrogated the demethylation activity of NO66 and because NO66 was unable to demethylate H3K4me3 in the absence of co-factors, we conclude that, like other JHDMs, NO66 is a co-factor-dependent JmjC histone demethylase. Several other known demethylases, including JMJD2A/2B/2C (KDM4A/4B/4C) and yeast Rph1 possess dual substrate specificity for both H3K9me and H3K36me; LSD1 (KDM1) has substrate specificity for both H3K4me and H3K9me (Allis et al, 2007). Interestingly, the dual substrate specificity we observe for NO66 for H3K4me and H3K36me is so far unique in that it targets these two markers of transcriptionally active chromatin.
The NO66 belongs to the ‘JmjC-domain only' subfamily and lacks other functional motifs that are present in other JHDMs, including PHD, tudor, bromo, C5HC2- and CXXC-zinc finger, FBOX, Bright/Arid, or TPR domains. These motifs are thought to mediate interactions between JmjC-containing proteins and histones or other proteins (Klose et al, 2006). Additional studies will need to delineate domains in NO66 that mediate such interactions.
In summary, our results provide, to the best of knowledge, the first evidence that a specific JmjC family histone demethylase, NO66, controls the activity of a ‘master' transcription factor that is essential for osteoblast differentiation and bone development. Loss- and gain-of-function experiments of NO66 in osteoblasts of mice should elucidate in greater detail the physiological role of NO66 during embryonic development and post-natally.
Materials and methods
Generation of cell lines stably transfected with an inducible Osx expression vector
Mouse C2C12 cells were stably transfected with pTet-off (Clontech) and selected on neomycin (G418). The G418 selected clones from each cell line were then stably transfected with pTRE-Flag–HA–Osx and pTK-hyg plasmids. Individual colonies were selected on both G418 and hygromycin, expanded and maintained in the presence of tetracycline (Tet). We verified that in several of these clones, Flag–HA–Osx was not expressed in the presence of Tet and was highly expressed in the absence of Tet, and furthermore, that these clones expressed osteoblast-specific genes after BMP-2 treatment similarly to the parental unstransfected cells (data not shown), indicating that these clones had maintained their potential to differentiate into osteoblasts. These clones were called C2-Osx and used for further experiments.
Purification and identification of NO66 as an Osx-interacting protein
C2-Osx cells were grown in the absence or presence of tetracycline and treated with BMP-2 (300 ng/ml) in DMEM containing 5% bovine serum albumin. Cells grown in the presence of tetracycline and BMP-2 served as control. Nuclear extracts were diluted in buffer A (50 mM Tris–Cl (pH 8), 150 mM NaCl, 0.5% NP-40, 1 mM PMSF and protease inhibitor cocktail) and immunoprecipitated with α-Flag antibody M2 agarose (Sigma) at 4°C followed by elution with 3 × FLAG peptide (200 μg/ml) in buffer A. Eluted samples were further immunoprecipitated with α-HA antibody agarose (Roche) at 30°C followed by elution with HA peptide (1 mg/ml) in buffer A, precipitation with cold acetone, and separation on a 4–15% gradient SDS–PAGE and Coomassie blue staining. Individual polypeptides were eluted from the gel and analysed by mass spectrometry.
Immunoprecipitation and western blot
Cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in buffer B (containing 50 mM Tris–Cl (pH 8), 150 mM NaCl, 1% NP-40) along with protease inhibitor cocktail. Total cell lysates were incubated with primary antibody for 2–3 h followed by protein A/G-Sepharose (Amersham Bioscience) for one additional hour at 4°C. Beads were extensively washed twice with buffer B then three times with buffer C (buffer B with 0.2% NP-40), boiled in 2 × SDS sample buffer, separated on SDS–PAGE, transferred to a nitrocellulose membrane and immunoblotted. Bound antibody was detected by Super Signal chemiluminescence reagent (Pierce).
GST pull down assay
Purified GST and GST–Osx (1–270) were incubated with 35S-labelled NO66 for 1 h at 4°C, washed with buffer C containing 1 mM PMSF and protease inhibitor cocktail and the complexes pulled down by glutathione Sepharose beads followed by SDS–PAGE. 35S-NO66 was prepared with the TNT T7 Quick coupled system (Promega). In the reciprocal experiment, purified GST and GST–NO66 bound to glutathione resins were mixed with purified His-S-tagged Osterix protein in the presence of 10 mg/ml BSA in PBS and incubated at 4°C for 2 h. After washing, resins were boiled in the presence of SDS sample buffer and polypeptides were separated on SDS–PAGE followed by western blotting using α-S-tagged antibody to detect Osterix.
Reporter assays, siRNA and shRNA transfections, and gene expression
For all reporter assays, HEK293T cells were grown in DMEM, transfected with 200 ng of reporter construct, 50–200 ng of expression vector, and 20 ng of pSV40/LacZ to control for transfection efficiency. Lysates were assayed for luciferase and β-galactosidase.
The C2C12 and MC3T3 cells were transfected with 100 nM stealth siRNAs (Invitrogen, USA) using lipofectamine 2000 (Invitrogen) for 48 h and then treated with BMP-2 for 24 h. For quantitative gene expression, total RNA was isolated using RNAeasy-Mini kit (Qiagen, Valencia, CA) and reverse-transcribed with random hexamer primers (Archive cDNA synthesis kit, Applied Biosystems). Complimentary DNA was used in triplicate for real-time PCR analysis using Taqman primer-probe. Representative qRT–PCR of two independent experiments is shown for which qPCR triplicate assays were performed. The experimental error of triplicate PCR assay is shown by error bar. Levels of RNA were normalized to those of GAPDH. Gene-specific primer-probes were obtained from Applied Biosystems. Sequences of siRNA are available on request.
Demethylation and immunofluorescence
Recombinant NO66 or mutant protein, which were affinity purified, was incubated with 5 μg calf thymus bulk histones (Sigma) in 50 μl demethylase buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 50 μM (NH4)2 Fe(SO4)2· 6(H2O), 1 mM μ-ketoglutarate, and 2 mM ascorbate). Reactions were carried out at 37°C for 3–7 h, terminated by addition of 2 × SDS-loading buffer, followed by separation on SDS–PAGE and immunoblotted with appropriate antibodies (α-H3K4me3, ab8580; α-H3K36me3, ab9050; α-H3, ab1791 from Abcam and α-H3K27me3, 07–449 from Millipore).
COS7 cells were seeded on top of coverslip in 24-well plate and transfected with 200 ng expression plasmids for 36 h. Cells were washed with PBS, fixed with 4% paraformaldehyde, permeabilized with 1% Triton X-100, and blocked with 10% BSA. The cover slips were incubated for 2 h at room temperature with α-myc antibody (9E10, Santa Cruz Biotechnology) and α-H3K4me3, washed and then incubated with secondary antibody (goat-α-rabbit Alexa 594 and goat-α-mouse Alexa 488, Molecular probes) for 1 h at room temperature. Cover slips were washed, mounted with antifade-Gold with DAPI (Molecular Probes), and analysed by fluorescence microscopy.
Knockdown of NO66 by shRNA and in vitro osteoblast differentiation assay
Pre-osteoblast MC3T3 cells were stably transfected with control non-specific and NO66 specific shRNAs (Open Biosystem, USA) using Arrest-in transfection reagent and selected on puromycin (2 μg/ml) for 10 days. Puromycin resistant cells were pooled and expanded in growth medium. Whole-cell lysates were used to analyse the levels of NO66 in both cell lines by immunoblotting with α-NO66 antibody. NO66-specific shRNA-expressing cells (NO66sh) and control non-specific shRNA-expressing cells (Nsh) were grown to confluency and maintained in differentiating medium (α-MEM with 7 mM β-glycerol-phosphate and 50 μg/ml ascorbic acid for several days). Cells were stained for alkaline phosphatase production using kit 86R (Sigma). Quantification of alkaline phosphatase activity was performed using Amplite colorimetric alkaline phosphatase assay kit (ABD Bioquest, USA) as per manufacturer's instructions. For extracellular mineralization, cells were stained with 1.4% Alizarin S Red (pH 4.2) for 10 min. Post-staining, plates were photographed by computer-assisted scanner. A subset of these cells during their differentiation was used for quantitative gene expression. Total RNA isolation, reverse-transcription and quantitative PCR were performed as described above.
Construction of NO66 and mutant expression plasmids
Expression vector pcDNA-NO66, which contains a full-length human NO66 with a myc-epitope at the N-terminus, was kindly provided by M Schmidt-Zachmann (Eilbracht et al, 2004). This plasmid was further modified to clone full-length NO66 cDNA (1–1923 bp) into EcoRI and XhoI sites of pcDNA3.1-myc to generate NO66 protein with myc and His tag at the C-terminus. Construction of NO66 mutants used conventional methods to clone into pcDNA3.1-myc whereas those for Osx mutants were described earlier (Zhang et al, 2008).
Expression and purification of recombinant proteins
Human NO66 cDNA was cloned into HT-FastBac vector (Invitrogen) to add a His tag at N-terminus and GST-FastBac vector to generate GST tag at N-terminus. The resulting construct was used to produce baculovirus and then infected to Sf9 cells. The recombinant NO66 was expressed in Sf9 cells and purified using Ni2+–NTA agarose (Qiagen) or glutathione-Sepharose 4B resin according to the manufacturer's protocol. His-S-tagged Osterix was cloned into pBACgus-2cp transfer plasmid (Novagen), which was then used to produce baculovirus.
Human NO66 cDNA and mouse Osx cDNA encoding amino-acids 1–270 were inserted into pGEX4T-1 and pGEX4T-2 (GE Healthcare), respectively. The GST fusion polypeptides were expressed in Escherichia coli (BL21, codon+) and purified using glutathione-Sepharose 4B resin (GE Healthcare) according to the manufacturer's protocol.
Chromatin immunoprecipitation and quantitative PCR
Sub-confluent cells were treated with or without BMP-2 for different time. The ChIP experiment was performed as described by Kouskouti et al (2004), with some modifications. Approximately 1.75 × 107 cells were treated with 1% formaldehyde in serum-free medium for 20 min followed by a treatment with 125 mM glycine. Cells were washed with cold PBS, re-suspended in 10 volumes of cell lysis buffer (25 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF, and protease inhibitor cocktail) and homogenized to release the nuclei that were then lysed with nuclear lysis buffer (50 mM HEPES (pH 7.9), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 0.5 mM PMSF, and protease inhibitor cocktail). Nuclear extracts were sonicated and cleared by centrifugation. Sonicated chromatin was diluted in lysis buffer without SDS to bring the final concentration of SDS to 0.05%. The antibodies specific to Osx, NO66, and rabbit IgG for control were first conjugated to magnetic coated protein G beads (Magnetic Dyna beads-Protein G, Invitrogen, CA), which were previously blocked with 0.5% BSA in PBS. After overnight immunoprecipitation at 4°C with rotation, immunoprecipitated complexes were collected using a magnetic stand. Washing, reversal of cross-links, and purification of DNA from immunocomplexes were done using EZ-ChIP kit (Millipore) as per manufacturer's instructions. For sequential ChIP experiments, chromatins prepared from C2-Osx cells were immunoprecipitated by HA–epitope antibody (ab9110, Abcam) and control IgG antibodies. Flag–HA–Osx-bound chromatin fragments were released from α-HA antibody with HA peptides (1 mg/ml, Roche Diagnostics, USA), which specifically eluted Flag–HA–Osx-bound chromatin, and then further re-immunoprecipitated with α-NO66 antibody in the presence of 50 μg/ml yeast tRNA and 5 mg/ml BSA. All other conditions were essentially same as described above.
Quantitative PCR reactions were performed with 500 nM of primers, 5 μl of 2 × SYBR green I PCR master mix (Applied Biosystems), 2 μl DNA in a total 10 μl reaction volume. Results were computed as percent antibody bound per input and data were shown after subtracting control IgG values. Representative ChIP assays of at least three independent experiments are shown for which qPCR triplicate assays were performed. The experimental error of triplicate PCR assay is shown by error bar. Primers used for qPCR were tested first with input DNA to analyse a linear range of amplification. Primer sequences are available on the request.
Generation of polyclonal rabbit anti-NO66 antibody
Rabbit α-NO66 antibody was generated against the peptide epitope (SYPEFVRVGDLPCDSVED), which is conserved at the C-terminus in mouse, rat, and human NO66 protein, and antiserum was affinity-purified on peptide-conjugated matrices (service provided by Open Biosystems, Huntsville, AL).
Supplementary Material
Supplementary Figures S1–10
Original scans
Review Process File
Acknowledgments
We thank Drs Ryuji Kobayashi and David Hawke for their technical help in performing peptide sequencing by mass spectrometry; Henry Adams for assistance with deconvolution microscopy; Lingna Zhang and Shelley Stephens for technical assistance. This study was funded by National Institutes of Health Grant R01 AR49072 (to BdC). KS is a recipient of the Rolanette and Berdon Lawrence Research Award of the Bone disease Program of Texas. We also acknowledge National Institutes of Health Grant CA16672 for DNA sequence analysis.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y (2007) New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 [DOI] [PubMed] [Google Scholar]
- Cheng SL, Shao JS, Cai J, Sierra OL, Towler DA (2008) Msx2 exerts bone anabolism via canonical Wnt signaling. J Biol Chem 283: 20505–20522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89: 747–754 [DOI] [PubMed] [Google Scholar]
- Eilbracht J, Reichenzeller M, Hergt M, Schnolzer M, Heid H, Stohr M, Franke WW, Schmidt-Zachmann MS (2004) NO66, a highly conserved dual location protein in the nucleolus and in a special type of synchronously replicating chromatin. Mol Biol Cell 15: 1816–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Hall BK (1997) Chondrogenic cell differentiation from membrane bone periostea. Anat Embryol (Berl) 196: 349–362 [DOI] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T (2006) Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev 20: 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB (2004) Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol 24: 9248–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C (2005) Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell 8: 727–738 [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449: 105–108 [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Kang JS, Alliston T, Delston R, Derynck R (2005) Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J 24: 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T (1994) Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol 127 (6 Pt 1): 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H (2005) NFAT and Osterix cooperatively regulate bone formation. Nat Med 11: 880–885 [DOI] [PubMed] [Google Scholar]
- Komori T (2006) Regulation of osteoblast differentiation by transcription factors. J Cell Biochem 99: 1233–1239 [DOI] [PubMed] [Google Scholar]
- Kouskouti A, Scheer E, Staub A, Tora L, Taliandis I (2004) Gene-Specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell 14: 175–182 [DOI] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T (2002) The many faces of histone lysine methylation. Curr Opin Cell Biol 14: 286–298 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R (2007) Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128: 877–887 [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- Nakashima K, de Crombrugghe B (2003) Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet 19: 458–466 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29 [DOI] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133: 3231–3244 [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, Westendorf JJ (2004) Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem 279: 41998–42007 [DOI] [PubMed] [Google Scholar]
- Shen J, Montecino M, Lian JB, Stein GS, Van Wijnen AJ, Stein JL (2002) Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. J Biol Chem 277: 20284–20292 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Tu Q, Valverde P, Chen J (2006) Osterix enhances proliferation and osteogenic potential of bone marrow stromal cells. Biochem Biophys Res Commun 341: 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN (2004) Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119: 555–566 [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481 [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125: 483–495 [DOI] [PubMed] [Google Scholar]
- Yang X, Karsenty G (2004) ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem 279: 47109–47114 [DOI] [PubMed] [Google Scholar]
- Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD, de Crombrugghe B (2008) Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc Natl Acad Sci USA 105: 6936–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ED, Demay MB, Gori F (2008) Wdr5 is essential for osteoblast differentiation. J Biol Chem 283: 7361–7367 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–10
Original scans
Review Process File