Abstract
The eukaryotic translation initiation factor eIF4E recognizes the mRNA cap, a key step in translation initiation. Here we have characterized eIF4E from the human parasite Schistosoma mansoni. Schistosome mRNAs have either the typical monomethylguanosine (m7G) or a trimethylguanosine (m2,2,7G) cap derived from spliced leader trans-splicing. Quantitative fluorescence titration analyses demonstrated that schistosome eIF4E has similar binding specificity for both caps. We present the first crystal structure of an eIF4E with similar binding specificity for m7G and m2,2,7G caps. The eIF4E·m7GpppG structure demonstrates that the schistosome protein binds monomethyl cap in a manner similar to that of single specificity eIF4Es and exhibits a structure similar to other known eIF4Es. The structure suggests an alternate orientation of a conserved, key Glu-90 in the cap-binding pocket that may contribute to dual binding specificity and a position for mRNA bound to eIF4E consistent with biochemical data. Comparison of NMR chemical shift perturbations in schistosome eIF4E on binding m7GpppG and m2,2,7GpppG identified key differences between the two complexes. Isothermal titration calorimetry demonstrated significant thermodynamics differences for the binding process with the two caps (m7G versus m2,2,7G). Overall the NMR and isothermal titration calorimetry data suggest the importance of intrinsic conformational flexibility in the schistosome eIF4E that enables binding to m2,2,7G cap.
Introduction
Eukaryotic initiation protein eIF4E2 is an essential translation factor that recognizes the mRNA cap (1–3). Recognition of the mRNA cap by eIF4E is the key and rate-limiting step in mRNA translation. The majority of translation in eukaryotic cells is cap-dependent; that is recruitment of mRNAs to the ribosome for translation is dependent on the interaction between eIF4E and the mRNA cap. eIF4E directly binds to the mRNA cap. However, for productive translation initiation to occur, eIF4E must interact with eIF4G. eIF4G acts as a bridge protein interacting with factors in the 40 S ribosomal subunit that facilitate ribosome recruitment to the mRNA. Increased expression of eIF4E is associated with a variety of cancers and cancer progression (4). Efforts in a number of laboratories are directed toward therapies against eIF4E in cancer, including the development of cap analogs (5–9).
The mRNA cap in most eukaryotes is m7GpppN (where N is A, C, G, or U). The cap contains a 5′–5′ triphosphate bridge with the first guanosine methylated at the N-7 position. However, spliced leader trans-splicing in metazoa adds a different cap to recipient mRNAs, a trimethylguanosine cap, m2,2,7GpppN (see Fig. 1A) (10–14). trans-splicing is present in a variety of parasitic nematodes and flatworms, and these organisms remain a significant health problem in many parts of the world, infecting upward of 2 billion people (15–17). Translation of these trans-spliced mRNAs is thought to require eIF4E recognition of the m2,2,7G cap to facilitate ribosomal recruitment (18–20). Vertebrate eIF4E has very low affinity for the trimethylguanosine cap in comparison with the monomethylguanosine cap, and association of the m2,2,7G cap with mammalian eIF4E seems to destabilize the overall structure of the protein (1, 9, 21). Approximately 70% of mRNAs in the nematode Caenorhabditis elegans undergo trans-splicing. Several isoforms of C. elegans eIF4E and an eIF4E from the parasitic nematode Ascaris suum recognize both monomethyl- and trimethylguanosine caps to a similar extent (19, 22–24). The unique ability of trans-splicing worms to interact efficiently with the m2,2,7G cap compared with vertebrate eIF4E represents a potential drug target against a diverse group of parasitic worms important in human and veterinary medicine.
FIGURE 1.
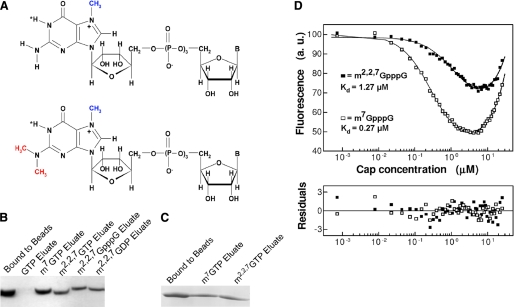
Schistosome eIF4E binds both m7G and m2,2,7G caps. A, chemical structures of m7GpppN (upper) and m2,2,7GpppN caps. The positive charge at the five-membered ring of the 7-methylguanosine moiety is localized at N-7 (67). B, purified, recombinant schistosome eIF4E is eluted from m7GTP-Sepharose with either m7G or m2,2,7G nucleotides. C, recombinant schistosome eIF4E binds to m2,2,7GTP-Sepharose and is eluted with either m7G or m2,2,7G nucleotides. D, quenching of intrinsic fluorescence of schistosome eIF4E on binding m7G or m2,2,7G dinucleotide cap and fitting residuals. The calculated dissociation constants are shown. The increase in fluorescence signal at higher concentrations of the cap analogs originates from the free cap molecules in the solution. The emission of cap has been explicitly taken into account in the numerical analysis. a.u., absorbance units.
Structural studies on mammalian, plant, and yeast eIF4E using NMR (25–29) and crystallography (5, 27, 30–34) have provided insight into the recognition of the m7GpppN cap by eIF4E. The eIF4E core resembles a “cupped hand” within which the cap-binding pocket residues are located. A key component of eIF4E cap-binding involves sandwiched stacking of the methylated guanine ring between two aromatic tryptophan residues at the top and bottom of the cupped hand that holds the guanine in place through interactions between the ring systems. Several other cap-binding proteins also involve stacking of the cap between aromatic residues as part of their mechanism of cap binding (35–37). The N-7 methyl group on the guanosine cap introduces a positive charge to this moiety that greatly enhances the stacking interaction and stabilizes the complex (25, 30). eIF4E does not effectively recognize unmethylated guanosine phosphate and requires more than one phosphate residue on the N-7 methylated guanosine (1). The mRNA cap is further stabilized inside the eIF4E cap-binding pocket by hydrogen bonds derived from the side chain carboxylate of a conserved glutamic acid, a backbone NH of one of the stacking tryptophans, and additional bonds derived from conserved eIF4E residues with the three phosphate groups and the ribose of the cap (30). van der Waals contacts also contribute to cap stabilization including contact from a third conserved tryptophan with the N-7 methyl group.
Despite this understanding of how eIF4E recognizes the monomethyl cap, the mechanism by which eIF4E proteins from trans-splicing organisms recognize and bind the alternate m2,2,7G cap is not known. Here we describe the cloning, expression, and characterization of the sole form of eIF4E in the human parasitic flatworm Schistosoma mansoni. Schistosomes infect ∼200 million people resulting in significant morbidity, and an estimated 800 million people are at risk of infection (16, 17). Schistosome eIF4E exhibits dual cap specificity with similar binding affinity for both m7GpppG and m2,2,7GpppG caps. As a first step in understanding the biochemical and biophysical basis for recognition of the m2,2,7G cap in some forms of eIF4E, we determined the high resolution crystal structure of schistosome eIF4E in complex with m7GpppG or m7GpppA. In addition, we used NMR, fluorescence titration, and isothermal titration calorimetry studies to obtain insights into the ability of schistosome eIF4E to bind the atypical m2,2,7G cap. These studies provide the basis for understanding recognition of the m2,2,7G cap by a diverse group of important parasitic worms and provide general insights into the key cap-binding protein eIF4E.
EXPERIMENTAL PROCEDURES
Cloning of the S. mansoni eIF4E
Sequences encoding the S. mansoni eIF4E were identified in public genomic and expressed sequence tag databases (38, 39). These sequences were used to design primers corresponding to the full eIF4E open reading frame (5′ = ATGACGGCTGTTGAGAGT and 3′ = CTAAATTTCATATTTTCCAGTAC). The primers were used to PCR amplify the open reading frame from cDNA prepared by random priming of adult schistosome RNA (40). The PCR product was initially cloned into a pET-30 Ek/LIC vector (Novagen, Madison, WI), and the sequence was confirmed.
Protein Expression and Purification
Initial protein expression and purification were carried out in RosettaTM 2(DE3) cells induced at 30 °C for 3 h with 0.4 mm isopropyl β-d-thiogalactoside. The protein was purified by nickel-nitrilotriacetic acid-agarose and used for cap analog-Sepharose chromatography as described previously (19). Protein for fluorescent titration studies was prepared from bacterial cultures induced with 0.2 mm isopropyl β-d-thiogalactoside at A600 = 0.7 and incubated overnight at 25 °C. The cells were collected by centrifugation at 4,000 × g, suspended in binding buffer (20 mm Hepes, pH 7.4, 100 mm KCl, 1 mm dithiothreitol, 1 mm EDTA), sonicated, and clarified by centrifugation at 16,000 × g. The supernatant was loaded onto m7GTP-Sepharose resin (GE Healthcare) and rotated at 4 °C for 2 h. The resin was washed with the binding buffer, and eIF4E was eluted with 2 m KCl in binding buffer. The eluted protein was applied to a Superdex 75 (GE Healthcare) column equilibrated in binding buffer, and monomeric protein was collected, analyzed by SDS-PAGE, and concentrated using an Amicon 10,000 molecular weight Ultra Centrifugal Filter Device (Millipore, Billerica, MA).
Cap Analog-Sepharose Assays
Sepharose affinity assays were carried out as described previously (19). Briefly ∼15 μg of protein in binding buffer (described above) was mixed with 20 μl of m7GTP-Sepharose resin (GE Healthcare) or m2,2,7GTP-Sepharose (22) pre-equilibrated in binding buffer. After incubation on a Nutator at 4 °C for 1 h, the resin was washed three times with 100 μl of binding buffer, and the protein was then eluted by addition of 20 μl of binding buffer with 200 μm cap analog. Eluted protein was analyzed by SDS-PAGE.
Fluorescence Titration
The fluorescence titration was performed using a FluoroMax-3 spectrophotometer (Horiba Ltd.) with full-length eIF4E at 20 °C and a protein concentration of 0.4 μm in binding buffer. The excitation wavelength was 280 nm (slit, 1 nm), and the emission wavelength was 335 nm (slit, 2 nm). The temperature was kept at 20 °C with a thermocouple inside a thermostated cuvette, and the sample was stirred magnetically. Assays were performed by addition of 1 μl of increasing concentrations of cap analog to 1400 μl of protein solution using an integration time of 30 s and a gap of 60 s. Dilution during the titration did not exceed 2.5%. Fluorescence intensity was corrected for the inner filter effect, and the emission of the free cap analogs was explicitly included in the numerical analysis. A theoretical curve for the fluorescence intensity as a function of the total ligand concentration was fitted to the experimental data points by means of non-linear, least squares method using Prism 3.02 (GraphPad Software) as described previously (9, 30, 41).
Protein Preparation for Crystallization
Because of the flexibility of the eIF4E N terminus, most eIF4E crystals have been produced and the structures have been solved using N-terminal truncated proteins (25, 27, 31). From eIF4E sequence alignments and limited trypsin digestions, we chose to truncate the schistosome protein by 22 residues at the N terminus to generate schistosome eIF4E-(23-203). This coding region was subcloned into pGEX6P-1 (GE Healthcare) using EcoRI and XhoI restriction sites and then transformed into Escherichia coli strain XA90. Protein expression was induced, the cells were collected and sonicated, and the supernatant was recovered as described above except that phosphate-buffered saline buffer was used. The cell lysate was incubated with Glutathione-Sepharose 4B (GE Healthcare) for 4 h at 4 °C with rotation. The resin was washed with phosphate-buffered saline buffer, and eIF4E was cleaved from the glutathione S-transferase fusion protein by incubation with PreScission Protease at 4 °C overnight. Eluted protein was further purified as described above on a Superdex 75 column.
Crystallization, Data Collection, and Refinement
Schistosome eIF4E-(23–203) was concentrated to 10 mg/ml (Amicon 10,000 molecular weight Ultra Centrifugal Filter Device) and then incubated with 1 mm 4E-BP peptide (SGSGRIIYDRKFLMECRNSPV, corresponding to residues 51–67 of the human 4E-BP1 sequence) and 0.5 mm m7GpppA or m7GpppG cap analogs at 4 °C for 1 h before crystallization setup using the hanging drop vapor diffusion method. Thin platelike crystals developed in 20% polyethylene glycol 4000, 0.2 m MgCl2, 100 mm MOPS, pH 6.0–6.5 at 4 °C within 1 week using 0.5 ml of 750 mm NaCl as the well solution. Before the data collection, the crystal was transferred into the equivalent mother solution containing 30% polyethylene glycol 4000 for 15 min and then flash-cooled in liquid nitrogen. Crystallographic data were collected using the mail-in data collection program at the Advanced Light Source beamline 4.2.2 at the Lawrence Berkley National Laboratory. The structure of schistosome eIF4E-(23-203) was solved by the molecular replacement method using human eIF4E (Protein Data Bank code 2V8W) as a model and the program PHASER in the CCP4 suite (42). Model building and manual refinement were performed in COOT (43) and REFMAC from the CCP4 suite (42), respectively. Figures were generated using Pymol (44).
Accession Numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank for schistosome eIF4E·m7GpppG·4E-BP complex (Protein Data Bank code 3HXI) and schistosome eIF4E·m7GpppA·4E-BP complex (Protein Data Bank code 3HXG).
NMR Sample Preparation
Full-length schistosome eIF4E protein was purified by a refolding method. The induction was done as described above, and the bacterial pellet was dissolved in denaturant solution (20 mm Hepes, pH 7.5, containing 6 m guanidine HCl, 1 mm dithiothreitol, 1 mm EDTA). The protein solution was then diluted 10-fold into refolding buffer (20 mm Hepes, pH 7.5, containing 1 m arginine, 300 mm NaCl, 1 mm dithiothreitol) and incubated at 4 °C for 2 h. Following centrifugation at 16,000 × g, the supernatant was dialyzed overnight against binding buffer and then mixed with m7GTP-Sepharose. Protein was eluted with 2 m KCl and further purified on Superdex 75 using NMR buffer (50 mm pH 7.4 phosphate, 100 mm KCl, 1 mm dithiothreitol, 1 mm EDTA).
Isotopically enriched schistosome eIF4E was prepared by growing cells in minimal M9 medium containing 15NH4Cl and/or [13C6]glucose. To prepare uniformly labeled schistosome eIF4E, host cells were grown in M9 minimum D2O medium supplemented with 1g/liter 2H,15N,13C-labeled ISOGRO® (ISOTECTM, Miamisburg, OH).
NMR Experiments and Data Processing
All NMR spectra were acquired on a 900-MHz Varian spectrometer using 0.6 mm triple labeled sample (2H,15N,13C) at a probe temperature of 25 °C. For the backbone assignment, standard three-dimensional resonance NMR experiments were conducted including HNCACB, HNCOCA, HNCOCACB, 15N NOESY-HSQC, and 15N-HSQC-NOESY-HSQC. For NMR chemical shift perturbation experiments, two-dimensional 15N HSQC were collected using 0.2 mm 15N-labeled sample supplemented with m7GpppG or m2,2,7GpppG cap analogs (45, 46) in a 1:1 ratio, leading to >90% of the protein in complex with cap according to the fluorescence binding constants. NMR data were processed using NMRPipe (43) and analyzed using CcpNmr (47).
Isothermal Titration Calorimetry
The N-terminal truncated protein (25 μm) used for crystallization studies in 20 mm Hepes, pH 7.4, 1 mm EDTA, 100 mm KCl was used in titration experiments carried out at 20 °C using a VP-ITC calorimeter (MicroCal Inc., Northampton, MA). Each titration experiment consisted of a 5-μl injection followed by 29 injections of 10 μl of 700 μm cap analog. All the data were processed using the single binding site model in Origin (Version 7.0, MicroCal Inc.). Control titration experiments were performed and subtracted (cap titrated into buffer) from the titrations for enthalpy changes due to dilution.
RESULTS
Cap Binding Specificity of Schistosome eIF4E
Sequence searches for eIF4E in comprehensive genomic and expressed sequence tag databases identified only a single isoform of eIF4E in S. mansoni or a second schistosome species, Schistosoma japonicum (38, 39, 48, 49). The presence of only a single eIF4E isoform is atypical for metazoa (50, 51). The schistosome eIF4E has 32% identity and 51% similarity with human eIF4E. Notably schistosome eIF4E is more highly divergent from human eIF4E than are nematode eIF4Es. Schistosome mRNAs exhibit two different types of mRNA caps, the typical m7GpppN cap and the m2,2,7GpppA cap added during spliced leader trans-splicing (Fig. 1A) (12). Although the exact percentage of schistosome mRNAs that are trans-spliced is not known, a rough estimate of ∼10% has been proposed (52, 53). Most eIF4E proteins have a lower affinity for trimethylguanosine caps compared with the monomethylguanosine form (30). The presence of only a single eIF4E isoform and the presence of two different mRNA caps in schistosomes prompted us to determine whether the S. mansoni eIF4E was capable of binding both monomethyl- and trimethylguanosine mRNA caps.
We first examined the cap specificity of recombinant schistosome eIF4E using a qualitative assay, cap analog-Sepharose chromatography (19). As shown in Fig. 1, B and C, purified schistosome eIF4E binds both monomethyl- and trimethylguanosine cap. To quantitatively investigate the binding affinity of schistosome eIF4E to cap analogs, a fluorescence titration assay was conducted. The equilibrium dissociation constants calculated for schistosome eIF4E are KD = 0.27 μm for m7GpppG and KD = 1.27 μm for m2,2,7GpppG (Fig. 1D). The affinity of schistosome eIF4E for m7GpppG is similar to that observed for truncated murine and full-length human eIF4E, KD = 0.14 and 0.10 μm, respectively (30). Schistosome eIF4E binds the m2,2,7GpppG cap with a slightly lower affinity (∼5-fold). Schistosome eIF4E binds the m2,2,7GpppA cap (the native schistosome trans-spliced cap) with a similar affinity (data not shown). Notably, previous studies on murine eIF4E have demonstrated a several hundred-fold lower affinity for the m2,2,7G compared with the m7G mononucleotide cap (30, 54). We conclude that the schistosome eIF4E protein binds both m7G and m2,2,7G cap and discriminates less against the m2,2,7G cap compared with other eukaryotic eIF4E proteins, consistent with its biological requirement of binding both caps.
Overall Crystal Structure of Schistosome eIF4E
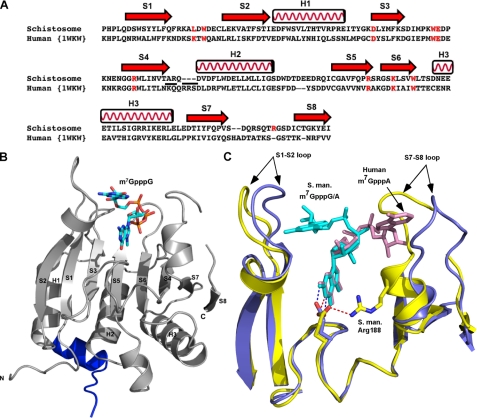
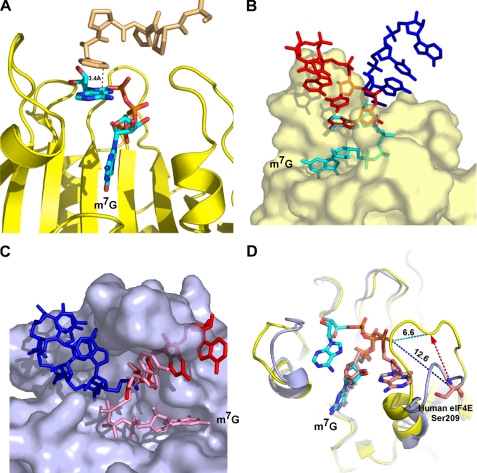
To understand the structural basis for the ability of schistosome eIF4E to bind both the m2,2,7G and m7G caps, we solved the crystal structure of a schistosome eIF4E·m7GpppG·4E-BP1 complex to 1.8-Å resolution (Table 1). The electron density is well defined except for residue Asp-196 and the side chains of Lys-39, Arg-70, Glu-71, Arg-145, Ser-174, and Asp-195. The overall structure of schistosome eIF4E is very similar to that of other eIF4Es (Fig. 2, B and C) (5, 25–27, 30, 33, 34, 55), containing eight β-strands and three α-helices that form a glove-shaped structure with the m7GpppG cap analog situated on the concave surface of the β-strands. The 4E-BP1 peptide binds to the opposite surface of the protein and forms interactions with schistosome eIF4E almost identical to those previously observed in the human eIF4E·4E-BP complex structure (56). Therefore, these are not discussed further here. The 4E-BP peptide was used to enable crystal formation. In comparison with the human eIF4E (Protein Data Bank code 1WKW), the schistosome eIF4E complex structure has a root mean square deviation of 0.66 Å. Although the overall structure of the schistosome and other eIF4Es are similar, there is a marked difference between the schistosome and human eIF4E (Protein Data Bank code 1WKW) in the position of two loops (S1-S2 loop and S7-S8 loop) (Fig. 2C). The different position for these loops appears to be related to the orientation of the second base of the cap in the schistosome eIF4E structure, which is very different in the two proteins (see below).
TABLE 1.
Crystal data and refinement statistics for schistosome eIF4E ternary complex (eIF4E·m7GpppG·4E-BP peptide)
| Data collection | m7GpppG | m7GpppA |
|---|---|---|
| Space group | P21212 | P21212 |
| Cell dimensions | ||
| a (Å) | 45.31 | 45.56 |
| b (Å) | 125.33 | 125.4 |
| c (Å) | 37.33 | 37.34 |
| α, β, γ (°) | α = β = γ = 90° | α = β = γ = 90° |
| Measured reflections | 91,837 | 104,640 |
| Resolutiona | 35.8-1.7 (1.8-1.7) | 41.7-2.0 (2.1-2.0) |
| Rsym or Rmerge | 0.082 (0.472) | 0.113 (0.275) |
| I/σ | 7.8 (2.0) | 6.6 (3.2) |
| Completeness (%) | 94.8 (96.2) | 98.3 (98.1) |
| Redundancy | 4.0 (3.0) | 3.7 (3.7) |
| Refinement | ||
| Resolution (Å) | 1.8 | 2.1 |
| Number of reflections | 17,407 | 13,064 |
| Rwork/Rfree | 23.8/28.9 | 23.6/29.1 |
| Number of atoms | ||
| Protein | 1,516 | 1,495 |
| Peptide | 129 | 123 |
| Ligand | 52 | 51 |
| Water | 112 | 114 |
| B factors (Å2) | ||
| Protein | 22.8 | 19.9 |
| Peptide | 26.4 | 23.3 |
| Ligand | 22.8 | 17.6 |
| Water | 29.8 | 25.6 |
| r.m.s.bdeviations | ||
| Bond length (Å) | 0.011 | 0.009 |
| Bond angles (°) | 1.348 | 1.222 |
| Ramachandran plot | ||
| Residues in most favorable regions (%) | 94.8 | 93.8 |
| Residues in disallowed regions (%) | 0 | 0 |
a Values in parentheses are for the highest resolution shell.
b Root mean square.
FIGURE 2.
Comparison of schistosome eIF4E with human eIF4E shows overall similarity in structure but differences in the orientation of the second base in the cap. A, structure-based sequence alignment of schistosome eIF4E with human eIF4E. The secondary structural elements were assigned based on the schistosome eIF4E structure. Residues involved in cap binding are colored in red. The underlined residues are substitutions and deletions discussed in the text. The alignment was prepared using the combinatorial extension algorithm (68) and an on-line server. B, the overall view of schistosome eIF4E structure. The 4E-BP peptide is colored blue. Three α-helices and eight β-strands are labeled. The m7GpppG cap is shown as sticks. C, superimposition of schistosome and human eIF4E complexes bound to the dinucleotide cap. Note the difference in orientation of the second cap base (marked by arrows) between schistosome eIF4E (yellow) and human eIF4E complex structure (Protein Data Bank code 1WKW) (blue). The S1-S2 and S7-S8 loops and caps are marked. The position of the key residue Glu-90 (schistosome, yellow; human, blue color) is also shown. The colored dashes (schistosome, red; human, blue) denote a hydrogen bond between the N2 amide from the N-7 methylated guanine base and the side chain of Glu. In addition, one hydrogen bond is shown between the side chain of Glu-90 to Nη of Arg-188 in the schistosome eIF4E complex structure. All the pictures were generated using Pymol (44). S. man., S. mansoni.
Schistosome eIF4E Cap-binding Site
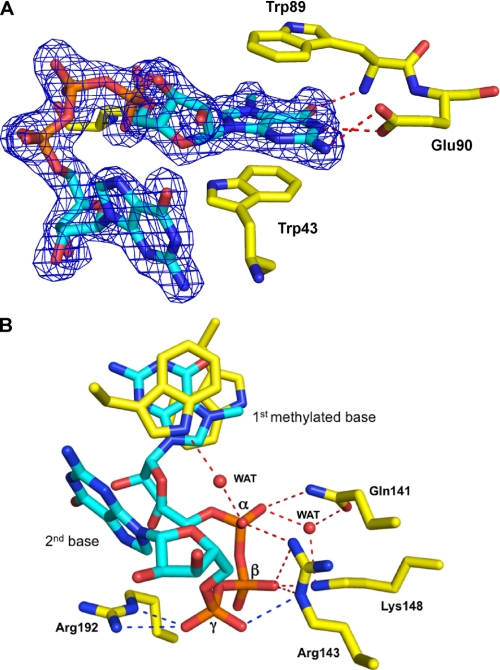
Overall the cap-binding pocket and interactions between the cap and residues in the pockets are very similar between schistosome and mammalian eIF4E. The electron density of the dinucleotide cap analog, m7GpppG, is well defined in our structure (Fig. 3A), allowing its position to be precisely determined. The residues involved in dinucleotide cap binding include Trp-43, Trp-89, Glu-90, Gln-141, Arg-143, Lys-148, and Arg-192. The major features that contribute to the interaction of the schistosome eIF4E with the cap include the following (Fig. 3 and supplemental Fig. 1): 1) cation-π stacking of the positive charged m7G moiety with two highly conserved aromatic Trp residues (Trp-43 and Trp-89) (Fig. 3A and supplemental Fig. 1); 2) two hydrogen bonds between the N-1 hydrogen and N2 hydrogen of the m7G moiety and the carboxyl group of conserved Glu-90 and one more between O6 of m7G and the backbone amide nitrogen of Trp-89 (Fig. 3A and supplemental Fig. 1); and 3) interaction between the three phosphates in the cap dinucleotide and residues (Gln-141, Arg-143, Lys-148, and Arg-192) that constitute the phosphate access slot as illustrated in Fig. 3B and supplemental Fig. 1. The presence of the N-1 H–O bond (2.93 Å) suggests that the cationic form of the m7G moiety is preferred. In the complex of m7GpppG, N2 hydrogen of the second base forms a weak hydrogen bond (3.1 Å) with the backbone oxygen of Leu-41. This bond is not present in the complex with m7GpppA because of the lack of a hydrogen donor in adenosine at a suitable position. This is consistent with a slightly higher affinity of mammalian eIF4E for m7GpppG versus m7GpppA and m7GpppC (30, 57). All of the cap-binding residues, except Leu-41 that interacts with the cap only through the backbone, are highly conserved in the eIF4E protein family (Fig. 5E).
FIGURE 3.
Cap density and interactions with schistosome eIF4E residues. A, the electron density map of m7GpppG cap bound to schistosome eIF4E illustrating stacking interactions with the first cap base (contour level at 1σ). B, cap-binding pocket of schistosome eIF4E in complex with m7GpppG illustrating the hydrogen bonding interactions with the cap phosphates. Two important water (WAT) molecules shown as spheres have interactions within the three phosphates in the cap-binding pocket. Colored dashed lines denote hydrogen bonding interactions, and unique schistosome hydrogen bonds are colored blue.
FIGURE 5.
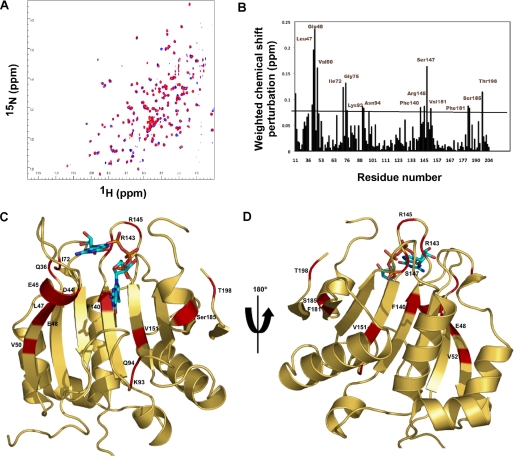
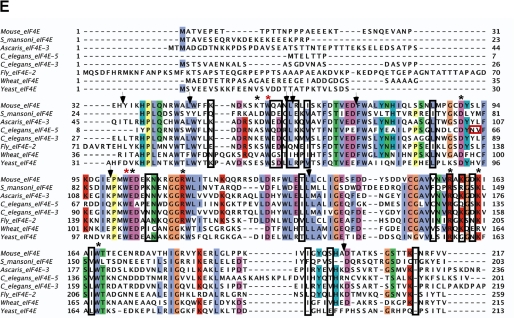
NMR-determined chemical shift perturbations on schistosome eIF4E binding to m2,2,7GpppG compared with m7GpppG. A, superposition of the 1H-15N HSQC spectrum of the schistosome eIF4E bound to m7GpppG (red) and bound to m2,2,7GpppG (blue). B, specific chemical shift perturbations on binding m2,2,7GpppG compared with m7GpppG. The reported chemical shift Δδ represents the weighted chemical shift by applying the Pythagorean theorem: Δδ{1H,15N} = {Δδ(1H)2 + 0.2 × Δδ(15N)2}½ where Δδ(1H) and Δδ(15N) are the chemical shift difference of the amide proton and nitrogen, respectively (28, 69). Amino acid residues with large chemical shifts are labeled. C and D, residues with major chemical shift perturbation on binding m2,2,7G compared with the m7G cap mapped onto the crystal structure of the m7GpppG·eIF4E complex. The backbone residues with major chemical shifts are colored red. Residues with major chemical shifts in the vicinity of the second base of the cap are shown in C, and those with major chemical shifts in the vicinity of the third phosphate of the cap are shown in D. E, Clustal sequence alignment of several eIF4E illustrating conserved residues, residues undergoing CSP, and mouse pepsin cleavage sites on cap binding. The black boxes denote residues with major chemical shift perturbation in our NMR experiments as described above. The vertical black arrows denote the specific pepsin cleavage sites based on the mammalian eIF4E (21). Starred residues are those involved in cap binding with the two key Trp residues in red. The red box indicates two key residues for C. elegans eIF4E reported to be important for m2,2,7G cap binding specificity (24). Note that C. elegans eIF4E-5 and Ascaris eIF4E-3 binds both types of caps, whereas mouse eIF4E3 and C. elegans eIF4E-3 are specific for m7G cap.
One observed difference in the geometry of the interatomic contacts within the schistosome eIF4E cap-binding center that might contribute to the similar specificity for the m7G and m2,2,7G forms of the mRNA 5′ cap is the variant conformation of the carboxylate of a conserved Glu-90. This carboxylate is turned by ∼80° compared with the orthologous Glu-103 carboxylate in the mammalian eIF4E (Fig. 2C). The result of this rotation is that Glu-90 forms only one hydrogen bond with m7G of the cap, whereas in human eIF4E, the equivalent residue Glu-103 forms two hydrogen bonds with both N-1 and N2 of the cap m7G. This almost perpendicular position of the schistosome Glu-90 carboxylate versus mammalian Glu-103 results from the interaction with Nη of Arg-188 (bond lengths of 2.97 and 3.29 Å in the m7GpppG and m7GpppA complexes, respectively) fixing the position of the Glu. In human eIF4E, there is no equivalent interaction as this region of the protein is moved out away from Glu-103 due to the positioning of the second base (Fig. 2C) (see below). Mutation of Glu-90 (Ala, Asp, or Arg) results in protein oligomerization suggesting that this residue contributes structurally to the apo form of the protein. Consequently direct analysis of the contribution of this residue to cap discrimination is difficult. The conformation of this conserved Glu and its contribution to m7G/m2,2,7G specificity in other eIF4E isoforms remains an open question.
Position of the Second Base of the Bound Cap Analog
Although the positions of the m7G moiety, the ribose, and the first phosphate group of the cap as well as the conserved eIF4E tryptophans align well between schistosome and mammalian eIF4E (Fig. 2C), the orientation of the second base in the schistosome structure differs from that observed in the mammalian eIF4E structure (Fig. 2C). In previous eIF4E structures with a dinucleotide cap, the second base of the cap was only well resolved in the human eIF4E complex structure (22, 23) apparently due to the flexibility of this nucleotide. In the human structure, the second base (m7GpppA) points to the C-terminal flexible loop and has some hydrogen bonding with the residues located in this loop. However, in the schistosome eIF4E complex structure, the orientation of the second base (m7GpppG) points to the N-terminal S1-S2 loop as shown in Fig. 2C. To test whether the orientation of the second base in the two structures was a consequence of the different purines at the second base (A versus G), the structure of schistosome eIF4E in complex with m7GpppA was also determined (supplemental Fig. 2). In the m7GpppA structure, the location of the second base also points to the N-terminal S1-S2 loop as observed for the m7GpppG complex.
The position of the second base in the schistosome eIF4E structure allows stacking of the base (A or G) onto a Phe (derived from vector sequence) from a symmetry-related molecule in the crystal as shown in Fig. 4A. This Phe may mimic the third base in an RNA and stabilize the position of the second base in our structure. The cap γ-phosphate in the schistosome eIF4E structure forms three hydrogen bonds with the side chain of Arg-143 and Arg-192 (Fig. 3B). These additional hydrogen bonds to the γ-phosphate in the schistosome eIF4E structure are consistent with the higher eIF4E binding affinity observed for m7GTP compared with m7GDP in mammalian eIF4E (30). Furthermore mammalian eIF4E binds the dinucleotide m7GpppG cap with lower affinity (∼20-fold) compared with m7GTP. The schistosome eIF4E has a similar affinity for the mono- and dinucleotide cap. Overall these data suggest that the second base is not likely to have extensive interactions with eIF4E protein and might suggest that the main interaction for the second base would be stacking with the third base of the RNA.
FIGURE 4.
Proposed mRNA path based on eIF4E complex crystal structure. A, a Phe derived from vector sequence in the symmetry-related molecule stacks with the second base, mimicking the third base of the RNA. The tan color denotes the model from a neighboring molecule. B, potential RNA paths based on the schistosome eIF4E complex crystal structure. C, potential RNA paths based on the human eIF4E complex crystal structure (Protein Data Bank code 1WKW). The two RNA orientations (red and blue) are based on a C-3 or C-2 endo ribose conformation with the extended RNAs in an idealized A-form conformation. D, illustration of potential distance change of human Ser-209 from the γ-phosphate of the cap based on the location of the schistosome S7-S8 loop. The red dashed arrow represents the movement of the loop based on the schistosome structure with the new location of the Ser-209 at the arrowhead. The dashed dark blue line represents the 12.6-Å distance of Ser-209 to the phosphate, and the light blue line represents the 6.6-Å distance to the phosphate with the altered location of the S7-S8 loop (yellow, schistosome; blue, human eIF4E).
Consequences of the Position of the Second Base
The orientation of the second cap base is associated with differences in the positions of the two loops (S1-S2 loop and S7-S8 loop) between the schistosome and human eIF4E structures. Specifically in the human structure loop S7-S8 is swung away from the rest of the protein due to the position of the second base of the cap (Fig. 2C). In the schistosome structure, the second base of the cap is positioned in a nearly opposite orientation that allows S7-S8 to move closer to the cap-binding pocket in a “clamp”-like position. Loop S1-S2 in the schistosome structure also shifts slightly (compared with the human structure) to accommodate the position of the second base. The orientation of the second base and its influence on loop S7-S8 likely does not play a role in cap binding specificity but would impact the path of the RNA.
To explore possible pathways that the mRNA may take, we modeled additional bases into the schistosome and human structures (Fig. 4, B and C). Although the structure of this part of the mRNA in different mRNAs may vary and is not known, we modeled the RNA using canonical A-form base stacking, the favored conformation for RNA, even in the single-stranded state (58). When the RNA is modeled onto the human structure so that the third base stacks directly on the second, the predicted RNA has significant steric clash with the protein in loop S7-S8 and the adjacent helix. Manual manipulation of the modeled mRNA revealed a position without steric clash, but this position required severe contortion of the RNA backbone between the second and third bases (Fig. 4C, blue RNA). In contrast, when the mRNA was modeled on the schistosome eIF4E structure so that the third base stacks directly on the second base, the path of the sugar phosphate backbone roughly parallels loop S1-S2. A single steric clash is seen between Phe-17 and the RNA. A second modeled position for the mRNA on the schistosome eIF4E structure in which the third base does not stack directly on the second base also allowed the mRNA to exit the protein without any steric clash (Fig. 4B). Both of these modeled positions on schistosome eIF4E are compatible with our structure and illustrate minimal direct interaction of the RNA with eIF4E. This is consistent with a variety of studies suggesting that eIF4E does not interact specifically with the body of the mRNA (59).
Importantly the position of the second base in the schistosome eIF4E structure is also consistent with data related to the phosphorylation of Ser-209 in mammalian eIF4E. Phosphorylation of Ser-209 can reduce eIF4E cap binding affinity (57, 60–62), suggesting that electrostatic repulsion between the cap phosphate and phosphorylated Ser-209 might reduce cap affinity (62). Ser-209 is located in a very flexible loop (S7-S8) in the eIF4E structure. In the human structure (Protein Data Bank code 1WKW), the distance between the Cα of Ser-209 to the cap phosphate is ∼12 Å, a distance likely too far for a significant repulsion interaction (Fig. 4D) (61). However, in the schistosome eIF4E structure, this flexible loop (S7-S8) is closer to the cap-binding pocket (6.6 Å; Fig. 4D) compared with that in the human eIF4E structure (Protein Data Bank code 1WKW). Thus, the human Ser-209 would be close enough to the phosphate backbone for repulsive electrostatic interactions to occur with the cap that could lead to a reduction in eIF4E affinity for the cap (Fig. 4D).
Chemical Shift Perturbation Associated with Different Caps
The determinants that enable some eIF4E proteins to have similar binding affinity for both m2,2,7G and m7G caps (schistosome and some nematode eIF4Es) whereas others have high selectivity for only m7G (mammalian, yeast, plant, etc.) remain unknown. The overall crystal structure and cap binding of schistosome eIF4E for m7GpppG is similar to that observed for other eIF4E proteins that have low affinity for the m2,2,7G cap. Thus, the general mechanism of cap binding and overall structure of the schistosome protein bound to m7GpppG is not different from other monospecific eIF4Es. However, modeling the m2,2,7G cap into the schistosome eIF4E structure indicates that the two additional methyl groups at N2 would likely lead to steric hindrance with the key Glu-90 residue. Our concerted efforts to obtain schistosome eIF4E crystals with a m2,2,7G cap were not successful. One possible reason for this is that the N2 of m7G cap forms crystal packing contacts with Asp-84 in a symmetry-related molecule. Thus, the methylation of N2 would likely disrupt crystal packing.
Therefore, to further explore the mechanism of schistosome m2,2,7G cap binding specificity, we undertook NMR chemical shift perturbation studies to compare schistosome eIF4E binding to the m7GpppG versus m2,2,7GpppG cap. NMR experiments could not be done for free protein because the free schistosome eIF4E protein was not stable at the requisite high concentration for NMR experiments. Superimposition of the 1H-15N HSQC spectrum of m7GpppG-bound (red) versus m2,2,7GpppG-bound (blue) schistosome eIF4E is shown in Fig. 5A, and a summary of the induced chemical shift perturbations with the different caps is provided in Fig. 5B. Surprisingly there are ∼15 residues with significant chemical shift perturbations (CSPs) that differ on schistosome eIF4E binding to the m2,2,7G versus the m7G cap (Fig. 5B). The only difference between the caps is the addition of two methyl groups at the N2 position (Fig. 1A), yet around 10% of the schistosome residues show major chemical shift differences between the m7G and m2,2,7G complexes. Residues with major CSPs were mapped onto the schistosome eIF4E crystal structure (Fig. 5, C–E). Most of these residues are distributed around the cap-binding pocket (Fig. 5C; Gln-36, Asp-44, Glu-45, Leu-47, Glu-48, Val-50, Lys-93, Asn-94, Phe-140, Arg-143, Arg-145, Ser-147, and Ser-185). The residues Gln-36, Asp-44, Glu-45, Leu-47, Glu-48, and Val-50, which are around Trp-43 and located in the S2 strands, also undergo major CSPs in the trimethylguanosine compared with the m7G cap-bound complex. Based on the cap binding mechanism recently suggested by Volpon et al. (28), this region is predicted to move like a hinge to lock the capped guanine initially and then hold the m7G moiety in the appropriate position to form the interaction with Trp-56. This suggests that this region plays a crucial role for enabling the m7G cap to enter the binding pocket. Significant CSPs for residues in this region were observed on binding m7GpppG compared with m2,2,7GpppG as shown in Fig. 5. One interpretation of these differences could be that the path or mechanisms through which the m7G and m2,2,7G caps enter into the binding pocket have discrete differences. Several residues (Ile-72, Leu-117, Val-151, Phe-181, and Thr-198) with major CSPs are present in a region distal from the cap-binding pocket (Fig. 5, C and D) indicating that long distance conformational changes also occur upon m2,2,7G cap binding. Overall these data suggest that some features of the overall binding mechanism and specific interactions within the binding pocket for each cap are different.
Isothermal Titration Calorimetry (ITC) Characterization of Schistosome eIF4E Interaction with Cap
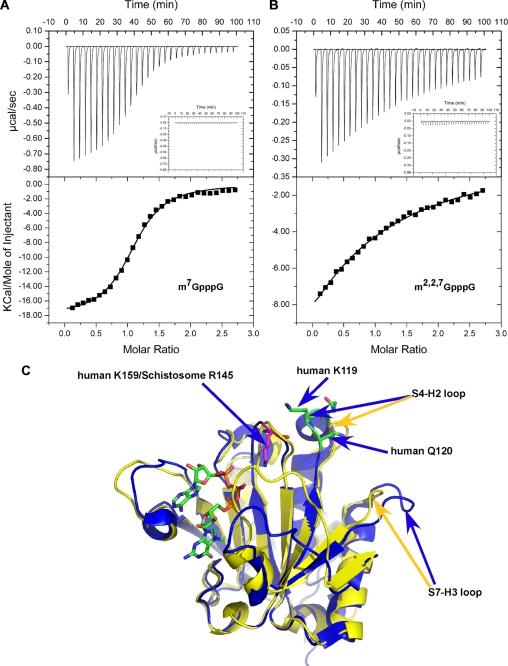
To probe thermodynamic parameters for schistosome eIF4E binding to the two caps, ITC was carried out. Binding affinity and thermodynamics parameters for schistosome eIF4E binding to m7G and m2,2,7G caps are shown in Table 2 and Fig. 6, A and B. The standard molar enthalpy change observed on binding m7GpppG and m2,2,7GpppG is similar for schistosome eIF4E (Table 2). The main enthalpy contributions to cap binding are from the cation-π stacking, hydrogen bonding (except for those involving the N2 amino group), and interactions of the phosphates with the positively charged residues. The similar enthalpy changes on binding the two caps do not account for the difference in binding free energy and binding affinity observed. The binding affinity is likely to be influenced by the different entropic changes observed suggesting that there are potential differences in the overall binding mechanism. The eIF4E cap binding affinity differences observed using ITC and fluorescence titration (Fig. 1D and Table 2) is likely due to methodological differences (63).
TABLE 2.
Summary of isothermal titration calorimetry measurements
| Ligand | KD | ΔH | TΔS | ΔG |
|---|---|---|---|---|
| μm | kcal/mol | kcal/mol | kcal/mol | |
| m7GTP | 0.58 ± 0.18 | −17.8 ± 0.1 | 9.4 ± 0.1 | 8.4 ± 0.2 |
| m2,2,7GTP | 11.2 ± 2.1 | −19.2 ± 0.3 | 12.5 ± 0.8 | 6.72 ± 0.06 |
| m7GpppG | 0.47 ± 0.01 | −16.8 ± 0.1 | 8.3 ± 0.3 | 8.48 ± 0.01 |
| m2,2,7GpppG | 13.5 ± 0.5 | −17.0 ± 1.0 | 10.5 ± 0.8 | 6.56 ± 0.03 |
FIGURE 6.
ITC analysis of schistosome eIF4E cap binding and conformational differences between schistosome and human eIF4E. A, m7GpppG titration. B, m2,2,7GpppG titration. The top panels show the raw data, and the bottom panels show the integrated data with the continuous lines representing the fit of the data to a single site binding model. The insets in the upper panels represent buffer injection alone. Similar values were obtained with m7GTP and m2,2,7GTP. C, structural comparison of human (blue) and schistosome (yellow) eIF4Es around H2-S4 loop.
DISCUSSION
Binding Specificity of Schistosome eIF4E and Translation
Cap analog-Sepharose chromatography demonstrates that schistosome eIF4E can bind both m7G and m2,2,7G caps. The fluorescent titration analyses demonstrate that 1) schistosome eIF4E recognizes both the m7G and m2,2,7G caps with similar affinity and that 2) the schistosome affinity for m7G cap is comparable to that of mammalian eIF4E (Fig. 1). However, the affinity of mammalian eIF4E for the m2,2,7GTP cap is several hundred-fold less than for the m7G (30). Thus, schistosome eIF4E has relatively high affinity for the m2,2,7G cap. Unlike other trans-splicing organisms examined (22, 23, 50, 64), only a single eIF4E isoform is identifiable in schistosomes. This suggests that this single eIF4E likely initiates translation for both non-trans-spliced (m7G) and trans-spliced mRNAs (m2,2,7G).
Overall Crystal Structure of Schistosome eIF4E·m7GpppG/A
Although the schistosome eIF4E has limited sequence identity with other eIF4Es, the overall structure in complex with either m7GpppG or m7GpppA is very similar to that observed for other eukaryotic eIF4Es (Fig. 2, B and C). The mode of binding m7G and the interactions within the cap-binding pocket are essentially identical to that observed for other forms of eIF4E (5, 25, 26, 28, 30, 33) (Fig. 3, A and B). One exception is different orientation of the conserved Glu-90 carboxylate in the cap-binding pocket that is stabilized by Arg-188. It remains to be determined whether this configuration and the orientation of the Glu-90 carboxylate play a role in the binding specificity.
Position of the Second Cap Base and RNA
A major difference in the schistosome eIF4E structure compared with human eIF4E bound to the dinucleotide m7GpppA is the position of two loops, S1-S2 and S7-S8 (Figs. 2C and 6C). The position of these loops appears to be a function of the different location of the second nucleotide in the cap in the schistosome compared with the human eIF4E structure (Fig. 2C). Several aspects of the position of the second nucleotide in the schistosome eIF4E structure are consistent with other data. Modeling of a longer RNA into the schistosome eIF4E structure indicates that the position of the second nucleotide leads to an unobstructed RNA path leaving the schistosome cap-binding pocket (Fig. 4B). In contrast, similar modeling of the human structure leads to steric hindrance with the protein or a highly contorted RNA (Fig. 4C).
The schistosome structure is also consistent with data showing that phosphorylation of Ser-209 in mammalian eIF4E can reduce the affinity of eIF4E for the cap (57, 60–62). In addition, mutation of Ser-209 to Glu decreases cap binding affinity of mammalian eIF4E (62), which is consistent with our structure. Although the role of Ser-209 phosphorylation remains unclear in eIF4E function and translation, the schistosome eIF4E crystal structure provides a reasonable model to explain how electrostatic repulsion between the human Ser-209 and cap might affect cap binding. In sum, the orientation of the second cap base in the schistosome structure is consistent with a likely RNA path and some functional data on cap binding affinity.
Comparison of Schistosome Binding to m7G Versus m2,2,7G Cap
Given that the structure and apparent mechanism of schistosome eIF4E binding to m7G cap is strikingly similar to mammalian eIF4E binding to m7G cap, what then enables the schistosome protein to bind to the structurally different m2,2,7G cap? To begin to address this, we used the NMR CSP technique to compare schistosome eIF4E binding to the m7G and m2,2,7G caps. The major CSP differences observed on binding the two different caps can be divided into three categories based on their location in the schistosome eIF4E complex structure with m7G cap: 1) residues in the vicinity of the two key Trp-43 and Trp-89 aromatics involved in stacking the first guanine of the cap (Fig. 5C), 2) residues in the vicinity of the phosphates in the cap-binding region of the protein (Fig. 5D), and 3) residues that are at a significant distance from the cap-binding pocket. Whereas residues in the first two categories likely are involved in direct interactions with the two different caps, residues at a significant distance from the cap-binding pocket may be involved in allosteric conformational and/or stability changes of the protein as a whole on binding the two different caps. These conformational change differences observed by NMR throughout the protein may reflect a difference in conformational dynamics, which contributes to differences in binding affinity as suggested by the thermodynamics studies (Fig. 6).
Recent studies have shown that murine eIF4E interaction with m2,2,7G versus m7G cap leads to differences in pepsin susceptibility within the protein (21). The appearance of new cleavage sites in the vicinity of Trp-56 (murine eIF4E) on interaction with m2,2,7G cap is consistent with our NMR chemical shift data (Fig. 5E). These data may indicate that the m2,2,7G moiety can enter the cap-binding slot even in mammalian eIF4E but that productive binding does not ensue (Fig. 5E). However, we also observed significant chemical shift perturbations for the residues located around Trp-102 and residues in the region that interact with the phosphates (Fig. 5, C and D). These perturbations differ from those observed in the pepsin cleavage studies on mammalian eIF4E. These differences are likely due to the selective binding affinity for the m2,2,7G cap and may suggest that the m2,2,7G cap does not have free access to the whole cap-binding slot in murine eIF4E.
The new schistosome m7G crystal structure described here suggests that the ability of this protein to bind both caps with similar affinity is not a result of major differences in the overall cap binding mechanism. Thus, all residues involved in binding the monomethyl cap are identical between mammalian and schistosome eIF4E. Therefore, based on the available data, we favor a model in which intrinsic protein flexibility or conformational changes likely play a crucial role in m2,2,7G cap binding. This hypothesis is supported by NMR and in part by our ITC data. Using data from our NMR chemical shift perturbation studies, mammalian eIF4E pepsin cleavage data on binding different cap analogs (21), and the mammalian eIF4E free protein structure (28), we suggest the following m2,2,7G cap binding mechanism model. When the schistosome eIF4E associates with the m2,2,7G cap, the stacking interaction between the guanine ring and the Trp (Trp-56 in human) induces a specific protein conformational change. This conformational change specifically promotes formation of a m2,2,7G cap-binding pocket, analogous to an “induced fit” mechanism. Thus, residues far away from the cap-binding pocket are also observed to have significant chemical shift perturbations. In mammalian eIF4E, association and interaction with the m2,2,7G cap may not lead to the requisite conformational changes and formation of the necessary m2,2,7G cap-binding pocket. As a consequence, mammalian eIF4E has a much lower binding affinity for m2,2,7G cap compared with m7G cap. This model suggests that intrinsic and specific conformational flexibility of the schistosome eIF4E plays the crucial role for m2,2,7G cap binding.
Steric hindrance of the cap due to the additional two methyl groups at the N2 position of the m2,2,7G cap has been suggested previously to likely be important in the reduced affinity of mammalian eIF4E in binding m2,2,7G cap (9, 21). Previous mutagenesis studies for C. elegans eIF4E-5, a form of eIF4E that can bind both types of caps, showed that two residue changes (N64Y/V65L) (Fig. 5E) led to a decrease in cap specificity for the m2,2,7G cap (24). It was suggested that these two residues could impact the width and depth of cap-binding slot. However, another nematode eIF4E (Ascaris eIF4E-3) has these Tyr-64/Leu-65 residues and readily binds the m2,2,7G cap (19). Furthermore our NMR chemical shift perturbation data show that more than 15 residues have major chemical shifts as shown in Fig. 5, A and B, indicating that relatively significant intrinsic conformational changes throughout the molecule are also likely to play a role in cap binding specificity rather than simply the dimensions of the cap-binding slot and steric considerations due to the additional methyl groups in the m2,2,7G cap.
Other Insights from the Schistosome eIF4E Structure on Cap Binding
The addition of a second base to an N-7 methylated nucleotide in general does not significantly increase mammalian eIF4E affinity for the cap (9). In fact, human eIF4E binding affinity for dinucleotide triphosphate cap (m7GpppG) is significantly lower than that observed for a mononucleotide (m7GTP) (9, 45) Interestingly, the schistosome eIF4E binding affinity for mono- and dinucleotide substrates is similar as determined by ITC (Table 2 and data not shown). Comparison of the schistosome and human 4E sequences (Fig. 2A) shows that two residues (Lys-119 and Gln-120) located in the S4-H2 loop are replaced by Ala-86 and Arg-87 in schistosome eIF4E. Mutation of Lys-119 or Gln-120 in the human eIF4E was reported to cause an increase in eIF4E binding affinity to cap analogs (65). In particular, the K119A mutation leads to a 10-fold increase in affinity for dinucleotide cap, whereas only a 3-fold increase is observed for a mononucleotide cap (66). An NMR structural analysis showed that the mutations in this region led to changes in the neighboring S5-S6 loop that directly interacts with the cap phosphates (28). From these data, Volpon et al. (28) suggested that the reduced interaction between these two loops (H2-S4 and S5-S6) might lead to greater conformational flexibility in the H2-S4 leading to the observed increase in cap binding.
The substitution of two residues (K119A/Q120R) and absence of three more residues in the schistosome (RRS) (Figs. 2A and 5E) H2-S4 loop results in a shorter H2 helix compared with human eIF4E. In addition, the schistosome H2-S4 loop is shifted away from the S5-S6 loop, and the S7-H3 loop is shifted inward (Fig. 6C). As a consequence, the interaction between these two loops might be stronger in the schistosome eIF4E enabling the S5-S6 loop to interact more directly with the cap phosphates. This may explain why the schistosome affinity for mono- and dinucleotide caps is similar. These changes may enable dinucleotide caps to more readily enter into the cap-binding slot in schistosome eIF4E. This might also explain why we were able to obtain a relatively high quality schistosome crystal with a dinucleotide cap.
Conclusion
We have identified and characterized the cap binding characteristics of the sole isoform of schistosome eIF4E. Several lines of data indicate that schistosome eIF4E is able to bind both m7G and m2,2,7G caps with similar affinity. We have determined the co-crystal structure of eIF4E with m7GpppG and m7GpppA. The schistosome eIF4E cap binding mechanism for m7G cap is very similar to that described for mammalian eIF4E. Our data demonstrate one potential difference in the cap-binding pocket and suggest a likely position for the RNA when the cap is bound by eIF4E consistent with biochemical data. We have also identified that significant conformational changes occur in eIF4E on binding m2,2,7G cap that provide insight into schistosome eIF4E binding to m2,2,7G compared with m7G cap. Interestingly analysis of schistosome eIF4E binding to m7G and m2,2,7G caps demonstrates significant thermodynamic differences. Although a detailed molecular mechanism for binding m2,2,7G cap is not yet available, our data indicate that major conformational differences occur on schistosome binding the two types of cap, and aspects of the binding mechanisms for each cap are likely to be different.
Mammalian eIF4E affinity for the m2,2,7G cap is several hundred-fold lower than observed for schistosome eIF4E (30, 54). The large difference in affinity for the m2,2,7G cap between schistosome and mammalian eIF4E suggests that this protein may represent a potential target for rational drug design against schistosomes (16, 17). Nematodes also have trans-splicing and eIF4E proteins with high affinity for the m2,2,7G cap. Nematodes infect upward of 2 billion people and are important agricultural pests (15, 17). A better understanding of the mechanisms of parasite eIF4E binding of the m2,2,7G cap might enable development of new compounds that are efficacious against a broad spectrum of important parasites.
Supplementary Material
Acknowledgments
We thank all members of the structural biology community at the University of Colorado Denver School of Medicine, University of Colorado Denver x-ray and NMR facility (supported in part by the University of Colorado Cancer Center), and Shaun Bevers at the University of Colorado Denver biophysical core without which this work would not have been possible. We thank Jay Nix at the Advanced Light Source at the Lawrence Berkeley National laboratory for eIF4E x-ray data collection and Ryszard Stolarski, Elan Eissenmesser, and members of the Davis laboratory for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI049558 and AI080805 (to R. E. D.), GM081346 and GM072560 (to J. K.), and GM080334 (to R. Z.). This work was also supported by Howard Hughes Medical Institute Grant 55005604 (to E. D.), Polish Ministry of Science and Higher Education Grant N301-035936 (to A. N. and E. D.), Warsaw University Grant BW 68/179203 (to M. J.-A.), and a National Science Support Project Grant PBZ-MNiSW-07/I/2007 (to E. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (codes 3HXI and 3HXG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- eIF4E
- eukaryotic initiation factor 4E
- m7G
- 7-methylguanosine
- m2,2,7G
- N2,N2,7-methylguanosine
- 4E-BP
- 4E-binding protein
- ITC
- isothermal titration calorimetry
- HSQC
- heteronuclear single quantum correlation
- MOPS
- 4-morpholinepropanesulfonic acid
- NOESY
- nuclear Overhauser effect spectroscopy
- CSP
- chemical shift perturbation.
REFERENCES
- 1.Gingras A. C., Raught B., Sonenberg N. (1999) Annu. Rev. Biochem. 68, 913–963 [DOI] [PubMed] [Google Scholar]
- 2.Pestova T. V., Lorsch J. R., Hellen C. U. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B., eds) pp. 87–128, Cold Spring Harbor Laboratory Press, Woodbury, NY [Google Scholar]
- 3.von der Haar T., Gross J. D., Wagner G., McCarthy J. E. (2004) Nat. Struct. Mol. Biol. 11, 503–511 [DOI] [PubMed] [Google Scholar]
- 4.Schneider R. J., Sonenberg N. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B., eds) pp. 401–432, Cold Spring Harbor Laboratory Press, Woodbury, NY [Google Scholar]
- 5.Brown C. J., McNae I., Fischer P. M., Walkinshaw M. D. (2007) J. Mol. Biol. 372, 7–15 [DOI] [PubMed] [Google Scholar]
- 6.Moerke N. J., Aktas H., Chen H., Cantel S., Reibarkh M. Y., Fahmy A., Gross J. D., Degterev A., Yuan J., Chorev M., Halperin J. A., Wagner G. (2007) Cell 128, 257–267 [DOI] [PubMed] [Google Scholar]
- 7.Pelletier J., Peltz S. W. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B., eds) pp 855–895, Cold Spring Harbor Laboratory Press, Woodbury, NY [Google Scholar]
- 8.Ghosh B., Benyumov A. O., Ghosh P., Jia Y., Avdulov S., Dahlberg P. S., Peterson M., Smith K., Polunovsky V. A., Bitterman P. B., Wagner C. R. (2009) ACS Chem. Biol. 4, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niedzwiecka A., Darzynkiewicz E., Stolarski R. (2004) Biochemistry 43, 13305–13317 [DOI] [PubMed] [Google Scholar]
- 10.Brehm K., Jensen K., Frosch M. (2000) J. Biol. Chem. 275, 38311–38318 [DOI] [PubMed] [Google Scholar]
- 11.Davis R. E., Singh H., Botka C., Hardwick C., Ashraf el Meanawy M., Villanueva J. (1994) J. Biol. Chem. 269, 20026–20030 [PubMed] [Google Scholar]
- 12.Rajkovic A., Davis R. E., Simonsen J. N., Rottman F. M. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8879–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maroney P. A., Hannon G. J., Nilsen T. W. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liou R. F., Blumenthal T. (1990) Mol. Cell. Biol. 10, 1764–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bethony J., Brooker S., Albonico M., Geiger S. M., Loukas A., Diemert D., Hotez P. J. (2006) Lancet 367, 1521–1532 [DOI] [PubMed] [Google Scholar]
- 16.Savioli L., Albonico M., Engels D., Montresor A. (2004) Parasitol. Int. 53, 103–113 [DOI] [PubMed] [Google Scholar]
- 17.Hotez P. J. (2008) Forgotten People, Forgotten Diseases: the Neglected Tropical Diseases and Their Impact on Global Health and Development, American Society for Microbiology (ASM) Press, Herndon, VA [Google Scholar]
- 18.Cheng G., Cohen L., Mikhli C., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., Davis R. E. (2007) Mol. Biochem. Parasitol. 153, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lall S., Friedman C. C., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., Davis R. E. (2004) J. Biol. Chem. 279, 45573–45585 [DOI] [PubMed] [Google Scholar]
- 20.Maroney P. A., Denker J. A., Darzynkiewicz E., Laneve R., Nilsen T. W. (1995) RNA 1, 714–723 [PMC free article] [PubMed] [Google Scholar]
- 21.Rutkowska-Wlodarczyk I., Stepinski J., Dadlez M., Darzynkiewicz E., Stolarski R., Niedzwiecka A. (2008) Biochemistry 47, 2710–2720 [DOI] [PubMed] [Google Scholar]
- 22.Jankowska-Anyszka M., Lamphear B. J., Aamodt E. J., Harrington T., Darzynkiewicz E., Stolarski R., Rhoads R. E. (1998) J. Biol. Chem. 273, 10538–10542 [DOI] [PubMed] [Google Scholar]
- 23.Keiper B. D., Lamphear B. J., Deshpande A. M., Jankowska-Anyszka M., Aamodt E. J., Blumenthal T., Rhoads R. E. (2000) J. Biol. Chem. 275, 10590–10596 [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi H., Dwyer D. S., Keiper B. D., Jankowska-Anyszka M., Darzynkiewicz E., Rhoads R. E. (2002) EMBO J. 21, 4680–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K. (1997) Cell 89, 951–961 [DOI] [PubMed] [Google Scholar]
- 26.Matsuo H., Li H., McGuire A. M., Fletcher C. M., Gingras A. C., Sonenberg N., Wagner G. (1997) Nat. Struct. Biol. 4, 717–724 [DOI] [PubMed] [Google Scholar]
- 27.Monzingo A. F., Dhaliwal S., Dutt-Chaudhuri A., Lyon A., Sadow J. H., Hoffman D. W., Robertus J. D., Browning K. S. (2007) Plant Physiol. 143, 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volpon L., Osborne M. J., Topisirovic I., Siddiqui N., Borden K. L. (2006) EMBO J. 25, 5138–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von der Haar T., Oku Y., Ptushkina M., Moerke N., Wagner G., Gross J. D., McCarthy J. E. (2006) J. Mol. Biol. 356, 982–992 [DOI] [PubMed] [Google Scholar]
- 30.Niedzwiecka A., Marcotrigiano J., Stepinski J., Jankowska-Anyszka M., Wyslouch-Cieszynska A., Dadlez M., Gingras A. C., Mak P., Darzynkiewicz E., Sonenberg N., Burley S. K., Stolarski R. (2002) J. Mol. Biol. 319, 615–635 [DOI] [PubMed] [Google Scholar]
- 31.Rosettani P., Knapp S., Vismara M. G., Rusconi L., Cameron A. D. (2007) J. Mol. Biol. 368, 691–705 [DOI] [PubMed] [Google Scholar]
- 32.Tomoo K., Matsushita Y., Fujisaki H., Abiko F., Shen X., Taniguchi T., Miyagawa H., Kitamura K., Miura K., Ishida T. (2005) Biochim. Biophys. Acta 1753, 191–208 [DOI] [PubMed] [Google Scholar]
- 33.Tomoo K., Shen X., Okabe K., Nozoe Y., Fukuhara S., Morino S., Ishida T., Taniguchi T., Hasegawa H., Terashima A., Sasaki M., Katsuya Y., Kitamura K., Miyoshi H., Ishikawa M., Miura K. (2002) Biochem. J. 362, 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomoo K., Shen X., Okabe K., Nozoe Y., Fukuhara S., Morino S., Sasaki M., Taniguchi T., Miyagawa H., Kitamura K., Miura K., Ishida T. (2003) J. Mol. Biol. 328, 365–383 [DOI] [PubMed] [Google Scholar]
- 35.Fechter P., Brownlee G. G. (2005) J. Gen. Virol. 86, 1239–1249 [DOI] [PubMed] [Google Scholar]
- 36.Gu M., Lima C. D. (2005) Curr. Opin. Struct. Biol. 15, 99–106 [DOI] [PubMed] [Google Scholar]
- 37.Quiocho F. A., Hu G., Gershon P. D. (2000) Curr. Opin. Struct. Biol. 10, 78–86 [DOI] [PubMed] [Google Scholar]
- 38.Berriman M., Haas B. J., LoVerde P. T., Wilson R. A., Dillon G. P., Cerqueira G. C., Mashiyama S. T., Al-Lazikani B., Andrade L. F., Ashton P. D., Aslett M. A., Bartholomeu D. C., Blandin G., Caffrey C. R., Coghlan A., Coulson R., Day T. A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J. A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D. A., Lacerda D., Macedo C. D., McVeigh P., Ning Z., Oliveira G., Overington J. P., Parkhill J., Pertea M., Pierce R. J., Protasio A. V., Quail M. A., Rajandream M. A., Rogers J., Sajid M., Salzberg S. L., Stanke M., Tivey A. R., White O., Williams D. L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C. M., Barrell B. G., El-Sayed N. M. (2009) Nature 460, 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verjovski-Almeida S., DeMarco R., Martins E. A., Guimarães P. E., Ojopi E. P., Paquola A. C., Piazza J. P., Nishiyama M. Y., Jr., Kitajima J. P., Adamson R. E., Ashton P. D., Bonaldo M. F., Coulson P. S., Dillon G. P., Farias L. P., Gregorio S. P., Ho P. L., Leite R. A., Malaquias L. C., Marques R. C., Miyasato P. A., Nascimento A. L., Ohlweiler F. P., Reis E. M., Ribeiro M. A., Sá R. G., Stukart G. C., Soares M. B., Gargioni C., Kawano T., Rodrigues V., Madeira A. M., Wilson R. A., Menck C. F., Setubal J. C., Leite L. C., Dias-Neto E. (2003) Nat. Genet. 35, 148–157 [DOI] [PubMed] [Google Scholar]
- 40.Cohen L. S., Mikhli C., Jiao X., Kiledjian M., Kunkel G., Davis R. E. (2005) Mol. Cell. Biol. 25, 8779–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niedzwiecka A., Stepinski J., Antosiewicz J. M., Darzynkiewicz E., Stolarski R. (2007) Methods Enzymol. 430, 209–245 [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 43.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 44.Delano W. L. (2002) The Pymol Molecular Graphics System, Delano Scientific, San Carlos, CA [Google Scholar]
- 45.Stepinski J., Bretner M., Jankowska M., Felczak K., Stolarski R., Wieczorek Z., Cai A. L., Rhoads R. E., Temeriusz A., Haber D., Darzynkiewicz E. (1995) Nucleosides Nucleotides 14, 717–721 [Google Scholar]
- 46.Jankowska M., Stepinski J., Stolarski R., Wieczorek Z., Temeriusz A., Haber D., Darzynkiewicz E. (1996) Collect. Czech. Chem. Commun. 61, S197–S202 [Google Scholar]
- 47.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 48.Liu F., Zhou Y., Wang Z. Q., Lu G., Zheng H., Brindley P. J., McManus D. P., Blair D., Zhang Q. H., Zhong Y., Wang S., Han Z. G., Chen Z. (2009) Nature 460, 345–35119606140 [Google Scholar]
- 49.Hu W., Yan Q., Shen D. K., Liu F., Zhu Z. D., Song H. D., Xu X. R., Wang Z. J., Rong Y. P., Zeng L. C., Wu J., Zhang X., Wang J. J., Xu X. N., Wang S. Y., Fu G., Zhang X. L., Wang Z. Q., Brindley P. J., McManus D. P., Xue C. L., Feng Z., Chen Z., Han Z. G. (2003) Nat. Genet. 35, 139–147 [DOI] [PubMed] [Google Scholar]
- 50.Joshi B., Lee K., Maeder D. L., Jagus R. (2005) BMC Evol. Biol. 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhoads R. E. (2009) J. Biol. Chem. 284, 16711–16715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davis R. E. (1996) Parasitol. Today 12, 33–40 [DOI] [PubMed] [Google Scholar]
- 53.Davis R. E., Hardwick C., Tavernier P., Hodgson S., Singh H. (1995) J. Biol. Chem. 270, 21813–21819 [DOI] [PubMed] [Google Scholar]
- 54.Yoffe Y., Zuberek J., Lerer A., Lewdorowicz M., Stepinski J., Altmann M., Darzynkiewicz E., Shapira M. (2006) Eukaryot. Cell 5, 1969–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross J. D., Moerke N. J., von der Haar T., Lugovskoy A. A., Sachs A. B., McCarthy J. E., Wagner G. (2003) Cell 115, 739–750 [DOI] [PubMed] [Google Scholar]
- 56.Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K. (1999) Mol. Cell 3, 707–716 [DOI] [PubMed] [Google Scholar]
- 57.Zuberek J., Wyslouch-Cieszynska A., Niedzwiecka A., Dadlez M., Stepinski J., Augustyniak W., Gingras A. C., Zhang Z., Burley S. K., Sonenberg N., Stolarski R., Darzynkiewicz E. (2003) RNA 9, 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seol Y., Skinner G. M., Visscher K., Buhot A., Halperin A. (2007) Phys. Rev. Lett. 98, 158103. [DOI] [PubMed] [Google Scholar]
- 59.Magee J., Warwicker J. (2005) Nucleic Acids Res. 33, 6694–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheper G. C., van Kollenburg B., Hu J., Luo Y., Goss D. J., Proud C. G. (2002) J. Biol. Chem. 277, 3303–3309 [DOI] [PubMed] [Google Scholar]
- 61.Sonenberg N., Dever T. E. (2003) Curr. Opin. Struct. Biol. 13, 56–63 [DOI] [PubMed] [Google Scholar]
- 62.Zuberek J., Jemielity J., Jablonowska A., Stepinski J., Dadlez M., Stolarski R., Darzynkiewicz E. (2004) Biochemistry 43, 5370–5379 [DOI] [PubMed] [Google Scholar]
- 63.Haq I., Ladbury J. E., Chowdhry B. Z., Jenkins T. C., Chaires J. B. (1997) J. Mol. Biol. 271, 244–257 [DOI] [PubMed] [Google Scholar]
- 64.Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., Allen J. E., Delcher A. L., Guiliano D. B., Miranda-Saavedra D., Angiuoli S. V., Creasy T., Amedeo P., Haas B., El-Sayed N. M., Wortman J. R., Feldblyum T., Tallon L., Schatz M., Shumway M., Koo H., Salzberg S. L., Schobel S., Pertea M., Pop M., White O., Barton G. J., Carlow C. K., Crawford M. J., Daub J., Dimmic M. W., Estes C. F., Foster J. M., Ganatra M., Gregory W. F., Johnson N. M., Jin J., Komuniecki R., Korf I., Kumar S., Laney S., Li B. W., Li W., Lindblom T. H., Lustigman S., Ma D., Maina C. V., Martin D. M., McCarter J. P., McReynolds L., Mitreva M., Nutman T. B., Parkinson J., Peregrín-Alvarez J. M., Poole C., Ren Q., Saunders L., Sluder A. E., Smith K., Stanke M., Unnasch T. R., Ware J., Wei A. D., Weil G., Williams D. J., Zhang Y., Williams S. A., Fraser-Liggett C., Slatko B., Blaxter M. L., Scott A. L. (2007) Science 317, 1756–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spivak-Kroizman T., Friedland D. E., De Staercke C., Gernert K. M., Goss D. J., Hagedorn C. H. (2002) FEBS Lett. 516, 9–14 [DOI] [PubMed] [Google Scholar]
- 66.Friedland D. E., Wooten W. N., LaVoy J. E., Hagedorn C. H., Goss D. J. (2005) Biochemistry 44, 4546–4550 [DOI] [PubMed] [Google Scholar]
- 67.Ruszczynska K., Kamienska-Trela K., Wojcik J., Stepinski J., Darzynkiewicz E., Stolarski R. (2003) Biophys. J. 85, 1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shindyalov I. N., Bourne P. E. (1998) Protein Eng. 11, 739–747 [DOI] [PubMed] [Google Scholar]
- 69.Grzesiek S., Stahl S. J., Wingfield P. T., Bax A. (1996) Biochemistry 35, 10256–10261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.