Abstract
The link between intra-abdominal adiposity and type II diabetes has been known for decades, and adipose tissue macrophage (ATM)-associated inflammation has recently been linked to insulin resistance. However, the mechanisms associated with ATM recruitment remain ill defined. Herein, we describe in vitro chemotaxis studies, in which adipocyte conditioned medium was used to stimulate macrophage migration. We demonstrate that tumor necrosis factor α and free fatty acids, key inflammatory stimuli involved in obesity-associated autocrine/paracrine inflammatory signaling, stimulate adipocyte expression and secretion of macrophage chemoattractants. Pharmacological studies showed that peroxisome proliferator-activated receptor γ agonists and glucocorticoids potently inhibit adipocyte- induced recruitment of macrophages. This latter effect was mediated by the glucocorticoid receptor, which led to decreased chemokine secretion and expression. In vivo results were quite comparable; treatment of high fat diet-fed mice with dexamethasone prevented ATM accumulation in epididymal fat. This decrease in ATM was most pronounced for the proinflammatory F4/80+, CD11b+, CD11c+ M-1-like ATM subset. Overall, our results elucidate a beneficial function of peroxisome proliferator-activated receptor γ activation and glucocorticoid receptor/glucocorticoids in adipose tissue and indicate that pharmacologic prevention of ATM accumulation could be beneficial.
Introduction
Obesity is associated with various metabolic abnormalities characterized as the metabolic syndrome or syndrome X (1, 2). These abnormalities are causally related to type 2 diabetes, nonalcoholic steatohepatitis, and coronary heart disease (1, 2). Recent findings indicate that the metabolic syndrome and its disease manifestations are linked to adipose tissue inflammation, which results from infiltration and accumulation of proinflammatory macrophages (3). Since numerous genetic manipulations of myeloid-derived cells in knock-out mice have clearly demonstrated the role of macrophage-mediated inflammation in insulin resistance/diabetes, strategies to interfere with macrophage migration could have clinical benefit (1, 2).
In inflammatory diseases, the secretion of specific chemoattractants, termed chemokines, mediates leukocyte recruitment. The chemokine family consists of numerous members (>50) that can be divided into two major subfamilies based on characteristic amino acid sequences (CCL versus CXCL). Specific monocyte/macrophage chemokines from the CCL subfamily have been identified, including CCL2 (MCP1 (monocyte chemoattractant protein 1)). Leukocyte recruitment is regulated by the expression of the corresponding chemokine receptor (CCR versus CXCR) on specific myeloid cell types. Chemokine and cytokine expression are tightly linked and are typically stimulated by proinflammatory cytokines (4, 5). Interestingly, in obese, insulin resistance states, such cytokines are elevated in adipose tissue, and this correlates with increased macrophage accumulation. MCP1 is one of the chemokines whose expression is increased in eWAT3 of obese mice; however, the role of the CCR2/MCP1 axis in mediating ATM recruitment is still unclear (1, 6–11).
Interestingly, a number of anti-inflammatory agents display insulin-sensitizing properties (12–14). In particular, insulin-sensitizing TZD PPARγ agonists, such as rosiglitazone, exert broad anti-inflammatory properties in macrophages (15). In addition to PPARγ agonists, GR, liver X receptor α, PPARα, and PPARβ/δ agonists can have anti-inflammatory effects in macrophages (16–20). Whether these nuclear hormone receptor agonists display similar anti-inflammatory properties directly in adipocytes is not fully understood.
In the current studies, we used an in vitro chemotaxis assay to investigate the mechanisms underlying macrophage recruitment by mature adipocytes (21). First, we demonstrate that proinflammatory stimuli, such as TNFα and free fatty acids (FFAs), increased the ability of adipocytes to release chemokines that induce chemotaxis. TZDs and glucocorticoids potently inhibited macrophage recruitment. The ability of glucocorticoids to interfere with chemotaxis was also observed in vivo, since treatment of obese mice with dexamethasone led to decreased ATM accumulation.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Aldosterone, corticosterone, dexamethasone, isobutylmethylxanthine, insulin LPS, PD98059, RU4868, spironolactone, and the free fatty acids (arachidonic, lauric, linoleic, myristic, and oleic acids) were from Sigma. AKT inhibitor VIII was from Calbiochem; rosiglitazone, T090137, GW501516, and Wy14643 were from Alexis. TNFα was from BIOSOURCE. Low endotoxin, free fatty acid-free BSA was from MP Biochemicals. MCP1-neutralizing antibody (2H5) and control IgG were from Biolegend.
Cell Culture
Mouse 3T3L1 cells were grown and maintained undifferentiated in DMEM (25 mm glucose) supplemented with 10% bovine calf serum (Hyclone) and differentiated to 3T3L1 adipocytes as described previously (12). Raw264.7 cells were maintained in 10% low endotoxin fetal bovine serum (Hyclone)/DMEM (25 mm glucose). Bone marrow-derived macrophages were obtained and cultured as described previously (22). Finally, THP1 cells were maintained in RPMI supplemented with 10% low endotoxin fetal bovine serum.
siRNA Electroporation
siRNAs were obtained from IDT. Sequences for GR and mineralocorticoid receptor (MR) siRNA duplexes were as described by Caprio et al. (23). Adipocytes at day +6 from differentiation protocol were collected and electroporated with siRNAs as described previously (GENE PULSER, Bio-Rad) (12). In the case of GR and MR knockdown, 24 h conditioned medium was prepared 36 h postelectroporation.
In Vitro Chemotaxis Assay
Mature 3T3L1 adipocytes, more than 99% of cells showing large lipids droplets (12 days after differentiation protocol initiation), were used for preparation of conditioned media. Treatment with the different drugs at the concentration indicated was performed in serum-free, 0.2% endotoxin- and FFA-free BSA/DMEM. Mature 3T3L1 and Raw264.7 cells were treated for 24 h, except when mentioned otherwise. Collected media were kept aliquoted and frozen at −20 °C until a chemotaxis assay or chemokine quantitation was performed. The FFA mixture (500 μm final concentration) consisted of a mixture of five different fatty acids (arachidonic, lauric, linoleic, myristic, and oleic acids), each one of them used at 100 μm. They were first suspended in 0.68% endotoxin- and FFA-free BSA/DMEM and exposed to mature adipocytes. After 24 h, cells were washed with phosphate-buffered saline, and, for each condition, exposed to 0.2% endotoxin- and FFA-free BSA/DMEM for another 24 h prior to collection of the media for the assays. For the migration per se, 200,000 Raw264.7 cells suspended in serum-free, 0.2% endotoxin- and FFA-free BSA/DMEM were used per condition. The Raw264.7 cells were placed in the upper chamber of an 8 μm polycarbonate filter (24-transwell format; Corning, Lowell, MA), whereas adipocyte conditioned medium was placed in the lower chamber. After 3 h of migration, which was determined to be the optimal period (i.e. within the linear phase of Raw264.7 cell accumulation (supplemental Fig. 1A)), cells were fixed in formalin and stained with 4′,6-diamidino-2-phenylindole. Cells that had not migrated and remained in the upper chamber were removed by gently swiping the filters with cotton tips. Raw264.7 cells found on the filter, in the lower chamber, were counted as cells having performed chemotaxis and quantified by fluorescence with Simple PCI imaging software (Compix Inc., Cranberry Township, PA). Macrophages were quantified from 5 fields/condition; each condition was performed in duplicate. Similar conditions were applied for the bone marrow-derived macrophage and peritoneal macrophage migration assays (24).
Animal Experiments
Male C57BL/6J mice (littermates) were obtained from Jackson Laboratories and housed four per cage under pathogen-free conditions. 3-Month-old male C57BL/6J mice were either fed a normal chow diet (13.5% fat; LabDiet) or a high fat diet (60% fat; Research Diet) for 20 weeks (8 mice/condition). Dexamethasone was suspended in drinking water at a concentration of 2 mg/liter (25, 26). Mouse body weight (g) and food and water intake (ml) were monitored weekly. Blood was withdrawn by tail vein bleeding on 5-h fasted mice (9 a.m. to 2 p.m.).
Immunohistochemistry and Adipocyte Size Quantification
Paraffin-embedded epididymal adipose tissue sections from 5 mice/group were incubated with MAC-2 antibody (Cedarlane) at a 1:3800 dilution at 4 °C. Subsequently, a biotinylated anti-rat secondary antibody (PharMingen) was used at a 1:100 dilution, followed by 1:500 horseradish peroxidase-Streptavidin (Jackson) and development in substratechromogen. Slides were counterstained with Mayer's hematoxylin and mounted with Vectashield mounting medium. Bright field photographs were taken of three representative fields per slide in a blinded fashion using a fluorescent microscope (Olympus MVX10, ×10 objective). We used the software Macnification (Orbicule BVBA) for quantification of adipocyte sizes (3).
Fluorescence-activated Cell Sorting (FACS) Analysis of Macrophages of Adipose Stromal Vascular Fraction
Epididymal fat pads were weighed, rinsed in phosphate-buffered saline, and then minced in FACS buffer (phosphate-buffered saline plus 1% BSA). Adipocytes and stromal vascular cells were prepared from collagenase digested adipose tissue as described previously (22). FACS analysis of stromal vascular cells for macrophage content and subtypes were performed as previously described (22). Estimation of macrophage subsets numbers/g of fat was performed as follows, number of cells = percentage of cells × number of live cells in SVF/mass of depot (g). Numbers obtained were subsequently represented as percentage of the highest subsets (i.e. F4/80+ in high fat diet (HFD)-untreated mice) (9, 22).
RNA Isolation and Quantitative PCR (QPCR)
Total RNA was extracted from cells or tissue with TRIzol (Invitrogen) reagent. 2 μg of total RNA was reverse-transcribed with using the ABI cDNA synthesis kit (Applied Biosciences). Primer sequences used in the PCRs were chosen based on the sequences available in GenBankTM and were designed to generate a PCR amplification product of 100–200 bp. Only primer pairs yielding unique amplification products without primer dimer formation were subsequently used for quantitative PCR assays. Primer sequences are available upon request. PCR was performed as described (3). The mRNA expression of all genes reported is normalized to the cylcophilin A gene expression.
Protein Isolation and Western Blots
Proteins from tissues or cell lysates were extracted with radioimmune precipitation buffer (Upstate) in the presence of phosphatase inhibitors and protease inhibitors (Roche Applied Science). 20 μg of proteins/lane were separated on a 10% polyacrylamide, precast SDS gel (Bio-Rad) followed by a transfer on polyvinylidene difluoride membrane (Immobilon, Millipore), and immunoblotting was performed as described (12, 27).
Deoxyglucose Uptake
3T3-L1 adipocytes were pretreated for 24 h in DMEM, 10% fetal bovine serum with the indicated drug concentrations; serum-starved for 1 h in DMEM containing 0.1% FFAs, endotoxin-free BSA; and stimulated with 0.17 nm insulin for 20 min at 37 °C. Glucose uptake was determined after the addition of 2-[3H]deoxyglucose (0.1 μCi, final concentration of 0.1 mm) in Krebs-Ringer-phosphate-HEPES buffer for 10 min at 37 °C. Deoxyglucose uptake was deducted from tritium counting normalized to total protein concentration (28).
Enzyme-linked Immunosorbent Assays
Corticosterone and MCP1 were directly quantified from cell tissue culture and white adipose tissue extracts according to the manufacturer's instructions (Neogen for corticosterone and BIOSOURCE for MCP1 and TNFα). Plasma insulin and adiponectin levels were measured by an enzyme-linked immunosorbent assay from ALPCO and B-Bridge, respectively.
Statistical Analysis
Data represent the means ± S.D. and were evaluated by Student's two-tailed t test or analysis of variance provided by VassarStats: Web Site for Statistical Computation. A p value cut-off of 0.05 was used to determine significance.
RESULTS
Adipocyte-secreted Factors Induce Macrophage Migration
We have studied the effects of proinflammatory signals on the ability of adipocytes to induce chemotaxis. Fig. 1 shows measurements of Raw264.7 monocyte/macrophage chemotaxis stimulated by conditioned medium (CM) harvested from 3T3L1 adipocytes that were treated with various stimuli.
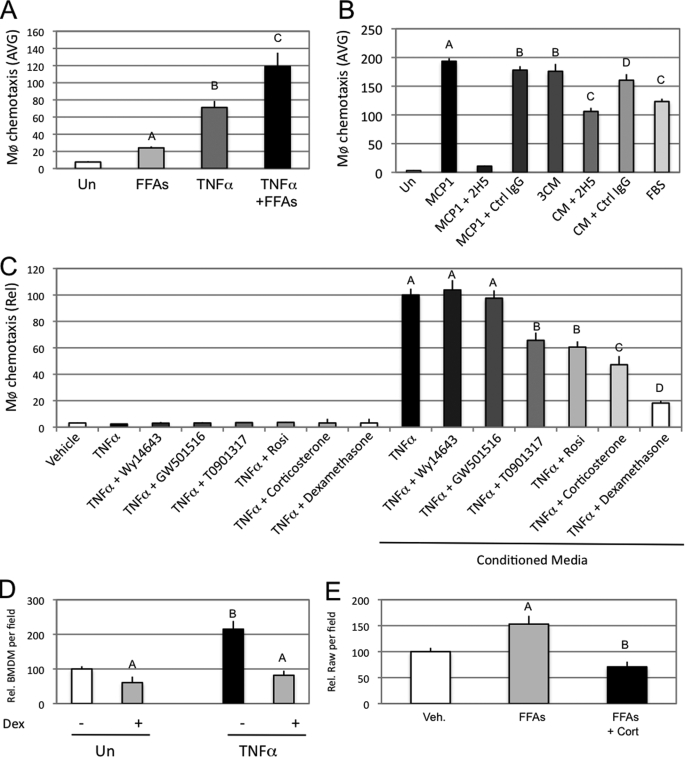
FIGURE 1.
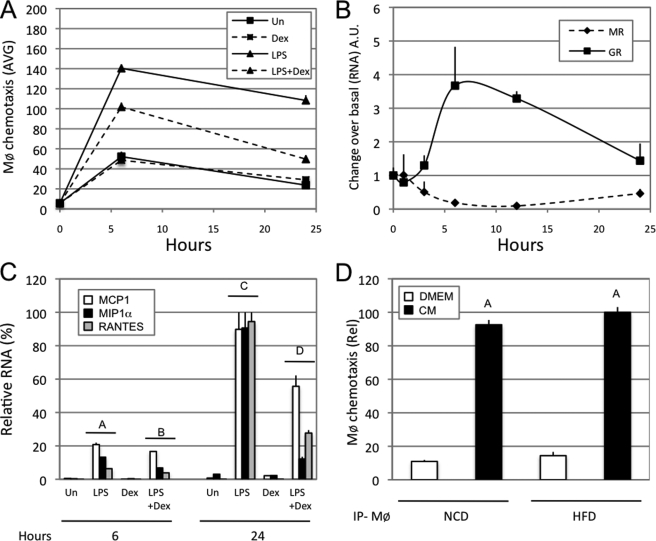
Adipocyte secreted factors induce macrophage migration. A, mature 3T3L1 adipocytes were pretreated for 24 h with an FFA mixture (500 μm final) and or TNFα (10 ng/ml final). Cells were subsequently washed and resuspended in serum-free medium for another 24 h. 3T3L1 conditioned media were then collected, and chemotaxis assays were performed with Raw264.7 macrophage cells. The graph represents the average (AVG) number of Raw264.7 cells counted per microscope field as having performed chemotaxis. B, chemotaxis assays were performed with THP1 cells in response to serum-free DMEM (Un), MCP1 (100 μg/ml), MCP1 and a MCP1 neutralizing antibody (2H5; 50 μg/ml), MCP1 and a control antibody (IgG; 50 μg/ml), 3T3L1 mature adipocyte CM, CM and an MCP1-neutralizing antibody (2H5; 50 μg/ml), CM and a control antibody (IgG; 50 μg/ml), and DMEM containing 10% fetal bovine serum (FBS). C, chemotaxis assays were performed with Raw264.7 cells on 3T3L1 mature adipocyte conditioned medium (serum-free), treated for 24 h with the indicated drugs (final concentrations: TNFα, 10 ng/ml; Wy14643, 10 μm; GW501516, 1 μm; T0901317, 5 μm; rosiglitazone (Rosi), 5 μm; corticosterone, 100 nm; dexamethasone, 100 nm). The left part of the graph represents the migration of Raw264.7 in response to non-conditioned medium, prior to incubation with 3T3L1 cells. The graph represents the relative (Rel) average number of Raw264.7 cells counted per microscope field as having performed chemotaxis. D, chemotaxis assays were performed with bone marrow-derived macrophage on 3T3L1 mature adipocyte conditioned media (serum-free), treated for 24 h with the indicated drugs (final concentrations: TNFα, 10 ng/ml; dexamethasone, 100 nm). E, chemotaxis assay was performed with Raw264.7 cells on 3T3L1 mature adipocyte conditioned media prepared from 24-h pretreated cells with vehicle (Veh) or an FFA mixture (500 μm final) in the presence or absence of 100 nm corticosterone (Cort). Conditioned media were collected 24 h after resuspension in serum-free media. Bars, average values ± S.D. of duplicates from at least three independent experiments. A–D above the bars show statistical groups (p < 0.05). BMDM, bone marrow-derived macrophages.
As shown in Fig. 1A, when 3T3L1 adipocytes were treated for 24 h with TNFα, the resulting adipocyte CM had a potent effect to stimulate macrophage chemotaxis. Treatment of the adipocytes with FFAs was also stimulatory, but less so than TNFα, whereas TNFα plus FFA treatment was synergistic on chemotaxis. THP1 human monocytes displayed comparable chemotactic responses when exposed to adipocyte CM (Fig. 1B). Macrophage chemotaxis was also strongly induced by MCP1 (100 μg/ml) and this effect was completely abolished by neutralizing anti-MCP1 antibody. Interestingly, preincubation of CM with the MCP1 neutralizing antibody inhibited chemotaxis by ∼40%. This partial inhibition indicates that secretion of MCP1 by adipocytes is one of the factors stimulating chemotaxis but that additional chemokines also play a role.
There are several nuclear hormone receptors that can mediate anti-inflammatory effects, and we assessed the ability of different nuclear hormone receptor agonists to inhibit adipocyte- mediated chemotaxis (Fig. 1C). The synthetic PPARγ and liver X receptor α agonists, rosiglitazone (5 μm) and T0901317 (5 μm), displayed substantial effects to inhibit CM-induced macrophage migration, but the effects of the GR agonists, corticosterone and dexamethasone, were more robust. The PPARα ligand, Wy14643 (10 μm), and the PPARδ/β ligand, GW501516 (1 μm), had no effect. Interestingly, in the absence of CM, none of the agonists had any effect on macrophage migration (Fig. 1C, left-hand bars). A similar chemotactic response was seen when bone marrow-derived macrophages were used instead of Raw264.7 cells (Fig. 1D). In addition to its ability to prevent TNFα-induced migration, corticosterone also inhibited the effects of FFAs (Fig. 1E). Hyperglycemia is a common feature of insulin resistance/diabetes, and glucose itself has been reported to be a proinflammatory stimulus (29). However, we did not detect any difference in macrophage recruitment by CM containing either 5 or 25 mm glucose (supplemental Fig. 1B).
Effects of CM on Signaling Pathways in Macrophages and Adipocytes
Chemokine receptors signal through molecules, such as extracellular signal-regulated kinase 1 and 2 (p42/p44) and Akt1, to induce cytoskeletal rearrangement, a prerequisite for chemotaxis (30–32). In Fig. 2A, we demonstrate that CM from 3T3L1 cells induces a rapid increase in phosphorylation of both Akt1 and extracellular signal-regulated kinase in Raw264.7 cells. This effect was more pronounced when CM was collected from adipocytes that had been pretreated with TNFα. Treatment of macrophages with a reversible Akt inhibitor but not an extracellular signal-regulated kinase inhibitor dose-dependently inhibited macrophage migration toward CM (Fig. 2B and supplemental Fig. 2A). Although a 30-min pretreatment of macrophages with the Akt inhibitor (10 μm) completely blocked Akt phosphorylation (supplemental Fig. 2B), macrophage migration was only partially prevented (Fig. 2B, right-hand bar). In contrast, chemotaxis was more strongly blocked when the Akt inhibitor was also added to the CM, indicating that the effects of the inhibitor are rapidly reversed after washing.
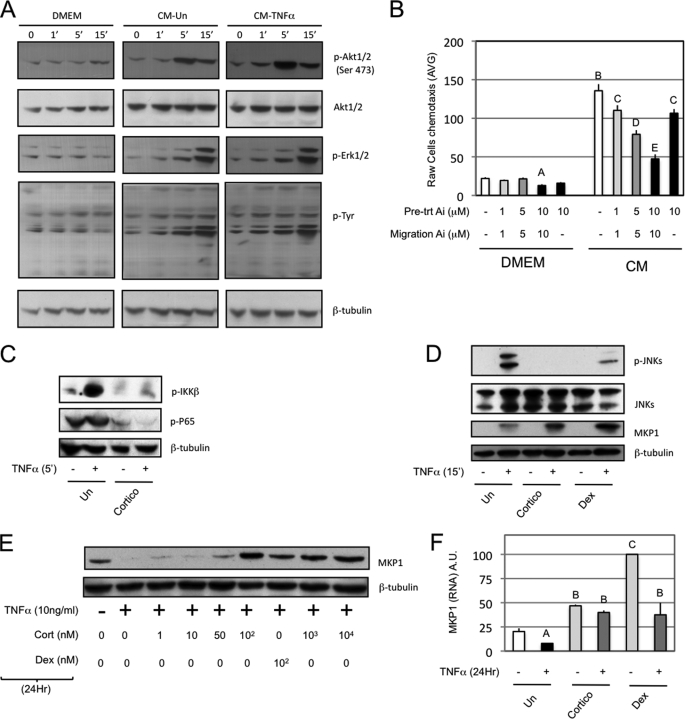
FIGURE 2.
Effects of CM on signaling pathways in macrophages and adipocytes. A, Western blots were performed on protein extracts from Raw264.7 cells treated for the indicated times with serum-free DMEM or mature 3T3L1 adipocyte conditioned media that were pretreated (CM-TNFα) or not (CM) with 10 ng/ml TNFα for 24 h prior to washes and resuspension for another 24 h in serum-free DMEM. B, a chemotaxis assay toward either serum-free DMEM or 24-h mature 3T3L1 adipocyte conditioned media (CM) was performed with Raw264.7. Akt inhibitor was used at the indicated concentrations as pretreatment (30 min prior to migration) and/or throughout the migration. C, Western blots were performed on protein extracts from mature adipocytes (3T3L1) that were pretreated with vehicle or corticosterone (100 nm, 24 h) and subsequently exposed to TNFα (10 ng/ml, 5 min). D, mature adipocytes (3T3L1) were pretreated with vehicle, corticosterone, or dexamethasone (100 nm, 24 h) and subsequently exposed to TNFα (10 ng/ml, 15 min) prior to protein extraction and Western blotting. E, Western blots were performed on 3T3L1 mature adipocytes treated for 24 h with the indicated drugs or vehicle in serum-free DMEM. F, QPCR was performed on 3T3L1 mature adipocytes RNA that were co-treated for 24 h with the indicated drugs (TNFα, 10 ng/ml; corticosterone and dexamethasone, 100 nm), and relative expression of MKP1 is normalized to glyceraldehyde-3-phosphate dehydrogenase expression. Bars, average values ± S.D. of duplicates from at least three independent experiments. A–E above the bars show statistical groups (p < 0.05). AVG, average; A.U., arbitrary units; p-, phosphorylated.
Iκ B kinase and Jun N-terminal kinase can link proinflammatory stimuli to chemokine expression (33, 34). Fig. 2, C and D, demonstrates that corticosterone and dexamethasone interfered with Iκ B kinase/NFκB and Jun N-terminal kinase activation in response to TNFα. Previous studies have shown that GR agonists can induce DUSP1/MKP1 (MAP kinase phosphatase 1), which can inhibit Jun N-terminal kinase phosphorylation (35, 36). Interestingly, short term treatment of 3T3L1 cells with TNFα (15 min) induced MKP1 expression, which was further increased by glucocorticoid pretreatment (Fig. 2D). On the other hand, 24-h TNFα treatment dramatically repressed MKP1 expression, which was reversed by glucocorticoid treatment at both the protein (Fig. 2E) and RNA level (Fig. 2F).
Macrophage Chemotaxis Correlates with Adipocyte Chemokine Expression and Secretion
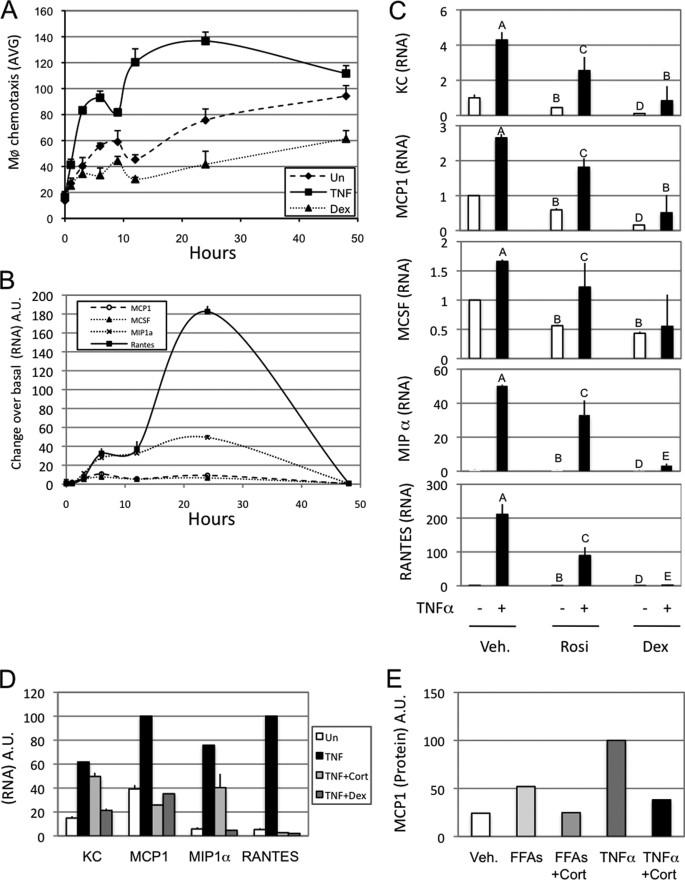
Adipocytes express several chemokines (34), and, in particular, increased MCP1 secretion has been linked to macrophage infiltration into adipose tissue (Fig. 1). Time course studies showed a striking parallel between the biphasic adipocyte expression pattern of four well described chemokines (macrophage colony stimulating factor, MCP1/CCL2, MIP1α (macrophage inflammatory protein 1α)/CCL3, and RANTES/CCL5) and the migration of Raw264.7 cells in response to CM (Fig. 3, B and A, respectively). Interestingly, the CCR1/5 agonists, RANTES and MIP1α, were more responsive to TNFα treatment compared with macrophage colony stimulating factor and MCP1. Rosiglitazone, corticosterone, and dexamethasone inhibited chemokine RNA expression (Fig. 3, C and D), and these effects were well correlated to the ability of these agents to inhibit chemotaxis (Fig. 3B). In addition, corticosterone inhibited FFA- or TNFα-induced MCP1 secretion into CM (Fig. 3E).
FIGURE 3.
Macrophages chemotaxis correlates with adipocyte chemokine expression and secretion. A, Raw264.7 cells were used for a chemotaxis assay on 3T3L1 media conditioned for the indicated times and treated with either vehicle (Un) or TNFα (10 ng/ml final) or dexamethasone (100 nm final). B, QPCR was performed on 3T3L1 cells treated with TNFα (10 ng/ml final) for the indicated times. C, QPCR was performed on 3T3L1 cells treated for 24 h with either vehicle (Veh.; DMSO), rosiglitazone (Rosi; 5 μm final), or Dex (100 nm final) in the presence (+) or absence (−) of TNFα (10 ng/ml final) for the indicated times. D, QPCR was performed on 3T3L1 cells treated for 24 h with vehicle (DMSO) and TNFα (10 ng/ml final) alone or in combination with corticosterone (Cort.; 100 nm) or dexamethasone (100 nm). E, MCP1 secretion was quantified from 24-h 3T3L1 mature adipocyte conditioned media. The FFA mixture was used at 500 μm, corticosterone (Cort.) was used at 100 nm, and TNFα was used at 10 ng/ml. Bars, average values ± S.D. of duplicates from at least three independent experiments. A–E above the bars show statistical groups (p < 0.05). AVG, average; A.U., arbitrary units; KC, keratinocyte-derived chemokine.
The Glucocorticoid Receptor (GR) Mediates the Repressive Effects of Glucocorticoids on Chemokine Expression and Macrophage Chemotaxis
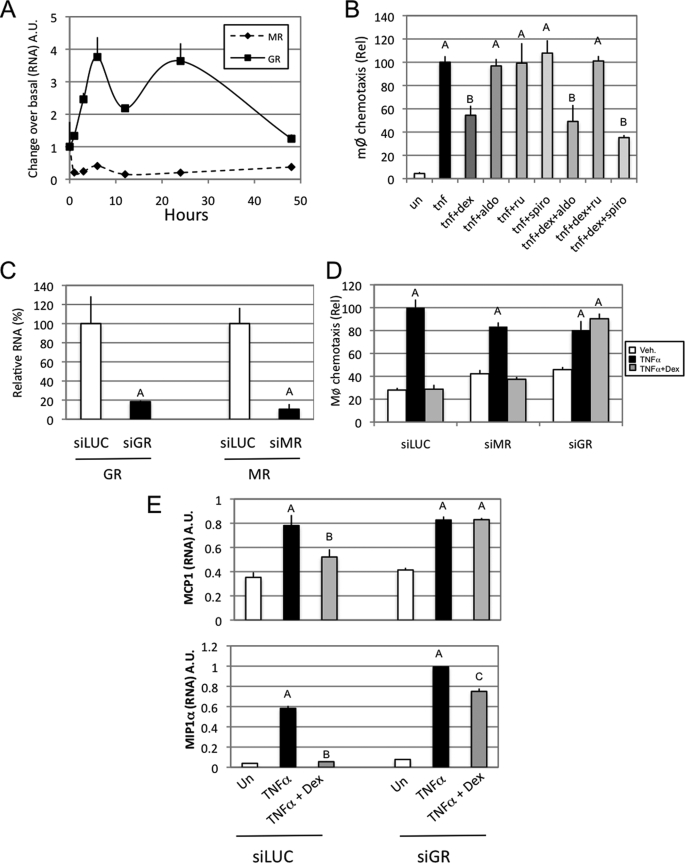
The MR can bind glucocorticoids with higher affinity than GR, and MR and GR are both expressed in 3T3L1 adipocytes. It has been demonstrated that MR mediates the adipogenic effects of glucocorticoids in vitro (23). Interestingly, time course studies showed that TNFα treatment decreases MR expression, whereas GR expression is markedly increased. This latter pattern is similar to the one observed for chemokine mRNA expression (Figs. 3B and 4A). To elucidate whether the MR or GR mediates the repressive effect of corticosterone and dexamethasone on macrophage chemotaxis, we treated adipocytes with synthetic agonists and antagonists for MR and GR (Fig. 4B) and then collected CM to measure chemotaxis. Aldosterone, a specific MR ligand, had no effect on either the stimulatory effect of TNFα or the repressive effect of dexamethasone on chemotaxis, and the specific MR antagonist spironolactone was also without effect. However, the GR antagonist RU486 blocked the anti-chemotactic effects of dexamethasone. We next used RNA interference to extend and confirm these findings. GR and MR siRNAs led to an 80 and 90% decrease of GR and MR mRNA, respectively (Fig. 4C), compared with scrambled siRNA. Studies with CM from these cells showed that the repressive effect of dexamethasone on TNFα-induced chemokine expression and macrophage migration was blocked by knockdown of GR but not MR (Fig. 4D). Measurements of chemokines (MCP1 and MIP1α) expression in knockdown adipocytes were consistent with the results for CM- induced chemotaxis (Fig. 4E). These results indicate that the adipocyte GR mediates the effects of glucocorticoids on chemokine expression and chemotaxis.
FIGURE 4.
The GR mediates the repressive effects of glucocorticoids on chemokine expression and macrophage chemotaxis. A, QPCR was performed on 3T3L1 cells treated with TNFα (10 ng/ml final) for the indicated times. B, a chemotaxis assay with Raw264.7 cells was performed on 24 h 3T3L1 mature adipocyte conditioned media treated with vehicle (un) or the indicated agents (final concentrations: TNFα, 10 ng/ml; dexamethasone (dex; 100 nm; aldosterone (aldo), 100 nm; RU4868 (ru), 1 μm; spironolactone (spiro), 1 μm). C, QPCR was performed on 3T3L1 cells (day 6 postdifferentiation) 36 h postelectroporation with siRNAs and 24 h incubation in serum-free DMEM. D, a chemotaxis assay with Raw264.7 cells was performed on 3T3L1 cell conditioned media, prepared from electroporated cells that were subsequently treated with either TNFα (10 ng/ml) alone or in the presence of dexamethasone (100 nm) for 24 h. E, QPCR was performed on 3T3L1 cells (day 6 postdifferentiation) 36 h postelectroporation and 24 h subsequent to incubation in serum-free DMEM with the indicated drugs (TNFα, 10 ng/ml; dexamethasone, 100 nm). Bars, average values ± S.D. of duplicates from at least three independent experiments. A–C above the bars show statistical groups (p < 0.05). A.U., arbitrary units.
Studies of Macrophage CM
Once macrophages accumulate, it is likely that they secrete additional chemokines, which further recruit ATMs in a feed-forward fashion. Consequently, we assessed the effects of CM harvested from Raw264.7 cells on chemotaxis of untreated Raw cells. We found that 6 and 24 h of LPS treatment increased the ability of Raw264.7 cell CM to stimulate chemotaxis, and this effect was inhibited by pretreatment with dexamethasone prior to CM collection (Fig. 5A). Similar to TNFα in 3T3L1 cells, LPS repressed MR expression and increased GR expression in the Raw264.7 cells (Fig. 5B). Furthermore, LPS enhanced and dexamethasone inhibited chemokine mRNA expression. Interestingly, CM-induced chemotaxis was maximal at 6 h of LPS treatment, whereas chemokine mRNA expression was maximal at 24 h (Fig. 5, A and C). These latter findings suggest the possibility that the Raw264.7 cells could contain a pool of presynthesized chemokines that are immediately released upon LPS treatment.
FIGURE 5.
Studies of macrophages CM. A, chemotaxis assays were performed with Raw264.7 cells prepared from Raw264.7 conditioned media incubated for 6 or 24 h with vehicle (Un), Dex (100 nm), and/or LPS (100 ng/ml) in serum-free DMEM. B, QPCR was performed on Raw264.7 cells treated with LPS (100 ng/ml final) in serum-free DMEM for the indicated times. C, QPCR was performed on Raw264.7 cells treated for the indicated times and reagents as described above. D, primary peritoneal macrophages (IP-MØ) elicited from mice fed a normal chow (NCD) or HFD were tested for migration toward adipocyte CM or control medium (DMEM). Bars, average values ± S.D. of duplicates from at least three independent experiments. A–D above the bars show statistical groups (p < 0.05). A.U., arbitrary units; Rel, relative.
Intrinsic Chemotactic Capacity of Macrophage from Chow and HFD Mice
These studies have shown the importance of chemotactic factors released from both adipocytes and macrophages as determinants of ultimate macrophage chemotaxis and migration. However, it remains possible that macrophages obtained from different groups of mice express differences in their intrinsic activity to respond to chemotactic signals. To assess this, we isolated peritoneal macrophages from normal chow fed mice versus HFD (16 weeks) mice and measured their chemotactic ability in response to adipocyte CM. As seen in Fig. 5D, the migration ability of these different macrophage populations was identical, indicating that extrinsic environmental cues, such as the mix of chemokines, rather than the intrinsic capacity of a macrophage to respond to a chemotactic signal is the more important factor determining macrophage migration and tissue accumulation.
In Vivo Dexamethasone Treatment Inhibits ATM Accumulation
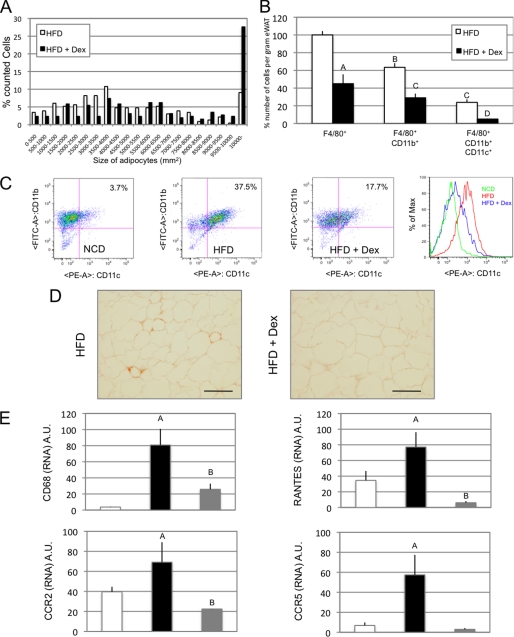
Since glucocorticoids decrease chemokine secretion and subsequent macrophage migration in vitro, we speculated that treatment of mice with dexamethasone would decrease ATM accumulation, even in the context of obesity. Accordingly, mice were placed on a 60% HFD along with dexamethasone treatment (120 ng/g of body weight/day). At the end of 20 weeks, total body weights were the same between groups, whereas epididymal fat pad weight was increased in the Dex-treated mice (Table 1 and supplemental Fig. 3). Quantification of adipocyte size showed an increase in average cell size in HFD Dex-treated animals due to a decrease in small adipocytes (<4000 μm2) and an increase in larger ones (>10,000 μm2) (Fig. 6A). This induction of adipocyte hypertrophy is in agreement with the reported lipogenic effects of chronic exposure to glucocorticoids (37). Stromal vascular fractions were prepared from the fat pads, and FACS analysis revealed a ∼50% reduction in the F4/80+ and F4/80+, CD11b+ macrophage fractions in the Dex-treated group (Fig. 6B). Interestingly, this decrease was even more pronounced (∼80%) in the triple positive F4/80+, CD11b+, and CD11c+ macrophage subset (Fig. 6, B and C) (3, 22, 38). This decrease in macrophage infiltration was confirmed by immunostaining of adipose tissue slices with MAC2, a macrophage-specific marker (Fig. 6D). The differences in macrophage recruitment were also quantified with QPCR for another macrophage-specific marker, CD68, the chemokine receptors CCR5 and CCR2, and the chemokine CCL5 (Fig. 6E). As can be seen, the expression of all of these inflammatory pathway genes was decreased in the Dex-treated mice (Fig. 6E).
TABLE 1.
Phyiological parameters
Total body weights; eWAT weights; average food, water, and dexamethasone intake; fasting blood glucose levels (FBG); insulin; FFAs; and adiponectin are represented for mice fed normal chow (NCD) or HFD in the absence or presence of Dex.
| NCD | HFD | HFD + Dex | |
|---|---|---|---|
| Total body weight (g) | 29.64 ± 0.66 | 50.35 ± 0.58a | 51.23 ± 1.92a |
| eWAT weight (g) | 0.66 ± 0.11 | 1.17 ± 0.14a | 2.29 ± 0.45ab |
| Food intake (g/mice/day) | 3.54 ± 0.16 | 2.70 ± 0.06a | 2.88 ± 0.02a |
| Water intake (ml/mice/day) | 5.58 ± 0.38 | 3.32 ± 0.14a | 4.06 ± 0.25ac |
| Dex intake (mg/g/day) | 0.10 ± 0.06 | ||
| FBG (m/dl) | 154.66 ± 10.47 | 205 ± 5.18a | 258.16 ± 14.36ab |
| Insulin (ng/ml) | 0.97 ± 0.05 | 9.19 ± 0.7a | 26.16 ± 8.05ab |
| FFA (mmol/liter) | 0.61 ± 0.08 | 0.57 ± 0.06 | 0.65 ± 0.03c |
| Adiponectin (mg/ml) | 19.386 ± 2.89 | 18.48 ± 0.83 | 24.93 ± 1.40b |
ap < 0.01 versus the normal chow group.
b p < 0.01, HFD + Dex group versus HFD group.
c p < 0.05, HFD + Dex group versus HFD group.
FIGURE 6.
In vivo dexamethasone inhibits ATM accumulation. A, adipocyte size areas were calculated from paraffin-embedded epididymal fat tissue sections. Data are presented as the percentage of cells counted falling within the indicated ranges of sizes for mice either fed an HFD or fed an HFD and treated with dexamethasone (HFD + Dex). B, FACS sorting was performed on epididymal fat stromal vascular cells sorted for F4/80, CD11b, and CD11c. Data represent relative cell numbers of the reported macrophage subtypes for HFD versus HFD + Dex mice. C, dot plot representation of CD11b versus CD11c relative expression for FACS data obtained from F4/80 gated macrophages. D, MAC2 immunostaining on paraffin-embedded epididymal fat tissue sections from HFD versus HFD + Dex mice. E, QPCR analysis of CD68, RANTES, CCR2, and CCR5 expression on epididymal fat from normal chow diet, HFD, and HFD + Dex mice. Bars, average values ± S.D. of duplicates from at least three independent experiments. A–D above the bars show statistical groups (p < 0.05). NCD, normal chow diet; A.U., arbitrary units.
Adipose Tissue Corticosterone Concentration and Inflammation
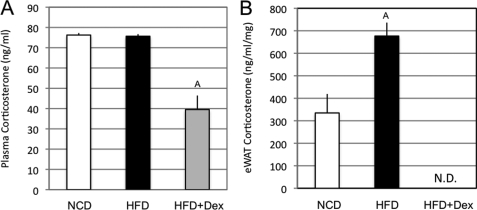
Glucocorticoids have potent diabetogenic properties, independent of their anti-inflammatory actions (39). Indeed, insulin resistance is a known effect of glucocorticoid treatment in humans and rodents. Consistent with this, our Dex-treated mice exhibited elevated glucose and insulin levels (Table 1). The independence of these diabetogenic properties from anti-inflammatory effects was also illustrated in 3T3L1 cells, because, despite interfering with TNFα signaling (Fig. 2, C and D), dexamethasone did not reverse the TNFα-induced cellular insulin resistance (supplemental Fig. 4, A and B). Glucocorticoid actions in target tissues can rely on local production. In various models of insulin resistance, omental fat glucocorticoid concentrations, unlike plasma, are elevated (37, 39, 40). Additionally, proinflammatory cytokines can increase glucocorticoid availability within adipose tissue, whereas anti-inflammatory agents, such as rosiglitazone, T0901317, and liver X receptor agonists lead to decreases (41, 42). In agreement with Masuzaki et al. (39), we observed that plasma corticosterone levels were not modified by HFD (Fig. 7A). However, dexamethasone treatment decreased circulating corticosterone by ∼50% (75.7 ng/ml HFD versus 39.5 ng/ml HFD + Dex), due to negative feedback on the hypothalamic-pituitary-adrenal axis (43). In contrast, in epididymal adipose tissue, the corticosterone concentration was >50% increased on HFD, whereas it was not detected in mice treated with dexamethasone (334.8 versus 676.7 ng/ml/mg protein in normal chow diet versus HFD) (Fig. 7B).
FIGURE 7.
Adipose tissue corticosterone concentration and inflammation. A, corticosterone was directly quantified from serum of mice fed a normal chow (NCD), HFD, or HFD + Dex. B, corticosterone was extracted and quantified from frozen eWAT of mice from the indicated groups. Bars, average values ± S.D. of duplicates from at least three independent experiments. A above the bars shows statistical groups (p < 0.05). N.D., not detected.
DISCUSSION
Chronic tissue inflammation is recognized as a key component of obesity- related insulin resistance (1, 2). It is also known that there is an increased content of macrophages in adipose tissue in obesity and that these proinflammatory macrophages are an important cause of the inflammation- induced insulin resistance (1, 2). Macrophages must undergo migration into tissues before they can accumulate, and the mechanisms underlying chemotaxis of macrophages into adipose tissue are poorly understood. In these studies, we have focused on the process of macrophage migration and, specifically, the ability of adipocytes to stimulate the process of macrophage chemotaxis. In these studies, we found that inflammatory stimuli, such as TNFα and FFAs, promote adipocyte secretion of chemokines into the medium, which, in turn, induces chemotaxis of macrophages. Treatment of adipocytes with anti-inflammatory nuclear hormone ligands suppresses chemokine expression, secretion, and macrophage migration. Among the chemokines studied, RANTES and MIP1α responded most dramatically to proinflammatory and anti-inflammatory signals. Of the receptors tested, GR agonism gave the most robust anti-inflammatory effects, followed by PPARγ activation. These studies directly show that the secretion of chemokines by adipocytes is under the control of pro- and anti-inflammatory signals and demonstrates the powerful effects of GR and PPARγ agonists on this process. In vivo treatment studies in mice confirm the potent effects of dexamethasone to inhibit macrophage accumulation in adipose tissue.
It is of interest that dexamethasone exerts the most potent effects of all compounds tested to inhibit chemokine expression, secretion, and macrophage chemotaxis and markedly restricts macrophage content of adipose tissue in the context of obesity and insulin resistance. If this were the only effect of dexamethasone, one would expect improved glucose tolerance and a reduction in insulin resistance. However, independent of their anti-inflammatory effects, GR agonists have well known prodiabetic effects, mediated through entirely different mechanisms. These diabetogenic effects include stimulation of the gluconeogenic program in hepatocytes as well as decreased insulin signaling and Glut4 expression in adipocytes and muscle cells (44, 45). If a selected GR agonist can be identified, lacking the diabetogenic effects, then such an agent could be a potent insulin-sensitizing compound.
These studies of in vitro chemotaxis clearly demonstrate that both adipocytes and macrophages secrete chemokines into the medium, which stimulates macrophage migration. Since there are multiple chemokines that reciprocally respond to proinflammatory and anti-inflammatory signals, the effects of the different chemokines on chemotaxis are likely to be combinatorial, with no single chemokine essential for the entire CM effects. Since both adipocytes and macrophages secrete chemokines, the results are consistent with a model in which stimuli, such as enlarging adipocytes, microhypoxia, ER stress, etc., activate inflammatory programs within adipocytes, leading to secretion of chemokines, which cause migration of macrophages. In turn, the recruited ATMs then release their own chemokines, which leads to a further increase in macrophage migration and even greater accumulation of ATMs in a feed-forward manner.
As illustrated through the expression of keratinocyte-derived chemokine, a known neutrophil chemoattractant, and RANTES, a macrophage/lymphocyte chemoattractant, adipocytes have the intrinsic property to recruit various leukocytes (i.e. neutrophils, lymphocytes, and macrophages). The early accumulation of neutrophils in eWAT of DIO mice was recently described; however, their involvement in insulin resistance is not established to date. Three recent publications demonstrate an increase in proinflammatory and/or a decrease in anti-inflammatory lymphocytes in eWAT of mice fed a high fat diet (46–48). These lymphocytes were identified in adipose tissue prior to M1 macrophage detection. Furthermore, Nishimura et al. (46) demonstrate that the interplay between lymphocytes and adipocytes leads to the secretion of factors that induce monocyte maturation and recruitment. It might hence be of particular interest to study the role of lymphocyte-specific cytokines, such as interferon-γ, on adipocytes and subsequent chemokine secretion. However, since adipocytes are clearly present before proinflammatory lymphocytes, they are probably the primary agent that relays the signal inducing leukocyte recruitment.
The adipose tissue expansion and adipocyte hypertrophy with obesity is accompanied by the accumulation of ATMs, and some have proposed a role for ATMs in mediating, or at least facilitating, the processes necessary for adipose tissue expansion (49). In the current studies, we find that dexamethasone treatment greatly decreases ATM content at the same time that it stimulates increased fat pad mass. This disconnect between ATM content and adipose tissue expansion is not consistent with a necessary role of ATMs in the processes of adipocyte hypertrophy and expansion, indicating that these events occur independently of macrophages.
Although MCP1 released by adipocytes and macrophages is clearly an important component in macrophage chemotaxis, it is interesting that the CCR5 ligands MIP1α and RANTES respond most robustly to inflammatory signals and are suppressed the most by the anti-inflammatory PPARγ ligand rosiglitazone and GR ligand dexamethasone. This is consistent with recent findings that the CD11c-positive M1-like macrophage subset expresses higher levels of CCR5 than do alternatively activated macrophages (50). The concept that the five chemokines we have measured work in a combinatorial, reinforcing fashion is supported by the fact that MCP1 alone induces substantial macrophage chemotaxis but that neutralization of MCP1 only abrogates a portion (∼40%) of the chemotactic effect of CM. Since the effects of chemokines on macrophage migration are combinatorial, an effective anti-inflammatory therapeutic for inflammation/insulin resistance should broadly inhibit chemokine secretion.
Supplementary Material
Acknowledgments
We thank Arezou Amidi and Jachelle Afrecio for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK033651, DK074868, and T32 DK007494 and by the Eunice Kennedy Shriver NICHD/National Institutes of Health through cooperative agreement U54 HD 012303-25 as part of the specialized Cooperative Centers Program in Reproduction and Infertility Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- eWAT
- epididymal white adipose tissue
- BSA
- bovine serum albumin
- CM
- conditioned medium
- Dex
- dexamethasone
- DMEM
- Dulbecco's modified Eagle's medium
- FFA
- free fatty acid
- GR
- glucocorticoid receptor
- HFD
- high fat diet
- LPS
- lipopolysaccharide
- MR
- mineralocorticoid receptor
- PPAR
- peroxisome proliferator-activated receptor
- RANTES
- regulated upon activation normal T-cell expressed and secreted
- TNFα
- tumor necrosis factor α
- TZD
- thiazolidinedione
- siRNA
- small interfering RNA
- ATM
- adipose tissue macrophage
- QPCR
- quantitative PCR
- FACS
- fluorescence-activated cell sorting.
REFERENCES
- 1.Schenk S., Saberi M., Olefsky J. M. (2008) J. Clin. Invest. 118, 2992–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Luca C., Olefsky J. M. (2008) FEBS Lett. 582, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patsouris D., Li P. P., Thapar D., Chapman J., Olefsky J. M., Neels J. G. (2008) Cell Metab. 8, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. (2004) Trends Immunol. 25, 677–686 [DOI] [PubMed] [Google Scholar]
- 5.Gerard C., Rollins B. J. (2001) Nat. Immunol. 2, 108–115 [DOI] [PubMed] [Google Scholar]
- 6.Harman-Boehm I., Blüher M., Redel H., Sion-Vardy N., Ovadia S., Avinoach E., Shai I., Klöting N., Stumvoll M., Bashan N., Rudich A. (2007) J. Clin. Endocrinol. Metab. 92, 2240–2247 [DOI] [PubMed] [Google Scholar]
- 7.Kamei N., Tobe K., Suzuki R., Ohsugi M., Watanabe T., Kubota N., Ohtsuka-Kowatari N., Kumagai K., Sakamoto K., Kobayashi M., Yamauchi T., Ueki K., Oishi Y., Nishimura S., Manabe I., Hashimoto H., Ohnishi Y., Ogata H., Tokuyama K., Tsunoda M., Ide T., Murakami K., Nagai R., Kadowaki T. (2006) J. Biol. Chem. 281, 26602–26614 [DOI] [PubMed] [Google Scholar]
- 8.Chavey C., Lazennec G., Lagarrigue S., Clapé C., Iankova I., Teyssier J., Annicotte J. S., Schmidt J., Mataki C., Yamamoto H., Sanches R., Guma A., Stich V., Vitkova M., Jardin-Watelet B., Renard E., Strieter R., Tuthill A., Hotamisligil G. S., Vidal-Puig A., Zorzano A., Langin D., Fajas L. (2009) Cell Metab. 9, 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inouye K. E., Shi H., Howard J. K., Daly C. H., Lord G. M., Rollins B. J., Flier J. S. (2007) Diabetes 56, 2242–2250 [DOI] [PubMed] [Google Scholar]
- 10.Kirk E. A., Sagawa Z. K., McDonald T. O., O'Brien K. D., Heinecke J. W. (2008) Diabetes 57, 1254–1261 [DOI] [PubMed] [Google Scholar]
- 11.Neels J. G., Badeanlou L., Hester K. D., Samad F. (2009) J. Biol. Chem. 284, 20692–20698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshizaki T., Milne J. C., Imamura T., Schenk S., Sonoda N., Babendure J. L., Lu J. C., Smith J. J., Jirousek M. R., Olefsky J. M. (2009) Mol. Cell Biol. 29, 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hundal R. S., Petersen K. F., Mayerson A. B., Randhawa P. S., Inzucchi S., Shoelson S. E., Shulman G. I. (2002) J. Clin. Invest. 109, 1321–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J. K., Kim Y. J., Fillmore J. J., Chen Y., Moore I., Lee J., Yuan M., Li Z. W., Karin M., Perret P., Shoelson S. E., Shulman G. I. (2001) J. Clin. Invest. 108, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pascual G., Fong A. L., Ogawa S., Gamliel A., Li A. C., Perissi V., Rose D. W., Willson T. M., Rosenfeld M. G., Glass C. K. (2005) Nature 437, 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glass C. K., Ogawa S. (2006) Nat. Rev. Immunol. 6, 44–55 [DOI] [PubMed] [Google Scholar]
- 17.Lehrke M., Pascual G., Glass C. K., Lazar M. A. (2005) Genes Dev. 19, 1737–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefebvre P., Chinetti G., Fruchart J. C., Staels B. (2006) J. Clin. Invest. 116, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharroubi I., Lee C. H., Hekerman P., Darville M. I., Evans R. M., Eizirik D. L., Cnop M. (2006) Diabetologia 49, 2350–2358 [DOI] [PubMed] [Google Scholar]
- 20.Barish G. D., Narkar V. A., Evans R. M. (2006) J. Clin. Invest. 116, 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu R., Kim C. S., Kwon B. S., Kawada T. (2006) Obesity 14, 1353–1362 [DOI] [PubMed] [Google Scholar]
- 22.Nguyen M. T., Favelyukis S., Nguyen A. K., Reichart D., Scott P. A., Jenn A., Liu-Bryan R., Glass C. K., Neels J. G., Olefsky J. M. (2007) J. Biol. Chem. 282, 35279–35292 [DOI] [PubMed] [Google Scholar]
- 23.Caprio M., Fève B., Claës A., Viengchareun S., Lombès M., Zennaro M. C. (2007) FASEB J. 21, 2185–2194 [DOI] [PubMed] [Google Scholar]
- 24.Solinas G., Vilcu C., Neels J. G., Bandyopadhyay G. K., Luo J. L., Naugler W., Grivennikov S., Wynshaw-Boris A., Scadeng M., Olefsky J. M., Karin M. (2007) Cell Metab. 6, 386–397 [DOI] [PubMed] [Google Scholar]
- 25.Wallerath T., Gödecke A., Molojavyi A., Li H., Schrader J., Förstermann U. (2004) Nitric Oxide 10, 36–41 [DOI] [PubMed] [Google Scholar]
- 26.Gunin A. G., Emelianov V. U., Tolmachev A. S. (2003) Eur. J. Obstet. Gynecol. Reprod. Biol. 107, 62–67 [DOI] [PubMed] [Google Scholar]
- 27.Nguyen M. T., Satoh H., Favelyukis S., Babendure J. L., Imamura T., Sbodio J. I., Zalevsky J., Dahiyat B. I., Chi N. W., Olefsky J. M. (2005) J. Biol. Chem. 280, 35361–35371 [DOI] [PubMed] [Google Scholar]
- 28.Liao W., Nguyen M. T., Yoshizaki T., Favelyukis S., Patsouris D., Imamura T., Verma I. M., Olefsky J. M. (2007) Am. J. Physiol. Endocrinol. Metab. 293, E219–E227 [DOI] [PubMed] [Google Scholar]
- 29.Takaishi H., Taniguchi T., Takahashi A., Ishikawa Y., Yokoyama M. (2003) Biochem. Biophys. Res. Commun. 305, 122–128 [DOI] [PubMed] [Google Scholar]
- 30.Zhang B., Ma Y., Guo H., Sun B., Niu R., Ying G., Zhang N. (2009) Eur. J. Immunol. 39, 894–901 [DOI] [PubMed] [Google Scholar]
- 31.Weiss-Haljiti C., Pasquali C., Ji H., Gillieron C., Chabert C., Curchod M. L., Hirsch E., Ridley A. J., Hooft van Huijsduijnen R., Camps M., Rommel C. (2004) J. Biol. Chem. 279, 43273–43284 [DOI] [PubMed] [Google Scholar]
- 32.Gevrey J. C., Isaac B. M., Cox D. (2005) J. Immunol. 175, 3737–3745 [DOI] [PubMed] [Google Scholar]
- 33.Takahashi K., Yamaguchi S., Shimoyama T., Seki H., Miyokawa K., Katsuta H., Tanaka T., Yoshimoto K., Ohno H., Nagamatsu S., Ishida H. (2008) Am. J. Physiol. Endocrinol. Metab. 294, E898–E909 [DOI] [PubMed] [Google Scholar]
- 34.Jiao P., Chen Q., Shah S., Du J., Tao B., Tzameli I., Yan W., Xu H. (2009) Diabetes 58, 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kassel O., Sancono A., Krätzschmar J., Kreft B., Stassen M., Cato A. C. (2001) EMBO J. 20, 7108–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Shepherd E. G., Nelin L. D. (2007) Nat. Rev. Immunol. 7, 202–212 [DOI] [PubMed] [Google Scholar]
- 37.Macfarlane D. P., Forbes S., Walker B. R. (2008) J. Endocrinol. 197, 189–204 [DOI] [PubMed] [Google Scholar]
- 38.Lumeng C. N., Bodzin J. L., Saltiel A. R. (2007) J. Clin. Invest. 117, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuzaki H., Paterson J., Shinyama H., Morton N. M., Mullins J. J., Seckl J. R., Flier J. S. (2001) Science 294, 2166–2170 [DOI] [PubMed] [Google Scholar]
- 40.Bujalska I. J., Hewitt K. N., Hauton D., Lavery G. G., Tomlinson J. W., Walker E. A., Stewart P. M. (2008) Endocrinology 149, 2584–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stulnig T. M., Oppermann U., Steffensen K. R., Schuster G. U., Gustafsson J. A. (2002) Diabetes 51, 2426–2433 [DOI] [PubMed] [Google Scholar]
- 42.Berger J., Tanen M., Elbrecht A., Hermanowski-Vosatka A., Moller D. E., Wright S. D., Thieringer R. (2001) J. Biol. Chem. 276, 12629–12635 [DOI] [PubMed] [Google Scholar]
- 43.Syed A. A., Weaver J. U. (2005) Obes. Res. 13, 1131–1133 [DOI] [PubMed] [Google Scholar]
- 44.Reynolds R. M., Walker B. R. (2003) Diabetes Obes. Metab. 5, 5–12 [DOI] [PubMed] [Google Scholar]
- 45.Andrews R. C., Walker B. R. (1999) Clin. Sci. 96, 513–523 [DOI] [PubMed] [Google Scholar]
- 46.Nishimura S., Manabe I., Nagasaki M., Eto K., Yamashita H., Ohsugi M., Otsu M., Hara K., Ueki K., Sugiura S., Yoshimura K., Kadowaki T., Nagai R. (2009) Nat. Med. 15, 914–920 [DOI] [PubMed] [Google Scholar]
- 47.Winer S., Chan Y., Paltser G., Truong D., Tsui H., Bahrami J., Dorfman R., Wang Y., Zielenski J., Mastronardi F., Maezawa Y., Drucker D. J., Engleman E., Winer D., Dosch H. M. (2009) Nat. Med. 15, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feuerer M., Herrero L., Cipolletta D., Naaz A., Wong J., Nayer A., Lee J., Goldfine A. B., Benoist C., Shoelson S., Mathis D. (2009) Nat. Med. 15, 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neels J. G., Olefsky J. M. (2006) J. Clin. Invest. 116, 33–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H., Ghosh S., Perrard X. D., Feng L., Garcia G. E., Perrard J. L., Sweeney J. F., Peterson L. E., Chan L., Smith C. W., Ballantyne C. M. (2007) Circulation 115, 1029–1038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.