Abstract
The widespread utilization of sugars by microbes is reflected in the diversity and multiplicity of cellular transporters used to acquire these compounds from the environment. The model bacterium Escherichia coli has numerous transporters that allow it to take up hexoses and pentoses, which recognize the more abundant pyranose forms of these sugars. Here we report the biochemical and structural characterization of a transporter protein YtfQ from E. coli that forms part of an uncharacterized ABC transporter system. Remarkably the crystal structure of this protein, solved to 1.2 Å using x-ray crystallography, revealed that YtfQ binds a single molecule of galactofuranose in its ligand binding pocket. Selective binding of galactofuranose over galactopyranose was also observed using NMR methods that determined the form of the sugar released from the protein. The pattern of expression of the ytfQRTyjfF operon encoding this transporter mirrors that of the high affinity galactopyranose transporter of E. coli, suggesting that this bacterium has evolved complementary transporters that enable it to use all the available galactose present during carbon limiting conditions.
Introduction
Microbes use a wide range of different carbohydrates in their metabolism and the biology of sugar utilization in model bacteria such as Escherichia coli is well understood (1). The preferred carbon source for growth in laboratory conditions is glucose, although in vivo this bacterium is likely to utilize simultaneously the wide range of mucin-derived sugars that are present at limiting concentrations in the gut (2–4). Given the important role of sugars as carbon sources, the mechanisms used to concentrate these compounds inside the bacterial cell have also been well-studied, and E. coli uses a range of different transporters to facilitate carbohydrate uptake including phosphotransferase system (PTS), ATP-binding cassette (ABC),5 and secondary transporters (5–7). E. coli can express multiple transporters for the same sugar, allowing the bacterium to utilize efficiently the widely varying concentrations of substrate available in vivo. For example, when galactose is abundant E. coli synthesizes a high capacity and low affinity secondary transporter for galactose (GalP), but when galactose is present at low concentrations (low to sub μm) it synthesizes a high affinity substrate-binding protein (SBP)-dependent ABC transporter, MglABCD, which functions as a scavenging system (8).
In the ABC transporters, the SBP component is soluble in the periplasm of Gram-negative bacteria and is essential for the initial capture of the substrate. The substrate is then delivered specifically to the cognate integral membrane transporter subunits and consequently the ligand binding properties of the SBP define those of the transporter as a whole (9). SBPs share a common overall structure comprising two globular α/β domains linked by a hinge (10). The ligand binding site is usually formed by residues from both domains and, via hinge movement, they close around the substrate when it is bound (11). In the absence of the transporter, SBPs usually bind their ligands tightly and, as off-rates are slow (12–14), can often co-crystallize with their ligand (13, 15).
While E. coli has multiple ABC transporters for sugars, the function of the ytfQRTyjfF operon, which encodes a predicted sugar ABC transporter, has not been previously studied. However, the promoter region for this operon contains two binding sites for the galactose repressor GalR (and GalS) upstream of the promoter at positions −97.5 and +57.5 relative to the predicted transcriptional start (see supplemental Fig. S1A) (16). There is also the additional presence of a weak site for the catabolite repressor protein (CRP) (17), located at a site centered at −41.5 bp relative to the putative transcriptional start, suggesting that this promoter is a type II CRP-activated promoter (18). Data from a number of microarray and proteomic studies suggest that expression of the ytfQ gene is induced during carbon limiting growth conditions and that the SBP component, YtfQ, accumulates in the periplasm consistent with the transporter functioning in uptake of an alternative carbon source during carbon limited growth (3, 4, 17, 20).
Using a strategy we have recently applied to the study of SBP proteins from uncharacterized tripartite ATP-independent periplasmic (TRAP) transporters (13, 21), we have examined the remaining ABC transporters in E. coli that contain an SBP and hence are likely to be involved in substrate uptake. Herein we report the biochemical and structural characterization of YtfQ, the SBP encoded in the ytfQRTyjfF operon and reveal a novel additional route for carbon scavenging via the utilization of rarer furanose forms of sugars present in vivo.
MATERIALS AND METHODS
Cloning, Expression, and Purification of YtfQ
The ytfQ gene was cloned from genomic DNA of E. coli K12 MG1665 following PCR amplification using KOD DNA polymerase (Novagen) with the following primers: YtfQF 5′-GGAATTCCATATGTGGAAACGCTTACTTATAGTC-3′ and YtfQR 5′-CCGCTCGAGATACCCCATATTTTTCTTC-3′. The blunt PCR product was cloned into a pZERO Blunt TOPO cloning vector (Invitrogen) and then excised on an NdeI/XhoI fragment. This was then subcloned into NdeI/XhoI-cut pET21b, and this final construct, pRSPH05, was transformed into BL21(DE3) pLysS (Stratagene) for protein expression. The sequence of the ytfQ gene in pRSPH05 was confirmed using DNA sequencing (MWG-Biotech). Expression of the native protein was optimal at 30 °C after 4 h of growth following induction with 1 mm isopropyl-1-thio-β-d-galactopyranoside. Expression was performed with cells grown either in Lennox broth (LB) or in M9 minimal medium with 1% glycerol. For purification of YtfQ, E. coli was grown in 2-liter baffled flasks in LB medium and inoculated from a 50-ml starter culture grown at 37 °C to an OD650 of 0.1. Cells were harvested by centrifugation at 5000 rpm in a SLC6000 rota at 4 °C for 15 min. The periplasmic fraction was isolated by osmotic shock (22) and Benzonase (Stratagene) added to the purified periplasm. The protein was purified using nickel affinity chromatography. Following protein loading, the Ni-nitrilotriacetic acid column was washed in five column volumes of 20 mm imidazole, and then YtfQ was eluted from the column with 100 mm imidazole. The protein was purified further using anion exchange chromatography on a DEAE-Sepharose column. Using an anion gradient from 0 to 200 mm NaCl, YtfQ eluted at ∼50 mm NaCl. Fractions were pooled and dialyzed into 50 mm Tris-HCl pH8 or 50 mm phosphate buffer as required. Protein levels were quantified using a NanoDrop spectrophotometer (Thermo Scientific). Total yields of ∼11 mg per liter of LB culture were achieved for YtfQ.
Crystallization, Data Collection, and Structure Solution
Crystallization trials employed a Mosquito nanoliter-dispensing robot with the sitting-drop, vapor-diffusion method in which 150 nl of solution containing YtfQ 10 mg/ml and 5 mm galactose was mixed with 150 nl of reservoir solutions from various commercially available crystallization screens. A crystal that diffracted well was identified from a single drop prepared with Pact D3 solution (23). This crystal was vitrified in a solution of mother liquor to which glycerol was added to a final concentration of 25% (v/v). Data were collected to a resolution of 1.2 Å on beamline ID23-1 at the ESRF, Grenoble and processed with Denzo and Scalepack (24). A Matthews coefficient of 2.1 Å3/Da was calculated suggesting the presence of one molecule in the asymmetric unit.
The structure was solved by molecular replacement using the program PHASER (25) and the coordinates for the d-ribose-binding protein from E. coli (PDB entry code 1DBP, Ref. 26) as the search model. ARP/wARP (27) built the initial model that was completed in iterative cycles of REFMAC5 (28) and COOT (29). The structure has been deposited with the PDB code 2VK2.
NMR Spectroscopy
After dialysis, 1.2 ml of YtfQ (64.7 μm bound to [13C1]galactose) was lyophilized. 1.2 ml of the dialysis buffer was also lyophilized. Both samples were redissolved in 600 μl of d6-DMSO (dimethyl sulfoxide-d6, 99.9 atom%, Aldrich) immediately prior to acquisition of an 1H-13C HSQC spectrum at 298 K. The total acquisition time was ∼50 min. A second HSQC spectrum of the YtfQ-containing sample was acquired 4 days later. Peak assignments are according to Hobley et al. (30). Spectra were recorded on a 700 MHz spectrometer (Bruker BioSpin) using Topspin 1.3 software.
Fluorescence Spectroscopy
All protein fluorescence experiments used a FluoroMax 3 fluorescence spectrometer (Jobin Yvon) with connecting water bath at 25 °C. YtfQ protein purified from M9-grown cells was used at a concentration of 0.19 μm in 50 mm Tris-HCl pH 8 and was excited at 295 nm with slit widths of 5 nm. Emission was monitored at 328 nm. To examine alternative ligands for YtfQ, potential ligands were added at 100 μm concentrations. To determine the Kd for ligand binding, the protein was titrated with increasing concentrations of the ligand and the corresponding fluorescence change was monitored in time acquisition mode. The cumulative fluorescence change was plotted in SigmaPlot, and the Kd was calculated from the hyperbolic fit of the binding curve.
Protein Analysis by Electrospray Mass Spectrometry
Samples of purified YtfQ protein were first dialyzed into 50 mm ammonium acetate pH 5 and were then diluted for analysis at a protein concentration of 5 μm. An API Qstar mass spectrometer using an Ionspray source was used for electrospray mass spectrometry (ES-MS). To monitor the mass of the protein with and without ligands, the protein was diluted into 3% methanol in 25 mm ammonium acetate pH 5. For analysis of YtfQ using denaturing conditions the protein was diluted into 1:1 acetonitrile-water containing 0.1% formic acid. Samples were infused at between 1 and 5 ml min−1 and were run within 1 h of preparation. Multiply charged protein signals were observed between 1800 and 3500 m/z under these conditions. Errors in the mass determinations are ∼1 in 104 over the range used; calibration was with caesium iodide and pheromone inhibitor peptide (sequence: ALILTLVS). The raw m/z data were deconvoluted to mass spectra using the Bayesian Protein Reconstruction routine in the BioAnalyst software provided by the manufacturer (Applied Biosystems). Ligands were added between a range of concentration of 0.7 μm and 700 μm.
Sequence Analysis and Bioinformatics
Sequence and genome contexts of ytfQ orthologues were obtained from the SEED (31) with additional manual curation. Sequences were aligned using ClustalX (32) followed by manual adjustment based on a structural superposition of the 4 structures (2DRI, 1RPJ, 1GCA, and 2KV2) and structural alignments using ESPript (33). Evidence for the expression of ytfQ was taken from proteomic and microarray experiments curated in the EchoBASE data base (34).
RESULTS
Purified YtfQ Protein Contains a Prebound Ligand
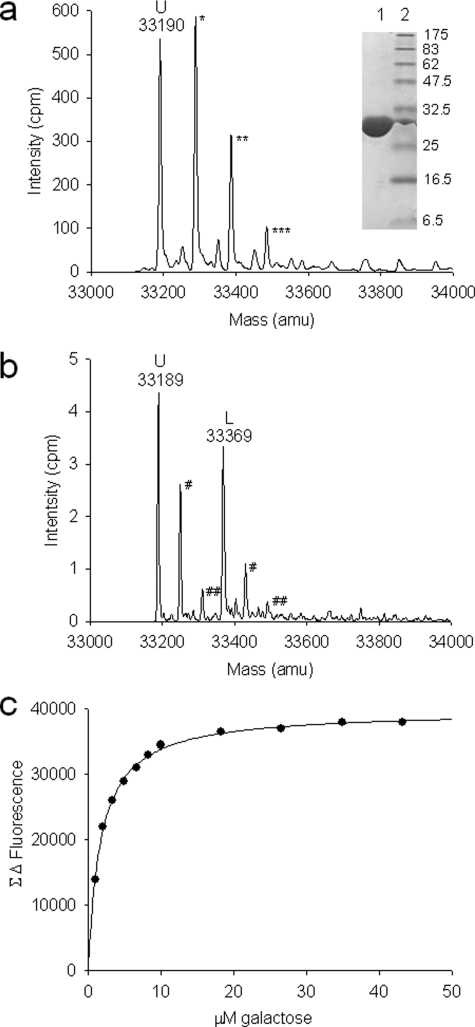
To study the function of YtfQ we expressed a C-terminally hexahistidine-tagged version in the E. coli periplasm which was purified to homogeneity using nickel affinity chromatography followed by anion exchange chromatography (Fig. 1, inset). We confirmed the identity of the purified protein using denaturing electrospray mass spectrometry (ES-MS) by determining its mass to be 33190 ± 3 mass units (Fig. 1a), which is consistent with the predicted mass of C-terminally tagged YtfQ protein following removal of its 21-amino acid signal peptide after export to the periplasm (33190 Da).
FIGURE 1.
Characterization of ligand binding to YtfQ. a, ES-MS spectrum of 5 μm YtfQ under denaturing conditions. The molecular mass of the unbound protein (U) is indicated at 33,190 atomic mass units (amu). Possible phosphate adducts, of 98 amu are indicated by asterisks. Inset, Coomassie-stained SDS-PAGE gel containing 2.4 μg of purified YtfQ (lane 1) and molecular weight markers (lane 2). b, ES-MS spectrum of 10 μm YtfQ under native conditions in 50 mm ammonium acetate pH5. The unbound protein (U) is the primary peak at the indicated mass of 33,189 with an additional ligand-bound species (L) at 33,369 amu. Acetate adducts are indicated with hashes. c, galactose binding to 0.19 μm YtfQ in 50 mm Tris-HCl, pH 8, as measured by tryptophan fluorescence spectroscopy.
Upon analysis of YtfQ in native conditions, in which SBP proteins often retain prebound ligands (14, 21), we saw an additional major species that accounts for about 40% of the protein signal (Fig. 1b). The peak mass of 33,369, being 180 mass units larger than that of the ligand-free protein (33,189 mass units in this sample), is consistent with that of a hexose-bound YtfQ. This suggests that the protein is partially saturated with a hexose ligand. Purification of the protein from M9 minimal medium resulted in a ligand-free protein, as judged using ES-MS (data not shown), suggesting that the 180-Da species was present in the LB growth medium.
Characterization of the Ligand Binding Properties of YtfQ
Given our bioinformatic analysis of YtfQ and the native ES-MS data, we assessed whether the M9-purified protein was able to bind a range of different sugars. We used tryptophan fluorescence spectroscopy, which exploits the large conformational changes that SBP proteins undergo upon ligand binding (Fig. 1c). The binding data is summarized in Table 1. Arabinose and galactose bound with the highest affinity, with Kd values in the range of 1–2 μm. Unusually the protein does not appear to bind glucose, which is notably different from other bacterial galactose transporters (35).
TABLE 1.
Dissociation constants (Kd) of 0.19 μm YtfQ for different sugars investigated by tryptophan fluorescence spectroscopy
The following compounds showed no change in fluorescence suggesting that they do not bind to the SBP: 1-phosphoglucose, 2-deoxyglucose, 6-phosphoglucose, altrose, fructose, glucosamine, glucuronate, lactose, lyxose, mannose, and sucrose. All titrations were performed at least three times. Standard deviations are reported to two significant figures.
| Ligand class | Ligand | Kd | S.D. |
|---|---|---|---|
| μm | μm | ||
| Hexose | Galactose | 1.7 | 0.30 |
| Talose | 25.7 | 2.5 | |
| Allose | 30.3 | 4.4 | |
| Glucose | NBa | ||
| Pentose | Arabinose | 1.3 | 0.24 |
| Ribose | 49.0 | 13 | |
| Other | Fucose (6-deoxy-l-galactose) | 6.2 | 0.54 |
| Methyl-β-galactoside | 145.4 | 21 | |
| Galacturonate | 220.9 | 22 |
a NB, no binding detected due to no apparent change in fluorescence upon addition of ligand.
Although arabinose and galactose bind most tightly to YtfQ, neither of the determined Kd values is particularly low for an ABC transporter. In fact, the SBP from the high-affinity ABC transporter for galactose, MglB, has a Kd around 10-fold lower than YtfQ (0.14–0.48 μm) (36, 37), while the corresponding SBP from the high-affinity ABC transporter for arabinose, AraF, also has higher affinity than YtfQ for this ligand (0.98 ± 0.33 μm) (12). While the micromolar affinities determined for YtfQ for arabinose and galactose do not suggest that these are the true ligands, the apparent regulation of the ytfQ operon via the galactose-responsive repressors GalR/GalS suggest that the transporter has some role in the transport of galactose or a similar molecule.
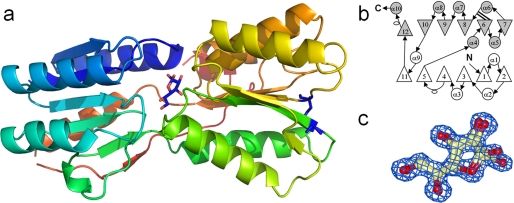
YtfQ Has a Type I SBP Structure with a Disulfide Bond
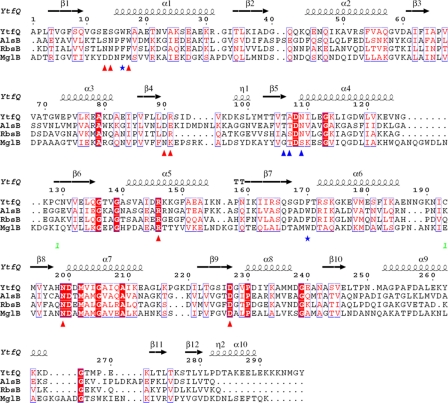
To help in determination of the physiological ligand for YtfQ, we solved its crystal structure to 1.2 Å resolution in the presence of galactose (Table 2 and Fig. 2a). The refined model contains residues 2–292 of the 297 residues in the mature protein. Like other SBPs, the protein has two α/β domains linked by a hinge region (38). Domain I is defined by residues 2–108 and 248–275 and contains 6 parallel β-strands with the order β2-β1-β3-β4-β5-β11 surrounded by 4 α-helices. Domain II is defined by residues 109–247 and 276–292 and has 6 β-strands surrounded by 6 α-helices. The order of the β-strands is β7-β6-β8-β9-β10-β12, with β12 running anti-parallel to the other 5 strands. The protein also contains a disulfide bond between residues Cys-129 and Cys-193 (Fig. 2a) which joins the start of the β7 and β6 strands in domain II. The topology of the protein is the type I class seen in SBPs (39) with additional β-strands (β11 and β12) and an additional α-helix (α10) (Fig. 2b). The protein is most similar at the sequence level (33% identity) and structural level, as judged using the DALI server (40), to the E. coli ribose-binding protein RbsB, and is also clearly homologous to the allose- (AlsB), galactopyranose- (MglB), and arabinose- (AraF) binding proteins from E. coli.
TABLE 2.
YtfQ x-ray data collection and refinement statistics
| Crystal 1 name | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 45.75, 51.54, 116.09 |
| a, b, g (°) | 90, 90, 90 |
| Resolution (Å) | 50.0-1.20 (1.23-1.20) |
| Rsym or Rmerge | 0.071 (0.35) |
| I/sI | 22 (1.5) |
| Completeness (%) | 91 (39) |
| Redundancy | 6.1 (1.8) |
| Refinement | |
| Resolution (Å) | 50.0 -1.2 |
| No. reflections | 74826 (3946) |
| Rwork/Rfree | 0.179 (0.190) |
| No. atoms | |
| Protein | 2241 |
| Ligand/ion | 24 |
| Water | 366 |
| B-factors | |
| Protein | 12.049 |
| Ligand/ion | 7.918 |
| Water | 27.364 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 1.206 |
FIGURE 2.
Crystal structure of YtfQ. a, schematic of the structure of YtfQ colored with rainbow coloring from the N terminus (blue) to the C terminus (yellow). The galactofuranose is shown in a stick representation. The single disulfide bond joining residues Cys-129 and Cys-193 is indicated in dark blue. b, topology diagram of YtfQ. The N-terminal domain is in white and the C-terminal domain in gray. Circles represent α-helices, triangles represent β-strands, and unfilled ovals are 310 helices. The double line from β6 indicates the position of the disulfide bond. c, structure of galactofuranose modeled into electron density contoured at 1.5 σ.
Presence of Galactofuranose in the Binding Site of YtfQ
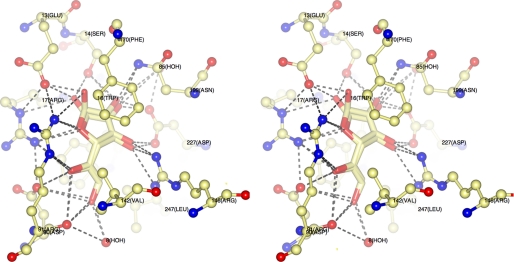
Our solution-based ligand binding experiments (Table 1) demonstrated that YtfQ binds galactose and this sugar was seen in the ligand binding site of YtfQ. However, unexpectedly it was bound as the rarer galactofuranose form of the sugar, rather than the more abundant galactopyranose form (Figs. 2, a and c and 3). Similar to RbsB and other monosaccharide SBPs, the ligand is trapped in a cleft between two structural α/β domains as indicated in Fig. 2a and is totally buried within the protein. The side chains of residues Ser-14, Arg-17, Asp-90, Arg-146, and Asn-199 form hydrogen bonds with the hydroxyl groups and the ring oxygen of the sugar molecule (Fig. 3). Further hydrogen bonds via a water molecule occur to the extended structure of the C6 position with residues Ala-107, Thr-106, Asn-109, and Asp-90 all co-coordinating this water. In addition aromatic residues, Trp-16 and Phe-170 provide hydrophobic surfaces packing against the nonpolar portions of both faces of the sugar. The high resolution of our structure resolved the presence of both the α-galactofuranose and the β-galactofuranose in the structure in an approximate 40:60 ratio, similar to the ratio seen in solution (41) (see in Figs. 2c and 3), suggesting that the protein is able to recognize both forms of the furanose and has no anomeric selectivity. For the α-furan and the β-furan forms, there is a hydrogen bond between the hydroxyl at C2 and the oxygen of the side chain of Ser-14. For β-furan there is a second hydrogen bond between the repositioned side-chain and the C1 hydroxyl in the sugar (see supplemental Figs. S2 and S3 for Ligplots).
FIGURE 3.
Stereo image of the binding site of YtfQ containing both α- and β-galactofuranose. The amino acids involved in coordinating the ligand are represented in thin stick format and are labeled, and the ligand is represented in thick stick format. Hydrogen bonds are indicated with dashed lines.
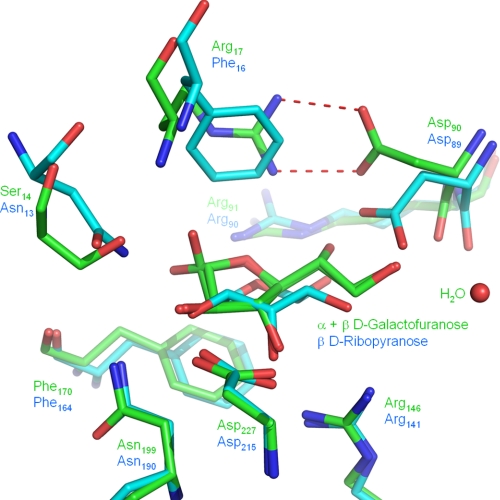
Closer comparison of the ligand binding site of YtfQ with that of RbsB (which binds ribopyranose), reveals that one side of the binding pocket is absolutely conserved between the two proteins (see the bottom side of Fig. 4). This pocket contains residues well-conserved in all the E. coli monosaccharide SBPs (Fig. 5). However, there are significant differences on the opposite side of the binding pocket (see the upper side of Fig. 4), the most striking change of which is at position 17 in YtfQ (position 16 in RbsB) where a phenylalanine in RbsB that stacks against the sugar ring is replaced by an arginine in YtfQ (Fig. 4). As a result, the aspartate residue at position 90 in YtfQ (Asp-89 in RbsB) flips out of the ligand binding site to form a salt bridge with Arg-17 (Fig. 4). This movement opens up the binding pocket to accommodate the more extended structure of the furanose form of the hexose.
FIGURE 4.
Superposition of the ligand binding sites of YtfQ and RbsB. The amino acid side chains involved in coordinating ribopyranose by RbsB (cyan) or galactofuranose by YtfQ (green) are labeled by their positions in the PDB files 2DRI and 2VK2, respectively. The coordination of galactofuranose also involves a water (red sphere).
FIGURE 5.
Multiple structure-based alignment of the amino acid sequences of E. coli YtfQ, AlsB, RbsB, and Salmonella typhimurium MglB proteins. Triangles indicate residues in contact with ligand in YtfQ (red direct, blue indirect). Blue stars indicate the aromatic residues that form part of the binding pocket. The cysteine residues (Cys-129 & Cys-193) that form the disulfide bond are labeled (green numeral 1).
Evidence for Selective Binding of Galactofuranose to YtfQ
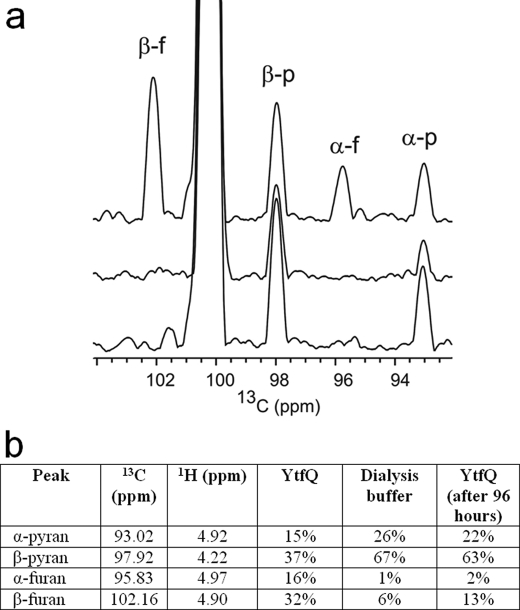
To verify that the unusual ligand observed in the crystal structure is relevant in solution, we used NMR, which is able to detect chemically the differences between the four galactose conformations (α-furanose, α-pyranose, β-furanose, and β-pyranose) in a sample of [13C1]galactose in one-dimensional 13C projections from a 1H-13C HSQC experiment (Fig. 6, lower trace) or in two-dimensional HSQC spectra (supplemental Fig. S4). The nature of the ligand bound to YtfQ was investigated by detecting the conformations of [13C1]galactose present immediately after release from YtfQ initiated by addition of d6-DMSO, a solvent in which exchange between the furanose and pyranose forms is very slow (42, 43) (Fig. 6a, upper). This trace is strongly enriched for the presence of both the α- and β-galactofuranose species. The sample was then left for 4 days and new HSQC spectra acquired (Fig. 6a, middle), which demonstrated the spontaneous rearrangement of the released galactose to the pyranose form to a ratio similar to that seen in the dialysis buffer used to remove free galactose from the galactose-bound protein (Fig. 6, a, lower and b).
FIGURE 6.
NMR analysis of ligands present in YtfQ. a, one-dimensional 13C projections of three 1H-13C HSQC experiments. The upper panel shows the [13C1]galactose after denaturation of YtfQ by addition of d6-DMSO. The middle panel shows the projection of the HSQC acquired 4 days later. The lower panel shows the projection of the HSQC acquired of the lyophilized dialysis buffer after re-dissolving in d6-DMSO. b, volume of the peaks in the corresponding two-dimensional spectra (supplemental Fig. S4). Peak assignments are according to those published previously (30). Peaks at the 13C chemical shifts of β-galactofuranose (β-f), α-galactofuranose (α-f), β-galactopyranose (β-p), and α-galactopyranose (α-p) are indicated. The large peak at ∼100.5 ppm arises from folding of the DMSO resonance.
As the YtfQ protein appears to be selective for the furanose form over the pyranose form, it is reasonable to reinterpret the binding data for galactose presented in Table 1. The determined Kd for galactose binding was 1.70 μm, but in aqueous solution the proportion of total galactose that is in the furanose form is small, being only 7.7% (41) Hence, the inferred affinity of YtfQ for galactofuranose, assuming that it is unable to recognize galactopyranose, is 0.13 μm, which is within the range for a typical SBP-dependent transporter.
DISCUSSION
In this work, which aims to determine the function of uncharacterized ABC transporters in the model bacterium E. coli K-12, we characterized the SBP of the ytfQRTyjfF operon. In silico analysis of the promoter region of this operon suggests a role for this transporter in galactose metabolism and the sequence of YtfQ is clearly related to other monosaccharide SBPs from E. coli (Fig. 5). Our initial characterization of the protein by ES-MS indicated that it was likely bound to a hexose and analysis of ligand binding by fluorescence spectroscopy demonstrated that both galactose and arabinose bind to the protein with low μm Kd values. This was unusual as E. coli has previously characterized ABC transporters for galactose and arabinose with higher affinities for both ligands. The structure of YtfQ provided the first clue as to the function of this protein, in that it revealed the presence of galactose bound to the protein in the rarer furanose form. Galactofuranose binding was confirmed in solution by measuring, via NMR methods, the ligand released from YtfQ.
The high resolution of the YtfQ structure, at 1.2 Å, the highest resolution structure for a monosaccharide SBP, allowed us to discern that both α- and β-galactofuranose were recognized by the protein. These two anomers are present at a ratio of ∼40% α and 60% β, which is similar to the ratio in a solution of free galactose (41). As well as a bound ligand, the structure revealed the presence of a disulfide bond in YtfQ that presumably stabilizes the structure after folding in the periplasm. While uncommon in SBPs, disulfides have been seen in a number of other structures including the E. coli LivJ and LivK SBPs, where they are likely to have similar structural roles (44, 45). The presence of a disulfide in YtfQ is consistent with the recent identification of YtfQ as a substrate for the Dsb proteins (46). In this study YtfQ was not detected in the periplasm of a dsbA strain suggesting that the formation of the disulfide is essential for the protein to fold correctly.
Examination of the binding pocket and comparison to RbsB revealed that relatively minor changes in the binding site have altered the ligand binding specificity of this protein. The presence of an arginine at position 17 which forms a salt bridge with Asp-90, appears to be unique to this subfamily of monosaccharide SBPs; orthologues with this arginine conserved are found in the proteobacteria and across the wider bacterial domain. While the ytfQRTyjfF operon is not genetically linked to any sugar metabolic genes in E. coli, its orthologues in Sinorhizobium meliloti and Mycobacterium smegmatis cluster in regions with sugar catabolic genes (see supplemental Fig. S1B). In M. smegmatis there are three additional genes present, which encode a likely pathway for the utilization of arabinose as a carbon source, while in S. meliloti the ytfQ operon is linked to a gene encoding a potential transcriptional regulator and in this organism expression of this ABC transporter is induced 40-fold in the presence of d-fucose and 33-fold in the presence of l-arabinose (47). Clearly these related transporters are sugar transporters, but their functions need further investigation to determine whether any recognize furanose forms. While YtfQ is able to recognize arabinose and its orthologue in M. smegmatis has a possible function in arabinose uptake, we believe that galactose is the more likely physiological substrate for this transporter in E. coli for the following three reasons. First, the regulation of expression of these genes via GalR/GalS suggests a specific function in galactose metabolism and the absence of AraC binding sites like those found upstream of araF does not support a role in arabinose metabolism. Secondly, the ligand that we identified that was prebound to the protein had a mass of 180 which is consistent with a hexose (galactose) and not a pentose (arabinose, with a mass of 150). However, it is notable that the ring structure of arabinofuranose is identical to that of galactofuranose and only differs in the C5 and C6 positions and so it is possible that we are also detecting binding of arabinofuranose in solution in the fluorescence spectroscopy studies. Finally in E. coli strains containing mutations in both the galP and mgl systems there is still a low level of residual galactose transport (35), which could be mediated via the YtfQ system. The deletion of the major galactopyranose transporters in this strain is very likely to reduce the level of induction of expression of the ytfQ operon via GalR/GalS and underestimate any contribution of the YtfQ system in this genetic background. The role of the transporter in vivo is currently under investigation.
Most monosaccharides in solution are present predominantly in their pyranose forms and have a small proportion of the sugar in the furanose form. With galactose this is around 8% of the total sugar and for other sugars this can rise to as much as 30% for fructose and altrose (48). In contrast, glucose is almost completely found in the pyranose form, which is presumably why YtfQ is unable to bind glucose, while it is recognized by the galactopyranose binding protein. All previous structures of monosaccharide SBPs from bacteria have contained the pyranose forms, for both hexoses and pentoses (19, 49–52). In addition, the structure of the sodium solute symporter (SSS) protein vSGLT from Vibrio parahemeolyticus also revealed that this galactose transporter binds the pyranose form of the sugar. We hypothesize that the YtfQ system is likely to function at the same time as the MglB system under carbon limiting conditions in the presence of galactose. As they recognize the furanose and pyranose forms of galactose, respectively, their combined use would allow E. coli to exploit maximally the available galactose. This might be important in the competitive environment of the human gut where mucin derived monosaccharides are important carbon sources for many components of the gut microflora. Also, given that other monosaccharides such as fructose have the furanose form present at up to 30% of the total sugar, it raises the question of whether other bacteria have evolved sugar transporters specific for the furanose forms and perhaps of a wider consideration of the role of furanose forms of sugars in biology more generally.
Supplementary Material
Acknowledgments
We thank the ESRF, Grenoble, for excellent data collection facilities, Berni Strongitharm and Andrew Leech for help with the ESI-MS, and Marcus Fischer for preparing LigPlot images. We would also like to thank Prof. Peter Henderson (University of Leeds) for access to reagents.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC) (to G. H. T.) and a BBSRC quota studentship (to R. S. P. H.).
The atomic coordinates and structure factors (code 2VK2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- ABC
- ATP-binding cassette
- SBP
- substrate-binding protein
- GalR
- galactose repressor
- PDB
- Protein Data Bank
- r.m.s.
- root mean-squared
- ES-MS
- electrospray mass spectrometry.
REFERENCES
- 1.Ingraham J. L., Neidhardt F. C. (1987) Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 2.Fabich A. J., Jones S. A., Chowdhury F. Z., Cernosek A., Anderson A., Smalley D., McHargue J. W., Hightower G. A., Smith J. T., Autieri S. M., Leatham M. P., Lins J. J., Allen R. L., Laux D. C., Cohen P. S., Conway T. (2008) Infect. Immun. 76, 1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franchini A. G., Egli T. (2006) Microbiology 152, 2111–2127 [DOI] [PubMed] [Google Scholar]
- 4.Liu M., Durfee T., Cabrera J. E., Zhao K., Jin D. J., Blattner F. R. (2005) J. Biol. Chem. 280, 15921–15927 [DOI] [PubMed] [Google Scholar]
- 5.Siebold C., Flükiger K., Beutler R., Erni B. (2001) FEBS Lett. 504, 104–111 [DOI] [PubMed] [Google Scholar]
- 6.Jahreis K., Pimentel-Schmitt E. F., Brückner R., Titgemeyer F. (2008) FEMS Microbiol. Rev. 32, 891–907 [DOI] [PubMed] [Google Scholar]
- 7.Saier M. H., Jr., Tran C. V., Barabote R. D. (2006) Nucleic Acids Res. 34, D181–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson D. B. (1974) J. Biol. Chem. 249, 553–558 [PubMed] [Google Scholar]
- 9.Davidson A. L., Dassa E., Orelle C., Chen J. (2008) Microbiol. Mol. Biol. Rev. 72, 317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quiocho F. A. (1990) Philos. Trans. R. Soc. Lond B Biol. Sci. 326, 341–351 [DOI] [PubMed] [Google Scholar]
- 11.Quiocho F. A., Ledvina P. S. (1996) Mol. Microbiol. 20, 17–25 [DOI] [PubMed] [Google Scholar]
- 12.Miller D. M., 3rd, Olson J. S., Pflugrath J. W., Quiocho F. A. (1983) J. Biol. Chem. 258, 13665–13672 [PubMed] [Google Scholar]
- 13.Müller A., Severi E., Mulligan C., Watts A. G., Kelly D. J., Wilson K. S., Wilkinson A. J., Thomas G. H. (2006) J. Biol. Chem. 281, 22212–22222 [DOI] [PubMed] [Google Scholar]
- 14.Thomas G. H., Southworth T., León-Kempis M. R., Leech A., Kelly D. J. (2006) Microbiology 152, 187–198 [DOI] [PubMed] [Google Scholar]
- 15.Müller A., Thomas G. H., Horler R., Brannigan J. A., Blagova E., Levdikov V. M., Fogg M. J., Wilson K. S., Wilkinson A. J. (2005) Mol. Microbiol. 57, 143–155 [DOI] [PubMed] [Google Scholar]
- 16.Bulyk M. L., McGuire A. M., Masuda N., Church G. M. (2004) Genome Res. 14, 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng D., Constantinidou C., Hobman J. L., Minchin S. D. (2004) Nucleic Acids Res. 32, 5874–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savery N. J., Lloyd G. S., Kainz M., Gaal T., Ross W., Ebright R. H., Gourse R. L., Busby S. J. (1998) EMBO J. 17, 3439–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Björkman A. J., Binnie R. A., Zhang H., Cole L. B., Hermodson M. A., Mowbray S. L. (1994) J. Biol. Chem. 269, 30206–30211 [PubMed] [Google Scholar]
- 20.Raman B., Nandakumar M. P., Muthuvijayan V., Marten M. R. (2005) Biotechnol. Bioeng. 92, 384–392 [DOI] [PubMed] [Google Scholar]
- 21.Severi E., Randle G., Kivlin P., Whitfield K., Young R., Moxon R., Kelly D., Hood D., Thomas G. H. (2005) Mol. Microbiol. 58, 1173–1185 [DOI] [PubMed] [Google Scholar]
- 22.Witholt B., Boekhout M., Brock M., Kingma J., Heerikhuizen H. V., Leij L. D. (1976) Anal. Biochem. 74, 160–170 [DOI] [PubMed] [Google Scholar]
- 23.Newman J., Egan D., Walter T. S., Meged R., Berry I., Ben Jelloul M., Sussman J. L., Stuart D. I., Perrakis A. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61, 1426–1431 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z., Minor W. (1997) Methods Enzmol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 26.Mowbray S. L., Cole L. B. (1992) J. Mol. Biol. 225, 155–175 [DOI] [PubMed] [Google Scholar]
- 27.Perrakis A., Harkiolaki M., Wilson K. S., Lamzin V. S. (2001) Acta Crystallogr. D. Biol Crystallogr. 57, 1445–1450 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Hobley P., Howarth O., Ibbett R. N. (1996) H-1 and C-13 NMR Shifts for Aldopyranose and Aldofuranose Monosaccharides: Conformational Analysis and Solvent Dependence. Magnetic Resonance in Chemistry Vol. 34, pp. 755–760, John Wiley and Sons; Ltd., New York, NY [Google Scholar]
- 31.Overbeek R., Begley T., Butler R. M., Choudhuri J. V., Chuang H. Y., Cohoon M., Crécy-Lagard V., Diaz N., Disz T., Edwards R., Fonstein M., Frank E. D., Gerdes S., Glass E. M., Goesmann A., Hanson A., Iwata-Reuyl D., Jensen R., Jamshidi N., Krause L., Kubal M., Larsen N., Linke B., McHardy A. C., Meyer F., Neuweger H., Olsen G., Olson R., Osterman A., Portnoy V., Pusch G. D., Rodionov D. A., Ruckert C., Steiner J., Stevens R., Thiele I., Vassieva O., Ye Y., Zagnitko O., Vonstein V. (2005) Nucleic Acids Res. 33, 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 34.Misra R. V., Horler R. S., Reindl W., Goryanin I. I., Thomas G. H. (2005) Nucleic Acids Res. 33, D329–D333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson P. J., Giddens R. A., Jones-Mortimer M. C. (1977) Biochem. J 162, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller D. M., 3rd, Olson J. S., Quiocho F. A. (1980) J. Biol. Chem. 255, 2465–2471 [PubMed] [Google Scholar]
- 37.Zukin R. S., Strange P. G., Heavey R., Koshland D. E. (1977) Biochemistry 16, 381–386 [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson A. J., Verschueren K. H. G. (2003) in ABC Proteins: From Bacteria to Man (Holland I. B., Cole S. P. C., Kuchler K., Higgins C. F. eds) pp. 187–207, Elsevier Science Ltd., London [Google Scholar]
- 39.Fukami-Kobayashi K., Tateno Y., Nishikawa K. (2003) Mol. Biol. Evol. 20, 267–277 [DOI] [PubMed] [Google Scholar]
- 40.Holm L., Kääriäïnen S., Wilton C., Plewczynski D. (2006) Curr. Protoc. Bioinformatics Chapter 5, Unit 5.5 [DOI] [PubMed] [Google Scholar]
- 41.Barlow J. N., Blanchard J. S. (2000) Carbohydr. Res. 328, 473–480 [DOI] [PubMed] [Google Scholar]
- 42.Jaseja M., Perlin A. S., Dias P. (1990) Magnetic Resonance in Chemistry Vol. 28, pp. 283–289, John Wiley and Sons, Ltd., New York [Google Scholar]
- 43.Franks F. (1987) Pure and Applied Chemistry Vol. 59, pp. 1189–1202, International Union of Pure and Applied Chemistry, Research Triangle Park, NC [Google Scholar]
- 44.Sack J. S., Saper M. A., Quiocho F. A. (1989) J. Mol. Biol. 206, 171–191 [DOI] [PubMed] [Google Scholar]
- 45.Magnusson U., Salopek-Sondi B., Luck L. A., Mowbray S. L. (2004) J. Biol. Chem. 279, 8747–8752 [DOI] [PubMed] [Google Scholar]
- 46.Vertommen D., Depuydt M., Pan J., Leverrier P., Knoops L., Szikora J. P., Messens J., Bardwell J. C., Collet J. F. (2008) Mol. Microbiol. 67, 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mauchline T. H., Fowler J. E., East A. K., Sartor A. L., Zaheer R., Hosie A. H., Poole P. S., Finan T. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17933–17938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins P., Ferrier R. (1995) Monosaccharides: Their Chemistry and Their Roles in Natural Products, Wiley [Google Scholar]
- 49.Chaudhuri B. N., Ko J., Park C., Jones T. A., Mowbray S. L. (1999) J. Mol. Biol. 286, 1519–1531 [DOI] [PubMed] [Google Scholar]
- 50.Quiocho F. A., Gilliland G. L., Miller D. M., Newcomer M. E. (1977) J. Supramol. Struct. 6, 503–518 [DOI] [PubMed] [Google Scholar]
- 51.Quiocho F. A., Vyas N. K. (1984) Nature 310, 381–386 [DOI] [PubMed] [Google Scholar]
- 52.Zou J. Y., Flocco M. M., Mowbray S. L. (1993) J. Mol. Biol. 233, 739–752 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.