Abstract
The removal of introns from pre-mRNA is performed by the spliceosome that stepwise assembles on the pre-mRNA before performing two catalytic steps. The spliceosome-associated CDC5L-SNEVPrp19-Pso4 complex is implicated in activation of the second catalytic step of pre-mRNA splicing, and one of its members, SNEVPrp19-Pso4, is also implicated in spliceosome assembly. To identify interaction partners of SNEVPrp19-Pso4, we have performed yeast two-hybrid screenings. Among the putative binding partners was a so far uncharacterized protein carrying two heterogeneous nuclear ribonucleoprotein K homology domains that we termed Blom7α. Blom7α is expressed in all tissues tested, and at least three splice variants exist. After confirming direct and physical interaction of SNEV and Blom7α, we investigated if it plays a functional role during pre-mRNA splicing. Indeed, Blom7α co-localizes and co-precipitates with splicing factors and pre-mRNA and is present in affinity-purified spliceosomes. More importantly, addition of Blom7α to HeLa nuclear extracts increased splicing activity in a dose-dependent manner. Furthermore, we tested if Blom7α influences splice site selection using two different minigene constructs. Indeed, both 5′- as well as 3′-site selection was altered upon Blom7α overexpression. Thus we suggest that Blom7α is a novel splicing factor of the K homology domain family that might be implicated in alternative splicing by helping to position the CDC5L-SNEVPrp19-Pso4 complex at the splice sites.
Pre-mRNA splicing is the removal of intronic, noncoding sequences from mRNA in a co-transcriptional catalytic process. This process is performed by the spliceosome, a large multiprotein machinery consisting of four small nuclear ribonucleoprotein particles (snRNPs2 U1, U2, U4-U6, and U5) and more than 100 different proteins that stepwise assemble on the pre-mRNA (1, 2).
One distinct subcomplex that is associated with the spliceosome is the human CDC5L-SNEVPrp19-Pso4 complex (3, 4) or its highly evolutionarily conserved counterpart in yeast, the NineTeen complex (5–8). The human core complex consists of SNEVPrp19-Pso4, CDC5L, PLRG1, SPF27(BCAS2), and Hsp73 (3, 4). Two to three additional proteins are considered as core members, depending on the study of either AD002 and β-catenin-like 1 (CTNNBL1) (4) or CCAP6 (3). Several reports highlight the importance of this complex for the second catalytic step, because immunodepletion or inhibition of the interaction between the two subunit members CDC5L and PLRG1 affects this step the most (3, 9, 10), which is in accordance with its presence in the C complex (11–13).
However, SNEVPrp19-Pso4 appears to associate with the spliceosome even prior to catalysis, because it has been identified in several pre-catalytic complexes as follows: in the A complex (14), the pre-catalytic BΔU1 complex, which can be immunopurified after 8 min of in vitro splicing reactions using antibodies to the U4-U6-specific 61-kDa protein (4); the B* complex, which is immunopurified from splicing reactions after 10 min using antibodies to SKIP and therefore might represent the activated spliceosome before catalysis (13); and the B complex, which can be immunopurified under native, low stringency conditions using glycerol gradient centrifugation and MS2-tagged MINX pre-mRNA for affinity purification (15). Because all of these complexes represent different steps in spliceosome activation, a pre-catalytic role for SNEVPrp19-Pso4 has been suggested. Consistently, the inhibiting SNEVPrp19-Pso4 self-interaction by peptides mimicking the self-interaction domain results in inhibition of spliceosome formation (16). This indicates that SNEVPrp19-Pso4 could form a scaffold that allows assembly of other splicing factors. Another possibility is that the E3 ligase activity of SNEVPrp19-Pso4 (17) and its interaction with the proteasome (18, 19) are necessary for spliceosome assembly. Indeed, the rearrangement of the yeast U4-U6-U5 tri-snRNP is dependent on ubiquitin (20).
We have previously found SNEVPrp19-Pso4 down-regulated during cellular senescence (21) to extend the replicative life span of human umbilical vein endothelial cells upon overexpression. Accordingly, we termed it Senescence Evasion factor (SNEV) (22). To merge the different names of SNEV that derive from the yeast homologue that were synonymously termed Pso4 as well as Prp19, we suggest here to use SNEVPrp19-Pso4. Knock-out of SNEVPrp19-Pso4 in mice is early embryonic lethal, whereas embryonic fibroblasts from heterozygous SNEVPrp19-Pso4+/− mice show reduced replicative life span (23) and hematopoietic progenitor defects in proliferation and self-renewal (24). This effect might be due to its additional function as a DNA repair factor (25, 26), especially in interstrand cross-link repair, where it interacts with WRN, the protein mutated in the premature progeroid Werner syndrome (27). Furthermore, SNEVPrp19-Pso4 itself is ubiquitinated upon DNA damage (28). SNEVPrp19-Pso4 has also been found in a protein complex different from the NineTeen complex, containing the xeroderma pigmentosa complementation group A-binding protein (XAB2)-hSYF1, involved in transcription-coupled nucleotide excision repair (29). Other members of this complex are hAquarius-intron-binding protein 6 (IBP160), hISY1, peptidylprolyl cis-trans-isomerase E (PPIE)-hCyp33, and a coiled-coil domain containing 16 CCDC16-OMCG1 (29). Finally, it is a regulator of adipocyte differentiation, as has been shown using mouse 3T3′L1 cells after adipogenic differentiation (30).
To further enhance our understanding of SNEVPrp19-Pso4, we performed yeast two-hybrid (Y2H) studies to detect novel interaction partners. Here we report the identification of a previously uncharacterized KH domain protein that we termed Bring lots of money 7α (Blom7α). Blom7α interacts with SNEVPrp19-Pso4 directly and physically, co-precipitates and co-localizes with other splicing factors, and is detected in affinity-purified, mature spliceosomes. Addition of recombinant Blom7α increased the activity of in vitro splicing reactions, whereas its overexpression in HeLa cells changes the splice site selection of reporter constructs, suggesting that Blom7α is a novel splicing factor involved in alternative 3′- and 5′-splice site selection.
MATERIALS AND METHODS
Y2H Assays
Directed Y2H assays were performed using the MATCHMAKER GAL4 Two-hybrid System.3 (Clontech) according to the manufacturer's guidelines. Therefore, the cDNAs of SNEVPrp19-Pso4 and SNEVPrp19-Pso4 deletion constructs were inserted into the bait vector pGBKT7 (Clontech) in-frame with the GAL4 DNA binding domain and a c-Myc epitope. As cDNA library for identification of novel putative interactors, the pACT2 plasmid-based human aorta cDNA library was used (Clontech). Blom7α and its deletion mutants were inserted into the vector pGADT7 (Clontech) providing the GAL4 activation domain and the hemagglutinin (HA) tag. All constructs were amplified in Escherichia coli and sequence-analyzed to confirm correct proteins. The constructs were co-transformed into Saccharomyces cerevisiae strain AH109 (Clontech) by LiAc co-transformation. We selected interaction-positive clones by growth on high stringency SD (synthetic dropout) medium (4× SD: −Trp−Leu−His−Ade; SD medium lacking four components: tryptophan, leucine, histidine, and adenine hemisulfate).
Sequence Analysis
For the determination of the sequence architecture and the prediction of molecular function of sequence segments, the protein sequences of the BLOM7 isoforms have been processed as described previously (31). In brief, more than 30 sequence-based methods for the prediction of nonglobular segments have been applied, including low complexity regions, post-translational modifications, subcellular translocations signals, as well as comparisons with six libraries of known sequence domains and repeats.
Recombinant Protein Expression, Purification, and Generation of Antibodies
His6-SNEV Prp19-Pso4 was expressed in Sf9 insect cells and affinity-purified on a Ni2+-NTA column as described recently (32). Blom7α-His6 was cloned into pET-30a(+) vector (Novagen) according to standard procedures using EcoRI and NotI restriction sites. After recombinant Blom7 expression using the overnight express autoinduction system 1 (Novagen), the Ni2+-NTA purified protein was used for generation of antibodies according to standard procedures.
Co-immunoprecipitation
CoIP was performed by incubation of in vitro translated HA-Blom7α (TnT-T7 coupled reticulocyte lysate system, Promega) and purified His6-SNEVPrp19-Pso4 for 1 h at 4 °C, followed by incubation with Ni2+-NTA-agarose beads (Qiagen, Germany) for 1.5 h at 4 °C. The beads were pre-equilibrated with TBST buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, and 0.1% v/v, Tween 20). The beads were washed five times with TBST buffer and eluted by boiling with SDS-PAGE sample buffer for 10 min. After SDS-PAGE, precipitated HA-Blom7α was detected using anti-HA antibody (Covance) and anti-mouse peroxidase conjugate as secondary antibody (Sigma).
Alternatively, for precipitation of endogenous proteins, protein A-agarose beads (Roche Diagnostics) coupled (coupling in 1× PBS, 1 h at room temperature) with mouse anti-Blom7α antibody and negative control beads coupled with mouse anti-HA antibody (Covance) were incubated with the nuclear extract (prepared as described in Ref. 33) in CoIP-buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.2% Triton X-100) for 2 h at 4 °C. The beads were washed four times with CoIP-buffer, and proteins were separated by SDS-PAGE. The detection of precipitated SNEV Prp19-Pso4 was done by Western blotting with a polyclonal anti-SNEV Prp19-Pso4 rabbit antibody (Prp19–866).
GST Pulldown Assay
GST-SNEVPrp19-Pso4 was expressed and purified as described (16). For the pulldown assay, equal amounts of the purified proteins (0.5 μg) were mixed together and incubated for 2 h at 4 °C in CoIP-buffer (200 mm NaCl, 25 mm Tris-HCl, pH 7.4, 0.5% Triton). Glutathione-Sepharose 4B beads (Amersham Biosciences), equilibrated with CoIP-buffer, were added, followed by incubation for 2 h at 4 °C. The beads were washed three times with CoIP-buffer, and proteins were eluted with SDS-PAGE sample buffer. After SDS-PAGE, co-precipitated Blom7α was detected using anti-His4 antibody (Qiagen, Germany).
Cell Culture
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 4 mm l-glutamine and 10% (v/v) fetal calf serum. HEK-293 (human embryonic kidney 293) cell line was grown in Dulbecco's modified Eagle's medium/Ham's medium supplemented with 4 mm l-glutamine and 10% fetal calf serum.
FRET Microscopy
SNEVPrp19-Pso4 and Blom7α inserted into pECFP-N1, pECFP-C1, pEYFP-N1, and pEYFP-C1, respectively, were transiently co-transformed into COS-1 and HEK-293 cells by LipofectamineTM 2000 (Invitrogen) according to the manufacturer's guidelines. After 24 h, FRET images from living cells were generated by the MicroFRET method as described (34). Photos were captured on a Nikon Diaphot TMD microscope with a cooled charge-coupled device camera (Kappa GmbH, Gleichen, Germany), with the yellow fluorescent protein (YFP), cyan fluorescent protein (CFP), and FRET filter sets (Omega Optical, Brattleboro, VT), under identical conditions and processed with Scion Image software version beta 4.0.2 (Scion, Frederick, MD). The images were aligned by pixel shifting abd inverted, and the background was subtracted. Images from the YFP and CFP filter sets were multiplied with their previously assessed correction factors (0.19 for YFP and 0.59 for CFP) and subtracted from the FRET filter set picture. The remaining signals were multiplied by 3 for better visualization, and they represent the corrected FRET.
Cell Staining and Immunofluorescence Analyses
HeLa cells were washed with PBS and fixed for 5 min in 3.7% (w/v) paraformaldehyde in CSK buffer (10 mm Pipes, pH 6.8, 10 mm NaCl, 300 mm sucrose, 3 mm MgCl2 and 2 mm EDTA) at room temperature. Permeabilization was performed with 1% Triton X-100 in PBS for 15 min at room temperature. Cells were incubated with primary antibodies diluted in PBS with 1% (v/v) goat serum for 1 h, washed three times with PBS for 10 min, incubated for 1 h with the appropriate secondary antibodies diluted in PBS with 1% goat serum, and washed three times for 10 min with PBS. Antibodies used were rabbit anti-SNEVPrp19-Pso4 antibody Prp19–867 and mouse anti-Blom7α. As secondary antibodies, TRITC-labeled anti-mouse antibody and fluorescein isothiocyanate-labeled anti-rabbit antibody (Jackson ImmunoResearch, West Grove, PA) were used. Microscopy and image analysis were performed using a Zeiss DeltaVision Restoration microscope as described (35).
Purification of Spliceosomes
Human spliceosome complexes were prepared as described (2). Briefly, a mixture of spliceosomal complexes was assembled on biotinylated, radioactively labeled RNA. As splicing substrate, adenovirus (AD1) transcript was used. The substrate was biotin-labeled and incubated under splicing conditions with HeLa nuclear extracts in 1-ml reactions at 30 °C for 1 h, forming both active spliceosomes and assembly intermediates. After incubation the samples were immediately loaded onto a 2.5 × 75-cm S-500 gel filtration column, and pooled fractions from the spliceosome peak were affinity-selected on streptavidin beads (36). Proteins bound to the beads were washed three times in wash buffer (100 mm NaCl, 20 mm Tris-HCl, pH 7.5) and then eluted in 0.3 ml of elution buffer (2% SDS, 20 mm Tris-HCl, pH 7.5, 20 mm dithiothreitol). Eluted proteins were precipitated with 1 ml of methanol together with 12 μg of slipper limpet glycogen carrier and finally resuspended in 50 μl of elution buffer. This procedure was repeated 12 times, and the resulting samples were pooled separately for each of the pre-mRNA substrates. Based on the staining with Coomassie Blue, we estimate that each fraction contained ∼6–10 μg of protein in total. For the background control, nuclear extract was incubated without labeled RNA, followed by gel filtration as described above. Beads were mixed with the fractions that corresponded to the ones that contained labeled RNA in the above-described experiment. Beads were washed, and the bound material was eluted as above.
In Vitro Splicing Assays
Nuclear extracts used in the splicing assays were obtained commercially from Computer Cell Culture Center (Mons, Belgium). Splicing assays were done using uniformly labeled, capped pre-mRNAs incubated with nuclear extract as described previously (37). Recombinant proteins were added to the splicing reactions. The adeno-pre- mRNA was transcribed from Sau3AI-digested plasmid pBSAd1 (38). The splicing reactions were loaded on a 10% polyacrylamide, 8 m urea denaturing gel and run in 1× TBE to separate the splicing products. When samples were to be used for the analysis of splicing complexes, the reactions were loaded onto a polyacrylamide-agarose composite gel (39) and run for about 5 h at 25 mA.
Splice Site Selection Assays
1 μg of expression plasmid was co-transfected into HeLa cells with 6 μg of the adenovirus E1A reporter plasmid pMTE1A (40) or with pDC20 (41, 42) in the presence of 20 μg of lipofectin (Invitrogen). The E1A gene plasmid pMTE1A used in the alternative splicing assays was described previously (40, 43–45). pMTE1A was kindly provided by J. Caceres and pdc20 by Pin Ouyang. The cells were grown to 60–75% confluence in 60-mm dishes; plasmid DNA was removed 12–16 h later, and Dulbecco's modified Eagle's medium containing 10% fetal calf serum was added for an additional 24 h. RNA was extracted using TRIzol (Invitrogen) for total RNA isolation. 5 μg of total RNA was analyzed by RT-PCR with Superscript II reverse transcriptase (Invitrogen) and GoTaq DNA polymerase (Promega). E1A mRNA detection was carried out with the exon 1 forward primer 5′-GTTTTCTCCTCCGAGCCGCTCCGA-3′ and the 5′-end-labeled exon 2 reverse primer 5′-CTCAGGCTCAGGTTCAGACACAGG-3′, and pDC20 primers were 5′-cgccaaacttgggggaagca-3′ (sense), 5′-cggaactgcctccaactatc-3′ (antisense 1), and 5′-ccagcatgcaagtactcaga-3′ (antisense 2). Amplified products separated by urea-PAGE were detected by autoradiography and quantitated by PhosphorImager analysis (BAS2000; Fujix) (42))
RESULTS
Identification of Putative Interaction Partners of SNEVPrp19-Pso4
To identify proteins interacting with the recently described human protein SNEVPrp19-Pso4, we have performed a two-hybrid library screening. After excluding that SNEVPrp19-Pso4-Gal4 fusion has toxic or autoactivating activities (data not shown), the bait was co-transformed with a human aorta tissue cDNA library (Clontech). Prey cDNAs were isolated and sequence-analyzed after selection on high stringency media and termed Blom2 to Blom7 (Table 1). These cDNAs coded for Exo70, a subunit of the exocyst complex, two collagen fragments (the fibrillar C-terminal regions of collagen α1(III) and collagen α2(I)), the Nω,Nω-dimethylarginine dimethylamino-hydrolase 2 (DDAH 2) that metabolizes Nω,Nω-dimethyl-l-arginine (l-ADDM) to citrulline and methylamines (46, 47), and mitochondrial enoyl-CoA hydratase (48), the enzyme catalyzing the second step in the β-oxidation. Additionally, Blom7 matched partially the sequence of the uncharacterized protein KIAA0907 suggesting an alternatively spliced variant.
TABLE 1.
Putative interaction partners of SNEVPrp19-Pso4 as identified by yeast two-hybrid screening against a human aorta cDNA library
| Clone | Protein identification | Gene name (amino acid where Y2H fusion protein starts) | Description |
|---|---|---|---|
| Blom2 | NP_000080 | Collagen α2(I) (1063) | Extracellular matrix component |
| Blom3 | NP_004083 | Enoyl-CoA hydratase (1) | Mitochondrial β-oxidation |
| Blom4 | NP_001013861 | hExo70 (1) | Subunit of octameric vesicle targeting complex (exocyst) |
| Blom5 | NP_000081 | Collagen α1(III) (1116) | Extracellular matrix component |
| Blom6 | NP_039268 | DDAH 2 (40) | Biogenesis of nitric oxide |
| Blom7 | AAM51855 | Similar to KIAA0907 (347) |
Blom7 Is a KH Domain Protein and Present as Different Splicing Variants
Because SNEVPrp19-Pso4 was found to be a nuclear protein (49), the two putative nuclear localization signals (pattern 7 at amino acids 457 and 551) of Blom7 encouraged us to further investigate this putative interaction partner. However, the cDNA that was contained in the library plasmid only partly matched the KIAA0907 data base entry suggesting an alternatively spliced variant of KIAA0907 with an alternative C terminus located to chromosome 1q22.
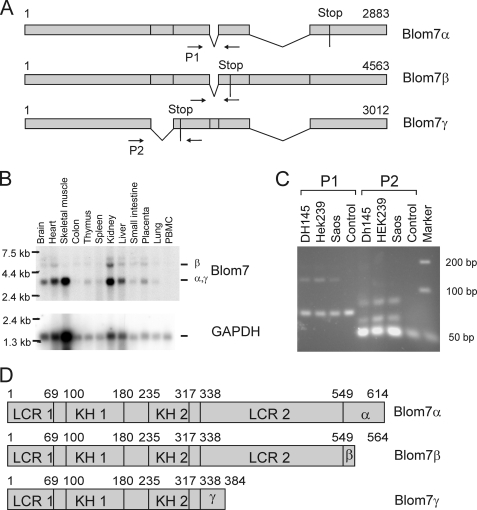
By comparison with expressed sequence tag (EST) data base 3 splicing isoforms were suggested (Fig. 1A). To confirm expression of Blom7 splicing variants, Northern blots using a full-length Blom7α probe were performed and show Blom7 mRNA expression in all tissues tested (Fig. 1C). A band visible at around 3 kb corresponds to Blom7α and -γ, which due to their small difference of 130 nucleotides cannot be distinguished on the multiple tissue Northern blot (Clontech). The 4.5-kb band corresponds to Blom7β, whereas the weak band at around 6 kb suggests an additional yet unknown variant. Furthermore, we used RT-PCR with primer pair P1 flanking the alternatively spliced sequences resulting in amplification of the expected fragments when using cDNA of HEK-293, Saos, or DH145 cells, whereas primer pair P2 detects at least one additional band, suggesting even more splicing variants than the three identified by our approach.
FIGURE 1.
Blom7 is a novel alternatively spliced protein. A, Blom7 isoforms as identified by RT-PCR and sequencing; arrows indicate primers for RT-PCRs. C, RT-PCR using two different primer sets to confirm simultaneous expression of different variants in three different cell lines. Whereas primer pair 1 (P1) shows the expected two bands, P2 shows three bands suggesting additional not yet identified splicing variants. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, multiple tissue Northern blots confirms ubiquitous expression of splicing variants in different tissues. D, protein sequence analysis suggests the presence of two KH domains (KH1 and -2), low complexity regions (LCR), and an alternative isoform-specific C terminus.
The three splicing variants were submitted to the GenBankTM data base as Blom7α (AAM51855), Blom7β (AAM51856) that is identical to KIAA0907, and Blom7γ (AAM51857). Putative homologous proteins to Blom7 are found in higher eukaryotes like chimpanzee (XP_524898.2), mouse (NP_083090.1), rat (XP_342278.3), dog (XP_851894.1), cow (XP_874767.2), chicken (NP_001026499.1), zebrafish (NP_997758.1), and Arabidopsis thaliana (NP_566850.3). No obvious homologue was found in Caenorhabditis elegans or Saccharomyces cerevisiae, whereas Schizosaccharomyces pombe contains a weakly similar protein (NP_593203.1).
The three corresponding protein variants and their domain structure are shown in Fig. 1D. All isoforms bear low complexity regions as well as two homologous domains to the KH domain, an RNA-binding motif (50). The second KH domain is similar to the one of splicing factor 1 (SF1) that recognizes specifically the intron branch point sequence (51). The three Blom7 variants differ only in their C terminus, a phenomenon that has also been described for other KH domain proteins (52). Proteins containing KH domains have been implicated in transcription, regulation of mRNA stability, translational silencing, mRNA localization, and of special interest in regard to its putative interaction with SNEVPrp19-Pso4, in mRNA splicing.
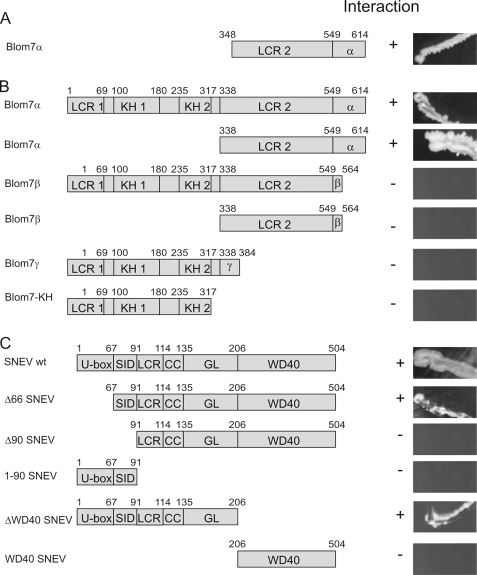
Blom7α-specific C Terminus and Self-interaction Domain of SNEVPrp19-Pso4 Are Necessary for Binding
The initial clone identified in our Y2H library screening contained only the C-terminal half of Blom7α (amino acids 348–614, as shown in Fig. 2A). To test whether full-length Blom7α still interacts with SNEVPrp19-Pso4 and whether other isoforms are interacting with SNEVPrp19-Pso4 as well, directed Y2H experiments were performed (Fig. 2B). Therefore, the cDNAs coding for the proteins and truncated forms were genetically fused to the GAL4 activation domain or to the GAL4 binding domain; however, prey and bait were not switched. After co-transformation of yeast strain AH109 with vectors containing Blom7α (or a deletion mutant -splicing variant) and SNEVPrp19-Pso4 (or a deletion mutant), interaction was tested by growth on high stringency selection media. No colonies were observed with either of the proteins and the respective second vector containing only the GAL4 activation or binding domain alone (data not shown). As indicated in Fig. 2B, the Blom7α specific C terminus was necessary for the interaction to full-length SNEVPrp19-Pso4. On the other side, the domain responsible for SNEVPrp19-Pso4 homo-oligomerization (SID), as identified previously (16), was necessary for mediating this interaction, although not sufficient, because the U-box together with the SID domain (1–90 SNEV) alone did not allow for colony formation. This suggests that either folding of the truncated protein is not correct or that homo-oligomerization of SNEVPrp19-Pso4 is necessary for Blom7α binding.
FIGURE 2.
Blom7α-specific C terminus and self-interaction domain of SNEVPrp19-Pso4 are necessary for interaction. A, C-terminal half of amino acid 347–614 were isolated from the human aorta cDNA library. B, Blom7α deletion mutants identify the α-specific C terminus as necessary for the interaction, whereas on the side of SNEVPrp19-Pso4. C, the self-interaction domain is necessary as determined by directed yeast two-hybrid analyses under high stringent conditions. U-box, UFD2-like motif with E3 ligase activity. SID, self-interaction domain; LCR, low complexity region; CC, coiled-coil domain, GL2, globular domain; LCR1, low complexity region APG-rich; KH1, -2, KH domain (RNA binding domain); LCR2, low complexity region PS-rich; α, α-specific C terminus; β, β-specific C terminus; γ, γ-specific C terminus.
SNEVPrp19-Pso4 Interacts Physically and Directly with Blom7α
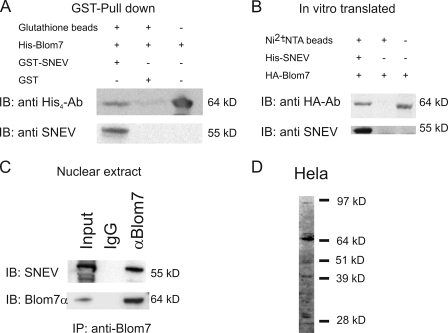
To confirm the interactions observed by the Y2H system, Blom7α was expressed as an HA tag fusion protein from the pGADT7 Y2H vector using rabbit reticulocyte lysate. His6-SNEVPrp19-Pso4 was purified from insect cells. Both recombinant proteins were incubated together with Ni2+-NTA beads and after washing separated by SDS-PAGE. Co-precipitated Blom7α was detected using anti-HA antibody, whereas no band was detectable when omitting SNEVPrp19-Pso4 (Fig. 3A).
FIGURE 3.
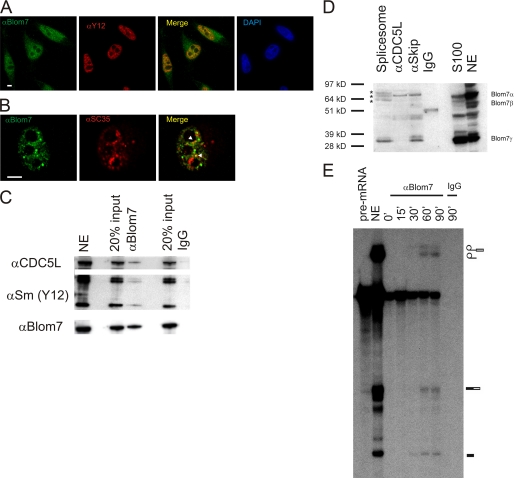
SNEVPrp19-Pso4 physically and directly interacts with Blom7α. A, GST pulldown experiments using bacterially expressed His6-tagged Blom7 and GST-SNEVPrp19-Pso4. His6-Blom7 is detected by Western blotting (IB) only when GST-SNEVPrp19-Pso4 is added to the beads. B, in vitro transcription coupled translation of HA-tagged Blom7 was incubated with insect cell expressed His6-SNEVPrp19-Pso4. Precipitation was performed using Ni2+-NTA-agarose beads and is dependent on the presence of His6-SNEVPrp19-Pso4 as detected by anti-HA antibody. C, immunoprecipitation of SNEVPrp19-Pso4 using beads covalently coupled to anti-Blom7 antibodies and detection on Western blots using anti-SNEVPrp19-Pso4 antibody. Whereas rabbit IgG coupled beads do not precipitate SNEVPrp19-Pso4, anti-Blom7 antibodies result in detection of SNEVPrp19-Pso4. D, Western blot using 20 μg of HeLa cell lysate was separated by SDS-PAGE and detected using Blom7 antibody.
To test if the interaction is direct and not bridged by factors present in reticulocyte lysates, we performed GST pulldown assays using GST-SNEVPrp19-Pso4 and His6-Blom7α recombinantly expressed and purified from E. coli. Indeed, GST-SNEVPrp19-Pso4 co-precipitated Blom7, whereas GST alone did not (Fig. 3B).
To test if nonrecombinant, endogenous proteins interact, Blom7 and SNEVPrp19-Pso4 were co-precipitated from nuclear extracts of HeLa cells. Beads covalently coupled with polyclonal anti-Blom7 mouse antibody were incubated with nuclear extracts. SNEVPrp19-Pso4 was detected after co-precipitation using anti-Blom7 antibody but not using pre-immune IgG (Fig. 3C). To test the specificity of our Blom7 antibody, we performed a Western blot on HeLa cell lysate showing a prominent band at the expected 64 kDa and additional bands that might represent the alternatively spliced isoforms (Fig. 3D).
Interaction of Blom7α and SNEVPrp19-Pso4 Occurs in the Nucleus
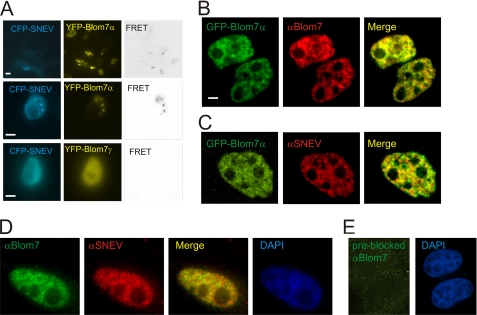
To visualize the cellular location of the Blom7α and SNEVPrp19-Pso4 interaction, we applied several microscopic techniques. After expression of ECFP and EYFP fusion proteins of SNEVPrp19-Pso4 and Blom7α in HEK-293 cells, we performed MicroFRET analysis on living cells (53). FRET signals localized to cell nuclei in dot-like structures (Fig. 4A), whereas EYFP Blom7γ (missing the α-specific C terminus) showed no interaction. As additional negative controls, we co-transfected SNEVPrp19-Pso4-ECFP together with unfused EYFP, and as positive control, an EYFP-ECFP fusion protein was used (data not shown).
FIGURE 4.
Blom7α co-localizes with SNEVPrp19-Pso4 and other splicing factors within the nucleus. A, FRET analysis was performed using ECFP-SNEVPrp19-Pso4 and EYFP-Blom7 constructs. Positive FRET signals were obtained in punctate structure in the nucleus after co-transfecting COS-1 and HEK-293 cells. B, EGFP-Blom7α is recognized by anti-Blom7 antibodies by indirect immunofluorescence microscopy after transfection of HeLa cells. C, co-localization of EGFP-Blom7α with endogenous SNEVPrp19-Pso4 stained by anti-SNEVPrp19-Pso4 antibody was observed. Bars represent 5 μm. D, co-localization of endogenous Blom7 with endogenous SNEVPrp19-Pso4 stained by anti-SNEVPrp19-Pso4 and anti-Blom7 antibody. DAPI, 4′,6-diamidino-2-phenylindole. E, Blom7 antibody pre-blocked with recombinant Blom7α does not give immunofluorescent signals above background.
Because our Blom7 antibody was raised against full-length Blom7α, the individual isoforms are not distinguished by immunofluorescence microscopy. Therefore, we overexpressed EGFP-Blom7α, which is well detected by our anti-Blom7 antibodies (Fig. 4B). Indeed, co-localization of EGFP-Blom7α and endogenous SNEVPrp19-Pso4 was observed within the nuclei in a speckle-like pattern (Fig. 4C).
Using both antibodies against both, Blom7α and SNEVPrp19-Pso4, co-localization within the cell nucleus is similarly observed, although the signal is more diffuse throughout the nucleoplasm (Fig. 4D). However, cytoplasmic staining was also observed to a low extent using the anti-Blom7 antibody.
Blom7α Is a Novel Subunit of the Spliceosome
To test if the interaction of Blom7 with SNEVPrp19-Pso4 occurs in the context of pre-mRNA splicing, we co-stained Blom7α with Sm proteins (Fig. 5A) and SC35 (Fig. 5B), two other canonical pre-mRNA splicing factors, and observed co-localization for Y12, whereas SC35 only shows 2–4 single co-localized dots per cell within the nuclei. Furthermore, anti-Blom7α antibodies co-precipitate Sm proteins detected by Y12 antibody as canonical pre-mRNA splicing factors, as well as the SNEVPrp19-Pso4 complex member CDC5L (Fig. 5, C and D).
FIGURE 5.
Blom7α associates with pre-mRNA splicing factors. A, co-localization of Blom7 is detected using antibodies against endogenous Sm proteins; B, SC35, arrowheads indicate two dots of co-localization. DAPI, 4′,6-diamidino-2-phenylindole. C, precipitation of Blom7 (lowest panel) from nuclear extract (NE) co-precipitates CDC5L as well as Sm proteins, whereas irrelevant IgG does not. 20% input of nuclear extracts that were added to anti-Blom7-Ab-protein A beads (αBlom7) or to IgG-loaded beads as negative control were separated by SDS-PAGE and analyzed by Western blotting. D, Blom7α is present in affinity-purified mature spliceosomes, in the CDC5L spliceosome-associated complex purified by anti-CDC5L or anti-Skip antibodies, and S100 extracts as probed by anti-Blom7 antibodies. Asterisks indicate bands of Blom7α that might have been post-translationally modified. E, anti-Blom7 antibody co-precipitates pre-mRNA and splicing intermediates from in vitro splicing reactions in a time-dependent manner further supporting that Blom7 is present in spliceosomes. In vitro splicing assays were separated on denaturing polyacrylamide gels after precipitation with anti-Blom7 antibody. Splicing reaction using HeLa nuclear extract (NE) without precipitation was loaded as positive control, and as negative control, irrelevant IgG was used.
In the opposite direction, we tested affinity-purified spliceosomes containing A, B, and C complexes assembled on biotinylated pre-mRNA as described previously (12) as well as anti-CDC5L and anti-SKIP immunoprecipitates. Indeed, Blom7α and smaller isoforms are detectable in the spliceosome as well as in the precipitates of anti-CDC5L and anti-SKIP (Fig. 5C), which is a SNEVPrp19-Pso4 complex-associated protein (4). Blom7α bands at 64 kDa were observed as well as a band at around 39 kDa representing Blom7γ. The purified spliceosome, however, shows a triple band at around 64 kDa that might represent post-translational modifications of Blom7α after integration into the spliceosome (Fig. 5D).
Finally, Blom7α also directly or indirectly associates with pre-mRNA. In time course immunoprecipitations of in vitro pre-mRNA splicing reactions using anti-Blom7 antibodies, Blom7 co-precipitated pre-mRNA immediately, whereas after 30 min splicing intermediates start to appear (Fig. 5E). Using preimmune serum as control, no pre-mRNA or splicing intermediates were precipitated.
Blom7α Modulates the Pre-mRNA Splicing Activity of Nuclear Extracts
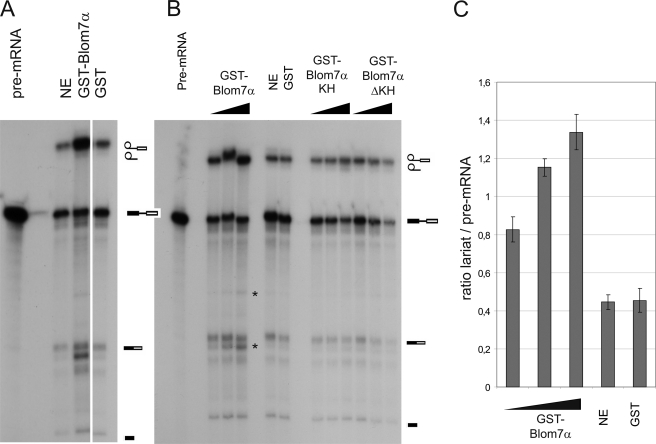
To test if Blom7 is also functionally involved in pre-mRNA splicing, we first tested if addition of recombinant GST-Blom7 expressed and purified from E. coli alters the in vitro splicing reaction. Surprisingly, addition of GST-Blom7α increased the ratio of pre-mRNA to lariat, suggesting acceleration of the splicing reaction (Fig. 6A) in a dose-dependent manner (Fig. 6, B and C), whereas neither GST alone nor the C-terminal nor the N-terminal half of Blom7α affected the splicing reaction as compared with nuclear extracts alone. Furthermore, two more bands appear to be specific for Blom7α overexpression (Fig. 6, A and B), suggesting that Blom7 might be involved in alternative splicing. However, it cannot be ruled out that these bands are unspecific and due to RNase activity, because no sequence analysis was performed.
FIGURE 6.
Blom7α is functionally involved in pre-mRNA splicing. A, addition of recombinant GST-Blom7α enhances in vitro splicing reactions as compared with standard splicing reactions or after addition of GST alone as control. B, this enhancement is dose-dependent, because increasing amounts of GST-Blom7α increase the ratio of lariat to pre-mRNA band as well as the joined exons. Asterisks indicate additional bands specific for Blom7α addition. C, lariat to pre-mRNA ratios were calculated from three independent experiments adding increasing amounts of recombinant Blom7α; untreated nuclear extracts as well as addition of GST were used as controls. NE, nuclear extract.
Blom7α Is Involved in Splice Site Selection
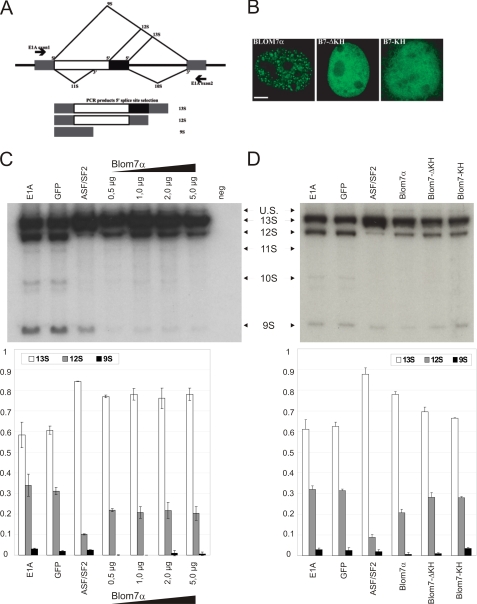
Because of the previous result and because KH domain proteins have been found to display alternative splicing activity (54), we also tested Blom7α in this regard. Therefore, we co-transfected EGFP-Blom7α together with an E1A splicing reporter minigene resulting in various splice isoforms to test for 5′-splice site selection (Fig. 7A). EGFP-Blom7α and its deletion mutants locate to the cell nuclei, but only the full-length protein shows a dotted distribution (Fig. 7B). As control, EGFP alone did not lead to alterations in the ratio of E1A splicing variants as compared with E1A-only transfected cells.
FIGURE 7.
Overexpression of Blom7α alters 5′-splice site selection of E1A minigene. A, scheme of the alternative splice products derived from of E1A minigene, showing the three most prevalent forms of 13S, 12S, and 9S. B, transfection of HeLa cells with EGFP-Blom7α shows nuclear localization, although a dot-like structure is only observed for full-length Blom7α. C, co-transfection of EGFP-Blom7α together with E1A minigene promotes formation of the 13S splice variant, whereas reducing 12S and 9S in comparison with the “E1A-only” and empty vector control. ASF-SF2 was used as positive control (n = 9). One representative gel of the alternative splicing analysis is shown. Neg, negative. D, neither the KH domains nor the C-terminal half (ΔKH) significantly alters splice site selection. U.S., unspliced
EGFP-Blom7α overexpression induces a significant increase in 13S and loss of the 9S isoform, whereas a less pronounced decrease in 12S is observed after transfection with increasing plasmid amounts (Fig. 7C), suggesting that full-length Blom7α is involved specifically in the selection of alternative 5′-splice sites and favoring proximal splicing of 13S over distal to 9S.
Neither the KH domains alone nor the C-terminal half alone resulted in a significantly altered splicing pattern (Fig. 7D) in three independent experiments using triplicates each time. ASF-SF2 served as a positive control for alternative 5′-splice site selection (40, 43, 45).
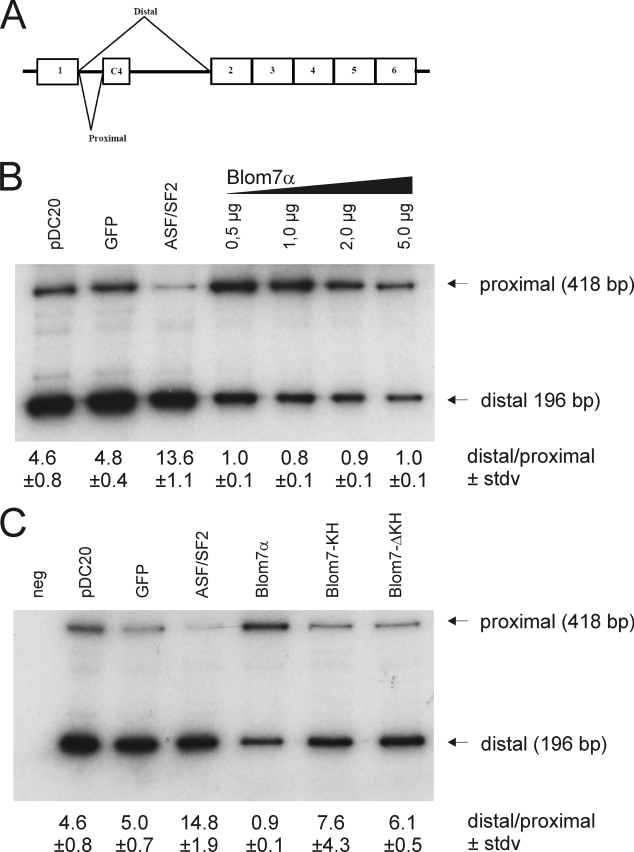
To test the effect of Blom7α on 3′-splice site selection, we additionally tested the pDC20 minigene construct harboring a calcitonin-dhfr chimeric pre-mRNA (41, 42) that gives rise to two different splicing isoforms (Fig. 8A). Overexpression of Blom7α markedly represses the proximal splicing, whereas EGFP alone does not alter the splice site selection in comparison with nontransfected cells (Fig. 8B). In contrast, ASF-SF2 shifts the balance to the proximally spliced variant as observed previously (41), indicating a specifically different function of Blom7α. Again, neither overexpression of the C- nor N-terminal half of Blom7α significantly alters splice site selection (Fig. 8C). Taken together these data suggest that Blom7α interacts with SNEVPrp19-Pso4 and is a novel pre-mRNA splicing factor also involved in regulating splice site selection.
FIGURE 8.
Blom7α also influences 3′-splice site selection of the pDC20 minigene. A, scheme of the alternative splice products deriving of pDC20 minigene, showing the two splicing variants (proximal and distal). B, representative gel of the co-transfection of Blom7α and pDC20. Whereas EGFP-ASF-SF2 increases the proximal splice variant, Blom7α favors selection of the distal splice site, and ratios of distal to proximal variants are shown below the lanes calculated from nine independent transfections. C, again, only full-length Blom7α alters the selection of splice sites, whereas neither the N- nor the C-terminal parts are sufficient to do so.
DISCUSSION
To identify novel SNEVPrp19-Pso4-interacting proteins, we performed a Y2H library screening. Besides its ability to interact with itself (16), we here identified several putative interacting proteins. Even if striking as odd interaction partners, a link between enoyl-coenzyme A hydratase and SNEVPrp19-Pso4 might indeed exist, because small interfering RNA-mediated SNEVPrp19-Pso4 knockdown increases β-oxidation slightly but significantly for 8% in differentiated murine 3T3-L1 preadipocytes (55). Similarly, an interaction of SNEVPrp19-Pso4 with collagens is possible, because splicing factors have been co-purified with collagens in a stable isotope labeling with amino acids in cell culture experiment aiming at identification of factors involved in cell adhesion (see supplemental material in Ref. 56).
However, we selected one so far uncharacterized candidate, Blom7α, for further analysis, due to putative nuclear localization signals and KH RNA-binding motifs. KH domains are a widespread RNA or single-stranded DNA-binding motif identified by sequence similarity searches to heterogeneous nuclear ribonucleoprotein K (hnRNP K) (57). They are highly conserved, found in archaea, bacteria, and eukaryotes (57, 58), and are involved in a variety of different RNA-processing steps.
Among the most prominent members of KH domain proteins is the fragile X syndrome protein FMR1 (59), where mutations in KH domains result in developmental defects (60), ribosomal protein S3 (61, 62), or the zipcode-binding protein 1 (ZBP-1), implicated in mRNA subcellular localization (63). Several KH domain proteins also interact with single-stranded DNAs, like DDP1 (64), far upstream element-binding protein (65), and hnRNP K (66, 67). A number of these proteins contain multiple KH domains, which are suggested to increase the usually weak RNA protein affinity of single KH domains (50).
Several KH domain proteins are also established pre-mRNA splicing factors, like hnRNP K itself (68), Nova proteins (54, 69, 70), Drosophila P-element somatic inhibitor protein (71, 72), KSRP (73), and splicing factor 1 (SF1), that specifically recognize the intron branch point sequence UACUAAC in the pre-mRNA transcripts during spliceosome assembly (51). Indeed, one of the KH domains of Blom7α is classified as SF1-type KH domain. Therefore, after confirming direct interaction with SNEVPrp19-Pso4, we tested if Blom7α also co-localizes and co-purifies with other splicing factors, although Blom7α escaped identification by the major proteomic spliceosome analyses conducted so far (3, 4, 11–13).
Because we indeed found association to the spliceosome, we tested if Blom7α might play a functional role in pre-mRNA splicing. Indeed, it accelerated in vitro pre-mRNA splicing using HeLa cell nuclear extracts. By immunodepletion of Blom7, however, we did not see a decrease in splicing activity, which might be due to residual Blom7 in the nuclear extract (data not shown).
KH domain proteins like Drosophila melanogaster Kep1 (74) and SF1 contribute to splice site selection (51, 75, 76). Therefore, we also tested if Blom7α is involved in regulating alternative splicing events using two different reporter minigene plasmids. Indeed, Blom7α overexpression regulates 5′-splice site selection of the E1A-containing cassette (43). The effect on 5′-splice site selection is similar to that of ASF-SF2. In contrast, the effect on 3′-splice site selection was opposite to ASF-SF2 favoring exon skipping in the calcitonin-dihydrofolate reductase chimeric minigene plasmid pDC20 (41).
Regulation of alternative splicing is supposed to mainly take place at the level of E and A complex assembly but is still possible in B complexes (77). We speculate that alteration of splice site selection by Blom7α might be influenced as early as in the E complex, because pre-mRNA is co-precipitated with Blom7 from the start of the splicing reactions. There, U1 snRNP binds to the 5′-splice site, and the branch site is recognized by SF1-mBBP, which interacts with U2AF65 (78). These, together with the smaller subunit of U2AF, U2AF35, and SR proteins, determine the 3′-splice site as “exon definition complex” (79) and bend the pre-mRNA (80).
Because Blom7β (KIAA0907) is supposed to bind to U2AF65 (81), according to a large scale Y2H interactome study (81), we hypothesize that Blom7α might bind intronic mRNA as well, because it only differs by 60 C-terminal amino acids from Blom7β. This binding might occur near the branch site due to SF1-like KH domain of Blom7, allowing for U2AF interaction and thus influencing 3′-splice site selection by altering the specificity of the exon definition complex.
The function of the interaction of Blom7α with SNEVPrp19-Pso4, however, is unclear. Blom7α might recruit SNEVPrp19-Pso4 to the E or A complex alone, where only the other members of the CDC5L-associated complex assemble on the SNEVPrp19-Pso4 homo-oligomer as a platform. Indeed, such a stepwise assembly seems possible because interfering with SNEVPrp19-Pso4 self-interaction interferes with spliceosome assembly (16).
Furthermore, in the A complex only SNEVPrp19-Pso4 and not CDC5L has been identified (14), whereas in the BΔU1 complex SNEVPrp19-Pso4 is joined by CDC5L together with CDC5L-SNEVPrp19-Pso4 complex-associated SKIP and hSyF3 (4). All members of the CDC5L-SNEVPrp19-Pso4 complex have been observed only in the B* complex (4). By addition of Blom7α to in vitro splicing reactions, this assembly might be enhanced, resulting in higher splicing activity in vitro.
By guiding the assembly of the spliceosome to specific sites on the pre-mRNA, this might further result in alternative selection of splice sites. However, several caveats to this hypothesis remain as follows: the interaction between Blom7 isoforms and U2AF65 has yet to be confirmed in the context of pre-mRNA splicing, and the kinetic behavior of Blom7 during spliceosomal activation and re-arrangement must be determined. Furthermore, the consensus sequence(s) to which the KH domains of Blom7α are binding have to be determined to understand if and how it is involved in exon definition. Finally, it will be of great interest to determine target mRNAs that are alternatively spliced by Blom7α and its isoforms.
Acknowledgments
We are grateful to Ursula Ryder, Christina Wolschner, Silvia Jakeli, Christian Kaisermayer, and Sabine Zehentner for excellent technical support. Plasmid E1A was kindly provided by Javier Caceres and pDC20 by Pin Ouyang.
This work was supported by the Austrian Science Fund Project S93-06, by Austrian Genome Research Grant 820982 (Noncoding RNAs), by a grant from Herzfelder-sche Familienstiftung (to R. G.-V.), by Wellcome Trust, and by Polymun Scientific.
- snRNP
- small nuclear ribonucleoprotein
- KH
- K homology
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- Y2H
- yeast two-hybrid
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- PBS
- phosphate-buffered saline
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- FRET
- fluorescence resonance energy transfer
- TRITC
- tetramethylrhodamine isothiocyanate
- EGFP
- enhanced green fluorescent protein
- RT
- reverse transcription
- CoIP
- co-immunoprecipitation
- YFP
- yellow fluorescent protein
- EYFP
- enhanced YFP
- SF1
- splicing factor 1
- CFP
- cyan fluorescent protein
- ECFP
- enhanced CFP
- Pipes
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Staley J. P., Guthrie C. (1998) Cell 92, 315–326 [DOI] [PubMed] [Google Scholar]
- 2.Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002) Genome Res. 12, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajuh P., Kuster B., Panov K., Zomerdijk J. C., Mann M., Lamond A. I. (2000) EMBO J. 19, 6569–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova O. V., Makarov E. M., Urlaub H., Will C. L., Gentzel M., Wilm M., Lührmann R. (2004) EMBO J. 23, 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarn W. Y., Hsu C. H., Huang K. T., Chen H. R., Kao H. Y., Lee K. R., Cheng S. C. (1994) EMBO J. 13, 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohi M. D., Gould K. L. (2002) RNA 8, 798–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Ren L., Hirsch J. P., Chazin W. J., Walz T., Gould K. L. (2005) Mol. Cell. Biol. 25, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng S. C., Tarn W. Y., Tsao T. Y., Abelson J. (1993) Mol. Cell. Biol. 13, 1876–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ajuh P., Lamond A. I. (2003) Nucleic Acids Res. 31, 6104–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajuh P., Sleeman J., Chusainow J., Lamond A. I. (2001) J. Biol. Chem. 276, 42370–42381 [DOI] [PubMed] [Google Scholar]
- 11.Bessonov S., Anokhina M., Will C. L., Urlaub H., Luhrmann R. (2008) Nature 452, 846–850 [DOI] [PubMed] [Google Scholar]
- 12.Neubauer G., King A., Rappsilber J., Calvio C., Watson M., Ajuh P., Sleeman J., Lamond A., Mann M. (1998) Nat. Genet. 20, 46–50 [DOI] [PubMed] [Google Scholar]
- 13.Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. (2002) Science 298, 2205–2208 [DOI] [PubMed] [Google Scholar]
- 14.Hartmuth K., Urlaub H., Vornlocher H. P., Will C. L., Gentzel M., Wilm M., Lührmann R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C. L., Kastner B., Stark H., Urlaub H., Lührmann R. (2006) Mol. Cell. Biol. 26, 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grillari J., Ajuh P., Stadler G., Löscher M., Voglauer R., Ernst W., Chusainow J., Eisenhaber F., Pokar M., Fortschegger K., Grey M., Lamond A. I., Katinger H. (2005) Nucleic Acids Res. 33, 6868–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K. I. (2001) J. Biol. Chem. 276, 33111–33120 [DOI] [PubMed] [Google Scholar]
- 18.Löscher M., Fortschegger K., Ritter G., Wostry M., Voglauer R., Schmid J. A., Watters S., Rivett A. J., Ajuh P., Lamond A. I., Katinger H., Grillari J. (2005) Biochem. J. 388, 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sihn C. R., Cho S. Y., Lee J. H., Lee T. R., Kim S. H. (2007) Biochem. Biophys. Res. Commun. 356, 175–180 [DOI] [PubMed] [Google Scholar]
- 20.Bellare P., Small E. C., Huang X., Wohlschlegel J. A., Staley J. P., Sontheimer E. J. (2008) Nat. Struct. Mol. Biol. 15, 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grillari J., Hohenwarter O., Grabherr R. M., Katinger H. (2000) Exp. Gerontol. 35, 187–197 [DOI] [PubMed] [Google Scholar]
- 22.Voglauer R., Chang M. W., Dampier B., Wieser M., Baumann K., Sterovsky T., Schreiber M., Katinger H., Grillari J. (2006) Exp. Cell Res. 312, 746–759 [DOI] [PubMed] [Google Scholar]
- 23.Fortschegger K., Wagner B., Voglauer R., Katinger H., Sibilia M., Grillari J. (2007) Mol. Cell. Biol. 27, 3123–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schraml E., Voglauer R., Fortschegger K., Sibilia M., Stelzer I., Grillari J., Schauenstein K. (2008) Stem Cells Dev. 17, 355–366 [DOI] [PubMed] [Google Scholar]
- 25.Mahajan K. N., Mitchell B. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10746–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck B. D., Park S. J., Lee Y. J., Roman Y., Hromas R. A., Lee S. H. (2008) J. Biol. Chem. 283, 9023–9030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang N., Kaur R., Lu X., Shen X., Li L., Legerski R. J. (2005) J. Biol. Chem. 280, 40559–40567 [DOI] [PubMed] [Google Scholar]
- 28.Lu X., Legerski R. J. (2007) Biochem. Biophys. Res. Commun. 354, 968–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuraoka I., Ito S., Wada T., Hayashida M., Lee L., Saijo M., Nakatsu Y., Matsumoto M., Matsunaga T., Handa H., Qin J., Nakatani Y., Tanaka K. (2008) J. Biol. Chem. 283, 940–950 [DOI] [PubMed] [Google Scholar]
- 30.Cho S. Y., Park P. J., Lee J. H., Kim J. J., Lee T. R. (2007) Biochem. Biophys. Res. Commun. 364, 844–849 [DOI] [PubMed] [Google Scholar]
- 31.Eisenhaber F. (2006) in Discovering Biomolecular Mechanisms with Computational Biology (Eisenharber F. ed) 1st Ed., pp. 39–54, Landes Biosciences, Georgetown, Washington D. C. [Google Scholar]
- 32.Böhm E., Grillari J., Voglauer R., Gross S., Ernst W., Ferko B., Kunert R., Katinger H., Borth N. (2005) J. Immunol. Methods 307, 13–23 [DOI] [PubMed] [Google Scholar]
- 33.Lamond A. I., Konarska M. M., Grabowski P. J., Sharp P. A. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youvan D. C., Coleman W. J., Silva C. M., Petersen J., Bylina E. J., Yang M. M. (1997) Biotech. Alia 1, 1–16 [Google Scholar]
- 35.Platani M., Goldberg I., Swedlow J. R., Lamond A. I. (2000) J. Cell Biol. 151, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calvio C., Neubauer G., Mann M., Lamond A. I. (1995) RNA 1, 724–733 [PMC free article] [PubMed] [Google Scholar]
- 37.Lamond A. I., Konarska M. M., Sharp P. A. (1987) Genes Dev. 1, 532–543 [DOI] [PubMed] [Google Scholar]
- 38.Konarska M. M., Sharp P. A. (1987) Cell 49, 763–774 [DOI] [PubMed] [Google Scholar]
- 39.Konarska M. M., Sharp P. A. (1986) Cell 46, 845–855 [DOI] [PubMed] [Google Scholar]
- 40.Zerler B., Moran B., Maruyama K., Moomaw J., Grodzicker T., Ruley H. E. (1986) Mol. Cell. Biol. 6, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai Y., Lee D., Yu T., Chasin L. A. (1999) Nucleic Acids Res. 27, 1126–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P., Lou P. J., Leu S., Ouyang P. (2002) Biochem. Biophys. Res. Commun. 294, 448–455 [DOI] [PubMed] [Google Scholar]
- 43.Cáceres J. F., Stamm S., Helfman D. M., Krainer A. R. (1994) Science 265, 1706–1709 [DOI] [PubMed] [Google Scholar]
- 44.van der Houven, van Oordt W., Diaz-Meco M. T., Lozano J., Krainer A. R., Moscat J., Cáceres J. F. (2000) J. Cell Biol. 149, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cazalla D., Newton K., Cáceres J. F. (2005) Mol. Cell. Biol. 25, 2969–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leiper J. M., Santa Maria J., Chubb A., MacAllister R. J., Charles I. G., Whitley G. S., Vallance P. (1999) Biochem. J. 343, 209–214 [PMC free article] [PubMed] [Google Scholar]
- 47.Ribas G., Neville M., Wixon J. L., Cheng J., Campbell R. D. (1999) J. Immunol. 163, 278–287 [PubMed] [Google Scholar]
- 48.Kanazawa M., Ohtake A., Abe H., Yamamoto S., Satoh Y., Takayanagi M., Niimi H., Mori M., Hashimoto T. (1993) Enzyme Protein 47, 9–13 [DOI] [PubMed] [Google Scholar]
- 49.Gotzmann J., Gerner C., Meissner M., Holzmann K., Grimm R., Mikulits W., Sauermann G. (2000) Exp. Cell Res. 261, 166–179 [DOI] [PubMed] [Google Scholar]
- 50.Valverde R., Edwards L., Regan L. (2008) FEBS J. 275, 2712–2726 [DOI] [PubMed] [Google Scholar]
- 51.Peled-Zehavi H., Berglund J. A., Rosbash M., Frankel A. D. (2001) Mol. Cell. Biol. 21, 5232–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J., Zhou L., Tonissen K., Tee R., Artzt K. (1999) J. Biol. Chem. 274, 29202–29210 [DOI] [PubMed] [Google Scholar]
- 53.Youvan D. C., Silva C. M., Bylina E. J., Coleman W. J., Dilworth M. R., Yang M. M. (1997) Biotech. Alia 3, 1–18 [Google Scholar]
- 54.Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B. J., Darnell R. B. (2006) Nature 444, 580–586 [DOI] [PubMed] [Google Scholar]
- 55.Cho S. Y., Shin E. S., Park P. J., Shin D. W., Chang H. K., Kim D., Lee H. H., Lee J. H., Kim S. H., Song M. J., Chang I. S., Lee O. S., Lee T. R. (2007) J. Biol. Chem. 282, 2456–2465 [DOI] [PubMed] [Google Scholar]
- 56.de Hoog C. L., Foster L. J., Mann M. (2004) Cell 117, 649–662 [DOI] [PubMed] [Google Scholar]
- 57.Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. (1993) Nucleic Acids Res. 21, 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grishin N. V. (2001) Nucleic Acids Res. 29, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson T. J., Rice P. M., Thompson J. D., Heringa J. (1993) Trends Biochem. Sci. 18, 331–333 [DOI] [PubMed] [Google Scholar]
- 60.Musco G., Stier G., Joseph C., Castiglione Morelli M. A., Nilges M., Gibson T. J., Pastore A. (1996) Cell 85, 237–245 [DOI] [PubMed] [Google Scholar]
- 61.Urlaub H., Kruft V., Bischof O., Müller E. C., Wittmann-Liebold B. (1995) EMBO J. 14, 4578–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., Lenardo M. J. (2007) Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 63.Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L., Singer R. H. (1997) Mol. Cell. Biol. 17, 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cortés A., Huertas D., Fanti L., Pimpinelli S., Marsellach F. X., Piña B., Azorín F. (1999) EMBO J. 18, 3820–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He L., Weber A., Levens D. (2000) Nucleic Acids Res. 28, 4558–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Backe P. H., Messias A. C., Ravelli R. B., Sattler M., Cusack S. (2005) Structure 13, 1055–1067 [DOI] [PubMed] [Google Scholar]
- 67.Braddock D. T., Baber J. L., Levens D., Clore G. M. (2002) EMBO J. 21, 3476–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Expert-Bezançon A., Le Caer J. P., Marie J. (2002) J. Biol. Chem. 277, 16614–16623 [DOI] [PubMed] [Google Scholar]
- 69.Jensen K. B., Dredge B. K., Stefani G., Zhong R., Buckanovich R. J., Okano H. J., Yang Y. Y., Darnell R. B. (2000) Neuron 25, 359–371 [DOI] [PubMed] [Google Scholar]
- 70.Ule J., Jensen K. B., Ruggiu M., Mele A., Ule A., Darnell R. B. (2003) Science 302, 1212–1215 [DOI] [PubMed] [Google Scholar]
- 71.Siebel C. W., Kanaar R., Rio D. C. (1994) Genes Dev. 8, 1713–1725 [DOI] [PubMed] [Google Scholar]
- 72.Chmiel N. H., Rio D. C., Doudna J. A. (2006) RNA 12, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Min H., Turck C. W., Nikolic J. M., Black D. L. (1997) Genes Dev. 11, 1023–1036 [DOI] [PubMed] [Google Scholar]
- 74.Robard C., Daviau A., Di Fruscio M. (2006) Biochem. J. 400, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Královicová J., Houngninou-Molango S., Krämer A., Vorechovsky I. (2004) Hum. Mol. Genet. 13, 3189–3202 [DOI] [PubMed] [Google Scholar]
- 76.Shitashige M., Naishiro Y., Idogawa M., Honda K., Ono M., Hirohashi S., Yamada T. (2007) Gastroenterology 132, 1039–1054 [DOI] [PubMed] [Google Scholar]
- 77.Chiara M. D., Reed R. (1995) Nature 375, 510–513 [DOI] [PubMed] [Google Scholar]
- 78.Berglund J. A., Abovich N., Rosbash M. (1998) Genes Dev. 12, 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matlin A. J., Clark F., Smith C. W. (2005) Nat. Rev. Mol. Cell Biol. 6, 386–398 [DOI] [PubMed] [Google Scholar]
- 80.Kent O. A., Reayi A., Foong L., Chilibeck K. A., MacMillan A. M. (2003) J. Biol. Chem. 278, 50572–50577 [DOI] [PubMed] [Google Scholar]
- 81.Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]