Abstract
The core domain of the tumor suppressor p53 has low thermodynamic stability, and many oncogenic mutations cause it to denature rapidly at body temperature. We made a series of core domain mutants that are significantly less or more stable than wild type to investigate effects of stability on the transcriptional activity and levels of native full-length p53 in H1299 mammalian cells. The levels of transcriptionally inactive native protein with inactivating mutations in the N-terminal transactivation domain correlated strongly with stability. The levels of transcriptionally active proteins, however, depended on both their stability and the transcriptional activity that leads to the feedback loop of proteolytic degradation via transcription of E3 ligases. A very highly stabilized quadruple mutant and an even more stable hexamutant were more active than wild-type p53 in terms of Bax transcription and apoptotic activity, and reached higher levels than wild type in cells. The increased activity did not result from increased overall stability but was due to a single known suppressor mutation, N239Y. It is possible that the low intrinsic stability of p53 is a means of keeping its level low in the cell by spontaneous denaturation, by a route additional to that of proteolytic degradation via E3 ligase pathways. Denatured p53 does accumulate in cells, and there are pathways for the proteolysis of denatured proteins.
Introduction
The p53 tumor suppressor protein regulates cell-cycle arrest, apoptosis, and DNA repair by transactivating a wide range of downstream genes in response to cellular stress (1). Upon activation through upstream stress signals, p53 levels rise rapidly via various mechanisms, such as post-translational modification, leading to transcription of downstream genes (2). The level of p53 protein in vivo is tightly regulated, and the basal level is kept low by rapid turnover through degradation, by ubiquitin-dependent proteolysis involving Hdm2 (3, 4) and other E3 ligases such as Pirh2 (5) and COP1 (6). Cellular stresses can lead to a decrease in Hdm2 or a disruption of interaction between Hdm2 and p53, for example, leading to an accumulation of p53 protein (7). Hdm2 is in turn transcriptionally induced by p53, thus forming an autoregulatory feedback loop that controls p53 levels (8, 9).
p53 contains intrinsically disordered N- and C-terminal domains, while its central core and its tetramerization domains are folded (10). The core domain (residues 94–312) dominates the overall functional stability of the full-length protein, being relatively unstable, with a melting temperature of only around 44 °C (11). In 50% of human cancers, p53 is inactivated by oncogenic mutations (12), which are mostly found in the DNA binding core domain of p53 (13). Some 30% of the mutations simply cause the protein to denature at body temperature, and these mutants have a half-life of a few minutes or less from spontaneous denaturation (13).
Other regulatory proteins involved in cell cycle control, such as p16INK4a, also have low thermodynamic stability (14). Therefore, it is possible that the low thermostability of p53 has the biological purpose of giving p53 a low in vivo half-life in the absence of the proteosomal degradation pathways. The difference in intrinsic stability between p53 and its more stable homologues, p63/p73, may also be important for their different in vivo functions (15–17).
The stabilized quadruple mutant M133L/V203A/N239Y/N268D core domain (QM3 or T-p53C, residues 94–312) was designed semirationally, and found to be more stable than the wild-type p53 core domain by 2.5 kcal/mol and fully functional in vitro (18). Its structure is identical to that of wild-type core apart from small local changes at the sites of mutation (19). The quadruple mutant has been used as a stabilized scaffold for determining the structures of hotspot-destabilizing mutations in the core domain (20) as well as for facilitating biophysical measurements with full-length p53 (21). Recently, the quadruple mutant has also been used in determining the quaternary structure of full-length p53 (22, 23).
We have designed a hexamutant, (HM) (M133L/V203A/Y236F/T253I/N239Y/N268D) (24), which is even more stable, based on the proposal that poorly hydrogen-bonded side chains of Tyr-236 and Thr-253 residues, buried in the core domain of p53, are replaced by Phe-236 and Ile-253, as found in the more stable paralogs p63 and p73 (25). The level of expression of the mutant p53 core domains in Escherichia coli, in the absence of Hdm2 and other E3 ligases feedback loops that are found in mammalian cells, depend on their thermodynamic stability (26). Destabilized mutants reach lower levels, since their rates of degradation are higher than that of stabilized mutants, although they all have the same rate of biosynthesis. Here, we examined the effects of thermodynamic stability of the core domain of full-length p53, by making use of the stabilized quadruple (QM) and HM, and expressing them in mammalian cells with active feedback loops involving Hdm2. We examined the levels of native p53 protein and the effects of stability on transcription and apoptosis.
MATERIALS AND METHODS
Cloning
Wild-type, QM, and HM full-length p53 were cloned into the pEGFP N1 vector using the SacII and XmaI restriction sites. pEGFP-N1 plasmid was a gift from Fiona Townsley.
Mammalian Cell Fluorescence Assay
H1299 cells were a gift from Carol Prives. The cells were maintained in RPMI medium with 10% fetal calf serum and penstrep antibiotics. For transient transfection, cells were seeded at 1.0 × 105 cells/6-well plate. After 24 h, cells were transfected with 1 μg of pEGFP-N1 p53 or mutant p53 plasmid. The medium was removed to harvest the cells. Cells on the plate surface were scraped into 250 μl of phosphate-buffered saline transferred into FACS tubes. The cells were then measured for fluorescence intensity using a Becton Dickinson FACScan. Cells were selected by setting the forward-scatter and the side-scatter range, and the emission intensity of each cell was measured at 530 and 585 nm. Experiments were carried out in triplicate.
Reporter Assay
H1299 cells were seeded at 1.0 × 104 cells per well in 96-well plates. After 24 h, the cells were co-transfected with 50 ng of pEGFP-N1 p53 or mutant p53 expression vector and 100 ng of Bax reporter luciferase plasmid (Gift from Moshe Oren) and 5 ng of CMV-Renilla plasmid, using Lipofectamine 2000 (Invitrogen) transfection reagent according to the manufacturer's instructions. Cells were assayed for luciferase and Renilla activity using a Dual-Glo luciferase assay kit (Promega) according to the manufacturer's instructions.
Western Blotting
H1299 cells were seeded at 1.0 × 105 cells per well in 6-well plates. After 24 h, the cells were transfected with 1 μg of pEGFP p53 or mutant p53 expression vector. Cells were harvested after 48 h by scraping into 300 μl of radioimmune precipitation assay buffer. Approximately 2 μg of cell lysate is run on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Invitrogen) and transferred onto a polyvinylidene difluoride membrane (Millipore). Quantification of cell lysate was done using a BCA kit (Thermo Scientific). Blots were incubated with a mouse anti-GFP primary antibody (1:2000) (Clontech) followed by an anti-mouse horseradish peroxidase secondary antibody (1:2000) (Santa Cruz Biotechnology) and assayed using ECL detection reagent (Amersham Biosciences). Mouse anti-γ-tubulin primary antibody (1:5000) (Sigma) was used for the loading control.
Caspase 3 Assay
H1299 cells were seeded at 1.0 × 105 cells per well in 6-well plates. After 24 h, the cells were transfected with 1 μg of pEGFP p53 or mutant p53 expression vector. Cells were assayed for caspase 3 activity using a caspase 3 assay kit (Promega), according to the manufacturer's instructions.
Cell Viability Assay
H1299 cells were seeded at 1.0 × 105 cells per well in 6-well plates. After 24 h, cells were transfected with 1 μg of pEGFP p53 or mutant p53 expression vector in triplicate wells. Cells were harvested after 24 h and pooled. 1.0 × 104 cells were sorted by GFP fluorescence into each well of a 96-well plate containing cell culture medium with 800 μg/ml of geneticin (Sigma) with a Moflo cell sorter. Cell viability was measured using a Celltitre-Glo Kit according to manufacturer's instructions (Promega).
RESULTS
Effects of Stability on Protein Levels
Full-length p53 mutants with a range of different stabilities (Fig. 1) were transiently expressed as a fusion protein with EGFP at the C terminus of p53 in H1299 cells. Fusion with EGFP does not affect the overall stability of p53, and EGFP fluoresces only when p53 is folded (26). This is based on the rapid protein-folding assay (27), in which an unfolded test protein fused to the N terminus of EGFP can interfere with the folding of EGFP. Only when the test protein is fully folded will proper formation of the EGFP chromophore occur and fluorescence be detected.
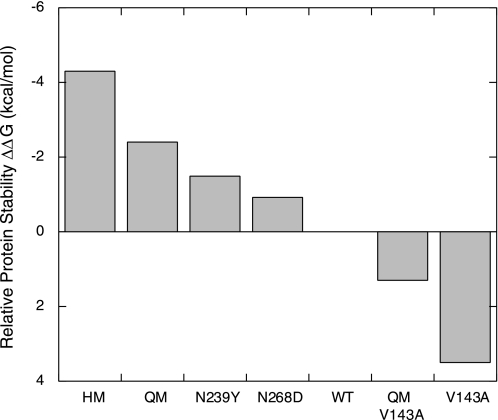
FIGURE 1.
Relative stabilities of p53 core domain (p53C) and its mutants. The thermodynamic stabilities of p53 core domains were measured as previously in vitro by equilibrium urea denaturation at 10 °C, at which temperature thermodynamic stability can be more accurately determined and compared (18, 20, 24, 29). Relative stabilities are given as ΔΔG values (kcal/mol) with respect to wild-type p53 core domain.
The steady-state levels of folded p53 were measured by flow cytometry, by monitoring the average fluorescence intensity of the green fluorescent cells, thus distinguishing transfected cells from non-transfected cells (Fig. 2) (28).
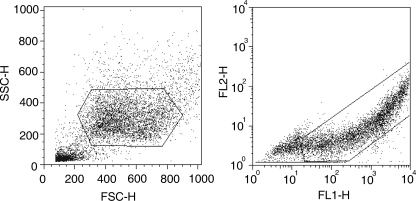
FIGURE 2.
FACs analysis for measuring cellular fluorescence. Left, cells with the right forward (FSC) and side scatter (SSC) were selected by gating. Right, EGFP-positive cells selected were measured for fluorescence emission through the FL1 (530 nm) and FL2 (583 nm) channels. This allowed p53-transfected cells to be distinguished from non-transfected ones. The fluoresence intensity of 10,000 individual cells was measured, and the average fluorescence intensity was recorded.
Levels of Transcriptionally Active p53 Over Time
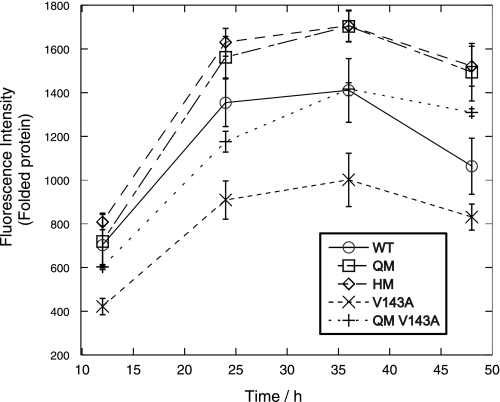
The levels of folded protein for transcriptionally active p53 mutants were also measured over a range of 48 h by EGFP fluorescence intensity (Fig. 3). We observed an accumulation in p53 levels to a maximum after 36 h followed by a decrease in signal. The levels of p53 correlated with thermodynamic stability. QM and HM, which have the highest stabilities, accumulated more rapidly than WT or V143A in the first 36 h and have the highest levels of folded protein over the 48-h period. V143A, which is a highly destabilized oncogenic mutant (29), had the lowest amount of folded protein over the time period. There was, however, little difference between QM and HM levels despite the higher stability in HM.
FIGURE 3.
Stabilized HM and QM mutants gave higher fluorescence (levels of folded p53) than wild-type p53 over a time course. EGFP-tagged transcriptionally active p53 mutants were transfected into H1299 cells, and the average fluorescence intensity was measured by FACS scan at different time points from 12 to 48 h after transfection. Higher thermodynamic stability led to higher accumulation over time. The QM-V143A control, which was similar to WT in terms of thermodynamic stability, had similar levels of folded protein to WT.
We also made a control, QM-V143A, which has a destabilizing V143A mutation, lowering its stability to slightly below that of WT p53 (Fig. 1). The levels of QM-V143A protein were very similar to WT p53 over the 48-h period, which suggested that the higher levels of protein observed for QM and HM was due to their higher stabilities rather than the effect of one or more of the stabilizing mutations.
The comparison of transcriptionally active p53 may reflect a more complex picture due to the induction of degradation and apoptosis, especially toward the later stages of the 48-h period. Differences in the extent of cell killing by the various mutants will be shown later.
Comparison with Transcriptionally Inactive p53 after 48 h
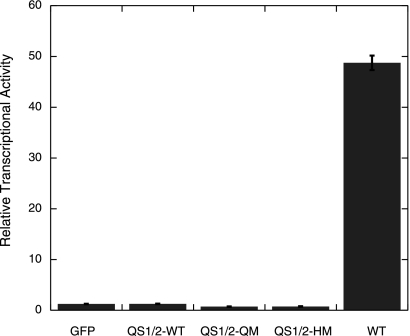
We made use of a transcriptionally inactive scaffold L22Q/W23S/W53Q/F54S (QS1/2) mutant (30) to rule out any self-induced degradation via the transcriptionally mediated Hdm2 feedback loop and to minimize apoptosis (Figs. 4 and 5). The QS1/2 mutations are in the natively unfolded N terminus domain of p53 and do not affect the overall thermodynamic stability of p53, which is governed by the core domain. The lack of cell killing in these transcriptionally inactive mutants also led to a more homogeneous population of GFP-positive cells, which allowed better comparison of fluorescence levels.
FIGURE 4.
Caspase 3 assay showing that QS1/2-WT, QS1/2-QM, and QS1/2-HM do not induce apoptosis. H1299 cells were transfected with EGFP-tagged p53 plasmid. Caspase 3 activity was measured after 24 h, and normalized for cell numbers using a cell viability kit in a parallel experiment.
FIGURE 5.
Transcriptional assay showing that the QS1/2-WT, QS1/2-QM and QS1/2-HM mutants are transcriptionally inactive. H1299 cells were co-transfected with EGFP-tagged p53, Bax-luciferase reporter plasmid and a CMV-Renilla plasmid, and luciferase activity was measured after 24 h.
The fluorescence levels between the transcriptionally inactive QS1/2 mutants showed a similar trend as the transcriptionally active ones (Fig. 6a). Levels of QS1/2-QM and QS1/2-HM were significantly higher than that of QS1/2-WT after 48 h, which suggests that higher stability does indeed lead to a higher accumulation of folded protein. QS1/2-V143A also yielded the smallest amount of protein. We also found that the fluorescence levels of QS1/2-WT, QS1/2-QM, and QS1/2-HM were higher than the transcriptionally active WT, QM, and HM, 48 h after transfection (Fig. 6a).
FIGURE 6.
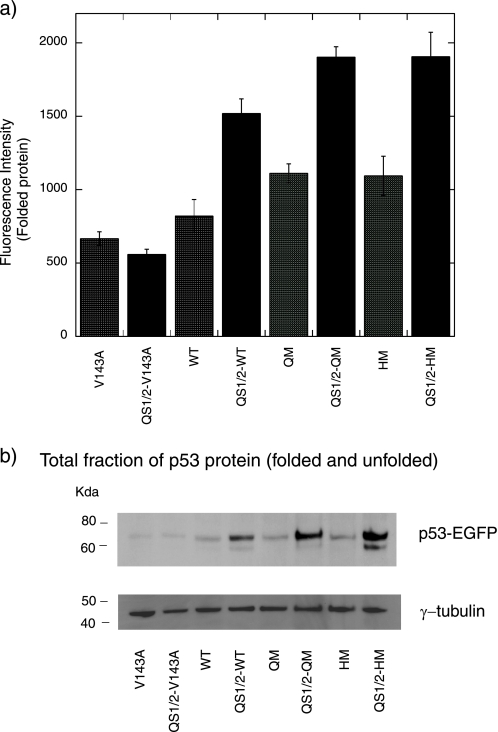
Higher stability of p53 leads to higher fluorescence (levels of folded p53) in both transcriptionally active and inactive (QS1/2) p53 mutants. a, EGFP-tagged p53 mutants were transfected into H1299 cells and the average fluorescence intensity was measured 48 h after transfection using a FACscan. Levels of folded p53 were correlated with intrinsic thermodynamic stability. Transcriptionally active mutants had lower levels of folded protein than transcriptionally inactive ones with the QS1/2 scaffold. b, Western blot showing the total fraction of p53 protein 48 h after transfection. The total fraction of p53 correlates well with thermodynamic stability of the transcriptionally inactive QS1/2 mutants.
Western blot analysis gave the total fraction of protein, which includes folded and unfolded protein (Fig. 6b). It can be seen that the total fraction of protein correlated well with the thermodynamic stability of the transcriptionally inactive QS1/2 mutants. The transcriptionally inactive QS1/2-WT, QS1/2-QM, and QS1/2-HM mutants were at higher levels in the total fraction than were their transcriptionally active counterparts because of activation of Hdm2 and the effects of cell death by the transcriptionally active mutants. No difference in levels was observed between V143A and QS1/2-V143A, since both mutants are inactive.
Higher Cell-killing Activity in QM and HM
We initially tried to measure the transcriptional activity of QM and HM and co-transfected p53 with a constitutive reporter plasmid, CMV-Renilla, to serve as a control for cell number. However, we found that Renilla activity was consistently lower for QM and HM than for WT after 24 h (results not shown). This led us to suspect that QM and HM were more potent in killing cells.
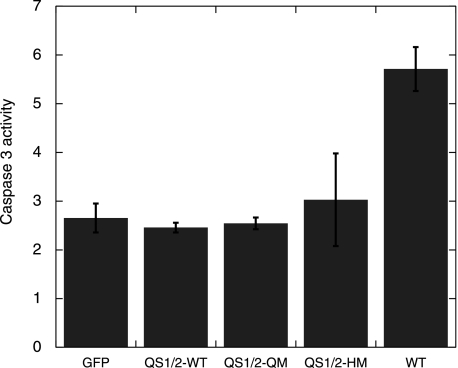
Our suspicion that QM and HM were more toxic persisted when we tried to select for EGFP-positive cells with geneticin. We observed that much fewer EGFP-positive cells transfected with QM and HM survived, as compared with wild-type, when viewed under the fluorescence microscope. We used a caspase 3 activity assay to show that QM and HM induced apoptosis and displayed higher activity than wild-type p53 (Fig. 7).
FIGURE 7.
The N239Y substitution leads to higher caspase 3 activity. H1299 cells were transfected with EGFP-tagged p53 plasmid. Caspase 3 activity was measured after 24 h, and normalized for cell numbers using a cell viability kit in a parallel experiment. Higher caspase activity could be traced back to a single N239Y mutation (N239Y mutation is also present in QM-V143A, QM, and HM). Comparison with the QM V143A and N268D controls showed that the higher caspase 3 activity in N239Y was not due to its higher thermodynamic stability.
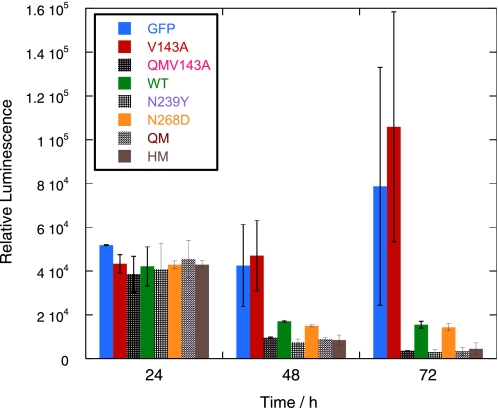
We then used a cell viability assay to measure the number of surviving cells after sorting the same number of EGFP-positive cells into each well of a 96-well plate to confirm our results. The use of cell sorting also ruled out proliferation of non-transfected cells. The number of viable cells transfected with HM and QM was significantly lower than with WT p53 after both 2 and 3 days (Fig. 8). Almost none of the cells transfected with QM and HM survived after 4 days (data not shown).
FIGURE 8.
Cell viability assay shows that the N239Y substitution contributes to lower cell viability. H1299 cells were transfected with EGFP-tagged p53 plasmid. 10,000 EGFP-positive cells were sorted into each well in a 96-well plate and allowed to proliferate. Cell viability was measured after 1 and 2 days. The results confirmed that N239Y led to higher cell death as shown by the caspase 3 assay.
Role of N239Y in Apoptosis
We investigated whether the higher stability of HM and QM was the cause of higher apoptosis levels seen in the caspase 3 and cell viability assays. The fact that QM-V143A was more active than WT in the caspase 3 and cell viability assays (Figs. 7 and 8) suggested that the increased stability of QM and HM per se was not the cause of the increase in cell death.
The oncogenic mutant V143A in the WT framework is inactive because of its low stability, which causes it to be unfolded at 37 °C. The V143A mutant was inactive in cell killing in both the caspase 3 assay (Fig. 7) and the cell viability assay (Fig. 8). Stabilizing the mutant with the additional four mutations from QM to give QM-V143A raised its stability to be about 1.3 kcal/mol less than WT (Fig. 1) but increased its activity in the caspase 3 assay to be close to that of the far more stable QM and HM. This finding implies that the higher caspase activity does not result from increased stability of the mutants, but most likely results from a specific effect of one or more of the four stabilizing mutations.
We thus studied two of the substitutions, N239Y and N268D, which are found at the surfaces of HM and QM. N239Y has a stabilizing effect of 1.49 kcal mol−1 while N268D has a stabilizing effect of around 0.92 kcal mol−1 (Fig. 1). The N239Y substitution, alone, led to higher apoptosis and cell death in the caspase 3 assay (Fig. 7) and cell viability assay (Fig. 8) whereas N268D caused no significant difference in p53 activity, compared with wild-type p53. Similar effects on apoptosis and cell death were seen for QM-V143A, QM, and HM, which contain the N239Y substitution (Figs. 7 and 8). These effects suggested that the N239Y substitution is the determining factor for the higher apoptotic activity seen in our results. It thus appears that increasing stability does not have any further observable effect on cell death in our assay.
Higher Cell-killing Activity of N239Y Is Transcriptionally Dependent
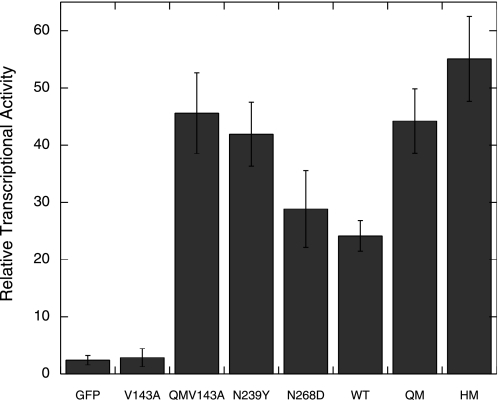
We repeated the transcription assay, but measured the activity at a shorter time point of 7 h to minimize cell death We observed that QM-V143A, N239Y, QM, and HM had higher transcriptional activities than WT and N268D (Fig. 9), which suggested that the N239Y mutation leads to higher transcription from the Bax promoter.
FIGURE 9.
N239Y mutation led to higher transcription from a Bax reporter plasmid. H1299 cells were co-transfected with EGFP-tagged p53, Bax-luciferase reporter plasmid and a CMV-Renilla plasmid. Luciferase activity was measured after 7 h. The higher transcriptional activity due to the N239Y mutation, suggest that the higher cell killing is transcriptionally dependent.
The V143A mutant did not have any transcriptional activity (Fig. 9), but its activity was fully restored to above WT levels in QM-V143A when the stabilizing QM scaffold is used.
We ruled out any transcription-independent pathway that could have led to the higher cell death observed in QM and HM by making use of the QS1/2 scaffold (Fig. 5). We also observed no difference in caspase 3 activities between QS1/2-WT, QS1/2-QM, and QS1/2-HM (Fig. 4).
DISCUSSION
Comparison of p53 Levels and the Role of Thermodynamic Stability in Cells
We determined the levels of p53 core domain mutants both when transcriptionally competent and when containing mutations in the transactivation domain that render them transcriptionally incompetent.
There was a correlation between the thermodynamic stability of the transcriptionally competent p53 mutants and the levels of folded protein in mammalian cells. To compare protein levels in the absence of cell death and self-induced degradation by up-regulation of Hdm2, we made use of the transcriptionally inactive QS1/2 scaffold. There were similar trends for both transcriptionally active and inactive QM and HM mutants, which accumulated to higher levels of folded protein than WT. However, the increase in stability from QM to HM led to a less significant increase in the fraction of folded protein.
The thermodynamic stability of p53 has been shown earlier to correlate with the rate of unfolding of protein in vitro at 37 °C (31). Our results strongly suggest that the intrinsic thermodynamic stability of p53 affects the rate of accumulation and degradation of folded protein in mammalian cells.
The in vivo level of a protein is related to its rate of synthesis and degradation, and p53 is known to be mainly regulated at its rate of degradation. It is likely that p53 can unfold spontaneously in vivo, in a manner similar to in vitro experiments (31). Stabilized p53 mutants, QM and HM, have slower rates of unfolding,4 and thus accumulate to higher levels of folded protein. This effect is observed even in the absence of the induction of Hdm2 and other E3 ligases, suggesting that spontaneous degradation occurs independently of these pathways.
p53, like many other cellular proteins, is degraded by the ubiquitin-dependent pathway, which involves the 26S proteosome (32). p53 degradation via Hdm2 ubiquitination involves recognition of the folded p53 core domain, in addition to its N terminus, by the Hdm2 protein (33, 34). p53 may also be degraded by a ubiquitin-independent mechanism, involving the 20S proteosome (35), known to degrade mainly unfolded proteins such as tau (36) and p21 (37). Denatured p53 does accumulate in cells, as detected by antibodies, and less stable mutants more so (38). Such spontaneously unfolded p53 may be targeted toward further proteolysis through the 20S proteosome pathway. p53 may have evolved to be unstable at body temperature so that the protein could have facile degradation via spontaneous unfolding when it is not needed. The rapid degradation of p53 by multiple mechanisms may facilitate the tight control of p53 levels.
Transcriptional Feedback Loop of p53
The QS1/2 scaffold used in our experiments rules out p53-induced degradation via the Hdm2 feedback loop and p53-induced apoptosis. The QS1/2 mutations have been studied separately as L22Q/W23S (QS1) (39) and W53Q/F54S (QS2) (40). These mutations affect the transactivation domains 1 and 2 of p53, respectively, both leading to reduced transcription and apoptosis. The four mutations (QS1/2) were later shown to abrogate synergistic transcription and apoptosis in H1299 cells (30), which we confirmed in our results.
The lack of transcription of Hdm2, and hence the absence of its regulatory feedback loop, led to the accumulation of higher levels of folded p53 protein when the QS1/2 scaffold was used. This suggests that the transcriptionally induced feedback loop is important in maintaining low levels of p53 in the cell.
Role of the Second Site Suppressor N239Y in Apoptosis
We observed higher transcriptional and apoptotic activities for QM and HM than WT p53. The increase in the level of transcription and apoptosis was traced to a single N239Y mutation, which was further confirmed by cell-viability assays, but we did not observe any further significant increase in activity from any higher intrinsic stability in our assay.
N239Y acts as a second site suppressor mutation, which is able to restore cancer hot-spot mutations such as G245S, found in loop L3 (41). N239Y increases the thermodynamic stability of the p53 core domain by about 1.5 kcal mol−1, which is able to restore wild type stability in G245S/N239Y (42). We observed that the N239Y mutation led to higher transcriptional and apoptotic activity in mammalian cells. It is possible that the surface mutation affects interactions with proteins that bind to p53. It has also been suggested that N239Y may alter DNA binding specificity as it is found close to the phosphodiester backbone of DNA when p53 is bound to DNA (41). p53 controls a vast number of genes in cell cycle arrest, apoptosis, and DNA repair in a DNA sequence-specific manner, and mutations in the DNA binding domain have been shown to lead to differential transcriptional and downstream biological responses (43). The higher activity of the N239Y mutant might explain why it has not been selected for in nature (19), as it is a more potent inducer of apoptosis. Future studies on this mutant may further elucidate how p53 chooses between cell cycle arrest and apoptosis, which is of interest for cancer therapy.
Rescue of Oncogenic V143A Mutant by QM Scaffold
The V143A oncogenic mutant shows neither DNA binding nor transcriptional activity at 37 °C, although these functions can be restored at lower temperatures (44). The mutation destabilizes p53 core domain by 3.5 kcal mol−1 (29). The transcriptional and apoptotic activity of V143A can be partially restored by individual N239Y and N268D mutations (41). Our results confirmed that V143A has no transcriptional or cell-killing activity at 37 °C and demonstrated that the destabilizing V143A mutation could be fully rescued in terms of transcriptional and apoptotic activity to above wild-type level by placing it in the QM-stabilizing scaffold. This presumably arises from the overall stabilizing effect of the QM scaffold, which contains both the N239Y and N268D mutations, and which contributes an increase of 2.5 kcal mol−1 to stability. This observation is in agreement with the crystal structure of QM-V143A, showing that the mutation creates an internal cavity but leaves the overall structure intact (20). The additional potency in inducing cell death comes from the N239Y substitution.
CONCLUSIONS
Our results show that the thermodynamic stability of p53 is correlated the level of folded protein in mammalian cells, but that increasing the intrinsic stability of p53 above that of WT appears to have no significant effect on transcriptional or apoptotic activity. It is possible that p53 has evolved to be marginally stable to allow the spontaneous denaturation of WT p53, without compromising its activity. This may serve as an alternative pathway for the tight control of p53 levels, independent of transcriptional feedback loops.
Acknowledgments
We thank Richard Grenfell for help with cell sorting and Dr. Fiona Townsley, Dr. Joel Kaar, and Caroline Blair for reading the manuscript. We are also grateful to Dr. Andreas Joerger for helpful discussions and Caroline Blair for help with molecular biology.
This work was supported in part by the Medical Research Council and Cancer Research UK and EC FP6 funding.
G. Jaggi and A. R. Fersht, unpublished in vitro data for QM.
- QM
- quadruple mutant
- HM
- hexamutant
- WT
- wild-type
- EGFP
- enhanced green fluorescent protein
- FACS
- fluorescent-activated cell sorter.
REFERENCES
- 1.Vogelstein B., Lane D., Levine A. J. (2000) Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 2.Brooks C. L., Gu W. (2003) Curr. Opin. Cell Biol. 15, 164–171 [DOI] [PubMed] [Google Scholar]
- 3.Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 4.Toledo F., Wahl G. M. (2006) Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 5.Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J. M., Lozano G., Hakem R., Benchimol S. (2003) Cell 112, 779–791 [DOI] [PubMed] [Google Scholar]
- 6.Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G. D., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. (2004) Nature 429, 86–92 [DOI] [PubMed] [Google Scholar]
- 7.Marine J. C., Lozano G. (2009) Cell Death Differ., in press [DOI] [PubMed] [Google Scholar]
- 8.Barak Y., Juven T., Haffner R., Oren M. (1993) EMBO J. 12, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X., Bayle J. H., Olson D., Levine A. J. (1993) Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 10.Joerger A. C., Fersht A. R. (2008) Annu. Rev. Biochem. 77, 557–582 [DOI] [PubMed] [Google Scholar]
- 11.Bullock A. N., Henckel J., DeDecker B. S., Johnson C. M., Nikolova P. V., Proctor M. R., Lane D. P., Fersht A. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14338–14342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olivier M., Eeles R., Hollstein M., Khan M. A., Harris C. C., Hainaut P. (2002) Hum. Mutat. 19, 607–614 [DOI] [PubMed] [Google Scholar]
- 13.Bullock A. N., Fersht A. R. (2001) Nat. Rev. Cancer 1, 68–76 [DOI] [PubMed] [Google Scholar]
- 14.Tang K. S., Guralnick B. J., Wang W. K., Fersht A. R., Itzhaki L. S. (1999) J. Mol. Biol. 285, 1869–1886 [DOI] [PubMed] [Google Scholar]
- 15.Klein C., Georges G., Künkele K. P., Huber R., Engh R. A., Hansen S. (2001) J. Biol. Chem. 276, 37390–37401 [DOI] [PubMed] [Google Scholar]
- 16.Levrero M., De Laurenzi V., Costanzo A., Gong J., Wang J. Y., Melino G. (2000) J. Cell Sci. 113, 1661–1670 [DOI] [PubMed] [Google Scholar]
- 17.Patel S., Bui T. T., Drake A. F., Fraternali F., Nikolova P. V. (2008) Biochemistry 47, 3235–3244 [DOI] [PubMed] [Google Scholar]
- 18.Nikolova P. V., Henckel J., Lane D. P., Fersht A. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14675–14680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joerger A. C., Allen M. D., Fersht A. R. (2004) J. Biol. Chem. 279, 1291–1296 [DOI] [PubMed] [Google Scholar]
- 20.Joerger A. C., Ang H. C., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15056–15061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ang H. C., Joerger A. C., Mayer S., Fersht A. R. (2006) J. Biol. Chem. 281, 21934–21941 [DOI] [PubMed] [Google Scholar]
- 22.Wells M., Tidow H., Rutherford T. J., Markwick P., Jensen M. R., Mylonas E., Svergun D. I., Blackledge M., Fersht A. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tidow H., Melero R., Mylonas E., Freund S. M., Grossmann J. G., Carazo J. M., Svergun D. I., Valle M., Fersht A. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12324–12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khoo K. H., Joerger A. C., Freund S. M., Fersht A. R. (2009) Protein Eng. Des. Sel. 22, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cañadillas J. M., Tidow H., Freund S. M., Rutherford T. J., Ang H. C., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2109–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer S., Rüdiger S., Ang H. C., Joerger A. C., Fersht A. R. (2007) J. Mol. Biol. 372, 268–276 [DOI] [PubMed] [Google Scholar]
- 27.Waldo G. S., Standish B. M., Berendzen J., Terwilliger T. C. (1999) Nat. Biotechnol. 17, 691–695 [DOI] [PubMed] [Google Scholar]
- 28.Mayer S. (2006) Effects of Mutations on p53 Level in Cells, PhD thesis, University of Cambridge [Google Scholar]
- 29.Bullock A. N., Henckel J., Fersht A. R. (2000) Oncogene 19, 1245–1256 [DOI] [PubMed] [Google Scholar]
- 30.Venot C., Maratrat M., Sierra V., Conseiller E., Debussche L. (1999) Oncogene 18, 2405–2410 [DOI] [PubMed] [Google Scholar]
- 31.Friedler A., Veprintsev D. B., Hansson L. O., Fersht A. R. (2003) J. Biol. Chem. 278, 24108–24112 [DOI] [PubMed] [Google Scholar]
- 32.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 33.Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Mol. Cell 23, 251–263 [DOI] [PubMed] [Google Scholar]
- 34.Yu G. W., Rudiger S., Veprintsev D., Freund S., Fernandez-Fernandez M. R., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asher G., Shaul Y. (2005) Cell Cycle 4, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 36.David D. C., Layfield R., Serpell L., Narain Y., Goedert M., Spillantini M. G. (2002) J. Neurochem. 83, 176–185 [DOI] [PubMed] [Google Scholar]
- 37.Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. (2003) Science 299, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Issaeva N., Friedler A., Bozko P., Wiman K. G., Fersht A. R., Selivanova G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13303–13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J., Zhou W., Jiang J., Chen X. (1998) J. Biol. Chem. 273, 13030–13036 [DOI] [PubMed] [Google Scholar]
- 40.Candau R., Scolnick D. M., Darpino P., Ying C. Y., Halazonetis T. D., Berger S. L. (1997) Oncogene 15, 807–816 [DOI] [PubMed] [Google Scholar]
- 41.Brachmann R. K., Yu K., Eby Y., Pavletich N. P., Boeke J. D. (1998) EMBO J. 17, 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikolova P. V., Wong K. B., DeDecker B., Henckel J., Fersht A. R. (2000) EMBO J. 19, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menendez D., Inga A., Resnick M. A. (2006) Mol. Cell Biol. 26, 2297–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W., Guo X. Y., Hu G. Y., Liu W. B., Shay J. W., Deisseroth A. B. (1994) EMBO J. 13, 2535–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]