Abstract
Solar ultraviolet (UV) A radiation is a well known trigger of signaling responses in human skin fibroblasts. One important consequence of this stress response is the increased expression of matrix metalloproteinase-1 (MMP-1), which causes extracellular protein degradation and thereby contributes to photoaging of human skin. In the present study we identify the proteasome as an integral part of the UVA-induced, intracellular signaling cascade in human dermal fibroblasts. UVA-induced singlet oxygen formation was accompanied by protein oxidation, the cross-linking of oxidized proteins, and an inhibition of the proteasomal system. This proteasomal inhibition subsequently led to an accumulation of c-Jun and phosphorylated c-Jun and activation of activator protein-1, i.e. transcription factors known to control MMP-1 expression. Increased transcription factor activation was also observed if the proteasome was inhibited by cross-linked proteins or lactacystin, indicating a general mechanism. Most importantly, inhibition of the proteasome was of functional relevance for UVA-induced MMP-1 expression, because overexpression of the proteasome or the protein repair enzyme methionine sulfoxide reductase prevented the UVA-induced induction of MMP-1. These studies show that an environmentally relevant stimulus can trigger a signaling pathway, which links intracellular and extracellular protein degradation. They also identify the proteasome as an integral part of the UVA stress response.
Solar ultraviolet A (UVA; 320–400 nm) radiation is a well known trigger of signaling responses in human dermal fibroblasts in vitro as well as in vivo in human skin (1–3). One important consequence of this stress response is the increased expression of matrix metalloproteinase-1 (MMP-1),8 which causes extracellular protein degradation and thereby contributes to photoaging of human skin. In fact, UVA-induced MMP-1 expression and the resulting increased degradation of collagen fibers, which occurs primarily in the upper part of the dermis, is thought to be a major reason for wrinkle formation in photoaged human skin (4–6).
The specific signaling steps involved in UVA radiation-induced MMP-1 expression have been extensively examined in recent years. These studies have identified UVA radiation-induced singlet oxygen formation as the initiating event (for review, see Ref. 7) that triggers a cascade of downstream steps which critically involve the activation of the transcription factor complex AP-1 and the subsequent increase in expression of MMP-1. This signaling model, however, includes a black box, because the precise signaling steps linking singlet oxygen with AP-1 activation are largely unknown.
In this regard it is of interest that skin fibroblasts in the upper part of the dermis, i.e. exactly the compartment where collagen degradation takes place, contain increased amounts of oxidized proteins (8). The functional relevance of protein oxidation in human skin is not known. Under normal conditions oxidized proteins are being degraded by the proteasome. The proteasome is located in the cytosol, nucleus, and attached to the endoplasmic reticulum and the cell membrane. The main body of this system is called 20 S “core” proteasome which is regulated by several peptides but also known to be able to degrade proteins without any regulator. It has been shown by many studies that proteasome activity decreases in aging tissues. A major reason for proteasome activity inhibition is protein aggregate formation which can occur as a consequence of protein oxidation.
In the present study we show that UVA radiation, through a singlet oxygen-mediated mechanism, causes protein oxidation and attenuation of proteasome activity in human skin fibroblasts. This then leads to the decreased degradation of intracellular proteins including constituents of the transcription factor complex AP-1 and ultimately to increased MMP-1 transcription. Our studies identify the proteasome as an integral part of the UVA stress response and link intracellular and extracellular protein degradation.
EXPERIMENTAL PROCEDURES
Cell Culture
Human foreskin fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% fetal calf serum in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. All experiments were performed with cells of passages 22–26. Cell viability was determined using MTT (3-(4,5- dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide) reduction by viable cells.
UVA Irradiation and Treatments of the Human Dermal Fibroblast Cells
The UVA irradiation source was a TL15W/05 lamp that emitted an energy spectrum in the UVA region (320–400 nm). The emitted dose was calculated using a UV-MAT dosimeter system. Before UVA irradiation, cells were washed twice with PBS to remove all medium, and during irradiation cells were held in PBS and maintained at 37 °C in a thermostatically controlled water bath. Control samples were subjected to the same conditions without irradiation. After irradiation, fresh medium was added, and the cells were incubated for a further 3 h before analysis. According to viability test results, 0–60 J/cm2 of UVA irradiation were chosen for the experiments. On average, 50 J/cm2 of UVA typically corresponds to one minimal erythemal dose in human skin (9). For lactacystin experiments cells were incubated with 20 μm lactacystin alone or with protein kinase inhibitors: 80 μm JNKi I (cell-permeable, Ref. 10), 100 μm JNKi II, SP600125 (11), 100 μm IκB kinase inhibitor (cell-permeable; Calbiochem) for 24 h. Chemical generation of 1O2 was achieved by incubation of cells with disodium 3,3′-(1,4-naphthylidene) dipropionate-1,4-endoperoxide (NDPO2) in PBS. NDPO2 was synthesized from NDP (Molecular Probes, Eugene, OR) by oxidation with H2O2/molybdate. Control experiments were performed with solutions of predecomposed (heated) NDPO2 containing the decomposition product, NDP.
In Vivo Irradiation Protocol
Healthy volunteers were irradiated at a dose of 120 J/cm2 in the buttock area. Biopsies were taken 1 h after irradiation from the irradiated area and, for comparison, from the contralateral nonirradiated area. The studies were approved by the ethical committee of the Heinrich-Heine-University, Düsseldorf. Biopsies were split into half and snap-frozen and stored at −80 °C until analysis. One part was used for immunostaining, whereas the other part of the biopsy was used for biochemical analysis.
Protein Carbonyl Measurement by Enzyme-linked Immunosorbent Assay
Protein carbonyl content was taken as a measure for protein oxidation. Carbonyls were determined in cell lysates (1 mg/ml in lysis buffer with 1 mm butylated hydroxytoluene) by an enzyme-linked immunosorbent assay as introduced by Buss et al. (12). Primary anti-DNP-rabbit-IgG antiserum (Sigma) and a secondary monoclonal anti-rabbit-peroxidase-conjugated IgG (Sigma) were used as detection system. Development was performed with o-phenylene diamine and H2O2. Absorbance was measured at 492 nm using a microplate reader (BioTek Instruments EL 340).
Proteasome Activity Analysis by Fluorometry
Cells were lysed in 1 mm dithiothreitol by vigorous shaking for 1 h at 4 °C. The lysates were centrifuged at 14,000 × g for 30 min to remove nonlysed cells, membranes, and nuclei. Supernatants were incubated in 225 mm Tris buffer (pH 7.8) containing 7.5 mm MgOAc, 7.5 mm MgCl2, 45 mm KCl, and 1 mm dithiothreitol. The fluorogenic peptide succinyl-LLVY-methyl coumarin was used as substrate at a concentration of 200 μm to measure chymotrypsin-like activity of the proteasome. After 30 min of incubation at 37 °C, methyl coumarin liberation was measured with fluorescence reader (360 nm excitation/485 nm emission) and calculated using free methyl coumarin as standards. To exclude other protease activities the selective proteasome inhibitor lactacystin with the final concentration of 20 μm was used in the reaction, and proteasome activity was calculated as the difference between the total activity and the remaining activity in the presence of lactacystin. For measurements in the isolated form of the proteasome, purification was made as described (13, 14).
Protein Carbonyl and Proteasome Content Observation by Immunohistochemistry in Tissue Samples
Immunostaining was performed on 5-μm-thick sections. Fixation was done as previously described (15). The following antibodies were used: rabbit polyclonal anti-DNP (Sigma, diluted 1:200) for protein carbonyls and rabbit polyclonal antibody to human 20 S proteasome (Biomol, diluted 1:100). For 4′,6-diamidino-2-phenylindole staining, 1 mg/ml of solution was used. Observations were made by fluorescence microscopy using an Olympus BX-60 transmission fluorescence microscope running standard software.
Quantitative PCR Analysis
Cells were lysed, and total RNA was isolated using the Qiagen RNeasy Mini kit following the manufacturer's protocol. The amount and purity of the extracted RNA were determined via spectrophotometry in a Smartspec 3000 (Bio-Rad). An iScript cDNA synthesis kit and 1 μg of RNA were used for cDNA synthesis. Quantitative real-time PCR was performed using a Bio-Rad iCycler 3.0 and the Bio-Rad IQ SYBR Green reaction mixture. The sequences of primers used in this work were as follows: human MMP-1 forward, CTGCTTACGAATTTGCCGAC; human MMP-1 reverse, GCAGCATCGATATGCTTCAC; human TIMP-1 forward, GATGGACTCTTGCACATCACT; human TIMP-1 reverse, TGGATAAACAGGGAAACACTG. As the housekeeping gene for relative mRNA quantification, 18 S rRNA was chosen; human 18 S rRNA forward, GGACATCTAAGGGCATCACA; human 18 S rRNA reverse, GGACATCTAAGGGCATCACA.
Immunoblot Analysis
Cells were lysed at 4 °C using 10 mm Tris HCl (pH 7.5) buffer containing 1 mm Pefabloc, 0.9% Nonidet P-40, 0.1% SDS. The protein concentrations of the supernatants were determined according to Lowry (16). 30 μg of total protein in reducing Laemmli buffer (0.25 M Tris (pH 6.8), 8% SDS, 40% glycerol, 0.03% bromphenol blue) were denatured at 95 °C for 5 min and applied to SDS-PAGE of 12% (w/v) acrylamide followed by electrophoresis and blotting onto nitrocellulose membrane according to standard procedures. Immunodetections were performed with the following antibodies at dilutions recommended by the suppliers: rabbit polyclonal c-Jun antibody, mouse monoclonal anti-phospho- (Pi-) c-Jun antibody raised against amino acids 56–69 of human c-Jun (Santa Cruz Biotechnology; Santa Cruz, CA), mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase antibody (Novus Biologicals). After exposure to peroxidase-coupled secondary antibodies (Calbiochem), membranes were developed using Lumi-Light Western blotting substrate (Cell Signaling).
AP-1 Binding Activity Analysis by Fluorescence Microscopy
The modulation of JNK/AP1 pathway was observed using Cignal AP1 Reporter Assay kit GFP (Superarray) according to the manufacturer's protocol. After reverse transfection of the cells for 16 h, cells were treated with 30 J/cm2 UVA or 20 μm lactacystin. GFP expression was analyzed via fluorescent microscopy using standard fluorescein isothiocyanate filters (excitation of 470 ± 20 nm and an emission filter of 515 nm).
Protein Aggregates and Proteolysis Measurement by Liquid Scintillation Counting
Cells were incubated with [35S]methionine/cysteine at the end activity of 100 μCi/ml in methionine and cysteine-free minimal essential Eagle's medium at 37 °C for 16 h. After washing non-incorporated medium with PBS, cells were irradiated with 30J/cm2 and incubated for 3 h in medium. For proteolysis measurement at the end of incubation, the medium was mixed with the equal volume of 20% trichloroacetic acid. Scintillation counting was performed with the acid-soluble supernatant after centrifugation at 14,000 × g for 15 min at 4 °C. The acid-soluble counts were calculated as (acid soluble sample counts/incorporated counts) × 100. For protein aggregate measurement, detergent solubility was taken as a measure. After collecting the medium for proteolysis measurement, cells were scraped, and pellets were resuspended in a detergent solution consisting of 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS in 10 mm Tris-HCl, 1 mm EDTA (pH 8) (17). After cell lysis at 4 °C for 15 min, samples were centrifuged at 13,000 × g for 10 min. Supernatants were counted as detergent-soluble proteins, and pellets, counted as detergent-insoluble proteins, were dissolved in 1 n NaOH and subjected to scintillation counting.
Statistics
Statistical analysis was performed using Prism 4 (GraphPad) software. For determination of statistical significances of differences, Student's t test and one-way ANOVA were performed followed by multiple comparisons using the Bonferroni's multiple comparison test. A p value of less than 0.05 was selected as the level of significance.
RESULTS
UVA Treatment Increases Protein Carbonyl Formation and Inhibits Proteasome Activity but Does Not Change Proteasome Content
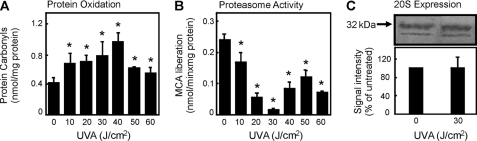
As an indicator of oxidized proteins, the amount of protein-bound carbonyls were measured after UVA treatment according to Buss et al. (12) with modifications by Voss et al. (18). The use of protein carbonyl groups as a biomarker of oxidative stress is widely accepted because of the relatively early formation and the stability of carbonylated proteins (19). The protein carbonyl content of cells was enhanced with increasing doses of UVA (Fig. 1A).
FIGURE 1.
Effect of UVA treatment on protein carbonyl formation and proteasome activity in dermal fibroblast cells. Cells were cultured and exposed to UVA as described under “Experimental Procedures.” Three hours later the cells were harvested, lysed, and analyzed for the protein carbonyl levels (A) by enzyme-linked immunosorbent assay (data are the means ± S.E.; n = 4; *, p < 0.05 versus nonirradiated, ANOVA, Bonferroni's multiple comparison test) or the 20 S proteasome activity (B) (data are the means ± S.E.; n = 4; *, p < 0.05 versus nonirradiated, ANOVA, Bonferroni's Multiple Comparison test). The same lysates were analyzed for the proteasome content (C) by an immunoblot (see the upper panel) and quantified amount (see the lower panel). Immunodetection of proteasomal subunits was by employing a polyclonal anti-proteasome antibody. MCA, 7-amino-4-methylcoumarin.
Protein oxidation appears to be lower at higher doses of UVA (50 and 60 J/cm2). More specifically, the amount of soluble protein carbonyls is lower. We believe that this is because of a masking of carbonyl moieties from detection via DNP hydrazine by reaction of carbonyls with several other oxidized components of the cells and by an enhanced formation of aggregated proteins.
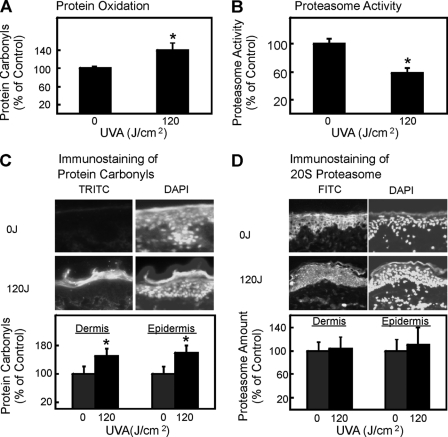
It was shown in earlier studies (20, 21) that the proteasome plays a major role in the degradation of oxidatively modified proteins. To test whether after UVA irradiation the proteasome is able to fulfill this task, we measured the proteasomal activity in fibroblast lysates after UVA treatment using the fluorogenic peptide succinyl-LLVY-methyl coumarin. Surprisingly, a clear decay of the proteasomal activity was detected, reaching a minimum at a dose of 30 J/cm2 (Fig. 1B). To further test whether this effect is because of an inhibition of the proteasome or due to a reduction of the presence of the proteasome, we determined the proteasome content by immunoblot. No change in the expression of the 20 S core proteasome subunits was observed (Fig. 1C). Therefore, we concluded that the reduction of proteasomal activity after UVA irradiation was because of inhibition of its catalytic activity. In this line we tested whether such effects also occur in vivo. Therefore, we next investigated protein carbonyls, proteasome activity, and proteasome content in sections of human skin biopsies exposed to UVA. Protein carbonyl content was increased, and proteasome activity was inhibited after UVA treatment (Fig. 2). Proteasome content and distribution was not changed because of UVA treatment (Fig. 2). 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to show the nuclei of the cells. In summary, we could see a clear increase of protein carbonyl formation in UVA-exposed human skin and no decline in proteasome amount (Fig. 2).
FIGURE 2.
Effect of UVA treatment on protein carbonyl formation and proteasome content in vivo. Human skin was UVA exposed in vivo, and biopsies were taken 1 h after irradiation or sham treatment. Biopsies were split into half. One part was used for biochemical analysis. After homogenization in PBS, the homogenate was analyzed by enzyme-linked immunosorbent assay for protein oxidation (A) or proteasome activity (B) (data are the means ± S.E.; controls in one experiment were set to 100%. n = 4; *, p < 0.05 versus nonirradiated, Student's t test). The other part was cut into 5-μm-thick sections and used for histochemical analysis. The protein carbonyl formation was measured after DNP hydrazine reaction using a rabbit polyclonal anti-DNP (dinitrophenyl) primary antibody and TRITC-labeled secondary antibody (C), whereas the proteasome content was detected using a rabbit polyclonal anti-20 S primary antibody and fluorescein isothiocyanate (FITC)-labeled secondary antibody (D). In both panels 4′,6-diamidino-2-phenylindole (DAPI) staining was used to identify cell nuclei. Quantification was done separately for the epidermal and dermal layers of the skin (data are the means ± S.E.; n = 4; *, p < 0.05 versus nonirradiated, ANOVA, Bonferroni's multiple comparison test).
Singlet Oxygen Formation after UVA Treatment
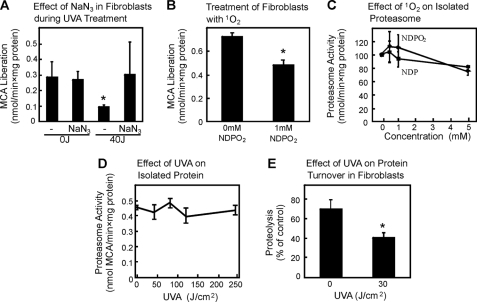
1O2 is known to be formed as a consequence of UVA irradiation, and the effects on protein oxidation and extracellular matrix changes can be correlated to 1O2 formation (22–24). To test the effect of 1O2 on proteasome inhibition after UVA, sodium azide (NaN3), a potent quencher of 1O2, was used in the model. NaN3 at 20 mm was shown to inhibit 1O2 formation caused by UVA irradiation (22, 25). Because NaN3 prevented the decline of the proteasome activity in our model (Fig. 3A), we concluded that 1O2 is the cause of the proteasomal inactivation. To confirm this we incubated fibroblasts with NDPO2 at millimolar concentrations, establishing pico-/low nanomolar steady-state concentrations of 1O2. As shown in Fig. 3B, proteasome activity was inhibited significantly by 1 mm NDPO2 in the cells (n = 3; p < 0.05). This was surprising because earlier studies by our group had demonstrated the resistance of the proteasome toward oxidative stress (13, 26, 27). Therefore, we tested whether the isolated proteasome is also susceptible toward 1O2. However, this was not the case (Fig. 3C). Our conclusion was that either 1O2 generates an unknown intermediate product in cells, affecting the proteasomal activity, or that UVA has a 1O2-independent effect on proteasomal activity. Because the direct exposure of isolated proteasome to UVA even at much higher doses did not cause any loss of activity (Fig. 3D), it seems to be most likely that UVA affects cellular structures which in turn inhibit the proteasome. It was shown by us (20, 21, 28) and others (29–33) that protein aggregates are potential factors inhibiting the proteasome after oxidative stress. If indirect inhibition of proteasomal activity occurs as a result of UVA-induced generation of cellular protein aggregates, the proteasomal activity should be impaired along with the total rate of cellular proteolysis. Total proteolysis was measured by metabolic radiolabeling of the intracellular protein pool, and we could clearly demonstrate an inhibition in overall protein degradation in cells after UVA (n = 3; p < 0.05) (Fig. 3E).
FIGURE 3.
UVA treatment of dermal fibroblast cells causes singlet oxygen formation and consequently inhibits proteolysis and proteasome activity. Fibroblasts were UVA treated as described under “Experimental Procedures” with the exception that NaN3 as a potential 1O2 quencher was present. Panel A demonstrates the proteasomal activity after UVA treatment. Data are the means ± S.E. (n = 3). *, p < 0.05 versus 0J/cm2, ANOVA, Bonferroni's multiple comparison test. To mimic the 1O2 generation of UVA, we used the singlet oxygen donor NDPO2 and tested the effect on the proteasomal activity (B). Data are the means ± S.E. (n = 3). *, p < 0.05 versus untreated, Student's t test. 20 S proteasome was isolated as described before (14), and the effects of NDPO2 were tested on its activity (C). Control experiments were performed with solutions of pre-decomposed NDPO2 containing the decomposition product, NDP. Data are the means ± S.E. (n = 3). In panel D the effect of UVA on the isolated 20 S proteasome was tested. UVA was used here in significantly higher doses compared with cell treatment. Data are the means ± S.E. (n = 3) ANOVA, Bonferroni's multiple comparison test. The effect of UVA on the overall protein degradation in fibroblasts after UVA irradiation was tested (E). The release of trichloroacetic acid-soluble radioactivity within 3 h from the intracellular protein pool was used as a measure of proteolysis. The data represent the means ± S.E. of three independent experiments, each with six independent measurements (*, p < 0.05 versus 0J/cm2, Student's t test). MCA, 7-amino-4-methylcoumarin.
Our conclusion from this series of experiments was that UVA generates 1O2. We then hypothesized that 1O2 may cause oxidative formation of protein aggregates that in turn inhibit the proteasome. We, therefore, measured formation of protein aggregates.
UVA-induced Protein Aggregate Formation as an Indicator of Severe Protein Oxidation
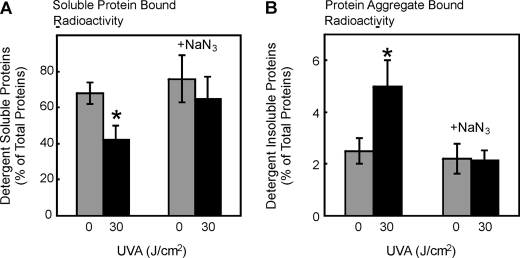
Because oxidatively modified proteins can form aggregates due to cross-linking and loss of solubility (34, 35), we determined the changes in protein detergent solubility to test for the formation of insoluble aggregates after UVA treatment. Cellular proteins were metabolically labeled with [35S]methionine/cysteine, and proteins which were soluble and insoluble in a detergent mixture were counted by liquid scintillation. In cells exposed to UVA at 30 J/cm2, the amount of detergent-soluble proteins was decreased by 35% (Fig. 4A) and detergent-insoluble proteins was increased 100% compared with nonirradiated cells (Fig. 4B) (n = 3; p < 0.05). Quenching singlet oxygen by the addition of azide ions completely abrogated UVA-induced loss of detergent-soluble protein (Fig. 4A) and UVA-induced increase in protein aggregate formation (Fig. 4B). This clearly indicates the formation of a fraction of highly cross-linked protein aggregates after a single exposure to UVA via singlet oxygen. To test whether the aggregate-induced inhibition of the proteasome is of any significance for cellular signaling and metabolism, we analyzed MMP-1 expression under these conditions.
FIGURE 4.
UVA treatment of dermal fibroblasts causes protein aggregation. Detergent solubility of cellular proteins was measured using liquid scintillation counting after [35S]methionine/cysteine labeling and UVA treatment in the absence or presence of the singlet oxygen quencher sodium azide (20 mm). The data represent the means ± S.E. of three independent experiments (*, p < 0.05 versus nonirradiated, ANOVA, Bonferroni's multiple comparison test). Detergent-soluble (A) proteins and detergent-insoluble aggregates (B) were determined without and after 30J/cm2 UVA treatment.
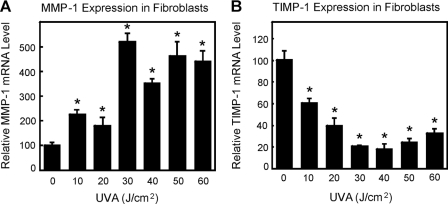
MMP-1 and TIMP-1 mRNA Levels after UVA Treatment
It is known that UVA changes MMP-1 and TIMP-1 expressions and that MMP-1 expression is elevated during skin aging. However, these changes of expression occur in a manner mechanistically not yet fully understood. Therefore, we tested whether the expression of MMP-1/TIMP-1 is changed in our fibroblast UVA exposure model. As demonstrated in Fig. 5, UVA irradiation led to significant increases in MMP-1 and decreases in TIMP-1 mRNA expressions (n = 3; p < 0.05). Therefore, we were able to mimic some of the in vivo effects of UVA. We next tested whether aggregate formation and the inhibition of the proteasome might play a role in the modulation of gene expression.
FIGURE 5.
UVA induced MMP-1 and TIMP-1 expressions in fibroblasts after UVA treatment. The dose dependence of MMP-1 mRNA (A) and TIMP-1 mRNA (B) expressions 3 h post-irradiation are shown (n = 3; *, p < 0.05 versus nonirradiated, ANOVA, Bonferroni's multiple comparison test. Data are the means ± S.E.).
Effects of Cross-linked Protein Aggregates on Proteasome Activity and MMP-1 mRNA Expression
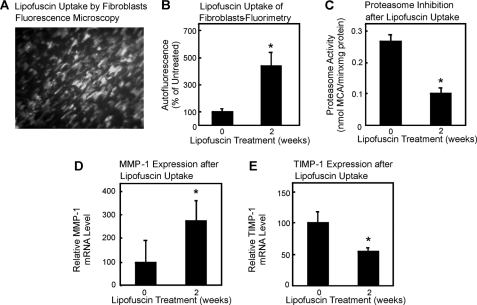
To test the role of proteasomal inhibition on gene expression, we decided to use a model of proteasome inhibition by protein aggregates without UVA as described by us before (29, 36, 37). As a model for cross-linked, oxidized proteins, we chose lipofuscin, a fluorescent pigment of aggregated polymers derived from cross-linked oxidation products of proteins and lipids accumulating during aging (28, 38). We prepared lipofuscin from isolated erythrocyte membranes exposed to UV as described in detail previously (28) and incubated dermal fibroblasts with this material. Because fibroblasts were described to readily incorporate such material (28), we measured the uptake of lipofuscin into the cells by fluorescence microscopy (Fig. 6A). Quantitation of the results revealed a 5-fold increase in autofluorescence (Fig. 6B), accompanied by a decline in proteasomal activity similar to that detected after UVA treatment (Fig. 6C). Most importantly, proteasome inhibition by protein aggregates was also accompanied by an increased expression of MMP-1 (Fig. 6D) and a decrease in TIMP-1 expression (Fig. 6E). Immunochemical detection of MMP-1 and TIMP-1 was unsuccessful, probably because of the fact that both are secreted proteins, the detection of which is hampered by lipofuscin, which was present in the cell culture media in this experimental setup. Therefore, the change of the MMP-1/TIMP-1 expression ratio in cells exposed to UVA appears to be a result of proteasome inactivation. To further support this hypothesis, we analyzed the effect of direct inhibition of the proteasome by inhibitor molecules on MMP-1 and TIMP-1 expression.
FIGURE 6.
Cross-linked protein aggregates inhibit the proteasome activity and cause an increase in MMP-1 mRNA expression and a decrease in TIMP-1 mRNA expression. Fibroblasts were treated with artificial lipofuscin as described under “Experimental Procedures.” Panel A shows the autofluorescence image after 2 weeks of incubation of fibroblasts with 0.6 mg of lipofuscin, whereas in panel B the quantification of autofluorescence using fluorometry with 360-nm excitation and 590-nm emission is demonstrated. Data are the means ± S.E., (n = 3). *, p < 0.05 versus untreated, Students t test. C, the proteasome activity without and with lipofuscin treatment of the cells was measured as described under “Experimental Procedures.” Panel D shows the relative MMP-1 mRNA level compared with 18 S rRNA levels, and panel E shows the relative TIMP-1 mRNA level compared with 18 S rRNA levels (n = 3, *, p < 0.05 versus untreated, Student's t test. Data are the means ± S.E.).
Effects of Proteasome Inhibition on MMP-1 and TIMP-1 Expressions
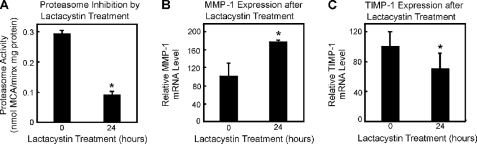
To determine whether direct proteasome inhibition affects MMP-1/TIMP-1 mRNA levels, we employed lactacystin, an irreversible inhibitor of the proteasome. Lactacystin binds to catalytic subunits of the 20 S proteasome and inhibits its protease activity (39). After the incubation of fibroblasts with 20 μm lactacystin for 24 h, the proteasome activity was inhibited more than 50% (Fig. 7A). The same concentration of lactacystin elicited significant elevation and decrease of MMP-1 and TIMP-1 mRNA steady-state levels, respectively (Fig. 7, B and C, p < 0.05). Therefore, proteasomal activity seems to be a crucial regulator of MMP-1/TIMP-1 mRNA levels.
FIGURE 7.
Proteasome inhibition increases MMP-1 mRNA levels. Fibroblasts were treated with lactacystin in panels A–C. Panel A shows the proteasome activity inhibition by 20 μm lactacystin. n = 3; *, p < 0.05 versus untreated. Panel B shows the MMP-1 mRNA expression without and with lactacystin. n = 3; *, p < 0.05 versus untreated, whereas panel C demonstrates the TIMP-1 mRNA expression without and with lactacystin. n = 3; *, p < 0.05 versus untreated. Student's t test. Data are the means ± S.E.
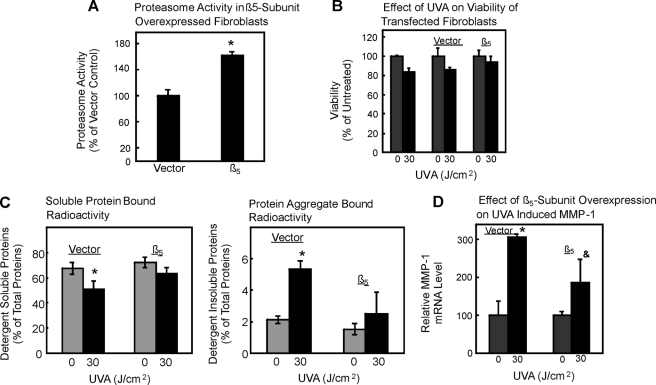
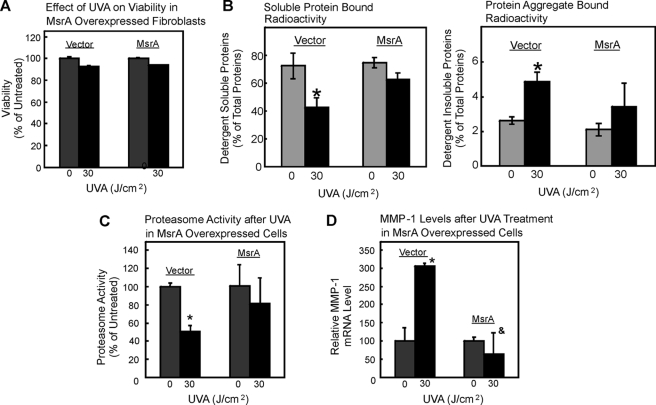
Thus, UVA should not be able to change mRNA levels to the same extent if high proteasomal activity is present in cells. Accordingly, we next studied the effect of UVA on fibroblasts with elevated proteasomal activity due to overexpression. WI38/T fibroblast cells were stably transfected with either an empty vector or a β5-subunit (40). Empty vector-transfected cells were used as controls for β5-overexpressing cells. Overexpression of the β5 proteasomal subunit was previously shown to increase cellular protease activities (40), and we also found a 60% increase in proteasome activity due to β5-subunit overexpression, as shown in Fig. 8A. Overexpression did not change the viability of the cells after UVA exposure (Fig. 8B). Exposure of vector-transfected cells to UVA at 30 J/cm2 caused a significant increase of protein aggregation (Fig. 8C, right panel) and of MMP-1 mRNA levels of 3-fold over control (i.e. nonirradiated vector) cells (n = 3; *, p < 0.05 versus nonirradiated) (Fig. 8D). In contrast, UVA-induced protein aggregation was blunted (Fig. 8C), and the change in MMP-1 mRNA levels was strongly attenuated (Fig. 8D) in β5-subunit-overexpressing cells exposed to 30 J/cm2 of UVA compared with nonirradiated cells. Moreover, mRNA levels were significantly lower in β5-subunit-overexpressing cells exposed to 30 J/cm2 of UVA relative to irradiated vector control cells (n = 3, &, p < 0.05 versus irradiated vector cells). These data clearly point to a crucial role of the proteasome in the regulation of MMP-1 mRNA levels. However, if oxidized proteins are formed during UVA irradiation, which in turn cross-link and inhibit the proteasome, it should be possible to block such a formation of oxidized cross-linked proteins as well as its consequences for proteasome activity and MMP-1 expression by introducing additional protein repair mechanisms into fibroblasts.
FIGURE 8.
Proteasome overexpression attenuates UVA-induced protein aggregation and rise in MMP-1 mRNA levels. A cell clone overexpressing the β5-subunit of the proteasome or the vector control was exposed to UVA. A, the proteasome activity after β5-subunit overexpression was measured; n = 3; *, p < 0.05 versus vector control. B, viability was measured under control conditions (gray columns) or after UVA treatment (black columns) in untransfected cells, vector control, and β5-overexpressing cells. C, protein aggregation (levels of detergent-insoluble aggregates, right panel) and levels of detergent-soluble proteins (left panel) were determined as in Fig. 4; n = 3; *, p < 0.05 versus nonirradiated. Panel D demonstrates the MMP-1 mRNA levels in vector control and β5-overexpressing cells after UVA treatment (black columns) or untreated cells (gray columns) after proteasome overexpression. n = 3; *, p < 0.05 versus nonirradiated vector cells; &, p < 0.05 versus irradiated vector cells, ANOVA, Bonferroni's multiple comparison test. Data are the means ± S.E.
MsrA Overexpression Protects Fibroblasts against UVA-induced Protein Aggregation, Proteasome Inhibition, and Elevation of MMP-1 Expression
MsrA contributes to cellular repair of oxidized proteins by reducing methionine sulfoxide to methionine (41). In addition to reconstituting previous activities of proteins that were targeted by an oxidant, the generation of reduced methionine serves the quenching of further oxidants, acting as an intramolecular antioxidative moiety, as proposed earlier (42). To test for the effects of MsrA on UVA-induced MMP-1 expression in fibroblasts, we used WI38/T cells stably overexpressing MsrA and exposed them to 30 J/cm2 of UVA. The employed cell lines were thoroughly characterized previously, and MsrA overexpression was established (43). No significant differences in survival were observed between control and MsrA-overexpressing cells (Fig. 9A). Overexpression of MsrA decreased UVA-induced loss of detergent-soluble protein and abrogated the UVA-induced increase in protein aggregate formation (Fig. 9B). Intriguingly, proteasome inhibition, which was seen in control cells after UVA, was not seen in MsrA-overexpressing cells (Fig. 9C), further indicating a crucial role of protein oxidation in causing proteasome inhibition. Moreover, MMP-1 mRNA expression was not increased (Fig. 9D) due to an enhanced protein repair after MsrA overexpression.
FIGURE 9.
Effect of MsrA overexpression on UVA-induced outcomes. MsrA-transfected cells were used for the experiment, and vector-transfected cells were used as controls. All experiments were performed in triplicate. *, p < 0.05, versus nonirradiated vector; &, p < 0.05, versus 30 J/cm2 irradiated vector, ANOVA, Bonferroni's multiple comparison test. Data are the means ± S.E. Panel A shows the effect of UVA on viability in vector and MsrA-transfected fibroblasts. B, protein aggregation (levels of detergent-insoluble aggregates, right panel) and levels of detergent-soluble proteins (left panel) were determined as in Fig. 4; n = 3; *, p < 0.05 versus nonirradiated. The proteasome activity after UVA treatment in vector and MsrA-overexpressing cells is demonstrated in panel C, whereas panel D shows MMP-1 mRNA levels after UVA treatment in vector and MsrA-overexpressing cells; &, p < 0.05 versus irradiated cells in the first and second columns.
Having established that UVA-induced changes in MMP-1 expression are because of oxidant (1O2) formation, the formation of oxidized, cross-linked proteins, and the inhibition of the proteasome, we next asked how decreased proteasomal activity relates to changes in mRNA levels.
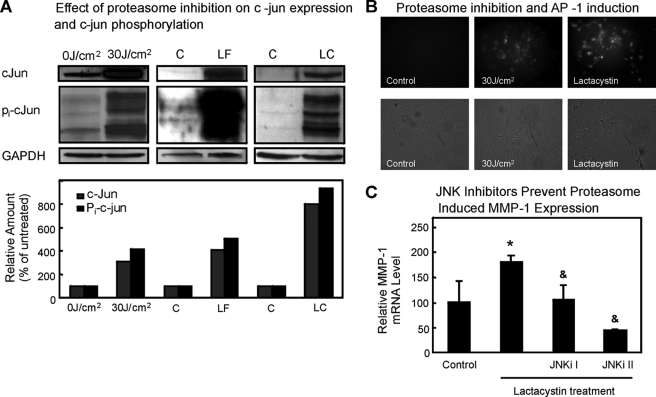
Proteasome Inhibition Leads to c-Jun and Pi-c-Jun Accumulation, Resulting in AP-1 Activation
UVA irradiation of human skin cells is known to lead to the activation of transcription factors AP-1 and NF-κB (44–46). Both transcription factors are related to the proteasomal pathway. Both the components of AP-1 factors (including c-Jun and c-Fos) (47–51) and the phosphorylated I-κB inhibitor of NF-κB (52, 53) are degraded by the proteasome. Active AP-1 is composed of phosphorylated c-Jun (Pi-c-Jun) and another protein (such as c-Fos). Therefore, we tested c-Jun and Pi-c-Jun protein levels in control cells and cells exposed to either UVA, lipofuscin, or lactacystin. Both protein levels of c-Jun and extent of c-Jun phosphorylation were significantly increased in all three models of proteasome inhibition compared with the controls (Fig. 10A). Because c-Jun is a protein involved in the formation of functional AP-1, an enhanced c-jun expression as well as c-Jun phosphorylation should cause an increase in AP-1 activity, i.e. its DNA binding and potency of stimulating transcriptional processes. We determined AP-1 activity using a Cignal reporter gene assay, employing GFP expression and detection thereof by fluorescence microscopy as read-out. As shown in Fig. 10B, AP-1 activity was increased in cells exposed to either 30 J/cm2 of UVA or to 20 μm lactacystin. Unfortunately, it was not possible to use lipofuscin for proteasome inhibition in this assay, as the autofluorescence of lipofuscin overlaps with GFP fluorescence spectra. To further confirm that c-Jun phosphorylation plays a crucial role in MMP-1 expression, we inhibited c-Jun N-terminal kinases (JNK) by incubation of cells with JNK inhibitors for 24 h to suppress the phosphorylation of c-Jun. The up-regulation of MMP-1 mRNA by proteasome inhibition was found to be attenuated by inhibition of JNK using a broad and rather unspecific inhibitor (SP600125 = JNKi II) and a specific peptide inhibitor of the kinases (JNKi I, Calbiochem) (Fig. 10C). On the other hand, use of NF-κB inhibitors (cell-permeable IκB kinase inhibitor, Calbiochem) did not show any effect (data not shown). These data demonstrate that inhibition of the proteasome is accompanied by an accumulation of c-Jun, activation (i.e. phosphorylation) of c-Jun by JNK, and formation of active AP-1. Therefore, we were able to establish a chain of events leading from UVA-induced singlet oxygen formation, via formation of oxidized cross-linked proteins, to proteasome inhibition, which in turn causes activation of the transcription factor AP-1, leading to an enhanced production of MMP-1 mRNA.
FIGURE 10.
C-Jun expression, phosphorylation, and AP-1 activation are results of proteasome inhibition. Fibroblasts were irradiated with 30 J/cm2, treated with lipofuscin (LF) or lactacystin (LC), or used without treatment (C) as described under “Experimental Procedures.” Cells were harvested and lysed, and proteins were analyzed by immunoblotting with anti-c-Jun, anti-Pi-c-Jun, and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies giving the products with the sizes of 40, 47, and 37 kDa, respectively (A). Immunoblotting for glyceraldehyde-3-phosphate dehydrogenase (bottom panel) showed equal loading of the proteins in each lane. The results of quantitative analysis of these immunoblots are depicted in the columns in the lower part of the panel. Amounts were quantified in relation to glyceraldehyde-3-phosphate dehydrogenase by densitometry. B, the Cignal AP1-GFP reporter assay measures the activation of AP-1 (see “Experimental Procedures”). After UVA and lactacystin treatments, bright field and fluorescent images were taken of the cultures. Clearly the green florescence in response to AP-1 activation after UVA and lactacystin is visible. The effect of JNK inhibitors (JNKi I and SP600125 or JNKi II) on lactacystin-induced MMP-1 mRNA level increase is shown in panel C; n = 3; *, p < 0.05 versus control cells without lactacystin treatment; &, p < 0.05 versus only lactacystin-treated cells, ANOVA, Bonferroni's multiple comparison test.
DISCUSSION
In this study we have found that UVA irradiation causes a loss in proteasome activity by way of protein oxidation followed by aggregate formation. The resulting proteasome inhibition plays a critical role in UVA-induced changes in MMP-1 expression levels.
Taking into account 1O2 formation after UVA irradiation (23, 25, 54), we tested for a role of singlet oxygen in UVA-induced proteasome inhibition employing NaN3 as a singlet oxygen quencher. Results suggest that 1O2 is indeed a mediator of UVA-induced proteasome inhibition in fibroblasts (Fig. 3). These data were confirmed in experiments employing 1O2 generated chemically by thermodecomposition of NDPO2 (Fig. 3). In contrast, neither NDPO2 nor UVA even at higher doses caused inhibition of proteolytic activity of isolated proteasome (Fig. 3). In this regard it is of interest that UVA irradiation causes an accumulation of highly oxidized and aggregated proteins. Therefore, proteasome inhibition in living cells might be explained by protein aggregates formed after severe oxidation (28, 34, 36, 37, 55). Evidence from several studies has indicated that photoaging and also intrinsic skin aging coincides with visible changes in the skin which are the results of collagen polymerization and degradation. These effects are also elicited by 1O2, which is generated in vivo under exposure to UVA and is known to mediate the UVA-induced enhancement of MMP-1 expression in the dermal layer of the skin (54, 56). In our hands exposure of up to 60 J/cm2 of UVA irradiation changes the expression levels of MMP-1 mRNA and TIMP-1 mRNA (Fig. 5), which is in agreement with earlier studies (12, 23, 24, 56–59). Furthermore, we demonstrate a direct chain of events and a connection between proteasome inhibition and mRNA expression. In line with this, an elevation of cellular proteasomal activity by overexpression of a proteasomal subunit blocked the induction of changes in mRNA levels (Fig. 8). Similarly, the reversion of oxidative protein damage by overexpression of MsrA interfered with UVA-induced changes in mRNA levels (Fig. 9).
The strategy employed was to enhance intrinsic defense against protein oxidation by elevating the levels of reduced methionine residues in proteins; protein carbonyls are no direct substrates of MsrA, but an overexpression of MsrA causes enhanced reduction of protein-bound methionine sulfoxide, which is generated during oxidative stress, to methionine. Methionine was proposed to act as an endogenous antioxidant in proteins protecting other amino acids in its vicinity from oxidation (42). MsrA is the only enzyme identified in human skin so far that is capable of repairing oxidative protein damage (60). It was demonstrated previously to be down-regulated during senescence in WI-38 fibroblasts and after chronic or high dose UVA treatment of keratinocytes (61, 62), and this decline was associated with an accumulation of oxidative protein damage. Furthermore, a crucial role of the Msr system in protecting against oxidative stress-induced cell death has been reported for several cell lines (43, 63–66).
There is ample evidence that UV radiation-induced increase in MMP-1 expression is caused by activation of transcription factors AP-1 and NF-κB (25, 45, 63, 67–70). The stimulation of AP-1 and NF-κB DNA binding activities by UVA was indeed demonstrated for human fibroblasts and keratinocytes to occur (44, 45). The transcription factor AP-1 is composed of members of the Jun and Fos families which bind to AP-1 sites (69). Here, UVA at 30 J/cm2 caused an increase in both expression and phosphorylation of c-Jun, as did lactacystin and lipofuscin, resulting in an enhanced AP-1 binding activity (Fig. 10). Jun phosphorylation is essential in lactacystin-induced MMP-1 mRNA expression, as confirmed in experiments with inhibited JNK (Fig. 10). NFκB did not affect the lactacystin-induced MMP-1 expression (data not shown).
We here demonstrate that protein aggregate formation is induced by (singlet oxygen-mediated) oxidation during UVA treatment, resulting in inhibition of the proteasome that is accompanied by phosphorylation of c-Jun and increased AP-1 activity. These effects were attenuated by MsrA and proteasome overexpression. Thus, the cause/effect relationship connecting UVA-induced protein oxidation and proteasomal dysfunction was established in four steps, i.e. (i) by demonstrating the formation of oxidized protein under exposure to UVA (Fig. 1A) and the loss of proteasomal activity (Fig. 1B) that was shown not to occur in the absence of cellular environment, including oxidizable protein (Fig. 3D); (ii) by imitating UVA effects on cellular protein oxidation and the proteasome by adding cross-linked protein to cells in the absence of UVA, which elicited the exact same effect as UVA in terms of inhibiting cellular proteasome (Fig. 6C) and causing changes in gene expression (Fig. 6, D and E); (iii) by enhancing the inherent resistance of cellular protein against oxidation by elevating levels of MsrA, a protein repair enzyme keeping up the levels of reduced methionine; (iv) by excluding that oxidants rather than oxidized proteins are the direct cause of proteasome dysfunction; neither oxidants nor UVA was capable of inhibiting isolated proteasome (Fig. 3). Intriguingly, with MMP-1 being a major protease of the extracellular matrix, the signaling cascade delineated in this work links the regulation of extracellular proteolytic events to intracellular proteolysis.
These results also provide novel insight into UVA- and singlet oxygen-induced signaling. It was hypothesized previously that singlet oxygen-induced oxidation of selected sensitive signaling proteins may result in signaling causing changes in gene expression (71–73). It is demonstrated here for the first time, however, that general protein oxidation rather than oxidation of selected proteins also contributes to the regulation of UVA-/singlet oxygen-induced signaling in that extensive protein oxidation causes attenuation of proteasome activity, resulting in a net accumulation of selected signaling proteins, including constituents of the transcription factor complex AP-1 that, in turn, controls MMP-1 expression. This introduces a novel level of specificity to oxidant-induced signaling as, rather than modulating signaling cascades by oxidation of specific oxidation-prone proteins, general oxidation causes a loss of degradation of selected proteins; selectivity is shifted from the level of oxidant/target interaction to that of an interaction between proteasome and its substrates. We propose a model of UVA- and singlet oxygen-induced signaling comprising both an immediate and a later response, the first being initiated at the level of oxidation of specific oxidation-prone signaling proteins and the latter being because of general overoxidation and accumulation of proteins with a consecutive proteasome inhibition.
It has previously been reported that photoaged as well as intrinsically aged skin contains increased the amounts of oxidized proteins within fibroblasts of the upper dermis. It should be noted that it is exactly this compartment of skin which shows stromal collagen breakdown in skin aging (8). The results described in this study are the first to indicate that the presence of oxidized proteins and collagen degradation in the upper dermis of (photo)aged skin are causally linked with each other and that prevention of protein oxidation represents a potential protective strategy regarding skin aging.
This work was supported by Henkel AG and CoKGaA, Düsseldorf, Germany.
- MMP-1
- matrix metalloproteinase-1
- AP-1
- activator protein-1
- DNP
- dinitrophenyl
- JNKi
- c-Jun NH2-terminal kinase inhibitor
- Msr
- methionine sulfoxide reductase
- NaN3
- sodium azide
- NDP
- disodium 3,3′-(1,4-naphthylidene) dipropionate
- NDPO2
- disodium 3,3′-(1,4-naphthylidene) dipropionate-1,4-endoperoxide
- 1O2
- singlet molecular oxygen
- Pi-c-Jun
- phospho-c-Jun
- TIMP-1
- tissue inhibitor of metalloproteinase-1
- GFP
- green fluorescent protein
- TRITC
- tetramethylrhodamine isothiocyanate
- PBS
- phosphate-buffered saline
- ANOVA
- analysis of variance.
REFERENCES
- 1.Tyrrell R. M. (1996) BioEssays 18, 139–148 [DOI] [PubMed] [Google Scholar]
- 2.Tyrrell R. M. (2004) Antioxid. Redox Signal. 6, 835–840 [DOI] [PubMed] [Google Scholar]
- 3.Scharffetter-Kochanek K., Wlaschek M., Brenneisen P., Schauen M., Blaudschun R., Wenk J. (1997) Biol. Chem. 378, 1247–1257 [PubMed] [Google Scholar]
- 4.Fisher G. J., Wang Z. Q., Datta S. C., Varani J., Kang S., Voorhees J. J. (1997) N. Engl. J. Med. 337, 1419–1428 [DOI] [PubMed] [Google Scholar]
- 5.Krutmann J., Gilchrest B. A. (2006) Skin Aging, pp. 33–44, Springer-Verlag New York Inc., New York [Google Scholar]
- 6.Rittie L., Fisher G. J., Voorhees J. J. (2006) Skin Aging, pp. 143–156, Springer-Verlag New York Inc., New York [Google Scholar]
- 7.Klotz L. O., Holbrook N. J., Sies H. (2001) Curr. Probl. Dermatol. 29, 95–113 [DOI] [PubMed] [Google Scholar]
- 8.Sander C. S., Chang H., Salzmann S., Müller C. S., Ekanayake-Mudiyanselage S., Elsner P., Thiele J. J. (2002) J. Invest. Dermatol. 118, 618–625 [DOI] [PubMed] [Google Scholar]
- 9.Luy H., Frenk E., Applegate L. A. (1994) Photodermatol. Photoimmunol. Photomed. 10, 126–133 [PubMed] [Google Scholar]
- 10.Bonny C., Oberson A., Negri S., Sauser C., Schorderet D. F. (2001) Diabetes 50, 77–82 [DOI] [PubMed] [Google Scholar]
- 11.Bennett B. L., Sasaki D. T., Murray B. W., O'Leary E. C., Sakata S. T., Xu W., Leisten J. C., Motiwala A., Pierce S., Satoh Y., Bhagwat S. S., Manning A. M., Anderson D. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13681–13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buss H., Chan T. P., Sluis K. B., Domigan N. M., Winterbourn C. C. (1997) Free Radic. Biol. Med. 23, 361–366 [DOI] [PubMed] [Google Scholar]
- 13.Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K. J., Grune T. (1998) Biochem. J. 335, 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hough R., Pratt G., Rechsteiner M. (1987) J. Biol. Chem. 262, 8303–8313 [PubMed] [Google Scholar]
- 15.Jung T., Engels M., Kaiser B., Poppek D., Grune T. (2006) Free Radic. Biol. Med. 40, 1303–1312 [DOI] [PubMed] [Google Scholar]
- 16.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 17.Demasi M., Davies K. J. A. (2003) FEBS Lett. 542, 89–94 [DOI] [PubMed] [Google Scholar]
- 18.Voss P., Horakova L., Jakstadt M., Kiekebusch D., Grune T. (2006) Free Radic. Res. 40, 673–683 [DOI] [PubMed] [Google Scholar]
- 19.Levine R. L., Williams J. A., Stadtman E. R., Shacter E. (1994) Methods Enzymol. 233, 346–357 [DOI] [PubMed] [Google Scholar]
- 20.Grune T., Reinheckel T., Joshi M., Davies K. J. A. (1995) J. Biol. Chem. 270, 2344–2351 [DOI] [PubMed] [Google Scholar]
- 21.Grune T., Reinheckel T., Davies K. J. A. (1996) J. Biol. Chem. 271, 15504–15509 [DOI] [PubMed] [Google Scholar]
- 22.Wlaschek M., Wenk J., Brenneisen P., Briviba K., Schwarz A., Sies H., Scharffetter-Kochanek K. (1997) FEBS Lett. 413, 239–242 [DOI] [PubMed] [Google Scholar]
- 23.Scharffetter K., Wlaschek M., Hogg A., Bolsen K., Schothorst A., Goerz G., Krieg T., Plewig G. (1991) Arch. Dermatol. Res. 283, 506–511 [DOI] [PubMed] [Google Scholar]
- 24.Dean R. T., Fu S., Stocker R., Davies M. J. (1997) Biochem. J. 324, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klotz L. O., Briviba K., Sies H. (1997) FEBS Lett. 408, 289–291 [DOI] [PubMed] [Google Scholar]
- 26.Grune T., Klotz L. O., Gieche J., Rudeck M., Sies H. (2001) Free Radic. Biol. Med. 30, 1243–1253 [DOI] [PubMed] [Google Scholar]
- 27.Reinheckel T., Ullrich O., Sitte N., Grune T. (2000) Arch. Biochem. Biophys. 377, 65–68 [DOI] [PubMed] [Google Scholar]
- 28.Sitte N., Huber M., Grune T., Ladhoff A., Doecke W. D., Von Zglinicki T., Davies K. J. (2000) FASEB J. 14, 1490–1498 [DOI] [PubMed] [Google Scholar]
- 29.Davies K. J., Shringarpure R. (2006) Neurology 66, S93–96 [DOI] [PubMed] [Google Scholar]
- 30.Powell S. R., Wang P., Divald A., Teichberg S., Haridas V., McCloskey T. W., Davies K. J., Katzeff H. (2005) Free Radic. Biol. Med. 38, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 31.Michalik A., Van Broeckhoven C. (2004) Neurobiol. Dis. 16, 202–211 [DOI] [PubMed] [Google Scholar]
- 32.Verhoef L. G., Lindsten K., Masucci M. G., Dantuma N. P. (2002) Hum. Mol. Genet. 11, 2689–2700 [DOI] [PubMed] [Google Scholar]
- 33.Ortega Z., Díaz-Hernández M., Lucas J. J. (2007) Cell Mol. Life Sci. 64, 2245–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grune T., Reinheckel T., Davies K. J. (1997) FASEB J. 11, 526–534 [PubMed] [Google Scholar]
- 35.McDuffee A. T., Senisterra G., Huntley S., Lepock J. R., Sekhar K. R., Meredith M. J., Borrelli M. J., Morrow J. D., Freeman M. L. (1997) J. Cell Physiol. 171, 143–151 [DOI] [PubMed] [Google Scholar]
- 36.Sommerburg O., Ullrich O., Sitte N., von Zglinicki D., Siems W., Grune T. (1998) Free Radic. Biol. Med. 24, 1369–1374 [DOI] [PubMed] [Google Scholar]
- 37.Sitte N., Merker K., von Zglinicki T., Grune T. (2000) Free Rad. Biol. Med. 28, 701–708 [DOI] [PubMed] [Google Scholar]
- 38.Grune T., Jung T., Merker K., Davies K. J. (2004) Int. J. Biochem. Cell Biol. 36, 2519–2530 [DOI] [PubMed] [Google Scholar]
- 39.Fenteany G., Schreiber S. L. (1998) J. Biol. Chem. 273, 8545–8548 [DOI] [PubMed] [Google Scholar]
- 40.Chondrogianni N., Tzavelas C., Pemberton A. J., Nezis I. P., Rivett A. J., Gonos E. S. (2005) J. Biol. Chem. 280, 11840–11850 [DOI] [PubMed] [Google Scholar]
- 41.Moskovitz J., Bar-Noy S., Williams W. M., Requena J., Berlett B. S., Stadtman E. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine R. L., Mosoni L., Berlett B. S., Stadtman E. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15036–15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picot C. R., Petropoulos I., Perichon M., Moreau M., Nizard C., Friguet B. (2005) Free Radic. Biol. Med. 39, 1332–1341 [DOI] [PubMed] [Google Scholar]
- 44.Djavaheri-Mergny M., Mergny J. L., Bertrand F., Santus R., Mazière C., Dubertret L., Mazière J. C. (1996) FEBS Lett. 384, 92–96 [DOI] [PubMed] [Google Scholar]
- 45.Vile G. F., Tanew-Ilitschew A., Tyrrell R. M. (1995) Photochem. Photobiol. 62, 463–468 [DOI] [PubMed] [Google Scholar]
- 46.Shaulian E., Schreiber M., Piu F., Beeche M., Wagner E. F., Karin M. (2000) Cell 103, 897–907 [DOI] [PubMed] [Google Scholar]
- 47.Jariel-Encontre I., Salvat C., Steff A. M., Pariat M., Acquaviva C., Furstoss O., Piechaczyk M. (1997) Mol. Biol. Rep. 24, 51–56 [DOI] [PubMed] [Google Scholar]
- 48.Salvat C., Aquaviva C., Jariel-Encontre I., Ferrara P., Pariat M., Steff A. M., Carillo S., Piechaczyk M. (1999) Mol. Biol. Rep. 26, 45–51 [DOI] [PubMed] [Google Scholar]
- 49.Jariel-Encontre I., Pariat M., Martin F., Carillo S., Salvat C., Piechaczyk M. (1995) J. Biol. Chem. 270, 11623–11627 [DOI] [PubMed] [Google Scholar]
- 50.Tsurumi C., Ishida N., Tamura T., Kakizuka A., Nishida E., Okumura E., Kishimoto T., Inagaki M., Okazaki K., Sagata N. (1995) Mol. Cell. Biol. 15, 5682–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basbous J., Jariel-Encontre I., Gomard T., Bossis G., Piechaczyk M. (2008) Biochimie 90, 296–305 [DOI] [PubMed] [Google Scholar]
- 52.Karin M., Ben-Neriah Y. (2000) Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 53.Palombella V. J., Rando O. J., Goldberg A. L., Maniatis T. (1994) Cell 78, 773–785 [DOI] [PubMed] [Google Scholar]
- 54.Scharffetter-Kochanek K., Wlaschek M., Briviba K., Sies H. (1993) FEBS Lett. 331, 304–306 [DOI] [PubMed] [Google Scholar]
- 55.Jung T., Bader N., Grune T. (2007) Ann. N.Y. Acad. Sci. 1119, 97–111 [DOI] [PubMed] [Google Scholar]
- 56.Naru E., Suzuki T., Moriyama M., Inomata K., Hayashi A., Arakane K., Kaji K. (2005) Br. J. Dermatol. 153, 6–12 [DOI] [PubMed] [Google Scholar]
- 57.Herrmann G., Wlaschek M., Lange T. S., Prenzel K., Goerz G., Scharffetter-Kochanek K. (1993) Exp. Dermatol. 2, 92–97 [DOI] [PubMed] [Google Scholar]
- 58.Polte T., Tyrrell R. M. (2004) Free Radic. Biol. Med. 36, 1566–1574 [DOI] [PubMed] [Google Scholar]
- 59.Hantke B., Lahmann C., Venzke K., Fischer T., Kocourek A., Windsor L. J., Bergemann J., Stäb F., Tschesche H. (2002) Photochem. Photobiol. Sci. 1, 826–833 [DOI] [PubMed] [Google Scholar]
- 60.Ogawa F., Sander C. S., Hansel A., Oehrl W., Kasperczyk H., Elsner P., Shimizu K., Heinemann S. H., Thiele J. J. (2006) J. Invest. Dermatol. 126, 1128–1134 [DOI] [PubMed] [Google Scholar]
- 61.Picot C. R., Moreau M., Juan M., Noblesse E., Nizard C., Petropoulos I., Friguet B. (2007) Exp. Gerontol. 42, 859–863 [DOI] [PubMed] [Google Scholar]
- 62.Picot C. R., Perichon M., Cintrat J. C., Friguet B., Petropoulos I. (2004) FEBS Lett. 558, 74–78 [DOI] [PubMed] [Google Scholar]
- 63.Fisher G. J., Datta S. C., Talwar H. S., Wang Z. Q., Varani J., Kang S., Voorhees J. J. (1996) Nature 379, 335–339 [DOI] [PubMed] [Google Scholar]
- 64.Yermolaieva O., Xu R., Schinstock C., Brot N., Weissbach H., Heinemann S. H., Hoshi T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kantorow M., Hawse J. R., Cowell T. L., Benhamed S., Pizarro G. O., Reddy V. N., Hejtmancik J. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9654–9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moskovitz J., Flescher E., Berlett B. S., Azare J., Poston J. M., Stadtman E. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14071–14075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X. Y., Bi Z. G. (2006) Chin. Med. J. 119, 827–831 [PubMed] [Google Scholar]
- 68.Wan Y. S., Wang Z. Q., Voorhees J., Fisher G. (2001) Cell. Signal. 13, 139–144 [DOI] [PubMed] [Google Scholar]
- 69.Angel P., Karin M. (1991) Biochim. Biophys. Acta 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 70.Klotz L. O., Pellieux C., Briviba K., Pierlot C., Aubry J. M., Sies H. (1999) Eur. J. Biochem. 260, 917–922 [DOI] [PubMed] [Google Scholar]
- 71.Klotz L. O. (2002) Biol. Chem. 383, 443–456 [DOI] [PubMed] [Google Scholar]
- 72.Klotz L. O., Kröncke K. D., Sies H. (2003) Photochem. Photobiol. Sci. 2, 88–94 [DOI] [PubMed] [Google Scholar]
- 73.von Montfort C., Sharov V. S., Metzger S., Schöneich C., Sies H., Klotz L. O. (2006) Biol. Chem. 387, 1399–1404 [DOI] [PubMed] [Google Scholar]