Abstract
Motivation: One of the most deadly cancer diagnoses is the carcinoma of unknown primary origin. Without the knowledge of the site of origin, treatment regimens are limited in their specificity and result in high mortality rates. Though supervised classification methods have been developed to predict the site of origin based on gene expression data, they require large numbers of previously classified tumors for training, in part because they do not account for sample heterogeneity, which limits their application to well-studied cancers.
Results: We present ISOLATE, a new statistical method that simultaneously predicts the primary site of origin of cancers and addresses sample heterogeneity, while taking advantage of new high-throughput sequencing technology that promises to bring higher accuracy and reproducibility to gene expression profiling experiments. ISOLATE makes predictions de novo, without having seen any training expression profiles of cancers with identified origin. Compared with previous methods, ISOLATE is able to predict the primary site of origin, de-convolve and remove the effect of sample heterogeneity and identify differentially expressed genes with higher accuracy, across both synthetic and clinical datasets. Methods such as ISOLATE are invaluable tools for clinicians faced with carcinomas of unknown primary origin.
Availability: ISOLATE is available for download at: http://morrislab.med.utoronto.ca/software
Contact: gerald.quon@utoronto.ca; quaid.morris@utoronto.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
While most cancerous tumors present at their cancer site of origin (CSO), ∼4% of all new tumors do not (American Cancer Society, 2001). Without knowledge of this site, treatment regimens are highly limited in their specificity and result in high mortality rates (Blaszyk et al., 2003; Shaw et al., 2007). In an effort to identify CSO, patients routinely undergo extensive clinical examination, radiology and histoimmunological testing (Hainsworth and Greco, 1993). However, these drastic interventions fail to identify the site of origin more than half of the time (Blaszyk et al., 2003).
Gene expression profiling provides a precise, high-resolution molecular fingerprint of a tumor that also offers insight into the underlying transcriptional activity that gave rise to its aberrant behavior (Liotta and Petricoin, 2000). To date, a number of supervised classification methods have been used to categorize tumors according to their site of origin based on gene expression profiles, including support vector machines (Ramaswamy et al., 2001; Su et al., 2001; Tothill et al., 2005), decision trees (Dennis et al., 2005; Shedden et al., 2003), K-nearest neighbors (Bridgewater et al., 2008; Giordano et al., 2001; Horlings et al., 2008), neural networks (Bloom et al., 2004) and others (Buckhaults et al., 2003; Dennis et al., 2002; Varadhachary et al., 2008).
These studies all share a similar three-step strategy: transcriptional profiles of many tumors with known sites of origin are used to identify individual marker genes whose expression levels discriminate cancers of different origin; then the expression levels of these marker genes in each tumor are used to train a classifier that is subsequently used to classify new tumors not previously labeled with a site of origin.
These microarray-based models have shown great diagnostic potential for identifying the site of origin of patients with carcinomas of unknown primary origin: accuracies of >80% were commonly reported for some types of carcinomas. However, because these methods are supervised classification methods, they require large amounts of transcriptionally profiled tumors with identified origin upon which to train. While this data may be available for mature tumors from common sites of origin, there are many less well-characterized cancers or poorly differentiated tumors that often have very little or no data available upon which to train. Reported prediction accuracy on these underrepresented tumors is little better than random performance (Ramaswamy et al., 2001; Shedden et al., 2003; Su et al., 2001). Classifier performance also depends critically on the CSO-specific marker genes identified in the preprocessing step, making downstream analysis highly sensitive to the marker set (Tothill et al., 2005). Unsupervised methods that neither rely on previously collected training data nor prescreen for marker genes are therefore of high value. However, to the best of our knowledge, such methods are not currently available.
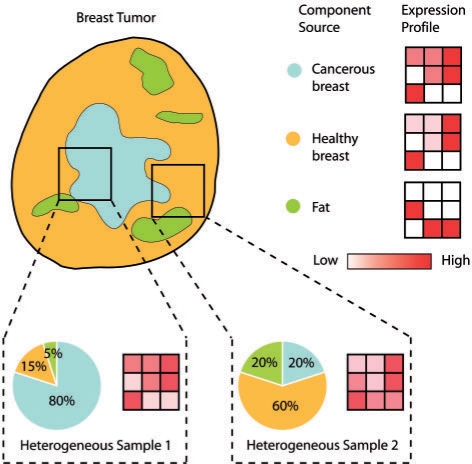
It is often of interest to not only identify the site of origin, but also to identify the genes differentially expressed in the cancer cells with respect to the site of origin. Ideally, the tumor expression profile can be directly compared with that of the CSO to identify those differentially expressed genes. However, tumors are not homogeneous masses of cancer cells, but are mixtures of cell populations with varying levels of heterogeneity, not only between tumors of the same cancer but also even across the samples from the same tumor (Dennis et al., 2005). Contaminating cell populations can include surrounding healthy tissues (such as the site of origin or the local site of a metastatic tumor) and supporting stroma (Masters and Lakhani, 2000). Sample heterogeneity contributes significantly to the large diversity of expression profiles observed even from similar tumor samples, and in many cases contaminating non-cancer cells can dominate the expression profile (Golub et al., 1999; Liotta and Petricoin, 2000; Reya et al., 2001), as illustrated in Figure 1. While methods exist for de-convolving heterogeneous expression profiles into their individual component profiles and inferring the so-called mixing proportions (also known as coefficients), based on techniques like Independent Components Analysis (ICA) (Hyvarinen, 2001; Lahdesmaki et al., 2005; Venet et al., 2001), they have been developed independently of the models for identifying CSO and therefore are currently applied as a preprocessing step.
Fig. 1.
Multiple samples taken from even the same tumor can be composed of different mixing proportions of component sources, giving rise to significantly different tumor expression profiles compared with the expression profile of the homogeneous cancer cell population. Methods for de-convolving sample heterogeneity and removing the contributions of non-cancerous cell populations to the measured expression profile aim to re-construct the expression profile of the homogeneous cancer cell population.
The advent and rapidly decreasing cost of high-throughput sequencing (HTS) methods for expression profiling promises much higher reproducibility and a wider dynamic range of detectable gene expression than microarrays (Marioni et al., 2008; Mortazavi et al., 2008). HTS methods are quickly becoming feasible for highly accurate characterization of the transcriptome profile of tumors. However, the digital counting of sequence tags in HTS methods leads to a different observation of noise process compared with the analog measurement of probe intensity in microarrays. This change requires an update to the statistical models used to analyze these data.
In this article, we present ISOLATE, a model for the Identification of Sites of Origin by LATEnt variables. ISOLATE is the first method that simultaneously identifies sites of origin in an unsupervised fashion and addresses sample heterogeneity using HTS cancer expression profiling. ISOLATE is designed to achieve three goals: identification of the site of origin from a set of profiled candidate sites, de-convolution of heterogeneous expression profiles into their individual components and identification of differentially expressed genes. We demonstrate on both synthetic and clinical datasets that ISOLATE achieves higher accuracy on all of these goals than a similar ICA-based unsupervised strategy that mirrors existing tools. The high accuracy levels achieved by ISOLATE demonstrate the feasibility of unsupervised methods for complementing traditional immunohistological and supervised classification models for identifying the site of origin of and characterizing tumors from carcinomas of unknown primary origin.
2 APPROACH OVERVIEW
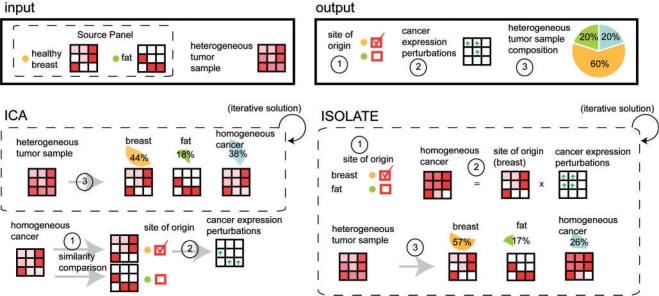
ISOLATE is designed to achieve three goals, illustrated in Figure 2: identification of the CSO, identification of the differentially expressed genes in the homogeneous cancer cell population and characterization of the cellular composition of each heterogeneous tumor sample (by estimation of the mixing proportions of their component cell populations). We compared ISOLATE with an ICA-based strategy that can be applied using existing tools to address the three challenges. In this study, we use Latent Dirichlet Allocation (LDA) (Blei et al., 2003) to implement the ICA strategy. LDA is an equivalent model to ICA (Shashanka et al., 2008) but with an observation noise model more appropriate for digital HTS data.
Fig. 2.
Outline of the ISOLATE and LDA methods for de-convolving cancer gene expression data and identifying the site of origin. The input to each method consists of the expression profile of a heterogeneous tumor sample(s), as well as the Source Panel representing previously profiled cell populations that may act either as contaminants or as candidate sites of origin. Each method performs three tasks: identification of the site of origin, identification of those genes differentially expressed in the cancer cells and characterization of the cellular composition of each heterogeneous sample by estimating the mixing proportions of each component cell population. The ICA-based strategy operates serially by first de-convolving the heterogeneous sample without constraining the cancer profile to be derived from the Source Panel, then predicts the site of origin and differentially expressed genes. This is in stark contrast to ISOLATE, which solves all three problems cooperatively.
Both ISOLATE and LDA model the expression profile of a heterogeneous tumor sample as a weighted mixture of expression profiles of ‘source’ cell populations (representing candidate sites of origin or contaminants), all of which have been previously characterized except for the homogeneous cancer cell population. The set of source cell populations are herein called the ‘Source Panel’. Candidate sites of origin and potential contaminating cells can be treated similarly in the context of LDA and ISOLATE and are hence both referred to as sources. ISOLATE differs from LDA in that it explicitly models the similarities in expression profile between the cancer cell population and the site of origin by representing the homogeneous cancer expression profile as a sparsely perturbed version of the profile of its site of origin. Tumor cells display functional, developmental and morphological similarities to their site of origin (Lobo et al., 2007; Sell and Pierce, 1994). This similarity is also reflected at the gene expression level, both between the primary tumor and the site of origin (Khan et al., 2001; Ross et al., 2000), and between the primary and metastatic tumor (D'Arrigo et al., 2005; Weigelt et al., 2003). By explicitly modeling the cancer cell expression profile as a perturbation of the site of origin profile, our model is a more precise representation of cancer that naturally leads to the identification of differentially expressed genes as those whose expression was perturbed to produce the tumor expression profile. ISOLATE then uses the estimate of the homogeneous cancer profile in conjunction with the Source Panel to decompose each tumor sample.
A key feature of ISOLATE is that it recognizes the interdependence of the solutions of all three goals and iteratively solves all of them simultaneously. In contrast, the ICA strategy first iteratively decomposes the tumor samples while estimating the profile of the homogeneous cancer cell population. Then it compares the estimated homogeneous cancer profile to the Source Panel and identifies the parent site as the most similar profile, and finally identifies differentially expressed genes by comparing the estimated cancer profile to that of the identified site of origin. This makes the ICA-based strategy for identifying the site of origin sensitive to imperfect de-convolution of the tumor expression profiles and often leads to misidentification of the site of origin as the surrounding tissue.
The following sections describe how we generated the synthetic datasets and collected and processed the clinical datasets that we used to test our model. We also describe statistical inference with ISOLATE and LDA.
3 METHODS
3.1 Synthetic data collection
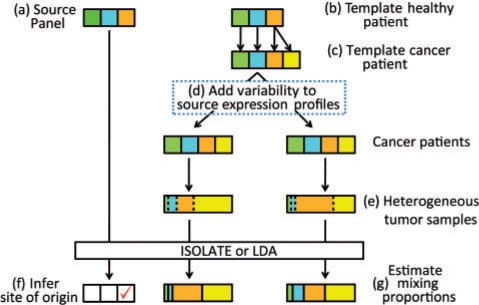
We measured the performance of ISOLATE on a comprehensive set of synthetic data for which the correct answer is known. Our strategy for generating data is shown in Figure 3 and summarized below, with more details provided in following sections. Each experiment is defined by five parameters: the number of genes whose expression is perturbed in the cancer cells, their multiplicative perturbation factor, the number of heterogeneous tumor samples profiled, the number of sources in the Source Panel and the level of biological variability of the expression profiles of the same sources between different cancer patients. First, using human kidney and liver data from Marioni et al. (2008), we generate expression profiles for each source, both for the (a) training profiles that make up the Source Panel, and (b) for a template healthy patient. From the template healthy patient, (c) we randomly select one component source as the site of origin, from which we perturb the expression profile to construct a cancer cell expression profile. The original template of healthy source expression profiles together with the cancer cell expression profile make up a template cancer patient, from which we (d) generate one or more unique cancer patients by adding variability to the template cancer patient independently for each cancer patient. (e) One heterogeneous tumor sample is generated from each individual using a unique set of mixing proportions to combine the source profiles of the cancer patient. Finally, we use the Source Panel and the heterogeneous tumor samples as input into the LDA and ISOLATE models, to (f) identify the CSO, (g) de-convolve the heterogeneity of each tumor sample, and identify differentially expressed genes.
Fig. 3.
Overall experimental strategy for generating the heterogeneous tumor samples from three sources (i.e. candidate sites of origin) to input into the LDA and ISOLATE models. The sources color-matched between the Source Panel and the template healthy patient differ only by technical variability in their expression profiles. Yellow represents cancer cells, while orange represents the site of origin.
3.1.1 Dataset
Human liver and kidney transcriptome profiling data from a single human male was obtained from Marioni et al. (2008) who sequenced each tissue seven times, split across two runs of an Illumina Genome Analyzer and at two concentrations, 1.5 pM and 3 pM. All reads were mapped to the genome using the Illumina ELAND algorithm, and only uniquely mapped reads were retained. A gene copy number is computed by counting the number of reads mapped to each known transcript, then computing the median number of copies for each gene over all of its respective transcripts. We discarded all genes for which there was not even one copy in all of the runs of both tissues, leaving 13 061 genes. Gene abundances (also called the expression profile) were computed from gene copy numbers by dividing each copy number by the sum of all gene copy numbers.
3.1.2 Generating a new source expression profile
We first applied a differential expression test (Lu et al., 2005) to identify the top 40% of all genes that were most likely to be constitutively expressed across all of the kidney and liver datasets and deemed these to be candidate house-keeping genes (Zhu et al., 2008). We then randomly selected two runs from either the kidney or liver datasets, and permuted the expression levels of their non-house-keeping genes randomly in the same order. One run is used in the Source Panel (Fig. 3a) as previously profiled abundances in LDA and ISOLATE, while the other is used in the template healthy individual (Fig. 3b).
3.1.3 Generating a cancer cell expression profile
To generate a cancer cell expression profile given the expression profile of the site of origin, the number of genes to perturb and their perturbation factor, we first randomly selected the set of genes to become differentially expressed, then randomly perturbed the abundances of each gene in that set either up or down (with equal probability), then renormalize the abundances to sum to 1 to make gene abundances correspond to parameters of a multinomial distribution. This cancer expression profile and the healthy tissue profiles in the template healthy individual combine to make the sources in the template cancer patient (Fig. 3c).
3.1.4 Generating a cancer patient from the template cancer patient
We use the template cancer patient to obtain a cancer patient profile by adding biological variability to each source expression profile. We represent biological variability by resampling the expression levels of a fraction of genes from the entire set of expression levels observed in that source's original expression profile. The expression profile is subsequently rescaled to sum to 1.
3.1.5 Generating a heterogeneous tumor sample from a cancer patient
We first determine what proportion of each source will compose the tumor sample, then we generate the sequence reads that are observed in the sample. For the tumor sample d, the mixing proportions of the sources θd are drawn from a Dirichlet distribution with parameters α = {αsc, αs1, αs2,…, αsS}, where sc indicates the cancer source. In our experiments, for all non-cancer sources i ≠ c, αsi = 1, and αsc = 3 by default. Larger values of αsc will result in tumor samples containing larger proportions of cancer cells. Once the mixing proportions θd are generated, for each transcript read to generate, we randomly select a source using the mixing proportions θd, then randomly select a transcript from which to generate a read using the multinomial distribution specified by the expression profile of the chosen source. Each tumor sample was generated with 1 675 078 reads, the average number of reads collected per experiment in Marioni et al. (2008), though the results were not sensitive to the total number of reads generated per tumor sample (data not shown).
3.2 Clinical data processing
Both the ISOLATE and LDA strategies require a fully profiled Source Panel and heterogeneous tumor samples, but owing to the current lack of such data available, we took advantage of the vast quantities of microarray data available and chose to digitize such datasets to make them compatible with our model. We downloaded a total of 93 tumor expression profiles from Su et al. (2001), consisting of 10 kidney, 6 liver, 24 lung, 23 ovary, 6 pancreatic and 24 prostate-originating tumors collected using Affymetrix U95a GeneChip arrays. Following the procedure of Su et al. (2001), for each tumor sample, raw intensity values were thresholded at 20. Mappings from the probe identifier to Ensembl gene identifiers were downloaded from the Affymetrix web site, and multiple probes matching the same Ensembl gene identifier were averaged together to produce a single measurement for each gene. The resulting array intensities were rounded to the nearest integer and treated as transcript counts from a HTS experiment.
As a Source Panel, we downloaded a separate set of microarray data collected using Affymetrix Human Genome U133A arrays (Su et al., 2004), giving us a healthy profile version of those same six tissues. Intensities for replicate array measurements were averaged together for each respective tissue, and using the provided annotation files, each probe was mapped to its respective Ensembl gene identifier, and multiple probes matching the same Ensembl gene identifier were averaged together.
The total set of common genes profiled in the Source Panel and the tumor profiles were 8667 genes. For these 8667 genes, their raw averaged intensities in each source of the Source Panel were divided by the total intensity measured to produce a proper multinomial distribution over the profiled genes.
3.3 Inference with the LDA model
The input to the LDA model (Blei et al., 2003), for both the synthetic and clinical datasets, consists of the expression profiles over all the genes in each source. Each profile is represented by a vector from the set {βs}s=1S that contains one vector for each of the S sources from the Source Panel. Also input to the LDA model are D sets of reads {td,n}d=1D originating from transcriptome profiling experiments of D heterogeneous tumor samples that each generate Nd reads. LDA estimates the expression profile βsc of all genes in the cancer source sc and performs de-convolution by inferring hidden variables {zd,n} (one for each read td,n) that indicate from which of the S + 1 sources (S from the Source Panel, and 1 from the cancer source) the transcript most likely originates. In doing so, LDA estimates the fraction of each cancer sample d (the mixing proportions), θd,s, coming from each of the S + 1 sources. The full model is specified below:
| (1) |
| (2) |
| (3) |
The model was trained using the same variational Expectation Maximization (EM) framework used in Blei et al. (2003) with 100 iterations, and rerun S times with random parameter initializations. The initialization that resulted in the highest log likelihood of the data is chosen. The model parameters estimated include α, θd for all tumor samples d, and cancer abundances βsc. Using the output of LDA, we predict the site of origin by choosing the source (from the Source Panel) whose expression profile has the least Kullback–Liebler divergence from the estimated cancer expression profile βsc. To rank genes in order of differential expression, we applied a two-class differential expression test (Lu et al., 2005) to compare the expression profile of the predicted site of origin against the set of reads the cancer cells are responsible for in each tumor sample (zd,n = sc), and sorted the genes based on the resulting P-value. Lastly, the mixing proportions (heterogeneity) of each sample d are estimated directly from the learned parameters of the model, θd.
3.4 Inference with the ISOLATE model
ISOLATE maintains the same probabilistic framework as LDA [Equations (1– 3)], but introduces the following key constraints on the learned parameters βsc, where β:,g is a column vector of abundances of gene g across all S non-cancer sources:
| (4) |
| (5) |
| (6) |
ω is a (S × 1)-dimensional parameter, where ωs = 1 denotes that source s is the site of origin. ρg is the estimated perturbation (multiplicative) factor that describes how much the cancer cells perturb the expression of gene g relative to the site of origin described by ω. Since we expect many genes to maintain similar expression levels to that of the CSO, we put a Gamma prior on ρg [Equation (5)], with mean E[ρg] = 1 to emphasize that we expect many genes to not have perturbed expression. ISOLATE uses the same variational EM framework as LDA (Blei et al., 2003) with 100 iterations, and rerun using S different initializations to test different candidate CSO. There is exactly one initialization per source s where the value ωs is set to 1, and the remaining entries set to 0. The initialization that resulted in the highest log likelihood of the data is chosen. To rank genes in order of differential expression, we sorted the genes based on the distance of ρg from the value 1. That is, the farther ρg is from 1, the more perturbed its abundance is from that of the site of origin. Finally, mixing proportions of each sample d is estimated directly from the learned parameters of the model, θd.
3.5 Performance metrics
The error rate in identifying the primary site of origin is the fraction of experiments in which the CSO was incorrectly identified. For the synthetic datasets, the reported error is averaged over the 20 datasets generated for each specific setting of the parameters. The model heterogeneity error is computed by averaging, over all tumor samples, the mean absolute error of the mixing proportions θd of the cancer cells and the true site of origin. We only measure the error with respect to these two sources because we found that almost all of the error in the mixing proportion estimates is from these two sources. Finally, we assess the error of the identification of differentially expressed genes for the synthetic datasets by using the ranks of the genes (in order of differential expression as defined by each model) and our knowledge of which genes are truly differentially expressed to compute an area under the receiver operator curve (ROC), where larger values correspond to higher accuracy. Error is computed as [1 − (Area under ROC)].
4 RESULTS
4.1 Synthetic datasets
We have evaluated the relative performance of ISOLATE and LDA as a function of realistic parameter settings to demonstrate their robustness to different conditions. We also compared naiveLu, a simple method for identifying differentially expressed genes by simply applying a differential expression test directly without accounting for sample heterogeneity. We do these comparisons because of the dearth of clinical data and the difficulties associated with defining gold standards therein. We are also able to query a larger variety of experimental conditions. In the absence of analytical estimates of performance, which are likely impossible due to the complexity of our models, these comparisons provide the best support for our claims of improved performance over LDA.
We varied the following parameters: the number of differentially expressed genes, the perturbation factor by which their expression levels are differentially expressed in cancer, the number of heterogeneous tumor samples, the number of sources in the Source Panel, and the (biological) variability between our profiled Source Panel and the corresponding profiles used to generate the tumor samples (see Section 3). This variability represents the expected differences between the normal source profiles in our Source Panel, which will likely come from different individuals, and the corresponding source profiles for the patient from which the tumor sample is drawn. Biological variability, which could represent either biological variation or technical noise, is a key parameter because it limits our ability to detect differentially expressed genes, as seen below. In the following, we vary only a single parameter from the default; the default parameters we use are 100 perturbed genes, 3 tumor samples, 10 sources, a perturbation factor of two, perturbation scale prior κ = 10 [see Equation (5)], and a biological variability of 0.16 (16%), which empirically leads to ∼14% of genes differentially expressed, as measured by Lu et al. (2005). This level of differential expression between simulated individuals is similar to reported variation between unrelated individuals (Sharma et al., 2005).
4.1.1 Identification of differentially expressed genes
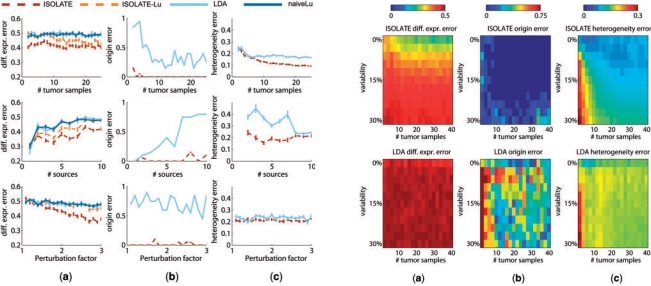
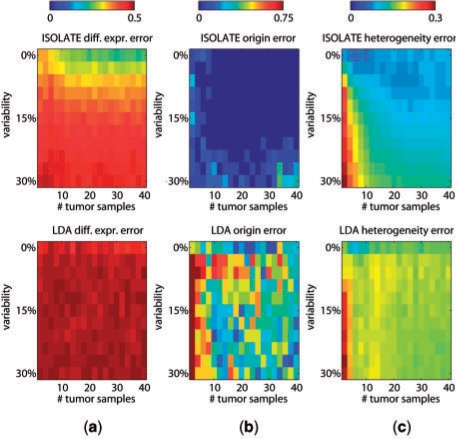
One of the principal objectives of identifying CSO is to identify genes that are differentially expressed in the cancer cell population with respect to healthy cells of the site of origin. We tested each models' ability to identify the differentially expressed genes, defined as those genes whose expressions were perturbed to differentiate the cancer source from the site of origin source, and measured the performance by the area under the ROC curve (see Section 3). Figure 4a demonstrates that ISOLATE consistently achieves higher accuracy at identifying differentially expressed genes than LDA across all three parameters at almost all settings. Surprisingly, the performance of both LDA and ISOLATE seem to stay constant despite increasing the number of tumor samples available. Figure 5a illustrates that beyond a variability level of 15%, increasing the amount of data does not improve ISOLATE performance. Because 15% is near the average variability between unrelated individuals (Sharma et al, 2005), this result suggests that ISOLATE's identification of differentially expressed genes can be improved by analyzing multiple tumor samples from the same individual but not necessarily by analyzing multiple samples from different individuals. Both ISOLATE and LDA improve performance as the perturbation factor increases—a direct result of its increasing differentiation from the site of origin source and hence easier de-convolution of sample heterogeneity—though ISOLATE improves at a much faster rate. The performance of naiveLu, which does not address heterogeneity, illustrates that de-convolution clearly improves the identification of differentially expressed genes. The ISOLATE performance gain over LDA is not just simply due to a difference in the specific method that ISOLATE uses to compute differential expression: we computed differential expression using the same method as for LDA (ISOLATE-Lu) and see that its performance is still better than LDA in many cases.
Fig. 4.
Performance of ISOLATE and LDA on synthetic datasets. Each row represents a different parameter tuned: the number of heterogeneous tumor samples, the number of sources (non-cancer) in the Source Panel and the perturbation factor of the differentially expressed genes (manipulating the number of perturbed genes within the range of 50–500 genes did not result in changes in performance to either model and are not shown). Each column represents a different performance metric applied to each dataset. (a) Differential expression error, (b) origin error and (c) heterogeneity error are as defined in Section 3. Two additional models are plotted in (a): ISOLATE-Lu is the performance achieved when applying the same method as ICA for identifying differentially expressed genes (Lu et al., 2005) to the output of the ISOLATE model, and naiveLu is the performance achieved when directly comparing the heterogeneous tumor expression profiles to the site of origin to identify differentially expressed genes, without accounting for sample heterogeneity.
Fig. 5.
Performance of ISOLATE and LDA on synthetic datasets under different biological variability conditions. Each column represents a different performance metric, and each row a different model (top, ISOLATE; bottom, LDA). Here, we co-vary the biological variability added to each tumor sample independently, and the number of tumor samples made available to each model. The performance metrics of (a) differential expression error, (b) origin error and (c) heterogeneity error are as defined in Section 3.
4.1.2 Identification of CSO
Figure 4b compares LDA and ISOLATE based on how often they are able to correctly identify the site of origin. ISOLATE consistently outperforms LDA across all datasets. Most importantly, while ISOLATE is robust against the number of sources in the Source Panel, the performance of LDA diminishes rapidly after six sources. This makes LDA and other ICA-like techniques impractical for considering many potential candidates for the CSO, an important feature given the potentially large set of candidate CSO to query. The difference between ISOLATE and LDA also illustrates the improvement in CSO identification, sometimes as staggeringly as 70%, achieved by solving for both cell population mixture coefficients and CSO simultaneously within the same framework. From Figure 5b, we see that ISOLATE achieves high accuracy at identifying CSO under even high variability conditions, while LDA accuracy varies quite widely even under low variability conditions. ISOLATE is therefore able to capture the underlying signal of the site of origin even despite large amounts of noise in the expression profiles. Most importantly, even when looking at very small sets of tumor samples, ISOLATE performs as well as it does with many more samples, an important feature given the cost of profiling tumors in a diagnostic setting.
4.1.3 Correction of sample heterogeneity
Figure 4c illustrates that when considering Source Panels containing fewer than 10 profiles, ISOLATE achieves better de-convolution of heterogeneity than LDA. However, their performance is nearly identical regardless of the perturbation factor applied to the differentially expressed genes, suggesting that the amount of tumor sample data is far more important for de-convolution than the difference in expression profiles of the cancer cells and the site of origin. As expected, as the number of tumor samples increases, both LDA and ISOLATE increase in performance. Figure 5c illustrates that ISOLATE achieves better accuracy for the same number of data points and level of variability, although it takes more samples for a given level of biological variability than LDA to achieve its maximum performance. Most of the performance loss in LDA appears to be due to confusion of the contributing expression signatures from the cancer cells and the site of origin source (Fig. 6 illustrates an example). This is a problem that ISOLATE is able to mitigate because of the constraints it places on the learned cancer expression profile.
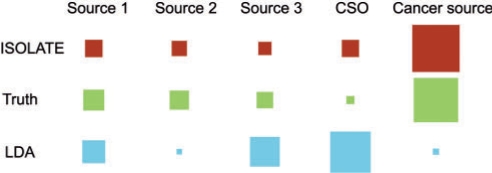
Fig. 6.
An example of heterogeneity error for a single tumor sample. Here, we illustrate a representative tumor sample whose mixing proportions of component sources were estimated by ISOLATE (top row) and LDA (bottom row), compared with the actual values (middle row). Each of the four columns on the left represent a source from the Source Panel (Sources 1–3, as well as the site of origin source), while the right-most fifth column represents the cancer cells. The area of each square is proportional to the fraction of the sample composed of that particular source. Whereas ISOLATE estimated mixing proportions fairly close to the truth, the LDA estimate of the CSO and cancer sources were quite erroneous.
4.2 Clinical dataset
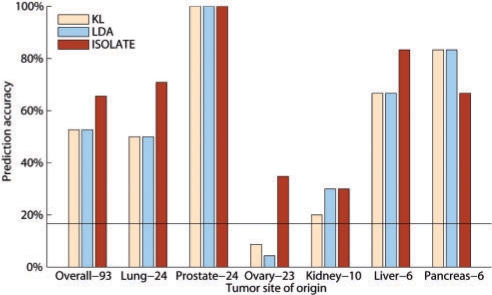
We used ISOLATE and LDA to predict the site of origin of 93 tumor samples from Su et al. (2001). Heterogeneity and differential expression error could not be measured on these datasets as the true values are not known. Each tumor, regardless of its site of origin, is predicted independently of all other tumors, to reproduce clinical diagnostic conditions. As a benchmark besides LDA, we constructed a predictor that chooses the source from the Source Panel whose Kullback-Leibler (KL)-divergence is least with respect to the tumor sample's expression profile (KL). To set the perturbation scaling prior κ of Equation (5), we tried several values of κ (100, 101, 102, 103, 104, 105), and performed 2-fold cross-validation by choosing the κ that maximized performance of half the data, in order to predict the other half of the dataset, and vice versa. The optimal value of κ was 105 for both halves of multiple splits of the data, and so was used to generate the results shown in Figure 7. Over the entire dataset of 93 tumor samples, ISOLATE achieves the highest performance of 65.59% accuracy, compared with 52.69% of both LDA and the KL measure. On a class-by-class basis, ISOLATE ties or performs better than LDA and ISOLATE in the larger classes, only performing worse when predicting tumors originating from the pancreas, the smallest class. Note that though previously reported performance of supervised classification methods is higher on some of these cancer types, ISOLATE achieves the observed performance considering each tumor separately without reference to any of the other tumor samples and without any training, mirroring clinical settings for the CSO identification of tumors underrepresented among previously profiled samples.
Fig. 7.
The performance of ISOLATE, LDA and another Kullback-Leibler (KL) divergence-based measure on the clinical dataset of 93 tumor samples. Each sample is predicted independently of all other samples in the dataset. The number of samples in each class is shown beside the class name, and classes are in decreasing order of size, from left to right, with the overall performance shown in the leftmost column. The black line shows random performance.
5 DISCUSSION
We have developed ISOLATE to provide a molecular diagnostic tool to aid in identifying the site of origin of tumors of poorly characterized cancers, situations in which classical supervised models perform poorly. ISOLATE simultaneously de-convolves tumor expression profiles, and identifies the CSO and genes differentially expressed in the cancer cells, three tasks that were previously solved independently. Our experiments detail the performance of ISOLATE under a wide range of realistic experimental conditions for synthetic and digitized clinical microarray data, showing that solving all three tasks simultaneously leads to greater predictive performance than solving them individually.
ISOLATE, unlike previous methods for classifying cancers of unknown primary origin, is an unsupervised classification algorithm, which provides it with several inherent advantages. It does not require a large training set of tumors of known primary origin, and in our clinical validation we only use data from a single tumor, a particularly important feature given the difficulty and cost of procuring many high-quality tumor samples in a diagnostic setting. Because it is an unsupervised algorithm, ISOLATE's performance is also less sensitive to the number of candidate sites of origin, as suggested by our synthetic data validation, in contrast to supervised learning methods that have difficulty with more than 10 classes (Su et al., 2001). By its construction, ISOLATE is also less prone to overfitting than supervised learning algorithms, and as such, does not require a prescreening stage to identify marker genes upon which cancers could be discriminated.
Despite our use of microarray data for our clinical validation, we recommend that ISOLATE be used exclusively with HTS expression profiles, which we believe support more accurate tumor diagnosis. HTS methods promise a substantial reduction in sample-to-sample variability, which our synthetic data-based validation shows limits the accuracy. Also, because HTS measurements are not probe-based, they are both less platform-specific, allowing easier integration of data from multiple labs, and less sensitive to polymorphisms in transcript sequence that are common in highly polymorphic cancer genomes. For these reasons, we have tailored ISOLATE's statistical model for HTS gene expression data.
The successful application of ISOLATE or other expression-based models will depend on the availability of expression profiles for a wide range of human tissues, in order to consider them as potential sites of origin. With the costs of high-throughput expression profiling dropping quickly and the number of studies using these technologies to profile tumors increases (Jones et al., 2008; Parsons et al., 2008), soon it will be practical to collect a compendium of expression data from many of the individual tissues of humans along with multiple tumor samples, as is currently available for microarrays (see, e.g. Su et al. (2004).
Molecular-based diagnostic tools for identifying cancer sites of origin represent an important class of tools that can potentially facilitate faster, more accurate diagnoses leading to the successful identification of primary sites. ISOLATE will be an invaluable tool for exploring new, uncharacterized cancers of unknown primary origin for which little expression data are available, or clinically ambiguous samples for which more traditional models cannot classify with high accuracy.
ACKNOWLEDGEMENTS
The authors thank Yoav Gilad for providing the processed read data from Marioni et al. (2008).
Funding: CFI/ORF equipment grants (to Q.M.); NSERC operating grant (to Q.M.); NSERC PGS Doctoral Scholarship (to G.Q., in part).
Conflict of Interest: none declared.
REFERENCES
- American Cancer Society. ACS Cancer Facts and Figures. 2001 ACS. Available at http://www.cancer.org/docroot/STT/content/STT_1x_2001_Facts_and_Figures.pdf.asp (last accessed date June 28, 2009) [Google Scholar]
- Bittner M, et al. Molecular classification of cutaneous malignant melaoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- Blaszyk H, et al. Cancer of unknown primary: clinicopathologic correlations. APMIS. 2003;111:1089–1094. doi: 10.1111/j.1600-0463.2003.apm1111203.x. [DOI] [PubMed] [Google Scholar]
- Blei DM, et al. Latent Dirichlet allocation. J. Mach. Learn. Res. 2003;3:993–1022. [Google Scholar]
- Bloom G, et al. Multi-platform, multi-site, microarray-based human tumor classification. Am. J. Pathol. 2004;164:9–16. doi: 10.1016/S0002-9440(10)63090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgewater J, et al. Gene expression profiling may improve diagnosis in patients with carcinoma of unknown primary. Br. J. Cancer. 2008;98:1425–1430. doi: 10.1038/sj.bjc.6604315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckhaults P, et al. Identifying tumor origin using a gene expression-based classification map. Cancer Res. 2003;63:4144–4149. [PubMed] [Google Scholar]
- D'Arrigo A, et al. Metastatic transcriptional pattern revealed by gene expression profiling in primary colorectal carcinoma. Int. J. Cancer. 2005;115:256–262. doi: 10.1002/ijc.20883. [DOI] [PubMed] [Google Scholar]
- Dennis JL, et al. Identification from public data of molecular markers of adenocarcinoma characteristic of the site of origin. Cancer Res. 2002;62:5999–6005. [PubMed] [Google Scholar]
- Dennis JL, et al. Markers of adenocarcinoma characteristic of the site of origin: development of a diagnostic algorithm. Clin. Cancer Res. 2005;11:3766–3772. doi: 10.1158/1078-0432.CCR-04-2236. [DOI] [PubMed] [Google Scholar]
- Giordano TJ, et al. Organ-specific molecular classification of primary lung, colon, and ovarian adenocarcinomas using gene expression profiles. Am. J. Pathol. 2001;159:1231–1238. doi: 10.1016/S0002-9440(10)62509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Hainsworth JD, Greco FA. Treatment of patients with cancer of an unknown primary site. N. Engl. J. Med. 1993;329:257–263. doi: 10.1056/NEJM199307223290407. [DOI] [PubMed] [Google Scholar]
- Horlings HM, et al. Gene expression profiling to identify the histogenetic origin of metastatic adenocarcinomas of unknown primary. J. Clin. Oncol. 2008;26:4435–4441. doi: 10.1200/JCO.2007.14.6969. [DOI] [PubMed] [Google Scholar]
- Hyvarinen A. Independent Component Analysis. John Wiley & Sons, Inc.; 2001. [Google Scholar]
- Jones S, et al. Core signalling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008 doi: 10.1126/science.1164368. [Epub ahead of print, doi: 10.1126/science.1164368, September 4, 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat. Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahdesmaki H, et al. In silico microdissection of microarray data from heterogeneous cell populations. BMC Bioinformatics. 2005;6:54. doi: 10.1186/1471-2105-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta L, Petricoin E. Molecular profiling of human cancer. Nat. Rev. Genet. 2000;1:48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- Lobo NA, et al. The biology of cancer stem cells. Annu. Rev. Dev. Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- Lu J, et al. Identifying differential expression in multiple SAGE libraries: an overdispersed log-linear model approach. BMC Bioinformatics. 2005;6:165. doi: 10.1186/1471-2105-6-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni J, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters JRW, Lakhani SR. How diagnosis with microarrays can help cancer patients. Nature. 2000;404:921. doi: 10.1038/35010139. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008 doi: 10.1126/science.1164382. [Epub ahead of print, doi: 10.1126/science.1164382, September 4, 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl Acad. Sci. USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Sell S, Pierce GB. Maturation arrest of stem cell differentiation is a common pathway for the cellular origin of teratocarcinomas and epithelial cancers. Lab. Invest. 1994;70:6–22. [PubMed] [Google Scholar]
- Sharma A, et al. Assessing natural variations in gene expression in humans by comparing with monozygotic twins using microarrays. Physiol. Genomics. 2005;21:117–123. doi: 10.1152/physiolgenomics.00228.2003. [DOI] [PubMed] [Google Scholar]
- Shashanka M.VS, et al. Probabilistic latent variable models as non-negative factorizations. Comput. Intell. Neurosci. 2008 doi: 10.1155/2008/947438. [Epub ahead of print, doi: 10.1155/2008/947438, May 11, 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PH, et al. A clinical review of the investigation and management of carcinoma of unknown primary in a single cancer network. Clin. Oncol. (R. Coll. Radiol.) 2007;19:87–95. doi: 10.1016/j.clon.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Shedden KA, et al. Accurate molecular classification of human cancers based on gene expression using a simple classifier with a pathological tree-based framework. Am. J. Pathol. 2003;163:1985–1995. doi: 10.1016/S0002-9440(10)63557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, et al. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothill RW, et al. An expression-based site of origin diagnostic method designed for clinical application to cancer of unknown origin. Cancer Res. 2005;65:4031–4040. doi: 10.1158/0008-5472.CAN-04-3617. [DOI] [PubMed] [Google Scholar]
- Varadhachary GR, et al. Molecular profiling of carcinoma of unknown primary and correlation with clinical evaluation. J. Clin. Oncol. 2008;26:4442–4448. doi: 10.1200/JCO.2007.14.4378. [DOI] [PubMed] [Google Scholar]
- Venet D, et al. Separation of samples into their constituents using gene expression data. Bioinformatics. 2001;17:S279–S287. doi: 10.1093/bioinformatics/17.suppl_1.s279. [DOI] [PubMed] [Google Scholar]
- Weigelt B, et al. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc. Natl Acad. Sci. USA. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, et al. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008;9:172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]