Abstract
The chloroplast enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) catalyzes the rate-limiting step of photosynthetic CO2 fixation. With a deeper understanding of its structure-function relationships and competitive inhibition by O2, it may be possible to engineer an increase in agricultural productivity and renewable energy. The chloroplast-encoded large subunits form the active site, but the nuclear-encoded small subunits can also influence catalytic efficiency and CO2/O2 specificity. To further define the role of the small subunit in Rubisco function, the 10 most conserved residues in all small subunits were substituted with alanine by transformation of a Chlamydomonas reinhardtii mutant that lacks the small subunit gene family. All the mutant strains were able to grow photosynthetically, indicating that none of the residues is essential for function. Three of the substitutions have little or no effect (S16A, P19A, and E92A), one primarily affects holoenzyme stability (L18A), and the remainder affect catalysis with or without some level of associated structural instability (Y32A, E43A, W73A, L78A, P79A, and F81A). Y32A and E43A cause decreases in CO2/O2 specificity. Based on the x-ray crystal structure of Chlamydomonas Rubisco, all but one (Glu-92) of the conserved residues are in contact with large subunits and cluster near the amino- or carboxyl-terminal ends of large subunit α-helix 8, which is a structural element of the α/β-barrel active site. Small subunit residues Glu-43 and Trp-73 identify a possible structural connection between active site α-helix 8 and the highly variable small subunit loop between β-strands A and B, which can also influence Rubisco CO2/O2 specificity.
In plants and green algae, ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco, EC 4.1.1.39)2 is comprised of eight chloroplast-encoded large subunits and eight nuclear-encoded small subunits (referred to as L8S8 Rubisco) (reviewed in Refs. 1–3). The ∼55-kDa large subunit contains a carboxyl-terminal α/β-barrel domain that, along with residues from the amino-terminal domain of a neighboring large subunit, forms the active site of the enzyme. Carboxylation of ribulose 1,5-bisphosphate (RuBP) initiates the rate-limiting step of photosynthetic CO2 fixation. However, O2 is mutually competitive with CO2, and oxygenation of RuBP is a nonessential side reaction that ultimately leads to the loss of CO2 in the photorespiratory pathway. Thus, net CO2 fixation is determined by the difference between the rates of carboxylation and oxygenation (4), which are ultimately determined by the Vmax values for carboxylation (Vc) and oxygenation (Vo), the Km values for CO2 (Kc) and O2 (Ko), and the concentrations of CO2 and O2 at the Rubisco active site. Another kinetic constant referred to as the CO2/O2 specificity factor (Ω) is equal to the catalytic efficiency of carboxylation (Vc/Kc) relative to the catalytic efficiency of oxygenation (Vo/Ko) (4). Because Ω is determined by the difference between the carboxylation and oxygenation free energies of activation at the rate-determining step of catalysis (5), changes in Ω may identify regions of Rubisco worthy of genetic engineering. There continues to be much interest in either engineering or selecting improvements in Rubisco as a means for increasing net CO2 fixation and the production of food, fiber, and renewable energy.
The catalytic mechanism of the large subunit active site has been studied extensively by employing directed mutagenesis of either the dimeric large subunit Rubisco enzyme of Rhodospirillum rubrum (referred to as L2 Rubisco) or the plant-like L8S8 Rubisco of Synechococcus expressed in Escherichia coli (reviewed in Refs. 6 and 7). Numerous x-ray crystal structures are available to serve as a basis for such studies (2). However, as of yet, this information has not been exploited for designing a better Rubisco or for explaining the variation in kinetic constants observed for Rubisco enzymes from different species (8–11). Much less is known about the role of the ∼15-kDa small subunit in Rubisco function (reviewed in Ref. 12). The small subunit is not in contact with any of the large subunit active site residues, but directed mutagenesis of prokaryotic and algal small subunits (13–15) or creation of hybrid enzymes comprised of large and small subunits from different species (16, 17) has indicated that the small subunit can also influence the value of Ω. Because there is greater divergence between eukaryotic and prokaryotic small subunits than between large subunits, one wonders whether the small subunits may, in part, be responsible for the higher Ω values of eukaryotic Rubisco enzymes (8).
The loop between β-strands A and B of the small subunit is the most variable structure among all Rubisco enzymes (12). It is comprised of 10 residues in prokaryotes and eukaryotic non-green algae, ∼22 residues in green plants, and 28 residues in green algae (see Fig. 1). The βA-βB loops of four small subunits, which reside at opposite ends of the octameric large subunit core, surround the opening of a solvent channel that passes through the holoenzyme (1, 2, 12). As a means for determining the significance of the longer loop in plant Rubisco, Bohnert and co-workers (28, 33) used in vitro transcription/translation and uptake into isolated pea chloroplasts to study the influence of engineered small subunits on the holoenzyme. This complex procedure was necessary because eukaryotic holoenzymes cannot be expressed in E. coli (34) and because the small subunit is coded by a family of rbcS genes in plants that cannot be eliminated and replaced with engineered copies (35). When the longer βA-βB loop of plants was engineered into the Synechococcus small subunit, this cyanobacterial small subunit was then able to assemble with plant large subunits in the isolated chloroplasts (33). Thus, the βA-βB loop appeared to be an assembly domain (33), but detailed analysis of Rubisco catalysis was difficult to perform because of the low yield of holoenzyme in the isolated chloroplast system (28, 33).
FIGURE 1.
Small subunit sequences aligned according to Rubisco x-ray crystal structures (18–23). The sequences are from C. reinhardtii (CR), Spinacea oleracea (SO), Nicotiana tabacum (NT), Pisum sativum (PS), Synechococcus (SY), Anabaena (AB), Alcaligenes eutrophus (AE), and Galdieria partita (GP). Residues that comprise α-helixes A and B are colored dark gray, and those that comprise β-strands A through F are colored light gray. Residues conserved in greater than 95% of all known small subunit sequences are colored red (12). Individual residues previously investigated by mutagenesis are colored blue (13–15, 24–32).
In the green alga Chlamydomonas reinhardtii, N54S, A57V, and C65S substitutions in the small subunit βA-βB loop were recovered as genetic suppressors of a temperature-conditional large subunit L290F mutant strain (15, 31). Whereas the L290F substitution caused a decrease in Ω and holoenzyme stability (36), the substitutions in the βA-βB loop returned Ω to nearly normal values and improved thermal stability of the holoenzyme (15, 31). When the rbcS gene family (comprised of linked rbcS1 and rbcS2 genes) of Chlamydomonas was deleted via an insertional mutagenesis procedure (37), it became possible to investigate the role of the βA-βB loop in greater detail, as well as the importance of the eukaryotic small subunit in general. Like all photosynthesis-deficient mutants of this model genetic organism, the Chlamydomonas rbcS deletion strain can be maintained with acetate as an alternative carbon source, and it can be restored to photosynthetic competency by transformation with either of the two rbcS genes (14, 37). Ala-scanning mutagenesis of conserved residues of the eukaryotic βA-βB loop showed that none was essential for holoenzyme assembly, but several affected catalysis, and an R71A substitution caused a decrease in Ω (14). Chlamydomonas Rubisco enzymes engineered to contain the smaller cyanobacterial or plant βA-βB loops could also assemble in vivo despite the loss of substantial structural interactions with the large subunit observed in x-ray crystal structures (38). Both chimeric enzymes had altered catalytic properties, and the enzyme with the cyanobacterial loop had a decreased Ω value. Most recently, the shorter plant loop, along with five substitutions of neighboring large subunit residues, has produced an enzyme with an increased Ω value and kinetic constants quite similar to those of the plant enzyme (11). Altogether, it appears that the small subunit βA-βB loop makes a greater contribution to Rubisco catalytic efficiency than to simply holding together large subunits in the holoenzyme.
Perhaps there are other structural features of the small subunit that contribute to Rubisco function. One approach for identifying such structural regions is to substitute highly conserved residues. Previous directed mutagenesis studies of prokaryotic Rubisco enzymes expressed in E. coli (see Fig. 1) identified a number of small subunit substitutions that affected carboxylase activity or holoenzyme stability (reviewed in Ref. 12). Because those studies were performed when relatively few rbcS sequences were available, focused on conserved regions rather than residues, and substituted a variety of similar or quite dissimilar residues, no specific structural region was studied in any greater detail. In the present study, the 10 most conserved small subunit residues were substituted with alanine in Chlamydomonas Rubisco. This eukaryotic system allows the effect of the substitutions to be assessed in vivo and in vitro. Analysis has indicated that the conserved small subunit residues may influence a single structural element that contributes to the Rubisco large subunit active site.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Cell wall-less C. reinhardtii strain rbcSΔ-T60-3 mt- was used as the host for rbcS nuclear gene transformation. It lacks photosynthesis and requires acetate for growth because of deletion of the 13-kb locus that contains rbcS1 and rbcS2 (37). All of the strains are maintained at 25 °C in darkness on medium containing 10 mm acetate and 1.5% Bacto agar (39). For biochemical analysis, the cells were grown in 250–500 ml of liquid acetate medium at 25 °C on a rotary shaker at 120 rpm in darkness.
Directed Mutagenesis and Transformation
Plasmid pSS1 (37), which contains the entire Chlamydomonas rbcS1 gene on a 5-kb EcoRI fragment, was used for site-directed mutagenesis and transformation. Mutagenesis was performed with synthetic oligonucleotides and a QuikChange kit from Stratagene (40). The mutant plasmids were propagated in E. coli XL1-Blue (Stratagene). To create the mutant substitutions, the sequences encoding Ser-16 (TCC), Leu-18 (CTG), Pro-19 (CCT), Tyr-32 (TAC), Glu-43 (GAG), Trp-73 (TGG), Leu-78 (CTG), Pro-79 (CCC), Phe-81 (TTC), and Glu-92 (GAG) were changed to the most common sequence that encodes alanine (GCC). PCR and restriction fragment analysis were used to screen for the presence of the mutations.
Transformation was performed by electroporation (41), and photosynthesis-competent colonies were selected on minimal medium (without acetate) in the light (80 microeinsteins/m2/s). The engineered rbcS1 gene from each mutant strain was then PCR-amplified and completely sequenced at the University of Nebraska DNA sequencing facility to confirm that only the intended mutation was present. The rbcS1 mutant strains were named S16A, L18A, P19A, Y32A, E43A, W73A, L78A, P79A, F81A, and E92A.
The unaltered pSS1 plasmid was also transformed into rbcSΔ-T60-3, and photosynthesis-competent colonies were recovered as described above. One of these strains, named SS1, was retained as a wild-type control strain. Previous studies have shown that there is no difference in growth phenotype or Rubisco function for Chlamydomonas strains containing either rbcS1 or rbcS2 alone (14).
Biochemical Analysis
Dark-grown cells were sonicated at 0 °C for 3 min in extraction buffer comprised of 50 mm Bicine (pH 8.0), 10 mm NaHCO3, 10 mm MgCl2, and 1 mm dithiothreitol. Cell debris was removed by centrifugation at 30,000 × g for 15 min, and the amount of protein in the supernatant was quantified (42). The cell extract was subjected to SDS-PAGE and Western blotting (43, 44) or fractionated on 10–30% sucrose gradients prepared in the same extraction buffer to isolate pure Rubisco holoenzyme (45). The thermal stability of purified Rubisco was determined as described previously (14, 46).
The Vc and Kc kinetic constants of purified and activated Rubisco were measured by the incorporation of acid-stable 14C from NaH14CO3 in the absence of O2. The ratio (R) of RuBP carboxylase activities measured in the absence and presence of O2 with varying concentrations of CO2 allows a direct determination of Ko according to the formula R = 1 + Kc[O2]/Ko(Kc + [CO2]) (36). The value of Ω (VcKo/VoKc) was determined as the ratio of the rates of carboxylase (vc) and oxygenase (vo) activities measured simultaneously with 88 μm [1-3H]RuBP (15.8 Ci/mol) and 2 mm NaH14CO3 (0.5 Ci/mol) in 30-min reactions at 25 °C according to the formula Ω = vc/vo·[CO2]/[O2] (47, 48). The [1-3H]RuBP and phosphoglycolate phosphatase used in the assays were synthesized/purified by standard methods (47, 49).
RESULTS
Recovery and Phenotypes of Mutants
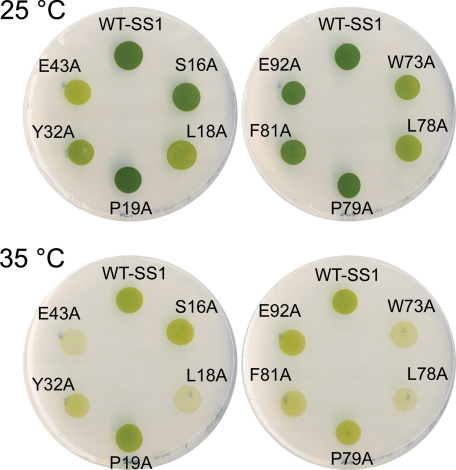
Considering that conserved residues in proteins are usually essential for structure or function, analysis of the most conserved small subunit residues may help to understand the role of the small subunit in Rubisco function. Previous analysis identified only 10 residues that are identical in greater than 95% of all Rubisco small subunit sequences (Fig. 1) (12). To assess the functionality of these residues, it seemed reasonable to substitute each with alanine. Because of its small side group and average ϕ and ψ torsion angles, alanine can mimic the removal of a side group without drastically affecting protein structure. The Chlamydomonas rbcS1 gene was subjected to directed mutagenesis to create the 10 mutant rbcS genes (S16A, L18A, P19A, Y32A, E43A, W73A, L78A, P79A, F81A, and E92A), and these genes were transformed into the Chlamydomonas rbcSΔ-T60-3 strain. Considering the high conservation of the engineered residues, it was surprising to find that all of these mutant genes restored photosynthetic growth to the rbcS knock-out strain. Thus, it was immediately apparent that none of these residues is essential for Rubisco function or assembly. Spot tests were performed (39) to better judge the phenotypes of the mutant strains (Fig. 2). Because nuclear transformation occurs by nonhomologous recombination in Chlamydomonas, there is concern that phenotypic variation may be the result of alterations in gene dosage or transcriptional regulation. To guard against these possibilities, the phenotypes of multiple, independent transformants were compared in spot tests prior to choosing a mutant strain for further analysis. All the mutants grew as well as wild type on acetate medium in darkness (data not shown). However, as shown in Fig. 2, five of the mutant strains (L18A, Y32A, E43A, W73A, and L78A) grew slower than the wild-type SS1 control strain under photosynthetic conditions at the normal growth temperature of 25 °C. Because temperature-conditional Rubisco mutants may be useful for selecting second site suppressor substitutions (50, 51), the phenotypes of the mutant strains were also checked at 35 °C (Fig. 2). At this restrictive temperature, the L18A, E43A, W73A, and L78A mutants failed to grow, and the Y32A and F81A mutants displayed substantial growth inhibition. The S16A, P19A, P79A, and E92A mutants had growth phenotypes comparable with the SS1 wild type.
FIGURE 2.
Photosynthetic growth phenotypes of wild-type SS1 and Rubisco small subunit mutants. Spot tests were performed by plating equal numbers of dark-grown cells on minimal medium in the light (80 microeinsteins/m2/s) at either the normal growth temperature of 25 °C or an elevated temperature of 35 °C. The wild-type SS1 control strain (WT-SS1) was created by transforming the rbcSΔ-T60-3 rbcS deletion mutant with the wild-type rbcS1 gene.
Analysis of Holoenzyme Stability
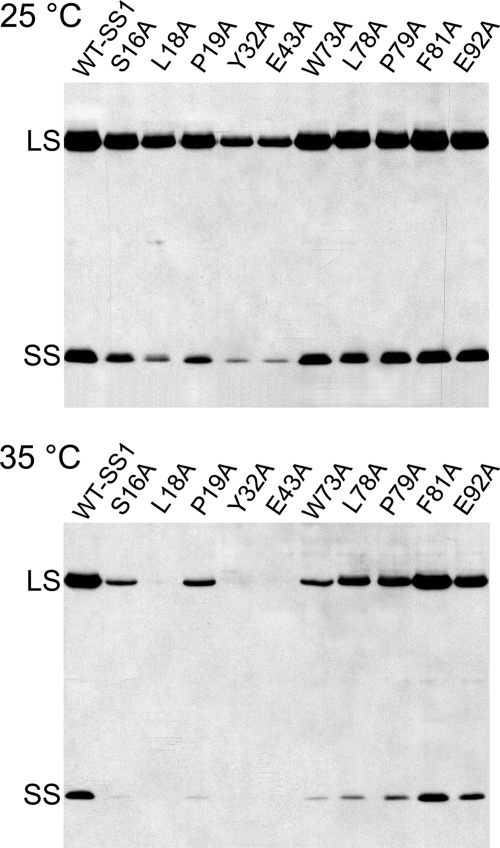
To see whether the growth phenotypes of the mutant strains (Fig. 2) reflected a decrease in the amount of Rubisco holoenzyme, extracts of cells grown at 25 and 35 °C in darkness were subjected to SDS-PAGE and Western analysis (Fig. 3). Because small subunits are rapidly degraded in the absence of large subunits (52), and large subunit synthesis is blocked at the level of translation in the absence of small subunits (37), the amount of subunits observed with SDS-PAGE reflects the amount of Rubisco holoenzyme in vivo. When grown at 25 °C, most of the mutant strains had nearly normal levels of Rubisco holoenzyme. The most obvious decreases in holoenzyme were observed for the L18A, Y32A, and E43A mutants (25 °C; Fig. 3), and these mutants also had reductions in growth when compared with wild type at 25 °C (Fig. 2). One generally suspects that the decreases in holoenzyme result from enhanced proteolysis of mutant enzymes altered in holoenzyme structure or stability. Mutants W73A and L78A also had reduced growth at 25 °C, but these mutants had normal levels of holoenzyme (25 °C; Fig. 3), indicating that the W73A and L78A Rubisco enzymes may be defective in net CO2 fixation. Furthermore, although mutants Y32A and E43A have similar amounts of holoenzyme (25 °C; Fig. 3), Y32A grows somewhat better than E43A under photosynthetic conditions at 25 °C (Fig. 2). When grown at 35 °C in darkness, mutants L18A, Y32A, and E43A lacked Rubisco holoenzyme (Fig. 2). However, other mutants that grew poorly if at all at 35 °C (W73A, L78A, and F81A) retained significant levels of holoenzyme, indicating that reduced growth at 35 °C may reflect alterations in Rubisco catalysis at this elevated temperature.
FIGURE 3.
Western blot analysis of total soluble proteins isolated from wild-type SS1 and Rubisco small subunit mutants. Protein extracts (60 μg/lane) of cells grown at either 25 or 35 °C in darkness were fractionated by gradient gel (7.5–15% polyacrylamide) electrophoresis (43), blotted to nitrocellulose, probed with rabbit anti-Chlamydomonas Rubisco immunoglobulin G (0.5 μg/ml) (38), and detected by enhanced chemiluminescence (Amersham Biosciences) (44). The Rubisco 55-kDa large subunit (LS) and 16-kDa small subunit (SS) are indicated.
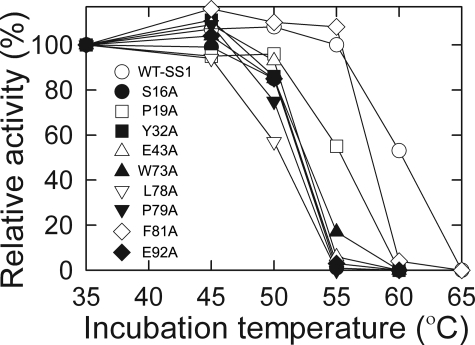
Because many of the mutant enzymes have reduced amounts of Rubisco when grown at the elevated temperature of 35 °C, structural stability of the mutant enzymes was further assessed by assaying thermal stability of purified Rubisco in vitro (14, 46) (Fig. 4). Although mutant L18A has substantial levels of Rubisco when grown at 25 °C (Fig. 3), assembled holoenzyme could not be isolated by sucrose gradient fractionation. Because of this presumed structural instability, L18A Rubisco was not studied further. All of the other mutant enzymes had decreased in vitro thermal stability relative to the SS1 wild type (Fig. 4), but, because this instability is detected at temperatures far above growth or enzyme assay temperatures, it is unlikely to affect the accurate determination of Rubisco kinetic constants.
FIGURE 4.
Thermal inactivation of Rubisco purified from wild-type SS1 and small subunit mutants (14, 46). Rubisco (5 μg) in 0.5 ml of 50 mm Bicine (pH 8.0), 10 mm NaH14CO3 (58 Ci/mol), 10 mm MgCl2, and 1 mm dithiothreitol was incubated at each temperature for 10 min. The samples were then cooled on ice for 5 min, and carboxylase activity was initiated at 25 °C by adding 20 μl of 10 mm RuBP. The reactions were terminated after 1 min with 0.5 ml of 3 m formic acid in methanol. The activities were normalized to the specific activities measured after the 35 °C incubation (WT-SS1, 1.6 μmol/min/mg; S16A, 1.5 μmol/min/mg; P19A, 1.6 μmol/min/mg; Y32A, 0.3 μmol/min/mg; E43A, 0.2 μmol/min/mg; W73A, 0.4 μmol/min/mg; L78A, 0.1 μmol/min/mg; P79A, 0.6 μmol/min/mg; F81A, 0.6 μmol/min/mg; E92A, 0.4 μmol/min/mg).
Analysis of Rubisco Function
As a first step for analyzing the catalytic properties of the mutant enzymes, Ω values, carboxylation specific activities, and the influence of O2 on carboxylation were determined (Table 1). The Y32A and E43A enzymes were found to have 8–10% decreases in Ω. Y32A caused an increase in O2 inhibition, but E43A caused a decrease. Because the ratio of carboxylase activities in the absence and presence of O2 is determined by only the Kc and Ko kinetic constants (36), one might assume that the decrease in Ω of the E43A enzyme results from the observed substantial decrease in carboxylation specific activity (assayed with saturating CO2 provided by 12.4 mm NaHCO3) (Table 1). The W73A, L78A, and F81A mutant enzymes also had decreases in O2 inhibition, but their Ω values were the same as that of the SS1 wild-type enzyme. The P79A enzyme had a wild-type Ω value and normal level of O2 inhibition, but, because its carboxylation specific activity was decreased ∼5-fold, further study of this enzyme was also warranted. Only three of the mutant enzymes (S16A, P19A, and E92A) had nearly normal catalytic properties in these initial assays (Table 1). Considering that the S16A, P19A, and E92A mutant strains also have wild-type phenotypes (Fig. 2) and normal levels of Rubisco holoenzyme (Fig. 3), they were not studied further.
TABLE 1.
Kinetic properties and oxygen inhibition of Rubisco purified from wild-type SS1 and small subunit mutants
The values are the means ± S.D. (n − 1) of three separate enzyme preparations.
| Enzyme | Ω(VcKo/VoKc) | RuBP carboxylase activity |
Ratio(A/B) | ||
|---|---|---|---|---|---|
| 100% N2(12.4 mmNaHCO3) | 100% N2(0.98 mmNaHCO3) (A) | 100% O2(0.98 mmNaHCO3) (B) | |||
| μmol CO2/h/mg of protein | |||||
| Wild-type SS1 | 60 ± 1 | 93 ± 4 | 32.4 ± 0.9 | 13.0 ± 0.4 | 2.5 |
| S16A | 59 ± 1 | 64 ± 3 | 23.3 ± 0.1 | 8.9 ± 0.1 | 2.6 |
| P19A | 59 ± 1 | 79 ± 4 | 28.4 ± 0.3 | 11.0 ± 0.1 | 2.6 |
| Y32A | 54 ± 2 | 11 ± 1 | 4.2 ± 0.1 | 1.4 ± 0.1 | 3.0 |
| E43A | 55 ± 2 | 3 ± 1 | 0.8 ± 0.1 | 0.5 ± 0.1 | 1.6 |
| W73A | 58 ± 1 | 22 ± 4 | 6.2 ± 1.0 | 3.8 ± 0.5 | 1.6 |
| L78A | 62 ± 2 | 4 ± 1 | 0.9 ± 0.1 | 0.6 ± 0.1 | 1.5 |
| P79A | 58 ± 1 | 19 ± 1 | 6.5 ± 0.9 | 2.7 ± 0.4 | 2.4 |
| F81A | 61 ± 2 | 18 ± 2 | 4.3 ± 0.7 | 2.5 ± 0.3 | 1.7 |
| E92A | 59 ± 1 | 54 ± 3 | 19.8 ± 1.0 | 7.3 ± 0.2 | 2.7 |
The Rubisco active sites are formed between large subunits, but initial studies indicated that at least six of the small subunit-directed mutant substitutions produced alterations in catalysis (Table 1). These mutant enzymes were analyzed in greater detail (Table 2). Although the Y32A and E43A enzymes both have decreases in the value of Ω, these decreases arise from different changes in kinetic constants. The Y32A enzyme has a beneficial increase in the Vc/Vo ratio, but this improvement is negated by a 27% decrease in Ko (and a decrease in the Ko/Kc ratio). In contrast, the E43A enzyme has an improved Ko/Kc because of a greater increase in Ko than in Kc, but its Ω value is decreased because of a substantial decrease in Vc and the Vc/Vo ratio. The W73A, L78A, and F81A mutant enzymes have kinetic properties much like those of the E43A enzyme (Table 2). In comparison with the wild-type enzyme, they all have decreases in Vc and Vc/Vo and increases in Kc, Ko, and Ko/Kc. However, in comparison with the E43A enzyme, the W73A, L78A, and F81A mutant enzymes have normal (or near normal) Ω values because of higher Vc and Vc/Vo values. The P79A mutant enzyme is quite different from the other mutant enzymes. It has only a 63% decrease in Vc and retains normal Kc, Ko, Vc/Vo, and Ω values. Nonetheless, the P79A mutant enzyme appears to be responsible for a decrease in the photoautotrophic growth of the P79A mutant strain, albeit less so than the negative effects on growth produced by the catalytically altered Y32A, E43A, W73A, L78A, and F81A mutant enzymes (Fig. 2). Altogether, it appears that the conserved small subunit residues can influence large subunit catalysis in several ways. A summary of mutant phenotypes and catalytic effects is presented in supplemental Table S1.
TABLE 2.
Kinetic properties of Rubisco purified from wild-type SS1 and small subunit mutants
| Enzyme | Ω(VcKo/VoKc)a,b | Vcb | Kcb | Kob | Vc/Kcc | Ko/Kcc | Vc/Voc |
|---|---|---|---|---|---|---|---|
| μmol/h/mg | μm CO2 | μm CO2 | |||||
| Wild-type | 60 ± 1 | 127 ± 13 | 31 ± 2 | 527 ± 20 | 4.1 | 17 | 3.5 |
| Y32A | 54 ± 2 | 23 ± 2 | 30 ± 2 | 383 ± 31 | 0.8 | 13 | 4.2 |
| E43A | 55 ± 2 | 10 ± 1 | 64 ± 3 | 1624 ± 309 | 0.2 | 25 | 2.2 |
| W73A | 58 ± 1 | 28 ± 8 | 46 ± 3 | 1159 ± 249 | 0.6 | 25 | 2.3 |
| L78A | 61 ± 2 | 16 ± 6 | 53 ± 7 | 974 ± 24 | 0.3 | 18 | 3.3 |
| P79A | 58 ± 1 | 47 ± 9 | 33 ± 3 | 529 ± 83 | 1.4 | 16 | 3.6 |
| F81A | 61 ± 2 | 47 ± 14 | 43 ± 2 | 1008 ± 69 | 1.1 | 23 | 2.6 |
a Values from Table I.
b The values are the means ± S.D. (n − 1) of three separate enzyme preparations.
c Calculated values.
Analysis of Rubisco Structure
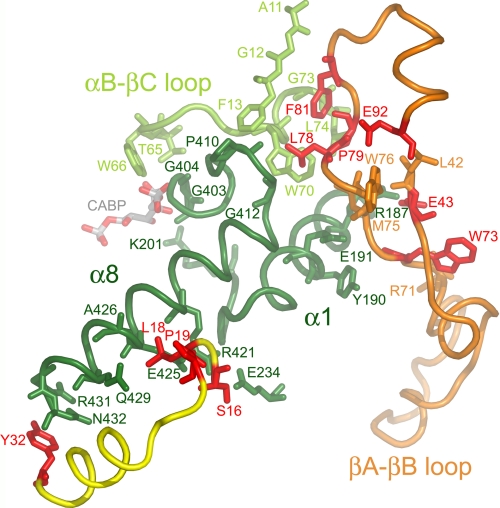
The small subunit residues analyzed in the present study were chosen because of their conservation in numerous small subunits (12). One might suppose that such highly conserved residues reside in the hydrophobic core of the small subunit and play some role in protein folding or assembly. However, analysis of the 1.4-Å x-ray crystal structure of Chlamydomonas Rubisco revealed that all but one (Glu-92) of the conserved residues are in contact with the Rubisco large subunit (Fig. 5 and supplemental Table 1), and as expected, the contacted large subunit residues are highly conserved (supplemental Table S2). Residues Ser-16, Leu-18, and Tyr-32 are in contact with the bottom half of large subunit α-helix 8 (Fig. 5), which is a structural element of the α/β-barrel that forms the active site. Leu-78 from a neighboring small subunit is in contact with the loop preceding large subunit α-helix 8 at the top of the α/β-barrel. Pro-79, Phe-81, and Glu-92 are closely associated with Leu-78 in this region (Fig. 5). Glu-43 and Trp-73 are in van der Waals contact with each other, and Glu-43 forms an ionic bond with Arg-187 in α-helix 1 of the large subunit α/β-barrel (Fig. 5). Considering that the E43A, W73A, L78A, and F81A mutant substitutions produce similar catalytic effects (Table 2), and the affected residues cluster near the top of α-helix 8 (Fig. 5), small subunit structure in this region may play a significant role in determining Rubisco function.
FIGURE 5.
Conserved small subunit residues (red) in the x-ray crystal structure of Chlamydomonas Rubisco (Protein Data Bank code 1GK8) (23). Looking down from the top of the Rubisco holoenzyme, the large subunit α/β-barrel is viewed from the side. Conserved small subunit residues from one small subunit (yellow ribbon) cluster near the carboxyl-terminal bottom (Tyr-32) and middle (Ser-16, Leu-18, and Pro-19) of large subunit α-helix 8 (dark green residues 413–432). Conserved small subunit residues Glu-43, Trp-73, Leu-78, Pro-79, Phe-81, and Glu-92 from a second small subunit (orange ribbon) cluster near the amino-terminal top of large subunit α-helix 8 or its preceding loop (dark green residues 403–412). Small subunit residues Glu-43 and Trp-73 are also in contact with α-helix 1 (dark green residues 182–194 and residues 195–201 following). Small subunit residues Leu-78, Pro-79, and Phe-81 are in contact with part of the loop between α-helix B and β-strand C from a neighboring large subunit (light-green residues 65–74). Large subunit active site residues Thr-65, Trp-66, Lys-201, Gly-403, and Gly-404 are in contact with the CABP transition state analog. A summary of contacts between the conserved small subunit residues and large subunit residues is provided in supplemental Table S1.
DISCUSSION
Analysis of hybrid (16, 17), chimeric (11, 38), and small subunit mutant enzymes (13–15, 31) has shown that Rubisco small subunits can influence both carboxylation catalytic efficiency and Ω. However, because small subunits do not interact directly with the large subunit active site (2), it remains difficult to determine the structural basis for this influence. A chimeric Chlamydomonas Rubisco enzyme containing the shorter small subunit βA-βB loop of Synechococcus has a decreased Ω value, but the x-ray crystal structure of the mutant enzyme failed to identify a direct structural path from the βA-βB loop to the large subunit active site (38).
It was interesting to find that the small subunit residues chosen for analysis in the present study, based solely on their high level of conservation (Fig. 1), associate in groups near α-helix 8 of the large subunit α/β-barrel (Fig. 5). Previous comparison of the x-ray crystal structure of L8S8 spinach Rubisco with the L2 structure of R. rubrum Rubisco identified shifts in α-helix 8 and its preceding loop (53). Because small subunit residues interact with these large subunit regions, it was concluded that the small subunit was responsible for the observed differences in conformation (53). Furthermore, because the loop before α-helix 8 contains two active site residues (Gly-403 and Gly-404) that bind to one of the phosphate groups of the CABP transition state analog (20, 23), it was also proposed (53) that small subunits might influence Rubisco catalysis via these interactions and account for the differences in Ω values between L2 (Ω ≈ 15) and L8S8 (Ω ≈ 80) Rubisco enzymes (8).
At the bottom of large subunit α-helix 8, the conserved small subunit residue Tyr-32 appears to shield large subunit residues Arg-431 and Asn-432 from solvent (Fig. 5). These residues are the last two at the carboxyl-terminal end of α-helix 8 at the bottom of the α/β-barrel. Nonetheless, the Y32A mutant enzyme has a 10% decrease in Ω (Table 2) in addition to an associated structural instability (Figs. 3 and 4). Because α-helixes are dipoles because of resonance at the peptide bond, perhaps the nature of small subunit residue 32 at the carboxyl-terminal end of large subunit α-helix 8 can influence the partial positive charge at the amino-terminal end of α-helix 8.
Of the three conserved small subunit residues that reside near the middle of large subunit α-helix 8 (Ser-16, Leu-18, and Pro-19) (Fig. 5), only Leu-18 has a significant effect on cell growth when substituted with alanine (Fig. 2). Because the side chain of Leu-18 is in van der Waals contact with three large subunit α-helix-8 residues (Glu-425, Ala-426, and Gln-429), the loss of these interactions may account for both the holoenzyme instability observed in L18A cells grown at 35 °C (Fig. 3) and the difficulty with purifying L18A mutant Rubisco. However, despite the fact that Ser-16 and Pro-19 are in contact with Leu-18, the S16A and P19A substitutions have only minor effects on carboxylation specific activity (Table 1) and in vitro holoenzyme thermal stability (Figs. 3 and 4). Perhaps less conservative substitutions at these residues would produce an effect. In previous studies with Synechococcus Rubisco expressed in E. coli, P19A and P19H substitutions also had no significant effect on carboxylation or holoenzyme assembly (13, 26). However, an S16D substitution disrupted holoenzyme stability (26), whereas an S16A substitution was reported to cause a decrease in Ω (13). In this latter study (13), an increased Ω value was also reported for a P20A substitution, but other Rubisco kinetic constants were not measured. Other substitutions were created in cyanobacterial small subunits at residues near to Ser-16, Leu-18, and Pro-19 (Fig. 1), but, except for one study (27), analysis of these mutant enzymes was incomplete. R10G and E13V substitutions blocked holoenzyme assembly (13, 25), and some substitutions at Thr-14, Tyr-17, and Leu-21 caused decreases in both carboxylation and holoenzyme stability (13, 26, 27). Perhaps Ser-16 and Pro-19 have provided some selective advantage during evolution that is not readily apparent from the completed biochemical analysis of Chlamydomonas Rubisco. Further study of the region defined by the conservative Ser-16, Leu-18, and Pro-19 residues may be necessary to determine whether this region is primarily responsible for holoenzyme stability, catalytic efficiency, or both.
The remainder of the conserved small subunit residues cluster near the top of α-helix 8 (Glu-43, Trp-73, Leu-78, Pro-79, Phe-81, and Glu-92) (Fig. 5). With respect to the evolution of Rubisco structure, it is interesting to note that these residues reside in a small subunit different from the one that comes in contact with the carboxyl-terminal end of large subunit α-helix 8. Although conserved Tyr-32 caps the carboxyl-terminal end of large subunit α-helix 8, small subunit Leu-78 is in contact with large subunit residues Pro-410 and Gly-412 just before the start of α-helix 8 at Asn-413 (Fig. 5). The L78A substitution does not alter Ω, but it does cause a decrease in Vc and increases in Kc and Ko (Table 2). The loss of interactions between small subunit Leu-78 and large subunit residues Pro-410 and Gly-412 may alter the structure of the loop preceding α-helix 8. Catalysis may be affected because this loop contains the active site residues Gly-403 and Gly-404, which interact with the CABP transition state analog (Fig. 5). Leu-78 and Phe-81 are not in direct contact with each other, but the L78A and F81A substitutions cause similar changes in catalytic properties (Table 2). The aromatic ring of Phe-81 is surrounded by large subunit residues Trp-70, Gly-73, and Leu-74 in the amino-terminal domain of a neighboring large subunit (Fig. 5). The loss of the Phe-81 side group might influence the position of Leu-78. However, the Leu-78 side group is also in van der Waals contact with large subunit Trp-70, and residues Trp-70, Gly-73, and Leu-74 are in the loop between large subunit α-helix B and β-strand C (αB-βC loop) that contains active site residues Thr-65 and Trp-66 (Fig. 5). It might be possible that L78A and F81A affect catalysis by altering the loop before α-helix 8, the amino-terminal-domain αB-βC loop, or both. Conserved small subunit residue Pro-79 is in contact with large subunit Trp-70 and small subunit residues Leu-78 and Phe-81, but the P79A substitution causes a relatively small decrease in only Vc. Perhaps a less conservative substitution would produce a different effect. In a previous study, when the homologous Pro-59 residue in the small subunit of Anabaena Rubisco was replaced with a larger and polar histidine (Fig. 1), holoenzyme assembly in E. coli was blocked (25). However, the Chlamydomonas L78A, P79A, and F81A mutant strains have normal amounts of Rubisco when grown at 25 °C, and the purified mutant enzymes have only a modest decrease in thermal stability in vitro (Fig. 4). The side chain of conserved residue Glu-92 resides at the surface of small subunit α-helix B (Figs. 1 and 5). Glu-92 is in contact with some of the same residues contacted by small subunit Leu-78, Pro-79, and Phe-81, but the E92A substitution causes only a small decrease in carboxylation (Table 2) with no effect on holoenzyme stability (Figs. 3 and 4). Altogether, it seems likely that the conserved Leu-78, Pro-79, Phe-81, and Glu-92 residues may have a greater role in function than in holoenzyme assembly or stability.
Whereas conserved small subunit residues Glu-43 and Trp-73 are also in the region near the top of large subunit α-helix 8, they are in direct contact with large subunit α-helix 1 of the α/β-barrel (Fig. 5). Glu-43 forms an ionic bond with large subunit Arg-187, and Trp-73 is in van der Waals contact with large subunit Tyr-190. The E43A and W73A enzymes both have decreases in Vc and increases in Kc and Ko, but the E43A enzyme also has a decreased Ω value (Table 2) and an associated structural instability in vivo (Fig. 3), which may result from the loss of the salt bridge to the large subunit. Analysis of small subunit Trp-55 and Trp-58 in Synechococcus Rubisco (which are homologous to Trp-73 and Trp-76 in Chlamydomonas; Fig. 1) indicated that W55F and W58F mutant enzymes also had a decrease in Vc, but Kc was not changed, and Ko was not measured (24, 26). When the residue homologous to Chlamydomonas Trp-73 was substituted with arginine in Anabaena Rubisco, the holoenzyme failed to assemble in E. coli (25). It was suggested, in part, that the structural instability might result from the additional arginine disrupting the ionic interaction between small subunit Glu-43 and large subunit Arg-187 (25). However, as demonstrated by the Chlamydomonas E43A mutant enzyme, this ionic bond is not essential for holoenzyme assembly. Substitutions were also created at Tyr-72 of the Chlamydomonas small subunit (32) and at the homologous plant and cyanobacterial tyrosine residues (26, 28) to assess Rubisco assembly/stability, but the catalytic properties of the mutant enzymes were not determined.
Although Glu-43 and Trp-73 are in contact with large subunit α-helix 1 and far from the other conserved residues that cluster at the top of large subunit α-helix 8 (Fig. 5), the E43A and W73A substitutions affect catalysis in much the same way as do the L78A and F81A small subunit substitutions (Table 2). All have similar decreases in Vc and increases in Kc and Ko. Furthermore, Glu-43 and Trp-73 may have a structural association with the other conserved small subunit residues near the amino-terminal end of α-helix 8. For example, Trp-73, Leu-78, and Pro-79 are all in contact with small subunit Met-75, which forms a hydrogen bond between its carbonyl oxygen and the OE2 of large subunit residue Glu-191. Glu-43 and Glu-92 are both in contact with small subunit Leu-42, which forms a hydrogen bond between its carbonyl oxygen and the amide nitrogen of small subunit Trp-76 (Fig. 5). Furthermore, Trp-73 is located at the base of the βA-βB loop (Fig. 5). In a previous study of the conserved residues unique to the βA-βB loops of plants and green algae (Fig. 1), an R71A substitution was found to decrease Ω (14). This mutant enzyme also has a decreased Vc and increased Kc and Ko values just like the E43A, W73A, L78A, and F81A mutant enzymes (Table 2). Arg-71 forms an ionic bond with large subunit residue Glu-223, but it is also in van der Waals contact with small subunit Trp-73 (Fig. 5). Perhaps the small subunit βA-βB loop can influence large subunit catalysis by a mechanism shared in common with small subunit Glu-43, Trp-73, Leu-78, and Phe-81.
None of the conserved small subunit residues is essential for Rubisco assembly or function, but many can influence large subunit catalytic efficiency (Table 2). Most of these residues cluster near the top or bottom of large subunit α-helix 8 (Fig. 5), supporting the idea that α-helix 8 may serve as a major structural element by which small subunits can influence large subunit catalysis (53). The conserved small subunit residues may be responsible for the differences in Ω observed between L2 and L8S8 Rubisco enzymes (8, 53). However, because they are conserved, they cannot be responsible for the differences in Ω observed among various L8S8 enzymes (8–10). Instead, divergent small subunit residues in the structural regions defined by the conserved residues may serve as targets for future investigation.
Supplementary Material
This work was supported in part by Grant DE-FG02-00ER15044 from the United States Department of Energy.
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- Rubisco
- ribulose 1,5-bisphosphate carboxylase/oxygenase
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- CABP
- 2-carboxy-d-arabinitol 1,5-bisphosphate
- RuBP
- ribulose 1,5-bisphosphate
- Ω
- CO2/O2 specificity factor.
REFERENCES
- 1.Spreitzer R. J., Salvucci M. E. (2002) Annu. Rev. Plant Biol. 53, 449–475 [DOI] [PubMed] [Google Scholar]
- 2.Andersson I., Backlund A. (2008) Plant Physiol. Biochem. 46, 275–291 [DOI] [PubMed] [Google Scholar]
- 3.Tabita F. R., Satagopan S., Hanson T. E., Kreel N. E., Scott S. S. (2008) J. Exp. Bot. 59, 1515–1524 [DOI] [PubMed] [Google Scholar]
- 4.Laing W. A., Ogren W. L., Hageman R. H. (1974) Plant Physiol. 54, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z., Spreitzer R. J. (1992) Photosynth. Res. 31, 157–164 [DOI] [PubMed] [Google Scholar]
- 6.Hartman F. C., Harpel M. R. (1994) Annu. Rev. Biochem. 63, 197–234 [DOI] [PubMed] [Google Scholar]
- 7.Cleland W. W., Andrews T. J., Gutteridge S., Hartman F. C., Lorimer G. H. (1998) Chem. Rev. 98, 549–562 [DOI] [PubMed] [Google Scholar]
- 8.Jordan D. B., Ogren W. L. (1981) Nature 291, 513–515 [Google Scholar]
- 9.Read B. A., Tabita F. R. (1994) Arch. Biochem. Biophys. 312, 210–218 [DOI] [PubMed] [Google Scholar]
- 10.Uemura K., Anwaruzzaman, Miyachi S., Yokota A. (1997) Biochem. Biophys. Res. Commun. 233, 568–571 [DOI] [PubMed] [Google Scholar]
- 11.Spreitzer R. J., Peddi S. R., Satagopan S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17225–17230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spreitzer R. J. (2003) Arch. Biochem. Biophys. 414, 141–149 [DOI] [PubMed] [Google Scholar]
- 13.Kostov R. V., Small C. L., McFadden B. A. (1997) Photosynth. Res. 54, 127–134 [Google Scholar]
- 14.Spreitzer R. J., Esquivel M. G., Du Y. C., McLaughlin P. D. (2001) Biochemistry 40, 5615–5621 [DOI] [PubMed] [Google Scholar]
- 15.Genkov T., Du Y. C., Spreitzer R. J. (2006) Arch. Biochem. Biophys. 451, 167–174 [DOI] [PubMed] [Google Scholar]
- 16.Read B. A., Tabita F. R. (1992) Biochemistry 31, 5553–5560 [DOI] [PubMed] [Google Scholar]
- 17.Kanevski I., Maliga P., Rhoades D. F., Gutteridge S. (1999) Plant Physiol. 119, 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman J., Gutteridge S. (1993) J. Biol. Chem. 268, 25876–25886 [PubMed] [Google Scholar]
- 19.Schreuder H. A., Knight S., Curmi P. M., Andersson I., Cascio D., Sweet R. M., Brändén C. I., Eisenberg D. (1993) Protein Sci. 2, 1136–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson I. (1996) J. Mol. Biol. 259, 160–174 [DOI] [PubMed] [Google Scholar]
- 21.Hansen S., Vollan V. B., Hough E., Andersen K. (1999) J. Mol. Biol. 288, 609–621 [DOI] [PubMed] [Google Scholar]
- 22.Sugawara H., Yamamoto H., Shibata N., Inoue T., Okada S., Miyake C., Yokota A., Kai Y. (1999) J. Biol. Chem. 274, 15655–15661 [DOI] [PubMed] [Google Scholar]
- 23.Taylor T. C., Backlund A., Bjorhall K., Spreitzer R. J., Andersson I. (2001) J. Biol. Chem. 276, 48159–48164 [DOI] [PubMed] [Google Scholar]
- 24.Voordouw G., De Vries P. A., Van den Berg W. A., De Clerck E. P. (1987) Eur. J. Biochem. 163, 591–598 [DOI] [PubMed] [Google Scholar]
- 25.Fitchen J. H., Knight S., Andersson I., Branden C. I., McIntosh L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5768–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee B., Berka R. M., Tabita F. R. (1991) J. Biol. Chem. 266, 7417–7422 [PubMed] [Google Scholar]
- 27.Paul K., Morell M. K., Andrews T. J. (1991) Biochemistry 30, 10019–10026 [DOI] [PubMed] [Google Scholar]
- 28.Flachmann R., Bohnert H. J. (1992) J. Biol. Chem. 267, 10576–10582 [PubMed] [Google Scholar]
- 29.Read B. A., Tabita F. R. (1992) Biochemistry 31, 519–525 [DOI] [PubMed] [Google Scholar]
- 30.Flachmann R., Zhu G., Jensen R. G., Bohnert H. J. (1997) Plant Physiol. 114, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Y. C., Hong S., Spreitzer R. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14206–14211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esquível M. G., Pinto T. S., Marín-Navarro J., Moreno J. (2006) Biochemistry 45, 5745–5753 [DOI] [PubMed] [Google Scholar]
- 33.Wasmann C. C., Ramage R. T., Bohnert H. J., Ostrem J. A. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cloney L. P., Bekkaoui D. R., Hemmingsen S. M. (1993) Plant Mol. Biol. 23, 1285–1290 [DOI] [PubMed] [Google Scholar]
- 35.Dhingra A., Portis A. R., Jr., Daniell H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6315–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z. X., Chastain C. J., Al-Abed S. R., Chollet R., Spreitzer R. J. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4696–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khrebtukova I., Spreitzer R. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13689–13693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karkehabadi S., Peddi S. R., Anwaruzzaman M., Taylor T. C., Cederlund A., Genkov T., Andersson I., Spreitzer R. J. (2005) Biochemistry 44, 9851–9861 [DOI] [PubMed] [Google Scholar]
- 39.Spreitzer R. J., Mets L. (1981) Plant Physiol. 67, 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papworth C., Bauer J. C., Braman J., Wright D. A. (1996) Strategies 9, 3–4 [Google Scholar]
- 41.Shimogawara K., Fujiwara S., Grossman A., Usuda H. (1998) Genetics 148, 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 43.Chua N. H. (1980) Methods Enzymol. 69, 434–446 [Google Scholar]
- 44.Thow G., Zhu G., Spreitzer R. J. (1994) Biochemistry 33, 5109–5114 [DOI] [PubMed] [Google Scholar]
- 45.Spreitzer R. J., Chastain C. J. (1987) Curr. Genet. 11, 611–616 [DOI] [PubMed] [Google Scholar]
- 46.Chen Z., Hong S., Spreitzer R. J. (1993) Plant Physiol. 101, 1189–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan D. B., Ogren W. L. (1981) Plant Physiol. 67, 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spreitzer R. J., Jordan D. B., Ogren W. L. (1982) FEBS Lett. 148, 117–121 [Google Scholar]
- 49.Kuehn G. D., Hsu T. C. (1978) Biochem. J. 175, 909–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong S., Spreitzer R. J. (1997) J. Biol. Chem. 272, 11114–11117 [DOI] [PubMed] [Google Scholar]
- 51.Du Y. C., Spreitzer R. J. (2000) J. Biol. Chem. 275, 19844–19847 [DOI] [PubMed] [Google Scholar]
- 52.Spreitzer R. J., Goldschmidt-Clermont M., Rahire M., Rochaix J. D. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 5460–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider G., Knight S., Andersson I., Brändén C. I., Lindqvist Y., Lundqvist T. (1990) EMBO J. 9, 2045–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.