Abstract
Deficiency of argininosuccinate lyase (ASL) causes argininosuccinic aciduria, an urea cycle defect that may present with a severe neonatal onset form or with a late onset phenotype. To date phenotype-genotype correlations are still not clear because biochemical assays of ASL activity correlate poorly with clinical severity in patients. We employed a yeast-based functional complementation assay to assess the pathogenicity of 12 missense ASL mutations, to establish genotype-phenotype correlations, and to screen for intragenic complementation. Rather than determining ASL enzyme activity directly, we have measured the growth rate in arginine-free medium of a yeast ASLnull strain transformed with individual mutant ASL alleles. Individual haploid strains were also mated to obtain diploid, “compound heterozygous” yeast. We show that the late onset phenotypes arise in patients because they harbor individual alleles retaining high residual enzymatic activity or because of intragenic complementation among different mutated alleles. In these cases complementation occurs because in the hybrid tetrameric enzyme at least one active site without mutations can be formed or because the differently mutated alleles can stabilize each other, resulting in partial recovery of enzymatic activity. Functional complementation in yeast is simple and reproducible and allows the analysis of large numbers of mutant alleles. Moreover, it can be easily adapted for the analysis of mutations in other genes involved in urea cycle disorders.
Argininosuccinic aciduria (ASAuria, MIM 207900)3 is an autosomal recessive disorder of the urea cycle caused by mutations of the ASL gene (hASL, MIM 608310), encoding argininosuccinate lyase (ASL; EC 4.3.2.1.) (1). This enzyme is ubiquitously expressed and catalyzes the reversible breakdown of argininosuccinate to arginine and fumarate. ASL belongs to a superfamily of hydrolases that includes adenylosuccinate lyase and fumarase, which share a homotetrameric structure and a similar catalytic mechanism. The tetrameric structure of ASL accounts for the phenomenon of intragenic complementation. This particular situation occurs when a multimeric protein is formed from subunits produced by differently mutated alleles of the same gene. On complementation, a partially functional hybrid protein is produced from the two distinct types of mutant subunits, neither of which individually has appreciable enzymatic activity (2).
ASL participates to the urea cycle, and in humans it is essential for ammonia detoxification, whereas in lower organisms it is required for the biosynthesis of arginine. Saccharomyces cerevisiae strains harboring a deletion of the homolog of human ASL (ARG4) cannot grow on media lacking arginine (3).
ASAuria is characterized by accumulation of argininosuccinic acid (ASA) in body fluids, and severe hyperammonaemia. The disease displays clinical heterogeneity with two main clinical phenotypes: the acute/neonatal onset form, with symptoms rapidly progressing to deep coma, apnea, and death (1), and the subacute/late onset type, which is diagnosed in infancy or childhood (4). Such patients may present simply with mental retardation or an epileptic disorder. In both types the diagnosis is established unambiguously by measuring plasma levels of ammonia (not always elevated in the late onset form), ASA, and its anhydrides by plasma amino acids assay (1). Over 40 mutations of the ASL gene have been reported, both amino acid substitutions and truncating variants, which are scattered throughout the gene (5, 6).
We have previously reported the identification of novel mutations of the ASL gene in a cohort of Italian patients (7). In this study we employed a yeast model to validate the pathogenicity of missense ASL mutations found in our cohort, to study the effects of different allelic combinations, and to establish possible genotype-phenotype correlations.
EXPERIMENTAL PROCEDURES
Patients and Mutation Analysis
Clinical diagnosis of ASAuria was based on the quantification of plasma ASA by amino acid chromatography, levels of serum ammonia, and determination of urinary orotic acid, during the acute phase of the disease.
Genomic DNA was extracted from peripheral blood leukocytes using the Puregene kit (Gentra). The entire coding region of the ASL gene was amplified from genomic DNA and sequenced as previously described (7).
Construction of Yeast Expression Vectors
Human total RNA was extracted and purified from about 106 cultured skin fibroblasts using the TRIzol kit (Invitrogen) according to the manufacturer's protocol. Synthesis of total cDNA was carried out using SuperScript II kit for reverse transcription (Invitrogen) with random hexamers provided by the manufacturer. A cDNA fragment containing the entire coding region of the human ASL gene (hASL) was then amplified using primers −4F (5′-CAACATGGCCTCGGAGAGT-3′) and 1400R (5′-AGGACCTAGGCCTGCTGTG-3′) and an Expand High Fidelity PCR system (Roche Applied Science). PCR conditions were as follows: 94 °C for 3 min, 35 cycles of 94 °C for 1 min, 55 °C for 1 min, 68 °C for 1 min and 30 s, and a final extension step of 7 min at 68 °C.
S. cerevisiae genomic DNA was extracted from the wild type strain BY4741 using standard rapid glass bead lysis and phenol-chloroform extraction as previously described (8). The coding region of the yeast homolog of argininosuccinate lyase (ARG4) was amplified from genomic DNA using primers −5F (5′-CAAACATGTCAGACGGCACT-3′) and 1422R (5′-TGAAATTCTTGCGCATAACG-3′), and PCR conditions were as above.
hASL and yeast ARG4 fragments were initially cloned into the yeast multi-copy expression vector pYES2.1/V5-His-TOPO (Invitrogen), containing the URA3 marker gene and a GAL1 promoter for the inducible expression of the inserted genes. The correctness and the reading frame of both plasmids were confirmed by direct sequencing.
Specific mutants were generated by site-directed mutagenesis using a QuikChange kit (Stratagene) according to the manufacturer's protocol; mismatched primers were designed with the assistance of the Stratagene Primer Design software (sequences of primers are available upon request). The correctness of each construct was verified by direct sequencing. A control plasmid was generated by cutting the pYES2.1/V5-His-TOPO-hASL vector with EcoRI (which cuts at position 799 related to the hASL ATG) and XbaI (which cuts the plasmid vector immediately after the hASL insert). After treatment with Klenow enzyme, to fill in the resulting overhangs, the fragment was recircularized by legation with T4 DNA ligase. The resulting construct encodes a truncated ASL protein, lacking 195 amino acids on the C terminus.
Each mutant was then cloned into the centromeric pFL vectors. The 2.3-kb-long expression cassette containing the GAL1 promoter, the full hASL cDNA, and the CYC1 transcription termination sequence was amplified from the pYES2.1/V5-His-TOPO-hASL construct using primers carrying adequate restriction sites (BamHIF, 5′-CTTCTGGATCCCTAGTACGGATTAGAAGCCGC-3′; and SphIR, 5′-CTCTTCGCATGCGCCGCAGCTTGCAAATTA-3′, with the restriction sites underlined) and an Expand High Fidelity PCR system (Roche Applied Science). PCR conditions were as follows: 94 °C for 3 min, 12 cycles of 94 °C for 1 min, 55 °C for 1 min, 68 °C for 2 min, and a final extension step of 7 min at 68 °C.
The PCR product was then digested with BamHI and SphI and cloned into the pFL38 and the pFL36 centromeric vectors (9) cut with the same enzymes. The two pFL vectors differ only for the selection marker (URA3 for pFL38 and LEU2 for pFL36). All of the inserts were then sequenced.
Yeast Strains, Media, and Transformations
BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and BY4742 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) wild type strains and the isogenic strains ΔARG4 Y00981 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YHR018c::kanMX4) and ΔARG4 Y10981 (MATα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0; YHR018c::kanMX4) were purchased from Euroscarf Consortium.
Yeast strains were cultured in rich medium (YPD) or synthetic minimal medium at 30 °C, as described (10). YPD (1% yeast extract, 2% peptone, 2% dextrose) was used to routinely maintain wild type and ΔARG4 strains. Synthetic minimal medium (0.17% yeast nitrogen base without amino acids, ammonium sulfate 0.5%, carbon source 2%) contained the necessary auxotrophic supplements and different carbon sources according to the specific experiment: 2% galactose, to achieve maximal expression of hASL, or 2% raffinose plus galactose 0.01% when minimal hASL expression was required. Growth media components were purchased from Difco. All of the yeast DNA transformations were performed with the polyethylene glycol-lithium acetate method as previously described (11).
Biochemical Assay and Western Blot Analysis
Yeast cultures were grown in synthetic minimal medium lacking uracil plus 2% galactose until exponential phase for biochemical assays or to A600 nm of ∼4 for protein extraction and immunoblot analyses. The cells were harvested and mechanically lysed using standard rapid glass bead lysis in a MagnaLyzer instrument (Roche Applied Science). ASL activity was assayed as described (7, 12) using a Jeol JLC 500/V amino acid analyzer, and it is expressed as nmol/h/mg of protein.
Proteins were precipitated with trichloroacetic acid and resuspended in Laemmli loading buffer (13). Equal aliquots of protein extracts were resolved by SDS-PAGE. A monoclonal mouse anti-ASL antibody (Santa Cruz), followed by peroxidase-labeled goat anti-mouse secondary antibody allowed detection by enhanced chemiluminescence (Amersham Pharmacia). The blots were stripped and reprobed with mouse monoclonal antibody raised against yeast Porin (Mitosciences).
Molecular Modeling
The images were generated as reported previously (7), using as templates for modeling the human ASL protein (Protein Data Bank code 1AOS) and Duck Delta Crystallin (Protein Data Bank code 1TJW), an ortholog of ASL, which is crystallized as a tetramer with its substrate.
RESULTS
We have analyzed 12 missense mutations. Seven were reported by Trevisson et al. (7), and two novel changes were identified in other ASAuria patients in the course of this study: c.217G>A (E73K) and c.890G>A (R297Q). Two mutations found in our cohort had been previously reported (V178M and R182Q) but had not been characterized extensively, whereas the Q286R mutation (found in Patient 1) has been widely studied (14) and was therefore included as a positive control. Table 1 summarizes the genotype and the phenotypic characteristics of patients included in this study.
TABLE 1.
ASL genotype and clinical phenotype of patients included in the study
| Patients | Mutations | Clinical phenotypea | Notes |
|---|---|---|---|
| 1 | Q286R/Q286R | N | |
| 2 | M382R (Del E6)b | N | |
| 3 | V178M/R186Q | N | |
| 4 | D31N/D31N | L | |
| 5 | D31N/D31N | L | Brother of Patient 4 |
| 6 | R113Q/R236W | N | |
| 7 | S184X/V335L | L | |
| 8 | S184X/V335L | L | Brother of Patient 7 |
| 9 | R182X/R456W | N | |
| 10 | R182Q/R297Q | L | |
| 11 | V178M T279fs_283Xc | L | |
| 12 | E73K/E73K | N | |
| 13 | R182X/R456W | N | Unrelated to Patient 9 |
| 14 | V178M/R236W | L |
a N, neonatal onset form; L, late onset form.
b Splicing mutation c.[IVS6 + 2T>G]; the effect on protein was deduced through a minigene functional splicing assay (7).
c Truncating mutation c.[839delG].
hASL Complements S. cerevisiae ARG4null Strain
The amino acid sequence of ASL is highly conserved throughout evolution; the human protein shares 56% of identity with the yeast polypeptide encoded by ARG4 gene (15). We checked whether the hASL gene could complement our haploid yeast null mutant ΔARG4, a derivative of the BY4741 strain that harbors a deletion of the ARG4 gene and consequently is auxotrophic for arginine (Table 2).
TABLE 2.
Summary of the characteristics of the mutations analyzed in this study
| Mutation | Class | Growth in minimal medium |
Protein stability (steady state levels) | Proposed effect on protein | |||
|---|---|---|---|---|---|---|---|
| Low copy vector |
High copy vector |
||||||
| 0.01% Gal | 2% Gal | 0.01% Gal | 2% Gal | ||||
| Q286Ra | I | − | − | − | − | +++ | Active site |
| R236Wb | I | − | − | − | − | +++ | Active site |
| R113Qb | I | − | − | − | − | +++ | Active site |
| R182Qc | I | − | − | − | − | −/+ | Interaction among subunits |
| E73Kd | I | − | − | − | − | − | Folding of individual subunit |
| C-terminal del | I | − | − | − | − | NTe | Truncated protein |
| D31Nb | II | − | − | − | ++ | +++ | Active site |
| R186Qb | II | − | − | − | ++ | −/+ | Interaction among subunits |
| R297Qd | II | − | − | − | ++ | +++ | Active site |
| M382Rb | II | − | − | +/− | +++ | + | Folding of individual subunit |
| R456Wb | II | − | ++ | ++++ | ++++ | ++ | Interaction among subunits |
| V335Lb | III | ++ | +++ | ++++ | +++++ | +++ | Not clear (distortion of the central domain of the enzyme?) |
| V178Mb | III | + | +++ | ++++ | +++++ | +++ | Not clear (distortion of the central domain of the enzyme?) |
The coding regions of the human ASL gene and of its S. cerevisiae homolog ARG4 were subcloned into the multi-copy, galactose-inducible yeast expression vector pYES2.1/V5-His-TOPO, carrying the URA3 gene as auxotrophic marker. This expression cassette, including the GAL1 promoter, the full coding region of the hASL gene, and the CYC1 transcription terminator, was also subcloned into the pFL38 (or pFL36) centromeric vector, carrying the same nutritional marker (URA3).
The human and yeast wild type constructs or the empty vector were transformed into ΔARG4. The cells were grown in synthetic minimal medium lacking uracil, containing 2% galactose for 48 h at 30 °C and then plated at serial dilutions in selective synthetic minimal medium plates, lacking uracil and arginine, containing either 2% galactose or 0.01% galactose plus 2% raffinose. Raffinose has no effect on the GAL1 promoter but provides the carbon source for yeast growth.
Under these conditions growth was indicative of functional complementation. Human ASL complemented the growth defect of deleted yeast strain when expressed from both plasmids, at both concentrations of inducer.
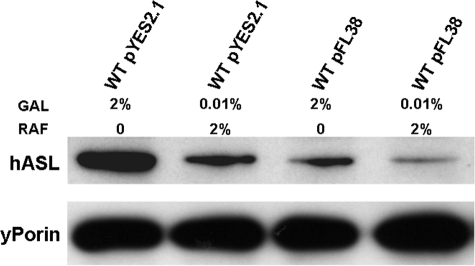
We then checked by immunoblot analysis the levels of the ASL protein under these different conditions (vector type versus concentration of the inductor). As shown in Fig. 1, the highest level of expression of the ASL protein is achieved using the multi-copy number pYES2.1/V5-His-TOPO vector in the presence of 2% galactose. Lower levels of protein were obtained with pYES2.1/V5-His-TOPO-hASL grown in the presence of 0.01% galactose or with pFL38-hASL in the presence of 2% galactose, whereas pFL38-hASL in 0.01% galactose yielded the lowest expression.
FIGURE 1.
ASL protein levels expressed from different plasmids (pFL38 versus pYES2.1-V5-His-TOPO) in the presence of either 2% galactose or 0.01% galactose (GAL) and 2% raffinose (RAF). WT, wild type.
All Missense Mutations Identified in Our Patients Are Pathogenic
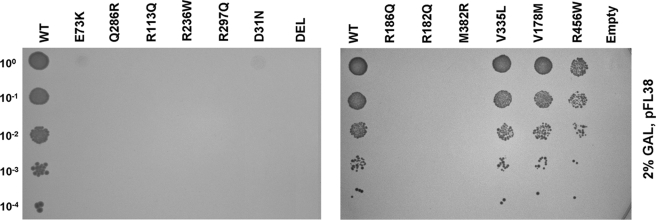
This functional complementation system was used to analyze the effects of missense mutations on ASL activity. Each of the 12 different missense mutations included in the study was introduced into the pFL38-hASL by site-specific mutagenesis. We also generated a further plasmid lacking 589 nucleotides on the 3′ region of the ASL gene to simulate truncating mutations. These plasmids were transformed into the null mutant strain ΔARG4, and we tested the ability of the resulting transformants to grow in the absence of arginine. Yeast cultures were grown as above in the presence of 2% galactose to induce ASL expression, and plates were incubated for 3 days at 30 °C. Under these conditions we observed that all mutant alleles analyzed abolish yeast growth in arginine-deficient medium, except for V178M, V335L, and R456W alleles, which retain residual growth (Fig. 2).
FIGURE 2.
Functional complementation of ΔARG4 yeast strain transformed with the centromeric pFL38 vector expressing the different mutated ASL alleles. Cells were grown in agar plates with synthetic minimal medium lacking uracil and arginine, and expression was induced with 2% galactose (GAL). The initial suspension at 1 unit of A600 nm/ml was diluted 1/10 four times. WT, wild type; DEL, ΔARG4 transformed with the truncated allele; Empty, ΔARG4 transformed with the empty vector.
We measured ASL enzymatic activity in strains expressing each of these three mutations and in controls transformed with the wild type human gene, grown in SM-URA medium with 2% galactose. Mutant alleles displayed a reduced enzymatic activity compared with the wild type: 14% for V178M and 17% for V335L, whereas we could not detect significant activity in the R456W mutant. Taken together, these data confirm the pathogenicity of all the missense mutations found in our patients.
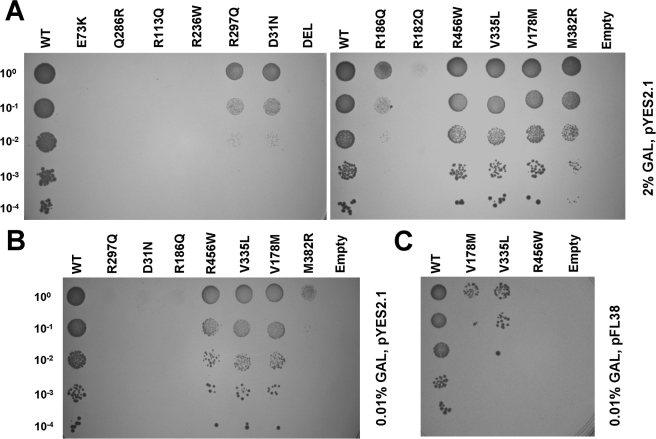
ASL Expression Levels Influence the Growth Phenotype of Some Mutations
We repeated the above functional complementation assay under conditions of maximal expression, using the mutagenized versions of the pYES2.1/V5-His-TOPO-hASL construct in the presence of high (2%) or low (0.01%) levels of the galactose inducer. Under conditions of maximal expression, we detected some growth with four alleles (D31N, R186Q, R297Q, and M382R) (Fig. 3A), whereas results obtained with low inducer concentrations (Fig. 3B) were similar to those of pFL38-hASL grown in 2% galactose (Fig. 2). This is consistent with the similar levels of expression of the ASL protein with these different experimental conditions (Fig. 1).
FIGURE 3.
A, functional complementation of ΔARG4 yeast strain transformed with the high copy pYES2.1-V5-His-TOPO vector expressing the different mutated ASL alleles. The cells were grown in plates with synthetic minimal medium lacking uracil and arginine, and expression was induced with 2% galactose (GAL). B, strains displaying residual growth in the above conditions were grown in 2% raffinose with 0.01% galactose to induce lower ASL expression. C, mutants allowing residual growth when expressed form the low copy pFL38 plasmid with 2% galactose were grown in 2% raffinose with 0.01% galactose to induce minimal ASL expression. Letter codes are as in Fig. 2.
In parallel, V178M, V335L, and R456W alleles were expressed from the pFL38 vector in the presence of only 0.01% galactose. In this case we detected a clear growth defect compared with the wild type (Fig. 3C).
These data allowed us to distinguish three classes of mutations: Class I, no growth under any condition: Q286R, R236W, R113Q, R182Q, E73K, and C-terminal deletion; Class II, no growth with minimal expression: D31N, R186Q, R297Q, M382R, and R456W; and Class III, growth with minimal expression, albeit reduced compared with wild type: V178M and V335L. All of the experiments were replicated using haploid strains of the opposite mating type and yielded overlapping results.
Screening for Intragenic Complementation Reveals That E73K Is Refractory to Complementation
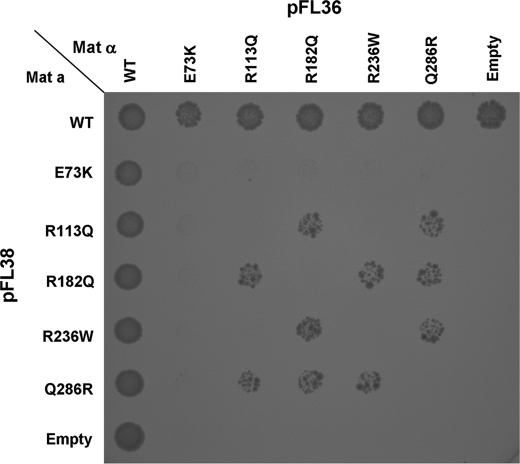
Intragenic complementation has been invoked to explain some of the cases with mild phenotypes (14). We devised a simple functional assay to verify the occurrence of intragenic complementation between different Class I ASL mutations. We chose these alleles because they do not grow under any of the conditions we tested. We transformed both the opposite mating types (MATa and MATα) ΔARG4 strains with each of the five human Class I alleles. The MATa ΔARG4 strain was transformed with the pFL38 (which expresses the URA3 selection marker) constructs and selected on synthetic minimal medium lacking uracil, whereas MATα ΔARG4 strain was transformed with the pFL36-derived constructs (obtained by cloning the pFL38 expression cassette in pFL36, which differs from pFL38 only for the presence of the LEU2 selection marker instead of URA3) and plated on synthetic minimal medium lacking leucine.
We mated the transformants on plates and tested their growth in synthetic minimal medium lacking uracil and leucine (to select for the presence of both plasmids), but also methionine and lysine (for which the MATa and the MATα strains are respectively auxotrophic), and arginine (to select for the occurrence of intragenic complementation). As expected, no growth was seen in “homozygous” diploid strains, whereas some allelic combinations were able to rescue the growth phenotype (Fig. 4). The E73K allele appeared to be refractory to complementation because no growth was detected in any of the different diploid strains harboring this allele; Q286R and R182Q could restore growth in all other allele combinations. The R113Q/R236W combination did not restore any growth in diploid strains; however, these alleles allowed intragenic complementation when associated with the Q286R or R182Q mutations (Fig. 4).
FIGURE 4.
Rapid screening for intragenic complementation. Haploid ΔARG4 yeast strain of both MATa (auxotrophic for methionine) and MATα (auxotrophic for lysine) mating types were transformed with different ASL mutated alleles cloned in either pFL38 (containing the URA3 selection marker) or pFL36 (identical to pFL38 except for the LEU2 selection marker). 10 μl from MATa and MATα cultures were spotted onto agar plates with synthetic minimal medium lacking uracil, leucine, methionine, and lysine; allowed to mate; and incubated at 30 °C for 3 days. These conditions allow growth only of the diploid strains harboring both plasmids but are not selective for arginine auxotrophy. Growth was observed in all spots (not shown), confirming that mating had occurred. The plates were then replicated on agar plates with synthetic minimal medium lacking uracil, leucine, methionine, lysine, and also arginine, which allow growth only of the diploid strains that recover some ASL activity by intragenic complementation. Note that no growth is observed in any of the “homozygous” diploid strains. Letter codes are as in Fig. 2.
Intragenic Complementation Accounts for the Mild Phenotype in Patient 10
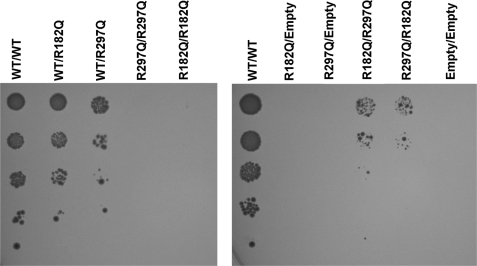
We investigated whether intragenic complementation could account for the mild phenotype displayed by Patient 10, who carried the R182Q and R297Q mutations. We mated MATa and MATα yeast transformed with each of these two alleles (cloned in the pFL38 or pFL36 plasmid as above). We detected growth in the “compound heterozygote” strain (R182Q/R297Q), whereas the “homozygote” strains (R182Q/R182Q or R297Q/R297Q) do not grow at all (Fig. 5), indicating that this combination of alleles results in partial rescue of the enzymatic activity of the tetramer, which is consistent with the milder phenotype observed in the patient. Interestingly, the lack of complementation of the R113Q/R236W combination correlates with the severe phenotype found in Patient 7, who harbored these two mutations (Fig. 4).
FIGURE 5.
Detailed analysis of intragenic complementation between the R182Q and R297Q mutated alleles. Diploid strains were obtained by mating haploid strain of different mating types transformed with individual mutants (expressed either from a pFL36 or pFL38 vector) and selected in agar plates lacking methionine and lysine, for which they are respectively auxotrophic. Diploid cells were grown as above and inoculated into agar plates with synthetic minimal medium lacking uracil, leucine, methionine, lysine, and arginine, diluting the initial suspension at 1 unit of A600 nm/ml. ASL expression was induced by 2% galactose. 10 μl of cultures were spotted onto plates, which were incubated at 30 °C for 3 days. Letter codes are as in Fig. 2.
Analysis of the V178M Allele in Different Diploid Combinations
The growth data and biochemical measurements obtained with the haploid strain indicate that the V178M allele retains significant residual activity, allowing growth of the corresponding transformant also when expressed at very low levels (Fig. 3C).
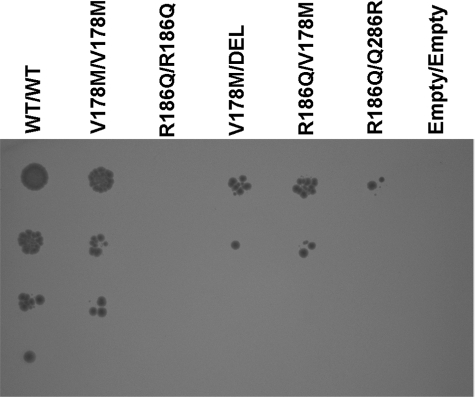
This mutation has been reported in association with both the neonatal and the late onset phenotypes in distinct genotypic combinations. In fact, it was previously associated to a mild phenotype when present in a homozygous state (6); we have found the same mutation in a compound heterozygous state with R236W (Patient 14) and with the truncating mutation c.839delG (Patient 11); both subjects presented with the late onset phenotype. However, we found also V178M in association with R186Q mutation in Patient 3 who apparently presented the severe phenotype (7). To explain this discrepancy, we hypothesized that the R186Q allele might have a sort of dominant negative effect, displaying an effect opposite to intragenic complementation.
We therefore analyzed the effects of V178M in compound heterozygosity with either the R186Q mutant or the truncating mutation. As a further control we studied the combination of R186Q and the high complementing Q286R allele. We crossed both the mating types of yeast transformed with the corresponding mutated constructs as above. Expression was induced with 0.01% galactose to increase the sensitivity of the assay. As shown in Fig. 6, the V178M mutation in the homozygous state displays a slightly reduced growth pattern compared with the wild type. There is a further reduction of growth when V178M is in combination with either the deletion mutant or R186Q. However, the growth of the V178M/DEL and V178M/R186Q combinations is similar, and we observed intragenic complementation with the R186Q/Q286R combination. These data reject the hypothesis of a dominant negative effect for the R186Q allele.
FIGURE 6.
Analysis of different genotypic combinations of the V178M, R186Q, and Q286R alleles. The experiments were performed as in Fig. 5 except that expression was induced with 0.01% galactose to increase sensitivity. The carbon source was 2% raffinose. Letter codes are as in Fig. 2.
Relationships between Mutation Type and ASL Protein Stability
The knowledge of the three-dimensional structure of the ASL enzyme allows predictions regarding the effect of individual mutants on protein structure and function. In a previous work we analyzed the consequences of some of the mutations included in the study (7). D31N, R113W, and R236W affect the active site of the enzyme (see Fig. 3B of Trevisson et al. (7)), and R186Q and R456W are predicted to alter the interaction between individual monomers (see Fig. 4 of Trevisson et al. (7)), whereas M382R is predicted to affect the folding of a peripheral loop of ASL. The effect of V335L is less clear. The substitution of valine 335 with a leucine is predicted to distort the central domain of the protein. Q286R was previously reported to affect the active site (2).
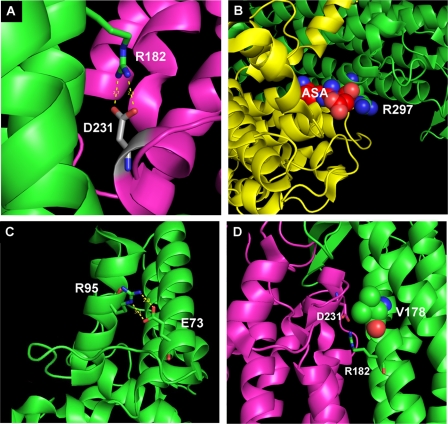
Analysis of the other mutations included in the study showed that arginine 182 is located in domain 2 of the protein, and it is critical for the interaction of two distinct subunits, forming hydrogen bonds with aspartate 231 of the opposite monomer (Fig. 7A). In this region a series of acidic and basic residues form a sort of “molecular zipper” that maintains the association of the monomers. Disruption of this interaction probably impairs the correct assembly of the tetramer. Arginine 297 is located close to the active site of the protein (Fig. 7B). Aspartate 73 is located on a peripheral α helix of domain 1 and forms hydrogen bonds with arginine 95 (Fig. 7C) of the same monomer. Residue 95 was found mutated in another ASAuria patient, and the R95C mutation was believed to impair the correct folding of the mutated protein, which is rapidly degraded (16, 17). Finally, valine 178 is located on the surface of the central domain of ASL (Fig. 7D). It does not appear to interact directly with residues on the opposite ASL monomer, and it is located close to both the active site and to arginine 182 (see above). Similar to what it has been hypothesized for the V335L mutation, the substitution with a larger residue like methionine may distort the structure of this portion of the molecule.
FIGURE 7.
A, interaction between arginine 182, located in helical bundle within the central domain of the protein, and aspartate 231 of the opposite monomer. B, representation arginine 297 located near the active site. C, interaction between aspartate 73 and arginine 95. D, position of the Val178 residue on domain 1 of the protein close to arginine 182.
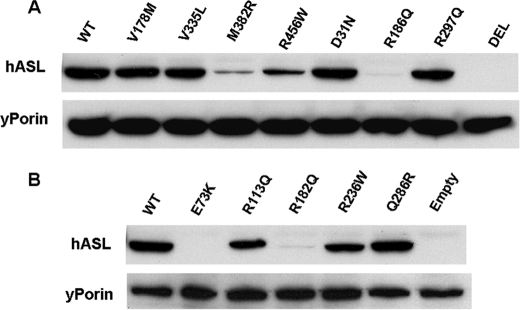
We then analyzed by immunoblot the levels of ASL protein expressed from the different mutant alleles cloned into pFL38 (Fig. 8). In this case yeast were grown in the presence of arginine (to allow growth of Class I mutants) and 2% galactose. Although there is no correlation between the mutation class and protein stability, we detected a clear relationship between the steady state levels of mutant ASL polypeptides and the type of mutation. In fact, all proteins with mutations affecting the active site of the enzyme were present at essentially normal levels. Conversely, mutations that by molecular modeling are predicted to affect the folding of individual subunits or the interaction between different monomers caused a considerable reduction of ASL protein levels (Table 2).
FIGURE 8.
Immunoblot analysis of different ASL mutants expressed from pFL38 vector induced by 2% galactose. Cells were grown in the presence of arginine to allow growth of yeast transformed with ASL alleles with virtually no residual activity. Porin was used as loading control. mRNA expression of the E73K allele was confirmed by reverse transcription-PCR (not shown). Letter codes are as in Fig. 2.
DISCUSSION
Although the genetic bases of ASAuria have been known for almost 20 years, genotype-phenotype correlations are still not well established, and the biochemical bases for its clinical heterogeneity are unclear. Environmental factors play an important role in determining the phenotype (18), and previous studies have shown a poor correlation between the clinical phenotype and residual ASL activity measured in cultured fibroblasts or in other tissues (1, 14, 19, 20). However, biochemical determination of ASL activity in cell lysates has a limited sensitivity, and the apparent absence of residual biochemical activity observed in fibroblasts of patients with the late onset phenotype has been explained invoking the lability of the mutant ASL protein, which may cause complete deterioration of the enzyme in a cell homogenate (6). Expression studies in COS cells have similar limitations, and the presence of basal ASL activity in these cells further complicates the analysis of alleles with low residual activity.
In vivo [14C]citrulline uptake studies are more sensitive and show a better correlation with the clinical phenotype (6), but they are complex and require a skin biopsy, which is an invasive procedure, especially in pediatric patients. Because patients are often compound heterozygotes for different mutations, direct measurements cannot discriminate the role of individual alleles.
We employed functional complementation of yeast deletion mutants as a tool to characterize the various mutant alleles identified in our patients. We evaluated growth in arginine-free medium rather than performing direct biochemical assays, because this method proved to be more sensitive to detect low levels of residual enzymatic activity. We also generated diploid strains to analyze intragenic complementation. In our hands yeast proved to be a useful model to study the effect of individual alleles, independently of the genetic background of the patient, and to test the effect of different allelic combinations.
The centromeric pFL38 vector provides a stable and reliable system to express the different mutant alleles in yeast (9), and the levels of expression that can be achieved are particularly suitable to study alleles with significant residual activity. However, low copy centromeric vectors may not achieve the higher expression levels required to study mutations with low residual activity. This is the main reason why we used the pYES2.1 high copy vector to study the alleles that did not display growth when expressed with pFL vectors. Even though high copy episomal vectors containing the 2μ element are less reliable than centromeric vectors because their copy number may fluctuate, they have been widely used, especially when expressing heterologous genes, including hASL, in S. cerevisiae (3, 21–23). Moreover, all experiments were repeated using strains of the opposite mating type and yielded the same results.
Our series of patients displayed both the acute neonatal and the late onset form. Our model shows that patients with the late onset form harbor either alleles, which retain significant residual activity or which allow the occurrence of intragenic complementation.
The V335L and V178M alleles found in patients with the late onset form encode for stable proteins, as shown by immunoblot analysis, and retain the highest residual activity, measured both by growth in SM-URA-ARG and by the direct biochemical assay. They probably can ensure sufficient ASL activity even when associated with a null allele.
However, we found V178M in association with R186Q in Patient 3, who had neonatal onset of symptoms (7). The yeast model excluded a dominant negative effect of R186Q. We therefore reviewed the clinical documentation of the patient. He presented after birth with a single acute severe episode of hyperammonaemia precipitated by an infection and with prolonged coma before being diagnosed with ASAuria. Although he was successfully treated, the damage to the central nervous system caused by the delayed diagnosis was devastating and permanent. Nevertheless, the subsequent clinical course was reminiscent of the late onset phenotype. In fact, despite undergoing stressful events that increased the turnover of endogenous proteins, such as infections or episodic fasting, he has never developed further episodes of acute hyperammonaemia, and the clinical course was uneventful. Moreover, he has never developed hypertransaminasemia and hepatomegaly, which are typically present in patients with the neonatal onset form, but they are rare in those with the late onset phenotype (6, 7). These data confirm the notion that environmental and/or iatrogenic factors play an important role in determining the outcome of ASAuria patients (18).
The D31N mutation was also associated with the late onset phenotypes (Patients 4 and 5). The two siblings harbored this mutation in the homozygous state (therefore excluding intragenic complementation). This is a Class II mutation that does retain some residual activity, albeit lower than other class II mutations as M382R and R456W, which were found in patients with the neonatal phenotype (Patients 2, 9, and 13). However, in these cases M382R and R456W were associated with a null allele.
One possible interpretation is that the modulation of ASL expression levels in vivo in critical tissues like the liver may determine the milder phenotype in these patients similarly to what we observed with the yeast growth phenotype. Alternatively the presence of two alleles with some residual activity may result in sufficient degradation of argininosuccinate to protect from the early onset disease. The D31N mutation has been reported in a single family; if further patients are reported, it will be interesting to compare their phenotypes.
Intragenic complementation accounts for the milder phenotype only in Patient 10. Our model shows that the coexpression of the R182Q and R297Q alleles found in this patient restores growth in yeast.
Complementation phenotypes do not arise with all mutant alleles, and several possible mechanisms have been proposed to explain the occurrence of this phenomenon (17). The first is dictated by the molecular symmetry of the ASL protein, in which the combination of distinct stable active site mutant monomers leads to the formation of at least one normal active with partial recovery of enzymatic activity (Fig. 9, A and B). The second occurs when a stable mutant subunit and an unstable mutant subunit form a hybrid protein with enough overall stability for catalysis to occur (24).
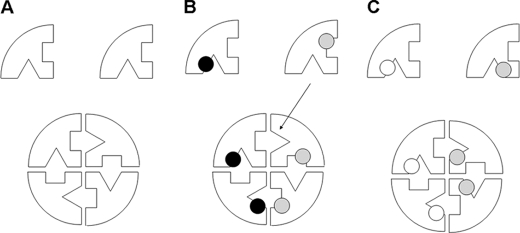
FIGURE 9.
Simplified representation of the mechanism by which two differently mutated monomers can produce a tetrameric protein with at least one normal active site. A, wild type protein. B, as in the case of the R113Q/Q286R combination, intragenic complementation occurs because one “normal” active site (arrow) can be formed. The dots represent the different mutations. C, as in the case of the R113Q/R236W combination, intragenic complementation does not occur because the mutations affect the same region of the monomer, and no “normal” active sites can be formed.
Our assays show that Q286R mutation in combination with either R113Q or R236W (all stable active site mutants) results in intragenic complementation, likely because of restoration of at least one normal active site in the tetramer. We did not detect intragenic complementation with the R113Q/R236W combination (interestingly this genotype was found in a patient with the severe phenotype). Arginine 113 and arginine 236 both participate in the formation of the active site but are located very close to each other in the quaternary structure of the protein (see Fig. 3B in Trevisson et al. (7)). In this case, lack of complementation can be explained by the fact that no wild type catalytic sites can be formed because the two mutations affect the same region of the monomer (Fig. 9C).
Intragenic complementation occurring with R182Q in combination with R113Q, R236W, or R297Q or with the R186Q/Q286R combination is an example of the second proposed mechanism of action, where a stable active site mutant stabilizes an unstable subunit. It should be noted that both R182Q and R186Q mutations are predicted to affect interaction between different subunits. In contrast, the E73K mutation, which is refractory to complementation, is predicted to disrupt the folding of a peripheral domain of the ASL monomer. The affected loop does not interact with other subunits and probably cannot be stabilized even in the presence of a stable mutant. Interestingly, we could not detect intragenic complementation also with the Q286R allele in combination with M382R (data not shown), the other mutant predicted to disrupt the folding of a peripheral domain of ASL (7).
In conclusion, our results show that to establish genotype-phenotype correlations in ASAuria, it is important to consider not only the role of individual alleles but also specific allelic combinations. Our model is particularly suitable to this purpose, and this type of analysis is simple, sensitive, and reproducible and overcomes the problems inherent to classical biochemical assays. Conceptually, it is similar to the indirect assay of radiolabeled citrulline uptake in fibroblasts, because both rely on an in vivo system. Functional complementation in yeast allows the simultaneous analysis of large numbers of mutant alleles and does not involve the complex procedures required to express and purify recombinant protein in Escherichia coli. Furthermore, it has higher sensitivity than the biochemical method, allowing the evaluation of mutations that retain low residual activity, undetectable by the classical biochemical assays, and it is particularly suitable to investigate intragenic complementation. This strategy can be easily adapted to study other urea cycle defects and in general most inborn errors of metabolism.
Acknowledgments
We are grateful to Mauro Naturale for technical assistance and to Prof. Iliana Ferrero and Paola Goffrini (Dept. of Genetics, University of Parma, Parma, Italy) for providing us with the pFL36 an pFL38 plasmids.
This work was supported by Telethon Italy Grant GGP06256 and by Fondazione Città della Speranza.
- ASAuria
- argininosuccinic aciduria
- ASL
- argininosuccinate lyase
- ASA
- argininosuccinic acid.
REFERENCES
- 1.Brusilov S. W., Horwich A. L. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. S., Valle D. eds) pp. 1909–1964, McGraw-Hill, New York [Google Scholar]
- 2.Turner M. A., Simpson A., McInnes R. R., Howell P. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9063–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa P., Cialkowski M., O'Brien W. E. (1991) J. Biol. Chem. 266, 5286–5290 [PubMed] [Google Scholar]
- 4.Kleijer W. J., Garritsen V. H., Linnebank M., Mooyer P., Huijmans J. G., Mustonen A., Simola K. O., Arslan-Kirchner M., Battini R., Briones P., Cardo E., Mandel H., Tschiedel E., Wanders R. J., Koch H. G. (2002) J. Inherit. Metab. Dis. 25, 399–410 [DOI] [PubMed] [Google Scholar]
- 5.Linnebank M., Homberger A., Rapp B., Winter C., Marquardt T., Harms E., Koch H. G. (2000) J. Inherit. Metab. Dis. 23, 308–312 [DOI] [PubMed] [Google Scholar]
- 6.Linnebank M., Tschiedel E., Häberle J., Linnebank A., Willenbring H., Kleijer W. J., Koch H. G. (2002) Hum. Genet. 111, 350–359 [DOI] [PubMed] [Google Scholar]
- 7.Trevisson E., Salviati L., Baldoin M. C., Toldo I., Casarin A., Sacconi S., Cesaro L., Basso G., Burlina A. B. (2007) Hum. Mutat. 28, 694–702 [DOI] [PubMed] [Google Scholar]
- 8.Dimmer K. S., Navoni F., Casarin A., Trevisson E., Endele S., Winterpacht A., Salviati L., Scorrano L. (2008) Hum. Mol. Genet. 17, 201–214 [DOI] [PubMed] [Google Scholar]
- 9.Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. (1991) Yeast 7, 609–615 [DOI] [PubMed] [Google Scholar]
- 10.Burke D., Dawson D., Stearns T. (2000) Methods in Yeast Genetics, pp. 113–114, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 11.Gietz R. D., Woods R. A. (2006) Methods Mol. Biol. 313, 107–120 [DOI] [PubMed] [Google Scholar]
- 12.Bastone A., Diomede L., Parini R., Carnevale F., Salmona M. (1990) Anal. Biochem. 191, 384–389 [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 14.Sampaleanu L. M., Vallée F., Thompson G. D., Howell P. L. (2001) Biochemistry 40, 15570–15580 [DOI] [PubMed] [Google Scholar]
- 15.O'Brien W. E., McInnes R., Kalumuck K., Adcock M. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 7211–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker D. C., McCloskey D. A., Simard L. R., McInnes R. R. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 9625–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valnot I., von Kleist-Retzow J. C., Barrientos A., Gorbatyuk M., Taanman J. W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., Rötig A. (2000) Hum. Mol. Genet. 9, 1245–1249 [DOI] [PubMed] [Google Scholar]
- 18.Lanpher B., Brunetti-Pierri N., Lee B. (2006) Nat. Rev. Genet. 7, 449–460 [DOI] [PubMed] [Google Scholar]
- 19.McInnes R. R., Shih V., Chilton S. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 4480–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker D. C., Christodoulou J., Craig H. J., Simard L. R., Ploder L., Howell P. L., McInnes R. R. (1997) J. Biol. Chem. 272, 6777–6783 [DOI] [PubMed] [Google Scholar]
- 21.Morillas M., Gómez-Puertas P., Rubí B., Clotet J., Ariño J., Valencia A., Hegardt F. G., Serra D., Asins G. (2002) J. Biol. Chem. 277, 11473–11480 [DOI] [PubMed] [Google Scholar]
- 22.Lodi T., Bove C., Fontanesi F., Viola A. M., Ferrero I. (2006) Biochem. Biophys. Res. Commun. 341, 810–815 [DOI] [PubMed] [Google Scholar]
- 23.Mollet J., Giurgea I., Schlemmer D., Dallner G., Chretien D., Delahodde A., Bacq D., de Lonlay P., Munnich A., Rötig A. (2007) J. Clin. Invest. 117, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu B., Paroutis P., Davidson A. R., Howell P. L. (2004) J. Biol. Chem. 279, 40972–40979 [DOI] [PubMed] [Google Scholar]