Abstract
Alternative splicing of 3′-terminal exons plays a critical role in gene expression by producing mRNA with distinct 3′-untranslated regions that regulate their fate and their expression. The Xenopus α-tropomyosin pre-mRNA possesses a composite internal/3′-terminal exon (exon 9A9′) that is differentially processed depending on the embryonic tissue. Exon 9A9′ is repressed in non-muscle tissue by the polypyrimidine tract binding protein, whereas it is selected as a 3′-terminal or internal exon in myotomal cells and adult striated muscles, respectively. We report here the identification of an intronic regulatory element, designated the upstream terminal exon enhancer (UTE), that is required for the specific usage of exon 9A9′ as a 3′-terminal exon in the myotome. We demonstrate that polypyrimidine tract binding protein prevents the activity of UTE in non-muscle cells, whereas a subclass of serine/arginine rich (SR) proteins promotes the selection of exon 9A9′ in a UTE-dependent way. Morpholino-targeted blocking of UTE in the embryo strongly reduced the inclusion of exon 9A9′ as a 3′-terminal exon in the endogenous mRNA, demonstrating the function of UTE under physiological circumstances. This strategy allowed us to reveal a splicing pathway that generates a mRNA with no in frame stop codon and whose steady-state level is translation-dependent. This result suggests that a non-stop decay mechanism participates in the strict control of the 3′-end processing of the α-tropomyosin pre-mRNA.
Introduction
Alternative splicing of 3′-terminal exons is a widespread mechanism in metazoans. Global in silico analysis showed that ∼20% of human transcript units contain alternative 3′-terminal exons (1, 2). This process contributes to the complexity of gene expression by not only expanding the proteome through the generation of protein isoforms with distinct carboxyl-terminal domains but also producing mRNAs that differ in their 3′-untranslated region. These sequences are pivotal for determining the behavior of mRNA, since they are known to regulate translation, stability, and localization. The importance of the 3′-untranslated region in mRNA physiology was recently underscored by the identification of microRNAs that control the translation or half-life of mRNAs through interactions with sequences present within the 3′-untranslated region (3). Mechanistically, the regulation of 3′-terminal exon processing is very complex because it requires a coordinated control of the splicing and cleavage/polyadenylation reactions, the latter being tightly interconnected to transcription termination (4). At present, the factors and the mechanisms involved in the coupling between cleavage/polyadenylation and splicing remain largely unknown.
Two classes of alternative 3′-terminal exons can be described based on the organization of the splice sites and the poly(A) site within the exon. The first class comprises exons that are delimited by a 3′ splice site and a poly(A) site. Competition for inclusion between two such exons results in the production of mature mRNA with one of two possible 3′-end termini. This class is described as skipped exons because the proximal exon of the pair is used as a terminal exon or entirely skipped (5). The second class includes exons with 3′ and 5′ splice sites followed closely by a poly(A) site. These exons, designated composite exon (5), can be used as terminal or internal exon, depending on the cellular context. In silico analysis showed that 10% of the transcription units within the human genome possess such composite exons (2). Bioinformatic studies also indicate that these exons are characterized by suboptimal 5′ splice sites and weak poly(A) sites (1, 6). Such an organization suggests that the control of the 5′ splice site or poly(A) site recognition is pivotal in regulating the processing of the composite exon as an internal or terminal exon, respectively.
The alternative processing of the IgM pre-mRNA is the best documented example of a composite exon regulation (reviewed in Ref. 7). In mature B cells (plasma cells), selection of the μs poly(A) site present within the composite Cμ4 exon produces mRNAs encoding the secreted Ig proteins, whereas non-mature B cells activate the use of the 5′ splice site associated with the splicing of downstream exons M1 and M2 generating mRNAs encoding the membrane-bound Ig proteins. It has been shown that the μs poly(A) site plays a key role in regulating the alternative processing of Ig M pre-mRNA (8). The fact that mutated Ig pre-mRNA as well as non-Ig pre-mRNA modified to contain a poly(A) site that competes with a 5′ splice are accurately regulated during B cell maturation (9, 10) strongly indicated that regulatory cis-acting sequences are not required to control the usage of the μs poly(A) site and suggests that the regulation is based on changes in general polyadenylation factors. Accordingly, it was shown that an artificial increase in CstF64 (cleavage stimulatory factor 64) in non-maturated B cells was sufficient to increase the use of the proximal poly(A) site (μs) (11), suggesting that the binding of CstF64 is crucial in the activation of the μs poly(A) site. However, the similar amount of CstF64 observed in established B cell and plasma cell lines that recapitulate the IgM splicing regulation (12) indicates that additional factors are required to regulate the binding of CstF64 during B cell maturation. Furthermore, extensive mutations of sequences surrounding the hexanucleotide AAUAAA of the μs poly(A) site do not impair regulation during B cell differentiation, implying that they are not contributing to the developmentally regulated recognition of the μs poly(A) site. More importantly, these mutation experiments also showed that it was very difficult to inactivate the μs poly(A) site, suggesting that additional regulatory sequences could influence recognition by the basic polyadenylation machinery (13).
We have developed the Xenopus laevis embryo as a true in vivo model to study the basis of tissue-specific splicing regulation (reviewed in Ref. 14). We are using this model and the α-fast tropomyosin (α-TM)4 pre-mRNA to study the regulation of alternative splicing of 3′-terminal exons in vivo. This α-TM pre-mRNA contains a composite exon designated exon 9A9′ that is differentially processed during Xenopus development. In embryonic myotomal cells, specific splicing of exon 9A9′ as a 3′-terminal exon by the selection of the αe poly(A) site present within this exon produces the α7 mRNA that encodes muscle α-TM (15) (see Fig. 1). In embryonic heart and adult striated muscle cells, α-TM is generated by the α2 mRNA isoform, which results from the activation of the internal 5′ splice site associated with the splicing to the downstream terminal exon 9B (16). α2 and α7 mRNAs differ only by their 3′-untranslated region, and the physiological function of this differential processing is at the moment unknown. In non-muscle cells, exon 9A9′ is skipped, and exon 9D is used as a 3′-terminal exon (16). Transient transgenesis of minigenes driven by tissue-specific promoters that recapitulate the specific use of exon 9A9′ in embryonic myotomal cells, and its repression in embryonic epidermal can be used to study the function and interplay of cis sequence elements within the α-TM pre-mRNA (17). Loss or gain of function of trans-acting factors and the consequences on the processing of pre-mRNA derived from minigenes or from the endogenous gene can also be apprehended (17, 18). Using this model, we showed in previous studies that xPTB, the Xenopus orthologue of the mammalian RNA-binding protein PTB1, represses the usage of the composite exon 9A9′ as a terminal exon in embryonic non-muscle cells. This repression requires several high affinity PTB binding sites that are present in the upstream intron between the distant branch site and the 3′ splice site. We also demonstrated that this effect is mediated by repressing both the splicing and cleavage/polyadenylation of exon 9A9′. In this study, we identify an intronic enhancer designated the upstream terminal exon enhancer (UTE) that is required for the specific use of the composite exon 9A9′ as a 3′-terminal exon in myotomal cells. Injection in the Xenopus embryos of antisense morpholino oligonucleotides that target this intronic element demonstrated its physiological relevance in the activation of the composite exon 9A9′ in the myotome and also revealed a splicing pathway producing a mRNA with no in frame stop codon. Targeted translation inhibition of this mRNA increases its steady-state level, suggesting that a non-stop decay mechanism (NSD) participates in the strict control of 3′-end processing of the endogenous α-TM pre-mRNA. We demonstrate in vivo that PTB prevents the activity of the UTE in non-muscle cells, whereas a subclass of SR proteins promotes the usage of exon 9A9′ through UTE.
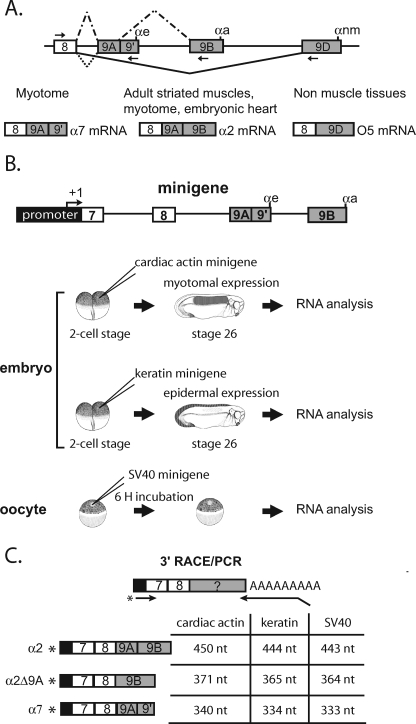
FIGURE 1.
Differential processing of the 3′-end of the Xenopus α-TM pre-mRNA. A, schematic diagram of the 3′-terminal region of the Xenopus α-TM pre-mRNA and of the different tissue-specific RNA processing patterns. Exons and introns are represented by boxes and horizontal lines, respectively. αe, αa, and αnm represent the poly(A) sites present within exons 9A9′, 9B, and 9D, respectively. The arrows represent the forward and reverse primers used to specifically RT-PCR-amplify the different RNA isoforms. The different RNA products in myotome, embryonic heart, adult striated muscle, and non-muscle tissues are indicated. B, diagrammed representation of the α-tropomyosin minigene and of the different in vivo expression assays using the Xenopus embryo and oocyte. C, schematic representation of the 3′-RACE/PCR assay. RNA was reverse transcribed with an oligo(dT) anchor primer. cDNA was further amplified with a reverse anchor primer and a radiolabeled forward primer that hybridize at the junction of the promoter and exon 7. Primers are indicated by arrows, and an asterisk marks the radiolabeling. The structure and size of all splicing products produced by the different minigenes are given.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The α-TM minigenes pBS-SV40.7-9B, pBSActin.7-9B, and pBSKeratin.7-9B have been described previously (17). The mutated minigenes were modified by PCR-mediated site-directed mutagenesis and cassette substitutions as described (17). The different oligonucleotides are given in the supplemental material (supplemental Table S1). For SR protein overexpression in oocytes, the open reading frames of X. laevis ASF/SF2, SC35, SRp20, and 9G8 and human SRp30c, SRp40, and SRp55 were amplified by PCR using specific upstream primers containing a BamHI (xASF/SF2, hSRp30c, and hSRp55) or EcoRV (all of the others) restriction sites and downstream primers containing a NotI restriction site (supplemental Table S1). The clone IMAGE p998E0212316Q3 and the plasmids pCGSRp30c, pcGSRp40, and pcGSRp55 (19) were used as templates for the amplification of xASF/SF2, SRp30c, SRp40, and SRp55, respectively. Xenopus tailbud RNA was used as template for reverse transcription (RT)-PCR amplification of xSRp20 and x9G8. Each amplimer was cloned into pGEM-T easy vector (Promega) and verified by DNA sequencing. The BamHI/NotI or EcoRV/NotI fragments were then cloned into pT7TS-V5 plasmid in frame with the carboxyl V5 epitope tag (17).
Xenopus Embryo Culture and Injection
X. laevis embryos were obtained by artificial fertilization of eggs from laboratory-reared females. Injections were carried out as described previously (17). For minigene injection, 250 pg of supercoiled DNA were injected in one blastomere at the two-cell stage, and the embryos were cultured until stage 26 according to the table of Nieuwkoop and Faber (20). Morpholino oligonucleotides (Mo) obtained from Gene Tools, LLC were injected into both blastomeres of two-cell stage Xenopus embryos. For the translational inhibition of xPTB, xSC35, and muscle α-TM, 20 ng/blastomere xPTB Mo (17), xSC35 Mo (5′-GAGGCCGACCGTAGCTCATGGAATC-3′), and TM Mo (5′-TTCTTGATGGCGTCCATGGCTGCTG-3′), respectively, were injected. To target UTE, 10 ng/blastomere UTE-1 Mo (5′-GATGAGGAATCAGACAGCGTGGAAG-3′) and UTE-2 Mo (5′-CATTGGCAAGAAACCACCATCCAGG-3′) were injected. A control Mo (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was used in all experiments.
Oocyte Culture and Injection
Injection and culture of oocytes were carried out as described previously (17). 1 ng of pBSV40.7-9B or pSV40.7-9BΔUTE minigenes was co-injected into the nucleus with different amounts of 150PY competitor RNA (17). For SR protein overexpression, different amounts of in vitro transcribed capped mRNA encoding V5-tagged SR proteins were injected in the cytoplasm 12 h before injection of the wild-type or ΔUTE minigenes into the nucleus.
In Vitro Transcription
150PY competitor RNA was transcribed from pGEM vector, linearized at a SalI site, using mMessage mMachine T7 kit (Ambion). The different recombinant V5-tagged SR proteins-capped mRNA were transcribed from pT7TS-V5 vector linearized at BamHI or SmaI, using the mMessage mMachine T7 kit (Ambion).
Antibodies and Western Blot Analysis
Western blot analysis was carried out as described previously (17). Muscle and non-muscle α TM were visualized by the Odyssey infrared imaging system (Li-COR Biosciences) using as primary antibodies the monoclonal anti-tropomyosin antibody clone TM311 (catalog number T2780, Sigma) and the anti-tropomyosin antibody (catalog number T3651, Sigma) diluted 1:500 and 1:200, respectively. xSC35, xPTB, and V5-tagged SR proteins were detected with the enhanced chemifluorescence technique (Amersham Biosciences) using xPTB antibodies (1:600) (17) and monoclonal V5 antibodies (Invitrogen).
Isolation of RNA, Analysis by RT-PCR, and RNase Protection Assay
RNA was isolated from embryos or oocytes as described previously (18). RNAs derived from minigenes were analyzed by a 3′-RACE/PCR assay (17). Briefly, 5 μg of RNA was used for the RT reaction using an oligo(dT) anchor as a primer, and one-sixth of the RT reaction was then used for the PCR amplification that was performed with 32P-end-labeled forward primers specific for each minigene (pBSSV40, pBSActin, and pBSKeratin) and a reverse PCR anchor primer. Endogenous transcripts were detected after RT using random primers and SuperScript II RT (Invitrogen) according to the manufacturer's instructions. One-seventh of the RT reaction was then used for the PCR amplification of the different α-TM transcripts using the 32P-labeled forward primer derived from exon 8 of the α-TM gene (5′-GAGTTTGCAGAGAGGACAGTA-3′) and reverse primers that specifically hybridized to exon 9′ (5′-TGGAAAGGGTACGGAGGTAAGC-3′), 9B (5′-AGGTTCCCAAGAAGAGACTG-3′), and 9D (5′-CGGAATTCCTGGCACTCAAGAGCAAG-3′). PCR amplification of the EF1-α transcript was done with a 32P-labeled forward primer (5′-GAGAGGGAAGCTGCTGAGATGG-3′) and a reverse primer (5′-CCACAGGGAGATGTCAATGGTA-3′). Following an initial denaturation at 94 °C for 3 min, DNA was amplified by 22 cycles (94 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s). Amplimers were separated on 4% acrylamide gel and quantified by PhosphorImager analysis (Amersham Biosciences). The riboprobes used for the detection of endogenous α-TM, EF1-α, and MLC1f transcripts and analyzing α-TM cleaved and uncleaved transcripts were described previously (15, 18). The riboprobe used to follow the splicing of exon 9A was PCR-amplified from the pSV40.7–9B construct using the forward (5′-CGGAATTCATTGAAGTCATGTCATTGAG-3′) and reverse (5′-CGGGATCCTCCTCTTTTGTGCCACC-3′) primers. The RNA probe for analyzing splicing and cleavage of exon 9A9′ was PCR-amplified using the forward (5′-CGGGATCCTACCTTGCCTTCCCTACTC-3′) and reverse (5′-CGGGATCCTCCTCTTTTGTGCCACC-3′) primers. The PCR products were digested with BamHI and EcoRI and cloned into pBS vector. The resulting plasmids were extensively digested with XbaI to assure a complete linearization. Riboprobes were prepared from 1 μg of linearized DNA templates in the presence of 20 μCi of [α-32P]UTP (800 Ci/mmol; PerkinElmer Life Sciences) and 9 units of T7 or T3 RNA polymerase (Promega). After an incubation of 30 min at 37 °C, the DNA templates were digested by the addition of 2 units of turbo-DNase (Ambion) and a further incubation for 20 min at 37 °C. The riboprobes were phenol/Sevag-treated, precipitated, and purified onto a 8% denaturing polyacrylamide/urea gel. The RNase protection assays were performed using 10 μg of total RNA and 5 × 104 cpm of probe per hybridization reaction as described previously (15). For each probe, 10 μg of yeast tRNA was used as background control. Single-strand RNAs were digested at 37 °C for 1 h with 30 μg/ml RNase A. The protected products were sized on a 5% polyacrylamide/urea gel and quantified with a PhosphorImager (GE Healthcare).
RESULTS
UTE, an Intronic Regulatory Element Required for the Splicing of Exon 9A9′ as a 3′-Terminal Exon
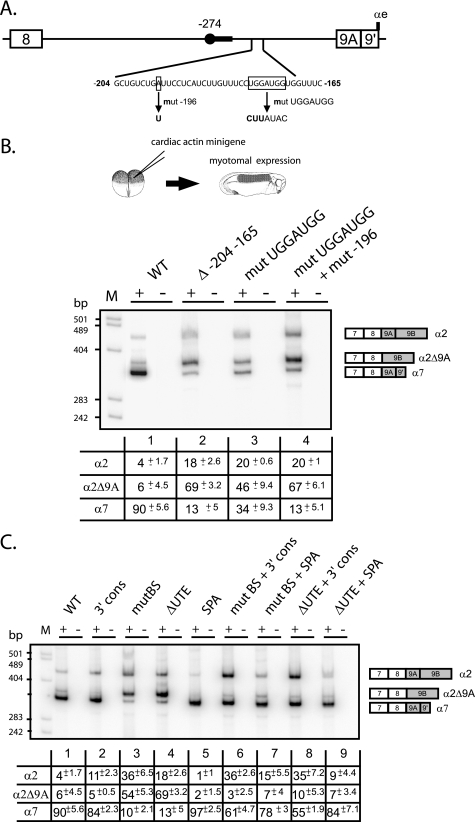
We previously demonstrated that the major branch point used in the myotome to splice exon 9A9′ as a 3′-terminal exon is localized 274 nucleotides upstream of the AG dinucleotide. Mutation of this branch site (mutBS; UUCUAAU to UUCUAUU) or of the associated 21-nucleotide-long pyrimidine tract resulted in a strong skipping of exon 9A9′ in a minigene-derived transcript expressed in the myotome (17). Several alternatively spliced exons associated with distal branch sites have been described, and in all cases, regulatory elements present between the branch site and the AG dinucleotide have been identified. Accordingly, we showed that several high affinity PTB binding sites distributed between the branch site and the dinucleotide AG were involved in PTB-mediated repression of exon 9A9′ usage in non-muscle cells (17, 18). To determine whether activating sequences were also present in this intronic region, mutant minigenes encompassing exon 7 to exon 9B (Fig. 1B) and with a series of 40 nucleotide deletions spanning the region between the distal branch site and the 3′ AG border were generated. The wild-type and mutant constructs driven by the cardiac actin promoter that targets expression in the embryonic myotomal cells were injected into two-cell stage embryos (Fig. 1B). The embryos were then cultured until tailbud stage (stage 26), and extracted mRNA was assayed by 3′-RACE/PCR as described previously (Fig. 1C) (17). One of the mutants (Δ−204−165) dramatically changed the splicing pattern from exon 9A9′ inclusion to exon 9A9′ skipping, producing an mRNA designated α2Δ9A, (Fig. 2B, compare lanes 1 and 2). No change of the splicing pattern was observed with minigenes driven by a keratin promoter that target expression in non-muscle cells in which exon 9A9′ is already strongly skipped (data not shown). An increase of α2 mRNA was also noticed, indicating that the deleted intronic sequence is required specifically for the splicing of exon 9A9′ as a 3′-terminal exon to produce α7 mRNA but not as an internal exon. Because intronic regulatory elements are often conserved between species, we undertook a comparative analysis of this 40-nucleotide element with the orthologous mammalian and avian intronic sequences upstream of exon 9A. A 7-nucleotide motif (UGGAUGG) located 178 nucleotides upstream of exon 9A in Xenopus was identified (Fig. 2A). This motif is present 94 nucleotides and 153 nucleotides upstream of exon 9A in chickens and humans, respectively. Mutation of this motif (UGGAUGG to CUUAUAC) resulted in a 2.6-fold decrease in terminal exon 9A9′ inclusion (α7 mRNA) relative to the wild-type construct (Fig. 2B, lane 3) and a concomitant increase in internal exon 9A splicing (α2 mRNA). However, the effect on exon 9A9′ inclusion was less marked than for the 40-nucleotide deletion, suggesting that additional motifs present within the deleted region were involved in the activation of exon 9A9′. Indeed, during the construction of the 7-nucleotide motif mutant, one of the clones contained a point mutation fortuitously introduced at position −196 (A→T). This double mutant behaved like the Δ−204−165 mutant (compare lanes 2 and 4). We conclude that an intronic sequence containing at least two motifs is required for the activation of exon 9A9′ as a 3′ terminal exon in muscle cells. This sequence was designated the UTE for upstream terminal exon enhancer.
FIGURE 2.
An intronic sequence upstream of exon 9A9′ contributes to its splicing as a 3′-terminal exon in muscle cells. A, schematic map of the intronic region upstream of exon 9A9′. The distant −274 branch site and its associated polypyrimidine tract are represented by a black circle and a black rectangle, respectively. The intronic sequence from −204 to −165 upstream of exon 9A9′ is shown. The generated mutations are shown below the wild-type sequence. B, the wild-type (WT) and the indicated mutant minigenes under the control of the cardiac actin promoter were injected into Xenopus embryos. The mRNA splicing pattern was assayed by 3′-RACE-PCR using a 32P-oligonucleotide specific to the cardiac actin minigene constructs in the presence (+) or absence (−) of reverse transcriptase. The structure of the amplified fragments is indicated on the right. Quantification was performed on a PhosphorImager to yield the proportion of each mRNA. The percentage of each mRNA is given in the table below a representative experiment as mean ± S.D. (n = 20 for the wild type construct and n = 3 for the mutants). C, the distant −274 branch site and UTE are functionally coupled. The Δ−204−165 mutation (ΔUTE) and the mutation of the distant nucleotide −274 branch site (mutBS) were associated with the 3′ cons mutant in which seven consecutive pyrimidine near the dinucleotide AG were introduced or to the SPA mutant in which a strong synthetic poly(A) site was substituted to the αe poly(A) site. The wild type and the indicated mutant minigenes under the control of the cardiac actin promoter were injected into Xenopus embryos. Samples were processed as for B. The percentage of each mRNA is given in the table below a representative experiment as mean ± S.D. (n = 20 for the wild-type construct, and n = 3 for the mutants).
We previously showed that exon 9A9′ is defined by a suboptimal 3′ splice site and a weak polyadenylation site. Accordingly, Hamon et al. (17) showed that the reinforcement of the 3′ splice site by the introduction of seven consecutive pyrimidines near the AG dinucleotide (mutant 3′ cons) or the substitution of the weak αe poly(A) site by a strong synthetic poly(A) site (mutant SPA) resulted in the constitutive usage of exon 9A9′ as a 3′-terminal exon in non-muscle cells. To study whether these mutations activate exon 9A9′ splicing through UTE and the distal nucleotide −274 branch site, several additional mutants were constructed in which the 3′ cons and SPA mutations were combined with the ΔUTE or branch site (mutBS) mutations. The different mutants were placed under the control of the cardiac actin promoter and injected into two-cell stage embryos. As already described by Hamon et al. (17), the single 3′ cons and SPA mutations only weakly changed the splicing pattern (Fig. 2C, compare lanes 2 and 5 with lane 1). A 2.75-fold increase in the production of α2 mRNA was observed with the 3′ cons mutant (lane 2), whereas the amount of α7 mRNA was slightly increased with the SPA mutant. As already described above, the single mutants mutBS and ΔUTE resulted in a strong skipping of exon 9A9′ (lanes 3 and 4). When the 3′ cons mutant was associated with the deletion of UTE or the mutation of the distal branch site, exon 9A9′ was used as a 3′-terminal exon or an internal exon to produce α7 and α2 mRNAs, respectively (lanes 6 and 8). Hence, the reinforcement of the 3′ splice site of exon 9A9′ is sufficient to define and splice this exon independently of UTE and the distal branch site. The association combination of ΔUTE or mutBS mutations with a strong poly(A) site mainly led to the inclusion of exon 9A9′ as a 3′-terminal exon (α7 mRNA) (lanes 7 and 9), demonstrating that the reinforcement of the poly(A) site is sufficient to define and splice exon 9A9′ as a 3′-terminal exon independently of UTE and the distal branch site (compare lanes 3 and 4, 6 and 8, and 7 and 9). Altogether, these results show that UTE is required to define exon 9A9′ as a 3′-terminal exon in the context of the wild-type pre-mRNA. It is also important to notice that the branch site and UTE mutants behave similarly when they are present alone or in combination with the 3′ cons or SPA mutations, strongly suggesting that UTE is required for the efficient usage of the distal branch site.
Blocking Access to UTE in Vivo Strongly Inhibits Endogenous α7 mRNA Production
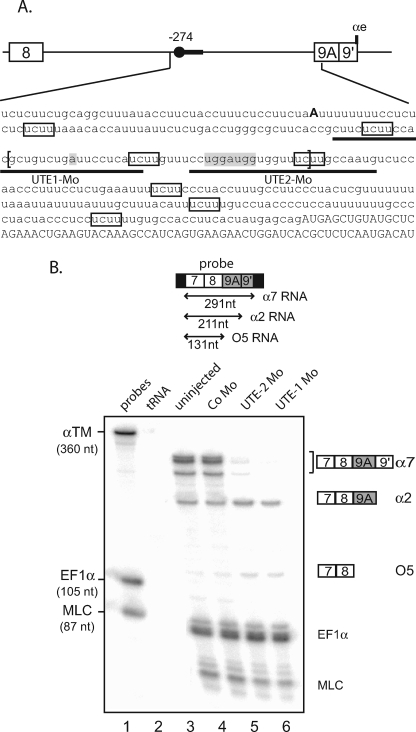
Mutational analysis of minigene reporters is a powerful approach to identify important cis-regulatory sequences. However, it is important to keep in mind that deletion or point mutation can also alter splicing by modifying the structure of the RNA or creating binding sites that change the composition of the RNP. Consequently, we decided to study the functional relevance of UTE by targeting it with antisense morpholino oligomers, the rationale being that such oligomers will block the binding of trans-acting factors that assemble onto UTE. In Xenopus embryo, the α-TM gene is expressed more strongly in the myotome than in the non-muscle tissues (16). Consequently, a whole embryo analysis reflects essentially the myotomal α-TM gene expression pattern. We took advantage of this feature to directly assay the relevance of UTE in the regulation of the endogenous α-TM pre-mRNA. Two contiguous 25-mer antisense morpholino oligonucleotides (UTE-1 Mo and UTE-2 Mo) complementary to UTE were designed (Fig. 3A). 10 ng/blastomere of UTE-1 Mo or UTE-2 Mo were injected into both blastomeres of two-cell stage Xenopus embryos. Embryos developed with normal morphology but presented altered myotomal contractions (data not shown). RNA isolated from tailbud stage embryos was assayed by an RNase protection assay using a probe spanning exons 7, 8, and 9A9′ that can distinguish between the three possible fates of exon 9A9′: use as a 3′-terminal exon (α7 mRNA), as an internal exon (α2 mRNA), and skipping. Two probes directed to EF1-α and MLC1f/3f were included in the assay as controls for RNA recovery and embryo staging (Fig. 3B). As already described, α7 and α2 mRNAs are strongly expressed in Xenopus embryos representing ∼75 and 20% of the α-TM transcripts, respectively, whereas the non-muscle isoform O5 is hardly detectable and accounts for less than 5% of the α-TM mRNA (lane 3). Injection of UTE-1 Mo or UTE-2 Mo resulted in a 35- and 15-fold decrease of α7 mRNA, respectively, whereas a slight increase in the steady-state level of α2 mRNA is observed. A 2–3-fold increase in the partially protected fragment corresponding to RNA in which exon 9A9′ is skipped was also observed. This result confirms the functional relevance of UTE as a myotomal specific enhancer of exon 9A9′ usage as a 3′-terminal exon. It also validates our in vivo minigene strategy to identify cis-regulatory elements.
FIGURE 3.
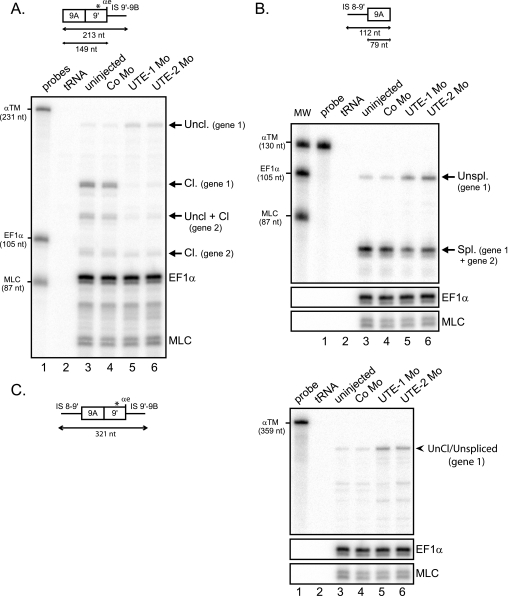
Morpholino-targeted inhibition of the endogenous UTE modifies the 3′-end processing of the α-TM pre-mRNA. A, partial sequence of the α-TM pre-mRNA upstream of exon 9A9′. Exon sequences are indicated in capital letters, and intron sequences are in lowercase letters. The distal −274 branch point is shown in boldface characters, and the high affinity PTB binding sites are framed. UTE is delimited by brackets, and the targeting positions of UTE-1 and UTE-2 Mo are underlined. B, uninjected embryos or embryos injected with control Mo (Co Mo) or UTE-1 Mo or UTE-2 Mo were analyzed by a RNase protection assay using the probe illustrated at the top. Sequences derived from vector are represented by black rectangles. EF1-α and MLC1/3 probes were included in the assay as control for RNA recovery and embryo staging, respectively. The identity or composition of each protected product is indicated on the right. α7 RNA produced two protected fragments because of three nucleotide differences within exon 9A9′ between the two α-tropomyosin genes.
UTE Repression Revealed a Splicing Pathway Producing an mRNA with No Stop Codon in Frame
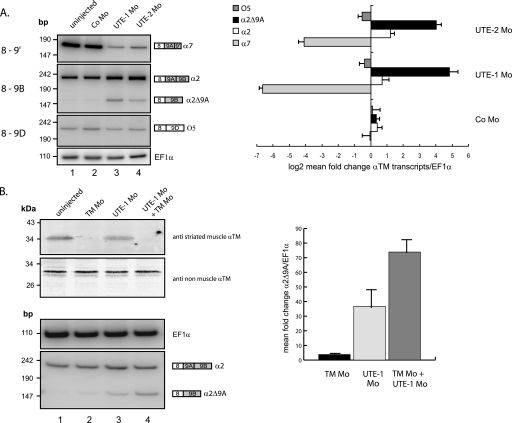
The quantification of the α-TM protected fragments normalized to EF1-α transcripts and uridine content indicated a 4.5-fold reduction of the α-TM transcripts in the UTE-1 Mo-injected embryos (Fig. 3B). Indeed, the decrease in α7 mRNA was not compensated by a symmetric increase in other mRNAs. The partially protected fragment corresponding to mRNA in which exon 9A9′ is skipped could potentially be generated from O5 mRNA but also mRNA in which exon 8 is spliced directly to exon 9B (α2Δ9A mRNA). To address this issue, we performed RT-PCR assays with a forward primer specific for exon 8 and reverse primers specific for exon 9′, 9B, and 9D (see Fig. 1). This analysis confirmed that the amount of α7 mRNA was strongly reduced in Mo-injected embryos (Fig. 4A, top), whereas the levels of α2 mRNA were slightly increased (middle). On the other hand, it showed that the steady-state level of O5 mRNA was not modified (bottom). With the primer specific for exon 9B, an additional PCR product, present in very low amount in uninjected embryos, was increased 43- and 17-fold in UTE-1 and UTE-2 Mo-injected embryos, respectively (middle; compare lanes 3 and 4 with lane 1, and see the bar diagram). Cloning and sequencing of the corresponding fragment showed that it corresponds to α2Δ9A mRNA in which exon 8 is spliced to the terminal exon 9B. This mRNA is characterized by the absence of an in frame stop codon (supplemental Fig. S1), suggesting that it could be a substrate for degradation via NSD, a mechanism that requires the translation of the mRNA (22). Such a feature could explain the overall decrease of α-TM mRNAs in Mo-injected embryos. To determine whether α2Δ9A mRNA is effectively a substrate for NSD, we analyzed its steady-state level when its translation was specifically inhibited (Fig. 4B). A morpholino oligomer (TM Mo) directed against the 5′ region encompassing the initiation codon of muscle α-TM mRNAs (α2, α7, and α2Δ9A mRNAs share the same initiation codon) was coinjected alone or along with UTE-1 Mo in Xenopus embryos. Embryos appeared to be morphologically normal until the tailbud stage, except that they presented neither myotomal contractions nor heart beating (data not shown). As expected, TM Mo resulted in a strong decrease of muscle α-TM protein, whereas the amount of non-muscle α-TM protein is unaffected (compare lanes 1 and 2), demonstrating that TM Mo efficiently inhibits the translation of muscle α-TM. This inhibition of translation was associated with a 3.8-fold increase in α2Δ9A mRNA, indicating that a low amount of α2Δ9A mRNA is physiologically produced during the processing of the α-TM pre-mRNA. UTE-1 Mo also generated a reduction in muscle α-TM protein, which is probably the consequence of the dramatic decrease of α7 mRNA and, as already described, generated a 37-fold increase in α2Δ9A mRNA. Co-injection of UTE-1 Mo and TM Mo resulted in an additional 2-fold stabilization. These results demonstrate that the degradation of α2Δ9A RNA is translation-dependent, suggesting that NSD contributes to the precise control of α-TM pre-mRNA processing.
FIGURE 4.
A, targeting the endogenous UTE generates α2Δ9A mRNA, a potential non-stop decay substrate. Endogenous α7, α2, and 05 RNAs were analyzed by RT-PCR using a 32P-labeled sense primer specific for exon 8 and reverse primers specific for exon 9′, 9B, and 9D, respectively. The PCR products were quantified by PhosphorImager analysis and normalized to EF1-α. The bar diagram shows the log2 mean -fold change ± S.D. of the different α-tropomyosin isoforms in morpholino-injected embryos relative to uninjected embryos (n = 3 independent experiments). B, reduction of the α2Δ9A mRNA is translation-dependent. A morpholino complementary to the region encompassing the start codon of the muscle α-tropomyosin mRNAs (TM Mo) was injected alone or in combination with UTE-1 Mo. The steady-state level of muscle α-tropomyosin was studied by Western blot with the monoclonal anti-tropomyosin antibody (clone TM311). Non-muscle tropomyosin was used as a loading control. Endogenous α2 and α2Δ9A RNAs were analyzed by RT-PCR using primers specific for exon 7 and 9B. The PCR products were quantified by PhosphorImager analysis and normalized to EF1-α. Data were presented as mean -fold change ± S.D. of α2Δ9A RNA in morpholino-injected embryos relative to uninjected embryos (n = 3 independent experiments).
UTE Promotes the Splicing and 3′-End Cleavage at the Proximal αe Poly(A) Site
Our results so far showed that the decrease in the accumulation of α-TM transcripts within the embryo after UTE inactivation is the consequence at least of splicing deregulation leading to the non-productive α2Δ9A mRNA. To study whether additional processes were affected by the inactivation of UTE we next monitored the splicing as well as the 3′-end cleavage of exon 9A9′ at the αe poly(A) site. Cleavage was assayed by RNase protection using a probe that covers exon 9A9′ and intronic sequences downstream of the αe poly(A) site (Fig. 5A, top). Because of three nucleotide differences between the two Xenopus α-TM genes within the sequence of exon 9′, the uncleaved and cleaved RNAs distributed in four protected fragments. As expected from the previous RNase protection assay (see Fig. 3B), UTE-Mo mediated inactivation resulted in a strong decrease in cleaved RNA (Fig. 5A, compare lane 3 with lanes 5 and 6). The amount of cleaved RNA produced from the gene copy that is fully complementary to the probe (gene 1) and normalized to EF1-α was reduced 21- and 13-fold in UTE-1 Mo- and UTE-2 Mo-injected embryos, respectively, which is consistent with the decrease of the α7 mRNA reported above (see Fig. 3B). More importantly, a 2.5–3-fold increase in uncleaved RNA derived from gene 1 was also observed. These results show that UTE is required for an efficient 3′-end cleavage of exon 9A9′. We next determined the consequence of UTE inactivation upon the splicing of exon 9A9′. A probe spanning the 3′ splice site junction of exon 9A was used in an RNase protection assay (Fig. 5B, top). This probe can identify spliced RNA derived from the two α-TM genes, whereas, because of several nucleotide differences within the intronic sequences upstream of exon 9A9′, only unspliced RNA derived from gene 1 can be visualized. UTE-1 Mo and UTE-2 Mo produced a 2-fold decrease in spliced RNA (Fig. 5B, compare lane 3 with lanes 5 and 6). Most of the spliced RNA probably corresponds to the 20% of α-TM transcripts in which exon 9A is spliced as an internal exon (α2 mRNA) (see Fig. 3B). A 3.5–5-fold increase in unspliced RNA was also observed, indicating that UTE is required to splice exon 9A9′ as a 3′-terminal exon.
FIGURE 5.
UTE activates the splicing and cleavage/polyadenylation of exon 9A9′. RNase protection analysis of RNA extracted from uninjected embryos or embryos injected with UTE-1 Mo, UTE-2 Mo, or control morpholino (Co Mo). The RNase protection probes are presented above. A, UTE activates the cleavage/polyadenylation of exon 9A9′. RNase protection was realized with an RNA probe extending beyond the αe poly(A) site, as presented at the top. The three-nucleotide difference within exon 9′ between the two α-TM genes is indicated by a star. The identity of uncleaved (Uncl.) and cleaved (Cl.) protected fragments is indicated on the right. B, UTE activates the splicing of exon 9A9′. RNase protection was realized with an RNA probe that covered the 3′ splice site of exon 9A9′, as presented at the top. The identity of unspliced and spliced fragments is shown on the right. C, unprocessed RNAs accumulate in UTE Mo-injected embryos. RNase protection was realized with a probe spanning the 3′ splice site and extending beyond the cleavage site of exon 9A9′, as shown on the left. The protected fragment corresponding to uncleaved/unspliced (Uncl./Unspl.) molecules is indicated on the right. A–C, the protected fragments were quantified by PhosphorImager analysis and normalized to uridine content and EF1-α. The α-TM, EF1-α, and MLC probes were used as molecular weight markers (MW).
Our results are consistent with a role of UTE in promoting 3′-end cleavage of exon 9A9′ associated, according to exon definition, with the reinforcement of the 3′ splice site that led to the definition of exon 9A9′ as a 3′-terminal exon and splicing. However, our RNase protection assays cannot rule out the possibility that uncleaved and unspliced RNA correspond to different molecules. To determine whether uncleaved and unspliced RNA molecules exist, we performed an RNase protection assay with a probe spanning the 3′ splice junction and the cleavage site of exon 9A9′ (Fig. 5C). Analysis of cleaved and spliced RNA products was again quite complex because of the small differences in the sequences of the two α-TM genes that produced additional partially protected fragments. However, it was possible to determine whether a full-length protected fragment is produced from gene 1, which is fully complementary to the probe. Such a fragment was observed in non-injected embryo, and a 5- and 4-fold increase was quantified in UTE-1 Mo- and UTE-2 Mo-injected embryos, respectively (Fig. 5C, compare lane 3 with lanes 5 and 6).
Altogether, these results are in accordance with a requirement of UTE to define exon 9A9′ as a 3′-terminal exon. In addition, the increase in pre-mRNA in which exon 9A9′ is still present indicates that the splicing of exon 8 to exon 9B in myotomal cells is less efficient than the splicing of exon 8 to exon 9A9′. This rise in unprocessed α-TM transcripts participates in the overall decrease in α-TM mRNAs.
xPTB Prevents the Activity of UTE in Non-muscle Cells
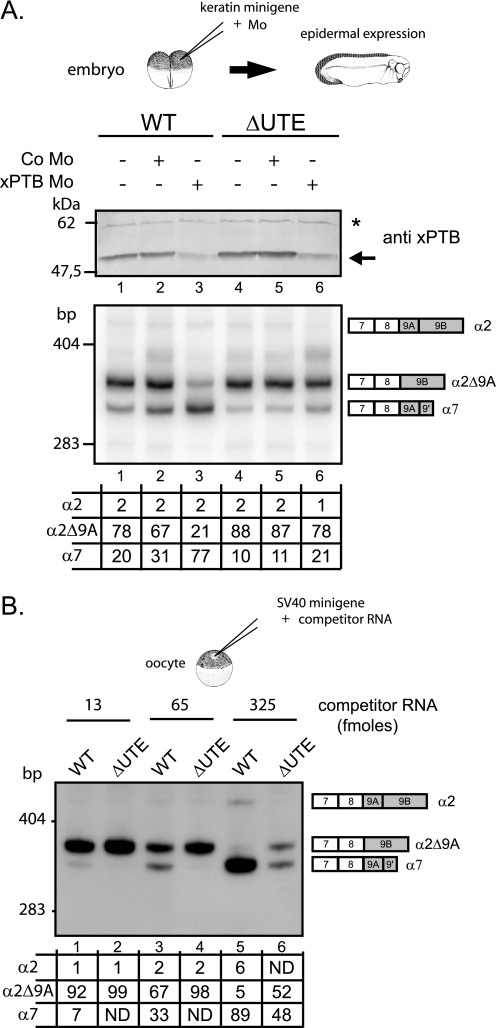
We previously demonstrated that xPTB was a key determinant in repressing exon 9A9′ in non-muscle cells. It was therefore important to study the relationship between xPTB and UTE. For that purpose, we used a strategy described previously (17), combining morpholino-mediated specific knockdown of xPTB and minigene injection into embryos. Morpholino antisense oligomers (xPTB Mo) directed against the 5′ region of xPTB mRNA or control nonspecific morpholino oligomers (Co Mo) were microinjected into both blastomeres of Xenopus embryos at the two-cell stage along with wild-type or ΔUTE mutant minigenes driven by the Xenopus keratin promoter that targets expression to epidermis (Fig. 6A, top). The xPTB Mo specificity was previously demonstrated by rescue experiments with a xPTB mRNA that was not targeted by the morpholino (17). Embryos were fixed at the tailbud stage, and extracted RNA and proteins were analyzed by RT-PCR analysis and Western blot, respectively. As expected, xPTB Mo microinjection resulted in a strong decrease of the amount of endogenous xPTB (Fig. 6A, top, lanes 3 and 6). The knockdown of xPTB resulted, as previously reported, in the derepression of exon 9A9′ in the embryos injected with the wild-type minigene (bottom, lane 3). In contrast, no change in the splicing pattern was observed in the embryos injected with the ΔUTE mutant minigenes (lane 6) demonstrating that xPTB prevents the activity of UTE in epidermis. We previously showed that the repression of exon 9A9′ was similarly controlled in Xenopus oocytes and embryonic epidermal cells and that the xPTB mediated repression can be relieved in oocytes by microinjecting RNA competitors containing four high affinity PTB binding sites (150PY RNAs) (17). As supplemental evidence of the relationship between UTE and PTB, we therefore tested the effects of such competitors upon the splicing pattern of RNA derived from wild type and ΔUTE mutant minigenes. Minigenes driven by the viral SV40 promoter that is functional in Xenopus oocytes were injected into oocyte nuclei along with increasing amount of competitors (Fig. 6B). As previously described, the competitors caused a complete derepression of exon 9A9′ in the context of the wild-type minigene pre-mRNA (compare lanes 1 and 5). However, only a partial derepression was observed with the mutant (compare lanes 2 and 6). These results demonstrate that xPTB represses exon 9A9′ usage in oocytes, in part, by antagonizing UTE activity, as happens in embryonic epidermal cells. Altogether, several important conclusions can be drawn from these experiments: (i) xPTB prevents the activity of UTE in non-muscle cells; (ii) UTE is functional in non-muscle cells when the PTB repression is relieved; (iii) UTE may interact with ubiquitously expressed factors when the amount of xPTB is low.
FIGURE 6.
xPTB prevents the activity of UTE in non muscle cells. A, mutant minigenes deleted for UTE (ΔUTE) and wild-type (WT) minigenes, driven by the keratin promoter, were injected in Xenopus embryo alone (−) or together with control morpholino (Co Mo) or xPTB-Mo. The steady-state level of xPTB was checked by Western blot using a xPTB antiserum. xPTB is indicated by an arrow, and the nonspecific cross-reacting band indicated by a star was used as a loading control. The mRNA splicing pattern was assayed by 3′-RACE-PCR using a 32P-oligonucleotide specific to the keratin minigene constructs. B, the wild-type and ΔUTE minigenes driven by the SV40 promoter were injected into oocyte nuclei alone or with increasing amounts of 150PY competitor RNA. The mRNA splicing pattern was assayed by 3′-RACE-PCR using a 32P-oligonucleotide specific to SV40 minigene constructs. A and B, quantification was performed on a PhosphorImager to yield the proportion of each product given in the table below. ND, not detected.
The SR Protein SC35 Activates Exon 9A9′ Usage through UTE
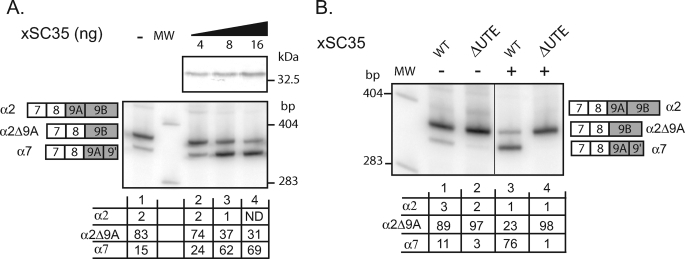
In view of the previous data and the established role of the ubiquitously expressed SR proteins in splicing regulation, we decided to study whether this class of RNA-binding protein might participate in exon 9A9′ activation through UTE. We first used the ESE Finder program (23) to examine the presence of potential SR proteins binding sites within UTE. Using the threshold values considered as being significant within the context of an ESE, ESE Finder predicted several binding sites for SC35, SRp40, and SRp55 (supplemental Fig. S1). The density of predicted binding sites was higher for SC35, and more importantly five and two SC35 binding sites of nine were absent in ΔUTE and mutUTE mutants, respectively. Since UTE is functional in Xenopus oocytes when repression exerted by xPTB is relieved, we decided to test whether SC35 activates the splicing of exon 9A9′ as a 3′-terminal exon in these cells. 4, 8, or 16 ng of in vitro transcribed mRNA encoding xSC35 tagged with a V5 epitope were injected into the cytoplasm of Xenopus oocytes, and 24 h later the wild type minigene was injected into the nucleus. Overexpression of xSC35 was obtained in oocytes, as determined by Western blot with V5 antibody (Fig. 7A, top). As described above, in the absence of xSC35 mRNA injection, processing of the wild-type minigene produced predominantly exon skipping (α2Δ9A mRNA) and a low level of exon 9A9′ usage (α7 mRNA) (Fig. 7A, bottom, lane 1). When xSC35 was overexpressed, exon 9A9′ usage was increased 4.6-fold (compare lane 1 with lane 3). To analyze whether this activation was dependent on UTE, the effects of xSC35 overexpression were also assayed using the ΔUTE minigene (Fig. 7B). Although the wild-type minigene showed a 7-fold activation of exon 9A9′ usage with xSC35 RNA injection (compare lane 1 with lane 3), no activation was observed with the ΔUTE construct. We therefore concluded that xSC35 activates the splicing of exon 9A9′ and that this activation is UTE-dependent. Additionally, the dose-dependent effects of xSC35 overexpression upon exon 9A9′ splicing strongly suggest a direct role.
FIGURE 7.
SC35 activates the usage of exon 9A9′ as a 3′-terminal exon through UTE. A, the wild-type minigene driven by the SV40 early promoter was injected into the nucleus of stage VI oocytes into which in vitro transcribed V5-tagged xSC35 RNA (4, 8, and 16 ng) had been previously injected into cytoplasm. V5-tagged xSC35 expression was monitored by Western blotting using a V5 antibody. B, the wild-type (WT) and ΔUTE mutant minigenes were injected into the nucleus of stage VI oocytes into which in vitro transcribed V5-tagged xSC35 RNA had been previously injected into cytoplasm (+) or not (−). A and B, total RNA was subjected to 3′-RACE-PCR using a 32P-oligonucleotide specific to the SV40 minigene reporter. Quantification was performed on a PhosphorImager to yield the proportion of each product given in the table below. MW, molecular weight markers.
In this study, we demonstrated that the repression activity of xPTB upon exon 9A9′ processing was mediated in part by UTE inactivation. This effect is probably direct, since UTE contains two high affinity PTB binding sites and recombinant PTB cross-linked to UTE (data not shown). Therefore, a possibility is that the use of exon 9A9′ as a 3′-terminal exon is regulated by a mechanism based on antagonistic binding of xSC35 and xPTB to UTE.
A Subclass of SR Proteins May Activate Exon 9A9′ Processing in a UTE-dependent Way
To test the physiological relevance of xSC35 in the activation of exon 9A9′ usage as a 3′-terminal exon in embryonic myotomal cells, we examined the effects of xSC35 depletion upon the processing of the endogenous α-TM pre-mRNA or of the pre-mRNA derived from the wild type minigene whose expression is targeted to myotomal cells. Morpholino-mediated translation inhibition of xSC35 within the embryo failed to modify the splicing pattern of both endogenous and minigene α-TM mRNA (data not shown). We therefore concluded that xSC35 inactivation within the embryo was not sufficient to prevent exon 9A9′ splicing as a 3′-terminal exon.
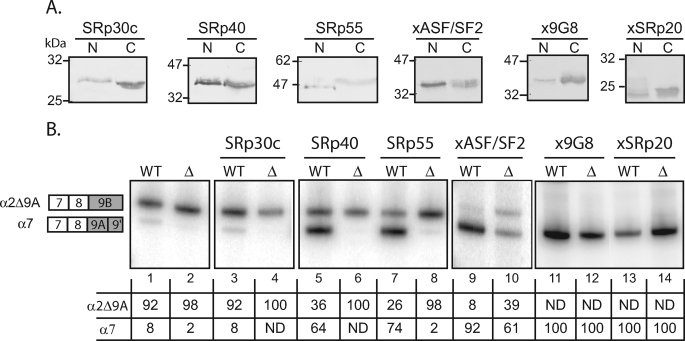
This result was not so surprising because it was reported that individual SR proteins may have redundant function in vivo. To test whether additional SR proteins may activate α7 mRNA production through UTE, we used the Xenopus oocyte model described above. In vitro transcribed mRNA encoding homologous or orthologous SR proteins tagged with a V5 epitope were injected in oocyte cytoplasm, and 24 h later minigenes were injected into the nucleus. The accumulation of the V5-tagged protein in the nucleus and cytoplasm was checked by Western blot (Fig. 8A), and the splicing pattern was assayed by RT-PCR, as already described (Fig. 8B). One of the SR proteins tested, SRp30c, did not activate exon 9A9′ usage (compare lanes 1 and 3), indicating that the switch to α7 mRNA is not the consequence of a general activation of exon 9A9′ splicing by SR proteins. The remaining SR proteins activated exon 9A9′ usage and can be divided into two classes. The first class corresponds to SR protein that activates exon 9A9′ usage through UTE and comprises SRp40 and SRp55 (compare lanes 5 and 6, 7 and 8, and 11 and 12). The second class corresponds to SR protein that activates exon 9A9′ usage independently of USE and comprises xASF/SF2, x9G8, and xSRp20 (compare lanes 9 and 10, 11 and 12, and 13 and 14). These results indicate that a subclass of SR proteins comprising SRp40, SRp55, and xSC35 is able to activate exon 9A9′ usage through UTE, suggesting that an activator complex comprising SR proteins is assembled onto UTE.
FIGURE 8.
A subclass of SR proteins activate exon 9A9′ splicing through UTE. The wild-type (WT) or ΔUTE mutant constructions were injected into the nucleus of stage VI oocyte alone or in presence of in vitro transcribed RNA encoding V5-tagged SR proteins. A, V5-tagged SR protein expression in cytoplasmic and nuclear fractions was monitored by Western blotting using a V5 antibody. B, total RNA was subjected to 3′-RACE-PCR using a 32P-oligonucleotide specific to the SV40 minigene reporter. The structure of the amplified fragments is indicated on the right. Quantification was performed on a PhosphorImager to yield the proportion of each mRNA given in the table below. ND, not detected.
DISCUSSION
PTB in Repression of the Composite Exon 9A9′
We previously showed that PTB was involved in the repression of exon 9A9′ usage as a 3′-terminal exon in non-muscle cells, by inhibiting both the splicing and cleavage/polyadenylation reactions (18). Our present data demonstrate that PTB is acting in part by preventing the activity of the intronic element UTE required to define the weak exon 9A9′ as a 3′-terminal exon. Several high affinity PTB binding sites are present within UTE, and UV-cross-linking demonstrated that PTB binds to this element. This result suggests that PTB may antagonize the binding of activator factors onto UTE, as previously demonstrated for the α-actinin and cardiac troponin pre-mRNA (24, 25). The observation that in the absence of UTE, a 2-fold increase in exon 9A9′ usage was still observed when PTB was down-regulated strongly suggests that in addition to precluding the assembly of an activator complex onto UTE, PTB directly inhibits the 3′ splice site and αe poly(A) site selection by the basal splicing and cleavage/polyadenylation machineries. Since numerous high affinity PTB binding sites are present between the −274 distal branch site and exon 9A9′, it is likely that PTB inhibits the splicing of exon 9A9′ by directly preventing the binding of U2AF65, as described for the mutually exclusive exon 6B of the β-tropomyosin pre-mRNA (21). It is also conceivable that PTB blocks the transition from an exon 9A9′ defined complex to a functional spliceosome, as demonstrated for the regulation of the alternative exon N1 of the c-src pre-mRNA (26). Regarding the cleavage/polyadenylation reaction, it could be envisioned that PTB directly inhibits this process by competing with CstF64 binding to the downstream uridine-rich region of αe poly(A), since it was demonstrated that PTB modulates the efficiency of polyadenylation by such a competition (27). However, the finding that a chimeric reporter β-globin minigene with an αe poly(A) site is not responsive to PTB repression is not in agreement with this hypothesis.5 Moreover, overexpression of CstF64 in Xenopus oocytes failed to activate exon 9A9′ usage, indicating that an increase in CstF64 is not sufficient to relieve the repression exerted on the αe poly(A) site.5 The morpholino-mediated inhibition of UTE in myotomal cells resulted in a strong decrease in exon 9A9′ usage associated with a direct splicing of exon 8 to exon 9B (α2Δ9A) RNA. This result differs from the splicing of exon 8 to exon 9D (O5 RNA) that occurs when PTB is overexpressed in myotomal cells (18). This discrepancy indicates that, in addition to inhibition of exon 9A9′ definition, PTB is either involved in the repression of exon 9B so that splicing of the distal 3′-terminal exon 9D is favored, or usage of exon 9D is activated by PTB.
Our data demonstrate that PTB-mediated repression of the proximal alternative 3′ exon 9A9′ is an important mechanism to ensure its complete skipping during 3′-end processing of the α-TM pre-mRNA. This observation led us to predict that PTB might be a major actor in the repression of proximal 3′-terminal exons in general. Interestingly, the study of the microRNA miR-124 down-regulation of PTB during neuronal differentiation uncovered several pre-mRNAs whose neural specific proximal 3′-terminal exon is repressed in non-neuronal cells by PTB (28). The switch to nPTB during neuronal differentiation alleviates this repression, allowing the use of these proximal 3′-terminal exons. It would be of interest to study whether the activation of the proximal exon is dependent on intronic regulatory sequences and whether PTB prevents the assembly of an activating complex onto these sequences.
UTE Function in the Activation of a 3′-Terminal Exon
The composite internal/3′-terminal exon 9A9′ is defined by suboptimal 3′ and 5′ splice sites and a weak poly(A) site (17). Informatic studies showed that such suboptimal signals are a general feature of this class of 3′-terminal exon, and it was proposed that it allows a dynamic competition between splicing and polyadenylation (1). The binding of CstF64 to the downstream uridine-rich element of a poly(A) site is a direct way to regulate a poly(A) site usage. For example, in the Drosophila female germ line, the RNA-binding protein SXL (sex-lethal) competes with CstF64 for binding to the uridine-rich element of the enhancer of rudimentary pre-mRNA and represses polyadenylation at the proximal poly(A) site (29). CstF64 binding is also pivotal in the choice of the proximal μs poly(A) site of the immunoglobulin pre-mRNA during B cell differentiation (11). However, the observation that the amounts of CstF64 are similar in established B cell and plasma cells lines, which recapitulate this splicing regulation (12), suggested that additional factor and auxiliary cis-sequences participate to the regulation of CstF64 binding (13). The intronic regulatory element (UTE) that we identified upstream of exon 9A9′ defines a new class of intronic splicing enhancer (ISE) that is specifically involved in the splicing activation of a composite exon as a 3′-terminal exon. The molecular mechanisms by which this element activates the splicing of exon 9A9′ as a terminal exon remain to be determined. This task is arduous using in vivo approaches because of the functional coupling between the cleavage/polyadenylation and splicing reactions (30) and the exon definition, in which activation of the 3′ splice site favors the recognition of the poly(A) site and the converse (31). In vitro systems should be appropriate to study both reactions independently; however, all approaches to develop such assays were unsuccessful,6 suggesting that coupling of both reactions is required.
Our present data, showing that UTE and the nucleotide −274 distal branch site are functionally linked, suggest a model in which UTE promotes the binding of U2snRNP to the distal branch site, which in turn might stabilize the binding of CPSF by a direct interaction (32). It is also possible that UTE controls the binding of U2AF65 to the polypyrimidine tract, which in turn recruits the cleavage factors CFIm through their respective RS domains (33). However, the fact that the mutation of UTE or its inhibition in vivo results in a weak but reproducible increase in the splicing of exon 9A as an internal exon strongly suggests that its primary function is to promote the assembly of an activating complex that increases the recognition of the weak αe poly(A) site. Then, in accordance with exon definition, this complex may stabilize proteins involved in the recognition of the 3′ splice site, such as U2AF65 and U2AF35, leading to the definition of exon 9A9′ as a 3′-terminal exon. Overexpression studies in the oocyte demonstrated that a subclass of SR proteins activate exon 9A9′ usage through UTE. Only a few studies have reported a relationship between SR proteins and the cleavage/polyadenylation machinery. It was shown that SRp20 participates in the definition of the proximal terminal exon of the CT/CGRP pre-mRNA through binding to a downstream intronic enhancer (34). More recently, it was shown that the binding of SR proteins to the negative regulator of splicing of the Rous sarcoma virus stimulates polyadenylation (35). Interestingly, this effect was dependent on an additional site that brought the negative regulator of splicing and the poly(A) site closer together, indicating that the distance between the bound SR protein and the poly(A) site is important. The authors anticipated that SR protein binding sites present upstream of suboptimal or alternative poly(A) sites might regulate their recognition. This is in accordance with the position of UTE within the α-TM pre-mRNA because it is only 300 nucleotides upstream of the αe poly(A) site. At present, it is not known how SR proteins might activate the cleavage/polyadenylation processes. It is known that SR protein can interact with U1snRNP (36), which in turn associates with the cleavage factor CFI (37). The SRm160 splicing coactivator is also a good candidate to mediate SR protein activity upon the cleavage/polyadenylation process. This factor that promotes transcript 3′-end cleavage (38) physically interacts with SR family proteins in vitro through their respective RS domains. Moreover, functional inactivation of SRm160 and individual SR protein in Caenorhabditis elegans resulted in specific phenotypes demonstrating functional interactions between these factors (39). To our knowledge, UTE is the first intronic enhancer present in the intron upstream of a regulated composite exon. This element defines, therefore, a new class of intronic enhancers, and it will be interesting to study whether such elements are present upstream of other regulated composite exons.
NSD and Alternative Splicing of 3′-Terminal Exons
In this study, the morpholino-mediated inhibition of UTE revealed that an endogenous α-TM mRNA (α2Δ9A) corresponding to the direct splicing of exon 8 to exon 9B was produced at a low level in Xenopus embryos. This result was not surprising because the generation of α2Δ9A RNA from α-TM minigenes showed that there is no intrinsic blockage of the splicing of both exons. The resulting endogenous mRNA does not possess an in-frame stop codon, making it a potential substrate for NSD. Stabilization of this mRNA after translation inhibition is in agreement with such a mechanism. There appears therefore to be no strict mechanism to preclude splicing of exon 8 to exon 9B; therefore, NSD participates in the strict control of the 3′-end processing of the endogenous α-TM pre-mRNA by degrading a non-productive mRNA species. NSD was characterized in yeast (40) and demonstrated to be present in mammalian cells (22). It was proposed that the use of 3′-end processing signals present in the coding region may be the basis of NSD selection. Our data add a new potential role for NSD, that of monitoring the accuracy of alternative 3′-end processing. Our data also showed that such a mechanism is active within the tissue of a living organism and contributes to the regulation of a complex gene regulation process. It is interesting to note that this process is not conserved through vertebrate phylogeny because in rats, splicing of exon 8 to exon 9B produces an mRNA that encodes a tropomyosin isoform (TMBr2) specifically expressed in the brain (41). This observation is in agreement with the fact that distinct alternative mRNA splicing participates in the speciation of organisms. In silico analyses showed that 20% of the composite exons do not contain stop codons in frame with upstream exons (2), indicating that these mRNAs are potential NSD substrates. Hence, NSD may participate in gene expression regulation in the same way as NMD (42) by down-regulating genes in specific tissues through the splicing of terminal exon devoid of in-frame stop codon.
Studying in Vivo the Mechanisms of Alternative Splicing and Polyadenylation
Since the pioneering work of Kole (43) showing that antisense oligonucleotides targeting the 3′ or 5′ splice sites can modify the splicing pathway of alternative exons, antisense oligonucleotides have become a promising therapeutic tool to redirect the splicing of aberrant mRNA by targeting splices sites and cis-regulatory elements present within exons (44). Such a strategy based on the skipping of the defective exons within the dystrophin pre-mRNA is currently in development as a treatment for Duschene muscular dystrophy (44). The feasibility of modulating the splicing pattern of the fibroblast growth factor receptor 1 by targeting intronic regulatory elements was also reported (45). The present data demonstrate for the first time that the splicing of a 3′-terminal exon can also be redirected by antisense oligonucleotides targeted at a cis-regulatory element. In addition to the potential of modifying splicing, antisense oligonucleotides might also be a powerful tool to study the complex interplay of regulatory sequences within an endogenous pre-mRNA in vivo. Indeed, simultaneously targeting regulatory elements upstream and downstream of the α-exon of the fibroblast growth factor receptor 1 demonstrated that these sequences act through a shared mechanism (45). In order to understand how UTE activates the usage of exon 9A9′, simultaneous injection of antisense oligonucleotides targeting UTE and the 3′ splice site or the αe poly(A) site of exon 9A9′ may provide useful information on the mechanisms involved and on their relationship with the basal splicing and cleavage/polyadenylation machineries. This approach, associated with a morpholino-mediated knockdown of the 5′ → 3′ exonuclease Xrn2 or the core component EXOSC9 of the exosome, should give access to intermediate RNA species. It would also be possible to identify additional cis-regulatory elements present within the endogenous α-tropomyosin pre-mRNA, using a systematic antisense oligonucleotide tiling strategy similar to the one successfully used to identify intronic cis-regulatory elements controlling the splicing of SMN2 exon 7 (46). The Xenopus embryo model is fully adapted for a true in vivo approach and should give new insights into the difficult task of deciphering the mechanisms that control the differential processing of 3′-terminal exons.
Supplementary Material
Acknowledgments
We thank Joelle Marie for the recombinant PTB protein and Adrian Krainer for the plasmids pCGSRp30c, pCGSRp40, and pCGSRp55. We thank Beverley Osborne for critically reading the manuscript, James L. Pearson for English correction of the manuscript and useful comments, and all of the members of the laboratory for helpful comments and suggestions.
This work was supported by grants from the Association Française contre les Myopathies (to S. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
C. Le Sommer and S. Hardy, unpublished data.
A. Méreau, unpublished data.
- α-TM
- α-fast tropomyosin
- UTE
- upstream terminal exon enhancer
- NSD
- non-stop decay mechanism
- RT
- reverse transcription
- Mo
- morpholino oligonucleotide(s)
- RACE
- rapid amplification of cDNA ends
- PTB
- polypyrimidine tract binding protein
- SR
- serine/arginine-rich.
REFERENCES
- 1.Tian B., Pan Z., Lee J. Y. (2007) Genome Res. 17, 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan J., Marr T. G. (2005) Genome Res. 15, 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proudfoot N. J., Furger A., Dye M. J. (2002) Cell 108, 501–512 [DOI] [PubMed] [Google Scholar]
- 5.Edwalds-Gilbert G., Veraldi K. L., Milcarek C. (1997) Nucleic Acids Res. 25, 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian B., Hu J., Zhang H., Lutz C. S. (2005) Nucleic Acids Res. 33, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson M. L. (2007) Immunol. Res. 37, 33–46 [DOI] [PubMed] [Google Scholar]
- 8.Peterson M. L., Gimmi E. R., Perry R. P. (1991) Mol. Cell. Biol. 11, 2324–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson M. L. (1994) Mol. Cell. Biol. 14, 7891–7898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seipelt R. L., Spear B. T., Snow E. C., Peterson M. L. (1998) Mol. Cell. Biol. 18, 1042–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takagaki Y., Seipelt R. L., Peterson M. L., Manley J. L. (1996) Cell 87, 941–952 [DOI] [PubMed] [Google Scholar]
- 12.Edwalds-Gilbert G., Milcarek C. (1995) Mol. Cell. Biol. 15, 6420–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson M. L., Bingham G. L., Cowan C. (2006) Mol. Cell. Biol. 26, 6762–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mereau A., Le Sommer C., Lerivray H., Lesimple M., Hardy S. (2007) Biol. Cell 99, 55–65 [DOI] [PubMed] [Google Scholar]
- 15.Hardy S., Hamon S., Cooper B., Mohun T., Thiébaud P. (1999) Mech. Dev. 87, 199–202 [DOI] [PubMed] [Google Scholar]
- 16.Hardy S., Fiszman M. Y., Osborne H. B., Thiebaud P. (1991) Eur. J. Biochem. 202, 431–440 [DOI] [PubMed] [Google Scholar]
- 17.Hamon S., Le Sommer C., Mereau A., Allo M. R., Hardy S. (2004) J. Biol. Chem. 279, 22166–22175 [DOI] [PubMed] [Google Scholar]
- 18.Le Sommer C., Lesimple M., Mereau A., Menoret S., Allo M. R., Hardy S. (2005) Mol. Cell. Biol. 25, 9595–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Screaton G. R., Caceres J. F., Mayeda A., Bell M. V., Plebanski M., Jackson D. G., Bell J. I., Krainer A. R. (1995) EMBO J. 14, 4336–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieuwkoop P., Faber J. (ds) (1956) Normal Table of Xenopus Laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilzed Egg till the End of Metamorphosis, North Holland Publishing Co., Amsterdam, The Netherlands [Google Scholar]
- 21.Sauliere J., Sureau A., Expert-Bezancon A., Marie J. (2006) Mol. Cell. Biol. 26, 8755–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frischmeyer P. A., van Hoof A., O'Donnell K., Guerrerio A. L., Parker R., Dietz H. C. (2002) Science 295, 2258–2261 [DOI] [PubMed] [Google Scholar]
- 23.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R. (2003) Nucleic Acids Res. 31, 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlet B. N., Logan P., Singh G., Cooper T. A. (2002) Mol. Cell 9, 649–658 [DOI] [PubMed] [Google Scholar]
- 25.Gromak N., Matlin A. J., Cooper T. A., Smith C. W. (2003) RNA 9, 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S., Kohlstaedt L. A., Damianov A., Rio D. C., Black D. L. (2008) Nat. Struct. Mol. Biol. 15, 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castelo-Branco P., Furger A., Wollerton M., Smith C., Moreira A., Proudfoot N. (2004) Mol. Cell. Biol. 24, 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007) Mol. Cell 27, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gawande B., Robida M. D., Rahn A., Singh R. (2006) EMBO J. 25, 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nesic D., Maquat L. E. (1994) Genes Dev. 8, 363–375 [DOI] [PubMed] [Google Scholar]
- 31.Niwa M., Berget S. M. (1991) Genes Dev. 5, 2086–2095 [DOI] [PubMed] [Google Scholar]
- 32.Kyburz A., Friedlein A., Langen H., Keller W. (2006) Mol. Cell 23, 195–205 [DOI] [PubMed] [Google Scholar]
- 33.Millevoi S., Loulergue C., Dettwiler S., Karaa S. Z., Keller W., Antoniou M., Vagner S. (2006) EMBO J. 25, 4854–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lou H., Neugebauer K. M., Gagel R. F., Berget S. M. (1998) Mol. Cell. Biol. 18, 4977–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciolek N. L., McNally M. T. (2007) J. Virol. 81, 11208–11217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Luhrmann R., Garcia-Blanco M. A., Manley J. L. (1994) Nature 368, 119–124 [DOI] [PubMed] [Google Scholar]
- 37.Awasthi S., Alwine J. C. (2003) RNA 9, 1400–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCracken S., Lambermon M., Blencowe B. J. (2002) Mol. Cell. Biol. 22, 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longman D., McGarvey T., McCracken S., Johnstone I. L., Blencowe B. J., Caceres J. F. (2001) Curr. Biol. 11, 1923–1933 [DOI] [PubMed] [Google Scholar]
- 40.van Hoof A., Frischmeyer P. A., Dietz H. C., Parker R. (2002) Science 295, 2262–2264 [DOI] [PubMed] [Google Scholar]
- 41.Lees-Miller J. P., Goodwin L. O., Helfman D. M. (1990) Mol. Cell. Biol. 10, 1729–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGlincy N. J., Smith C. W. (2008) Trends Biochem. Sci. 33, 385–393 [DOI] [PubMed] [Google Scholar]
- 43.Kole R. (1998) in Applied Antisense Oligonucleotides Technology (Stein C. A., Krieg A. M. eds) pp. 451– 469, Wiley-Liss, Inc., New York [Google Scholar]
- 44.Sazani P., Kole R. (2003) J. Clin. Invest. 112, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bruno I. G., Jin W., Cote G. J. (2004) Hum. Mol. Genet. 13, 2409–2420 [DOI] [PubMed] [Google Scholar]
- 46.Hua Y., Vickers T. A., Okunola H. L., Bennett C. F., Krainer A. R. (2008) Am. J. Hum. Genet. 82, 834–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.