Abstract
Misincorporation of amino acids in proteins expressed in Escherichia coli has been well documented but not in proteins expressed in mammalian cells under normal recombinant protein production conditions. Here we report for the first time that Ser can be incorporated at Asn positions in proteins expressed in Chinese hamster ovary cells. This misincorporation was discovered as a result of intact mass measurement, peptide mapping analysis, and tandem mass spectroscopy sequencing. Our analyses showed that the substitution was not related to specific protein molecules or DNA codons and was not site-specific. We believe that the incorporation of Ser at sites coded for Asn was due to mischarging of tRNAAsn rather than to codon misreading. The rationale for substitution of Asn by Ser and not by other amino acids is also discussed. Further investigation indicated that the substitution was due to the starvation for Asn in the cell culture medium and that the substitution could be limited by using the Asn-rich feed. These observations demonstrate that the quality of expressed proteins should be closely monitored when altering cell culture conditions.
Introduction
Many recombinant proteins have been approved as therapeutic drugs by the Food and Drug Administration, and many more are undergoing clinical trial (1). For economic and practical reasons, considerable effort has been made to increase product yield and process efficiency for proteins made in mammalian cell culture. Nowadays, large amounts of proteins can be expressed efficiently in optimized expression systems, with yields from bioreactors having improved more than 100-fold during the past two decades (2). Yields as high as 10 g/liter have been reported for production of monoclonal antibodies in CHO2 cells (3). These yields are due mainly to improvements in host cell engineering, cell line selection, and culture medium optimization (4). However, it is well known that overexpressing recombinant proteins can lead to nutritional stresses in the host cells and that these stresses can markedly increase the frequency of random translational errors, resulting in a heterogeneous mixture of proteins (5–11). A variety of translational errors have been observed during overexpression of proteins in Escherichia coli, including frame shifts, premature truncation, read-through, leaky stop codons, and amino acid misincorporation (12–16). Nevertheless, there are few such reports for proteins made in mammalian cells, and it is commonly believed that the fidelity of translation in mammalian cells is higher (8, 17). Here we report for the first time that misincorporation, namely of Ser for Asn, can occur in proteins overexpressed in CHO cells under normal recombinant protein production conditions. Further investigation showed that supplementation of the medium with Asn can overcome this problem. Our work demonstrates that protein products should be closely monitored for misincorporation, for example, by molecular mass determination and peptide mapping during optimization of culture conditions.
EXPERIMENTAL PROCEDURES
Protein Expression
Proteins were expressed in CHO cells cultured in 125-ml, 1-liter, and 3-liter shake flasks or 5-liter bioreactors. The cultures were grown in proprietary medium supplemented with feeds containing additives described under “Investigation of the cause of the Asn → Ser substitution section” and Table 2. The process was carried out at a temperature of 35–37 °C or at 37 °C with a subsequent change to 28 °C on the third day. For bioreactors, the pH was maintained at neutrality by the addition of 1 m sodium carbonate and by sparging with CO2. Dissolved oxygen was controlled at 30% air saturation. Cell viability was determined by the trypan blue exclusion method, and viable cell density was determined using a Cedex instrument (Innovatis, Bielefeld, Germany). The conditioned medium samples were harvested at the indicated time points.
TABLE 2.
Summary results of investigations into the causes of the Asn → Ser substitution
| Sample information |
Estimated degree of Asn → Ser substitution | |||||||
|---|---|---|---|---|---|---|---|---|
| Row no. | Cell line | SF/BRa | Temperature | Medium | Feedb | Day | Titerc | |
| ° C | mg/liter | |||||||
| 1 | mAb A-1 | BR | 37/28 | M1 | F1 | 14 | 80 | Not detected |
| 2 | CL 24-13 | SF | 37/28 | M1 | F1 | 16 | 566 | 2 |
| 3 | CL 24-13 | SF | 36 | M2 | F2 + hydrolysates | 16 | 821 | 6 |
| 4 | CL 24-34 | SF | 36 | M2 | F2 + hydrolysates | 16 | 689 | 7 |
| 5 | CL 24-13 | BR | 35 | M2 | F4 + hydrolysates | 14 | 2582 | 6 |
| 6 | CL 29-13 | SF | 35 | M2 | F4 | 14 | 546 | 2 |
| 7 | CL 29-13 | BR | 35 | M2 | F4 + hydrolysates | 14 | 1032 | 10 |
| 8 | CL 13-21 (progeny CL 24-13) | BR | 35 | M2 | F4 + hydrolysates | 12 | 2887 | 8 |
| 9 | CL 9-05 (progeny CL 17-14) | BR | 36.5 | M2 | F4 + hydrolysates | 7 | 500 | 2 |
| 10 | CL 9-05 (progeny CL 17-14) | BR | 36.5 | M2 | F4 + hydrolysates | 14 | 2057 | 4 |
| 11 | CL 13-01 (progeny CL 24-13) | BR | 36.5 | M2 | F4 + hydrolysates | 7 | 600 | 8 |
| 12 | CL 13-01 (progeny CL 24-13) | BR | 36.5 | M2 | F4 + hydrolysates | 14 | 1377 | 6 |
| 13 | CL 15-29 (progeny CL 24-34) | BR | 36.5 | M2 | F4 + hydrolysates | 14 | 4162 | 2 |
| 14 | CL 13-21 (progeny CL 24-13) | BR | 35 | M2 | F4 | 14 | 2497 | 5 |
| 15 | CL 13-21 (progeny CL 24-13) | BR | 35 | M3 | F5 + hydrolysates | 14 | 1128 | Not detected |
| 16 | CL 15-29 (progeny CL 24-34) | BR | 35 | M3 | F5 + hydrolysates | 14 | 1312 | Not detected |
| 17 | CL 13-21 (progeny CL 24-13) | BR | 35 | M2 | F4 + hydrolysates | 14 | 3274 | 4 |
| 18 | CL 13-21 (progeny CL 24-13) | BR | 35 | M2 | F4 + hydrolysates + Asn | 14 | 2991 | Not detected |
| 19 | Antibody B | BR | 35 | M2 | F4 + hydrolysates | 14 | 2073 | 3 |
| 20 | Fusion Protein C | BR | 35 | M3 | F3 | 14 | 894 | 2 |
a BR = bioreactor; SF = shake flask.
b F1, F2, F3, F4, and F5 contain 2.4, 1.5, 0.0, 1.5, and 4.5 g/liter of Asn.H2O, respectively.
c Titer of the expressed protein.
Protein Purification
Antibody A and its mutants were purified by chromatography using HiTrap rProtein A FF GE followed by size exclusion chromatography on a Superdex 200 column (GE Healthcare). Antibody B was purified on protein A-Sepharose followed by anion exchange chromatography on TMAE-Fractogel (EM Merck). Purification of fusion protein C was the same as for antibody A except that the protein was further purified by a hydrophobic interaction chromatography on a phenyl-Sepharose column (Amersham Biosciences) before the size exclusion chromatography.
Antibody A Binding Assay
The ability of monoclonal antibody A and its mutants to bind to antigen protein was tested in an ELISA assay. Individual wells in a 96-well plate were coated overnight using the antigen protein at 2 μg/ml. Antibody A and its mutants were titrated into the plate at concentrations from 5 μg/ml to 24 pg/ml and then incubated for 1 h. Binding of antibody A or its mutant to the antigen protein was detected by subsequent binding of horseradish peroxidase anti-human IgG and reaction with tetramethylbenzidine.
Deglycosylation of Proteins
N-Linked glycans were removed from the proteins with peptide N-glycosidase F. About 1 μl of peptide N-glycosidase F (2.5 milliunits/μl, Prozyme) was added to 25 μl of a solution containing about 40 μg protein, after which the solution was incubated at 37 °C overnight.
Intact Mass Measurement
N-Deglycosylated proteins were reduced with 40 mm dithiothreitol in phosphate-buffered saline, pH 7.6, containing 4 m urea. The samples were then analyzed on an LC-MS system composed of an HPLC solvent delivery system (2695 Alliance Separations Module), a 2487 dual wavelength UV detector, and an LCT mass spectrometer (Waters Corp.). A Vydac C4 cartridge was used for desalting. Molecular masses were obtained by deconvolution of raw mass spectra using the MaxEnt 1 program embedded in MaxLynx 4.0 software (Waters Corp.).
Endo-Lys-C Peptide Mapping
Proteins were reduced and alkylated essentially as described in Wen et al. (18). The reduced and alkylated protein was digested with 10% (w/w) of endo-Lys-C (Roche) in 2 m urea, 0.12 m Tris-HCl, pH 8.0, for 8 h at room temperature. Portions of this solution were analyzed on an LC-MS system described above. Peptides from the digest were eluted from a 1.0-mm × 25-cm YMC C18 column (Waters Corp.) with a 185-min water, acetonitrile gradient (0–70% acetonitrile) containing 0.03% trifluoroacetic acid at a flow rate of 0.07 ml/min at 30 °C.
Identification of Peptides by Mass Spectrometry
Peak components on the peptide maps were identified using MassLynx 4.0 software (Waters Corp.). MS/MS spectra were acquired using information-dependent acquisition on a nano-flow LC-MS/MS system composed of a nano-flow HPLC (Dionex, Sunnyvale, CA) and a QSTAR XL mass spectrometer (Applied Biosystems, Foster City CA). The HPLC was equipped with a 0.3-mm × 1-mm Pepmap C18-trap column for desalting and a 0.075-mm × 150-mm, 100-Å, Pepmap C18 column for separation. Peptides were eluted with a 70-min linear gradient (0–50% acetonitrile) containing 0.1% formic acid at a flow rate of 0.2 μl/min. The nanoelectrospray was generated with a nanoelectrospray ionization source (Sciex) using a Picoview needle (15-μm inner diameter; New Objectives) maintained at a voltage of 1700 V. MS/MS spectra were in the m/z range 50–2000, and the collision energy setting was optimized for broader sequence coverage.
Spiking Experiments
Both the wild type and mutant protein samples were diluted to 0.50 mg/ml with phosphate-buffered saline. The concentration of the protein was calculated from its UV absorbance at 280 nm using a calculated extinction coefficient (A280 (1 mg/ml) = 1.4 ml/mg·m). To determine the detection limit of the Asn → Ser substitution, different amounts (0.025–25%) of a monoclonal antibody (mAb) with Asn → Ser mutations at residue 163 in the light chain and residue 392 in the heavy chain were spiked into the wild type mAb, using lowest-volume microsyringes. For analysis by intact mass measurement, an aliquot containing 100 pmol of the protein was injected; for analysis by peptide mapping, 250 pmol was injected. The samples were analyzed in triplicate in each case; the sample with the lowest concentration of Asn → Ser mutant spike was run first followed by increasingly higher concentrations of spike. Between sample runs, the column was cleaned by injecting water and running the gradient. For intact mass measurement data, the amount of the Asn → Ser substitution was calculated by comparison of the heights of peaks of MH10+10, MH15+15, and MH16+16 ions from the light chain. For peptide mapping data, the amount of the substitution was estimated from peak heights of the combined mass spectra for the predicted peptide and for the corresponding Asn → Ser-containing peptide, after subtracting background noise.
RESULTS
Discovery of Low Mass (−27 Da) Components
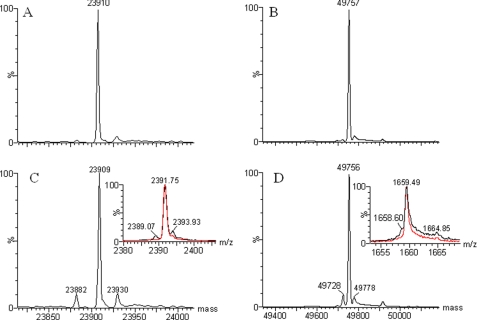
Antibody A is a humanized monoclonal antibody raised against a human protein. To ensure that a cell line selected for production of clinical study materials does not produce a protein with unexpected mutations or modifications, we analyzed samples of antibody A made from candidate cell lines using mass spectrometry. Unexpectedly, in addition to the expected light chain mass (observed = 23,909 Da; calculated = 23,910.8 Da) and heavy chain mass (observed = 49,756 Da; calculated = 49,757.2 Da), two minor components with masses of 23,882 and 49,728 Da were detected in the respective chains of some of the candidate cell line samples. The masses of these species were about −27 Da lower than those calculated for the light chain and the heavy chain, respectively. The amount of the “−27 Da” component in each candidate cell line sample varied. Fig. 1, C and D, shows the mass spectra of candidate cell line CL 24-34 as an example of one of the worst cases. The −27-Da components were absent when the antibody was produced in a low-expressing cell line (mAb A-1) as shown in Fig. 1, A and B.
FIGURE 1.
Deconvoluted mass spectra of the reduced antibody A. A, shown is the light chain of the mAb A-1. B, shown is the heavy chain of the mAb A-1. C, shown is the light chain of a batch from candidate cell line CL 24-34. Inset, shown is the MH1010+ peak of the light chain. D, shown is the heavy chain of a batch from candidate cell line CL 24-34. Inset, shown is the MH3030+ peak of the heavy chain. The calculated molecular mass for the light chain is 23,910.8 Da; that for the heavy chain is 49,757.2 Da.
Identity of the Low Mass (−27 Da) Components
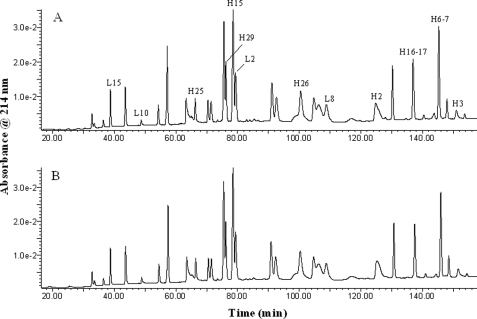
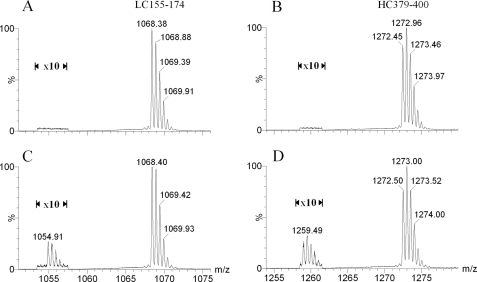
To identify the −27-Da species seen for both the light chain and the heavy chain of the antibody, we carried out endo-Lys-C and endo-Asp-N peptide mapping of the reduced antibody A using an LC-MS system. The sequence coverage of the combined endo-Lys-C and endo-Asp-N peptide maps for antibody A is 100% for both the light and heavy chains (data for the endo-Asp-N peptide maps are not shown). Fig. 2 shows the HPLC profile, monitored at 214 nm, for endo-Lys-C peptide digests of samples of the mAb A-1 and of cell line candidate CL 24-34. Comparison of the UV profiles and total ion chromatograms for the two samples did not reveal any obvious differences between samples. However, when we carefully compared the mass spectral profiles peak by peak, we observed low levels of extra components in the map of the CL 24-34 sample, specifically components whose molecular masses were all 27 Da lower than the nearby peptides having the expected masses. For example, Fig. 3 shows mass spectra of a light chain peptide, LC 155–174, and of a heavy chain peptide, HC 379–400, from samples CL 24-34 and mAb A-1. There are four possible changes that could produce a peptide with a molecular mass of 27 Da lower than predicted: Arg → Glu, Gln → Thr, Lys → Thr, or Asn → Ser. We found that the −27-Da components were detected only when the corresponding normal peptides contained an Asn residue (see Fig. 2 and Table 1) and, therefore, concluded that the −27-Da components detected in candidate cell line samples must be due to an Asn → Ser substitution. To confirm this, several of the −27-Da-containing peptides were subjected to MS/MS sequencing analysis along with their “normal” corresponding peptides as controls.
FIGURE 2.
UV profile with monitoring at 214 nm for endo-Lys-C peptide maps of the reduced antibody A. A, shown is mAb A-1. B, shown is a batch from candidate cell line CL 24-34.
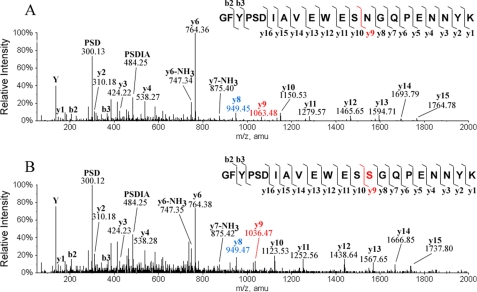
FIGURE 3.
Mass spectra of peptide LC 155–174 (A and C) and HC 379–400 (B and D) of antibody A. Only peaks in the doubly charged state are shown. A and B, shown are mAb A-1. C and D, shown is a batch from candidate cell line CL 24-34. The intensities of peaks near m/z 1055 and 1260 were increased 10-fold for easy viewing and comparison.
TABLE 1.
Predicted endo-Lys-C peptides for antibody A (huP2D10v2) and summary results of Asn → Ser substitution analysis
| Endo-LysC peptide (residues) | Number of Asn residues in the peptide | Amount of Asn → Sera |
|---|---|---|
| % | ||
| Antibody A, light chain | ||
| L1 (1–33) | 0 | |
| L2 (34–44) | 1 (CDR1) | 1.4 |
| L3 (45–55) | 0 | |
| L4 (56–79) | 1 (CDR2) | 1.5 |
| L5, L6, L7, L6 (80–131) | 0 | |
| L8 (132–150) | 2 | 3.0 |
| L9 (151–154) | 0 | |
| L10 (155–174) | 2 | 3.1 |
| L11, L12, L13, L14 (175–212) | 0 | |
| L15 (213–219) | 1 | 1.6 |
| Antibody A, heavy chain | ||
| H1, 1–43 | 0 | |
| H2, 44–76 | 1 | 1.9 |
| H3, 77–129 | 2 | 4.0b |
| H4, H5 (130–155) | 0 | |
| H6–7, 156–218 | 4 | 6.7 |
| H8, H9, H10, H11 (219–230) | 0 | |
| H12, H13, H14 (231–282) | 0 | |
| H15, 283–296 | 2 | 2.4b |
| H16–17, 297–325 | 2 | 2.9 |
| H18, H19 (326–330) | 0 | |
| H20, 331–334 | 1 | 1.4b |
| H21, H22, H23, H24 (335–368) | 0 | |
| H25, 369–378 | 1 | 1.4 |
| H26, 379–400 | 3 | 4.6 |
| H27, H28 (401–422) | 0 | |
| H29, 423–447 | 2 | 2.8 |
| H30, 448–454 | 0 | |
a The amount of the substitution was estimated from peak heights of the combined mass spectra from the extracted ion chromatograms of the predicted peptide and the corresponding peptide containing the Asn → Ser substitution.
b Amounts are estimated from the endo-Asp-N peptide map of the protein (data are not shown).
During MS/MS sequencing analysis, the peptide is fragmented by collision-induced dissociation, which breaks backbone amide bonds generating a series of fragment ions, mainly C-terminal y ions and N-terminal b ions in this case. The identity of an amino acid at each position in a peptide can be determined by the difference in the m/z values of two adjacent y or b ions. For example, a difference of 114 atomic mass units between adjacent y ions (or b ions) indicates an Asn at that position, and a difference of 87 atomic mass units indicates a Ser.
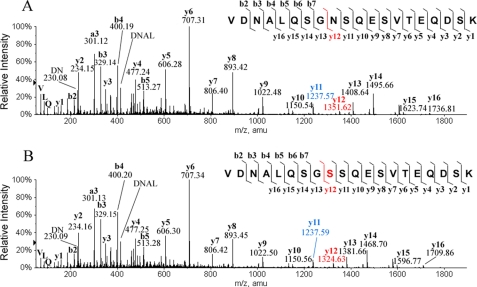
Fig. 4A shows the MS/MS spectrum of the predicted peptide LC 155–174 (i.e. a light chain peptide containing predicted residues 155–174). There are two Asn residues in peptide LC 155–174, at positions 157 and 163, based on the predicted sequence. As shown in Fig. 4A, the differences in m/z values between y12 (1351.62) and y11 (1237.57) and between b3 (329.14) and b2 (230.08) are each 114.04, confirming Asn at the both positions in this peptide as predicted. LC-MS peptide mapping detected two −27-Da components related to peptide LC 155–174; one co-eluted with the predicted wild type peptide, and the other eluted slightly later. Fig. 4B shows a MS/MS spectrum for the −27-Da component that eluted slightly later. As one can see in this spectrum, the difference of m/z values between b3 (329.15) and b2 (230.09) is 114.04, confirming an Asn at position 157. However, the difference of m/z values between y12 (1324.63) and y11 (1237.59) is 87.03, indicating that a Ser is at the position 163, not the predicted Asn. The differences in m/z values for the other adjacent ions in this peptide are all as expected, which confirms that the sequence of the rest of the peptide is the same as the wild type. The MS/MS sequencing analysis of the −27-Da component that co-eluted with the wild type peptide showed that a Ser replaced Asn-157 in the peptide but not Asn-163 (data not shown). Similarly, there are three −27-Da components related to peptide HC 379–400 (residues 379–400 in the heavy chain). Fig. 5 shows the MS/MS spectrum of one of three peptides along with the corresponding wild type peptide HC 379–400; Asn is at position 392 in the wild type peptide as predicted (Fig. 5A; the difference in m/z values for y9 (1063.48) and y8 (949.45) is 114.04), but Ser is at this position in the −27-Da-containing peptide (Fig. 5B; the difference in m/z values for y9 (1036.47) and y8 (949.47) is 87.03). MS/MS sequencing of the other two −27-Da-containing peptides demonstrated that the −27-Da peptide was also because of an Asn → Ser substitution (data not shown) antibody A contains 7 Asn residues in the light chain and 18 in the heavy chain. A combination of endo-Lys-C and endo-Asp-N peptide mapping detected Asn → Ser substitutions at all the predicted Asn positions, including the predicted N-glycosylation site Asn-305 in the Fc region. Table 1 summarizes the results. The Asn → Ser substitution occurred randomly in the antibody polypeptide chains, with about 1–2% at each Asn position in the CL 24-34 sample; no preferred site was observed. In addition, we detected by peptide mapping Asn → Ser substitution in the samples made in many candidate cell lines, the level varying from 0.3 to 2% per Asn position.
FIGURE 4.
MS/MS spectra of peptide LC 155–174 of antibody A. A, shown is the wild type peptide. B, shown is the peptide with an Asn → Ser substitution. The sequence of the peptide, the fragmentation pattern, and the detected fragment ions are shown at the top of each panel. y ions contain the C-terminal region of the peptide, and b ions contain the N-terminal region of the peptide. Calculated m/z values are y11 = 1237.55 and y12 = 1351.60 when residue 163 is Asn, and y11 = 1237.55 and y12 = 1324.59 when residue 163 in Ser.
FIGURE 5.
MS/MS spectra of peptide 379–400 of antibody A. A, shown is the wild type peptide. B, shown is the peptide with an Asn → Ser substitution. The sequence of the peptide, the fragmentation pattern, and the detected fragment ions are shown at the top of each panel. y ions contain the C-terminal region of the peptide, and b ions contain the N-terminal region of the peptide. Calculated m/z values are y8 = 949.44 and y9 = 1063.48 when residue 387 is Asn, and y8 = 949.44 and y9 = 1036.47 when residue 387 in Ser.
Limits of Detection of Asn → Ser Substitutions Using Intact Mass Measurement and Peptide Mapping
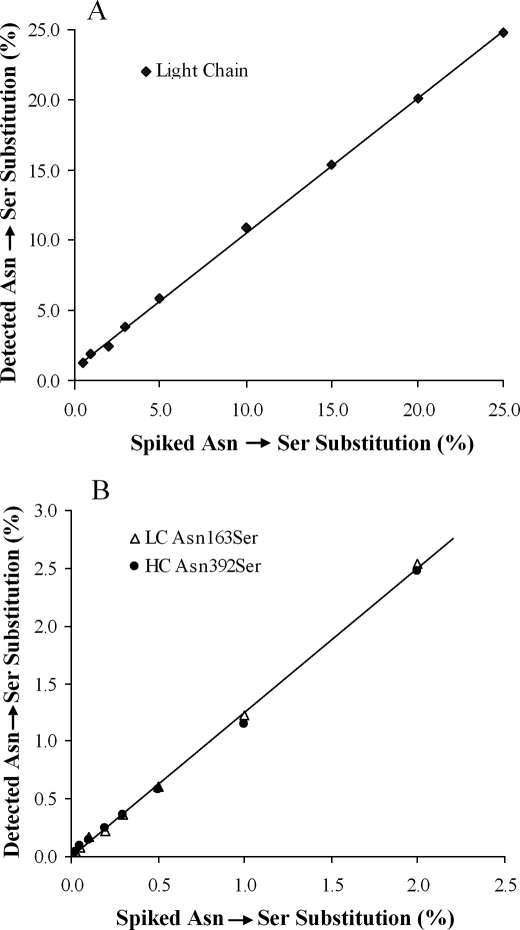
To determine the limits of detection for the intact mass measurement and peptide mapping methods, spiking experiments were carried out using a mutant of antibody A, L163/H392, in which residue Asn-163 in the light chain and residue Asn-392 in the heavy chain had been mutated to Ser. The wild type protein (mAb A-1) was then spiked with the mutant, and the lowest levels of detection were determined. These experiments showed that ≥0.5% of an Asn → Ser substitution in the light chain (Fig. 6A) or ≥2% in the heavy chain (data not shown) of antibody A could be detected by intact mass measurement using 100 pmol of the reduced protein, and that using endo-Lys-C peptide mapping and 250 pmol of the reduced protein, ≥0.025% of an Asn → Ser substitution in a peptide could be detected. The mass spectrometric response to substitution in a peptide is linear and the intercept is very close to zero (Fig. 6B). However, the observed amounts of the mutant peptides on a particular peptide map are about 15–20% higher than spiked values, which is partly caused by better ionization (∼12% better, data not shown) of the Ser mutant peptide compared with its wild type form and partly caused by partial saturation of the wild type peptide in the electrospray ionization-time of flight mass spectrometer used in this study. This work also showed that ≥2% of the mutant is required for quantification of the Asn → Ser substitution in the intact light chain or ≥0.1% in a peptide. The degree of substitution from intact mass measurement for the heavy chain cannot be used for estimating the overall amount of substitution because the mutant and wild type forms of the heavy chain are poorly resolved.
FIGURE 6.
Plots of the detected amounts of the Asn → Ser substitution in monoclonal antibody samples that were spiked with varying amounts of a monoclonal antibody with Asn → Ser mutations at residue 163 in the light chain and at residue 392 in the heavy chain. A, shown is intact mass measurement for the light chain of antibody A. B, shown is peptide mapping of antibody A. LC, light chain; HC, heavy chain.
Investigation of the Cause of the Asn → Ser Substitution
As mentioned above, the Asn → Ser substitution was observed in samples made from all candidate cell lines that had been produced by independent transfections (rows 3, 4, and 6 in Table 2). Furthermore, substitution was detected at all Asn positions in the protein and at similar levels (Table 1). It, therefore, seemed that the substitution was not due to a particular cell line. To confirm this and to find out whether the Asn → Ser substitution was due to variations in cell culture conditions in the shake flasks, where pH, oxygen sparge rate, etc., were not tightly controlled, we expressed antibody A from different cell lines in bioreactors before and after amplification. As shown in Table 2, the Asn → Ser substitution was also observed in samples made in bioreactors regardless of which cell line was used (rows 5, 7, 10, and 13) whether or not amplification was done (rows 5 versus 8) and whether the cells were harvested at days 7 or 14 (rows 9 versus 10 and rows 11 versus 12). Although we found that the amount of the substitution varies between cell lines, no trend was found; the protein made from cell line CL 24-34 (row 4) had the highest level of substitution of all the samples made in shake flasks, but the protein made from the same cell line in a bioreactor had a very low level of the substitution (row 13). On the other hand, the samples from cell line CL 24-13 had similar amounts of the substitution regardless of whether they were made in a shake flask or a bioreactor (rows 3 and 8). Furthermore, the level of the substitution in one sample harvested at day 7 was lower than that from day 14 (row 9 versus 10) but higher at day 7 compared at day 14 in another sample (rows 11 versus 12). Therefore, we concluded that the Asn → Ser substitution is not due to the cell lines used.
Because we had not observed the Asn → Ser substitution in any recombinant proteins made previously and had not seen any reports of this problem in the literature, we thought that the Asn → Ser substitution might be antibody A molecule-specific. To test the idea, we made two other proteins under the same culture conditions; antibody B (raised against a very different target antigen) and fusion protein C. Once again endo-Lys-C peptide mapping of the two samples revealed the Asn → Ser substitution in many Asn-containing peptides for both proteins. The substitution level was about 0.4% at each Asn position for antibody B and about 0.2% for fusion protein C (supplemental Table 1 and supplemental Figs. 1 and 2). Thus, the Asn → Ser substitution is not protein molecule-specific. These results suggested that cell culture conditions might be responsible for the Asn → Ser substitution.
We tested various culture conditions using different in-house-made media (M1, M2, and M3) and feeds (F1, F2, F3, F4, and F5 with or without hydrolysates and Asn). Representative results are shown in Table 2. Our experiments showed that the substitution could occur in proteins made in medium M1, M2, or M3 that contain 0.2–0.9 g/liter l-Asn monohydrate. The Asn → Ser substitution was not detected in the mAb A-1 but was observed in protein made from a high-expressing cell line grown in the same medium (M1) with the same feed (F1), which contains 2.4 g/liter l-Asn (rows 1 versus 2). Although it seemed that the Asn → Ser substitution was related to the higher product titer in the cell culture, our studies did not show a linear relationship between the level of the Asn → Ser substitution and the product titers in bioreactors, at least not within our testing range (1–4.2 g/liter, rows 7, 8, 10, 12–14 in Table 2). When M1 and M2 media were tested using different feeds, the substitution was not observed when the culture was augmented with Asn at a high concentration, i.e. in culture with optimized feed containing additional l-Asn (F5 contains 4.5 g/liter Asn, rows 15 and 16 in Table 2), or when l-Asn was added to an optimized feed (row 18 in Table 2). Further experiments showed that the results were reproducible and that the strategy of adding l-Asn in the feed could be scaled up for a 200-liter bioreactor. Thus, we demonstrated that Asn → Ser substitution was caused by starvation for l-Asn in the cell culture medium.
Binding of an Asn → Ser Mutant of Antibody A
Antibody A has two Asn residues in the complementarity determining region of the light chain. To test whether the Asn → Ser substitution would affect the function of the antibody, an antibody A mutant was made in which Asn-35 in the light chain was mutated to Ser (LC-N35S). The binding affinity of the LC-N35S to the antigen protein was tested and compared with the wild type antibody A. As shown in supplemental Fig. 3, no significant difference was detected in the binding affinity between the LC-N35S mutant and wild type antibody A.
DISCUSSION
We have discovered that Ser can be misincorporated randomly at Asn positions when proteins are expressed at very high levels in CHO cells. Using our methods, <1% of an Asn → Ser substitution in a 20-kDa protein can be detected by mass measurement of the intact protein, and <0.03% of the substitution can be detected in a peptide, e.g. by peptide mapping analyses. Further investigation showed that the substitution was due to starvation for Asn and that adding extra Asn to culture medium can overcome the problem.
Random amino acid substitutions in proteins occur naturally. The frequency of the errors under subnormal growth conditions occurs at a rate of about 4 × 10−4 to 5 × 10−5 per codon, or 0.005–0.04% per site (12). However, the error frequency can be 20–150-fold higher under conditions of stress (10). The fidelity of protein biosynthesis depends on accurate codon-anticodon interaction between mRNA and tRNA and also on the specific attachment of amino acids to their cognate tRNA species. Random amino acid substitution can be caused by either an anticodon-codon mismatch, i.e. misreading, or the use of an erroneously charged tRNA, i.e. mischarging or misacylation (12).
We believe that the misincorporation of Ser at Asn positions was due to mischarging of tRNAAsn rather than misreading for three reasons. First of all, it has been shown that most misreading errors occur at the third position of the codon, which has a weaker interaction with its cognate base than do bases at the first and second positions (20, 21). So far, all Asn starvation experiments have shown that the Asn codons AAU and AAC were misread as lysine, AAA and AAG, errors at the third position of the codon (22, 23). Second, Asn starvation studies have also shown (24, 25) that the frequency of misreading of the Asn codon, AAU, is much higher (2–10-fold) than for the AAC codon, whereas we see no codon preference even though about two-thirds of the Asn codons in our proteins are AAU. Third, mischarging, as seen here, is not codon-related but results from the attachment of a different but structurally related amino acid to its cognate tRNA; misincorporation can be prevented by supplementing the culture medium with the cognate amino acid, Asn, in this case (11, 26).
Acylation of tRNA is a two-step reaction: the amino acid is activated by ATP to form an aminoacyl-adenylate intermediate (aa-AMP), and then the intermediate reacts with the tRNA to produce aminoacyl-tRNA and AMP. Acylation of tRNAs is controlled by aminoacyl-tRNA synthetases (aminoacyl-RSs) which not only catalyze the reactions but also employ proofreading or editing mechanisms at various steps during the catalytic cycle to ensure that cognate amino acids are attached to the appropriate tRNAs. Complete discrimination of amino acids by aminoacyl-RSs is achieved through different strategies. First of all, aminoacyl-RSs have to recognize their cognate amino acids in intracellular pools of amino acids. About half of the aminoacyl-RSs can easily recognize their cognate amino acids based on structural or chemical and physical properties, such as size, charge, hydrophobicity, and space orientation as these enzymes produce conjugates with molecular geometries for cognate amino acids and unfavorable geometries for non-cognate amino acids. However, in the case of substrates with close structural and chemical similarities, the enzymes use proofreading or editing strategies during the process of tRNA aminoacylation to ensure high accuracy of protein biosynthesis. Fidelity mechanisms of the aminoacyl-RSs with known editing functions have been reviewed thoroughly recently by Mascarenhas et al. (27) and Splan et al. (28). To ensure fidelity, misactivated aminoacyl-adenylates and misaminoacylated tRNAs can be hydrolyzed in respective pre-transfer editing and post-transfer editing processes. Some non-cognate amino acids are edited at the synthetic active site, and others are edited at a separate editing site in the aminoacyl-RS. Hydrolysis of misacylated tRNA can occur in a distal hydrolytic active site or in an editing domain in an aminoacyl-RS (29–32). The mechanism for hydrolysis of non-cognate aminoacyl-adenylates is not completely understood yet, but several mechanisms have been proposed including hydrolysis in a distinct editing active site after translocation to it (33, 34), hydrolysis within the synthetic active site (35, 36), and selective release into the cellular milieu (36, 37). Recent studies have shown that editing-like reactions can occur within the synthetic active site in glutaminyl-RS (a class I aminoacyl-RS) (35), prolyl-RS (a class II aminoacyl-RS) (37), and seryl-RS (a class II aminoacyl-RS) (38). Asparaginyl-RS, a class IIb aminoacyl-RS, does not possess an editing domain (39), and so far no editing activity for asparaginyl-RS has been reported, which suggests that asparaginyl-RS can select its cognate substrates from a pool of competitive substrates in a very efficient initial binding step based on structure and physical and chemical properties.
It is probably true that when Asn is in good supply for protein biosynthesis, asparaginyl-RS has the highest binding affinity for Asn and the lowest hydrolysis rate for asparaginyl-adenylate compared with non-cognate aminoacyl-adenylates. However, when Asn is not present in sufficient amounts, other amino acids, especially ones with the most similar structures and physical and chemical properties, will be misactivated so that biosynthesis can continue. The misincorporation of norvaline, a non-protein amino acid, at leucine positions in recombinant human hemoglobin when the ratio of norvaline to leucine in culture medium is high is a good example (11).
Why is it that Ser rather than other amino acids is misincorporated at Asn positions? Both Asn and Ser are neutral, polar amino acids, with similar physical properties. Although aspartic acid is the closest structural analog for Asn, it does not bind to the synthetic active site in asparaginyl-RS because Glu-225 in asparaginyl-RS has a dual role of positive recognition of Asn and discrimination against the negatively charged aspartic acid side chain (39). Calculated binding energies also suggest that Ser will compete with Asn for formation of an activated intermediate with asparaginyl-RS (40). In addition, both asparaginyl-RS and seryl-RS are class II aminoacyl-RSs that share many structural features. For example, both are homodimers and lack an editing domain, and their size and catalytic domains are similar (41, 42). The structure of the asparaginyl-tRNA synthetase-ATP complex has exactly the same configuration of three divalent cations as does the seryl-RS-ATP complex (39), indicating that the structure of Ser is similar to that of Asn. Overall, the slightly smaller size of Ser and similar physical and chemical natures of Ser and Asn seem to result in a higher binding affinity of Ser at the synthetic active site and slower rate of hydrolysis of seryl-AMP in asparaginyl-RS than for other amino acids assuming that the same pre-transfer editing exists in asparaginyl-tRNA synthase as in seryl-RS (38). Thus, tRNAAsn appears to have been misacylated by Ser but not other amino acids when there is a shortage of Asn during protein biosynthesis.
Based on our results we propose a model for Ser misincorporation at Asn positions. Asparagine is a nonessential amino acid that can be supplied by biosynthesis in CHO cells under normal biosynthesis conditions. Whenever CHO cells need Asn, the extent of aminoacylation of tRNAAsn decreases, and the activity of Asn synthetase for production of Asn amino acid increases (43, 44). Thus, cells regulate themselves to meet their requirement for Asn. Asparaginyl-RS has the highest selectivity for Asn and does not need significant editing activities as do other aminoacyl-RSs. Asparaginyl-RS excludes non-cognate amino acids that are larger than Asn, e.g. Gln, Glu, Ile, Leu, Val, Met, etc. It does not bind smaller amino acids (Ala, Pro, and Gly) whose side chains cannot form hydrogen bonds with residues in its synthetic active site (39), it does not bind Asp because Glu-225 in the synthetic active site repels the negatively charged side chain of Asp (39), it cannot bind Cys tightly because it cannot provide the zinc-thiolate interaction required for binding Cys to cysteinyl-RS (45), and it does not bind well with Thr because the methyl group of the side chain of Thr is bulky and hydrophobic and because the geometry of the side chain is very different from Asn. Ser is the only amino acid that is both smaller than Asn and has some similar hydrogen-bonding properties. Thus, Ser can bind to the active site in asparaginyl-RS relatively more tightly than can other non-cognate amino acids. However, the binding affinity of Ser to asparaginyl-RS will be much weaker than that of Asn as the hydroxyl group of its side chain can form only one hydrogen bond with a residue in the synthetic active site of asparaginyl-RS, whereas Asn can form two hydrogen bonds (with Glu-225 and Arg-368) (39). In addition, non-cognate-adenylates may be hydrolyzed at the synthetic active site of asparaginyl-RS, as has been reported for seryl-RS (38).
No miscorporation at Asn positions was observed when cells grew at a normal growth rate or when a sufficient amount of Asn was supplied in the culture medium. However, if the rate of Asn biosynthesis is not high enough to meet the need for overexpression of a foreign protein, the cells become starved for Asn, which can increase the rate of formation of misacylated adenylates and mischarged tRNAAsn. In this case the result is detectable Ser misincorporation at Asn positions in the protein, as Ser is the closest analog to Asn, and it is available. This problem can be corrected simply by adding adequate amounts of Asn to the cell culture medium. It is interesting that we did not detect any substitution of Asn by Lys, as noted by others (5, 22, 24) in E. coli under conditions of extreme Asn starvation. Perhaps it is because in all Asn starvation studies reported so far, the relevant asparaginyl-RS was inactivated by using temperature-sensitive asparaginyl-RS mutants. Thus, no t-RNAAsn mischarging could have occurred, only misreading. Furthermore, Stanner et al. (47) have shown that asparaginyl-RS in CHO cells is much more sensitive to amino acid starvation than are other aminoacyl-RSs.
The large scale synthesis of recombinant proteins always involves overexpression of the product, which can lead to nutritional stresses in the production cells and create imbalances in the charged tRNA supply, resulting in a marked increase in the frequency of random translational errors. To date, most published data dealing with charged tRNA imbalances in overexpression systems used for making recombinant proteins refer to E. coli. Our finding of the Asn → Ser substitution is the first such report for proteins overexpressed in CHO cells under normal recombinant protein production conditions. This should not be a total surprise as the pathways of protein synthesis in prokaryotes and eukaryotes are similar, although the quality control for protein biosynthesis in eukaryotes is tighter than that in prokaryotes (17).
Although little is known about the possible deleterious effects in humans due to minor amounts of erroneously synthesized recombinant protein therapeutics, abnormal bioactivities and undesirable immune responses could be problems. For instance, diseases caused by editing-defective aminoacyl-RSs are well known (48–50); single Asn → Ser mutations have caused the loss in binding capacity in proteins (51–53), diminished enzymatic activity (54), and changed protein folding structure (55) and have been associated with diseases (46, 56, 57). We observed only a minor decrease of the binding affinity when Asn-35 was mutated to Ser in the complementarity determining region of the light chain of antibody A (supplemental Fig. 3), but the mutation could be immunogenic in vivo. Therefore, steps need to be taken both to identify and eliminate such potential errors when developing expression strategies for producing recombinant proteins used in human therapies, e.g. when limiting Asn in the culture media and feeds (19) to gain maximum growth rates and to reduce the amount of ammonia in CHO cell cultures. Random misincorporation is difficult to detect and quantify by conventional analytical methods, especially when the cognate and erroneous residues belong to similar chemical groups, as they manifest themselves as a heterogeneous mixture of proteins, each having slightly different chemical and physical properties from the other. Because removal of erroneously synthesized molecules after production is likely to be extremely difficult, it is better to prevent their formation in the first place, i.e. by careful monitoring for such errors when changing protein expression conditions using modern analytical tools as we have demonstrated here.
Supplementary Material
Acknowledgments
We thank Holly Prentice, Donald Bennett III, Konrad Miatkowski, and Yen-Ming Hsu for contributions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- CHO
- Chinese hamster ovary
- aminoacyl-RS
- aminoacyl-tRNA synthetases
- LC
- liquid chromatography
- MS
- mass spectrometry
- HPLC
- high pressure liquid chromatography
- mAb
- monoclonal antibody.
REFERENCES
- 1.Reichert J. M., Rosensweig C. J., Faden L. B., Dewitz M. C. (2005) Nat. Biotechnol. 23, 1073–1078 [DOI] [PubMed] [Google Scholar]
- 2.Wurm F. M. (2004) Nat. Biotechnol. 22, 1393–1398 [DOI] [PubMed] [Google Scholar]
- 3.Low D., O'Leary R., Pujar N. S. (2007) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 848, 48–63 [DOI] [PubMed] [Google Scholar]
- 4.Birch J. R., Racher A. J. (2006) Adv. Drug Deliv. Rev. 58, 671–685 [DOI] [PubMed] [Google Scholar]
- 5.Parker J., Pollard J. W., Friesen J. D., Stanners C. P. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberger R. F. (1991) Mutat. Res. 256, 255–262 [DOI] [PubMed] [Google Scholar]
- 7.Scorer C. A., Carrier M. J., Rosenberger R. F. (1991) Nucleic Acids Res. 19, 3511–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos M. A., Tuite M. F. (1993) Trends Biotechnol. 11, 500–505 [DOI] [PubMed] [Google Scholar]
- 9.Roserberg R. F., Holliday R. (1993) Trends Biotechnol. 11, 498–499 [DOI] [PubMed] [Google Scholar]
- 10.Rosenberger R. F. (1994) Dev. Biol. Stand. 83, 21–26 [PubMed] [Google Scholar]
- 11.Apostol I., Levine J., Lippincott J., Leach J., Hess E., Glascock C. B., Weickert M. J., Blackmore R. (1997) J. Biol. Chem. 272, 28980–28988 [DOI] [PubMed] [Google Scholar]
- 12.Parker J. (1989) Microbiol. Rev. 53, 273–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spanjaard R. A., Chen K., Walker J. R., van Duin J. (1990) Nucleic Acids Res. 18, 5031–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinkmann U., Mattes R. E., Buckel P. (1989) Gene 85, 109–114 [DOI] [PubMed] [Google Scholar]
- 15.Seetharam R., Heeren R. A., Wong E. Y., Braford S. R., Klein B. K., Aykent S., Kotts C. E., Mathis K. J., Bishop B. F., Jennings M. J., Smith C. E., Siegel N. R. (1988) Biochem. Biophys. Res. Commun. 155, 518–523 [DOI] [PubMed] [Google Scholar]
- 16.Calderone T. L., Stevens R. D., Oas T. G. (1996) J. Mol. Biol. 262, 407–412 [DOI] [PubMed] [Google Scholar]
- 17.Ibba M., Söll D. (1999) Science 286, 1893–1897 [DOI] [PubMed] [Google Scholar]
- 18.Wen D., Wildes C. P., Silvian L., Walus L., Mi S., Lee D. H., Meier W., Pepinsky R. B. (2005) Biochemistry 44, 16491–16501 [DOI] [PubMed] [Google Scholar]
- 19.Seewöster T., Lehmann J. (1995) Appl. Microbiol. Biotechnol. 44, 344–350 [DOI] [PubMed] [Google Scholar]
- 20.Lagerkvist U. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 1759–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lustig F., Elias P., Axberg T., Samuelsson T., Tittawella I., Lagerkvist U. (1981) J. Biol. Chem. 256, 2635–2643 [PubMed] [Google Scholar]
- 22.Parker J., Friesen J. D. (1980) Mol. Gen. Genet. 177, 439–445 [DOI] [PubMed] [Google Scholar]
- 23.Parker J., Johnston T. C., Borgia P. T., Holtz G., Remaut E., Fiers W. (1983) J. Biol. Chem. 258, 10007–10012 [PubMed] [Google Scholar]
- 24.Johnston T. C., Borgia P. T., Parker J. (1984) Mol. Gen. Genet. 195, 459–465 [DOI] [PubMed] [Google Scholar]
- 25.Precup J., Parker J. (1987) J. Biol. Chem. 262, 11351–11355 [PubMed] [Google Scholar]
- 26.Barker D. G., Bruton C. J. (1979) J. Mol. Biol. 133, 217–231 [DOI] [PubMed] [Google Scholar]
- 27.Mascarenhas A. P., An S., Rosen A. E., Martinis S. A., Musier-Forsyth K. (2009) in Protein Engineering-Nucleic Acids and Molecular Biology Series 22 (Köhrer C., RajBhandary U. L. eds) 1st Ed., pp. 155– 203, Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 28.Splan K. E., Musier-Forsyth K., Boniecki M. T., Martinis S. A. (2008) Methods 44, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silvian L. F., Wang J., Steitz T. A. (1999) Science 285, 1074–1077 [PubMed] [Google Scholar]
- 30.Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) Nat. Struct. Mol. Biol. 12, 923–930 [DOI] [PubMed] [Google Scholar]
- 31.An S., Musier-Forsyth K. (2005) J. Biol. Chem. 280, 34465–34472 [DOI] [PubMed] [Google Scholar]
- 32.Ruan B., Söll D. (2005) J. Biol. Chem. 280, 25887–25891 [DOI] [PubMed] [Google Scholar]
- 33.Lincecum T. L., Jr., Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., Van Den Eynde W., Link A., Van Calenbergh S., Grøtli M., Martinis S. A., Cusack S. (2003) Mol. Cell 11, 951–963 [DOI] [PubMed] [Google Scholar]
- 34.Nomanbhoy T. K., Hendrickson T. L., Schimmel P. (1999) Mol. Cell 4, 519–528 [DOI] [PubMed] [Google Scholar]
- 35.Gruic-Sovulj I., Uter N., Bullock T., Perona J. J. (2005) J. Biol. Chem. 280, 23978–23986 [DOI] [PubMed] [Google Scholar]
- 36.Splan K. E., Ignatov M. E., Musier-Forsyth K. (2008) J. Biol. Chem. 283, 7128–7134 [DOI] [PubMed] [Google Scholar]
- 37.Hati S., Ziervogel B., Sternjohn J., Wong F. C., Nagan M. C., Rosen A. E., Siliciano P. G., Chihade J. W., Musier-Forsyth K. (2006) J. Biol. Chem. 281, 27862–27872 [DOI] [PubMed] [Google Scholar]
- 38.Gruic-Sovulj I., Rokov-Plavec J., Weygand-Durasevic I. (2007) FEBS Lett. 581, 5110–5114 [DOI] [PubMed] [Google Scholar]
- 39.Berthet-Colominas C., Seignovert L, Härtlein M., Grotli M., Cusack S., Leberman R. (1998) EMBO J. 17, 2947–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClendon C. L., Vaidehi N., Kam V. W., Zhang D., Goddard W. A., 3rd (2006) Protein Eng. Des. Sel. 19, 195–203 [DOI] [PubMed] [Google Scholar]
- 41.Cusack S. (1995) Nat. Struct. Biol. 2, 824–831 [DOI] [PubMed] [Google Scholar]
- 42.Sankaranarayanan R., Moras D. (2001) Acta Biochim. Pol. 48, 323–335 [PubMed] [Google Scholar]
- 43.Andrulis I. L., Hatfield G. W., Arfin S. M. (1979) J. Biol. Chem. 254, 10629–10633 [PMC free article] [PubMed] [Google Scholar]
- 44.Arfin S. M., Simpson D. R., Chiang C. S., Andrulis I. L., Hatfield G. W. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 2367–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newberry K. J., Hou Y. M., Perona J. J. (2002) EMBO J. 21, 2778–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishitobi M., Miyoshi Y., Hasegawa S., Egawa C., Tamaki Y., Monden M., Noguchi S. (2003) Cancer Lett. 200, 1–7 [DOI] [PubMed] [Google Scholar]
- 47.Stanners C. P., Wightman T. M., Harkins J. L. (1978) J. Cell. Physiol. 95, 125–137 [DOI] [PubMed] [Google Scholar]
- 48.Lee J. W., Beebe K., Nangle L. A., Jang J., Longo-Guess C. M., Cook S. A., Davisson M. T., Sundberg J. P., Schimmel P., Ackerman S. L. (2006) Nature 443, 50–55 [DOI] [PubMed] [Google Scholar]
- 49.Schimmel P. (2008) Protein Sci. 17, 1643–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park S. G., Schimmel P., Kim S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11043–11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pöschl E., Fox J. W., Block D., Mayer U., Timpl R. (1994) EMBO J. 13, 3741–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jnaoui K., Minet M., Michiels T. (2002) J. Virol. 76, 8138–8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevens C., Lin Y., Sanchez M., Amin E., Copson E., White H., Durston V., Eccles D. M., Hupp T. (2007) J. Biol. Chem. 282, 13791–13803 [DOI] [PubMed] [Google Scholar]
- 54.Kaye E. M., Shalish C., Livermore J., Taylor H. A., Stevenson R. E., Breakefield X. O. (1997) J. Child Neurol. 12, 242–247 [DOI] [PubMed] [Google Scholar]
- 55.George E., Huisman T. H., Yang K. G., Kutlari F., Wilson J. B., Kutlar A., Storming T. A., Gonzales-Redondo J. M., Faridah K., Khalid A. K. (1989) Med. J. Malaysia 44, 259–262 [PubMed] [Google Scholar]
- 56.Walz R., Castro R. M., Landemberger M. C., Velasco T. R., Terra-Bustamante V. C., Bastos A. C., Bianchin M., Wichert-Ana L., Araújo D., Alexandre V., Jr., Santos A. C., Machado H. R., Carlotti C. G., Jr., Brentani R. R., Martins V. R., Sakamoto A. C. (2004) Neurology 63, 557–560 [DOI] [PubMed] [Google Scholar]
- 57.Klannemark M., Suurinkeroinen L., Orho-Melander M., Groop L., Taskinen M. R. (2000) Diabet. Med. 17, 599–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.