Abstract
Prostaglandin E2 (PGE2) is an arachidonic acid metabolite that counters transforming growth factor-β-induced fibroblast activation via E prostanoid 2 (EP2) receptor binding. Phosphatase and tensin homologue on chromosome 10 (PTEN) is a lipid phosphatase that, by antagonizing the phosphoinositol 3-kinase (PI3K) pathway, also inhibits fibroblast activation. Here, we show that PTEN directly regulates PGE2 inhibition of fibroblast activation by augmenting EP2 receptor expression. The increase in collagen production and α-smooth muscle actin expression observed in fibroblasts in which PTEN is deficient was resistant to the usual suppressive effects of PGE2. This was due to marked down-regulation of EP2, a Gs protein-coupled receptor (GPCR) that mediates the inhibitory actions of this prostanoid via cAMP. pten−/− or PTEN-inhibited fibroblasts in which the PI3K pathway was blocked demonstrated a restoration of EP2 receptor expression, due to augmented gene transcription and mRNA instability. Importantly, restoration of the balance between PI3K and PTEN reestablished the inhibitory effect of PGE2 on fibroblast activation. No such influence of PTEN was observed on alternative E prostanoid GPCRs. Moreover, our studies identified a positive feedback loop in which cAMP signaling enhanced EP2 receptor expression, independent of PTEN. Therefore, our findings indicate that PTEN regulates the antifibrotic effects of PGE2 by a specific and permissive effect on EP2 receptor expression. Further, our data imply that cAMP signaling circumvents EP2 down-regulation in pten-deficient cells to restore EP2 receptor expression. This is the first description, to our knowledge, of PI3K/PTEN balance directing GPCR expression, and provides a novel mechanism for cellular effects of PTEN.
Introduction
Prostaglandin E2 (PGE2)2 is a cyclooxygenase-derived metabolite of arachidonic acid that influences a spectrum of important biological processes including pain, inflammation, and fibrosis. PGE2 effects may be cell-specific; for example, PGE2 promotes migration of epithelial cells (1, 2), but it inhibits migration of fibroblasts (3, 4). These seemingly paradoxical actions of PGE2 reflect the distinct responses resulting from ligation of a diverse repertoire of E prostanoid (EP) receptors on the cell surface. The four G protein-coupled (GPCR) EP receptors, termed EP1–4, signal by mobilizing intracellular calcium (EP1), increasing intracellular cAMP (EP2 and 4), or decreasing intracellular cAMP (EP3) (5). We have shown previously that PGE2 is capable, in lung fibroblasts, of tempering the profibrotic, activating effects induced by TGF-β (e.g. α-smooth muscle actin (α-SMA) and collagen I expression) (6) as well as the migratory effects of fibroblast growth factor (3). These effects are mediated by intracellular cAMP subsequent to binding of EP2, the most abundant of the fibroblast EP receptors.
Phosphatase and tensin homologue on chromosome 10 (PTEN) is a dual specificity lipid and protein phosphatase, but its primary action involves dephosphorylation of the D3 position on the inositol ring of phosphatidylinositol 3,4,5-trisphosphate (7), a major intracellular lipid second messenger that influences diverse cellular responses by recruiting pleckstrin homology domain-containing proteins to the cell membrane and is formed via the catalytic actions of PI3K following cell surface receptor activation. PTEN, as a direct antagonist of PI3K signaling, thus serves as a major brake in growth factor signaling. PTEN was first identified as a tumor suppressor, being mutated or deleted in a variety of malignant tumors (8, 9) and hamartomatous disorders (10). More recently, we and others have identified PTEN as a major down-regulator of fibroblast function (3, 11, 12), with loss of PTEN expression resulting in activation and migration of fibroblasts (11). Interestingly, we have also observed that PGE2 enhances PTEN catalytic activity and that the antimigratory effects of PGE2 depend on PTEN (3). This is in line with subsequent observations that another GPCR, the sphingosine 1-phosphate-2 receptor, regulates PTEN activity to inhibit cell migration (13). In this article, we show that PTEN also regulates EP receptor expression on fibroblasts, which we believe represents the first report of this phosphatase regulating GPCR expression. Specifically, we demonstrate that unopposed PI3K/Akt signaling results in significant down-regulation of the EP2 receptor and that blockade of this pathway restores functional EP2 signaling in PTEN-deficient cells. Moreover, we identify cAMP as a second novel regulator of EP2 expression, thereby placing cAMP both upstream and downstream of EP2. These data reveal bidirectional interplay between PTEN and PGE2/EP2 signaling that comprises a positive feedback loop, thereby amplifying the antifibrotic effects of each molecule.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
IMR-90 fibroblasts, derived from the lungs of a normal diploid human female fetus, were from the Coriell Institute for Medical Research (Camden, NJ). Murine (C57BL/6) embryonic fibroblasts were from ATCC (Rockville, MD) and pten−/− murine embryonic fibroblasts (14) were a kind gift from Drs. V. Stambolic and T. Mak (University Health Network, Ontario). Cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum, antibiotics, and Hepes. PGE2 was from Cayman Chemical (Ann Arbor, MI). Human recombinant TGF-β1 was from Roche Diagnostics (Mannheim, Germany). Bisperoxo(pyridine-2-carboxyl)oxovanadate (bpV(pic)) was from EMD Biosciences. The EP4-specific agonist ONO-AE1-329 was kindly provided by ONO Pharmaceuticals (Osaka, Japan). Akt inhibitor X (10-(4°-(N-diethylamino)butyl)-2-chlorophenoxazine HCl) was from Calbiochem and was demonstrated in preliminary studies to inhibit Akt phosphorylation and activity by ∼90% in pten−/− cells at a concentration of 10 μm (data not shown).
Antibodies
Antibody to α-SMA was obtained from Dako Corporation (clone 1A4, 1:5000; Carpinteria, CA). β-Tubulin and GAPDH antibodies (1:1000) were from Upstate (Billerica, MA). Total Akt (1:1000) antibody was from Cell Signaling Technologies (Beverly, MA). Type I collagen antibody (H-197) was from Santa Cruz Biotechnology (1:1000, Santa Cruz, CA).
Quantitative Real Time-PCR
Quantitative real time-PCR was performed on an Applied Biosystems (Foster City, CA) 7300 real time PCR machine as we have reported previously (15). Relative quantitation was based on the ΔΔCT method. All primers and probes were from Integrated DNA Technologies (Coralville, IA). Sequences are shown in supplemental Table S1.
Western Blotting
Western blot analysis under reducing conditions was performed on whole cell lysates as described previously (11). Densitometry of visualized bands was performed using ImageJ software (version 1.31; National Institutes of Health).
PTEN siRNA
PTEN and control siRNA were purchased from Cell Signaling Technology as a kit (catalogue no. 6250) and were used according to the manufacturer's instructions. Confirmatory experiments were performed in IMR-90 cells stably transfected with retrovirus encoding PTEN short hairpin RNA that targets human PTEN (16), resulting in functional abrogation of PTEN activity (Fig. S1).
Measurement of Intracellular cAMP Production
Intracellular production of cAMP was measured as described previously (3) utilizing a commercially available enzyme immunoassay kit according to the manufacturer's instructions (Assay Designs, Ann Arbor, MI).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5.01 (GraphPad Software, La Jolla, CA). In all cases, data are presented as mean ± S.D. Unless otherwise noted, all experiments were performed a minimum of three times. Differences between groups were evaluated using Student's t test. For multiple comparisons, one-way analysis of variance with Bonferroni's post test analysis was used. Data were considered significant at p < 0.05.
RESULTS
PTEN Is Necessary for PGE2 Inhibition of Fibroblast Activation
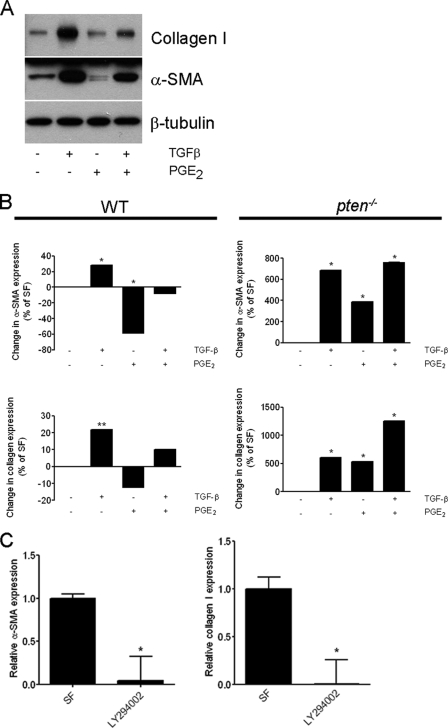
We demonstrated previously that PTEN is necessary for the antimigratory effects of PGE2 (3). Because PTEN also negatively regulates collagen synthesis and myofibroblast differentiation (11), we asked whether PTEN played an intermediary role in antifibrotic PGE2 effects. Normal fetal human lung fibroblasts (IMR-90) were incubated with TGF-β1 (2 ng/ml) in the presence or absence of PGE2 (1 μm) for 24 h. Western blot analysis confirmed our previous work (6) that TGF-β induced α-SMA and collagen I expression in human lung fibroblasts, and both responses were inhibited in the presence of PGE2 (Fig. 1A). Similar induction of α-SMA and collagen I was observed in IMR-90 cells treated with the relatively specific PTEN inhibitor, bpV(pic) (17) (data not shown). In embryonic C57BL/6 murine fibroblasts, we observed a consistent pattern of TGF-β-induced α-SMA expression by Western blotting (Fig. S2) and of α-SMA and collagen I expression by quantitative real time PCR (Fig. S3). PGE2 also blocks basal α-SMA and collagen I expression (18). However, in pten−/− cells, which possess a constitutively activated phenotype (11), PGE2 failed to block basal α-SMA and collagen I and instead resulted in substantial enhancement of both proteins (Fig. 1B). Further, in the presence of TGF-β, PGE2 caused a synergistic increase in collagen I expression in pten−/− cells, indicating that PTEN activity is necessary to transduce antifibrotic signals from PGE2.
FIGURE 1.
A, PGE2 inhibits TGF-β-induced α-SMA and collagen I expression in IMR-90 cells. Results are representative of three separate experiments. B, quantitative PCR (±S.D.) of pten−/− embryonic fibroblasts for α-SMA and collagen I (Col I). *, p < 0.0001 compared with the control condition. Results are representative of three separate experiments. C, quantitative PCR expression (±S.D.) of α-SMA (upper panel) and collagen I (lower panel) in pten−/− fibroblasts under serum-free (SF) conditions or following incubation with various concentrations of LY294002 (left graphs) or with wortmannin (right graphs). *, p < 0.0001 compared with the serum-free condition. Results are pooled from four separate experiments on independently generated RNA samples.
PTEN is the primary physiologic antagonist of PI3K activity. To ascertain whether elevated basal α-SMA and collagen I gene expression could be diminished by blockade of PI3K activity, pten−/− cells were incubated with the PI3K inhibitor LY294002 (200 μm) for 24 h. As shown in Fig. 1C, both α-SMA and collagen I gene expression were significantly reduced following PI3K inhibition. Likewise, PTEN reconstitution in pten−/− cells by transfection with full-length murine PTEN similarly decreased α-SMA and collagen I expression compared with cells transfected with the empty vector, although this did not reach statistical significance (Fig. S4).
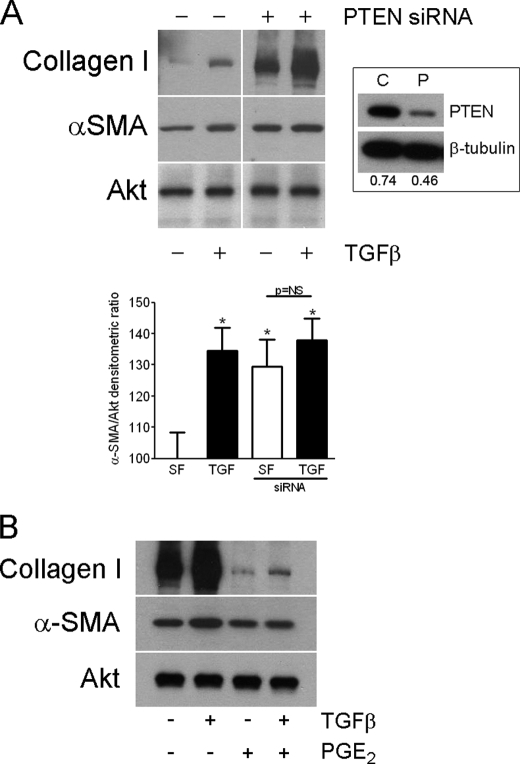
To that ensure these data were not confounded by the transgenic nature of the pten−/− cells, we inhibited PTEN in IMR-90 cells using siRNA. Such treatment resulted in a ∼50% reduction in PTEN expression by Western blotting compared with control siRNA (Fig. 2A, inset). Moreover, these cells demonstrated augmented α-SMA and collagen I expression (Fig. 2A), confirming our previous findings (11). Also consistent with our prior findings (11), we found that PTEN-silenced cells no longer responded to TGF-β stimulation with augmented α-SMA expression above their base line, although collagen I expression could be enhanced further. Unlike pten−/− cells, however, we observed that PGE2 was still capable of inhibiting TGF-β-induced α-SMA and collagen I expression (Fig. 2B), which was likely due to the incomplete silencing of PTEN. These data therefore suggest that the degree of PTEN inhibition or reconstitution dictates cellular phenotype. Indeed, a dose-dependent effect of PTEN can be observed in the progression of prostate carcinoma (19).
FIGURE 2.
A, PTEN inhibition in normal human lung fibroblasts augments α-SMA and collagen I expression. Akt was probed as a protein-loading control. Blot is representative of three separate experiments. Inset shows PTEN expression in IMR-90 cells treated with control (C) or PTEN (P) siRNA. Numbers under lanes refer to densitometric ratio of PTEN to β-tubulin. The bar graph represents the densitometric ratio of α-SMA to Akt depicted as percentage increase over the serum-free (SF) condition ± S.D. The ratio was pooled from three separate blots. *p < 0.05. NS, not significant. B, PGE2 inhibits α-SMA and collagen I expression in PTEN siRNA-treated IMR-90 lung fibroblasts. The blot is representative of three separate experiments.
Inability of EP2 to Signal Appropriately in the Absence of PTEN
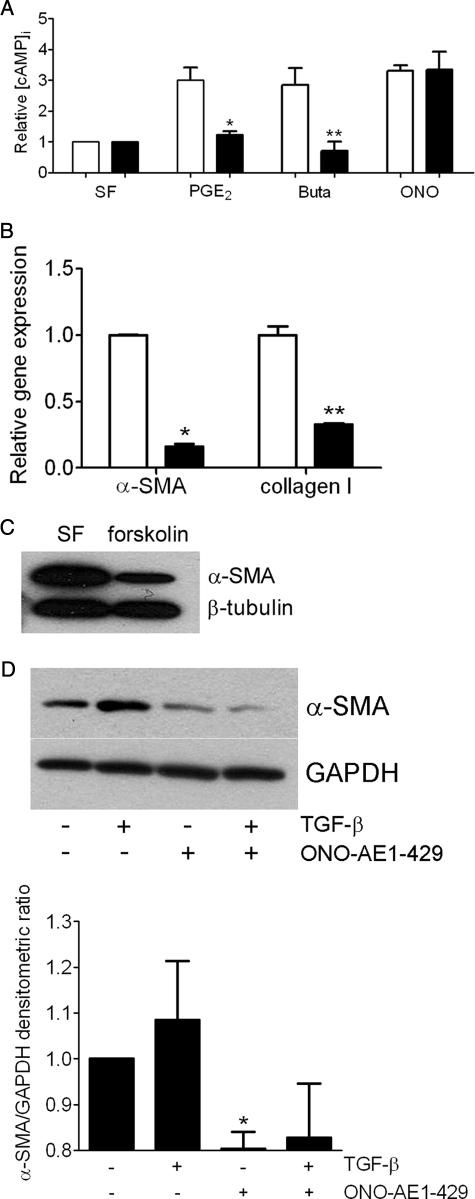
PGE2 exerts antifibrotic and antimigratory effects via EP2 (3, 4, 6), a GPCR that activates adenylate cyclase and augments intracellular cAMP (5). Thus, we asked whether PGE2 or a specific EP2 receptor agonist enhanced intracellular cAMP in pten−/− fibroblasts compared with WT fibroblasts. Unexpectedly, we observed that intracellular cAMP levels failed to increase in pten−/− embryonic fibroblasts in response to either PGE2 or the EP2-specific agonist butaprost whereas WT cells demonstrated an intact response (Fig. 3A). In contrast, both pten−/− and WT cells stimulated with a specific agonist (ONO-AE1-329) (20) of the alternate Gs-coupled EP receptor, EP4, demonstrated similar increases in intracellular cAMP concentration (Fig. 3A), implying that PTEN regulation of EP receptor signaling is restricted to EP2 and that Gs coupling to adenylate cyclase is intact within pten−/− cells.
FIGURE 3.
A, impaired EP2,but not EP4, receptor signaling in pten−/− cells. WT (open bars) or pten−/− (closed bars) cells were incubated under serum-free (SF) conditions or in the presence of PGE2, the EP2-specific agonist butaprost (Buta), or the EP4-specific agonist ONO-AE1-329 (ONO) for 30 min prior to assaying intracellular cAMP concentration. *, p < 0.01 compared with WT cells treated with PGE2. **, p < 0.05 compared with WT cells treated with butaprost. Data are representative of three independent experiments for WT cells and four independent experiments for pten−/− cells ± S.D. B, increased cAMP expression down-modulates α-SMA and collagen I gene expression by quantitative PCR in pten−/− cells. pten−/− cells were untreated (open bars) or treated with forskolin (closed bars) for 24 h. *p < 0.0001, **p = 0.0005 compared with the untreated conditions. Data are representative of three separate experiments. C, forskolin treatment inhibits α-SMA expression by Western blotting in pten−/− cells. pten−/− cells were untreated (SF) or treated with forskolin for 24 h. The membrane was reprobed for β-tubulin as a loading control. The blot is representative of data from three separate experiments. D, specific EP4 stimulation decreases basal and TGF-β-induced α-SMA expression by Western blotting in pten−/− cells. pten−/− cells were treated in the presence or absence of TGF-β ± ONO-AE1-329 for 24 h. The membrane was reprobed for GAPDH as a loading control. The blot is representative of three separate experiments. The graph shows densitometric ratios of α-SMA to GAPDH pooled from three independent experiments. *, p < 0.005.
To ascertain whether stimulating cAMP independent of the PGE2/EP2 axis in pten−/− cells exerted functional effects on α-SMA and collagen I expression, pten−/− cells were treated with forskolin (200 μm) for 24 h. Utilizing quantitative real time PCR, we observed a significant inhibition of gene expression for both parameters of fibroblast activation (Fig. 3B). Consistently, pten−/− cells treated with forskolin also revealed a decrease in α-SMA protein expression (Fig. 3C). Moreover, we also found that direct stimulation of pten−/− cells with the ONO-AE1-329 compound decreased basal and TGF-β-stimulated α-SMA protein expression (Fig. 3D), implying that although EP2 is the preferential receptor under normal conditions, EP4 is capable of mediating PGE2 antifibrotic effects in fibroblasts under conditions where EP2 is down-regulated. In aggregate, these data signify that pten−/− cells possess functional and activatable adenylate cyclase, which, when stimulated directly by forskolin or via EP4, results in intact downstream signaling to suppress the activated phenotype.
EP Receptor Gene Expression Is Influenced by the PI3K/Akt/PTEN Axis
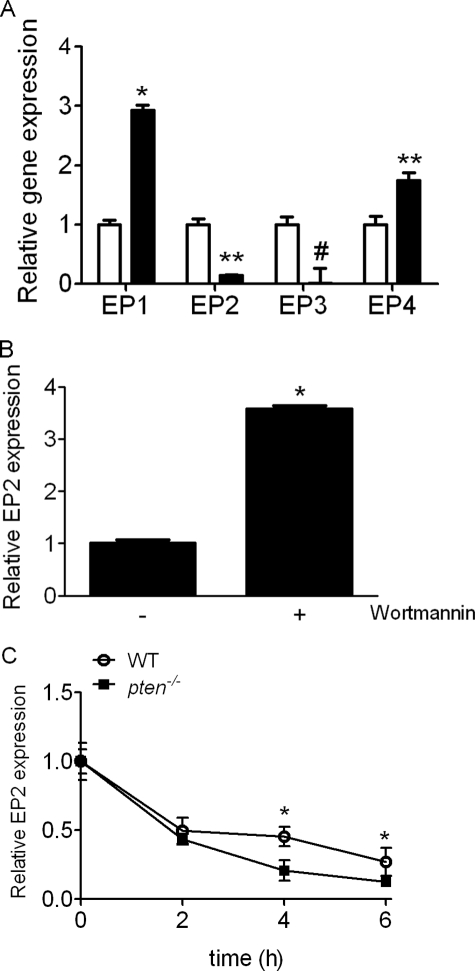
Given that we observed no defect in cAMP generation in pten−/− cells treated with an EP4 agonist, we next questioned whether the defect in EP2 receptor signaling in pten−/− cells was primarily due to differences in receptor gene expression. Indeed, we found a markedly different mRNA expression profile of EP receptors between pten−/− and WT cells, with pten−/− cells demonstrating significantly lower levels of the EP2 gene (p < 0.005) and EP3 gene (p < 0.05) and significantly higher amounts of the EP1 gene (p < 0.0001) and EP4 gene (p = 0.007) than WT cells (Fig. 4A), supporting a novel regulatory effect of PTEN.
FIGURE 4.
A, EP2 receptor gene expression is significantly reduced in pten−/− cells (closed bars) compared with WT cells (open bars) by quantitative PCR. *, p < 0.0001; **, p < 0.005; #, p < 0.05; compared with WT condition. Error bars indicate ±S.D. B, PI3K blockade with wortmannin (50 nm) up-regulates EP2 receptor gene expression in pten−/− cells. Results are representative of triplicate studies. *, p < 0.0001. C, EP2 mRNA stability in actinomycin-treated pten−/− versus WT fibroblasts. Results are pooled from five independently performed experiments. *, p < 0.05.
On pten−/− cells, significantly decreased EP3 expression and significantly augmented EP4 expression would both be expected to increase intracellular cAMP in response to PGE2. However, the reductions in cAMP levels observed in pten−/− cells could only be explained by the down-regulation of EP2 expression and signaling; therefore, we focused further mechanistic study specifically on EP2 receptor gene regulation by PTEN. Concordantly, IMR-90 cells treated with the PTEN inhibitor bpV(pic) had significantly diminished EP2 receptor gene expression compared with untreated cells (Fig. S5), albeit to a lesser degree than that seen in pten−/− cells. That reduced EP2 gene expression in PTEN-deficient/inhibited cells was due in part to unopposed PI3K signaling was evidenced by significant increases in EP2 gene expression in pten−/− cells treated with the PI3K inhibitor wortmannin (50 nm; Fig. 4B).
We next assessed EP2 mRNA turnover in both cell lines in the presence of the transcriptional inhibitor actinomycin D. EP2 mRNA expression declined at similar rates in both WT and pten−/− cells for up to 2 h following actinomycin D treatment. However, pten−/− EP2 gene expression declined at a modestly but statistically significantly accelerated rate through 6 h following actinomycin D (Fig. 4C). In aggregate, these data suggest that reductions in EP2 mRNA expression in pten−/− compared with WT cells is due to a combination of reduced gene transcription and mRNA stability.
Akt Inhibition Restores Functional EP2 Receptor Expression in pten−/− Cells
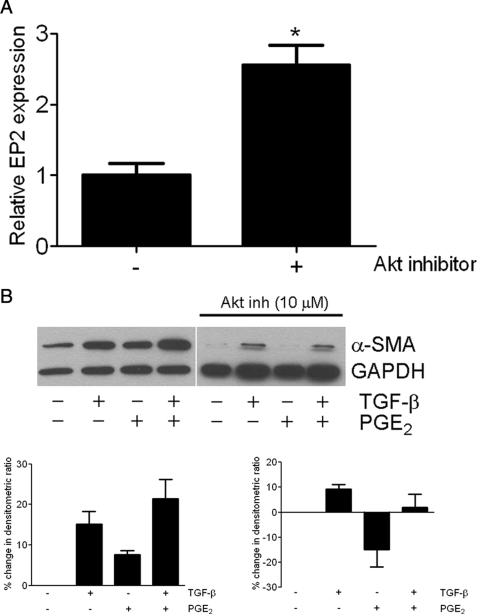
To ascertain whether decreased EP2 expression in pten−/− cells resulted from unchecked Akt activity, pten−/− fibroblasts were treated in the presence or absence of Akt inhibitor X (10 μm) for 24 h. Consistent with our findings with a PI3K inhibitor, Akt inhibition likewise resulted in a significant increase in EP2 expression (Fig. 5A). To determine whether this increase in EP2 was sufficient to confer enhanced PGE2 effects, pten−/− cells, in the presence or absence of Akt inhibitor, were stimulated with both TGF-β and PGE2. Akt inhibition in pten−/− cells indeed restored the ability of PGE2 to inhibit α-SMA expression by Western blotting (Fig. 5B).
FIGURE 5.
A, in the absence of PTEN, Akt inhibition augments EP2 receptor gene expression. pten−/− cells were treated with Akt inhibitor X (10 μm) for 24 h. Results are pooled from four independent experiments. *, p < 0.0001. B, restoration of EP2 by Akt inhibition (Akt inh) results in functional PGE2 signaling. Serum-starved pten−/− cells were stimulated with TGF-β ± PGE2 in the absence or presence of Akt inhibitor. The graph shows the densitometric ratio of α-SMA to GAPDH (used as a loading control) for each condition, expressed as the percentage change compared with the untreated condition. Densitometry data are pooled from four separate blots. Error bars indicate ±S.D.
Positive Feedback between PGE2 and EP2 Receptor Is Regulated by PI3K/Akt Signaling
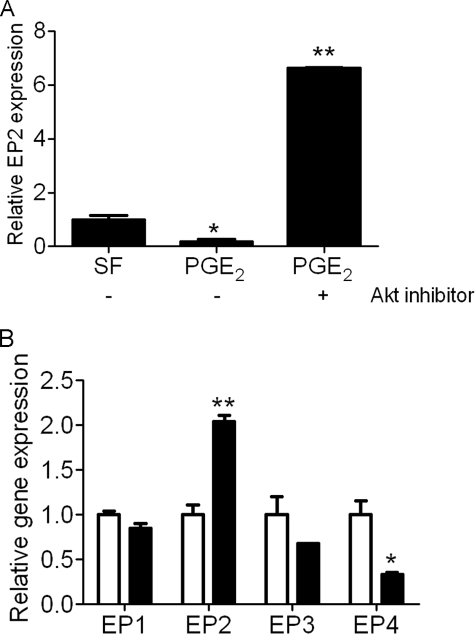
Our previous findings that PGE2 activated PTEN (3) and the current findings that PI3K/Akt blockade enhanced EP2 expression and PGE2 signaling suggest the possible existence of a positive feedback mechanism. We therefore sought to determine whether PGE2/EP2/cAMP signaling promoted further increases in EP2 receptor expression as a possible synergistic mechanism that supports the antifibrotic effects of PGE2. pten−/− cells treated with PGE2 demonstrated a further decrease in EP2 expression (Fig. 6A). However, PGE2 significantly augmented EP2 expression in pten−/− cells in the presence of the Akt inhibitor (Fig. 6A). These data demonstrate the importance of PTEN opposition to Akt in maintaining a normal positive feedback loop between PGE2 and EP2. To confirm the reliance of this feedback loop on cAMP, pten−/− cells were treated with or without forskolin for 24 h; in this setting, we found a significant increase in EP2 receptor gene expression in treated cells compared with untreated cells (Fig. 6B). Moreover, we found that expression of EP1 and EP3 genes was relatively unchanged in pten−/− cells in the presence of forskolin, whereas forskolin treatment resulted in a significant decrease in EP4 expression (Fig. 6B).
FIGURE 6.
A, PGE2-EP2 mRNA positive feedback loop is regulated by the PI3K/Akt/PTEN axis. pten−/− cells were untreated (SF) or treated with PGE2 for 24 h in the absence or presence of Akt inhibitor. *, p < 0.0001 compared with the SF condition. B, forskolin effects on EP receptor gene expression in pten−/− cells. Cells were untreated (open bars) or treated with forskolin (closed bars) for 24 h, and receptor gene expression was monitored by quantitative PCR. *, p < 0.05; **, p < 0.005 compared with the untreated condition. Error bars indicate ±S.D.
DISCUSSION
In this article, we demonstrate that PTEN acts as novel regulator of EP2 gene expression and functional receptor activity. Normally, PGE2 is capable of preventing α-SMA and collagen I induction in TGF-β-stimulated fibroblasts via intracellular increases in cAMP elicited by ligation of EP2 (6). We now show that unopposed PI3K/Akt activity following loss or inhibition of PTEN culminates in down-regulation of EP2 receptor expression, which correlates with fibroblast activation. Somewhat unexpectedly, we also found that pten−/− cells stimulated with PGE2 demonstrate enhanced fibroblast activation, which may be due to signaling via alternative EP receptors, such as EP1. In agreement with this possibility, our data demonstrate a 3-fold increase in EP1 expression in pten−/− cells compared with WT. Coupled with the data that PGE2 possesses almost 2-fold greater affinity for EP1 than EP2 (21), this is a reasonable possibility. Importantly, we demonstrate that circumventing EP2 by direct stimulation of adenylate cyclase-dependent cAMP production in pten−/− cells not only antagonizes the basal enhancement of α-SMA and collagen I expression, but also restores functional EP2 activity. Taken together, our data suggest that the PI3K/Akt/PTEN axis is instrumental in regulating EP2 receptor expression and activity on fibroblasts. Furthermore, our data provide what we believe to be the first description of PTEN as a regulator of GPCR expression.
A relationship between GPCRs and PTEN has been described previously. Sanchez et al. (13) recently showed that the sphingosine 1-phosphate receptor, a GPCR that binds the multifunctional lipid second messenger sphingosine 1-phosphate, activates PTEN and inhibits fibroblast migration. Similarly, we reported that PTEN activity is enhanced in fibroblasts following EP2 receptor ligation due to tyrosine phosphorylation of PTEN (3). In another study, inhibition of the GPCR EP1 enhanced PTEN phosphorylation, although the specific residue(s) were not identified (22). Moreover, PI3K inhibition in this system resulted in abrogation of the neuroprotective effects of EP1 blockade. Thus, it is clear that GPCR signaling modulates PTEN expression, phosphorylation, or activity in various systems. However, our data are the first to demonstrate the directionally reciprocal relationship. Specifically, our data show that PTEN modulates EP2 receptor expression, resulting in alteration of cellular responses. It is noteworthy that pten−/− fibroblasts responded to EP4 receptor stimulation by increasing intracellular cAMP levels to the same degree as WT cells, suggesting that the relationship between PTEN and EP2 appears to be relatively specific.
Our data also demonstrate that direct activation of adenylate cyclase enhanced EP2 receptor expression in pten−/− cells. This represents a second regulatory control on EP2 receptor expression in our system, although the mechanism by which this occurs is as yet unknown. The EP2 promoter possesses a half-site cAMP-response element (23), although a corresponding TATA box (necessary for robust cAMP-driven transcription) (24) is not present. This suggests that cAMP might directly stimulate further EP2 expression. In support of this possibility, Oh et al. (25) have recently reported that cyclooxygenase-2-dependent PGE2 production by C33A cervical carcinoma cells exposed to the human papilloma virus E5 signals in an autocrine fashion, via EP4-mediated cAMP production, to enhance EP4 receptor expression. Further, they also noted an increase in EP2 expression; these observations contrast with our data that EP2 expression is enhanced following forskolin stimulation but that EP4 gene levels are depressed. We speculate that this discrepancy relates to the use of the C33A cell line in that study, which is known to harbor a mutated and hypofunctioning retinoblastoma (pRb) gene that renders the cell deficient in pRb. It is clear that the PI3K/PTEN pathway relies upon functioning pRb protein for cell cycle control in tumor cells (26). Moreover, the retinoblastoma protein GRK3 is known to phosphorylate and desensitize the GPCR CRF1 (27), suggesting a possible regulatory relationship between pRb and GPCRs. Further, cyclooxygenase-2 expression, the rate-limiting step in the generation of PGE2, is substantially up-regulated in retinoblastoma tumors (28) suggesting that the C33A line may be predisposed to enhanced expression of EP2 and EP4.
Our data suggest that PTEN impacts not only EP receptor expression, but also cAMP-regulated effects following EP2 receptor binding by PGE2. Although the mechanism behind this observation is not readily apparent, a number of possibilities exist. First, it is conceivable that PTEN may also regulate the expression of various molecules necessary for the generation of intracellular cAMP, such as the Gαs subunit or adenylate cyclase itself. This seems unlikely given the equivalent production of cAMP in pten−/− cells treated with the EP4 receptor agonist ONO-AE1-129. Alternatively, PTEN may act downstream of cAMP production by regulating the expression of cAMP-dependent signaling pathways controlled via protein kinase A or by exchange proteins activated by cAMP (Epac) or by dephosphorylating downstream effectors of these pathways. Indeed, PTEN possesses both lipid and protein phosphatase activities, although the protein target(s) of PTEN are largely unknown.
PGE2 is a pleiotropic lipid second messenger that induces quite disparate responses depending on the cell tested and the receptor(s) expressed. Indeed, although PGE2 dampens fibrotic responses (6, 29), it also promotes tumorigenesis. For example, PGE2 has been shown to induce, via EP2, vascular endothelial growth factor expression in the prostate cancer cell line PC-3 (30). Interestingly, PC-3 cells are PTEN-null by virtue of biallelic deletion of PTEN (31); hence, these results appear to contradict our observations. However, a second prostate cancer cell line, LNCaP, which is also null for PTEN, was found to express no EP2 receptor (30) and in fact demonstrated substantially higher basal levels of phosphorylated Akt than the PC-3 cell line (32). This observation is in agreement with our own data demonstrating that Akt activity is an important mediator of EP2 receptor regulation and reinforces the importance of PI3K/PTEN balance in regulation of EP2 receptor expression. It also stands to reason that other components of the cellular context seem to influence the nature of the PI3K-EP2 relationship, which may account for the discrepant findings between PC3 and LNCaP cells.
The mechanism by which unopposed PI3K/Akt activity in PTEN-deficient cells results in suppression of EP2 is not yet elucidated. The 5′-untranslated region of the human EP2 gene contains a transcription initiation site located within a cluster of transcriptional regulatory motifs spanning a ∼600-bp sequence from approximately −1600 to −1000 bp (relative to the translation start site), and included in this region are a number of potential activating protein-2 (AP-2) binding sequences (33). Notably, recent evidence suggests that nerve growth factor-induced human umbilical vein endothelial cell matrix metalloproteinase-2 production and Matrigel invasion are mediated via PI3K/Akt-dependent AP-2 activity (34). Similarly, extracellular matrix fibronectin, signaling via α5β1 integrins, activates PI3K/Akt to enhance AP-2 binding to the EP4 promoter (35). Our data are compatible with these studies but suggest that the PI3K/Akt/PTEN axis more broadly regulates GPCR expression than was recognized previously.
In summary, our data show a previously undescribed relationship between PTEN and GPCR expression. Moreover, we show that the PI3K/Akt/PTEN axis specifically regulates EP2, with loss of PTEN activity or unopposed Akt activity resulting in significant suppression of EP2 expression and signaling. Finally, our data suggest that interventions designed to block Akt activity or to directly stimulate cAMP production in fibroblasts restores functional EP2 receptor signaling.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL085083 (to E. S. W.), HL094311 (to M. P.-G.), AI065543 (to B. B. M.), and HL083385 (to C. D. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
- PGE2
- prostaglandin E2
- α-SMA
- α-smooth muscle actin
- bpV(pic)
- bisperoxo(pyridine-2-carboxyl)oxovanadate
- EP
- E prostanoid
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GPCR
- G protein-coupled receptor
- PI3K
- phosphoinositol 3-kinase
- PTEN
- phosphatase and tensin homologue on chromosome 10
- siRNA
- small interfering RNA
- TGF
- transforming growth factor
- WT
- wild type.
REFERENCES
- 1.Buchanan F. G., Wang D., Bargiacchi F., DuBois R. N. (2003)J. Biol. Chem. 278,35451–35457 [DOI] [PubMed] [Google Scholar]
- 2.Sheng H., Shao J., Washington M. K., DuBois R. N. (2001)J. Biol. Chem. 276,18075–18081 [DOI] [PubMed] [Google Scholar]
- 3.White E. S., Atrasz R. G., Dickie E. G., Aronoff D. M., Stambolic V., Mak T. W., Moore B. B., Peters-Golden M. (2005)Am. J. Respir. Cell Mol. Biol. 32,135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohyama T., Ertl R. F., Valenti V., Spurzem J., Kawamoto M., Nakamura Y., Veys T., Allegra L., Romberger D., Rennard S. I. (2001)Am. J. Physiol. Lung Cell Mol. Physiol. 281,L1257–L1263 [DOI] [PubMed] [Google Scholar]
- 5.Narumiya S., Sugimoto Y., Ushikubi F. (1999)Physiol. Rev. 79,1193–1226 [DOI] [PubMed] [Google Scholar]
- 6.Kolodsick J. E., Peters-Golden M., Larios J., Toews G. B., Thannickal V. J., Moore B. B. (2003)Am. J. Respir. Cell Mol. Biol. 29,537–544 [DOI] [PubMed] [Google Scholar]
- 7.Maehama T., Dixon J. E. (1998)J. Biol. Chem. 273,13375–13378 [DOI] [PubMed] [Google Scholar]
- 8.Li D. M., Sun H. (1997)Cancer Res. 57,2124–2129 [PubMed] [Google Scholar]
- 9.Li D. M., Sun H. (1998)Proc. Natl. Acad. Sci. U.S.A. 95,15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng C., Peacocke M. (1998)Nat. Genet. 19,223. [DOI] [PubMed] [Google Scholar]
- 11.White E. S., Atrasz R. G., Hu B., Phan S. H., Stambolic V., Mak T. W., Hogaboam C. M., Flaherty K. R., Martinez F. J., Kontos C. D., Toews G. B. ( 2006) Am. J. Respir. Crit. Care Med. 173, 112– 121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pap T., Franz J. K., Hummel K. M., Jeisy E., Gay R., Gay S. (2000)Arthritis Res. 2,59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez T., Thangada S., Wu M. T., Kontos C. D., Wu D., Wu H., Hla T. (2005)Proc. Natl. Acad. Sci. U.S.A. 102,4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stambolic V., Suzuki A., de la Pompa J. L., Brothers G. M., Mirtsos C., Sasaki T., Ruland J., Penninger J. M., Siderovski D. P., Mak T. W. (1998)Cell 95,29–39 [DOI] [PubMed] [Google Scholar]
- 15.White E. S., Thannickal V. J., Carskadon S. L., Dickie E. G., Livant D. L., Markwart S., Toews G. B., Arenberg D. A. (2003)Am. J. Respir Crit. Care Med. 168,436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hjelmeland A. B., Hjelmeland M. D., Shi Q., Hart J. L., Bigner D. D., Wang X. F., Kontos C. D., Rich J. N. (2005)Cancer Res. 65,11276–11281 [DOI] [PubMed] [Google Scholar]
- 17.Schmid A. C., Byrne R. D., Vilar R., Woscholski R. (2004)FEBS Lett. 566,35–38 [DOI] [PubMed] [Google Scholar]
- 18.Thomas P. E., Peters-Golden M., White E. S., Thannickal V. J., Moore B. B. (2007)Am. J. Physiol. Lung Cell Mol. Physiol. 293,L417–L428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trotman L. C., Niki M., Dotan Z. A., Koutcher J. A., Di Cristofano A., Xiao A., Khoo A. S., Roy-Burman P., Greenberg N. M., Van Dyke T., Cordon-Cardo C., Pandolfi P. P. (2003)PLoS Biol. 1,E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mutoh M., Watanabe K., Kitamura T., Shoji Y., Takahashi M., Kawamori T., Tani K., Kobayashi M., Maruyama T., Kobayashi K., Ohuchida S., Sugimoto Y., Narumiya S., Sugimura T., Wakabayashi K. ( 2002) Cancer Res. 62, 28– 32 [PubMed] [Google Scholar]
- 21.Sugimoto Y., Narumiya S. (2007)J. Biol. Chem. 282,11613–11617 [DOI] [PubMed] [Google Scholar]
- 22.Zhou P., Qian L., Chou T., Iadecola C. (2008)Neurobiol. Dis. 29,543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Odom D. T., Koo S. H., Conkright M. D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J. R., Emerson B., Hogenesch J. B., Unterman T., Young R. A., Montminy M. (2005)Proc. Natl. Acad. Sci. U.S.A. 102,4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conkright M. D., Guzmán E., Flechner L., Su A. I., Hogenesch J. B., Montminy M. (2003)Mol. Cell 11,1101–1108 [DOI] [PubMed] [Google Scholar]
- 25.Oh J. M., Kim S. H., Lee Y. I., Seo M., Kim S. Y., Song Y. S., Kim W. H., Juhnn Y. S. (2009)Carcinogenesis 30,141–149 [DOI] [PubMed] [Google Scholar]
- 26.Paramio J. M., Navarro M., Segrelles C., Gómez-Casero E., Jorcano J. L. (1999)Oncogene 18,7462–7468 [DOI] [PubMed] [Google Scholar]
- 27.Dautzenberg F. M., Wille S., Braun S., Hauger R. L. (2002)Biochem. Biophys. Res. Commun. 298,303–308 [DOI] [PubMed] [Google Scholar]
- 28.Souza Filho J. P., Martins M. C., Correa Z. M., Odashiro A. N., Antecka E., Coutinho A. B., Macedo C. R., Vianna R. N., Burnier M. N., Jr. (2006)Am. J. Ophthalmol. 142,625–631 [DOI] [PubMed] [Google Scholar]
- 29.Wilborn J., Crofford L. J., Burdick M. D., Kunkel S. L., Strieter R. M., Peters-Golden M. (1995)J. Clin. Invest. 95,1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Klein R. D. (2007)Mol. Carcinog. 46,912–923 [DOI] [PubMed] [Google Scholar]
- 31.McMenamin M. E., Soung P., Perera S., Kaplan I., Loda M., Sellers W. R. (1999)Cancer Res. 59,4291–4296 [PubMed] [Google Scholar]
- 32.Chen X., Thakkar H., Tyan F., Gim S., Robinson H., Lee C., Pandey S. K., Nwokorie C., Onwudiwe N., Srivastava R. K. (2001)Oncogene 20,6073–6083 [DOI] [PubMed] [Google Scholar]
- 33.Smock S. L., Pan L. C., Castleberry T. A., Lu B., Mather R. J., Owen T. A. (1999)Gene 237,393–402 [DOI] [PubMed] [Google Scholar]
- 34.Park M. J., Kwak H. J., Lee H. C., Yoo D. H., Park I. C., Kim M. S., Lee S. H., Rhee C. H., Hong S. I. (2007)J. Biol. Chem. 282,30485–30496 [DOI] [PubMed] [Google Scholar]
- 35.Han S., Ritzenthaler J. D., Wingerd B., Rivera H. N., Roman J. (2007)J. Biol. Chem. 282,7961–7972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.