Abstract
The impact of globalization on the emergence and spread of pathogens is an important veterinary and public health issue. Staphylococcus aureus is a notorious human pathogen associated with serious nosocomial and community-acquired infections. In addition, S. aureus is a major cause of animal diseases including skeletal infections of poultry, which are a large economic burden on the global broiler chicken industry. Here, we provide evidence that the majority of S. aureus isolates from broiler chickens are the descendants of a single human-to-poultry host jump that occurred approximately 38 years ago (range, 30 to 63 years ago) by a subtype of the worldwide human ST5 clonal lineage unique to Poland. In contrast to human subtypes of the ST5 radiation, which demonstrate strong geographic clustering, the poultry ST5 clade was distributed in different continents, consistent with wide dissemination via the global poultry industry distribution network. The poultry ST5 clade has undergone genetic diversification from its human progenitor strain by acquisition of novel mobile genetic elements from an avian-specific accessory gene pool, and by the inactivation of several proteins important for human disease pathogenesis. These genetic events have resulted in enhanced resistance to killing by chicken heterophils, reflecting avian host-adaptive evolution. Taken together, we have determined the evolutionary history of a major new animal pathogen that has undergone rapid avian host adaptation and intercontinental dissemination. These data provide a new paradigm for the impact of human activities on the emergence of animal pathogens.
Keywords: evolution, host adaptation, pathogen, phylogeography, globalization
Globalization and the trend toward the integration of trade of crops and livestock could have a major impact on the emergence and dissemination of pathogens. Shifts in agricultural practice result in opportunities for pathogens to expand into new host species and to spread rapidly to new territories. For example, the epidemics of bovine spongiform encephalitis (1) and the foot and mouth disease epidemic (2) were caused by changing agricultural practices providing new opportunities for transmission, including the use of meat and bone meal in cattle feed, and the long-distance transport of livestock, respectively.
The broiler poultry industry has been transformed within the last 50 years from a market dominated by smallholder chicken farms to a multibillion dollar industry controlled by a handful of multinational companies who supply a limited number of breeding lines to a global market (3, 4). Infectious diseases of chicken flocks are a major economic burden on the industry. In particular, Staphylococcus aureus is associated with several infections of poultry including septic arthritis, subdermal abscesses (i.e., “bumble foot”), and gangrenous dermatitis (5). In the 1970s, a new form of S. aureus infection of broiler poultry known as bacterial chondronecrosis with osteomyelitis (BCO) was described (6). Since then, BCO has increased in frequency to become a leading cause of lameness in the broiler chicken industry (7, 8). The reasons for the emergence and subsequent increase in incidence of BCO among chickens are unknown.
Results and Discussion
The Majority of S. aureus Isolates from Poultry Belong to a Single Clonal Complex (CC5) Usually Associated with Humans.
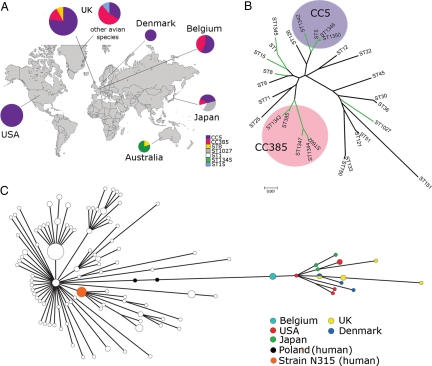
To examine the population genetics of S. aureus strains infecting farmed birds, we carried out multi-locus sequence typing (MLST) of 57 S. aureus isolates, including 48 isolates from healthy and diseased poultry, in 8 countries on 4 continents isolated in the past 54 years, in addition to 9 isolates from different species of reared game and wild birds [supporting information (SI) Table S1]. Remarkably, the majority of all avian isolates (n = 35; 61%) including 32 (67%) from broiler chickens, belonged to a single sequence type (ST), ST5, or its single locus variants (ST1342, ST1346, and ST1350), including isolates from all countries examined except Australia (Fig. 1). The clonal complex CC5 (which includes related haplotypes differing at a small number of loci) is one of the most successful human-associated lineages of S. aureus, characterized by its global distribution and frequent emergence of methicillin-resistant strains (9). Of the non-CC5 isolates identified, which included isolates from broiler chickens, reared bird species such as pheasant and partridge, and a wild buzzard, 11 belonged to an unrelated clonal complex, CC385, which has not been identified previously among human or mammalian strains. The lack of association of CC385 with humans or other mammals and its wide distribution among an array of reared and wild bird species in different countries suggests that the CC385 lineage has had a long-term avian host restriction (Table S1). Of the remaining 11 isolates, 4 from Australia belonged to a novel ST1345, 3 Japanese isolates were ST1027, 2 were of ST8, and single isolates were of ST1 and ST15, all being related or identical to STs previously identified among human S. aureus isolates (Fig. 1 A and B).
Fig. 1.
The majority of poultry S. aureus isolates evolved by a single host jump from humans followed by wide dissemination. (A) Geographical distribution of poultry and other avian S. aureus STs identified in countries from 4 different continents. Pie chart diameter is proportional to the number of isolates from each location. (B) Neighbor joining tree with bootstrapping consensus inferred from 500 replicates was constructed using concatenated sequences of S. aureus STs from birds (green branches) and representatives of the major clonal complexes of human and animal origin in the S. aureus species (black branches). The 2 major avian-associated clonal complexes, CC5 and CC385, are indicated in blue and red, respectively. (C) Minimum spanning tree based on 185 BiPs in 19 representative poultry ST5 isolates compared with 135 S. aureus ST5 human isolates of global origin analyzed in a previous study (9). Circle size is proportional to haplotype frequency and line length is proportional to the number of mutational steps between haplotypes. Colored nodes indicate the geographical origin of the poultry isolates, closely-related human isolates from Poland, and the human strain N315.
The Poultry ST5 Clade Is the Result of a Single, Recent Human-To-Poultry Host Jump.
Recently, a high-resolution analysis of the phylogenetic structure of the human ST5 clonal radiation was carried out by mutation discovery at 108 loci (46 kb), resulting in the identification of at least 14 distinct lineages within the ST5 radiation (9). To trace the evolutionary origin of the poultry ST5 strains, we carried out mutation discovery at the same genetic loci among a representative selection of 19 poultry ST5 isolates, as previously described, and identified 29 novel bi-allelic polymorphisms (BiPs) (9). These polymorphisms, along with 156 informative BiPs previously identified among human ST5 isolates (9), were used to construct a minimum spanning tree, which included the 19 poultry isolates and 135 isolates of human origin (Fig. 1C). The phylogenetic analysis indicates that all the poultry isolates are closely related to each other and do not fall into separate clades for diseased and asymptomatic birds, suggesting that S. aureus poultry infections are caused by resident commensal strains (Fig. 1C). Further, the poultry ST5 clade belongs to an ST5 sub-lineage that includes human strains that were circulating in Poland in the 1990s (Fig. 1C). Importantly, examination of the distribution of SNPs within the lineage indicates that the Polish human isolates are basal to all isolates in the poultry ST5 clade (Fig. 1C). Nubel et al. (9) recently demonstrated that ST5 sub-lineages demonstrate phylogeographic clustering, including many that are confined to single countries, indicating that intercontinental spread and fixation in local populations by human S. aureus is very rare. For example, the widespread clone represented by Polish human isolates MR1 and PL72 that was predominant in Polish hospitals in the 1990s (10) has not been identified elsewhere in the world.
To investigate the date of the most recent common ancestor of the ST5 poultry clade and the Polish human ST5 strains (MR1 and PL72), we carried out a Bayesian phylogenetic analysis of human and poultry strains sampled at different time points (ranging from 1976 to 2006), and estimated that a host switch occurred 38 years ago with a 95% CI of 30 to 63 years (Fig. S1). The great diversity of geographically isolated subtypes of the CC5 lineage colonizing humans indicates a long-term association of the CC5 lineage with its human host (9) and implies that the common poultry S. aureus clade evolved as a result of a human-to-poultry host jump by a S. aureus strain originating in or near Poland. Further, in common with bovine-adapted strains of S. aureus (11), poultry ST5 strains have lost the function of several genes involved in human disease pathogenesis, whereas the majority of pseudogenes that accumulated in the ancestral lineage of human MR1 and poultry ED98 have not been implicated in direct human host-pathogen interactions (Table S2). This is consistent with a strain adapting to a recent host switch after long-term association with humans. Taken together, these data strongly suggest that the direction of the host jump was from humans to poultry, leading to the emergence of a new strain of S. aureus with a tropism for birds. The alternative scenario of a poultry-to-human host jump could be explained only by multiple host species jumps, including an initial jump from humans into an unknown non-human intermediate host species, which occurred since the most recent common ancestor of the CC5 lineage, followed by a second jump into poultry resulting in the poultry ST5 clade. In turn, a final jump back into humans would be required, leading to wide dissemination in Polish hospitals (10).
The Poultry ST5 Clade Has Undergone Rapid Intercontinental Dissemination.
The distribution of the poultry ST5 clade in countries in 3 different continents strongly contrasts with the geographical isolation of the closely related human ST5 isolates to Poland (Fig. 1C), indicating that frequent intercontinental spread of the poultry strains has occurred. This finding is consistent with the globalized nature of the poultry industry, whereby a very small number of breeder companies distribute large numbers of live broiler chickens via a global distribution network. The lack of ST5 poultry isolates identified in Australia is consistent with stringent regulations controlling the import of livestock to the island (see http://www.daff.gov.au/).
The Poultry ST5 Clade Has Acquired Novel Mobile Genetic Elements from an Avian-Specific Accessory Gene Pool.
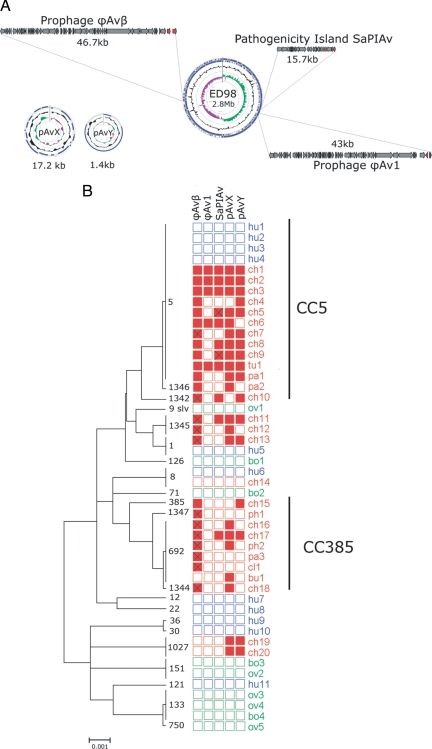
The poultry clade forms a long branch in the minimum spanning tree constructed by using ST5 polymorphism data (Fig. 1C), indicating a large number of mutational steps relative to the ST5 clonal radiation. To investigate the genetic basis for the adaptation of the ST5 poultry clade to its new host, we determined the whole genome sequence for a representative poultry ST5 clade strain ED98, isolated from a chicken with BCO in Ireland (12), and compared it with the genome of the basal human strain from Poland, MR1 (10) (Table S3). We discovered that the poultry strain ED98 has acquired 5 novel mobile genetic elements (MGEs) not identified among human S. aureus isolate sequences identified to date, including 2 prophages, 2 plasmids, and a staphylococcal pathogenicity island (SaPI; Fig. 2A and Table S4). PCR screening of a panel of 48 S. aureus isolates from poultry, reared and wild birds, humans, cows, and sheep representing the breadth of diversity within the species revealed the wide distribution of these MGEs among avian S. aureus isolates and their complete absence among isolates of human or mammalian animal origin (Fig. 2B). In particular, ϕAvβ, a novel member of the β-converting phage family, lacks the immune evasion cluster of genes which are central to the human niche-specific activity of most β-converting phages (13). Instead of the immune evasion cluster, ϕAvβ contains genes encoding a novel ornithine cyclodeaminase and a putative novel protease that may be involved in avian niche-specific activities. ϕAvβ-related sequences were detected in all 13 avian strains of the CC5 poultry clade, 8 of 9 strains of the avian-specific CC385 lineage, the ST1345 clone specific for Australia, and a Japanese ST1 strain, implying frequent horizontal gene transfer of ϕAvβ between S. aureus strains from different avian species and of distinct clonal origin (Fig. 2B). In addition, pAvX, a 17-kb plasmid encoding a thiol protease (ScpA), previously implicated in the pathogenesis of poultry S. aureus infections (14, 15), and a putative lysophospholipase, was present in 11 of 13 avian CC5 strains examined and in 5 of 9 CC385 strains (Fig. 2). A second plasmid (pAvY), 1.4 kb in size, encoding 2 hypothetical proteins of unknown function, was distributed among 11 of 13 avian CC5 strains examined and in 2 of 9 CC385 isolates (Fig. 2). A novel member of the SaPI family (SaPIAv) that encodes 2 putative novel virulence genes at the region typically occupied by superantigen genes in SaPIs (Table S4) (16) was found in 9 of 13 avian ST5 isolates and in 2 isolates from unrelated lineages (ST1345 and CC385). Finally, a second prophage ϕAv1 encoding an allele of the ear gene (SAAV_0875), which encodes a protein with putative β-lactamase activity (16), was found among 5 of 13 poultry ST5 isolates examined but not among other avian isolates or isolates of human or ruminant origin (Fig. 2B). The identification of several novel MGEs that are widely distributed among avian S. aureus isolates from distinct clonal lineages and are not associated with human or ruminant strains indicates the presence of an avian niche-specific accessory gene pool for S. aureus. The maintenance of such a gene pool implies that it confers an important selective advantage to bacteria residing in the niche and that horizontal gene acquisition of MGE has played a fundamental role in the avian host adaptation of the poultry ST5 clade. Of note, we found avian-specific MGEs among poultry isolates other than CC385 or CC5 that share identical or closely related STs with human strains in the MLST database. These data suggest that, in common with the ST5 poultry clade, other poultry S. aureus strains have recently switched from a human to a poultry host and are undergoing host adaptation, at least in part through acquisition of avian-specific MGEs. These findings also suggest that the CC5 sub-lineage that includes the poultry strains and human isolates from Poland may be particularly proficient at acquiring MGEs, leading to the emergence of the successful poultry pathogen and the human methicillin-resistant S. aureus clone predominant in Polish hospitals in the 1990s.
Fig. 2.
S. aureus avian isolates of distinct clonal lineage share a common accessory gene pool. (A) Novel MGEs identified in the whole-genome sequence of the ST5 poultry isolate ED98. The ED98 genome and plasmid maps represent ORFs (blue arrows on outer circle), GC content (middle circle), and GC skew (inner circle; GC+ and GC- skew in green and purple, respectively). Prophage and SaPI ORFs are represented by arrows, and red arrows denote genes putatively involved in virulence and/or host specificity. (B) The distribution of novel MGEs identified in the ED98 genome sequence among a panel of diverse isolates of avian, human, bovine, and ovine origin. Neighbor joining tree with bootstrapping consensus inferred from 500 replicates was constructed using concatenated sequences of S. aureus STs from different host species, including representatives of the major clonal complexes identified in the S. aureus species. Labels at branch tips denote STs of strains. A filled box indicates presence of an MGE and an empty box its absence as determined by MGE-specific PCR. A crossed box indicates the presence of a related but non-identical genetic element (determined by successful amplification in 1 of 2 PCRs). Red, blue, and green boxes denote strains of avian, human, and ruminant (bovine or ovine) origin, respectively. Strain prefixes denote the host species origin of the strains including ch, broiler chicken; ph, farmed pheasant; tu, farmed turkey; pa, farmed partridge; cl, poultry layer; bu, wild buzzard; hu, human; bo, cattle; and ov, sheep, respectively.
The Poultry ST5 Clade Has Diversified from Its Human Progenitor Strain by the Loss of Function of Genes Involved in Human Disease Pathogenesis.
In addition to the acquisition of genes by the poultry ST5 clade, examination of the genome sequence of poultry strain ED98 and comparison with its human basal strain MR1 revealed 197 SNPs. Of note, 10 pseudogenes have arisen in ED98 since the most recent common ancestor with the human strain MR1, including several that would influence host-pathogen interactions (Table S2). For example, mutations leading to loss of function have occurred in genes encoding a putative surface-exposed lipoprotein, a protease SspA involved in the maturation of SspB that contributes to blood clot formation (17), and asp1 required for the surface expression of SraP, a cell wall-associated protein involved in the pathogenesis of infective endocarditis (18). In addition, a mutation leading to a premature stop codon in the gene encoding the major staphylococcal protein A (SpA) has occurred in strain ED98. An identical spA nonsense mutation was found in all poultry ST5 strains examined, indicating that it happened early in the evolution of the poultry ST5 clade. SpA has multiple roles in the pathogenesis of human infections, including adherence to lung epithelium via the TNFR1 receptor (19), binding to human platelets through its interaction with von Willebrand factor (20, 21), and inhibition of opsonophagocytosis resulting from non-specific binding of the Fc region of IgG (22, 23). Of note, the avian equivalent of human IgG is IgY, which has a structurally distinct Fc region that does not bind to SpA (24, 25), suggesting that SpA would not contribute to the inhibition of opsonophagocytosis in birds.
The Poultry ST5 Strain ED98 Exhibits Enhanced Resistance to Killing by Chicken Heterophils Compared with the Human Basal Strain MR1.
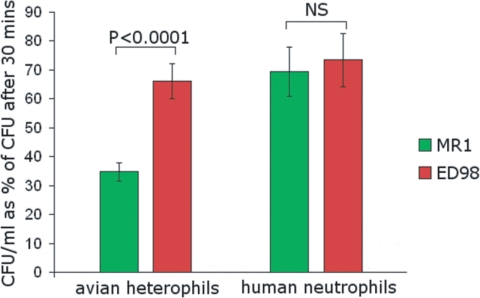
Taken together, we have discovered considerable genetic diversification of the poultry ST5 subtype compared with its basal human strain, which could be (at least in part) the result of host-adaptive genetic events that increase the fitness of S. aureus in its poultry niche. Inasmuch as neutrophils (and the avian equivalent heterophils) are a critical component of the innate immune system of humans and birds, and the primary cellular defense against S. aureus infections, we tested this hypothesis by comparing the ability of the poultry ED98 strain and its human basal strain MR1 to survive cellular killing by avian heterophils or human neutrophils. Remarkably, poultry ED98 had very enhanced resistance to killing by avian heterophils compared with the closely related human MR1 strain (Fig. 3). In contrast, there was no significant difference between the strains in resistance to phagocytic killing by human neutrophils. These data indicate that genetic differences identified in the ED98 genome compared with the human MR1 strain contribute to avian host-specific innate immune avoidance, reflecting avian host-adaptive evolution. In particular, we speculate that the acquisition of MGEs that are specific to the avian niche may play a key role in resistance to killing by chicken heterophils, perhaps through expression of novel virulence factors that interfere with heterophil function.
Fig. 3.
Poultry S. aureus strain ED98 exhibits increased resistance to killing by avian neutrophils (heterophils) in comparison with the human basal S. aureus strain MR1. Survival of poultry strain ED98 and human strain MR1 after opsonization with avian or human plasma followed by incubation with avian heterophil samples (n = 17) or human neutrophils (n = 13), measured by percentage reduction in colony count in the coculture compared with the colony count of the bacteria alone. Error bars indicate SE.
Concluding Comments.
The increase in prevalence of skeletal infections of poultry to become a major cause of lameness in the industry correlates with the emergence and wide dissemination of the ST5 poultry subtype. Of note, a previous study reported that the ST5 clonal complex is associated with increased frequency of hematogenous infections of humans, including osteomyelitis (26), consistent with the largely skeletal tropism of infections caused by the poultry ST5 clade. The reduction in the biodiversity of chickens in the broiler poultry industry such that a very small number of breeding lines are supplied for global consumption promotes the spread of opportunistic pathogens that are resident on those birds and could select for infectious diseases to which those animals are genetically predisposed. Our data highlight the importance of regular screening of the normal flora of livestock to identify newly emerging bacterial pathogens and suggest that decolonization of resident S. aureus from fledgling stocks may be an effective means to reducing the burden of staphylococcal infections.
Although the potential impact of globalization on the emergence of new pathogens is a major public concern, our understanding of the evolutionary basis for such events is lacking. Here we provide a unique paradigm of a major bacterial pathogen of humans that has switched to a poultry host, presumably as a consequence of an increase in human-poultry contact during broiler breeding programs or production. Adaptation through extensive acquisition of MGEs from other S. aureus isolates colonizing poultry, and loss of gene function, has been followed by intercontinental dissemination. The globalization of the food industry has resulted in a massive increase in the transport of livestock worldwide, but the potential consequences of this expansion for pathogen emergence and transmission to previously unaffected geographical regions have not been well examined. Taken together, these data provide broad new insights into bacterial host adaptation and the influence of human activities on pathogen emergence.
Materials and Methods
Bacterial Strains and Growth Conditions.
A total of 77 S. aureus isolates from human, avian, bovine, and ovine hosts were examined (Table S1). Of the isolates, 48 were from healthy or diseased broiler chickens in 8 countries in 4 continents isolated during the past 54 years, 9 were from different species of reared birds (pheasant, partridge, turkey, poultry layer) and a wild buzzard in Scotland. Ruminant isolates (4 bovine and 5 ovine) were isolated in 9 different countries, and 11 isolates were obtained from human infections in 4 countries (Table S1). Bacteria were grown to stationary phase in trypticase soy broth (TSB) at 37 °C for 16 h with shaking. For exponential phase, 200 μL of stationary-phase culture was added to 40 mL fresh TSB and grown to an OD600 value of 0.6.
Genomic DNA Extraction.
DNA was isolated from 1 mL volume of stationary-phase S. aureus TSB culture by using a bacterial genomic DNA purification kit (Edge Biosystems) according to the manufacturer's instructions, with addition of lysostaphin (5 mg mL−1; AMBI Products) to the cell lysis step.
Genome Sequencing, Annotation, and Comparative Analysis.
Whole-genome sequencing by a combination of 454 pyro-sequencing and directed sequencing of PCR products representing gap regions, followed by assembly, and annotation of the S. aureus poultry strain ED98, was carried out before deposition of the sequence in the NCBI GenBank database (see SI Materials and Methods for further detail). Genome sequencing of human strain MR1 was performed with an Illumina/Solexa genome analyzer, and sequence reads were aligned against the ED98 sequence to identify SNPs and large regions of difference (see SI Materials and Methods for further detail).
Pseudogene Identification.
ORFs displaying evidence of frame shifts or mutations leading to a premature stop codon were identified using information from the BLAST-extend-repraze (BER) search performed in the J. Craig Venter Institute annotation pipeline, manually checked, and confirmed by directed sequencing. The distribution of pseudogenes was examined in 18 additional poultry ST5 isolates and the human strains MR1 and N315, the latter of which had been analyzed for the presence of pseudogenes in a previous study (27) (see SI Materials and Methods for further detail).
PCR Detection of MGEs.
The presence of novel MGEs among a panel of avian, human, bovine, and ovine isolates was determined by PCR using MGE-specific primers (Table S5). PCR was performed in 50-μL reaction volumes containing approximately 10 ng of genomic DNA, 200 nM of each primer, 1U of GoTaq DNA polymerase (Invitrogen), 1× PCR buffer, 1.5 mM MgCl2 (Promega) and 0.2 mM deoxynucleoside triphosphates (Promega). Reactions were performed in a Biometra TGradient with an initial 2-min denaturation at 95 °C, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min or 4 min (depending on predicted product size with each primer pair), with a final incubation at 72 °C for 5 min.
MLST and Mutation Discovery.
MLST was carried out by PCR amplification and sequencing of internal fragments of 7 housekeeping genes as described previously (28). Mutation discovery was performed as described previously (9). Briefly, internal regions of 108 gene loci corresponding to a total of 46 kb were amplified by PCR, and PCR products polymorphic to those of the reference N315 genome were detected by denaturing HPLC and sequenced.
Phylogenetic Analysis.
Reconstruction of evolutionary relationships was carried out by using the MEGA 4 package (29). Concatenated MLST sequence data were used to construct trees using the neighbor-joining method (30) with bootstrapping consensus inferred from 500 replicates (31). For reconstruction of relationships among ST5 strains, informative BiPs identified by mutation discovery were used to construct a minimum spanning tree by using the Bionumerics 5.1 program (Applied Maths).
To date the predicted host switch from humans to poultry, a Bayesian phylogenetic analysis of human and poultry strains sampled at different time points (ranging from 1976 to 2006) was performed to estimate the date of their most recent common ancestor (see SI Materials and Methods for further detail).
Neutrophil/Heterophil Bacteriocidal Assay.
For preparation of heterophils, the method was adapted from Brooks et al. (32). Heparin anticoagulated (10 IU/mL) whole blood was centrifuged at 350 × g and the plasma phase removed. The pelleted cells were resuspended in 1.5 × HBSS without divalent cations (HBSS-/-) containing 1% (wt/vol) methylcellulose and 1% (vol/vol) FBS. The cells were centrifuged at 25 × g for 15 min and the extended buffy layer aspirated, washed once in HBSS-/-, and resuspended in reaction buffer [HBSS-/- with 1% (vol/vol) FBS] before applying to Histopaque density gradients (1.077 and 1.119). The gradients were centrifuged for 20 min at 225 × g. The top 2 layers and mono-nuclear cell band were aspirated and the pellet resuspended in reaction buffer, before washing once in reaction buffer and re-suspending in lysis buffer [distilled water with 0.87% (wt/vol) NH4Cl and 0.1% (wt/vol) KHCO3]. The cells were then washed twice in reaction buffer before being resuspended in HBSS with divalent cations (HBSS+/+) at 10 × 106/mL, resulting in an average yield of 106 heterophils per sample. Human neutrophils were extracted as previously described (33).
An overnight TSB culture of S. aureus was subcultured in TSB for 2 to 3 h until the bacteria were in early exponential phase (OD600 of 0.1) and diluted by a factor of 1:1,000 in HBSS+/+ with 10% pooled plasma. Bacterial suspension (225 μL) and 25 μL neutrophils or heterophils were added to an Eppendorf tube and incubated at 41 °C (normal chicken body temperature) or 37 °C (human body temperature) for 30 min. Simultaneously, bacterial suspension was incubated with 25 μL of HBSS+/+ without neutrophils or heterophils. The neutrophils or heterophils were then lysed with 0.1% Triton X-100, and dilutions of 10−2 and 10−4 made in HBSS+/+. One hundred microliters of neat, 10−2, and 10−4 solutions were plated in duplicate on trypticase soy agar plates and incubated at 37 °C for 14 to 16 h before determination of colony counts. Killing of bacteria was expressed as the percent reduction in colony count in the co-culture compared with the colony count of the bacteria alone.
Supplementary Material
Acknowledgments.
We are grateful to J. Rodgers, G. Foster, D. Vancraeynest, W. Grubb, D. Bertolatti, S. Takeuchi, L. Guardabassi, J. Lindsay, J. Penades, T. Tollersrud, H. de Lencastre, J. Maurer, and M. Maier for providing isolates; and O. Moncayo, L. Farrell, A. Weller, and members of the sequencing labs at the University of Edinburgh (Genepool) and Robert Koch Institute for technical assistance. We thank G. Somerville for his contribution to sequence interpretation, the J. Craig Venter Institute for assistance with annotation, and P. Hocking and G. Robertson for supply of chicken blood samples. This work was funded by Grant BB/D521222/1 from the Biotechnology and Biological Sciences Research Council (to J.R.F.) and the Royal Society (to J.R.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. CP001781, CP001782, CP001783, and CP001784 (ED98 genome sequence, chromosome and plasmids, respectively); and ACZQ00000000 (MR1 Whole Genome Shotgun Project)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0909285106/DCSupplemental.
References
- 1.Nathanson N, Wilesmith J, Griot C. Bovine spongiform encephalopathy (BSE): Causes and consequences of a common source epidemic. Am J Epidemiol. 1997;145:959–969. doi: 10.1093/oxfordjournals.aje.a009064. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson NM, Donnelly CA, Anderson RM. The foot-and-mouth epidemic in Great Britain: Pattern of spread and impact of interventions. Science. 2001;292:1155–1160. doi: 10.1126/science.1061020. [DOI] [PubMed] [Google Scholar]
- 3.Boyd W, Watts M. Agro-industrial just-in-time: The chicken industry and postwar American capitalism. In: Goodman D, Watts M, editors. Globalising Food: Agrarian Questions and Global Restructuring. London: Routledge; 1997. pp. 192–225. [Google Scholar]
- 4.Manning L, Baines RN. Globalisation: A study of the poultry-meat supply chain. Br Food J. 2004;106:819–836. [Google Scholar]
- 5.Smyth JA, McNamee PT. Staphylococci, streptococci and enterococci, Poultry Diseases. In: Jordan F, Pattison M, Alexander D, Faragher T, editors. 6th Ed. Amsterdam: Elsevier; 2001. pp. 191–199. [Google Scholar]
- 6.Nairn ME, Watson ARA. Leg weakness in poultry - a clinical and pathological characterisation. Aust Vet J. 1972;48:645–656. doi: 10.1111/j.1751-0813.1972.tb09237.x. [DOI] [PubMed] [Google Scholar]
- 7.McNamee PT, et al. Study of leg weakness in two commercial broiler flocks. Vet Rec. 1998;143:131–135. doi: 10.1136/vr.143.5.131. [DOI] [PubMed] [Google Scholar]
- 8.McNamee PT, Smyth JA. Bacterial chondronecrosis with osteomyelitis ('femoral head necrosis') of broiler chickens: A review. Avian Pathol. 2000;29:253–270. doi: 10.1080/03079450050118386. [DOI] [PubMed] [Google Scholar]
- 9.Nubel U, et al. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2008;105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leski T, et al. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J Clin Microbiol. 1998;36:3532–3539. doi: 10.1128/jcm.36.12.3532-3539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodgers JD, et al. Comparison of Staphylococcus aureus recovered from personnel in a poultry hatchery and in broiler parent farms with those isolated from skeletal disease in broilers. Vet Microbiol. 1999;69:189–198. doi: 10.1016/s0378-1135(99)00112-1. [DOI] [PubMed] [Google Scholar]
- 13.van Wamel WJB, et al. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J Bacteriol. 2006;188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi S, Kinoshita T, Kaidoh T, Hashizume N. Purification and characterization of protease produced by Staphylococcus aureus isolated from a diseased chicken. Vet Microbiol. 1999;67:195–202. doi: 10.1016/s0378-1135(99)00034-6. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi S, et al. Structural gene and strain specificity of a novel cysteine protease produced by Staphylococcus aureus isolated from a diseased chicken. Vet Microbiol. 2002;89:201–210. doi: 10.1016/s0378-1135(02)00171-2. [DOI] [PubMed] [Google Scholar]
- 16.Yarwood JM, et al. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J Biol Chem. 2002;277:13138–13147. doi: 10.1074/jbc.M111661200. [DOI] [PubMed] [Google Scholar]
- 17.Massimi I, et al. Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease of Staphylococcus aureus. J Biol Chem. 2002;277:41770–41777. doi: 10.1074/jbc.M207162200. [DOI] [PubMed] [Google Scholar]
- 18.Siboo IR, Chaffin DO, Rubens CE, Sullam PM. Characterization of the accessory sec system of Staphylococcus aureus. J Bacteriol. 2008;190:6188–6196. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez MI, et al. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J Biol Chem. 2006;281:20190–20196. doi: 10.1074/jbc.M601956200. [DOI] [PubMed] [Google Scholar]
- 20.Hartleib J, et al. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood. 2000;96:2149–2156. [PubMed] [Google Scholar]
- 21.O'Seaghdha M, et al. Staphylococcus aureus protein A binding to von Willebrand factor A1 domain is mediated by conserved IgG binding regions. Febs J. 2006;273:4831–4841. doi: 10.1111/j.1742-4658.2006.05482.x. [DOI] [PubMed] [Google Scholar]
- 22.Gemmell C, et al. Susceptibility to opsonophagocytosis of protein A, α-haemolysin and β-toxin deficient mutants of Staphylococcus aureus isolated by allele-replacement. J Zentralbl Bakteriol. 1991;21(Suppl) [Google Scholar]
- 23.Foster TJ. Immune evasion by staphylococci. Nat Rev Micro. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 24.Barkas T, Watson CM. Induction of an Fc conformational change by binding of antigen: The generation of protein A-reactive sites in chicken immunoglobulin. Immunology. 1979;36:557–561. [PMC free article] [PubMed] [Google Scholar]
- 25.Warr GW, Magor KE, Higgins DA. IgY: Clues to the origins of modern antibodies. Immunol Today. 1995;16:392–398. doi: 10.1016/0167-5699(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 26.Fowler VG, et al. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. J Infect Dis. 2007;196:738–747. doi: 10.1086/520088. [DOI] [PubMed] [Google Scholar]
- 27.Lerat E, Ochman H. Recognizing the pseudogenes in bacterial genomes. Nucl Acids Res. 2005;33:3125–3132. doi: 10.1093/nar/gki631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright MC, et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Brooks RL, Bounous DI, Andresen CB. Functional comparison of avian heterophils with human and canine neutrophils. Comp Haematol Int. 1996;6:153. [Google Scholar]
- 33.Haslett C, et al. Modulation of multiple neutrophil functions by preparative methods or trace concentrations of bacterial lipopolysaccharide. Am J Pathol. 1985;119:101–110. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.