Abstract
Helicobacter pylori infection is associated with gastric adenocarcinoma in some humans, especially those that develop an antecedent condition, chronic atrophic gastritis (ChAG). Gastric epithelial progenitors (GEPs) in transgenic gnotobiotic mice with a ChAG-like phenotype harbor intracellular collections of H. pylori. To characterize H. pylori adaptations to ChAG, we sequenced the genomes of 24 isolates obtained from 6 individuals, each sampled over a 4-year interval, as they did or did not progress from normal gastric histology to ChAG and/or adenocarcinoma. H. pylori populations within study participants were largely clonal and remarkably stable regardless of disease state. GeneChip studies of the responses of a cultured mouse gastric stem cell-like line (mGEPs) to infection with sequenced strains yielded a 695-member dataset of transcripts that are (i) differentially expressed after infection with ChAG-associated isolates, but not with a “normal” or a heat-killed ChAG isolate, and (ii) enriched in genes and gene functions associated with tumorigenesis in general and gastric carcinogenesis in specific cases. Transcriptional profiling of a ChAG strain during mGEP infection disclosed a set of responses, including up-regulation of hopZ, an adhesin belonging to a family of outer membrane proteins. Expression profiles of wild-type and ΔhopZ strains revealed a number of pH-regulated genes modulated by HopZ, including hopP, which binds sialylated glycans produced by GEPs in vivo. Genetic inactivation of hopZ produced a fitness defect in the stomachs of gnotobiotic transgenic mice but not in wild-type littermates. This study illustrates an approach for identifying GEP responses specific to ChAG-associated H. Pylori strains and bacterial genes important for survival in a model of the ChAG gastric ecosystem.
Helicobacter pylori establishes a lifelong infection in most of its human hosts. The majority of colonized individuals remain asymptomatic and may even benefit from harboring this bacterium; for example, evidence is accumulating that colonization may offer protection against gastroesophageal reflux disease (1) and esophageal cancer (2, 3) as well as asthma and allergies (4). However, a minority of hosts go on to develop severe gastric pathology, including peptic ulcer disease and gastric cancer.
The mechanisms that link H. pylori infection and gastric cancer, specifically adenocarcinoma, are largely ill-defined. The risk for developing cancer is greater if an individual develops chronic atrophic gastritis (ChAG),3 a histopathologic state characterized in part by loss of acid-producing parietal cells. Several observations have suggested the possibility that this organism may interact directly with gastric stem cells, creating a dangerous liaison that could influence tumorigenesis. First, we have used a germ-free transgenic mouse model of ChAG where parietal cells are eliminated using an attenuated diphtheria toxin A fragment (tox176) expressed under the control of parietal cell-specific regulatory elements from the mouse Atpb4 gene to show that loss of this gastric epithelial lineage is associated with amplification of gastric stem cells expressing NeuAcα2,3Galβ1,4-containing glycan receptors recognized by H. pylori adhesins (5). H. pylori colonization of these germ-free transgenic mice results in invasion of a subset of gastric epithelial progenitors (GEPs), which harbor small communities of intracellular bacteria weeks to months after a single gavage of bacteria into the stomachs of these animals. This internalization exhibits cellular specificity; it does not occur in the differentiated descendants of GEPs, notably NeuAcα2,3Galβ1,4-expressing, mucus-producing gastric pit cells. The ability to establish residency within GEPs may have implications for tumorigenesis as stem cells are long-lived and have been postulated to be the site of origin for many types of neoplasms (cancer stem cell hypothesis) (6). Second, studies of human gastric biopsies with pre-neoplastic as well as neoplastic changes have revealed intracellular H. pylori (7). Third, we have identified notable differences between H. pylori strains isolated from a single host who progressed from ChAG to gastric adenocarcinoma that may be relevant to initiation and/or progression of carcinogenesis. Both ChAG and cancer-associated strains were able to initially colonize the stomachs of germ-free Atbp4-tox176 mice as indicated by an IgM response, but the ChAG-associated strain was able to maintain a more persistent infection (8). However, the cancer-associated strain was more adapted to an intracellular habitat in GEPs. Using a mouse GEP cell line (mGEPs) with a transcriptome that resembles that of laser capture microdissected, Atp4b-tox176-derived GEPs, and that can support attachment, internalization, and intracellular survival of H. pylori, we noted that the cancer strain had a greater invasive capacity (8). Whole genome transcriptional profiling of mGEP and bacterial responses to infection disclosed isolate-specific and progenitor cell-specific molecular phenotypes (a non-progenitor-like mouse gastric epithelial cell line was used as a control in these infection experiments) (8). Compared with the ChAG-associated strain, the cancer strain exhibited increased expression of bacterial ketol-acid reductoisomerase upon mGEP infection, suggesting that it is more able to overcome its requirements for valine and isoleucine by establishing residency within stem cells (8). Infection with the cancer strain also prompted transcriptional changes in mGEP polyamine biosynthetic pathways indicative of augmented production of this class of compounds, which are known to stimulate growth of a variety of bacterial as well as mammalian cell lineages (9, 10). Moreover, one of these affected genes, ornithine decarboxylase, exhibits increased expression in gastric adenocarcinomas compared with tissue without metaplasia (11). Finally, infection of mGEPs with the cancer strain resulted in lower levels of expression of several tumor suppressors, including kangai1, whose reduced expression correlates with poorer prognosis in human gastric cancers (12). Together, these comparative studies of strains, isolated from the same host as they progressed from ChAG to adenocarcinoma, suggest an evolving bacterial-progenitor cell interaction that not only provides a microhabitat for H. pylori in gastric stem cells but also affects the risk for malignant transformation.
The ChAG and cancer strains were obtained from a single individual enrolled in a population-based study, performed a decade ago, that explored the prevalence of peptic ulcer disease in individuals who lived in two northern Swedish cities, Kalix and Haparanda. A subset of enrollees in this “Kalixanda” study (13, 14) had esophagogastroduodenoscopy performed at two time points, separated by four years. In this report we now extend our studies of the genomic adaptations of H. pylori to development of ChAG and its interactions with mGEPs. We have done so by sequencing isolates recovered from the body (corpus region) of the stomachs of additional Kalixanda enrollees. Four isolates, two recovered at the time of each endoscopy, were characterized from (i) each of two individuals who maintained normal histology over the four-year interval, (ii) one participant who progressed from normal gastric histology to high grade ChAG, (iii) one person who presented with mild ChAG and then progressed over four years to severe ChAG, (iv) another who had progressed from moderate atrophy to high grade atrophy, and (v) the single patient who had completed the progression from ChAG to adenocarcinoma. Gene and SNP content were compared in the sequenced microbial genomes, and the resulting datasets were used to determine the extent of genome-wide diversity and whether the genomes cluster according to host and/or host pathologic status. A subset of 6 strains was culled from this collection of 24 isolates, and together with HPAG1, a previously sequenced ChAG strain from another independent clinical study (15), were used for GeneChip-based functional genomics analyses to define a shared mGEP response to ChAG-associated isolates. Finally, whole genome transcriptional profiling of the HPAG1 strain grown under various pH conditions in vitro and during its infection of mGEPs was used to identify an outer membrane bacterial protein that plays an important role in colonization of Atpb4-tox176 mice.
EXPERIMENTAL PROCEDURES
Bacterial Strains
H. pylori strain HPAG1 was obtained from a patient with ChAG in a Swedish case-control study of gastric cancer (15). Strains recovered from participants enrolled in the Kalixanda study are listed in Table 1. Bacteria were grown under microaerophilic conditions for 48–72 h at 37 °C on brain heart infusion (BHI) agar plates supplemented with 10% calf blood, vancomycin (6 μg/ml), trimethoprim (5 μg/ml), and amphotericin B (8 μg/ml). For liquid culture, bacteria were grown in Brucella or BHI broth supplemented with 10% fetal calf serum (Sigma) and 1% IsoVitaleX (BD Biosciences), pH 7.0.
TABLE 1.
List of H. pylori strains sequenced and histologic features of the corpus region of their host gastric habitat
| Patient ID | Agea/gender | Gastric histopathology | t = 0 | t = 4 years |
|---|---|---|---|---|
| 1259 | 67/male | Normal to normal | HPKX_1259_NL0C1 | HPKX_1259_NL4C1 |
| HPKX_1259_NL0C2 | HPKX_1259_NL4C2 | |||
| 1379 | 70/female | Normal to normal | HPKX_1379_NL0C1 | HPKX_1379_NL4C1 |
| HPKX_1379_NL0C2 | HPKX_1379_NL4C2 | |||
| 345 | 65/male | Normal (0) to high grade (3) atrophy | HPKX_345_NL0C1 | HPKX_345_AG4C1 |
| HPKX_345_NL0C2 | HPKX_345_AG4C2 | |||
| 1039 | 68/male | Slight atrophy (grade 1) to high grade (3) atrophy | HPKX_1039_AG0C1 | HPKX_1039_AG4C1 |
| HPKX_1039_AG0C2 | HPKX_1039_AG4C2 | |||
| 1172 | 70/male | Moderate atrophy (grade 2) to high grade (3) atrophy | HPKX_1172_AG0C1 | HPKX_1172_AG4C1 |
| HPKX_1172_AG0C2 | HPKX_1172_AG4C2 | |||
| 438 | 75/male | Moderate atrophy (grade 2) to gastric cancer | HPKX_438_AG0C1 | HPKX_438_CA4C1 |
| HPKX_438_AG0C2 | HPKX_438_CA4C2 |
a Age at initial endoscopy.
Construction of a hopZ Mutant Strain
The hopZ gene was amplified from strain HPAG1 using primers HopZ-F (GTGAAAAACACCGGCGAATTGA) and HopZ-R (CGGAGTTGAAAAAGCTGGATTTGAT), cloned into pCR®4TOPO® (Invitrogen), and then subcloned into pGEM®-5Zf(+) (Promega) using SpeI and NotI sites. hopZ was disrupted by insertion of the kanamycin resistance cassette of pJMK30 (59) into a HindIII site. The hopZ::KanR construct was electroporated into the recipient HPAG1 strain. Bacteria were plated on BHI agar plates, grown for 3 days, harvested, resuspended in PBS, and then plated on BHI agar containing 50 mg/ml kanamycin to select for transformants. Correct insertion of the kanamycin cassette was confirmed by PCR, sequencing, and Southern blot hybridization.
Genome Sequencing
Genomic DNA was prepared according to Oh et al. (39). Standard Illumina genomic sequencing protocols were employed. In general, one lane of an eight-lane flow cell in an Illumina GA-I or GA-II sequencer was used for each strain. However, an additional lane of sequencing was done for genomes with poor assemblies (total assembly length <1 Mbp or N50 contig length <1500 bp).
Genome Assembly
The Velvet assembler (17) was used for sequence assembly. All combinations of k (the word length for velveth) between 19 and 31 and minimum coverage (the -cov_cutoff option for velvetg) between 6 and 40 were tested. The best combination was manually chosen for each data set to maximize N50 contig length, total assembly length, and k in that order. A high coverage cutoff (the -max_coverage option for velvetg) was chosen manually from a weighted histogram of the number of nucleotides versus coverage reported by Velvet, generated as described in the appendix of the Velvet manual (the high coverage cutoff was chosen by visual inspection to include only the largest peak in the coverage histogram).
Assembled Data Operational Gene Units (OGUs) Calling
Genes were predicted using Glimmer in each assembled data set. All predicted coding sequences <300 nucleotides were filtered out (this reduced the false positive rate and increased the false negative rate only mildly). These genes were added to the pan-genome generated from published genome sequences and the nr data base as described in the “Comparisons of H. pylori Genomes” section under “Results and Discussion.” This entire set of genes was binned (clustered) using the CD-HIT program with default parameters into OGUs. If any predicted gene from an assembled genome was present in a given OGU (cluster), that OGU was called as present within that genome. The method used for “raw read” OGU calling (as opposed to “assembled data” OGU calling) is described in the “Comparisons of H. pylori Genomes” section below. The pan-genome used for the raw read analysis encompased all Glimmer-predicted genes, including those <300 bp.
Functional Analysis
All sequences in all OGUs predicted for each genome using the raw read method were aligned to the COG or KEGG data base using BLASTX. Each OGU was assigned the union of the COG categories or KEGG pathways matching its constituent sequences (with few (<5%) exceptions, all sequences within a single OGU matched the same COG category or KEGG pathway). Comparisons of fractional representation of COG and KEGG pathways between different gene sets (i.e. core versus variable genes) were done using a χ2 goodness-of-fit test; categories or pathways with less than 5 genes in either “core” gene sets (i.e. represented in all sequenced H. pylori genomes) or “variable” gene sets (not represented in all genomes) were excluded from the χ2 test. A post hoc Z-test on residuals was used to determine which categories or pathways contributed to significant differences, with a cutoff set at 3 S.D., corresponding to an unadjusted type I error rate of 0.0013.
SNP Calling
HPAG1 was used as a reference genome. Each assembled genome was aligned using BLASTN against HPAG1. Only the first alignment reported by BLASTN was used for each assembled contig. The position of each SNP (based on the HPAG1 genome sequence) was recorded. A gap was counted as 1 SNP. The SNPs for each assembled genome was then represented as a vector of 1 (SNP present) or 0 (no SNP or no alignment) with 1,596,366 elements (the length of the HPAG1 genome). SNP rates were calculated for each pair of assembled genomes by counting the number of sequence differences only at positions in the HPAG1 reference genome where the both genomes had contigs that aligned. The total number of sequence differences in overlapping regions was divided by the total length of overlapping alignments, giving a rate of SNPs per aligned base pair.
Positive Selection
For a given gene, using the fully sequenced HPAG1 genome as a reference, a set of all orthologous sequences from the Illumina data sets was collected. Orthology was defined as a reciprocal best BLAST hit; i.e. using the HPAG1 sequence for a gene, the best BLASTN hit from an assembled genome was extracted and compared back to the HPAG1 genome using BLASTN and BLASTX. Only sequences that mapped back to the original HPAG1 gene sequence were used. The set of orthologs for a given gene was analyzed for positive selection according to Chen et al. (54) with minor modifications. Specifically, each ortholog set was aligned as translated protein sequence using ClustalW (60); the alignment was imposed on the DNA sequences. Aligned DNA sequences were trimmed to the shortest sequence in the data set (so there were no gaps at the ends of the alignments). The GENECONV program (61) was used to detect recombination; alignments that had a p value of less than 0.05 were split into fragments based on the recombination breakpoints identified by GENECONV. Each alignment or fragment alignment was then used to infer a maximum likelihood tree using the DNAML program from PHYLIP, then tested for positive selection using the M1 and M2 models of CODEML from the PAML package (Version 4) (62). M1 is the null model that does not include positive selection. M2 is the test model that includes positive selection. An unadjusted p value cutoff of 0.05 was used for each gene.
Infection of mGEP Cells with H. pylori Strains and Isolation of Cellular RNA
mGEP cells (8) were seeded (passage number 4–7) at 4 × 105 cells per T75 flask in 12–15 ml of RPMI 1640 medium (Sigma, pH 7.1–7.2) supplemented with 10% fetal bovine serum (Hyclone) and grown for 3 days at 37 °C to 70% confluency. Medium was then removed. H. pylori strains, which had been grown to log phase A600 ≈ 0.8 in Brucella broth plus 5% fetal bovine serum and 1% IsoVitaleX, were spun down, washed in PBS, resuspended in fresh cell culture medium, and added to the flasks (1.6 × 107–5 × 109 bacteria/flask). After a 24-h incubation at 37 °C, residual medium and non-attached bacteria were washed off from mGEPs using PBS (2 times; 25 °C), and cells were harvested by trypsinization (5 min at 37 °C; 0.05% trypsin (Sigma); 0.02% EDTA). After neutralization with ice-cold medium and a PBS wash, cells were flash-frozen in liquid nitrogen. As controls, mGEP cells alone were incubated for 24 h in culture medium under the same conditions and underwent identical treatments as the infected samples. Alternatively, bacteria were heat-killed by incubation at 95–100 °C for 5 min. The heat-treated cultures were subcultured on BHI plates at 37 °C for 96 h to prove that no viable organisms remained. mGEP infections with heat-killed bacteria were carried out as described above.
All infections were performed in triplicate/strain. Total cellular RNA was then extracted using the RNeasy mini-prep kit (Qiagen), and contaminating genomic DNA was removed using the DNA-free kit (Ambion) as described by the manufacturer.
GeneChip Analysis of the Responses of mGEP Cells to Infection with H. pylori Strains
Biotinylated and fragmented cRNAs prepared from total cellular mGEP RNA were hybridized to Moe430_2 Affymetrix Gene Chips. Raw data were normalized with RMA (63). Z-score-based p values were calculated for each gene based on the mean and S.D. of the gene across all experiments in a set of 12 control microarrays of uninfected mGEPs. FDR was estimated using the Benjamini and Hochberg procedure (64).
Studies of Wild-type and Mutant Strains of HPAG1
Growth Assay
Overnight liquid cultures of HPAG1 and the isogenic ΔhopZ mutant were grown to A600 of 0.5–0.7. Bacteria were harvested by centrifugation and resuspended in 50 ml of BHI broth, pH 7.0, supplemented with 10% fetal calf serum and 1% IsovitaleX to yield a starting A600 ≈ 0.05. Growth was subsequently monitored at 37 °C over a 50-h period.
Acid Exposure and RNA Isolation
Overnight liquid cultures of HPAG1 and HPAG1ΔhopZ (A600 = 0.5–0.7) were harvested by centrifugation and resuspended in 50 ml of BHI broth, pH 7.0, supplemented with 10% fetal calf serum and 1% IsovitaleX to yield a starting A600 of 0.05. Cultures were grown for 25 h to mid-log phase (A600 ≈ 0.5). At this point hydrochloric acid was added to half of the culture to lower the pH to 5.0, whereas the other half remained at pH 7.0. Aliquots (4 ml) were collected from both these cultures after 1 h of growth, yielding 2 samples for RNA isolation (pH 5.0 and 7.0). Samples were immediately placed in 2 volumes of RNAprotect bacteria reagent (Qiagen). The solution was mixed by vortexing for 10 s, incubated at room temperature for 5 min, and then centrifuged (3500 × g for 15 min at 22 °C). The resulting pellet was stored at −80 °C until RNA was isolated. The acid exposure experiment was done in triplicate.
Total RNA was isolated using the RNeasy Mini kit (Qiagen). Cell pellets were resuspended in 200 μl of TE buffer (10 mm Tris, 1 mm EDTA, pH 8.0) containing 1 mg/ml lysozyme (specific activity, 50,000 units/mg; Sigma), incubated in room temperature for 20 min, and vortexed every 3 min. Genomic DNA was removed by on-column DNase digestion using RNase-Free DNase Set (Qiagen) as described by the manufacturer.
GeneChip Analysis
To analyze the responses of the wild-type HPAG1 and ΔhopZ isogenic strains to low pH in vitro, cDNA targets were prepared from 8–10-μg aliquots of each bacterial RNA sample using protocols described in the Escherichia coli Antisense Genome Array manual (Affymetrix). To analyze the bacterial transcriptome after a 24-h infection of mGEP cells, we used 25–43 mg of total RNA; each sample contained 8–10 mg of bacterial RNA based on quantification of the mammalian/bacterial 18 S to 16 S rRNA ratio using RNA 6000 Pico LabChips and a 2100 Bioanalyzer (Agilent Technologies). RNA was reverse-transcribed using random primers and Superscript-II reverse transcriptase (Invitrogen), and the RNA template was removed by incubation with 0.25 n NaOH for 30 min at 65 °C. The cDNA product was isolated (QiaQuick Spin columns; Qiagen), fragmented using DNase-I (Amersham Biosciences), and biotinylated (Enzo-BioArray Terminal Labeling kit). Standard Affymetrix protocols were used for hybridization of each cDNA target to a custom Affymetrix H. pylori HPAG1 GeneChip (Hp-AG78a520172F).
GeneChip data were normalized with RMA. FDR was estimated using the Benjamini and Hochberg procedure (64). Functional enrichment was calculated using a hypergeometric distribution. COG functions and GO terms were assigned to genes by BLAST (for COG, eval <10E-10 using STRING data base Version 7.1) and with hmmpfam (for GO terms using TIGRFAM Version 8.0 and PFAM Version 23).
Bacterial Invasion Assay
mGEP cells were seeded in 24-well plates (Corning; 1 × 105 cells/well) and cultured in RPMI 1640 medium (Sigma) supplemented with 10% fetal bovine serum (Hyclone) for 24 h under an atmosphere of 5% CO2, 95% air at 37 °C. Wild-type HPAG1 and the ΔhopZ mutant strain were grown overnight, separately, in Brucella medium, pH 7.0 (supplemented with 10% fetal calf serum and 1% IsovitaleX), harvested by centrifugation, and resuspended in RPMI1640 medium with 10% fetal bovine serum. 2.5 × 108 cfu of wild-type HPAG1, and an equivalent amount of the isogenic ΔhopZ mutant was added to each well at 37 °C. Three hours later, cells were washed with PBS (4 times; 25 °C) to remove non-attached bacteria and then incubated with fresh medium for 21 h at 37 °C. Cells were washed with PBS (3 times; 25 °C), fixed in 4% paraformaldehyde for 20 min, washed with PBS (3 times; 25 °C), incubated with PBS containing 0.2% bovine serum albumin (blocking buffer) for 15 min, and attached extracellular bacteria marked with rabbit polyclonal antibodies to H. pylori surface proteins (DAKO, 1:1000 dilution; overnight incubation in blocking buffer at 4 °C). Bound antibodies were incubated with biotin-tagged anti-rabbit Ig (1:500; 1 h at 25 °C). After washing (3× in PBS, 5 min/cycle at 25 °C) to remove unbound antibodies, mGEP cells in some wells were permeabilized (1% saponin (Sigma), 3% bovine serum albumin in PBS; 15 min at 37 °C). Antibodies to H. pylori were added (DAKO; 1 h at 25 °C, 1:1000 dilution) to label both extra- and intracellular bacteria, and bacteria were visualized with streptavidin-Alexa-Fluor 350 (blue) and Cy3 (red)-tagged donkey anti-rabbit Ig (1:500; 1 h at 25 °C). Intracellular bacteria were defined as those that were only stained with Cy3 and not with Alexa-Fluor 350. Alexa-Fluor 488-phalloidin (Molecular Probes; 1:500) was used to label actin filaments.
In Vivo Competition Experiments
All experiments involving mice used protocols approved by the Washington University Animal Studies Committee. Germ-free Atbp4-tox176 mice were maintained in plastic gnotobiotic isolators (65) and given an autoclaved chow diet (B & K Universal, East Yorkshire, UK) ad libitum. HPAG1 and ΔhopZ, strains were grown in Brucella broth to mid-log phase (A600 = 0.6). Equal amounts of each strain were mixed and then inoculated with a single gavage of 107 cfu into the stomachs of germ-free 9–13-week-old Atbp4-tox176 animals. Mice were killed 5 weeks later. Each stomach was removed and divided in half along the cephalocaudal axis, and one-half was homogenized in 0.5 ml of PBS. Serial dilutions of the homogenates were plated on BHI agar to assay for cfu.
RESULTS AND DISCUSSION
Selecting H. pylori Strains for Identification of a Shared mGEP Response to Infection in the Setting of ChAG
18–50 H. pylori isolates were recovered from each of the 6 individuals listed in Table 1. Histopathological features of their gastric mucosal biopsies were classified using the updated Sydney system (16) by a pathologist who was blinded to the identity of each patient. It is important to emphasize that according to then prevailing Swedish medical practices and the human studies committee-approved study protocol, ChAG diagnosed at the initial endoscopy was not viewed as an indication for required H. pylori eradication.
Random amplified polymorphic DNA assays of H. pylori isolate DNAs yielded results consistent with infection with a dominant strain for each host. Based on these findings, we randomly selected two isolates per time point (t = 0 and 4 years) from the set of strains available from each individual. All isolates were recovered from the corpus region of the stomach. We collected shotgun genome sequence data from the 24 isolates using massively parallel Illumina GA-I and GA-II DNA sequencers (total dataset of 4,442,560,380 bp; 72–155-fold coverage/genome; supplemental Table S1).
Comparisons of H. pylori Genomes
The program Velvet (17) was used to generate consistent, high quality genome assemblies from the shotgun sequence data (supplemental Table S1). To do so, we created an interactive hybrid pipeline that performed a parallel parameter search with Velvet on a high performance computer cluster, presented a web-based graphical interface for users to configure the parameter search and select optimal assembly parameters, and performed a final assembly and contig filtering based on user-identified parameters (see “Experimental Procedures”).
We evaluated two gene-calling methods, one based on a more traditional approach of assembly followed by gene calling using the program Glimmer (18), and one based on raw (unassembled) Illumina reads. The first method used Velvet-assembled data. Genes were predicted using Glimmer in each assembly. All predicted coding sequences less than 300 bp were filtered out (this reduced the false positive rate with only a mild increase in the false negative rate). The entire set of genes was then binned using the program CD-HIT (20) with default parameters to generate OGUs, analogous to the OTU concept used in 16 S rRNA gene-based metagenomic analyses of microbial community membership. The presence or absence of a given OGU was then determined for each genome (see “Experimental Procedures”). For the second method a reference H. pylori“pan-genome” was created from the annotated genes of all fully sequenced H. pylori genomes as well as from all sequences in the NCBI non-redundant data base that were identified with the H. pylori organism tag. Also, genes predicted by Glimmer in the Velvet-assembled genomes, including short genes <300 bp, were added to the pan-genome for this “raw read method.” All raw reads for each genome were mapped to this pan-genome using BLAT (19) with default parameters. The pan-genome was then clustered using CD-HIT. The total number of raw reads mapping to a given OGU was then used as the score for that OGU. A null cutoff score was calculated by dividing the total number of reads by the total length of OGU representatives (as determined by CD-HIT); this cutoff represents the expected number of reads per OGU normalized by length if reads were randomly selected from all OGU representatives. OGUs with scores less than this cutoff were called “absent,” and those above were called “present.”
We found that both methods performed well. Using the previously sequenced HPAG1 genome for validation, the raw read method had slightly higher false positive and false negative rates (4.2% false positive rate and 7% false negative rate) but had the advantage of performing well even for short (<300 bp) genes. In contrast, the assembled data method had a very low false positive rate and comparable false negative rate when excluding short genes (0% false positive, 6.5% false negative). Based on these observations and our desire not to remove short genes, we used the raw read method for all subsequent analysis.
Data generated from just one lane of an 8-lane Illumina flow cell (≥4–5 million 36-nucleotide-long reads) were sufficient for nearly full-length genome assembly of the H. pylori strains: i.e. 92% (22/24) of the single lane data sets were assembled with an average total assembly of 1.58 Mbp and an N50 contig length of 12,348 bp (supplemental Table S1; data for strain HPKX_1172_AG0C2 represent the combined data from two lanes). Using the finished genome sequence of the ChAG-associated H. pylori strain HPAG1 for a reference sequencing control, we estimated that our average nucleotide error rate after assembly was 30–50 per 1.5 Mbp genome (i.e. 1 error per 30,000–50,000 bp, corresponding to a Phred quality of 45–47; supplemental Fig. S1).
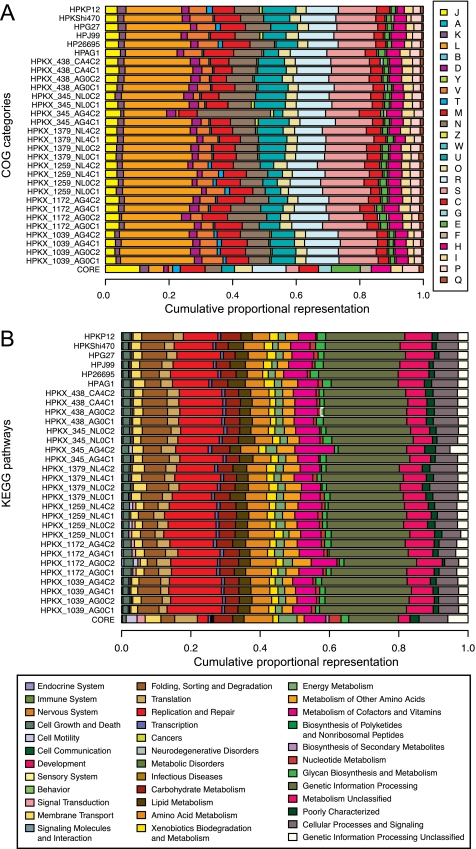
Using the raw read method, we identified a total of 4563 OGUs in the H. pylori pan-genome, with 1073 of these conserved in all strains (the “core” genome). We subsequently performed a functional analysis of OGUs using COG categories, KEGG pathways, and the raw read dataset (Fig. 1). There were significant differences in the relative representation of functions in the core versus variable genomes (p < 0.0001, χ2 test). For example, nearly all metabolism COG categories (C, E, F, G, H, and P) and category J (translation, ribosomal structure, and biogenesis) were significantly enriched in the set of core genes (Z-score > 3, post-hoc Z-score test); categories E, F, and G were also significantly depleted in the set of variable genes (Z-score < −3), consistent with the need for conserved basic metabolic and growth functions in the core genome. Only one COG category, L (replication, recombination, and repair), was significantly enriched in the set of variable genes and depleted in the set of core genes, consistent with previous observations that restriction-modification systems are variably represented among H. pylori strains (21–23). Similar trends were seen when using KEGG pathways (i.e. enrichment of basic metabolic functions in the core genome and enrichment of replication and repair in the variable genome). Interestingly, there were no significant differences in functional classes represented by the variable genes in the individual genomes.
FIGURE 1.
COG and KEGG functional analyses of identified genes in the sequenced genomes of Kalixanda H. pylori strains. The relative representation of each COG category or KEGG pathway in the core genome (OGUs shared in all genomes) and in variable genes in each genome is graphed as a stacked bar chart. A, COG categories. Codes for functional annotations: J, translation, ribosomal structure and biogenesis; A, RNA processing and modification; K, transcription; L, replication, recombination, and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; Y, nuclear structure; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; Z, cytoskeleton; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; R, general function prediction only; S, function unknown; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism. B, KEGG pathways (level 2). COG labels and KEGG pathways shown in the legend for each panel are labeled by color and in the same vertical order as shown in the bar graph.
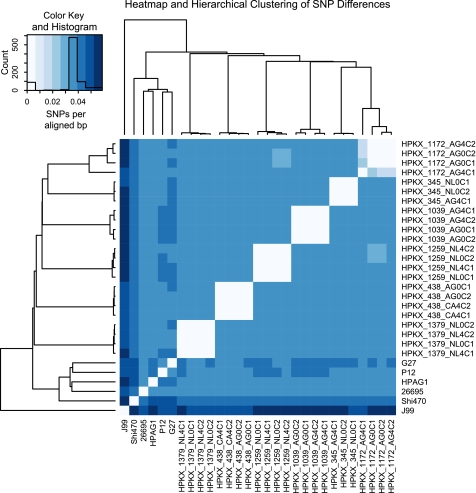
Using BLASTN, we subsequently compared the nucleotide sequences of different genome assemblies against each other and against the reference finished HPAG1 genome. This allowed us to call the number of SNPs between genomes. In general, strains from the same patient were very close to each other, with SNPs in the range of noise (<100; supplemental Table S2). In contrast, strains recovered from different patients were all approximately equidistant from each other, with 30,000–50,000 SNPs, a value similar to the SNP distance between 6 previously reported completely sequenced (finished) H. pylori genomes from patients with different gastric pathologies living in different countries (HPAG1, J99, 26695, G27, P12, and Shi470) (supplemental Table S2).
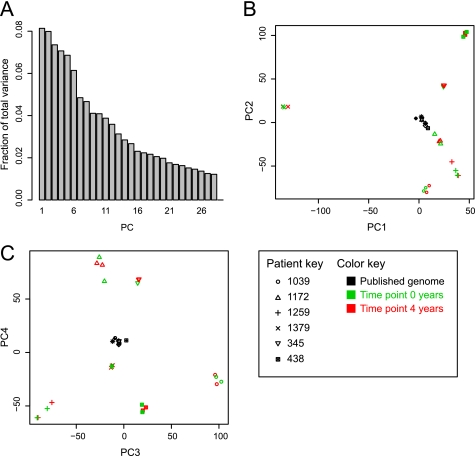
Clustering was examined using principal components analysis (Fig. 2) and hierarchical clustering (Fig. 3). Both techniques gave the same results. There was no obvious clustering by disease state or any consistent changes between different sampling times. There was also no consistent difference in isolates taken from patients experiencing similar gastric pathology (i.e. two patients had normal pathology at both time points, and two patients had ChAG at both time points; H. pylori strains isolated from these four patients showed no clustering beyond clustering by host).
FIGURE 2.
Principal components analysis of SNP differences between H. pylori strains. Each genome was represented by a binary vector consisting of all SNP positions relative to HPAG1. A principal components analysis was run on the set of all these vectors. A, a scree plot of fraction of variance explained by each principal component (PC). B, a biplot of PC1 and PC2. C, a biplot of PC3 and PC4. Previously published H. pylori genome sequences are colored black. Green symbols represent isolates taken at time point 0 years. Red indicates isolates from time point 4 years. Isolates from different patients are represented by different symbols, as indicated.
FIGURE 3.
Hierarchical clustering of SNP differences between H. pylori strains. SNP rates (SNPs/aligned bp) between different sequenced strains were interpreted as a distance matrix. Hierarchical clustering was done on this symmetric matrix of SNP rates. Color in the central heatmap represents SNP rate as shown in the legend at the top left. A tree based on Euclidean distance is shown at the top and left of the heatmap. Superimposed on the color scale at the top left is a histogram in black of the number of cells with the indicated SNP rate. G27, P12, HPAG1, 26695, Shi470, and J99 are previously published sequenced H. pylori genomes used as reference.
Thus, genome-wide analysis of multiple isolates from multiple patients with variable gastric pathology demonstrated that H. pylori populations within individuals are largely clonal, whereas different isolates from different individuals appear to be essentially unrelated regardless of disease state. Furthermore, our results revealed that the H. pylori population within a given individual is remarkably stable over a period of four years, consistent with the concept of host-pathogen co-adaptation throughout the lifetime of a persistent infection (24–26). These results are also consistent with other reports that H. pylori is very diverse, highly recombinogenic, clonally descendent within individuals or closely related individuals (27–29) and slowly divergent over time within a host (30).
Functional Genomic Studies of the mGEP Response to Infection
Because the extent of genome-wide variation precluded our ability to readily identify specific genes and functional properties associated with development of ChAG, we tested whether such common properties exist by determining whether ChAG-linked strains evoked a shared transcriptional response after infection of mGEPs. Six of the 24 sequenced Kalixanda strains were used for these analyses: i.e. (i) the set of strains recovered from patient 345 when he had normal gastric histology and then 4 years later when he had developed high grade atrophy, (ii) the set of strains from patient 1039 as he progressed from mild atrophy to high grade atrophy, and (iii) the set of strains from patient 438 with moderate atrophy who later progressed to gastric adenocarcinoma. HPAG1 was also used in this analysis, with heat-killed HPAG1 cells employed as a control.
The mGEP cell line was established from FVB/N transgenic mice that express SV40 T antigen under the control of the same Atbp4 transcriptional regulatory elements that were used to direct expression of tox176 (see the Introduction). Atbp4-TAg expression results in entrapment/amplification of GEPs because of a block in the differentiation of oligo-potential pre-parietal progenitors to mature parietal cells. Electron microscopy and immunohistochemical studies of the cloned mGEP cell line showed that cells have the morphologic features of GEPs present in the normal mouse stomach and express a number of biomarkers that are enriched in GEPs harvested by laser capture microdissection from Atbp4-tox176 transgenic mice (8).
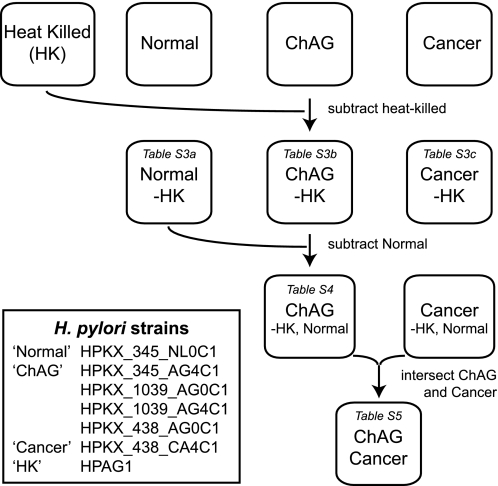
Low passage number mGEP cells were grown to 70% confluency. 107–5 × 109 cfu from a log phase culture of a given H. pylori strain were introduced into the culture medium, and infection was allowed to proceed for 24 h (n = 3 mGEP infections in separate culture flasks/strain). After washing away non-attached bacteria, RNA was prepared from host cells, and cRNA targets generated from these RNA preparations were hybridized to mouse Moe430_2 GeneChips. Uninfected mGEPs served as reference controls. The resulting GeneChip transcriptional profiles of mGEP responses were placed into three groups: normal (one strain), “ChAG” (five strains including HPAG1), and “gastric adenocarcinoma” (one strain). To identify differentially expressed genes in each group, Z-score-based p values were calculated on a gene-by-gene basis by comparing mean expression levels to a set of 12 control GeneChips obtained from uninfected mGEP cells (31, 32). An FDR of 0.5% was chosen as a threshold for significance. For each group, we subtracted any genes that were also found to be differentially expressed when mGEP cells were infected with heat-killed HPAG1 (n = 3 infections) (Fig. 4).
FIGURE 4.
Flowchart of bioinformatic analysis that identified a shared mGEP transcriptional response to ChAG-associated H. pylori isolates and a shared response to both ChAG and cancer strains.
Supplemental Table S3, A–C, lists the differentially expressed genes for each of the three groups. We defined a ChAG-associated signature mGEP response by identifying 695 transcripts that were differentially expressed upon infection with all ChAG strains but not with the strain associated with normal gastric histology or heat-killed HPAG1 (Fig. 4 and supplemental Table S4). 434 transcripts from this 695-member ChAG-associated response were also differentially expressed after infecting mGEPs with the cancer-associated H. pylori isolate (Fig. 4 and supplemental Table S5).
Ingenuity Pathways Analysis software was used to identify biological categories, plus signaling and metabolic pathways, which were significantly over-represented in the 695-member ChAG-associated signature response of mGEPs. The top four statistically significant biological categories were cell death, cancer, cellular function and maintenance, and cellular growth and proliferation (see supplemental Table S6, A and B for a list of genes in these overrepresented categories and pathways). Importantly, these functions and pathways were also statistically significantly overrepresented among the 434 genes present in the shared ChAG-cancer response (supplemental Table S7, A and B). A number of the most highly regulated genes in the shared 434-member ChAG-cancer mGEP response are linked to gastric cancer. The most highly induced Ingenuity Pathways Analysis-annotated mGEP gene in the dataset is Serpine-1 (22-fold up-regulated). This member of the urokinase activator system is also induced in human gastric adenocarcinoma cells upon infection with cag-positive H. pylori strains (33). In humans with gastric cancer, increased levels of Serpine-1 are associated with a poor clinical outcome plus lymph node and vascular invasion (34). Several protein tyrosine phosphatases (Ptpre, Ptpn14, and Ptpn11) were also represented in this group of significantly up-regulated genes. When H. pylori CagA is introduced into gastric epithelial cells, it undergoes Src-dependent tyrosine phosphorylation and activates the host cell SHP-2 phosphatase encoded by Ptpn11. A single nucleotide polymorphism in Ptpn11 (SNP JST057927, G to A) is associated with increased risk for progression to ChAG and gastric cancer (35). Intriguingly, Ptpn11, in contrast to other PTP family members that act as tumor suppressors, is now believed to be a proto-oncogene (36); its up-regulation in our dataset would be consistent with a role in malignant transformation of progenitor cells upon infection with ChAG and cancer-associated strains. Other notable induced genes in the “shared ChAG-cancer” mGEP response include Sod2 (superoxide dismutase 2), whose level is increased human gastric cancers (37), as well as genes associated with neuroendocrine differentiation (Gadd45B and enolase 2) (38). Neuroendocrine differentiation is a histopathologic feature that appears in a number of epithelial cell cancers, including those arising from the prostate, and heralds a more aggressive phase. Finally, among the down-regulated genes in the shared ChAG-cancer mGEP response, is Cdkn2c, a known tumor suppressor.
In summary, our results reveal a common mGEP response to isolates associated with ChAG and gastric cancer. This response is significantly enriched in genes associated with tumorigenesis in general and gastric carcinogenesis in the specific cases discussed above, suggesting that the influence of H. pylori isolates on progenitor cell biology may mediate progression from ChAG to gastric cancer.
The Bacterial Perspective; Functional Genomics Studies of the Response of a ChAG-associated Strain to Infection of mGEPs
We selected HPAG1 as a model to analyze a ChAG strain response to mGEP infection for several reasons; (i) its highly efficient colonization of the stomachs of Atpb4-tox176 gnotobiotic mice and its known ability to establish intracellular residency in their GEPs (5), (ii) its complete genome sequence had been defined, and (iii) it evoked an mGEP transcriptional response that was shared with the other ChAG isolates. Therefore, RNA was prepared from bacteria harvested from mGEP cultures infected for 24 h. In addition, RNA was isolated from HPAG1 harvested after a 24-h incubation in cell culture medium alone (n = 3 parallel incubations with and without mGEPs). cDNA targets were generated from each RNA preparation, and each cDNA preparation hybridized to separate custom Affymetrix GeneChips that contain probesets to 1530 of the 1536 chromosomal protein-coding genes known or predicted to be present in the HPAG1 genome (39).
274 genes satisfied our selection criteria for being differentially expressed upon mGEP infection (unpaired t test, FDR 5%) (see supplemental Table S8 for a list, including -fold differences in expression). The dataset included genes enriched (p value < 0.001 from a hypergeometric distribution) for the GO terms “iron ion binding” and “metal ion binding” and for the COG term “pyruvate:ferredoxin oxidoreductase and related 2- oxoacid:ferredoxin oxidoreductases, gamma subunit.” We were also intrigued to find hopZ in the dataset (2.2-fold up-regulated, p value = 6.02e-05). H. pylori expresses numerous outer membrane proteins that function as porins and/or adhesins; several have been implicated as being important for attachment to gastric epithelial cells, including the Lewisb blood group antigen binding adhesin, BabA2, which recognizes Fucα1,2Galβ1,3[Fucα1,4]GlcNAcβ epitopes produced by differentiated pit cells in the majority of humans (40), and the sialic acid binding adhesin, SabA (also known as HopP), which recognizes NeuAcα2,3Galβ1,4 epitopes (41) produced by GEPs (42).
Previous in vitro DNA microarray studies of non-ChAG H. pylori isolates have demonstrated that that the bacterial response to acid involves down-regulation of outer membrane proteins; notably, Hop family members that function mainly as adhesins (39, 43–45). HopZ has been shown to be important for bacterial attachment to a gastric adenocarcinoma-derived epithelial cell line (AGS) in vitro, although its receptor is not known (46, 47). Introducing a null allele of hopZ does not affect the ability of the bacterium to establish residency in the stomachs of mice or guinea pigs with normal acid-producing capacity (47, 48).
To understand the role of HopZ in the context of ChAG, we generated a hopZ knock-out mutant in strain HPAG1. Loss of HopZ did not result in a detectable growth phenotype when the wild-type and isogenic ΔhopZ strains were incubated in standard culture medium at pH 7.0 (supplemental Fig. S2). We subsequently used our custom HPAG1 GeneChips to compare their transcriptomes at pH 7 and 5. Fifty-nine genes in HPAG1 and 169 genes in the ΔhopZ mutant satisfied our criteria for being differentially expressed (either up- or down-regulated) at pH 7.0 versus pH 5.0 (supplemental Fig. S3 and Table S9, A and B). Moreover, comparison of the wild-type and ΔhopZ datasets revealed 85 genes that were uniquely up-regulated and 66 genes that were uniquely down-regulated in ΔhopZ cells upon switching from pH 7.0 to 5.0 (i.e. the wild-type strain did not exhibit these changes with the same pH shift) (supplemental Fig. S3 and Table S10).
Several of the pH-regulated, hopZ-dependent genes encoded outer membrane proteins whose expression was lower at pH 7.0. They include HopP (SabA HPAG1_0709), which as noted above, binds sialylated glycans produced by GEPs in vivo and HPAG1_0636, which encodes α1,3-fucosyltransferase. Variation of surface antigen expression (e.g. surface Lewis (Le) antigens) is a mechanism deployed by H. pylori to adapt to changes in its gastric habitat (49–51), including those associated with development of ChAG in our Swedish population (52).
We asked whether these outer membrane proteins were under positive selection in the set of previously published fully sequenced H. pylori strains and in the strains we sequenced. hopA, hopP, (sabA), and hopZ were all defined as being under positive selection (see supplemental Table S11 and “Experimental Procedures” for criteria used). As a control, six housekeeping genes (atpA, efp, mutY, ppa, trpC, and ureI) (53) and two virulence factors (cagA, vacA) were also examined. Because efp, mutY, trpC, cagA, and vacA satisfied our criteria for being under positive selection (supplemental Table S11), we extended this survey further to 20 randomly chosen genes from the HPAG1 genome. Remarkably, 5 of these 20 were also defined as being under positive selection. Thus, 8 (31%) of the 26-member control set of genes not expected to be under positive selection had significant evidence for this phenomenon, a much higher proportion than the 1–4% typically found in large surveys of positive selection (54–57), even after controlling for possible intragenic recombination (supplemental Table S11). A recent population bottleneck can give a signal of positive selection in many genes simultaneously, regardless of the actual selective pressures acting on those genes. Given previous data and our results indicating that each patient had been originally colonized by a single H. pylori strain, this high rate of positive selection appears to be largely because of a severe population size reduction occurring at the time of initial colonization, although we cannot completely exclude the possibility that hopA, hopP, and hopZ also evolve under positive selection.
An in Vivo Phenotype Produced by HopZ Deficiency
Loss of HopZ did not produce an appreciable effect on the ability of the HPAG1 strain to bind and invade cultured mGEPs. Invasion was scored using a multilabel immunohistochemical assay where H. pylori antibodies, labeled with one fluorescent tag, are used to stain infected mGEPs followed by the addition of the cell-permeabilizing agent saponin plus the same H. pylori antibody but labeled with a second tag (supplemental Fig. S4). Moreover, profiling with GeneChips revealed no mouse genes that satisfied our criteria for having significant differences in expression in mGEPs infected with the wild-type versus ΔhopZ strain (unpaired t test, FDR 5%). With the exception of hopZ itself, there were only three bacterial transcripts whose expression was scored as significantly different after H. pylori GeneChip-based comparisons of the two strains 24 h after mGEP infection at pH 7.1–7.2 (n = 3 separate mGEP infections/strain); they encoded the outer membrane protein HorF (1.5-fold down-regulated in the mutant compared with the wild-type) and two putative type III restriction enzymes (HPAG1_1328, 12.5-fold up-regulated and HPAG1_1329, 14.5-fold up-regulated).
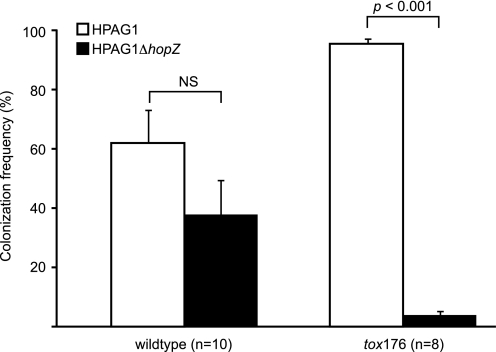
Loss of HopZ markedly reduced the fitness of H. pylori in the stomachs of parietal-cell deficient Atbp4-tox176 mice but not in their normal nontransgenic littermates. This phenotype was identified after inoculating germ-free mice with a single gavage of equal numbers (107 cfu) of the wild-type and ΔhopZ strains. When animals were killed 5 weeks later, all (10/10) of the nontransgenic mice contained both strains in their stomachs, and there was no statistically significant difference in their relative proportions (62% of the H. pylori population was composed of wild-type HPAG1; p > 0.05; Student's t test). In contrast, 2 of the 8 Atpb4-tox176 mice only retained the wild-type strain at the time of their sacrifice; in the remaining 6 parietal cell-deficient transgenic animals, HPAG1 was the dominant strain representing an average 96% of the population (p < 0.001; Student's t test) (Fig. 5). Control experiments involving a single gavage of 107 cfu of either strain alone did not produce statistically significant differences in (i) the efficiency of colonization in non-transgenic versus transgenic animals (92% (11/12) of non-transgenic mice were colonized with HPAG1 versus 93% (14/15) with the ΔhopZ mutant; 100% (12/12) of Atbp4-tox176 mice were colonized with wild-type HPAG1 and 79% (15/19) with ΔhopZ) or (ii) the density of colonization (range, 3.2–9.5 × 103 cfu/stomach) (data not shown).
FIGURE 5.
Competition experiments involving wild-type and isogenic ΔhopZ strains of H. pylori HPAG1 introduced into the stomachs of gnotobiotic Atbp4-tox176 transgenic and non-transgenic littermates. 9–13-Week-old mice each received a single gavage of 107 cfu of an equal mixture of HPAG1 and ΔhopZ. Animals were killed 5 weeks later, and the proportional representation of the two strains was defined in gastric contents by plating onto selective medium. Mean values ± S.E. for the percent representation of the wild-type and mutant strains in the gastric microbiota are plotted.
Our findings suggest possible mechanisms that could underlie the reduced fitness. As noted above, hopZ is up-regulated in response to interactions with mGEPs at pH 7 and positively regulates expression of genes encoding several adhesins, including HopP/SabA, which binds sialylated carbohydrate epitopes produced by GEPs in Atpb4-tox176 mice (but not by cultured mGEPs; data not shown). hopZ is a member of a large family of hop genes that share extensive homology at the 5′ and 3′ termini; this shared sequence identity allows for frequent recombination that in turn could modulate the adherence properties of a colonizing strain of bacteria (58). Reduction in the expression of HopP/SabA in combination with other Hop family adhesins whose expression is regulated by HopZ would be expected to affect the ability of the bacterium to bind to gastric stem cells and their descendants as well as to the mucus slime layer that overlies the gastric epithelium. Because loss of acid producing capacity removes a barrier to colonization of the stomach, changes in gastric microbial ecology would be expected to increase competition with other microbes for nutrients in this ecosystem and, together with reduced adherence, could have deleterious effects on the fitness of the bacterium as it seeks to maintain itself in extracellular habitats or to enter more protected intracellular milieus.
Prospectus
The mechanisms by which infection of the human stomach by H. pylori can lead to adenocarcinoma in a subset of hosts need to be elucidated; those at risk have to be identified and should be treated, whereas those not at risk may derive benefit from persistent colonization (see the Introduction). In the current report, we have continued our analysis of the hypothesis that interactions between H. pylori and a subset of gastric stem cells in the setting of ChAG may influence tumorigenesis. The liaison between this bacterium and this host cell population is envisioned to benefit the bacterium because of the nutrient foundation that the progenitor cell provides (e.g. amino acids that it cannot synthesize) and the safe haven for persistence that it creates. In this scenario, the bacterium can affect host progenitor cells in part by regulating expression of GEP genes involved in the control of proliferative activity and/or tumorigenesis.
The present study illustrates one approach to the very challenging problem of characterizing the co-evolution of H. pylori genomes and host GEP responses during progression to ChAG and gastric cancer. Our initial supposition was that deep draft assemblies of H. pylori genomes, generated from isolates obtained over a 4-year interval from patients as they progress from normal gastric histology to ChAG, or from mild to more severe ChAG, and/or from ChAG to cancer, where each individual serves as his/her own control and where multiple individuals with the same histopathologic classification of their gastric mucosa are included, would be useful in identifying shared features in the genomes of isolates associated with a given stage in the evolution of host pathology versus its precursor state. Moreover, isolates from subjects living in the same region of the world who maintain normal gastric histology could be used to “filter-out” H. pylori genetic changes occurring as a result of continued adaptation to its stomach habitat independent of progression to a more pathologic state. As illustrated, this type of approach identified genomic variations that clustered by host rather than disease state and were too numerous between hosts, even those with shared histopathology, to serve as the foundation for developing compelling, testable hypotheses about functional alterations in the bacterium that are associated with disease progression. The number of SNPs between strains isolated at different time points from the same host as ChAG developed was ∼1000-fold lower than the number of SNPs that occurred between hosts with ChAG, highlighting the relative stability of the organism's genome within a person when sampled over a 4-year interval. It is possible that a comparison of strains recovered from the same host over a longer interval (>20 as opposed to 4 years) could circumvent this problem and identify genes and genomic changes linked to disease pathogenesis.
mGEP cells provided us with another approach for identifying shared features among ChAG isolates recovered from different hosts; namely, their ability to evoke shared progenitor cell transcriptional responses. They also allowed us to define differential bacterial transcriptional responses to GEP infection and to determine whether these differentially regulated genes are under positive selection. A downstream step in this analytic pipeline was to examine the effects of disrupting these bacterial genes in competitive fitness assays based on colonization of germ-free Atpb4-tox176 and their nontransgenic littermates with isogenic wild-type and mutant strains. An additional arm of the analysis not illustrated in this report would be to introduce these wild-type and mutant strains into more complex model human gastric microbiotas/microbiomes composed of cultured (and sequenced) representatives of the microbial community present in the stomachs of H. pylori-infected patients with ChAG. The hoped-for outcome of these efforts is new bacterial and host cell biomarkers associated with increased risk for disease progression and new therapeutic guidelines and targets.
Supplementary Material
Acknowledgments
We are indebted to Maria Karlsson and David O'Donnell for gnotobiotic husbandry, Jessica Hoisington-Lopez for technical assistance with genome sequencing, and Lars Agreus, Tom Storskrubb, Jukka Ronkainen, Pertii Aro, and Nicholas J. Talley for coordinating the clinical study and collecting the strains described in this report. We thank Scott Hultgren for support during the course of these studies.
This work was supported by National Institutes of Health Grants DK58529, DK52574, DK081620, and DK64540 and the Swedish Cancer Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S11 and Figs. S1–S4.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) SRP001104.
The eChip data sets reported in this paper have been submitted to the Gene Expression Omnibus (GEO) database under accession number GSE16440.
- ChAG
- chronic atrophic gastritis
- GEP
- gastric epithelial progenitor
- BHI
- brain heart infusion
- PBS
- phosphate-buffered saline
- OGU
- operational gene unit
- contig
- group of overlapping nucleotide sequences
- cfu
- colony-forming units
- SNP
- single nucleotide polymorphism
- FDR
- false discovery rate.
REFERENCES
- 1.Holtmann G., Maldonado-Lopez E., Haag S. (2004) J. Gastroenterol. 39, 1027–1034 [DOI] [PubMed] [Google Scholar]
- 2.Ye W., Held M., Lagergren J., Engstrand L., Blot W. J., McLaughlin J. K., Nyrén O. (2004) J. Natl. Cancer Inst. 96, 388–396 [DOI] [PubMed] [Google Scholar]
- 3.Rokkas T., Pistiolas D., Sechopoulos P., Robotis I., Margantinis G. (2007) Clin. Gastroenterol. Hepatol. 5, 1413–1417 [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Blaser M. J. (2007) Arch. Intern. Med. 167, 821–827 [DOI] [PubMed] [Google Scholar]
- 5.Oh J. D., Karam S. M., Gordon J. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5186–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reya T., Morrison S. J., Clarke M. F., Weissman I. L. (2001) Nature 414, 105–111 [DOI] [PubMed] [Google Scholar]
- 7.Necchi V., Candusso M. E., Tava F., Luinetti O., Ventura U., Fiocca R., Ricci V., Solcia E. (2007) Gastroenterology 132, 1009–1023 [DOI] [PubMed] [Google Scholar]
- 8.Giannakis M., Chen S. L., Karam S. M., Engstrand L., Gordon J. I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4358–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida M., Kashiwagi K., Shigemasa A., Taniguchi S., Yamamoto K., Makinoshima H., Ishihama A., Igarashi K. (2004) J. Biol. Chem. 279, 46008–46013 [DOI] [PubMed] [Google Scholar]
- 10.Gerner E. W., Meyskens F. L., Jr. (2004) Nat. Rev. Cancer 4, 781–792 [DOI] [PubMed] [Google Scholar]
- 11.Miao X. P., Li J. S., Li H. Y., Zeng S. P., Zhao Y., Zeng J. Z. (2007) World J. Gastroenterol. 13, 2867–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H. S., Lee H. K., Kim H. S., Yang H. K., Kim W. H. (2003) J. Pathol. 200, 39–46 [DOI] [PubMed] [Google Scholar]
- 13.Aro P., Ronkainen J., Storskrubb T., Bolling-Sternevald E., Carlsson R., Johansson S. E., Vieth M., Stolte M., Engstrand L., Talley N. J., Agréus L. (2004) Scand. J. Gastroenterol. 39, 1280–1288 [DOI] [PubMed] [Google Scholar]
- 14.Storskrubb T., Aro P., Ronkainen J., Vieth M., Stolte M., Wreiber K., Engstrand L., Nyhlin H., Bolling-Sternevald E., Talley N. J., Agréus L. (2005) Scand. J. Gastroenterol. 40, 302–311 [DOI] [PubMed] [Google Scholar]
- 15.Enroth H., Kraaz W., Engstrand L., Nyrén O., Rohan T. (2000) Cancer Epidemiol. Biomarkers Prev. 9, 981–985 [PubMed] [Google Scholar]
- 16.Dixon M. F., Genta R. M., Yardley J. H., Correa P. (1996) Am. J. Surg. Pathol. 20, 1161–1181 [DOI] [PubMed] [Google Scholar]
- 17.Zerbino D. R., Birney E. (2008) Genome Res. 18, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. (1999) Nucleic Acids Res. 27, 4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent W. J. (2002) Genome Res. 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Godzik A. (2006) Bioinformatics 22, 1658–1659 [DOI] [PubMed] [Google Scholar]
- 21.Xu Q., Morgan R. D., Roberts R. J., Blaser M. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9671–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salama N., Guillemin K., McDaniel T. K., Sherlock G., Tompkins L., Falkow S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14668–14673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aras R. A., Small A. J., Ando T., Blaser M. J. (2002) Nucleic Acids Res. 30, 5391–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaser M. J., Parsonnet J. (1994) J. Clin. Invest. 94, 4–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaser M. J., Kirschner D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8359–8364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaser M. J., Kirschner D. (2007) Nature 449, 843–849 [DOI] [PubMed] [Google Scholar]
- 27.Suerbaum S., Smith J. M., Bapumia K., Morelli G., Smith N. H., Kunstmann E., Dyrek I., Achtman M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12619–12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner K. M., Hanage W. P., Fraser C., Connor T. R., Spratt B. G. (2007) BMC Microbiol. 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen R. J., Xerry J. (2003) J. Med. Microbiol. 52, 515–524 [DOI] [PubMed] [Google Scholar]
- 30.Lundin A., Björkholm B., Kupershmidt I., Unemo M., Nilsson P., Andersson D. I., Engstrand L. (2005) Infect. Immun. 73, 4818–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dwyer D. J., Kohanski M. A., Hayete B., Collins J. J. (2007) Mol. Syst. Biol. 3, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosgrove E. J., Zhou Y., Gardner T. S., Kolaczyk E. D. (2008) Bioinformatics 24, 2482–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keates A. C., Tummala S., Peek R. M., Jr., Csizmadia E., Kunzli B., Becker K., Correa P., Romero-Gallo J., Piazuelo M. B., Sheth S., Kelly C. P., Robson S. C., Keates S. (2008) Infect. Immun. 76, 3992–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Binder B. R., Mihaly J. (2008) Immunol. Lett. 118, 116–124 [DOI] [PubMed] [Google Scholar]
- 35.Hamajima N., Naito M., Kondo T., Goto Y. (2006) Cancer Sci. 97, 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan G., Kalaitzidis D., Neel B. G. (2008) Cancer Metastasis Rev. 27, 179–192 [DOI] [PubMed] [Google Scholar]
- 37.Takeno A., Takemasa I., Doki Y., Yamasaki M., Miyata H., Takiguchi S., Fujiwara Y., Matsubara K., Monden M. (2008) Br. J. Cancer 99, 1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baudin E., Gigliotti A., Ducreux M., Ropers J., Comoy E., Sabourin J. C., Bidart J. M., Cailleux A. F., Bonacci R., Ruffié P., Schlumberger M. (1998) Br. J. Cancer 78, 1102–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh J. D., Kling-Bäckhed H., Giannakis M., Xu J., Fulton R. S., Fulton L. A., Cordum H. S., Wang C., Elliott G., Edwards J., Mardis E. R., Engstrand L. G., Gordon J. I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9999–10004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilver D., Arnqvist A., Ogren J., Frick I. M., Kersulyte D., Incecik E. T., Berg D. E., Covacci A., Engstrand L., Borén T. (1998) Science 279, 373–377 [DOI] [PubMed] [Google Scholar]
- 41.Mahdavi J., Sondén B., Hurtig M., Olfat F. O., Forsberg L., Roche N., Angstrom J., Larsson T., Teneberg S., Karlsson K. A., Altraja S., Wadström T., Kersulyte D., Berg D. E., Dubois A., Petersson C., Magnusson K. E., Norberg T., Lindh F., Lundskog B. B., Arnqvist A., Hammarström L., Borén T. (2002) Science 297, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syder A. J., Guruge J. L., Li Q., Hu Y., Oleksiewicz C. M., Lorenz R. G., Karam S. M., Falk P. G., Gordon J. I. (1999) Mol. Cell. 3, 263–274 [DOI] [PubMed] [Google Scholar]
- 43.Ang S., Lee C. Z., Peck K., Sindici M., Matrubutham U., Gleeson M. A., Wang J. T. (2001) Infect. Immun. 69, 1679–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merrell D. S., Goodrich M. L., Otto G., Tompkins L. S., Falkow S. (2003) Infect. Immun. 71, 3529–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bury-Moné S., Thiberge J. M., Contreras M., Maitournam A., Labigne A., De Reuse H. (2004) Mol. Microbiol. 53, 623–638 [DOI] [PubMed] [Google Scholar]
- 46.Peck B., Ortkamp M., Diehl K. D., Hundt E., Knapp B. (1999) Nucleic Acids Res. 27, 3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaoka Y., Kita M., Kodama T., Imamura S., Ohno T., Sawai N., Ishimaru A., Imanishi J., Graham D. Y. (2002) Gastroenterology 123, 1992–2004 [DOI] [PubMed] [Google Scholar]
- 48.de Jonge R., Durrani Z., Rijpkema S. G., Kuipers E. J., van Vliet A. H., Kusters J. G. (2004) J. Med. Microbiol. 53, 375–379 [DOI] [PubMed] [Google Scholar]
- 49.Appelmelk B. J., Martin S. L., Monteiro M. A., Clayton C. A., McColm A. A., Zheng P., Verboom T., Maaskant J. J., van den Eijnden D. H., Hokke C. H., Perry M. B., Vandenbroucke-Grauls C. M., Kusters J. G. (1999) Infect. Immun. 67, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G., Rasko D. A., Sherburne R., Taylor D. E. (1999) Mol. Microbiol. 31, 1265–1274 [DOI] [PubMed] [Google Scholar]
- 51.Appelmelk B. J., Martino M. C., Veenhof E., Monteiro M. A., Maaskant J. J., Negrini R., Lindh F., Perry M., Del Giudice G., Vandenbroucke-Grauls C. M. (2000) Infect. Immun. 68, 5928–5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skoglund A., Bäckhed H. K., Nilsson C., Björkholm B., Normark S., Engstrand L. (2009) PLoS One 4, e5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aanensen D. M., Spratt B. G. (2005) Nucleic Acids Res. 33, W728–W733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen S. L., Hung C. S., Xu J., Reigstad C. S., Magrini V., Sabo A., Blasiar D., Bieri T., Meyer R. R., Ozersky P., Armstrong J. R., Fulton R. S., Latreille J. P., Spieth J., Hooton T. M., Mardis E. R., Hultgren S. J., Gordon J. I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5977–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petersen L., Bollback J. P., Dimmic M., Hubisz M., Nielsen R. (2007) Genome Res. 17, 1336–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davids W., Gamieldien J., Liberles D. A., Hide W. (2002) J. Mol. Evol. 54, 458–464 [DOI] [PubMed] [Google Scholar]
- 57.Roth C., Betts M. J., Steffansson P., Saelensminde G., Liberles D. A. (2005) Nucleic Acids Res. 33, D495–D497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bäckström A., Lundberg C., Kersulyte D., Berg D. E., Borén T., Arnqvist A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16923–16928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferrero R. L., Cussac V., Courcoux P., Labigne A. (1992) J. Bacteriol. 174, 4212–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawyer S. (1989) Mol. Biol. Evol. 6, 526–538 [DOI] [PubMed] [Google Scholar]
- 62.Yang Z. (2007) Mol. Biol. Evol. 24, 1586–1591 [DOI] [PubMed] [Google Scholar]
- 63.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 [Google Scholar]
- 65.Hooper L. V., Mills J. C., Roth K. A., Stappenbeck T. S., Wong M. H., Gordon J. I. (2002) Methods in Microbiology: Molecular Cellular Microbiology, Vol. 31, pp. 559–589, Elsevier Ltd. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.