Abstract
The Forkhead box M1 (FoxM1) transcription factor is critical for expression of the genes essential for G1/S transition and mitotic progression. To explore the cell cycle regulation of FoxM1, we examined the phosphorylation profile of FoxM1. Here, we show that the phosphorylated status and the activity of FoxM1 increase as cells progress from S to G2/M phases. Moreover, dephosphorylation of FoxM1 coincides with exit from mitosis. Using mass spectrometry, we have identified a new conserved phosphorylation site (Ser-251) within the forkhead domain of FoxM1. Disruption of Ser-251 inhibits phosphorylation of FoxM1 and dramatically decreases its transcriptional activity. We demonstrate that the Ser-251 residue is required for CDK1-dependent phosphorylation of FoxM1 as well as its interaction with the coactivator CREB-binding protein (CBP). Interestingly, the transcriptional activity of the S251A mutant protein remains responsive to activation by overexpressed Polo-like kinase 1 (PLK1). Cells expressing the S251A mutant exhibit reduced expression of the G2/M phase genes and impaired mitotic progression. Our results demonstrate that the transcriptional activity of FoxM1 is controlled in a cell cycle-dependent fashion by temporally regulated phosphorylation and dephosphorylation events, and that the phosphorylation at Ser-251 is critical for the activation of FoxM1.
Transitions of the eukaryotic cell cycle are orchestrated by multiple protein kinases and by the transcriptional control of cell cycle regulators (1–3). Perturbations in the cell cycle process result in abnormal cell division and proliferation, the hallmark of cancer (4). Progression through the G1/S and G2/M phases of the cell cycle is regulated by CDK2-cyclin E or A and CDK1-cyclin B kinase, respectively (3). In addition, the activity of other mitotic kinases such as Polo-like kinase 1 (PLK1)3 must be maintained for proper mitotic progression (2, 5, 6). Previous studies demonstrated that the polo-box domain in PLK1 acts as a phosphopeptide-binding domain and targets PLK1 to its substrates that have been “prime” or phosphorylated by CDK1 (7). The polo-box domain-mediated PLK1 recruitment is responsible for both temporal and spatial regulation of PLK1 substrates (2, 7).
The mammalian Forkhead box (Fox) proteins belong to a large family of transcription factors consisting of more than 50 proteins that share homology in the winged helix DNA-binding domain (8, 9). Within this extensive family, FoxM1 is a proliferation specific transcription factor (10–14). Expression of Foxm1 is detected only in proliferating cells and is extinguished when cells undergo terminal differentiation and exit the cell cycle (10, 12, 13). Transcription of the Foxm1 locus results in three distinct splice variants that are almost identical in sequence but differ by the addition of two small exons: Foxm1b (HFH-11B) contains no additional exons, whereas the Foxm1c (Trident, WIN, or MPP2) and Foxm1a (HFH-11A) isoforms contain one and two additional exons, respectively (11–13, 15). Of these three variants, Foxm1b and Foxm1c are transcriptionally active (13, 16).
Previous studies demonstrated that FoxM1 orchestrates the transcription of genes that are essential for cell cycle progression and cell proliferation. During G1/S progression, FoxM1 is required for the transcriptional activation of JNK1, KIS, SKP2, and CKS1 (17–19). FoxM1 also controls transcription of a subset of G2/M-specific genes, which are essential regulators of mitosis and chromosomal segregation, such as cyclin B, Cdc25B, Aurora B, PLK1, Survivin, CENP-A, CENP-B, and CENP-F (14, 19–21). Silencing of FoxM1 expression using small interfering RNA results in diminished S-phase cell population, G2/M arrest, chromosome missegregation, and polyploidization (18–21). Foxm1−/−-deficient mouse embryonic fibroblasts display not only polyploidy genotype but also premature senescence, suggesting that Foxm1 is required for the maintenance of chromosomal integrity. Consistent with its important role in cell cycle progression, overexpressed Foxm1b protein has been demonstrated to promote tumor cell proliferation and progression in different mouse models (22–25). The association between the elevated expression level of FoxM1 and tumorigenesis, as well as unfavorable prognosis, has also been observed in a number of human malignancies, including hepatocellular carcinoma, prostate cancer, lung cancer, colorectal cancer, breast cancer, and gastric cancer (13, 22, 23, 25–28). Those observations implicated the important role of FoxM1b as a transcription factor in tumorigenesis. Thus, exploring how the function of FoxM1b is regulated in the cell might have significant impact on the design of anti-cancer therapeutics.
Expression of the FoxM1b protein is barely detectable in quiescent cells. During re-entry into the cell cycle, FoxM1b is expressed at late G1/early S phase and sustained throughout G2 phase and mitosis (10, 13). It has been shown that the transcriptional activity of FoxM1b is dependent upon the activation of the Ras-mitogen-activated protein kinase (MAPK) signaling pathway, which activates FoxM1b through cyclin-CDKs (29). The C-terminal transcriptional activation domain of FoxM1b contains a cyclin-binding motif (LXL motif), and both CDK1 and CDK2 were shown to associate with FoxM1b. Mutation of the cyclin-binding motif eliminates the binding of both CDK1 and CDK2 (29). Further analysis reveals that the Leu-641 residue within an LXL motif is required for the recruitment of the cyclin-CDK complex, and the Thr-596 residue is a critical CDK1 phosphorylation site within the activation domain of FoxM1b. CDK-dependent phosphorylation stimulates the FoxM1b transcriptional activity, which correlates with binding to the CREB-binding protein (CBP), the transcriptional coactivator. Mutation of Thr-596 abolishes binding to CBP and inhibits FoxM1b transcriptional activity (29).
We and others have characterized an autorepression domain in the N-terminal region between residues 1 and 232 of FoxM1b (16, 30, 31). Deletion of the N-terminal repression domain renders FoxM1b constitutively active and independent of cyclin-CDK. The N-terminal repression domain inhibits FoxM1b transcriptional activity presumably by binding to the C-terminal activation domain. It is postulated that cyclin-CDK-mediated phosphorylation activates FoxM1b by disrupting the interaction of the N-terminal repression domain with the C-terminal activation domain (16, 30, 31). These observations indicated a critical role of cyclin-CDK phosphorylation in the cell cycle-regulated activation of FoxM1b. However, the mechanism of temporal activation of a subset genes by FoxM1b at the G1/S and G2/M phases remains unknown.
In this study, we investigated the phosphorylation pattern of the endogenous FoxM1 protein during progression through the cell cycle. We show that the endogenous FoxM1 protein maintains a hypophosphorylation state at the G1/S boundary, exhibits increased phosphorylation status from S phase to G2/M transition, and reaches its maximal phosphorylation status at mitosis. FoxM1 is dephosphorylated upon the completion of mitosis. The transcriptional activity of FoxM1b increases through the cell cycle and coincides with the phosphorylation status of FoxM1. Consistent with a recent study (32), we observed that PLK1 interacts with FoxM1 in M phase, phosphorylates FoxM1b on Ser-724 at the C terminus, and stimulates its transcriptional activity. Furthermore, we identified a novel, conserved phosphorylation site at Ser-251 within the forkhead box DNA binding domain of hyperphosphorylated FoxM1b using tandem mass spectrometry (liquid chromatography-MS/MS). Mutation of Ser-251 to alanine interferes with neither the subcellular localization nor the DNA binding ability of FoxM1b. However, the S251A mutant exhibits deficiency in undergoing phosphorylation by CDK1, remains predominantly hypophosphorylated and shows significantly diminished transcriptional activity. Interestingly, the transcriptional activity of the S251A mutant protein fails to be activated by CDK1, but remains responsive to PLK1 stimulation. Our results demonstrate that the transcriptional activity of FoxM1b is temporally regulated through phosphorylation modifications by multiple protein kinases during progression of the cell cycle. Moreover, we show that phosphorylation of Ser-251 in the forkhead box domain of FoxM1b is a critical event for activation of its transcriptional activity at the G2/M phases by CDK1.
MATERIALS AND METHODS
Cell Culture, Transfections, Synchronization, and Flow Cytometry Analysis
Human osteosarcoma U2OS cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (HyClone Laboratories Inc.) and 100 units of penicillin/streptomycin at 37 °C with 5% CO2. Cells were transfected with plasmid DNA using LipofectamineTM 2000 (Invitrogen) in serum-free tissue culture medium following the manufacturer's protocol. Four hours after transfection, cells were fed with complete Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cell cycle synchronization was performed by double thymidine block. U2OS cells were arrested at G1/S transition for 17 h with 2.5 mm thymidine (Sigma) with a 7-h release interval. Arrested cells were released into fresh medium to follow cell cycle progression. To analyze cell cycle progression, flow cytometry analyses were carried out using a Beckman Coulter EPICS Elite ESP apparatus, and data were analyzed by FCSPress software (Ray Hicks, FCSPress, Cambridge, UK). To isolate mitotic cells, we used the mitotic shake-off method. Briefly, U2OS cells were arrested by 300 ng/ml of nocodazole (Sigma) treatment for 16 h. The partially detached nocodazole-arrested cells were shaken-off and washed three times with phosphate-buffered saline before being plated to allow re-entry into the cell cycle.
Plasmids and Mutagenesis
The pCMV T7-FoxM1b expression vector, 6×FoxM1 TATA luciferase reporter plasmid, and −749 bp Aurora B promoter-luciferase construct are described previously (19, 29). Human wild type and K82R kinase-dead PLK1 plasmids were kindly provided by David V. Hansen (Stanford University). Site-directed mutagenesis was performed by using the QuikChange® site-directed mutagenesis kit (Stratagene), and the FoxM1b point mutations were verified by sequencing (University of Illinois, Chicago, DNA Sequencing Facility).
Antibodies
The following antibodies were used for immunoblotting and immunoprecipitation: mouse monoclonal T7 tag (Novagen, Madison, WI); mouse monoclonal Plk1 (F8), mouse monoclonal CBP (C1), normal mouse IgG, and normal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal Plk1 (pT210), mouse monoclonal cyclin B1 (GNS-11) (BD Biosciences); mouse monoclonal MPM2, rabbit anti-phosphohistone H3 (Ser-10) (Upstate, Lake Placid, NY); rabbit phospho-Rb (Ser-795) (Cell Signaling Technology, Danvers, MA); mouse anti-α-tubulin (T6074), mouse anti-β-actin (A5441) (Sigma); and rabbit polyclonal anti-FoxM1 antibody was generated as described previously (19).
Immunoprecipitation, in Vitro Dephosphorylation Assay, and Western Blot Analysis
All procedures including lyses, clarification, and immunoprecipitation were performed at 4 °C. Whole cell extracts were prepared using the Nonidet P-40 lysis buffer as described (29). For lysis, cells were incubated for 20 min in ice-cold Nonidet P-40 lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 5 mm EDTA, 5 mm EGTA, 1% Nonidet P-40, 5% glycerol, 10 mm β-glycerolphosphate, and 1 mm Na3VO4). Lysates were clarified by centrifugation at 13,000 × g for 20 min, and supernatants were collected. Immunoprecipitations were carried out with 500 μg of total protein extract from each sample. Cell lysates were first pre-cleared by incubation with 20 μl of protein A-Sepharose beads (50% slurry) (Amersham Biosciences) for 1 h. The supernatants were then incubated with 2 μg of antibodies and protein A-Sepharose beads overnight with gentle rocking at 4 °C. The beads were washed three times with lysis buffer, and bound proteins were eluted from the beads in SDS sample buffer. Coimmunoprecipitated proteins were then detected by Western blot analysis. For the dephosphorylation assay using λ-phosphatase, FoxM1 immunoprecipitates were washed twice in lysis buffer in the absence of phosphatase inhibitors and resuspended in phosphatase buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 0.1 mm EGTA, 2 mm dithiothreitol, and 0.01% Brij 35). Parallel samples were incubated with or without 100 units of λ-phosphatase (New England Biolabs, Ipswich, MA) at 30 °C for 30 min.
Luciferase Reporter Assays
U2OS cells were plated in a 24-well plate at a density of 105 cells/well. Cells were transfected using 1 μl of Lipofectamine 2000 with 0.1 μg of either pCMV FoxM1b expression construct or pCMV empty vector, 0.2 μg of either 6×FoxM1 TATA luciferase reporter or the −749 bp Aurora B promoter luciferase reporter, and 1 ng of pCMV-Renilla luciferase, which serve as an internal control. Twenty-six hours after transfection, cells were harvested and extracts were prepared for dual luciferase assay (Promega, Madison, WI) as described previously. The luciferase levels were normalized to Renilla luciferase activity (19, 29). All experiments were performed in triplicate, and repeated twice.
FoxM1b Phosphopeptide Analysis by Mass Spectrometric Analysis
A vector expressing T7-FoxM1b was coexpressed with either wild type or kinase-dead PLK1 plasmid in U2OS cells in five 15-cm dishes. T7-FoxM1b was immunoprecipitated by T7 tag antibody-conjugated beads (Novagen). The immunoprecipitated proteins were resolved in 7% SDS-PAGE. The hyper- and hypophosphorylated forms of the FoxM1b protein were visualized by Coomassie Blue staining. The gel bands were excised, washed, reduced with 10 mm dithiothreitol (Sigma), and alkylated with 55 mm iodoacetamide (Sigma) followed by digestion with modified sequencing grade trypsin (Promega). The extracted tryptic peptides were analyzed by data-dependent reversed-phase microcapillary liquid chromatography-tandem mass spectrometry using a LTQ linear ion trap mass spectrometer (ThermoFisher Scientific). MS/MS spectra were searched against the concatenated target and decoy (reversed) Swiss-Prot protein data base using Sequest (Proteomics Browser, ThermoFisher Scientific) with differential modifications for Ser/Thr/Tyr phosphorylation (+79.97) and the sample processing artifacts for Met oxidation (+15.99) and Cys alkylation (+57.02). Phosphorylated and unphosphorylated peptide sequences were identified if they initially passed the following Sequest scoring thresholds: 1+ ions, Xcorr ≥ 2.0, Sf ≥ 0.4, p ≥ 5; 2+ ions, Xcorr ≥ 2.0, Sf ≥ 0.4, p ≥ 5; 3+ ions, Xcorr ≥ 2.60, Sf ≥ 0.4, p ≥ 5 against the target protein data base. Passing MS/MS spectra that represented the phosphorylated forms in the FoxM1b sequence were manually inspected to be sure that all b- and y-fragment ions aligned with the assigned sequence and modification sites. Determination of the exact sites of phosphorylation was aided using GraphMod software (Proteomics Browser).
In Vitro Phosphorylation of FoxM1b
For the in vitro kinase assays, 500 μg of protein lysates from U2OS cells transfected with T7-FoxM1b wild type or S251A mutant plasmids were coimmunoprecipitated overnight with T7 tag antibody and protein A-Sepharose beads. The immunoprecipitates were washed three times with lysis buffer and twice with kinase buffer. To start the kinase reaction, 20 μl of kinase buffer supplemented with 50 μm ATP, 5 μCi of [γ-32P]ATP, and 5 μg of recombinant cyclin B1-CDK1 proteins was added to the beads, and samples were incubated at 30 °C for 30 min. Reactions were terminated by adding an equal volume of 2× Laemmli sample buffer (Bio-Rad). The samples were separated on SDS-PAGE and analyzed by Coomassie Blue staining and then dried prior to autoradiography.
Retroviral Vector Cloning and Generation of T7-tagged Wild Type and S251A FoxM1 Stable Clones
T7-FoxM1b wild type and S251A mutant constructs were subcloned from pCMV T7-FoxM1b and pCMV T7-FoxM1bS251A vectors into a pLPCX-puro vector, respectively. Retrovirus stocks were prepared by transfecting 24 μg of the vectors into the Amphotropic 293 cells in a 15-cm plate using the calcium phosphate method. Viral supernatants were collected at 48 and 72 h post-transfection, filtered, concentrated, and stored at −80 °C until use (pLPCX-puro vector and Amphotropic 293 cells were kindly provided by Chia-Chen Chen, University of Illinois, Chicago). U2OS cells were transduced by concentrated viral supernatant in Dulbecco's modified Eagle's medium and 10 μg/ml Polybrene. Forty-eight hours after transduction, cells were selected with 1 μg/ml puromycin.
McKay Assay
A modified McKay assay was performed with the double-stranded FoxM1 binding site oligonucleotide (from 6×FoxM1 TATA-luciferase reporter gene) (29, 33, 34). Briefly, U2OS cells, transfected with a vector expressing either T7-FoxM1b wild type or S251A mutant protein, were lysed in 1 ml of McKay binding buffer (10% glycerol, 5 mm EDTA, 20 mm Tris, pH 7.2, 100 mm NaCl, 0.1% Nonidet P-40 and protease inhibitors). Cell debris was removed from the supernatant by spinning down at 14,000 × g for 10 min at 4 °C, and protein concentrations were determined by the Bradford method with the Bio-Rad protein assay reagent (Bio-Rad). Three hundred micrograms of cell extract was pre-cleared by incubation with 20 μl of protein A-Sepharose beads (50% slurry) (Amersham Biosciences) for 40 min and then incubated with 500,000 cpm of 32P-labeled FoxM1 binding site oligonucleotide, 35 μl of T7 tag antibody-conjugated beads (Novagen), and 1 μg/μl of poly(dI/dC) overnight with gentle rocking at 4 °C. The beads were washed three times with the McKay washing buffer (2% glycerol, 5 mm EDTA, 20 mm Tris, pH 7.2, 100 mm NaCl, 0.1% Nonidet P-40 and protease inhibitors), and resuspended in 100 μl of TE buffer (10 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0). Five micrograms of proteinase K were then added and samples were incubated for 30 min at 37 °C. The 32P-labeled FoxM1 binding site oligonucleotides were purified using the QIAquick Nucleotide Removal kit (Qiagen), eluted with 20 μl of ddH2O and resolved on 5% non-denaturing acrylamide gel. For competition experiments, a 100-fold excess of cold probe was preincubated with extract for 10 min prior to the binding reaction. After electrophoresis, the gel was dried and exposed to x-ray film.
Chromatin Immunoprecipitation Assay
U2OS, U2OS-WT, and U2OS-251A cell lines were used to perform chromatin immunoprecipitation (ChIP) assay as described (39) with additional modification. Briefly, 8 × 106 cells were cross-linked in situ by addition of 37% formaldehyde (Fisher Scientific) to a final concentration of 1% (w/v) and incubated at room temperature for 10 min with gentle swirling. The cross-linking reaction was terminated by the addition of 125 mm glycine for 5 min. Cells were washed twice in ice-cold phosphate-buffered saline, scraped, and collected from the plate with 1 ml of ice-cold phosphate-buffered saline containing protease inhibitors (Roche Applied Science). After spinning down at 2,000 × g for 5 min, the cell pellet was resuspended in 0.8 ml of SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1) and placed on ice for 10 min. The extract was sonicated to 200–1000 bp using a microtip and the remaining insoluble material was removed by centrifugation at 4 °C for 10 min. After measuring the protein concentration of the lysate, 500 μg of lysate was diluted with 9 volumes of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris, pH 8.0, 167 mm NaCl) and precleared with protein A-agarose beads followed by immunoprecipitation. Each precleared ChIP sample was incubated with 35 μl of either T7 tag antibody-conjugated beads (Novagen) or protein A-agarose beads for 12 to 16 h at 4 °C with rotation and washed according to the Upstate Cell ChIP assay protocol (catalog number 17–295; Lake Placid, NY). Cross-links were resolved on all samples, including 20% input, by addition of 100 μl of TE (1 mm EDTA, 10 mm Tris, pH 7.4) containing 10 μg of RNase A and then incubated for 15 min at 25 °C. Ten micrograms of proteinase K and 4 μl of 5 m NaCl were than added, and samples were digested for 16 h at 65 °C. DNA was purified using the PCR purification kit (Qiagen) and eluted with 50 μl of ddH2O. The total input was diluted 1:10, and 2 μl of samples and diluted input were used for standard PCR. The primer sequences and annealing temperature for the ChIP assay are as follow: Aurora B −1045F, 5′-AAG AAA GGA GAA GAG TGC TGG ACA G-3′, and Aurora B −741R, 5′-TCC GTT GTT TGC TCA TTT GCC-3′ (Ta, 56 °C); Survivin −1152F, 5′-GAG GCA GGA GAA TCG CTT GAA C-3′, and Survivin −831R, 5′-GGG CAT CAC ATC CAC TCA CTT C-3′ (Ta, 62 °C).
Real Time Quantitative RT-PCR
Total RNA from double thymidine-synchronized U2OS cells was prepared and subjected to quantitative RT-PCR as described (19). The following forward (F) and reverse (R) primer sequences and annealing temperature (Ta) were used to amplify and measure the amount of human mRNA by real time RT-PCR: Plk1 −946F, 5′-ATC ACC TGC CTG ACC ATT CCA C-3′ and Plk1 −1045R, 5′-TCT CCA AGC CTT TAT TGA GGA CTG-3′ (Ta, 56 °C); Aurora B −829F, 5′-TCA CAC AAC GAG ACC TAT CGC C-3′, and Aurora B −936R, 5′-GGG GTT ATG CCT GAG CAG TTT G-3′ (Ta, 56 °C); Survivin-F, 5′-TCA AGG ACC ACC GCA TCT CTA-3′, Survivin-R, 5′-TGA AGC AGA AGA AAC ACT GGG C-3′ (Ta, 61 °C); and CENPA-F, 5′-CTT CCT CCC ATC AAC ACA GTC G-3′, and CENPA-R, 5′-TGC TTC TGC TGC CTC TTG TAG G-3′ (Ta, 54 °C). The real time RT-PCR RNA levels were normalized to human cyclophilin mRNA levels as described (19). All experiments were performed in triplicate, and repeated twice.
Statistical Analysis
Statistical significance was calculated by the Student's t test using the Prism statistics software. Statistically significant changes (p values < 0.05) are indicated with asterisks.
RESULTS
Identification of a Novel Conserved Phosphorylation Site in the Forkhead Box Domain of FoxM1b That Is Critical for Its Transcriptional Activity
FoxM1 is a cell cycle-regulated transcription factor that participates in the expression of genes required for both G1 and M phase progression (14, 19, 20). It is phosphorylated in M phase and degraded in the early G1 phase (29, 31, 35, 36). However, the mechanisms that are involved in its activation are poorly understood. Previous studies identified Thr-596 as a CDK1 phosphorylation site that is important for the activity of FoxM1b (Thr-611 in FoxM1c (37)). To elucidate how FoxM1 is controlled by phosphorylation, we investigated the potential phosphorylation sites in FoxM1 that might be important for its activation in the M phase. First, U2OS cells were synchronized at the G1/S boundary by double thymidine block, and then released into fresh medium to allow the cells to reenter the cell cycle. The cells were collected at the indicated time points for analysis (Fig. 1). A flow cytometric analysis identified the distribution of cells in different phases of the cell cycle (supplemental Fig. S1). As shown by Western blot analysis in Fig. 1A (top panel), several slower migrating forms of FoxM1 were initially detected at 4 and 8 h (S phase) after release from the G1/S block and accumulated in G2 and M phases. The slower migrating bands were indeed phosphorylated FoxM1, because when immunoprecipitated FoxM1 from 0 (G1/S) and 16 h (M phase) cells were treated with phosphatase (λ-phosphatase), the slower migrating forms of FoxM1 in the 16-h time point were converted to the faster migrating form, which co-migrated with the form of FoxM1 from the 0-h time point (Fig. 1B). This result suggests that FoxM1 is initially phosphorylated at S phase and reaches its maximal phosphorylation status at G2 and M phases.
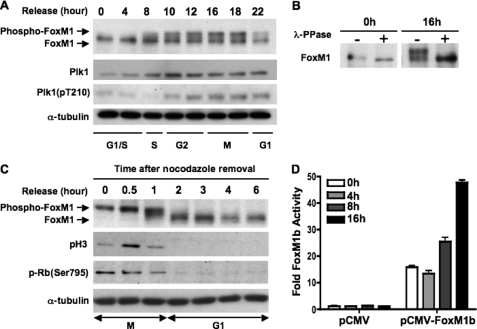
FIGURE 1.
FoxM1 is hyperphosphorylated during G2 and M phase and becomes dephosphorylated upon cells completing mitosis. A, phosphorylation profile of endogenous FoxM1 through th S-G2-M phases of the cell cycle. U2OS cells were synchronized at the G1/S boundary by a double thymidine block, released into fresh medium, and harvested at the indicated time points (hours). Cell extracts were prepared and immunoblotted for FoxM1, Plk1, active form Plk1 (pT210-Plk1), and α-tubulin. B, equal amounts of G1/S phase (0 h) or G2/M phase (16 h) cell extracts from A were immunoprecipitated using anti-FoxM1 antibody. Immunoprecipitated endogenous FoxM1 was treated with or without phosphatase (λ-PPase) followed by Western blot analysis. C, mitotic cells were collected as described under “Materials and Methods” and replated in fresh medium to allow re-entry into the cell cycle. Cell extracts were collected at various time points as indicated and immunoblotted for FoxM1, phospho-Histone H3, phospho-Rb(Ser-795), and α-tubulin. D, U2OS cells were synchronized by double thymidine block. At the release intervals, cells were cotransfected with the 6×FoxM1-TATA-luciferase reporter and a vector expressing FoxM1b. Cells were harvested at the indicated time points and analyzed by the dual luciferase assay as described under “Materials and Methods.” Results are presented as fold-induction relative to G1/S (0 h) cells transfected with a pCMV empty vector. Error bars represent S.D.
Reversible phosphorylation is one of the most common mechanisms for rapid alterations of transcription factor activity (38–40). To determine the fate of the phosphorylated FoxM1 in M phase, we analyzed the mitotic U2OS cells released from nocodazole arrest. The mitotic cells were collected by the mitotic shake-off method and replated in fresh media to allow progression through M phase. Cells were harvested at various time points after release, and the extracts were analyzed by Western blot for FoxM1. 30 min to 2 h after release, the slower migrating forms of FoxM1 were converted to the faster migrating forms (Fig. 1C, top panel), indicating dephosphorylation. The dephosphorylation of FoxM1 was gradual, with the appearance of “intermediate” migrating forms (at 1 h post-release) before the appearance of the fastest migrating form of FoxM1 (at 2 h post-release). At the 2-h time point, FoxM1 exhibited mainly the hypophosphorylated form. The mitosis-specific phosphorylation of histone H3 (pH3 Ser-10) and phospho-Rb (Ser-795) were barely detectable at the 2-h time point, suggesting that FoxM1 is dephosphorylated when cells complete mitosis.
To determine whether the transcriptional activity of FoxM1b correlated with its phosphorylation status, U2OS cells were synchronized by double thymidine block and cotransfected with the 6×FoxM1 TATA-luciferase reporter plasmid and pCMV or pCMV-FoxM1b expression vectors at the release interval. Cells were harvested and extracts were analyzed by the luciferase assay at the 4-, 8-, and 16-h time points after release from the G1/S block. This assay demonstrated that FoxM1b has basal transcriptional activity at the G1/S boundary and reaches its maximal activity at the G2 and M phases, coinciding with its increased phosphorylation status (Fig. 1D).
Consistent with a recent study (32), we observed that the increased phosphorylation status of FoxM1 coincided with the expression and activation of PLK1 (Fig. 1A, middle panel). PLK1 is a key regulator of mitotic progression. It has been shown that the protein level of PLK1, its phosphorylation status, and kinase activity are tightly regulated through cell cycle progression (2). Interestingly, the C-terminal region of FoxM1b contains PLK1 consensus motifs, (E/D-X-(S/T)-Q (41), around Ser-623 and Ser-724. Therefore, PLK1 could phosphorylate FoxM1b to stimulate its transcriptional activity. Indeed, FoxM1b is phosphorylated by wild type but not kinase-dead PLK1 and its transcriptional activity is significantly induced by PLK1 (Fig. 2A). Coimmunoprecipitation experiments demonstrated that endogenous PLK1 interacts with exogenous T7-tagged FoxM1b (T7-FoxM1b) in vivo (Fig. 2B). To determine whether the association between endogenous FoxM1 and PLK1 is cell cycle regulated, cell extracts, prepared as in Fig. 1A, were subjected to immunoprecipitation assay. The interaction between FoxM1 and PLK1 was detected at the 16- and 18-h time points (M phase) during which the kinase activity of PLK1 was also the highest (Fig. 2C, upper panel). Moreover, the FoxM1 signals in the input lanes suggested that PLK1 associated with phosphorylated FoxM1 (Fig. 2C, lower panel). To further demonstrate whether PLK1 interacted with phosphorylated FoxM1, cells were transfected with a vector expressing the T7-FoxM1b protein, and cell extracts were immunoprecipitated with monoclonal PLK1 antibody or normal mouse IgG. The immunoprecipitates were treated with or without phosphatase (λ-phosphatase), and T7-FoxM1b was detected by immunoblotting with T7 tag antibody. The result shows that endogenous PLK1 associated with phosphorylated T7-FoxM1b (Fig. 2D). Together, these results indicate that the association between FoxM1 and PLK1 is temporally and tightly regulated during cell cycle progression. Therefore, we considered the possibility that PLK1 phosphorylates FoxM1 and modulates its activity in the M phase. To investigate PLK1-dependent phosphorylation of FoxM1, we cotransfected U2OS cells with a T7-FoxM1b expression vector and a vector expressing wild type (WT) or kinase-dead (KD) PLK1. Extracts were immunoprecipitated with a monoclonal antibody against the T7 tag to purify the T7-FoxM1b protein. Expression of PLK1 converted T7-FoxM1b to the phosphorylated form, whereas the KD PLK1 had very little effect (Fig. 2A). These two different forms of FoxM1b bands were subjected to phosphopeptide analyses using tandem mass spectrometry (supplemental Fig. S2). The phosphosites that were predominantly found in the phosphorylated form of T7-FoxM1b were obtained from the PLK1-cotransfected sample, but not from the KD PLK1-cotransfected sample, are summarized in Fig. 3A. One of these phosphorylation sites, Ser-623, was indeed a PLK1 consensus site. However, the others were not PLK1 consensus sites. Phosphorylation at Thr-596, which is the CDK1-phosphorylation site, was expected because PLK1 is known to activate cyclin B-Cdk1 (2, 6, 42). It has been reported that PLK1 could directly phosphorylate FoxM1 at Ser-715 and Ser-724 for full activation and proper mitotic progression (32). Although we identified a PLK1 phosphorylation motif around Ser-724 and conducted experiments that demonstrated that PLK1 could phosphorylate FoxM1 at 2 sites (including Ser-724 residue) within the 688–748 fragment of FoxM1b in vitro (data not shown), no phosphopeptide containing Ser-724 was detected by our in vivo mass spectrometry analysis. The basis for this disparity is unclear. We did observe the phosphorylation of Ser-715 in both WT and KD PLK1-cotransfected samples but there was no significant difference in the number of phosphopeptides between these two samples (supplemental Fig. S2).
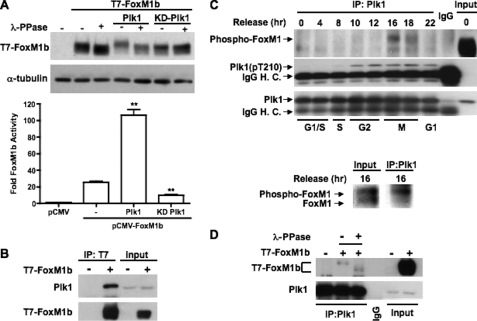
FIGURE 2.
Plk1 hyperphosphorylates FoxM1 and stimulates its transcriptional activity. A, U2OS cells were cotransfected with vectors expressing T7-FoxM1b and either WT or kinase-dead (KD) Plk1. Upper panel, cell extracts were treated or not with λ-phosphatase (PPase) and analyzed by immunoblotting using a monoclonal antibody against T7 tag. Lower panel, cell extracts were used in a dual luciferase assay. Results are expressed as fold-induction over the pCMV control. ± SD. **, p < 0.01. B, cells were transiently transfected with pCMV-T7-FoxM1b, and cell extracts were immunoprecipitated (IP) with a monoclonal antibody recognizing T7 tag. Endogenous Plk1 in the immunoprecipitates was detected by Western blot and using a monoclonal antibody specific to Plk1. C, cell extracts prepared as described in the legend to Fig. 1A were used for coimmunoprecipitation to determine the association of endogenous FoxM1 and Plk1 during cell cycle progression. Lysates from different time points were immunoprecipitated with monoclonal antibody against Plk1 or normal mouse IgG followed by immunoblotting with anti-FoxM1, anti-pT210 Plk1, and anti-Plk1 antibody. Lower panel, Western blot of FoxM1 from the input and the immunoprecipitates from the 16-h time point extract is shown. D, cell extracts prepared as in B were subjected to coimmunoprecipitation with monoclonal Plk1 antibody or normal mouse IgG. The immunoprecipitates were treated or not with λ-phosphatase, and the T7-FoxM1b protein was detected by Western blot analysis. Detection of Plk1 demonstrated that an equal amount of Plk1 protein was pulled down in all lanes.
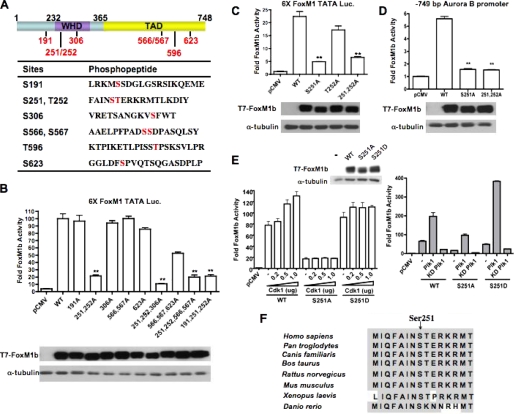
FIGURE 3.
Identification of in vivo phosphorylation sites within the hyperphosphorylated FoxM1b. A, a schematic representation of the FoxM1b protein indicating the N terminus (1–232), wing-helix domain (WHD), and transcriptional activation domain (TAD). The phosphorylated serine and threonine residues as revealed by mass spectrometry are shown in red. The table lists the sequences of peptides surrounding the phosphorylated serine and/or threonine residues. B and C, Ser-251 is critical for FoxM1b transcriptional activity. Vectors expressing T7-FoxM1b WT or mutant proteins in which the indicated residues have been mutated to alanine were cotransfected into U2OS cells with the 6×FoxM1-TATA-luciferase reporter construct. Cell extracts were analyzed by dual luciferase assay and immunoblotting. Western blot shows the expression levels of T7-FoxM1b WT and the mutant proteins. D, cells were cotransfected with a vector expressing T7-FoxM1b WT, S251A, or S251A/T252A mutant proteins and a luciferase reporter construct containing −749 bp human Aurora B promoter. The Western blot shows the expression of T7-FoxM1b WT and mutant proteins. The asterisks in panels B, C, D indicate statistically significant changes, with p values calculated by the Student t test. **, p < 0.01. E, left, cells were cotransfected with a vector expressing T7-FoxM1b WT, S251A, or S251D, an increasing amount of pCMV-Cdk1 plasmid, and the 6×FoxM1-TATA luciferase reporter construct. Cell extracts were analyzed by dual luciferase assay. Right, cells were cotransfected with a vector expressing T7-FoxM1b WT, S251A, or S251D mutant, a vector expressing wild type or kinase-dead Plk1, and the 6×FoxM1-TATA luciferase reporter construct. Western blot shows the expression levels of T7-FoxM1b WT and the mutant proteins. F, comparison of the amino acid sequence surrounding Ser-251 of FoxM1b in FoxM1 orthologues.
To investigate the impact of the other phosphorylation sites (Fig. 3A) on the transcriptional activity of FoxM1b, we mutated the threonine or serine residues to alanine by site-directed mutagenesis. The transcriptional activities of the different FoxM1b mutants were measured by a dual luciferase assay using the 6×FoxM1 TATA-luciferase reporter. Surprisingly, mutation of Ser-623, a potential PLK1 phosphorylation site, had only a marginal effect on the transcriptional activity of FoxM1b. However, a significant reduction (∼80%) of transcriptional activity was observed in the S251A/T252A mutant that harbor substitutions of Ser-251 and Thr-252. Moreover, the transcriptional activities of the other mutants were dramatically decreased when they also contained mutations at Ser-251 and Thr-252 (Fig. 3B). Further analyses revealed that Ser-251 is critical, as the S251A mutant, but not the T252A mutant, exhibited significantly diminished transcriptional activity (Fig. 3, C and D). Taken together, these results suggest that Ser-251 plays an important role in transcriptional activity of FoxM1b. Moreover, phylogenetic analysis revealed that Ser-251 is conserved in all FoxM1 orthologues characterized, thus implying that regulation of FoxM1b is also conserved (Fig. 3F).
We have shown that the transcriptional activity of FoxM1b can be stimulated by CDK1 and PLK1 (see Ref. 29 and Fig. 2A). A previous study demonstrated that PLK1 targets its substrates through the polo-box domain, which recognizes the “priming” phosphorylation generated by CDK1 kinase (7). Moreover, a recent study also suggested that the phosphorylation of FoxM1b by CDK1 serves as priming factor to recruit PLK1 to FoxM1b and stimulate its transcriptional activity (32). To demonstrate whether CDK1 and/or PLK1 could activate the FoxM1b mutant proteins, we cotransfected a vector expressing the wild type, S251A mutant, or S251D phosphomimetic mutant FoxM1b protein with CDK1 or PLK1 expressing plasmids into U2OS cells and measured FoxM1b transcriptional activity. We observed that transcriptional activity of the S251A mutant did not respond to CDK1 stimulation, whereas CDK1 induces the transcriptional activity of wild type FoxM1b in a dose-dependent manner. Interestingly, PLK1 could still stimulate transcriptional activity of the S251A mutant protein. (PLK1 also caused a shift in the mobility of the S251A mutant (see supplemental Fig. S3).) Moreover, activity of the S251D mutant is more responsive to PLK1 stimulation compared with the wild type FoxM1 (Fig. 3E). The lack of activation of the S251A mutant by CDK1 suggests that Ser-251 might be critical for CDK1-dependent phosphorylation.
Effect of S251A Mutation on Nuclear Localization and DNA Binding Ability of FoxM1b
To elucidate the transcription deficiency phenotype of the S251A mutant, we first investigated whether Ser-251 is required for nuclear localization of FoxM1b. The U2OS cells constitutively expressing T7-FoxM1b wild type or S251A mutant protein were grown on chamber slides and synchronized by double thymidine block. The G1/S-arrested cells were released into fresh medium for 10 h to allow them to reach the G2 phase, fixed, and subjected to immunostaining with a monoclonal antibody against the T7 tag. To identify the nucleus, cells were counterstained with 4′,6-diamidino-2-phenylindole. We observed that both the wild type and S251A mutant proteins were localized in the nucleus at G2 phase (Fig. 4B). The S251A mutant protein, like the wild type, was found to be nuclear in all phases (data not shown). Therefore, Ser-251 is not required for nuclear localization of FoxM1b.
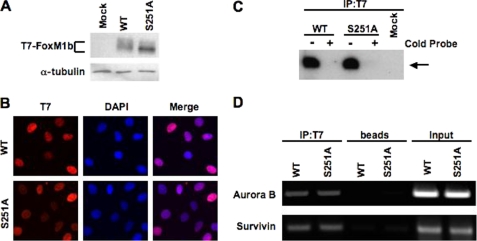
FIGURE 4.
Mutation in Ser-251 affects neither the nuclear localization nor the DNA binding of FoxM1b. A, mutation of Ser-251 generates mainly the hypophosphorylated form of FoxM1b. U2OS cells were transfected with a vector expressing T7-FoxM1b WT or S251A mutant proteins, and cell extracts were subjected to immunoblotting with T7-tag monoclonal antibody. B, U2OS cells constitutively expressing the T7-FoxM1b WT or S251A mutant proteins were synchronized at the G1/S boundary and released into fresh medium for 10 h to allow them reach the G2 phase. Cells were fixed and cellular localization of T7-FoxM1b WT or S251A mutant proteins was revealed by immunofluorescence staining with T7-tag antibody (red). Cells were stained also with 4′,6-diamidino-2-phenylindole (DAPI) (blue) to identify the nuclei. C, McKay assay was performed with 32P-labeled FoxM1 binding site oligonucleotide and cells extracts containing exogenous T7-FoxM1b WT or S251A mutant proteins. The T7-FoxM1b protein-DNA complex was immunoprecipitated with monoclonal antibody specific to T7 tag; the bound DNA were purified, resolved by electrophoresis on a nondenatured 5% polyacrylamide gel, and detected by autoradiography as described under “Materials and Methods.” For DNA competitions, 100-fold molar excess of cold probe was used as competitor to show binding specificity. D, promoter binding ability of the T7-FoxM1b S251A mutant determined by ChIP assay. U2OS cells constitutively expressing T7-FoxM1b WT or S251A mutant proteins were cross-linked and sonicated. The chromatin fragments were immunoprecipitated (IP) with T7 tag monoclonal antibody followed by PCR using the primers specific to human Aurora B (−1045/−741) or Survivin (−1152/−831) promoters.
Ser-251, which is required for the transcriptional activity of FoxM1b, lies within the winged helix DNA binding domain of FoxM1b. Therefore, it is possible that the mutation of Ser-251 inhibits the transcriptional activity of FoxM1b by impairing its DNA binding activity. However, based on structural modeling done with the winged helix DNA binding domain of FoxA3 (also known as HNF-3γ) as template, this residue is not expected to make contact with DNA (not shown) (8). To determine the effects of mutation on the DNA binding activity, we expressed T7-FoxM1b wild type or S251A mutant protein in U2OS cells (Fig. 4A). Interestingly, the mutant protein exhibited a faster mobility in SDS-polyacrylamide gel electrophoresis, indicating a deficiency in phosphorylation. To compare the DNA binding activity, the extracts were subjected to a modified McKay assay using a 32P-labeled FoxM1 consensus binding site oligonucleotide probe (29, 33, 34). To confirm specific binding, an unlabeled DNA probe was included as competitor. The protein-DNA complexes were immunoprecipitated using T7 tag antibody, and the DNA was purified and resolved on a 5% nondenaturing polyacrylamide gel and visualized by autoradiography. As shown in Fig. 4C, mutation of Ser-251 did not significantly affect the DNA binding ability of FoxM1b. In additional experiments, we did not detect any difference in the off-rates of DNA binding by FoxM1b wild type and the S251A mutant (data not shown). Furthermore, to determine the promoter binding efficiency of the mutant protein in vivo, we conducted ChIP experiments in cells constitutively expressing wild type or S251A mutant protein. Previously, we reported that FoxM1 is recruited to the promoters of human Aurora B and Survivin to promote their transcription (19). As shown in Fig. 4D, we observed a similar occupancy of FoxM1b wild type and S251A mutant protein on human Aurora B and Survivin promoters. We therefore conclude that the Ser-251 to Ala mutation does not significantly alter the DNA binding activity of FoxM1b. However, we did not rule out the possibility that the mutant FoxM1b has a different binding specificity.
Ser-251 Is Required for Efficient Binding of FoxM1b to the Transcriptional Coactivator CBP
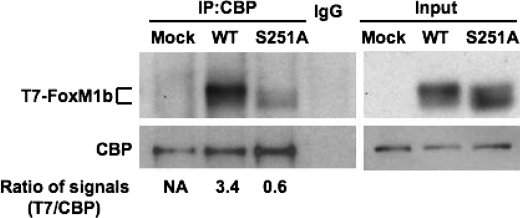
Because we did not detect any significant impairment in DNA binding and in nuclear localization of S251A mutant, we considered the possibility that the mutant might be defective in binding to the transcriptional activation partners. Previous studies identified CBP as a major transcriptional partner of FoxM1b. It was shown that CBP associates with the cyclin-CDK-phosphorylated form of FoxM1b to stimulate FoxM1b-activated transcription (29). It is noteworthy that cyclin-Cdk phosphorylates FoxM1 at Thr-596 and not at Ser-251. Nevertheless, we compared the interaction between the wild type and mutant FoxM1b with CBP. U2OS cells were transfected with plasmids expressing T7-FoxM1b wild type or S251A mutant protein. Cell extracts were subjected to immunoprecipitation with a monoclonal antibody against CBP. The immunoprecipitates were examined by Western blot analysis using the T7 tag antibody to detect the presence of FoxM1b. As expected, the FoxM1b wild type protein co-immunoprecipitated with CBP (Fig. 5). However, co-immunoprecipitation of the S251A mutant protein was significantly impaired compared with that of the wild type. The impaired interaction with CBP could potentially explain the reduced transcriptional activity of the S251A mutant. These observations also suggest that Ser-251 directly or indirectly participates in the interaction of FoxM1b with CBP.
FIGURE 5.
Ser-251 of FoxM1b is required for its efficient association with the co-activator CBP. U2OS cells were transfected with T7-FoxM1b WT or S251A expression plasmids. Endogenous CBP was immunoprecipitated with a monoclonal CBP antibody and the immunocomplexes were subjected to Western blot analysis with antibodies specific to T7 tag or CBP. The relative ratio of the T7-FoxM1b signal compared with immunoprecipitated (IP) CBP signal in the immunocomplexes was determined. NA, not analyzed.
Ser-251 Is Necessary for CDK1 Phosphorylation of FoxM1b
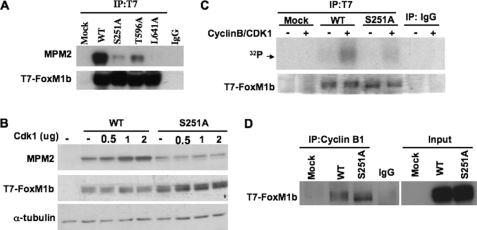
Binding of CBP to FoxM1b requires phosphorylation of FoxM1b by CDK1/CDK2. Mutation in the CDK1/CDK2 phosphorylation site (Thr-596) leads to inhibition of association between FoxM1b and CBP (29). Moreover, expression of a dominant-negative CDK1 diminishes the interaction between CBP and FoxM1b (29). It has been demonstrated that the FoxM1 protein is immunoreactive with the MPM2 monoclonal antibody, which recognizes a CDK-related epitope containing phosphoserine or phosphothreonine followed by proline (phospho(S/T)P) (29, 43, 44). Because the S251A mutant is impaired in binding to CBP and its transcriptional activity fails to respond to CDK1 stimulation (Figs. 3E and 5), we determined whether it is deficient in cyclin-CDK-dependent phosphorylation using the monoclonal MPM2 antibody. Cells were transfected with a vector expressing T7-FoxM1b wild type, S251A mutant, T596A mutant, or L641A mutant proteins. The T596A mutant is deficient in CDK-dependent phosphorylation, and the L641A mutant is deficient in the binding of cyclin-CDK complex (29). The wild type and FoxM1b mutant proteins were immunoprecipitated with T7 tag antibody and immunoblotted with MPM2 monoclonal antibody. The MPM2 signal was barely detected in S251A and L641A mutants and was reduced in T596A mutant proteins compared with that in the wild type T7-FoxM1b (Fig. 6A). To further investigate the deficiency in CDK1-dependent phosphorylation, we co-expressed increasing levels of CDK1 with the wild type or S251A mutant protein. Expression of CDK1 increased MPM2 signals in the wild type, but not in the S251A mutant, in a dose-dependent manner (Fig. 6B). The observation was further confirmed by in vitro kinase assays. T7-FoxM1b wild type and S251A mutant proteins were immunoprecipitated from transfected cell lysates and then subjected to in vitro phosphorylation using [γ-32P]ATP and recombinant cyclin B-CDK1 kinase. Cyclin B-CDK1 phosphorylated the T7-FoxM1b wild type protein at a significantly higher level compared with the S251A mutant protein (Fig. 6C). As pointed out before, Ser-251 of FoxM1b is not a CDK-phosphorylation site, as it lacks the (S/T)-P motif. We considered the possibility that Ser-251 might play a role in the interaction between FoxM1b and the cyclin B-CDK1 complex. To determine whether the S251A mutant protein was deficient in binding to the cyclin B-CDK1 complex, we performed immunoprecipitation experiments with protein extracts prepared from cells transfected with T7-FoxM1b wild type or S251A mutant constructs. Endogenous cyclin B1 was immunoprecipitated, and the FoxM1b protein was detected by Western blot with T7 tag antibody. Surprisingly, the S251A mutant protein interacted with the endogenous cyclin B1 protein as efficiently as the T7-FoxM1b wild type protein (Fig. 6D). Therefore, Ser-251 of FoxM1b is not required for binding to cyclin B-CDK1, but it is required for cyclin B-CDK1-dependent phosphorylation.
FIGURE 6.
Ser-251 is necessary for Cdk1-dependent phosphorylation of FoxM1b. A, U2OS cells were transfected with a vector expressing T7-FoxM1b WT, S251A, T596A, or L641A mutant proteins. The exogenous T7-FoxM1b proteins were immunoprecipitated with T7 tag antibody and immunoblotted with a monoclonal antibody against the MPM2 (mitotic protein monoclonal 2) epitope or T7 tag. B, Cdk1 increases MPM2 signals on T7-FoxM1b WT but not S251A mutant protein in vivo. A vector expressing T7-FoxM1b WT or S251A was cotransfected with an increasing amount of Cdk1 plasmids into cells. Cell extracts were subjected to Western blot analysis with MPM2 antibody. C, Cdk1 phosphorylates T7-FoxM1b WT but not S251A mutant in vitro. Cells were transfected with T7-FoxM1b WT or S251A expressing plasmids, and cell extracts were immunoprecipitated with either normal mouse IgG or T7 tag antibody. The immunocomplexes were incubated with [γ-32P]ATP in the presence or absence of recombinant cyclin B-CDK1 for the kinase reaction. Following the kinase reaction, samples were subject to Coomassie Blue staining and autoradiography. D, endogenous cyclin B1 was immunoprecipitated from cell extracts prepared as in C and the immunocomplexes were analyzed by Western blot analysis using T7 tag antibody.
Expression of S251A Mutant Protein Fails to Stimulate the G2/M Target Genes of FoxM1b and Impairs M Phase Progression
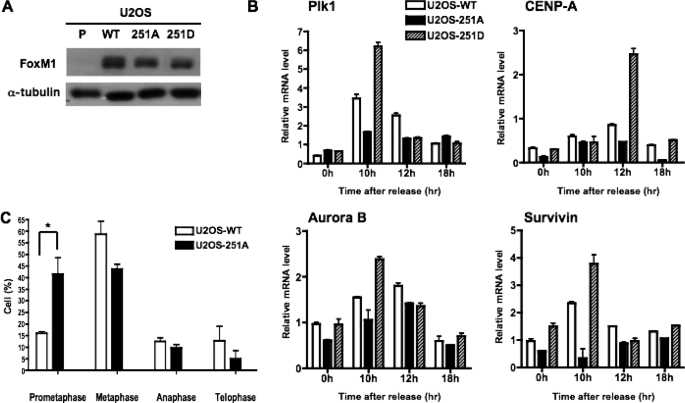
To further investigate the role of Ser-251 in the function of FoxM1b, we generated stable cell lines by infecting U2OS cells with retrovirus expressing the T7-FoxM1b wild type, S251A, or the S251D proteins. After selection, the expression of FoxM1b in U2OS-WT, U2OS-251A, U2OS-251D, and parental U2OS cells were analyzed by Western blot analysis. The wild type and mutant FoxM1b proteins were expressed at comparable levels (Fig. 7A).
FIGURE 7.
Expression of T7-FoxM1b S251A mutant protein fails to stimulate the G2/M target genes and impairs mitotic progression. A, cell extracts were prepared from parental U2OS cells (P), and cells constitutively expressing T7-FoxM1b WT (U2OS-WT), S251A (U2OS-251A), or S251D mutant proteins (U2OS-251D). The expression level of WT and mutant proteins in stable cell lines was determined by immunoblotting with FoxM1 antibody. B, U2OS-WT, U2OS-251A, and U2OS-251D cells were synchronized at the G1/S phase by double thymidine block and then released into fresh media. Following release, cells were harvested at the indicated time points for real time RT-PCR analysis. Total RNA was isolated, and the mRNA levels of G2/M target genes were analyzed by quantitative real time RT-PCR analysis using primers specific to PLK1, Aurora B, Survivin, and CENP-A. The experiments were repeated twice in triplicate. C, U2OS-251A cells exhibit mitotic delay. U2OS cells stably expressing either WT or S251A mutant FoxM1 were synchronized by nocodazole treatment. The mitotic cells were collected, replated with fresh medium on chamber slides for 40 min, fixed, and then subjected to immunostaining with α-tubulin, FoxM1, and 4′,6-diamidino-2-phenylindole. Cells in each stage of mitosis were counted and presented in the bar graph. The experiments were repeated twice in triplicate and 10 random fields were counted per sample. *, p < 0.05.
Because the phosphorylation status and the activity of FoxM1b are significantly increased in G2 and M phases, we investigated whether the S251A or S251D proteins could stimulate the expression of FoxM1 target genes that are activated in G2/M phases using U2OS-WT, U2OS-251A, and U2OS-251D cells. The cell lines were synchronized to the G1/S boundary by double thymidine block and then released into fresh medium. At different time points following the release, cells were harvested and total RNA was prepared followed by quantitative real time RT-PCR assays to measure the mRNA levels of the FoxM1 target genes, including PLK1, Aurora B, Survivin, and CENP-A. The expression of the wild type or the phosphomimetic S251D FoxM1b caused a significant increase in G2/M expression of these FoxM1 target genes. In contrast, the phosphorylation-defective S251A mutant was deficient in stimulating expression of its target genes (Fig. 7B). Next, we investigated whether the cells expressing the S251A mutant were defective also in mitotic progression. The U2OS-WT and U2OS-251A cells were synchronized and arrested at prometaphase (early mitosis) by nocodazole treatment. The nocodazole-arrested cells were collected by the mitotic shake-off method and released into fresh medium. Forty minutes after release, cells were fixed and stained with α-tubulin antibody to visualize microtubule assembly. The cells were counterstained with 4′,6-diamidino-2-phenylindole to follow M phase progression. The majority of the wild type FoxM1b expressing cells were found to progress into metaphase, whereas the S251A mutant expressing cells were mainly found in prometaphase (Fig. 7C). These results suggest that the expression of the S251A mutant protein delays progression through early metaphase.
DISCUSSION
The work presented here provides insight into the mechanisms that activate FoxM1b during the G2 and M phases of the cell cycle. Deregulation of FoxM1b has been implicated in cell transformation, tumor development, and metastasis (18, 22, 23, 25, 26, 45–49). Therefore, an understanding of the mechanisms that control the activation of FoxM1b is significant in designing therapeutic interventions. In this study, we identified a highly conserved phosphorylation site within the forkhead box DNA binding domain of FoxM1b that controls CDK1-dependent phosphorylation of FoxM1b without affecting its DNA binding ability. Disruption of that conserved site precludes phosphorylation of FoxM1b by cyclin-CDK1, generates a hypophosphorylated FoxM1b, and significantly diminishes its binding to the coactivator protein CBP. Moreover, we show that this conserved phosphorylation site is essential for the transcriptional activation of mitotic genes by FoxM1b.
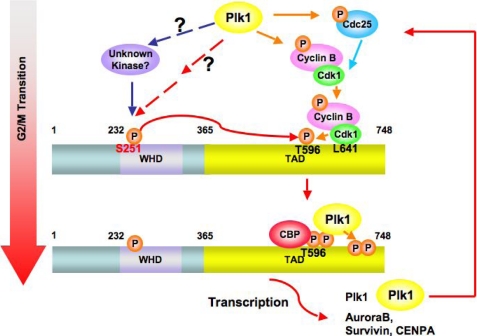
FoxM1b is initially phosphorylated at the S phase and becomes hyperphosphorylated in G2 and M phases of the cell cycle (Fig. 1A). Previous studies indicated that cyclin-CDK1 is one of the major kinases that phosphorylates FoxM1b (29). Because there appears to be mainly one functional CDK-phosphorylation site (Thr-596; we did not rule out the possibility that phosphorylation at this site opens up other CDK sites in FoxM1b), we considered other G2/M kinases that might be involved in generating the hyperphosphorylated FoxM1b. We observed that activation of PLK1 coincides with the appearance of the hyperphosphorylated form of FoxM1b (Fig. 1A). FoxM1b possesses at least two potential PLK1 phosphorylation sites (data not shown). Coexpression of PLK1 with FoxM1b increases the slower migrating hyperphosphorylated form of FoxM1b and stimulates FoxM1b transcriptional activity (Fig. 2A). Moreover, the association between PLK1 and FoxM1 mainly occurs in the M phase (Fig. 2C). These observations prompted us to analyze the phosphorylation of FoxM1b by PLK1. Indeed, we identified one of the PLK1 sites to be phosphorylated by PLK1. However, mutation of that site had only a marginal effect on the transcriptional activity of FoxM1b. A recent report described the identification of two residues (Ser-715 and Ser-724) on FoxM1 that are phosphorylated by PLK1 (32). In our hands, we did not observe the phosphorylation of Ser-724 in the mass spectrometry assay, although we did identify the PLK1 phosphorylation motif around Ser-724 through bioinformatic analysis and observed that Ser-724 could be phosphorylated by PLK1 in vitro (data not shown). The mutagenesis assay also suggested a marginal effect of Ser-724 phosphorylation on FoxM1 activity (data not shown). We did observe the phosphorylation on Ser-715 but in both wild type- and kinase-dead PLK1-cotransfected samples. The percentage of Ser-715 phosphopeptides is slightly more in the FoxM1 sample cotransfected with wild type PLK1 expressing vector but the difference was not significant. The potential discrepancy between Fu et al. (32) and our studies could be that we cotransfected a T7-FoxM1b expressing vector with wild type or kinase-dead Plk1 constructs into cells and then analyzed the hyper- and hypophosphorylation forms of FoxM1 by mass spectrometry assay. Fu and colleagues (32) use recombinant FoxM1 protein purified from bacteria to study the phosphorylation sites generated in vitro by PLK1. We also performed an in vitro PLK1 kinase assay and found that the C-terminal FoxM1 recombinant protein (residue 688–748) was highly phosphorylated by PLK1. We observed at least two major phosphorylation sites within this region (data not shown), which are consistent with the sites identified by Fu et al. (32). Furthermore, in our analysis we observed that PLK1 also indirectly phosphorylates FoxM1b. For example, there was an increase in the phosphorylation (phospho-Thr-596) of FoxM1b by expression of PLK1, an event mediated by cyclin-CDK1. CDK1 is known to be activated by PLK1 (2, 6, 42). Also, we identified a functionally important phosphorylation, stimulated by PLK1, at Ser-251 that appears to be phosphorylated by an unknown kinase. Our preliminary data suggests that this site might be phosphorylated by CK2 kinase. Although CK2 plays important roles in G2/M progression (50–52), it is not known to be activated by PLK1. Nevertheless, the mechanism by which PLK1 increases phosphorylation at Ser-251 remains unclear.
We demonstrate that Ser-251 is critical for transcriptional activation by FoxM1b. It is required for interaction with CBP, which was shown to be a transcriptional coactivator of FoxM1b. It has been shown that the transcriptional activity and interaction with CBP are mainly regulated by phosphorylation at Thr-596, which is mediated by the cyclin-CDK1 kinase (29). Fu and colleagues (32) also report that PLK1 interacts with FoxM1 in a cyclin-CDK1 phosphorylation-dependent manner to stimulate and maintain FoxM1 activity during mitotic progression. We observed that the CDK1-dependent phosphorylation of FoxM1b is impaired by the mutation of Ser-251 to Ala, suggesting that phosphorylation of Ser-251 is required for further cyclin-CDK1-mediated phosphorylation of FoxM1. The S251A mutant binds cyclin B-CDK1 at a comparable level to the wild type FoxM1b, but fails to undergo CDK1-dependent phosphorylation. Thus, there are other regulatory steps that control activation of FoxM1b at the G2/M transition by cyclin B-CDK1. Moreover, we observed the interaction between FoxM1 and PLK1 in M phase but not G2 phase (Fig. 2C). Taken together, we hypothesized that the CDK1-phosphorylation site (Thr-596) in FoxM1b is masked by other interactions, and phosphorylation at Ser-251 helps to unmask the site, allowing phosphorylation by already bound cyclin B-CDK1. This CDK1-dependent phosphorylation of FoxM1 will serve as the priming factor to recruit not only its coactivator CBP, but also PLK1. That is, the hyperphosphorylated form of FoxM1 is generated and its activity is promoted and maintained for proper mitotic progression (Fig. 8). Thus, it would appear that Plk1 and an unknown mediator (CK2 kinase?) collaborate to activate the phosphorylation of FoxM1b by cyclin B-CDK1. Interestingly, a collaboration between PLK1 and CK2 was observed in the inactivation of the Wee1A kinase in G2/M phases of the cell cycle (42, 53).
FIGURE 8.
A model for the activation of FoxM1 regulated by sequential phosphorylation during G2 and M phase progression. First, during the G2/M transition, Ser-251 within the winged helix DNA binding domain (WHD) of FoxM1 is phosphorylated by an unknown kinase, which might be activated by Plk1. Phosphorylation at Ser-251 is required for Cdk1-mediated phosphorylation on the C terminus of FoxM1. The Cdk1-dependent phosphorylation serves as a priming factor to recruit Plk1 onto FoxM1 at M phase (33). The positive feedback loop between FoxM1 and Plk1 maintains the hyperphosphorylation status and maximal activity of FoxM1 during the progression of mitosis.
Furthermore, although we demonstrated that the S251A mutant is deficient in phosphorylation and activation by CDK1, transcriptional activity of the S251A mutant remains responsive to stimulation by overexpressed PLK1. Those observations suggested that the phosphorylation of Ser-251 controls FoxM1b transcriptional activity through CDK1-dependent phosphorylation. However, overexpressed PLK1 could overcome the requirement for CDK1-dependent phosphorylation of FoxM1b, which is consistent with the observations of Fu et al. (32), who suggested that PLK1 activates FoxM1 downstream of CDK1.
The region between residues 1 and 232 of FoxM1b has a potent inhibitory function. Elimination of that region stimulates transcriptional activity of FoxM1b by about 20-fold (16, 30, 31). Interestingly, the N-terminal deletion mutant of FoxM1b, lacking residues between 1 and 232, is constitutively active independent of phosphorylation by cyclin-CDK1. For example, a dominant-negative CDK1 or other inhibitors of CDK1, which would inhibit FoxM1b-activated transcription, had very little inhibitory effect on the transcriptional activity of the deletion mutant lacking the N-terminal repressor domain (30, 31). These observations suggested a model in which cyclin-CDK1-mediated phosphorylation stimulates transcriptional activity of FoxM1 by overcoming inhibition by the N-terminal repressor domain (30, 31). The N-terminal fragment of FoxM1b could interact with the C-terminal activation domain to inhibit its own activity, when expressed in trans. Therefore, we think that the N-terminal repression domain (alone or in conjunction with other protein(s)) plays a role in masking the activating phosphorylation sites, thereby blocking interaction of FoxM1b with coactivators, such as CBP. CBP interacts with CREB when the latter is phosphorylated by cAMP-activated protein kinase (54, 55). Therefore, it is possible that the sequential phosphorylation of FoxM1b first by an unknown kinase followed by cyclin-CDK1 functions as a switch that promotes phosphorylation by PLK1 and recruitment of coactivators for transcriptional activation by FoxM1b.
FoxM1b has been implicated also in G1/S progression. Depletion of FoxM1b delays progression into S phase. The delay in progression has been linked to reduced expression of several genes that are expressed in the G1 phase and are targets of FoxM1b. However, it is not clear whether the transcriptional activity of FoxM1 in G1 phase also depends upon cyclin-CDK-mediated phosphorylation. Previous studies provided evidence for association between FoxM1b and cyclin E-CDK2 (29, 31, 37). Therefore, it is possible that cyclin E-CDK2 phosphorylates FoxM1 to stimulate expression of the G1 target genes. In that case, we have to assume that the cyclin E-CDK2-mediated activation of FoxM1 is inefficient because very little hyperphosphorylated FoxM1 is observed in the late G1 or G1/S phases (only the intermediate phosphorylation form of FoxM1 is observed) (Fig. 1). On the other hand, FoxM1 is dephosphorylated, degraded in early G1 phase (see Fig. 1 and Refs. 35 and 36), and synthesized in the late G1 phase (10, 13). It is possible that the newly synthesized FoxM1 activates expression of the G1/S target genes (SKP2, CKS1, KIS, and JNK1) through a mechanism that is distinct from the G2/M target genes. Clearly, further work will be necessary to understand how phosphorylation of FoxM1 orchestrates the expression of genes that are critical for G1/S and G2/M progression.
Supplementary Material
Acknowledgments
We thank Dr. Chih-Chiun Chen for helpful discussions. We also thank Dr. K. L. Hagen for flow cytometry analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA 124488 and CA 77637 (to P. R.) from the United States Public Health Service, and NIH Grants AG021842 (to L. F. L.), DK44525, and DK068503 (to A. L. T.).
This work is dedicated to the memory of Dr. Robert H. Costa who unfortunately passed away after a heroic fight with pancreatic cancer.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Plk1
- Polo-like kinase 1
- CREB
- cAMP-response element-binding protein
- ChIP
- chromatin immunoprecipitation
- CBP
- CREB-binding protein
- MS
- mass spectrometry
- RT
- reverse transcriptase
- WT
- wild type
- KD
- kinase-dead
- pCMV
- plasmid containing cytomegalovirus promoter.
REFERENCES
- 1.Nigg E. A. (2001) Nat. Rev. Mol. Cell Biol. 2, 21–32 [DOI] [PubMed] [Google Scholar]
- 2.Barr F. A., Silljé H. H., Nigg E. A. (2004) Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 3.Pines J. (1999) Nat. Cell Biol. 1, E73–E79 [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 5.Takaki T., Trenz K., Costanzo V., Petronczki M. (2008) Curr. Opin. Cell Biol. 20, 650–660 [DOI] [PubMed] [Google Scholar]
- 6.van Vugt M. A., Medema R. H. (2005) Oncogene 24, 2844–2859 [DOI] [PubMed] [Google Scholar]
- 7.Elia A. E., Cantley L. C., Yaffe M. B. (2003) Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 8.Clark K. L., Halay E. D., Lai E., Burley S. K. (1993) Nature 364, 412–420 [DOI] [PubMed] [Google Scholar]
- 9.Kaestner K. H., Knochel W., Martinez D. E. (2000) Genes Dev. 14, 142–146 [PubMed] [Google Scholar]
- 10.Korver W., Roose J., Clevers H. (1997) Nucleic Acids Res. 25, 1715–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lüscher-Firzlaff J. M., Westendorf J. M., Zwicker J., Burkhardt H., Henriksson M., Müller R., Pirollet F., Lüscher B. (1999) Oncogene 18, 5620–5630 [DOI] [PubMed] [Google Scholar]
- 12.Yao K. M., Sha M., Lu Z., Wong G. G. (1997) J. Biol. Chem. 272, 19827–19836 [DOI] [PubMed] [Google Scholar]
- 13.Ye H., Kelly T. F., Samadani U., Lim L., Rubio S., Overdier D. G., Roebuck K. A., Costa R. H. (1997) Mol. Cell. Biol. 17, 1626–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa R. H. (2005) Nat. Cell Biol. 7, 108–110 [DOI] [PubMed] [Google Scholar]
- 15.Korver W., Roose J., Heinen K., Weghuis D. O., de Bruijn D., van Kessel A. G., Clevers H. (1997) Genomics 46, 435–442 [DOI] [PubMed] [Google Scholar]
- 16.Wierstra I., Alves J. (2006) Biol. Chem. 387, 963–976 [DOI] [PubMed] [Google Scholar]
- 17.Petrovic V., Costa R. H., Lau L. F., Raychaudhuri P., Tyner A. L. (2008) J. Biol. Chem. 283, 453–460 [DOI] [PubMed] [Google Scholar]
- 18.Wang I. C., Chen Y. J., Hughes D. E., Ackerson T., Major M. L., Kalinichenko V. V., Costa R. H., Raychaudhuri P., Tyner A. L., Lau L. F. (2008) J. Biol. Chem. 283, 20770–20778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang I. C., Chen Y. J., Hughes D., Petrovic V., Major M. L., Park H. J., Tan Y., Ackerson T., Costa R. H. (2005) Mol. Cell. Biol. 25, 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. (2005) Nat. Cell Biol. 7, 126–136 [DOI] [PubMed] [Google Scholar]
- 21.Wonsey D. R., Follettie M. T. (2005) Cancer Res. 65, 5181–5189 [DOI] [PubMed] [Google Scholar]
- 22.Kalin T. V., Wang I. C., Ackerson T. J., Major M. L., Detrisac C. J., Kalinichenko V. V., Lyubimov A., Costa R. H. (2006) Cancer Res. 66, 1712–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim I. M., Ackerson T., Ramakrishna S., Tretiakova M., Wang I. C., Kalin T. V., Major M. L., Gusarova G. A., Yoder H. M., Costa R. H., Kalinichenko V. V. (2006) Cancer Res. 66, 2153–2161 [DOI] [PubMed] [Google Scholar]
- 24.Kalinichenko V. V., Major M. L., Wang X., Petrovic V., Kuechle J., Yoder H. M., Dennewitz M. B., Shin B., Datta A., Raychaudhuri P., Costa R. H. (2004) Genes Dev. 18, 830–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida Y., Wang I. C., Yoder H. M., Davidson N. O., Costa R. H. (2007) Gastroenterology 132, 1420–1431 [DOI] [PubMed] [Google Scholar]
- 26.Pilarsky C., Wenzig M., Specht T., Saeger H. D., Grützmann R. (2004) Neoplasia 6, 744–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q., Zhang N., Jia Z., Le X., Dai B., Wei D., Huang S., Tan D., Xie K. (2009) Cancer Res. 69, 3501–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bektas N., Haaf A., Veeck J., Wild P. J., Lüscher-Firzlaff J., Hartmann A., Knüchel R., Dahl E. (2008) BMC Cancer 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Major M. L., Lepe R., Costa R. H. (2004) Mol. Cell. Biol. 24, 2649–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park H. J., Wang Z., Costa R. H., Tyner A., Lau L. F., Raychaudhuri P. (2008) Oncogene 27, 1696–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laoukili J., Alvarez M., Meijer L. A., Stahl M., Mohammed S., Kleij L., Heck A. J., Medema R. H. (2008) Mol. Cell. Biol. 28, 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Z., Malureanu L., Huang J., Wang W., Li H., van Deursen J. M., Tindal D. J., Chen J. (2008) Nat. Cell Biol. 10, 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman W. H., Biade S., Zilfou J. T., Chen J., Murphy M. (2002) J. Biol. Chem. 277, 3247–3257 [DOI] [PubMed] [Google Scholar]
- 34.Lin T., Chao C., Saito S., Mazur S. J., Murphy M. E., Appella E., Xu Y. (2005) Nat. Cell Biol. 7, 165–171 [DOI] [PubMed] [Google Scholar]
- 35.Park H. J., Costa R. H., Lau L. F., Tyner A. L., Raychaudhuri P. (2008) Mol. Cell. Biol. 28, 5162–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laoukili J., Alvarez-Fernandez M., Stahl M., Medema R. H. (2008) Cell Cycle 7, 2720–2726 [DOI] [PubMed] [Google Scholar]
- 37.Lüscher-Firzlaff J. M., Lilischkis R., Lüscher B. (2006) FEBS Lett. 580, 1716–1722 [DOI] [PubMed] [Google Scholar]
- 38.Hunter T., Karin M. (1992) Cell 70, 375–387 [DOI] [PubMed] [Google Scholar]
- 39.Hunter T. (2007) Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 40.Whitmarsh A. J., Davis R. J. (2000) Cell Mol. Life Sci. 57, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakajima H., Toyoshima-Morimoto F., Taniguchi E., Nishida E. (2003) J. Biol. Chem. 278, 25277–25280 [DOI] [PubMed] [Google Scholar]
- 42.Watanabe N., Arai H., Nishihara Y., Taniguchi M., Watanabe N., Hunter T., Osada H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4419–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung T. W., Lin S. S., Tsang A. C., Tong C. S., Ching J. C., Leung W. Y., Gimlich R., Wong G. G., Yao K. M. (2001) FEBS Lett. 507, 59–66 [DOI] [PubMed] [Google Scholar]
- 44.Westendorf J. M., Rao P. N., Gerace L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang I. C., Meliton L., Tretiakova M., Costa R. H., Kalinichenko V. V., Kalin T. V. (2008) Oncogene 27, 4137–4149 [DOI] [PubMed] [Google Scholar]
- 46.Wang Z., Banerjee S., Kong D., Li Y., Sarkar F. H. (2007) Cancer Res. 67, 8293–8300 [DOI] [PubMed] [Google Scholar]
- 47.Chandran U. R., Ma C., Dhir R., Bisceglia M., Lyons-Weiler M., Liang W., Michalopoulos G., Becich M., Monzon F. A. (2007) BMC Cancer 7, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Boom J., Wolter M., Kuick R., Misek D. E., Youkilis A. S., Wechsler D. S., Sommer C., Reifenberger G., Hanash S. M. (2003) Am. J. Pathol. 163, 1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai B., Kang S. H., Gong W., Liu M., Aldape K. D., Sawaya R., Huang S. (2007) Oncogene 26, 6212–6219 [DOI] [PubMed] [Google Scholar]
- 50.Yde C. W., Olsen B. B., Meek D., Watanabe N., Guerra B. (2008) Oncogene 27, 4986–4997 [DOI] [PubMed] [Google Scholar]
- 51.Poole A., Poore T., Bandhakavi S., McCann R. O., Hanna D. E., Glover C. V. (2005) Mol. Cell. Biochem. 274, 163–170 [DOI] [PubMed] [Google Scholar]
- 52.Barz T., Ackermann K., Dubois G., Eils R., Pyerin W. (2003) J. Cell Sci. 116, 1563–1577 [DOI] [PubMed] [Google Scholar]
- 53.Watanabe N., Arai H., Iwasaki J., Shiina M., Ogata K., Hunter T., Osada H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11663–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arias J., Alberts A. S., Brindle P., Claret F. X., Smeal T., Karin M., Feramisco J., Montminy M. (1994) Nature 370, 226–229 [DOI] [PubMed] [Google Scholar]
- 55.Kwok R. P., Lundblad J. R., Chrivia J. C., Richards J. P., Bächinger H. P., Brennan R. G., Roberts S. G., Green M. R., Goodman R. H. (1994) Nature 370, 223–226 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.