Abstract
The post-translational import of nucleus-encoded preproteins into chloroplasts occurs through multimeric translocons in the outer (Toc) and inner (Tic) membranes. The high fidelity of the protein import process is maintained by specific recognition of the transit peptide of preproteins by the coordinate activities of two homologous GTPase Toc receptors, Toc34 and Toc159. Structural and biochemical studies suggest that dimerization of the Toc receptors functions as a component of the mechanism to control access of preproteins to the membrane translocation channel of the translocon. We show that specific mutations that disrupted receptor dimerization in vitro reduced the rate of protein import in transgenic Arabidopsis compared with the wild type receptor. The mutations did not affect the GTPase activities of the receptors. Interestingly, these mutations did not decrease the initial preprotein binding at the receptors, but they reduced the efficiency of the transition from preprotein binding to membrane translocation. These data indicate that dimerization of receptors has a direct role in protein import and support a hypothesis in which receptor-receptor interactions participate in the initiation of membrane translocation of chloroplast preproteins as part of the molecular mechanism of GTP-regulated protein import.
Introduction
The Toc2 (translocon at the outer membrane of chloroplasts) and Tic (translocon at the inner membrane of chloroplasts) complexes constitute the major pathway for the import of nucleus-encoded proteins into chloroplasts (1, 2). The Toc translocon contains two surface-exposed membrane receptors, Toc159 and Toc34, that recognize the transit peptides of newly synthesized chloroplast preproteins and initiate protein import by promoting the insertion of preproteins into the Toc channel (3, 4). Toc75, a β-barrel membrane protein, is the primary component of the protein-conducting channel (5, 6). The transition from preprotein binding to membrane translocation represents the committed step in the import process, and this key step is controlled by the intrinsic GTPase activity of Toc159 and Toc34.
Considerable biochemical evidence has accumulated to support a role for the Toc receptors as GTP-dependent regulators of the import reaction. Nonhydrolyzable GTP analogs significantly inhibit preprotein import into isolated chloroplasts, demonstrating that a GTPase cycle plays an important role in receptor function (7, 8). Furthermore, genetic studies demonstrate that members of the Toc159 and Toc34 family members are essential in Arabidopsis (9–12). Although it is clear that the Toc GTPase receptors control transit peptide recognition and the initial stages of membrane translocation, the molecular mechanism that couples GTPase activity with receptor function remains undefined. One clue to a potential mechanism of receptor function has been revealed by the x-ray crystal structure of pea Toc34 (psToc34) (13). The structure revealed the existence of a Toc34 homodimer, with amino acid side chains from each monomer interacting with bound GDP in the nucleotide binding site on the reciprocal monomer. Subsequent direct binding studies support the ability of psToc34 and the Arabidopsis Toc34 ortholog, atToc33, to form homodimers (14–19). Key residues involved in Toc34 dimerization are conserved in the GTPase domain of Toc159, and biochemical studies also demonstrate that Toc34 interacts with Toc159 via their GTPase domains (20, 21). These studies have led to models in which changes in receptor homo- or heterodimerization in response to nucleotide binding/hydrolysis correspond to the molecular switch from preprotein binding to membrane translocation (3, 4, 22). These models include proposals that changes in Toc receptor interactions represent a molecular gate to the translocon channel or that receptor dimerization functions as a mechanism to hand off preproteins from one receptor to another during the initial stages of import.
The observation that residues from one monomer interact with a bound nucleotide in the GTP binding pocket of the reciprocal monomer in the psToc34 dimer suggests that the two subunits might act as reciprocal GTPase-activating proteins (GAPs) (13). This hypothesis is attractive because it provides a link between receptor dimerization and the GTP-controlled events in the import reaction. Several reports have addressed this hypothesis by examining mutants in which arginine 133 in psToc34 or the comparable residue in atToc33 (arginine 130) was converted to alanine. Arg130/Arg133 of one monomer makes contact with the β-phosphate of bound GDP in the other monomer in the crystal structure; this residue was originally proposed to act as an “arginine finger” to promote γ-phosphate hydrolysis in a manner similar to the GAPs of other small GTPases. Although all of these studies demonstrate that Arg130/Arg133 disrupts homo- and heterodimerization (15–18, 23), they yield data both for and against a role for dimerization in activating GTP hydrolysis. Additional studies investigating the relationship between nucleotide binding and dimerization indicate that the nucleotide-bound state of the receptors does not appear to have a major impact on either homo- or heterodimerization of the receptors (16, 19, 20). Taken together, the in vitro studies provide equivocal data on the role of dimerization in Toc GTPase function. Furthermore, there is little evidence to demonstrate that receptor-receptor interactions play a role in the structure or functional dynamics of Toc complexes in chloroplasts despite the prevalent role of receptor dimerization in models of Toc receptor function.
To address the physiological role of Toc receptor dimerization in protein import, we introduced amino acid substitutions within atToc33 that are predicted to affect dimer formation. We identified mutants that reduced receptor dimerization but had no detectable effects on atToc33 GTPase activity. These mutants were introduced into the Arabidopsis atToc33 null mutant, ppi1 (plastid protein import mutant 1), and their effects on protein import and chloroplast biogenesis were assessed both in vivo and in vitro. These studies support a role for dimerization in Toc receptor function and suggest that receptor-receptor interactions play a key role in the initiation of membrane translocation at Toc complexes.
EXPERIMENTAL PROCEDURES
atToc33 Mutant Constructs
To generate atToc33-R130A, the atToc33 coding region was amplified in two fragments using PCR. Fragment one was amplified using sense (5′-GAAATTAATACGACTCACTATAGGGG-3′) and antisense (5′-CCCAAGCTTGAGCTCATCGACTGCATACACATCCAAACGATCA-3′) primers. Fragment two was amplified using sense (5′-CATGGAGCTCGATAAGCAAGTTGTTATAG-3′) and antisense (5′-TTATGCTAGTTATTGCTCAG-3′) primers. The PCR fragments were inserted into the NcoI and SalI sites of pET21d-Tev:atToc33FL in a triple ligation to yield pET21d:atToc33-R130A.Tev.His. Other atToc33 mutations were introduced into pET21d-Tev:atToc33FL using the DpnI-mediated mutagenesis technique (24). AtToc33-F67A was amplified with sense (5′-CAATCCTTCAGCTTGGGCAGGACTGACACGGAC-3′) and antisense (5′-GTCCGTGTCAGTCCTGCCCAAGCTGAAGGATTG-3′) primers. AtToc33-D127A was amplified with sense (5′-ATCACTCTATACACAGCCAAACGATCAACATA-3′) and antisense (5′-TATGTTGATCGTTTGGCTGTGTATAGAGTCGAT-3′) primers. AtToc33-Y129A was amplified with sense (5′-TAGCTCATCGACTCTAGCCACATCCAAACGATC-3′) and antisense (5′-GATCGTTTGGATGTGGCTAGAGTCGATGAGCTA-3′) primers. AtToc33-L134A was amplified with sense (5′-AACAACTTGCTTATCTGCCTCATCGACTCTATA-3′) and antisense (5′-TATAGAGTCGATGAGGCAGATAAGCAAGTTGTT-3′) primers. For constructs with no C-terminal tag, atToc33 or the appropriate mutant was amplified from the full-length constructs using the sense (5′-GGAGCCATGGGGTCTCTCGTTCGTG-3′) and antisense (5′-CGGTCGACTTAAAGTGGCTTTCCACTTGT-3′) primers.
To generate the atToc33G constructs corresponding to the GTP-binding domains of the receptor, sequences corresponding to amino acids 1–256 of atToc33 or the appropriate mutant were amplified from the full-length constructs using the sense primer, 5′-GGAGCCATGGGGTCTCTCGTTCGTG-3′, in combination with one of the following antisense primers: 5′-AGGTCGACTTACTTTCCTTTATCATCAGAG-3′ for constructs with no C-terminal tag, 5′-GATGAGAAGCTTTCCTTTATCATCAGAG-3′ for constructs encoding a C-terminal His6 tag, or 5′-GCAAGCTTTCTTTCCTTTATCATCAGA-3′ for constructs encoding a C-terminal Tev protease cleavage site and a His6 tag. The PCR fragments were inserted into double digested pET21d:atToc33 plasmids to generate the final expression plasmids. For expression of atToc33.Tev.His in Arabidopsis, the coding regions of wild type or mutant genes were cloned into the EcoRI/BamHI sites of the binary vector construct pSMB-CaMV35S:atToc33.Tev.His.
In Vitro Translation and Purification of Escherichia coli-expressed Proteins
All [35S]methionine-labeled in vitro translation products were generated in a coupled transcription-translation system containing reticulocyte lysate according to the manufacturer's instructions (Promega, TnT coupled reticulocyte lysate system). For E. coli expression, all constructs containing His6 tags were transformed into E. coli strain BL21(DE3), and expression was induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 15 °C for 16 h after the culture had reached A600 = 0.6. Overexpressed proteins were purified using Ni-NTA chromatography (Novagen) under nondenaturing conditions at 4 °C. Proteins for GTPase assays were used immediately. Otherwise, protein fractions were stored in HMK buffer (50 mm Hepes-KOH, pH 7.5, 5 mm MgCl2, 25 mm KOAc) containing 10% glycerol. Protein concentration was measured by the Bradford method (25).
GTP Hydrolysis Assays
GTP hydrolysis by atToc33 wild type and atToc33 mutants was assayed using the phosphate release assay as described previously (23). Reactions were carried out in GBS (20 mm Tricine-KOH, pH 7.65, 1 mm MgCl2, 50 mm NaCl, 1 mm β-mercaptoethanol), 25 nm [γ-32P]GTP, and 25 μm GTP in a final volume of 25 μl at 25 °C for 20 min. Three independent experiments were performed, and each data point was counted in duplicate. To examine preprotein stimulation of GTP hydrolysis activity, hydrolysis experiments were performed as described above in the presence of a 10-fold molar excess of the E. coli-expressed precursor protein of the small subunit of Rubisco (preSSU).
Solid Phase Binding Assays
Direct interactions of Toc wild type or mutant receptors were measured using a solid phase binding assay as described previously (20) with E. coli-expressed proteins as the bait and in vitro translated 35S-labeled proteins as the prey. Bound [35S]methionine-labeled proteins were detected in dried gels using a Fuji Fla-5000 phosphorimaging device and Multi Gauge V2.02 software for quantification. Bound [35S]methionine-labeled protein is presented as the percent of added total [35S]methionine-labeled protein. Purified recombinant CRABP-His10 was a gift from Dr. L. Gierasch, University of Massachusetts, Amherst, MA.
Arabidopsis Transformation and Screening of Transgenic Plants
The pSMB-CaMV35S:atToc33.Tev.His plasmids were introduced into Arabidopsis plants using Agrobacterium tumefaciens (LBA4404)-mediated transformation by the floral dip method (26, 27). Transformants were sown on soil and selected using 100 μg/ml Basta (glufosinate ammonium). The presence of the transgenes was confirmed by PCR of genomic DNA using transgene-specific primers (sense primer, 5′-GGAGCCATGGGGTCTCTCGTTCGTG-3′; antisense primer, 5′-TAGTTTGCGTATATTTTG-3′). To determine the genetic background of transformed ppi1 plants, PCR-based identification was conducted as described previously (28).
Protein Extraction and Immunoblotting
Protein was extracted directly in SDS-PAGE sample buffer from total aboveground tissue of the plants indicated. Samples corresponding to equivalent amounts of total protein were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and subjected to immunoblotting with antisera to the indicated proteins. Immunoblotting was performed as described previously (29) using chemiluminescence detection. Antisera to atToc159, atToc75, atToc33, and atToc34 were described previously (10).
Chloroplast Isolation and Protein Import Assays
Arabidopsis thaliana seedlings were grown on agar plates containing 0.5× Murashige and Skoog growth medium containing 1% sucrose under long day conditions (18 h day:6 h night) at 22 °C for 10 days. Chloroplasts were isolated as described previously (30) and quantified by measuring the chlorophyll content according to Arnon (31).
Protein import was performed with [35 S]preSSU using chloroplasts corresponding to 10 μg of chlorophyll in a total volume of 100 μl of import buffer (330 mm sorbitol, 50 mm Hepes-KOH, pH 7.5, 25 mm KOAc, 5 mm MgOAc) as described previously (20, 30). When necessary, import reactions were divided into two equivalent fractions for protease treatment. One-half was treated with 100 μg/ml thermolysin on ice for 30 min, and the reactions were stopped by adding ice-cold HS buffer (50 mm Hepes-KOH, pH 7.5, 330 mm sorbitol) containing 50 mm EDTA. Kinetic analysis of import was performed using various concentrations of [35S]methionine-labeled E. coli-expressed and 6 m urea-denatured preSSU for 10 min at 23 °C (32). All samples in the kinetic analysis contained a final concentration of 0.2 m urea after dilution of the [35S]preSSU into the import reaction. After the import reaction, the chloroplasts were analyzed by SDS-PAGE and phosphorimaging. Kinetic constants were calculated using nonlinear fitting of Michaelis-Menten data with GraphPad Prism software (GraphPad Software, Inc.).
For preprotein binding reactions or early import intermediate formation, the in vitro translated [35S]preSSU was dialyzed against HS buffer to remove nucleotides (33). Intact chloroplasts corresponding to 35 μg of chlorophyll were used in 100 μl of import buffer. Chloroplasts were held for a dark adaptation for 10 min at 23 °C prior to performing the assays to deplete internal ATP. To uncouple ATP generation in chloroplasts, nigericin (final concentration, 800 nm) was added to the samples. Where indicated, 10 mm glycerate was added to deplete internal ATP in chloroplasts during the dark adaptation (7). Energy-depleted chloroplasts were incubated in the presence of various concentrations of ATP. In the case of samples with no added ATP, apyrase (2 units/100 μl) was added instead of ATP before the addition of the energy-depleted preSSU in vitro translation product. Assays to measure preprotein binding, the formation of early import intermediates, and analysis of the kinetics of preSSU import were conducted as described previously (30). All samples from chloroplast binding and import reactions were resolved by 14% SDS-PAGE and analyzed by phosphorimaging (Fuji Fla-5000). For quantification, Multi Gauge V2.02 software was used.
Cross-linking and Co-immunoprecipitation
Chloroplasts (200 μg of chlorophyll) from preprotein import reactions were reisolated, washed with HS buffer, and resuspended in 100 μl of 50 mm Hepes-NaOH, pH 7.5. To catalyze disulfide bond formation, 0.1 mm copper (II)-1,10-phenanthroline (CuP) was added and incubated at room temperature for 5 min. Iodoacetamide (final concentration, 10 mm) was added to quench the cross-linking reaction. Recovered chloroplasts were washed once with HS buffer. Co-immunoprecipitations under native and denaturing conditions were performed as described previously (30) using chloroplasts corresponding to 150 μg of chlorophyll.
RESULTS
Generation and Analysis of atToc33 Dimerization Mutants
The psToc34 GTPase domain formed a dimer with a 2-fold axis of symmetry in the x-ray crystal structure (13). Five residues contribute side chains that form major noncovalent bonds with the other monomer at the dimer interface (Table 1). The list of residues includes Arg133, which has been shown previously to disrupt dimerization (17, 18, 23). We reasoned that substitutions of the corresponding amino acids with alanine in atToc33 would destabilize the dimer interface and receptor-receptor interactions. To minimize effects on the folding or overall structure of atToc33, we substituted each residue individually. We established three initial criteria to identify substitutions that specifically affect dimerization with minimal disruption of other known receptor activities. The three criteria were: 1) reduction of receptor dimerization, 2) normal receptor GTPase activity, and 3) normal preprotein interaction as assayed by stimulation of GTPase activity.
TABLE 1.
Comparison of amino acids involved in homodimer formation in Arabidopsis Toc33 (atToc33) and pea Toc34 (psToc34)
| Amino acids substituted |
Interactions on opposite monomer |
Type of interaction | ||
|---|---|---|---|---|
| atToc33 | Corresponding amino acids in psToc34 | atToc33 | Corresponding amino acids in psToc34 | |
| Arg130 | Arg133 | Gly46 | Gly49 | Hydrophobic contact |
| Arg130 | Arg133 | Ser65 | Ser68 | Hydrogen bonding |
| Arg130 | Arg133 | GDP | GDP | Hydrogen bonding |
| Phe67 | Phe70 | Phe174 | Phe177 | Hydrophobic contact |
| Asp127 | Asp130 | Arg125 | Arg128 | Hydrogen bonding |
| Tyr129 | Tyr132 | Pro66 | Pro69 | Hydrophobic contact |
| Tyr129 | Tyr132 | GDP | GDP | Hydrophobic contact |
| Leu134 | Leu137 | Gly98 | Gly101 | Hydrophobic contact |
| Leu134 | Leu137 | Leu134 | Leu137 | Hydrophobic contact |
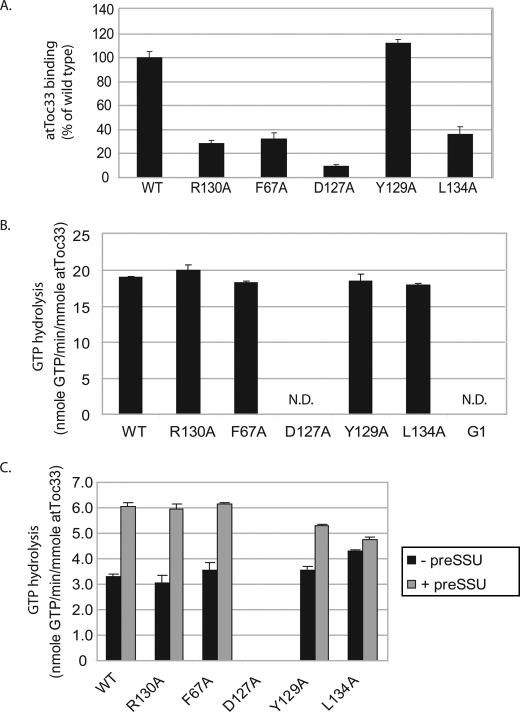
We expressed wild type and mutated atToc33 proteins as 29-kDa fragments containing C-terminal His6 tags and lacking their C-terminal transmembrane segments (atToc33G-His6). All of the proteins were expressed in E. coli and purified from soluble extracts using Ni-NTA affinity chromatography (data not shown). To examine the effects on dimerization, we performed a solid phase binding assay using 0.3 nmol of immobilized His6-tagged wild type and mutant proteins as the bait and in vitro translated wild type [35S]atToc33 as the prey. This assay had been used previously to investigate atToc33 homo- and heterodimerization (17, 20). In this initial screen, all mutants except atToc33-Y129A showed significantly decreased dimer formation as compared with wild type atToc33 (Fig. 1A).
FIGURE 1.
Biochemical characteristics of atToc33 dimerization mutants. A, analysis of receptor dimerization. E. coli-expressed and purified atToc33G-His6 wild type or the indicated mutant proteins (0.3 nmol) were immobilized on Ni-NTA-agarose and incubated with in vitro translated 35S-labeled full-length atToc33 (WT) lacking a C-terminal tag. The amount of bound receptor was quantified by phosphorimaging of SDS-PAGE-resolved samples. B, GTP hydrolysis by atToc33 (WT) and the indicated atToc33 mutants. G1 corresponds to atToc33-G45R/K49N/S50R, a receptor shown previously to lack GTPase activity (35). The concentration of total GTP is 10 μm. C, stimulation of GTPase activity of the atToc33 receptors by preprotein. The graph shows unstimulated GTP hydrolysis and stimulated hydrolysis in the presence of a 10-fold molar excess of preSSU. The concentration of total GTP is 1 μm. N.D., not detected. Error bars correspond to S.D.
Our second priority was to identify the substitutions that did not affect basal GTPase activity. Although dimerization originally was proposed to affect GTPase activity directly (13, 34), subsequent studies demonstrated that dimerization is not required for GTP hydrolysis by atToc33 (15–17). We examined basal GTP hydrolysis and preprotein-stimulated GTP hydrolysis (Fig. 1, B and C). AtToc33G-His6 wild type and mutant proteins were incubated with [γ-32P]GTP under substrate-saturating conditions (23), and hydrolysis was measured as released γ- phosphate. The assay was performed under conditions in which the total GTP concentration (10 μm) was saturating (23). Consequently, the measured GTPase activity corresponds to Vmax for hydrolysis. A previously characterized GTPase mutant, atToc33-G1, which lacks nucleotide binding and hydrolytic activity, was used as a negative control (35, 36). atToc33-G1 harbors three point mutations in the consensus G1 motif (P-loop) of the conserved GTP-binding domain. All of the alanine substitution proteins retained normal GTPase activity relative to wild type atToc33 with the exception of atToc33-D127A (Fig. 1B). atToc33-D127A did not exhibit measurable GTPase activity, suggesting that its ability to bind and hydrolyze GTP was significantly impaired.
Previous studies demonstrated that preprotein binding stimulates GTP hydrolysis on the Toc GTPase receptors (37, 38). We tested the stimulation of GTP hydrolysis by atToc33 constructs in the presence of preSSU. In this assay, the concentration of [γ-32P]GTP was reduced to 1 μm to maximize sensitivity. All of the proteins, except atToc33-L134A and atToc33-D127A, exhibited preprotein-stimulated hydrolysis at levels indistinguishable from wild type atToc33 (Fig. 1C). Preprotein stimulation of atToc33-L134A GTPase activity was only ∼10% above that of basal activity as compared with the ∼2-fold stimulation observed with the other proteins.
Analysis of the results shown in Fig. 1 indicated that atToc33-R130A and atToc33-F67A were the only two proteins that met all three of our initial criteria. The two receptor mutants appeared to disrupt dimerization without significantly affecting basal and stimulated GTPase activity. The results with atToc33-R130A are consistent with previously published findings (15, 17). Arg130 (Arg133 in psToc34) as well as several amino acids in the other monomer makes contact with the GDP molecule at the dimer interface (Table 1). Phe67 (Phe70 in psToc34) interacts with Phe174 at the other monomer via hydrophobic contacts (13) (Table 1). On the basis of these results, we focused our attention on these two proteins for further study.
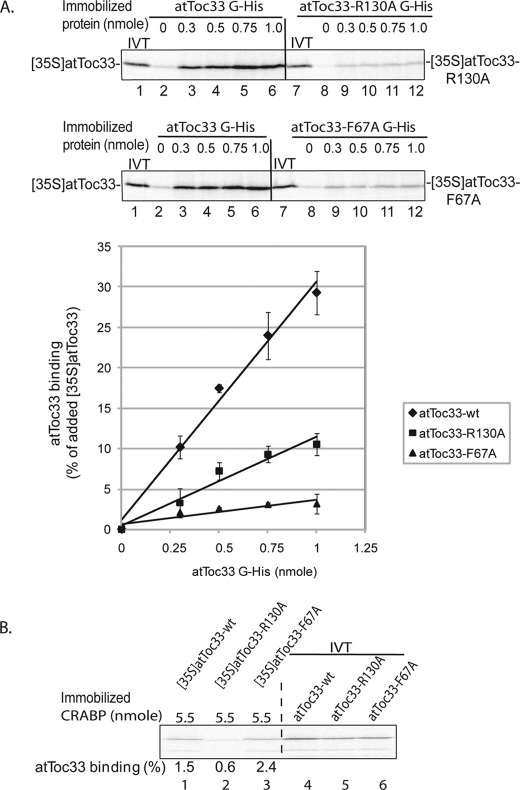
We performed a more detailed study of the biochemical characteristics of atToc33-R130A and atToc33-F67A. We tested the concentration dependence of homodimerization using the atToc33-R130A and atToc33-F67A proteins as both the prey and bait in the pulldown assay. A maximum of 35% of the [35S]atToc33 bound to atToc33G-His6 at the highest concentration tested (Fig. 2A). In contrast, ∼10% of the added [35S]atToc33-R130A and 4% of the added [35S]atToc33-F67A bound to atToc33G-R130A-His6 and atToc33G-F67A-His6, respectively (Fig. 2A). These results confirm that atToc33-R130A and atToc33-F67A are significantly deficient in homodimerization compared with wild type protein. Furthermore, the results indicate that mutations in both monomers have a more significant effect on dimerization than if only one monomer was mutated (compare Figs. 1A and 2A).
FIGURE 2.
Homodimerization of atToc33-R130A and atToc33-F67A. A, increasing concentrations of immobilized atToc33G-His6, atToc33G-R130A-His6, or atToc33G-F67A-His6 were incubated with full-length in vitro translated [35S]atToc33, [35S]atToc33-R130A, or [35S]atToc33-F67A, respectively. 10% of the in vitro translation (IVT) products added to each reaction is indicated (lanes 1 and 7). The graph presents quantitation of bound proteins from triplicate experiments corresponding to the representative data shown in the upper panels. B, increasing concentrations of immobilized CRABP-His10 were incubated with full-length in vitro translated [35S]atToc33, [35S]atToc33-R130A, or [35S]atToc33-F67A as a control for nonspecific binding. Bound proteins were analyzed by SDS-PAGE and quantified using phosphorimaging. The error bars in both panels correspond to S.D.
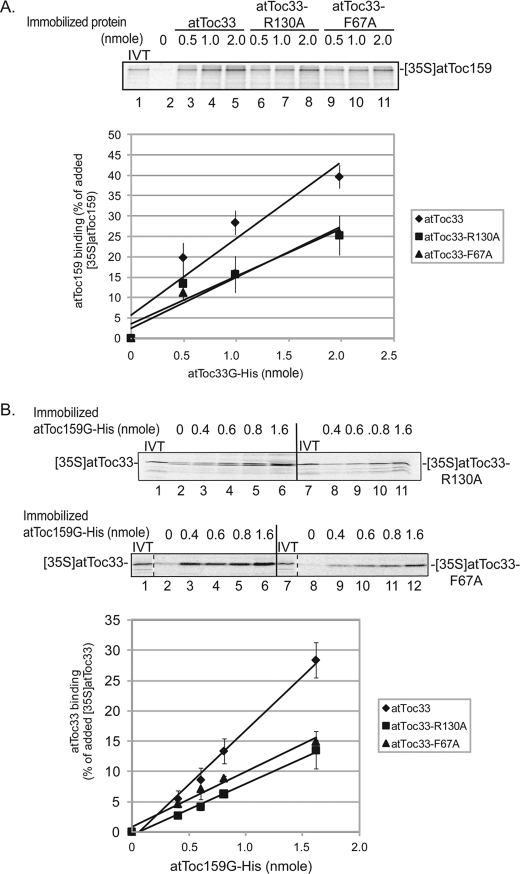
We also examined the impact of the mutations on heterotypic interactions with atToc159. In vitro binding studies have demonstrated that Toc34 and Toc159 family members can interact also via their GTP-binding domains (14, 17, 19–21, 39). In the case of atToc159 binding, [35S]atToc159 also bound more efficiently to immobilized atToc33G-His6 (maximum of 40% of total [35S]atToc159 added) than to atToc33-R130A-His6 (26% maximum) and atToc33-F67A-His6 (27% maximum) (Fig. 3A). To confirm these results, we preformed the converse experiment using immobilized atToc159G-His6 as the bait and in vitro translated [35S]atToc33 proteins as the prey in the assay. As shown Fig. 3B, a maximum of 30% of wild type [35S]atToc33 was pulled down with the highest levels of immobilized atToc159G-His6 (Fig. 3B). In contrast, the binding was reduced to ∼13–15% of added [35S]atToc33-R130A or [35S]atToc33-F67A (Fig. 3B). This result suggests that heterodimerization of atToc33 and atToc159 also involves Arg130 and Phe67. However, the single point mutations appear to have less of an effect on the heterotypic interaction than on the homotypic interaction. This probably reflects the fact that residues in only one of two binding partners are mutated, a situation that might be less destabilizing to binding. Taken together, these results demonstrate that Arg130 and Phe67 play roles in both homotypic and heterotypic interactions.
FIGURE 3.
Binding of atToc33-R130A and atToc33-F67A to atToc159. A, increasing concentrations of immobilized atToc33G-His6, atToc33G-R130A-His6, or atToc33G-F67A-His6 were incubated with full-length in vitro translated [35S]atToc159. 10% of the in vitro translation (IVT) products added to each reaction is indicated in lane 1. B, increasing concentrations of immobilized atToc159G-His6 were incubated with full-length in vitro translated [35S]atToc33, [35S]atToc33-R130A, or [35S]atToc33-F67A. 10% of the in vitro translation products added to each reaction is indicated in lanes 1 and 7. The graphs correspond to the quantitative analysis of bound proteins from triplicate experiments of the representative data shown in the upper panels in both A and B. The error bars correspond to S.D.
None of the 35S-labeled proteins exhibited significant binding to Ni-NTA beads in the absence of immobilized protein, confirming that both wild type and mutant atToc33 had directly bound to partner proteins (Figs. 2A and 3B, lanes 2 and 8, and Fig. 3A, lane 2). Furthermore, the in vitro translated proteins did not bind to an unrelated immobilized protein, CRABP-His10, at levels of immobilized protein that were 2 to 5 times the maximum level used in the receptor-receptor binding assays (Fig. 2B). These controls indicate that the binding interactions were specific.
Assessment of Dimer Formation within Toc Complexes
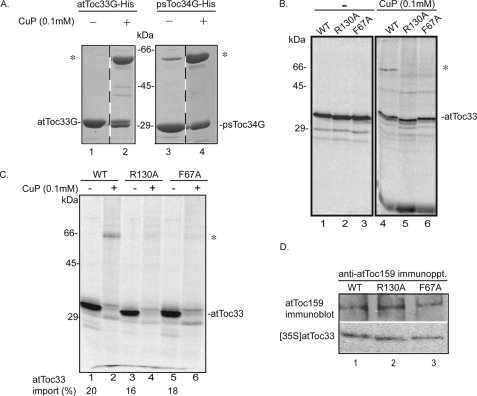
We wished to establish that the effects of the atToc33-R130A and atToc33-F67A substitutions on dimerization could be detected within Toc complexes in chloroplasts. To this end, we developed a method to covalently capture receptor dimers by inducing intersubunit disulfide formation. AtToc33 contains two cysteine residues, one at position 152 and the second at position 213. In the psToc34 dimer crystal structure, the cysteines at the position comparable with Cys213 (Cys215) are closely apposed in the longest loops of each monomer (13). PsToc34 contains only this one cysteine (Cys215). This suggested that this cysteine pair could be used to capture the dimer under oxidizing conditions. As a first step, we examined the interactions of the recombinant receptors in solution using a zero-length covalent cross-linking strategy based on oxidation by the reagent CuP (40, 41). We incubated E. coli-expressed atToc33 and psToc34 with CuP and assessed covalent dimer formation by SDS-PAGE under nonreducing conditions. Fig. 4A shows that both the atToc33 and psToc34 dimers can be captured using CuP oxidation (compare lanes 1 and 3 with 2 and 4). The fact that psToc34 contains only one cysteine indicates that Cys215 (psToc34) participates in the covalent disulfide linkage. We assume that the dimer observed in atToc33 involves the comparable cysteine pair (Cys213). Titration of CuP at a concentration range of 0.1 to 2.5 mm or at varying incubation times had no significant effect on the degree of dimer detected by the cross-linking approach (data not shown).
FIGURE 4.
In organello visualization of atToc33 dimerization. A, E. coli-expressed atToc33 and psToc34 were cross-linked with CuP to promote disulfide formation. B, in vitro translated [35S]atToc33, [35S]atToc33-R130A, or [35S]atToc33-F67A was incubated in the presence or absence of CuP in the absence of chloroplasts. C, in vitro translated [35S]atToc33, [35S]atToc33-R130A, or [35S]atToc33-F67A was inserted into isolated ppi1 chloroplasts. After reisolation, the chloroplasts were incubated with CuP for 5 min at room temperature. Samples recovered from cross-linking reactions were analyzed by nonreducing SDS-PAGE. D, chloroplasts from the insertion reactions were detergent-solubilized, and proteins were immunoprecipitated with atToc159 antiserum. All samples were analyzed by SDS-PAGE under reducing or nonreducing conditions as indicated. Phosphorimaging or immunoblotting was used to detect [35S]atToc33 or atToc159, respectively. An asterisk indicates the position of the atToc33 dimer in A–C.
As an additional control, we tested whether the dimer could be detected in in vitro translated [35S]atToc33, [35S]atToc33-R130A, and [35S]atToc33-F67A in the absence of chloroplasts (Fig. 4B). A band corresponding to an atToc33 dimer (∼66 kDa) was detectable only in wild type atToc33 but not in the two mutants (Fig. 4B, compare lanes 4–6, asterisk) in CuP-treated samples. These data provide additional evidence that the mutations disrupt homodimer formation.
The ability to capture the atToc33 dimer by disulfide formation suggested that we could apply this approach to assess dimer formation in native Toc complexes. To this end, we imported in vitro translated [35S]atToc33 into isolated ppi1 chloroplasts that lack atToc33 to promote insertion and assembly into native Toc complexes. The import efficiency of [35S]atToc33, [35S]atToc33-R130A, and [35S]atToc33-F67A in ppi1 chloroplasts was similar (Fig. 4C, compare lanes 1, 3, and 5). Immunoprecipitation of Toc complexes from the untreated insertion reactions using anti-atToc159 serum demonstrated that the wild type and mutant [35S]atToc33 proteins were indeed incorporated into Toc complexes (Fig. 4D).
The chloroplasts were reisolated after the insertion reaction and treated with CuP. Oxidation was allowed to continue for 5 min in 0.1 mm CuP to minimize possible nonspecific cross-linking (40, 41). Following CuP treatment, a band with mobility identical to that of the atToc33 dimer was observed in ppi1 chloroplasts containing newly imported [35S]atToc33 (Fig. 4C, lane 2, asterisk). No cross-linking product was observed in chloroplasts containing imported [35S]atToc33-R130A or [35S]atToc33-F67A (Fig. 4C, lanes 4 and 6). These results indicate that the mutations inhibit the interaction between receptors within Toc complexes. Together, the solid phase binding and the in organello cross-linking data indicate that atToc33-R130A or atToc33-F67A significantly reduce homodimerization compared with the wild type receptors.
We also attempted to use the CuP method to detect atToc33-atToc159 heterodimers. However, cross-linking reactions with in vitro translated [35S]atToc33 and [35S]atToc159 yielded very high molecular weight complexes that could not be resolved by standard methods, and therefore we were unable to detect a product that could be identified as an atToc33-atToc159 cross-link (data not shown). Similar results were also obtained with isolated chloroplasts. Consequently, we could not use this assay to assess heterodimerization within Toc complexes.
Phenotypic Analysis of Wild Type and Mutant Plants
The primary goal of this study was to assess the role of receptor dimerization on the function of the Toc complex. Toward this aim, we reconstituted the atToc33-R130A or atToc33-F67A receptors into Toc complexes in vivo by expressing the mutant proteins in the ppi1 mutant of Arabidopsis and assessed the effects on protein import into isolated chloroplasts. ppi1 seedlings exhibit a pale yellow phenotype because of the lack of atToc33 expression (28). Although ppi1 plants contain a second Toc34 isoform with partially overlapping functions (atToc34), the atTOC34 gene is expressed at very low levels in young seedlings. Consequently, chloroplasts from ppi1 seedlings exhibit significantly reduced levels of preprotein import (42–44) because of the lack of atToc33 receptor. On the basis of these data, we reasoned that the import characteristics of chloroplasts from ppi1 seedlings expressing atToc33-R130A or atToc33-F67A would reflect the effects of the mutations on the mechanism of receptor function.
We stably transformed wild type (Col) and ppi1 plants (28, 44) with mutant and wild type atToc33 constructs. The cDNA coding region (894 bp) of the wild type and mutant atToc33 constructs with Tev protease recognition sites and His6 tags at their C termini was cloned into the binary vector, pSMB, under the control of the 35S promoter of CaMV35S. Transformation by floral dipping of wild type or ppi1 plants with these constructs renders the transformants resistant to the herbicide Basta. Independent Basta-resistant transformants were segregated to select for homozygous ppi1 plants carrying the CaMV35S::atToc33.Tev.His transgenes. At least three independent lines from each transformation were examined. No significant differences in the phenotypes were observed between the lines from each transformation (data not shown). Therefore, we selected one representative line from each group for further analysis.
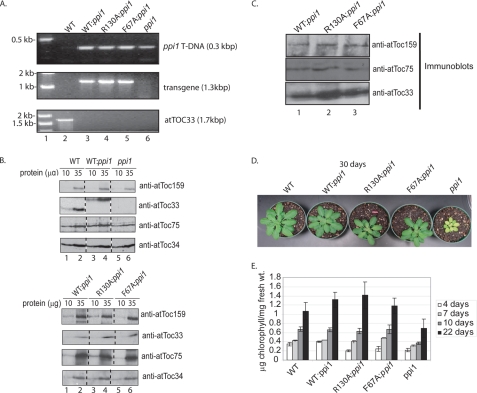
The genotypes of representative ppi1 plants transformed with CaMV35S::atToc33.Tev.His (WT:ppi1), CaMV35S::atToc33-R130A.Tev.His (R130A:ppi1), and CaMV35S::atToc33-F67A.Tev.His (F67A:ppi1) were confirmed by PCR-based genotyping (Fig. 5A) and DNA sequencing (data not shown). Immunoblots of serial dilutions of extracts from these plants indicate that atToc33 receptor expression in WT:ppi1, R130A:ppi1, and F67A:ppi1 is similar and that all three proteins are expressed at levels only moderately higher than authentic atToc33 in wild type (WT) plants (Fig. 5C). The SDS-PAGE mobility of the atToc33 transgenic proteins is reduced compared with the endogenous wild type protein because of the addition of the C-terminal Tev-His tags (Fig. 5B, compare lanes 1 and 2 with 3 and 4 in the second panel from the top).
FIGURE 5.
Genotypic and phenotypic analysis of transgenic plants. A, the genotypes of WT and ppi1 plants transformed with atToc33.Tev.His (WT:ppi1), atToc33-R130A.Tev.His (R130A:ppi1), and atToc33-F67A.Tev.His (F67A:ppi1) were confirmed by PCR analysis of genomic DNA from plants with primer sets specific for the transgene (see “Experimental Procedures”). B, immunoblot analysis of levels of protein expression in isolated chloroplasts from 10-day-old plants grown on agar plates. Serial dilutions of chloroplast samples were immunoblotted with the affinity-purified antibodies as indicated. C, detergent-solubilized chloroplasts from WT:ppi1, R130A:ppi1, and F67A:ppi1 plants were subjected to immunoprecipitation with anti-atToc159 serum. The total fractions from the immunoprecipitates were resolved by SDS-PAGE and immunoblotted with affinity-purified antibody as indicated on each panel. D, visual phenotype of atToc33 transgenic plants from fully matured 30-day-old plants grown in soil. E, change in chlorophyll content of ppi1 transformants during development. The data in the graph represent the mean of three independent experiments at 4, 7, 10, and 22 days after germination. The error bars correspond to S.D.
To ensure that expression of the mutants did not disrupt expression of other Toc components, we examined the levels of atToc159, atToc75, and atToc34 (a second Toc34 homolog) by immunoblotting. All three proteins were expressed at levels similar to those in wild type plants. To examine Toc complex assembly, co-immunoprecipitation was conducted in all three transgenic Arabidopsis lines (Fig. 5B). Anti-atToc159 serum co-immunoprecipitated atToc33 and atToc75 from detergent-soluble extracts in all cases, demonstrating that the mutations did not detectably affect receptor targeting or Toc complex assembly.
Visual analysis of the transgenic lines indicated that all constructs for the most part were able to complement the pale phenotype of ppi1 plants (Fig. 5D). The chlorophyll accumulation was similar in native wild type, WT:ppi1, R130A:ppi1, and F67A:ppi1 plants at 22 days after germination. The R130A:ppi1 and F67A:ppi1 plants showed a lower level of chlorophyll at the earliest time point (Fig. 5E, 4 days). The R130A:ppi1 and F67A:ppi1 plants had fully recovered and were indistinguishable from the wild type controls by 7 days post-germination. Previously, atToc33 expression had been shown to correlate with rapid growth and development in green tissues (28, 43). The effects of the mutation on early development are consistent with a defect in protein import in these plants. The demand for protein import is highest in seedlings because of the rapid development and increased division of chloroplasts in young tissues. In contrast, ppi1 plants exhibited ∼50–60% of the levels of chlorophyll at all developmental stages compared with native wild type plants (Fig. 5E) (28, 43). These data are consistent with the visual phenotypes of the transgenic plants, and they indicate that the atToc33 mutant receptors at least partially complement the absence of atToc33 in ppi1 plants. No phenotypes were observed in wild type plants transformed with either of the atToc33 mutants (data not shown).
Protein Import in R130A:ppi1 and F67A:ppi1 Chloroplasts
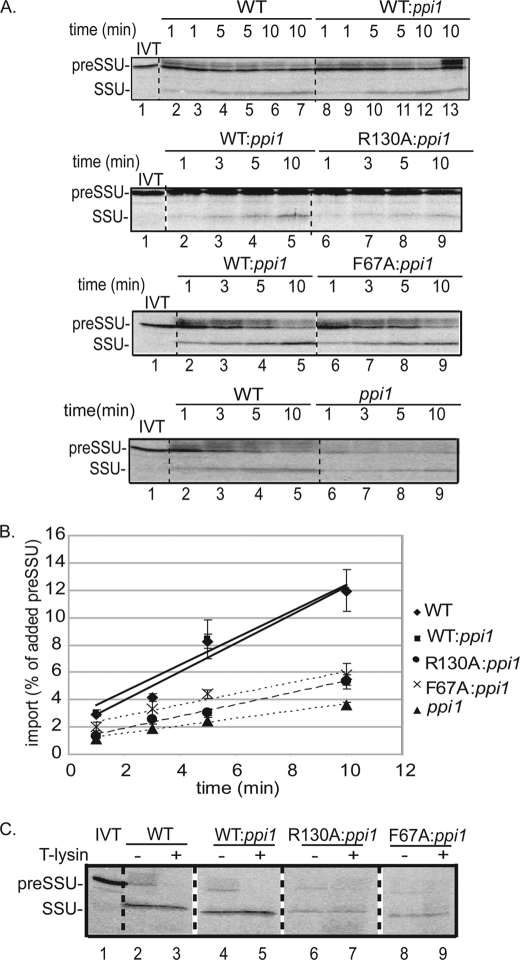
To address the question of whether dimerization affects protein import, we compared preprotein import rates using isolated chloroplasts from wild type, WT:ppi1, R130A:ppi1, and F67A:ppi1 chloroplasts. The import rate was determined by incubating isolated chloroplasts with [35S]preSSU and measuring the accumulation of the mature protein (SSU) over time. As shown in Fig. 6, the rate of protein import was linear over a 10-min time course for all chloroplasts tested. The import rate of WT:ppi1 chloroplasts was indistinguishable from that of native wild type chloroplasts (Fig. 6, A and B). PreSSU import in R130A:ppi1 chloroplasts was 46% of the rate observed in wild type chloroplasts under these conditions (Fig. 6, A and B). Similarly, the import rate of F67A:ppi1 chloroplasts was only 50% that of wild type chloroplasts at the 10-min time point (Fig. 6, A and B). Thermolysin treatment of the chloroplasts following the import reaction removed bound preSSU but did not digest the mature form of SSU (Fig. 6C), which indicates that processed SSU represents completely imported protein and not a partially imported substrate. These data support the hypothesis that dimerization of the Toc GTPase receptors participates in the protein import reaction.
FIGURE 6.
atToc33-R130A and atToc33-F67A chloroplasts exhibit a decreased rate of protein import. A, isolated chloroplasts from the indicated plants were incubated with [35S]preSSU in the presence of 1 mm GTP and 2 mm ATP at 23 °C for the indicated times. B, quantitation of the data from triplicate experiments is presented in the graph. Lane 1 in each panel represents 20% of the in vitro translation (IVT) products added to each reaction. C, chloroplasts from a [35S]preSSU 10-min import reaction in A were treated with 100 μg/ml thermolysin on ice for 30 min to discriminate between bound and fully imported proteins. The position of [35S]preSSU (20 kDa) and imported mature SSU (13.5 kDa) are indicated at the left of each panel. Dashed lines indicate that the illustrations were generated from different regions of the same SDS-polyacrylamide gel using samples from the same experiment. The error bars correspond to S.D.
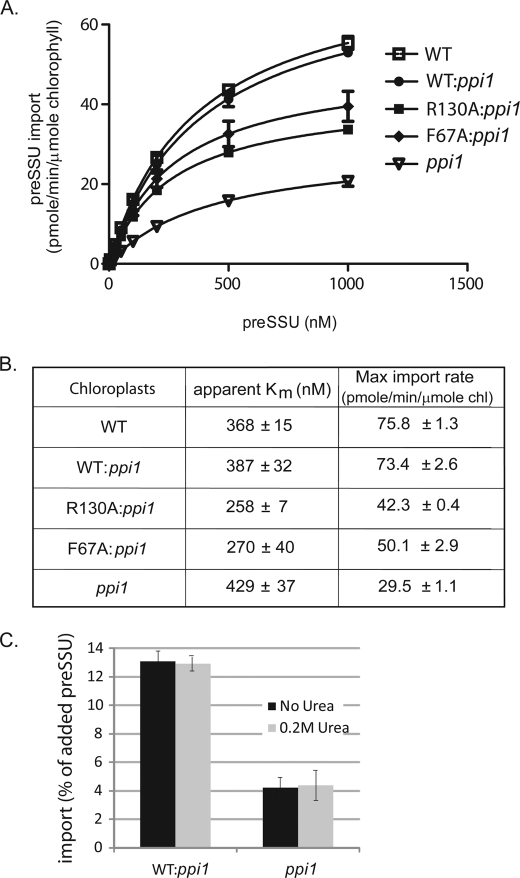
To examine in more detail which step in import was affected in R130A:ppi1 and F67A:ppi1, we performed a kinetic analysis of import (30). Isolated chloroplasts were incubated with increasing concentrations of E. coli-expressed and urea-denatured [35S]preSSU for 10 min at 23 °C. The final concentration of urea was held constant at 0.2 m after preSSU was diluted into each import reaction. This concentration of urea does not reduce the levels of import into wild type or ppi1 chloroplasts (Fig. 7C) (30, 32). Fig. 7A shows the import saturation curves for native wild type, ppi1, WT:ppi1, R130A:ppi1, and F67A:ppi1 chloroplasts. We calculated kinetic constants from these data using nonlinear fitting of Michaelis-Menten curves (Fig. 7B). Native wild type and WT:ppi1 chloroplasts had indistinguishable import rate constants (apparent Km) and maximum import rates, indicating that the activity of atToc33.Tev.His is indistinguishable from that of native atToc33. Native wild type and WT:ppi1 chloroplasts had slightly higher apparent Km values for import (368 ± 15 and 387 ± 32 nm, respectively) compared with R130A:ppi1 chloroplasts (258 ± 7 nm) (Fig. 7B). F67A:ppi1 chloroplasts had an apparent Km for import (270 ± 40 nm) similar to that of R130A:ppi1chloroplasts (Fig. 7B). In contrast, the maximum import rates for native wild type and WT:ppi1 chloroplasts (75.8 ± 1.3 and 73.4 ± 2.6 pmol/min/μmol chlorophyll, respectively) were ∼1.5 times higher than that of R130A:ppi1 chloroplasts (42.3 ± 0.4 pmol/min/μmol chlorophyll) or of F67A:ppi1 chloroplasts (50.1 ± 2.9 pmol/min/μmol chlorophyll), respectively (Fig. 7B). These data are consistent with the import data in Fig. 6. The apparent Km data suggest that the affinity of preSSU for R130A:ppi1 and F67A:ppi1 chloroplasts is slightly increased compared with wild type chloroplasts. In contrast, the maximal preprotein import is significantly decreased in the mutant chloroplasts. On the basis of these observations, we concluded that disruption of dimerization does not decrease preprotein binding at Toc complexes but decreases the efficiency of membrane translocation of the preprotein.
FIGURE 7.
Comparison of import kinetics of WT:ppi1 with R130A:ppi1 or F67A:ppi1 chloroplasts. A, increasing amounts of E. coli-expressed and urea-denatured [35S]preSSU were imported into isolated chloroplasts under standard conditions for 10 min. Quantification of the data from triplicate experiments is presented as a Michaelis-Menten plot. B, the maximum import velocity and apparent Km for import were calculated by nonlinear fitting of the data in A, where v = rate of preSSU import and [S] = total concentration of preSSU. C, [35S]preSSU import into wild type and ppi1 chloroplasts was performed under standard conditions in the presence or absence of 0.2 m urea to ensure that protein import was not affected by the addition of urea. The error bars correspond to S.D.
To investigate the hypothesis that the atToc33-R130A and atToc33-F67A mutations do not disrupt initial preprotein binding but reduce the efficiency of membrane translocation, we examined the energetics of protein import. Protein import into chloroplasts can be divided into at least three distinct stages in vitro on the basis of energetic requirements (29, 45, 46). The first stage of import involves a reversible, energy-independent interaction of preproteins with receptors of the Toc complex. At low concentrations of ATP and GTP (≤100 μm), the preprotein partially inserts across the outer envelope membrane via the Toc channel. At higher concentrations of ATP (≥1 mm) the protein translocates fully across both envelope membranes into the stroma.
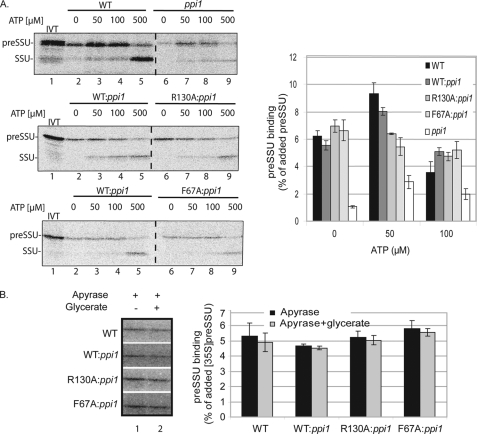
We first examined the levels of preprotein binding in the absence of ATP or GTP. Chloroplasts were treated with nigericin and incubated for 10 min in the dark to deplete internal ATP and inhibit ATP synthesis (45, 47). In addition, the in vitro translated [35S]preSSU was dialyzed to remove nucleoside triphosphates (33). The chloroplasts were incubated with preprotein and varying concentrations of ATP for 10 min at 23 °C in the dark. After incubation, the chloroplasts were reisolated and analyzed by SDS-PAGE and phosphorimaging. The R130A:ppi1 or F67A:ppi1 chloroplasts exhibited similar levels of preprotein binding compared with wild type and WT:ppi1 chloroplasts in the absence of added energy (Fig. 8A, 0 μm ATP). In native wild type and WT:ppi1 chloroplasts, preSSU binding increased with the addition of 50 μm ATP (Fig. 8A). This represents the early import intermediate that has partially inserted across the outer membrane. The level of early import intermediate at 50 μm ATP was higher in wild type and WT:ppi1 chloroplasts compared with R130A:ppi1 or F67A:ppi1 (Fig. 8A, lane 3 and graph). In fact, the levels of preprotein associated with R130A:ppi1 or F67A:ppi1 chloroplasts did not increase with the addition of low levels of ATP (Fig. 8A, graph), indicating that formation of the early intermediate was inhibited in these chloroplasts. At concentrations of ATP exceeding 100 μm, the amount of bound preSSU decreased with a concomitant increase in mature, imported SSU (Fig. 8A, lanes 4, 5, 8, and 9 in each panel and graph).
FIGURE 8.
Examination of the energetics of preSSU binding and import in R130A:ppi1 and F67A:ppi1 chloroplasts. A, ATP dependence of preSSU import into isolated wild type, ppi1, WT:ppi1, R130A:ppi1, or F67A:ppi1 chloroplasts. Energy-depleted chloroplasts equivalent to 35 μg of chlorophyll were incubated with [35S]preSSU in the presence of increasing amounts of ATP, as indicated, at 23 °C for 10 min. Quantification of the data from triplicate experiments is presented in the graph. Lane 1 in the upper panel shows 10% of the in vitro translation (IVT) products added to each reaction. Lane 1 in the bottom two panels shows 20% of the in vitro translation products added to each reaction. B, nigericin- and apyrase-treated chloroplasts were incubated for 5 min in either the presence or absence of 10 mm glycerate. Quantification of the data from triplicate experiments is presented in the graph. All samples were analyzed by SDS-PAGE and phosphorimaging. Dashed lines indicate that the illustrations were generated from different regions of the same SDS-polyacrylamide gel using samples from the same experiment. The error bars correspond to S.D.
To confirm that the initial binding was not affected by the mutations, isolated chloroplasts were incubated with apyrase and 10 mm glycerate to ensure that both external and internal ATP, respectively, were depleted (7, 49). Preprotein binding was similar in wild type, WT:ppi1, R130A:ppi1, and F67A:ppi1 under all conditions tested (Fig. 8B), which is consistent with the conclusion that preprotein binding is not decreased by the mutations. Taken together with the kinetic data, these results suggest that disrupted dimerization affects the transition of preSSU from binding at the surface to energy-dependent insertion into the protein-conducting channel of Toc complexes.
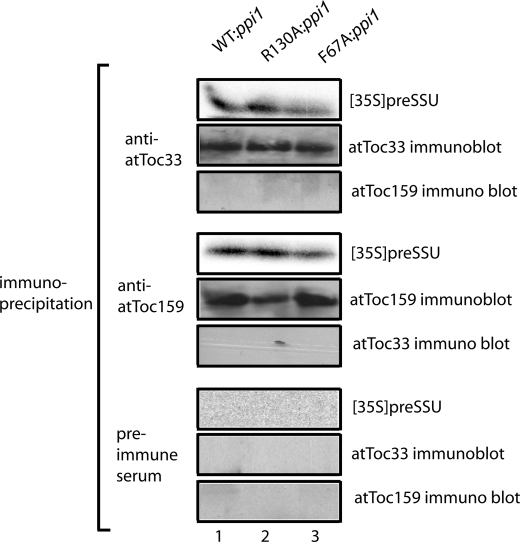
Previously, it was demonstrated that atToc33 and atToc159 both interact with preproteins during the binding step of import (29, 30), suggesting that the two receptors cooperatively participation in preprotein recognition. To examine receptor participation during preprotein binding in the dimerization mutants, we used the cysteine cross-linking reagent CuP to cross-link bound preSSU to isolated, energy-depleted chloroplasts. Consistent with the similarity in levels of preprotein binding (Fig. 8), bound preSSU co-immunoprecipitated with either atToc159 or atToc33 in WT:ppi1, R130A:ppi1, and F67A:ppi1 chloroplasts (Fig. 9). These results are consistent with the previous conclusions that both receptors participate in the initial recognition of preproteins (29, 30) but suggest that the disruption of receptor dimerization does not inhibit these interactions.
FIGURE 9.
Binding of preSSU to Toc translocons is mediated by both atToc159 and atToc33. A, energy-depleted, in vitro translated [35S]preSSU was incubated with isolated chloroplasts from WT:ppi1, R130A:ppi1, or F67A:ppi1 plants in the absence of energy. The samples were treated with 0.1 mm CuP for 5 min. After the cross-linking reactions were quenched, the chloroplasts were detergent-solubilized, and proteins were immunoprecipitated with anti-atToc159 serum, atToc33 affinity-purified antibodies, or preimmune serum. The immunoprecipitates were analyzed by SDS-PAGE and phosphorimaging to detect co-immunoprecipitated [35S]preSSU or by immunoblotting to detect atToc159 or atToc33.
DISCUSSION
We studied the dimerization of atToc33 using a combination of biochemical and in vivo approaches in an effort to test the physiological role of receptor-receptor interactions in protein import into chloroplasts. The original dimeric crystal structure of psToc34 and subsequent direct binding data led to the hypothesis that homotypic or heterotypic interactions between Toc34 and Toc159 function as a key part of Toc complex activity (13, 14, 19, 34). We have shown that alanine substitution mutants, atToc33-R130A and atToc33-F67A, disrupt the dimerization of Toc receptors in binding studies with purified recombinant proteins (Figs. 1–3) and in studies using zero-length covalent cross-linking in native Toc complexes (Fig. 4). Several groups have demonstrated that the atToc33-R130A and psToc34-R133A mutations disrupt dimerization relative to wild type atToc33 in vitro (15, 17, 18, 23). Our findings are consistent with previous studies demonstrating that the atToc33-R130A mutation dramatically reduces homodimerization and has a lesser, but significant effect on the interaction of atToc33 with atToc159 (15–17).
We investigated the effects of atToc33-R130A and atToc33-F67A on protein import and chloroplast biogenesis by expressing the proteins in the ppi1 mutant of Arabidopsis that lacks atToc33 expression (28). Arabidopsis contains another Toc34 homolog, atToc34. This receptor is expressed at low levels throughout the plant, and its function at least partially overlaps that of atToc33 (12, 28). The relatively mild pale phenotype of ppi1 is attributed to the low level of atToc34 expression in this mutant. Although the presence of atToc34 does result in a basal level of protein import in the absence of atToc33, our results clearly demonstrated a distinct difference between ppi1 plants and ppi1 plants expressing atToc33-R130A and atToc33-F67A. These plants provided a system to assess the effects of the mutations on protein import directly. The import rates in wild type and R130A:ppi1 and F67A:ppi1 chloroplasts confirmed that the import capacity was decreased by the mutations (Fig. 6). The analysis of import kinetics (Fig. 7) and binding studies with isolated chloroplasts (Fig. 8) demonstrated that the initial binding of preproteins to the Toc complex was not decreased. In fact, the apparent Km of preprotein import in both R130A:ppi1 and F67A:ppi1 chloroplasts was slightly decreased, reflecting a detectable increase in the binding affinity of the mutant chloroplasts. In contrast, the maximum import rates of the mutant chloroplasts were significantly lower than in wild type chloroplasts (Fig. 7), and examination of the energetics of import indicated that the initial insertion of preprotein in the Toc channel (i.e. formation of the early import intermediate) was decreased by the mutations (Fig. 8). These observations suggest that receptor dimerization is important in promoting the preprotein from its initial binding at the Toc receptors to its insertion in the Toc channel.
The ability to cross-link bound preSSU to both atToc159 and atToc33 in wild type, R130A:ppi1, and F67A:ppi1 chloroplasts (Fig. 9) is consistent with the hypothesis that the two receptors participate together in preprotein recognition. Previous direct and label transfer cross-linking approaches have demonstrated that both receptors are in close contact with transit peptides during initial binding to Toc complexes (30, 50). However, the fact that atToc33-R130A and atToc33-F67A do not decrease the levels of preprotein binding (Fig. 8) or the apparent affinity of the import apparatus for preproteins (Fig. 7) suggests that receptor dimerization might not be required for the interaction of the two receptors with preproteins at this step. The decrease in the overall import capacity (Fig. 7) and the formation of the early import intermediate (Fig. 8) suggest that receptor-receptor interactions are important in initiating the transition from preprotein binding to membrane translocation. This is consistent with elements of models for Toc receptor function in which receptor-receptor dimerization triggers the transfer of preprotein to the Toc channel (22).
Although the R130A and F67A mutations significantly decreased both homodimer and heterodimer formation in vitro, we were only able to detect a defect in homodimer formation in chloroplasts because of technical limitations of the cross-linking assay. Therefore, we were unable to distinguish the roles of homotypic or heterotypic interactions in initiating membrane translocation. It should be noted that previous studies had indicated that Toc34-Toc159 interactions are important for the targeting and assembly of Toc159 into Toc complexes (20, 21), and a previous study using a split ubiquitin system demonstrated that the atToc33 and atToc159 GTPase domains can interact in planta (14). Considering that there were no detectable defects in Toc complex assembly in R130A:ppi1 and F67A:ppi1 chloroplasts (Figs. 4D and 5C), it is likely that the residual heterotypic binding in the mutants is sufficient to facilitate Toc159 targeting. It also is possible that the effects of the atToc33 mutations on atToc159-atToc33 interactions are insufficient to disrupt receptor function within Toc complexes.
Receptor dimerization was proposed to stimulate the GTPase activity of Toc34 based on the crystal structure of psToc34 (13, 34). However, analysis of the psToc34-R133A and atToc33-R130A mutants gave varying results regarding the role of dimerization in receptor GTPase activity. Yeh et al. (18) provide evidence that atToc33 dimers have modestly increased GTPase activity relative to monomers (∼1.5 fold stimulation). In contrast, several other studies have concluded that the GTPase activities of psToc34 or atToc33 are not stimulated by dimerization (15, 17, 23). Consistent with these observations, atToc33-R130A and wild type atToc33 monomers exhibited similar GTPase activity in our assays. Furthermore, we identified two additional, independent mutants (atToc33-F67A and atToc33-L134A) with reduced dimerization. These mutations exhibited no detectable effects on basal GTPase activity (Fig. 1), providing additional evidence that dimerization is not linked directly to GTPase activity.
Koenig et al. (15) noted that the 1.5-fold stimulation of GTP hydrolysis observed in dimer versus monomer preparations (15, 16, 18) is very modest compared with that promoted by the typical GAPs of other GTPases. GAPs generally exhibit greater than 10-fold GTPase stimulation (51–53). Thus, arginine 130 of atToc33 appears not to be an arginine finger under the conditions assayed. Koenig et al. (16) suggested that another unknown protein could bind to the dimer and induce conformational changes that would trigger arginine 130 participation in GTP hydrolysis. Our studies do not rule out this possibility.
Our data suggest that dimerization is critical for the initiation of preprotein translocation across the outer membrane (i.e. transfer of the transit peptide from the receptors to the Toc channel). Although the mechanism by which dimerization participates in the translocation reaction remains to be defined, our data are consistent with models in which transient dimerization of receptors is involved during the transfer of preprotein from the receptors to the channel (4, 22). Koenig et al. (16) proposed that an exchange between Toc159-Toc34 and Toc34-Toc34 dimerization might be important for the switch to trigger translocation. Other studies have suggested that the transit peptides of preproteins play a key role in the GTPase activation event (22, 23, 48), and it has been suggested that transit peptide binding may promote dimerization (22). It is possible that dimerization might be important in transmitting the conformational changes in the receptor dimer that are necessary for GTPase-stimulated dissociation of the transit peptide and result in preprotein insertion in the Toc channel. Both receptors interact directly with preproteins (Fig. 9) (29, 30). Thus, it is reasonable to propose that the two receptors would dimerize to coordinate preprotein transfer. Finally, it cannot be ruled out that Toc34 might function as a stable dimer within the Toc complex. Disruption of this interaction would thereby disrupt translocon function. More detailed structural analysis of Toc receptor complexes with and without bound transit peptides will provide more clues to the role of dimerization in promoting translocation.
Acknowledgment
We thank Dr. Caleb Rounds for expert technical assistance in generating the transgenic plants.
This work was supported, in whole or in part, by National Institutes of Health Grant GM61893 (to D. J. S.). This work was also supported by a Gilgut Fellowship in Plant Biology (to J. L.).
- Toc
- translocon at the outer membrane of chloroplasts
- Tic
- translocon at the inner membrane of chloroplasts
- GAP
- GTPase-activating protein
- Ni-NTA
- nickel-nitrilotriacetic acid
- CuP
- copper (II)-1,10-phenanthroline
- Rubisco
- ribulose-1,5-bisphosphate carboxylase/oxygenase
- preSSU
- precursor to the small subunit of Rubisco
- SSU
- mature form of the small subunit of Rubisco
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CRABP
- cellular retinoic acid-binding protein
- Tev
- tobacco etch virus
- CaMV
- cauliflower mosaic virus
- WT
- wild type.
REFERENCES
- 1.Inaba T., Schnell D. J. (2008) Biochem. J. 413, 15–28 [DOI] [PubMed] [Google Scholar]
- 2.Jarvis P. (2008) New Phytol. 179, 257–285 [DOI] [PubMed] [Google Scholar]
- 3.Rounds C., Wang F., Schnell D. J. (2007) in The Enzymes: Molecular Machines Involved in Protein Transport across Cellular Membranes (Tamanoi F., Dalbey R. E., Koehler C. M. eds) Vol. 25, pp. 407–430, Academic Press, Orlando, FL [Google Scholar]
- 4.Li H. M., Kesavulu M. M., Su P. H., Yeh Y. H., Hsiao C. D. (2007) J. Biomed. Sci. 14, 505–508 [DOI] [PubMed] [Google Scholar]
- 5.Hinnah S. C., Hill K., Wagner R., Schlicher T., Soll J. (1997) EMBO J. 16, 7351–7360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinnah S. C., Wagner R., Sveshnikova N., Harrer R., Soll J. (2002) Biophys. J. 83, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue H., Akita M. (2008) J. Biol. Chem. 283, 7491–7502 [DOI] [PubMed] [Google Scholar]
- 8.Young M. E., Keegstra K., Froehlich J. E. (1999) Plant Physiol. 121, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. (2000) Nature 403, 203–207 [DOI] [PubMed] [Google Scholar]
- 10.Ivanova Y., Smith M. D., Chen K., Schnell D. J. (2004) Mol. Biol. Cell 15, 3379–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubis S., Patel R., Combe J., Bédard J., Kovacheva S., Lilley K., Biehl A., Leister D., Rios G., Koncz C., Jarvis P. (2004) Plant Cell 16, 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constan D., Patel R., Keegstra K., Jarvis P. (2004) Plant J. 38, 93–106 [DOI] [PubMed] [Google Scholar]
- 13.Sun Y. J., Forouhar F., Li Hm H. M., Tu S. L., Yeh Y. H., Kao S., Shr H. L., Chou C. C., Chen C., Hsiao C. D. (2002) Nat. Struct. Biol. 9, 95–100 [DOI] [PubMed] [Google Scholar]
- 14.Rahim G., Bischof S., Kessler F., Agne B. (2009) J. Exp. Bot. 60, 257–267 [DOI] [PubMed] [Google Scholar]
- 15.Koenig P., Oreb M., Höfle A., Kaltofen S., Rippe K., Sinning I., Schleiff E., Tews I. (2008) Structure 16, 585–596 [DOI] [PubMed] [Google Scholar]
- 16.Koenig P., Oreb M., Rippe K., Muhle-Goll C., Sinning I., Schleiff E., Tews I. (2008) J. Biol. Chem. 283, 23104–23112 [DOI] [PubMed] [Google Scholar]
- 17.Weibel P., Hiltbrunner A., Brand L., Kessler F. (2003) J. Biol. Chem. 278, 37321–37329 [DOI] [PubMed] [Google Scholar]
- 18.Yeh Y. H., Kesavulu M. M., Li H. M., Wu S. Z., Sun Y. J., Konozy E. H., Hsiao C. D. (2007) J. Biol. Chem. 282, 13845–13853 [DOI] [PubMed] [Google Scholar]
- 19.Jelic M., Soll J., Schleiff E. (2003) Biochemistry 42, 5906–5916 [DOI] [PubMed] [Google Scholar]
- 20.Smith M. D., Hiltbrunner A., Kessler F., Schnell D. J. (2002) J. Cell Biol. 159, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer J., Hiltbrunner A., Weibel P., Vidi P. A., Alvarez-Huerta M., Smith M. D., Schnell D. J., Kessler F. (2002) J. Cell Biol. 159, 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker T., Jelic M., Vojta A., Radunz A., Soll J., Schleiff E. (2004) EMBO J. 23, 520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddick L. E., Vaughn M. D., Wright S. J., Campbell I. M., Bruce B. D. (2007) J. Biol. Chem. 282, 11410–11426 [DOI] [PubMed] [Google Scholar]
- 24.Fisher C. L., Pei G. K. (1997) BioTechniques 23, 570–571, 574 [DOI] [PubMed] [Google Scholar]
- 25.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 26.Desfeux C., Clough S. J., Bent A. F. (2000) Plant Physiol. 123, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 28.Jarvis P., Chen L. J., Li H., Peto C. A., Fankhauser C., Chory J. (1998) Science 282, 100–103 [DOI] [PubMed] [Google Scholar]
- 29.Ma Y., Kouranov A., LaSala S., Schnell D. J. (1996) J. Cell Biol. 134, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F., Agne B., Kessler F., Schnell D. J. (2008) J. Cell Biol. 183, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnon D. I. (1949) Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnell D. J., Blobel G. (1993) J. Cell Biol. 120, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agne B., Infanger S., Wang F., Hofstetter V., Rahim G., Martin M., Lee D. W., Hwang I., Schnell D., Kessler F. (2009) J. Biol. Chem. 284, 8670–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessler F., Schnell D. J. (2002) Nat. Struct. Biol. 9, 81–83 [DOI] [PubMed] [Google Scholar]
- 35.Wallas T. R., Smith M. D., Sanchez-Nieto S., Schnell D. J. (2003) J. Biol. Chem. 278, 44289–44297 [DOI] [PubMed] [Google Scholar]
- 36.Chen D., Schnell D. J. (1997) J. Biol. Chem. 272, 6614–6620 [DOI] [PubMed] [Google Scholar]
- 37.Jelic M., Sveshnikova N., Motzkus M., Hörth P., Soll J., Schleiff E. (2002) Biol. Chem. 383, 1875–1883 [DOI] [PubMed] [Google Scholar]
- 38.Schleiff E., Soll J., Küchler M., Kühlbrandt W., Harrer R. (2003) J. Cell Biol. 160, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiltbrunner A., Bauer J., Vidi P. A., Infanger S., Weibel P., Hohwy M., Kessler F. (2001) J. Cell Biol. 154, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobashi K. (1968) Biochim. Biophys. Acta 158, 239–245 [DOI] [PubMed] [Google Scholar]
- 41.Dabney-Smith C., van Den Wijngaard P. W., Treece Y., Vredenberg W. J., Bruce B. D. (1999) J. Biol. Chem. 274, 32351–32359 [DOI] [PubMed] [Google Scholar]
- 42.Gutensohn M., Pahnke S., Kolukisaoglu U., Schulz B., Schierhorn A., Voigt A., Hust B., Rollwitz I., Stöckel J., Geimer S., Albrecht V., Flügge U. I., Klösgen R. B. (2004) Mol. Genet. Genomics 272, 379–396 [DOI] [PubMed] [Google Scholar]
- 43.Gutensohn M., Schulz B., Nicolay P., Flügge U. I. (2000) Plant J. 23, 771–783 [DOI] [PubMed] [Google Scholar]
- 44.Kubis S., Baldwin A., Patel R., Razzaq A., Dupree P., Lilley K., Kurth J., Leister D., Jarvis P. (2003) Plant Cell 15, 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theg S. M., Bauerle C., Olsen L. J., Selman B. R., Keegstra K. (1989) J. Biol. Chem. 264, 6730–6736 [PubMed] [Google Scholar]
- 46.Inoue H., Ratnayake R. M., Nonami H., Akita M. (2008) Plant Physiol. Biochem. 72, 2926–2935 [DOI] [PubMed] [Google Scholar]
- 47.Olsen L. J., Keegstra K. (1992) J. Biol. Chem. 267, 433–439 [PubMed] [Google Scholar]
- 48.Schleiff E., Soll J., Sveshnikova N., Tien R., Wright S., Dabney-Smith C., Subramanian C., Bruce B. D. (2002) Biochemistry 41, 1934–1946 [DOI] [PubMed] [Google Scholar]
- 49.Olsen L. J., Theg S. M., Selman B. R., Keegstra K. (1989) J. Biol. Chem. 264, 6724–6729 [PubMed] [Google Scholar]
- 50.Kouranov A., Schnell D. J. (1997) J. Cell Biol. 139, 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wittinghofer A., Gierschik P. (2000) Biol. Chem. 381, 355. [DOI] [PubMed] [Google Scholar]
- 52.Haas A. K., Fuchs E., Kopajtich R., Barr F. A. (2005) Nat. Cell Biol. 7, 887–893 [DOI] [PubMed] [Google Scholar]
- 53.Bourne H. R., Sanders D. A., McCormick F. (1991) Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]