Abstract
Opsins are the universal photoreceptor molecules of all visual systems in the animal kingdom. They can change their conformation from a resting state to a signalling state upon light absorption, which activates the G protein, thereby resulting in a signalling cascade that produces physiological responses. This process of capturing a photon and transforming it into a physiological response is known as phototransduction. Recent cloning techniques have revealed the rich and diverse nature of these molecules, found in organisms ranging from jellyfish to humans, functioning in visual and non-visual phototransduction systems and photoisomerases. Here we describe the diversity of these proteins and their role in phototransduction. Then we explore the molecular properties of opsins, by analysing site-directed mutants, strategically designed by phylogenetic comparison. This site-directed mutant approach led us to identify many key features in the evolution of the photoreceptor molecules. In particular, we will discuss the evolution of the counterion, the reduction of agonist binding to the receptor, and the molecular properties that characterize rod opsins apart from cone opsins. We will show how the advances in molecular biology and biophysics have given us insights into how evolution works at the molecular level.
Keywords: opsin, retinal, rods, cones, phototransduction, evolution
1. Introduction
Light is potentially the most important signal for living organisms, as most of the life on Earth ultimately depends on light energy. Many animals utilize light cues to regulate biological processes, including vision and circadian clock regulation. In humans, a substantial part of the brain is dedicated to processing visual information (Wandell et al. 2007). The mechanisms of vision have inspired great scientific interest, and today vision is one of the best characterized biological signal transduction systems.
Rhodopsins are highly diversified proteins that present researchers with the perfect material to study the way evolution takes place at the molecular level. This review takes a look at our understanding of the molecular evolution of opsins and phototransduction. Hopefully, it presents an exciting picture of new discoveries that may aid the understanding of evolution in general.
(a). New technologies and new discoveries
Technological advances in recent years have greatly contributed to our understanding of the molecular mechanism and evolution of vision. Early works on rhodopsin were conducted on protein purified from animal retinas. This approach is still useful and important today because it is the only way to ensure the physiological conditions surrounding the protein, as it has become clear that functional characteristics of a membrane protein are greatly influenced by its membrane environment. However, now we can obtain opsins from cultured cells by transiently expressing their genes. Although this way of obtaining opsin has the underlying difficulty that it will often be slightly different from the native protein because the membrane conditions differ, it gives researchers the opportunity to play with the protein. Traditionally, researchers were limited to exploring the naturally occurring variations of certain proteins. However, recent advances in molecular biology have allowed researchers to selectively modify a protein and study it. Biologists can now induce site-directed mutations in genes, create truncated proteins, fuse different proteins that would help us study them or even combine two parts of different proteins into a new protein (called a chimera). These techniques allow us to study the protein function in detail.
In the year 2000, the first rhodopsin crystal structure was solved, providing the world with the first G-protein-coupled receptor (GPCR) structure (Palczewski et al. 2000). Since then, many crystal structures of opsins have become available revealing the structure of opsins at different states. A crystal structure provides us with an atomic model of the protein, containing the spatial information of the amino acids that constitute the protein. Having an atomic model allows us to identify the interactions between the amino acids, which confer important biochemical and biophysical properties to molecules.
Another crucial advancement is the ever-increasing sequence data from different opsin genes. The ease of cloning and sequencing has allowed researchers to explore the eyes of different organisms, revealing underlying similarities and differences in their molecular machinery. Comparing opsin sequences can reveal residues crucial for the function of the protein. Moreover, phylogeny permits us to explore their phylogenetic relationships, that is to say, it helps us to understand how different genes evolved.
Finally, with genetic engineering techniques, we can create animals with the genes that we desire. We can introduce modified genes to test the effect of these proteins in vivo. With this technique, we can create deduced ancestral sequences that are not seen anymore in the present, and test our hypotheses.
(b). Light sensing and signal transduction
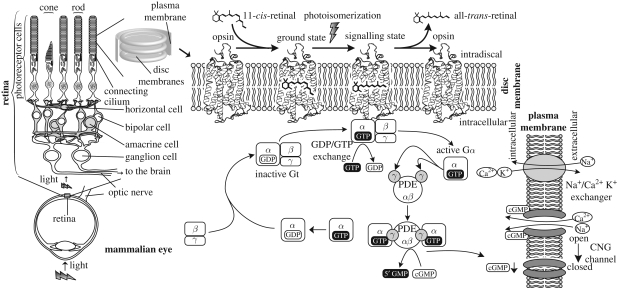
In order to understand how the molecular components that allow us to sense light and transform that light signal into electric signals evolved, we first need to understand the molecular machinery at work. This section presents the basic concepts of light sensing and signal transduction. Figure 1 summarizes the signal flow in the vertebrate phototransduction system.
Figure 1.
A diagram showing the mechanism of phototransduction in mammalian eyes. Light is captured by two specialized morphologically distinct photoreceptor cells derived from neurons: rods and cones that have the same molecular mechanism. Opsins in these cells absorb photons and form a signalling state, which can bind to and activate the G protein by catalysing the exchange of GDP to GTP. The GTP-bound Gα dissociates from Gβγ exposing its active site. Activated Gα binds to its effector, PDE (cyclic nucleotide phosphodiesterase), and activates it. PDE breaks the phosphodiester bond of cGMP producing 5′GMP, and the decrease in the concentration of cGMP causes CNG (cyclic nucleotide gated) channels to close, which creates a hyperpolarization response in the photoreceptor cells. Light-activated rhodopsin is thermally unstable and the chromophore eventually detaches from the opsin. The hyperpolarization of the membrane potential of the photoreceptor cell modulates the release of neurotransmitters to downstream cells. The light signal is transmitted through different cells, finally reaching ganglion cells which form the optic nerve and project to the brain.
(i). Molecular machinery of light sensing
Even though there are a variety of visual systems throughout the animal kingdom, all the visual systems known to date share certain striking similarities in their components. It appears that the underlying molecular machinery of the visual systems is common to all living organisms that possess the ability to see. The first step in vision is light sensing, and rhodopsin is the molecule that absorbs light and thus ‘senses’ light. Light absorption induces changes in the molecular structure of rhodopsin that allow it to activate another molecule, the G protein, which mediates an enzymatic signalling cascade that eventually generates an electrical response in the photoreceptor cell. The signal received from rhodopsin is amplified at this stage since one rhodopsin molecule can activate many G proteins. The downstream signalling cascade depends on the G protein subtype, because different G proteins can act through different pathways.
Throughout this review, we will refer to bovine rhodopsin, as it is the best characterized of all the opsins, and one of the best characterized membrane proteins. The availability of large amounts of rhodopsin obtained from bovine retinas made bovine rhodopsin the photoreceptor molecule of choice for researchers. The amino acid numbering system refers to the amino acid positions of bovine rhodopsin.
(ii). Rhodopsin
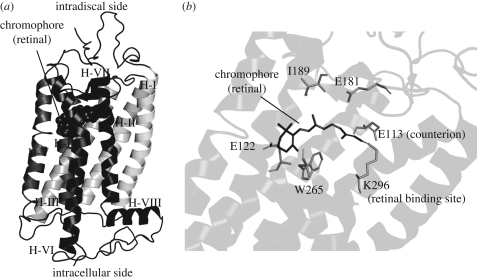
Rhodopsin is a membrane protein that consists of two parts: the apoprotein, termed opsin, and the prosthetic group chromophore, whose presence is responsible for the colour of the compound. Rhodopsin is folded into a characteristic seven transmembrane helical structure, with the N-terminus in the lumen of the disc membrane (topologically the extracellular side) and the C-terminus on the cytoplasmic side. The chromophore is covalently bound to a lysine residue at helix 7 (H7) (figure 2) through a Schiff base linkage, which can be protonated or unprotonated depending on the environment that a particular opsin provides. The protonation of the Schiff base causes delocalization of π electrons, which results in a red shift in the absorbance of the compound allowing it to absorb visible light. In the case of bovine rhodopsin, the chromophore is protonated, and thus it absorbs visible light.
Figure 2.
Molecular structure of rhodopsin. (a) Cartoon representation of the atomic model of rhodopsin, consisting of seven transmembrane helices, an eighth helix at the intracellular side and parallel to the membrane, and the chromophore shown as spheres. (b) A close-up of the structure of the chromophore and the spacial location of some of the amino acids that characterize rhodopsin, discussed in this article. Molecular graphics representations were created using PyMol (DeLano Scientific LLC).
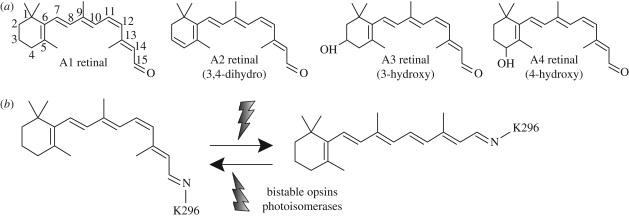
The chromophore: retinal. The chromophore moiety is a vitamin A-based retinaldehyde, either retinal (A1), 3,4-dehydroretinal (A2), 3-hydroxyretinal (A3) or 4-hydroxyretinal (A4). A1 retinal is the most common chromophore for both vertebrates and invertebrates. A2 is commonly encountered in vertebrates such as fish, amphibia and reptiles. The A2 retinal generally causes a red shift in the absorbance maxima of the retinal/opsin complex, which is sometimes called porphylopsin (derived from purple) as opposed to the A1 retinal/opsin complex, which is generally called rhodopsin (derived from rose). Freshwater fish often switch from A1 to A2 retinal to adapt to their light environment. There are also reports of seasonal variations in the A1 and A2 retinal content of some fish and in the crayfish (Suzuki & Eguchi 1987). A3 retinal is commonly observed in many insects, and the A3 retinal/opsin complex is sometimes called xanthopsin (derived from yellow). A4 retinal has been observed in the firefly squid, which seems to use A1, A2 and A4 retinals to create photoreceptor molecules of different absorbance maxima and achieve colour vision (Seidou et al. 1990). Although there are several names for a retinal-based photoreceptor molecule based on its chromophore and its absorbance maxima, rhodopsin is used as a generic term to describe all the visual pigments.
The retinal can take the form of many isomers such as all-trans, 13-cis, 11-cis or 9-cis, etc. but in the dark most opsins preferentially bind 11-cis-retinal as a chromophore. The chromophore is the key component of the light sensor; not only does the retinal absorb light, but it also changes its conformation upon light absorption. The retinal is light-isomerized from the 11-cis to the all-trans form upon light absorption (figure 3). The photoisomerization efficiency of retinal is less than 20 per cent in any solution. However, in rhodopsin the surrounding amino acids provide the necessary environment to achieve an exceptionally high photoisomerization efficiency of approximately 65 per cent, i.e. two out of three photons will cause the isomerization, rendering animals with an extremely sensitive light sensing device (Dartnall 1968).
Figure 3.
(a) Differences in A1, A2, A3 and A4 retinals used by opsins. (b) Photoisomerization of the retinal. In opsins, the retinal is covalently bound to a lysine located at H7, and the isomerization is sterospecific from 11-cis to all-trans. Some opsins are bistable and the photoisomerized all-trans-retinal can be reconverted to 11-cis-retinal by the absorption of a second photon. Also there are photoisomerases, opsins that bind all-trans-retinal and form 11-cis-retinal.
The apoprotein: opsin. The opsins are average sized proteins of 30–60 kDa, formed from about 355 amino acids that act as a shell which modifies the physico-chemical properties of the chromophore. One of the functions of the opsin is to provide the necessary environment for the absorption of light at a particular wavelength. Therefore, by providing a different opsin to the retinal, organisms can sense light of different wavelengths or colours. Small changes near the chromophore are enough to change its absorbance maxima. In addition to this spectral tuning, there are properties common to all opsins. As previously mentioned, an opsin must enhance the retinal's isomerization efficiency upon light absorption in order to create a sensitive light sensor. The chromophore is tightly bound to the opsin, and its isomerization causes structural changes in the opsin that allow rhodopsin to activate the G protein. The opsin provides an interface that binds to and subsequently interacts with the G protein, transmitting the light signal by activating the G protein (Emeis et al. 1982; Morizumi et al. 2003). Light-activated rhodopsin can activate hundreds of G proteins and thus the light signal is amplified at this stage. To ensure that rhodopsin does not continuously activate the G protein, a specialized mechanism quickly inactivates the light-activated rhodopsin and thus the signal is terminated.
(iii). The G protein
The G protein (guanine nucleotide-binding protein) or transducin is the signal transducing molecule that mediates and relays the signal from the light sensing rhodopsin. It is the molecule that transduces light stimuli into more familiar chemical signals for the cell. All ocular systems function through the signalling cascade initiated by a G protein. The G protein is a heterotrimeric protein that is activated by the exchange of guanine nucleotide, GDP to GTP, induced by rhodopsin. When the inactive Gαβγ is activated by rhodopsin, it changes its GDP to a GTP, which allows it to dissociate into two molecules: the GTP-bound α subunit and the βγ complex. Phototransduction acts primarily through the α subunit, Gα. Separation of the subunits exposes the active site of Gα, allowing it to act on its effector enzyme. Gα has an intrinsic GTPase activity and the subunits remain active until Gα hydrolyses its GTP to GDP. The intrinsic GTPase activity is too slow to account for the inactivation, and activated Gα usually requires GAP (GTPase activating proteins) to rapidly hydrolyse its GTP and terminate the signal. GDP-bound Gα binds to Gβγ once again and together they hide their active sites, effectively suppressing their activity. Different opsin families are coupled to specific types of G proteins that produce different responses. As an example, the phototransduction signalling cascade of the vertebrate visual pigments is depicted in figure 1.
2. The Opsins
(a). The big picture
GPCRs are heptahelical transmembrane proteins and they constitute the biggest family of cell membrane receptors. Based on sequence homology, it is clear that opsins belong to the family-A (or rhodopsin-like superfamily) GPCRs. Rhodopsin is the best characterized GPCR to date, and it is used as a template to understand other GPCRs.
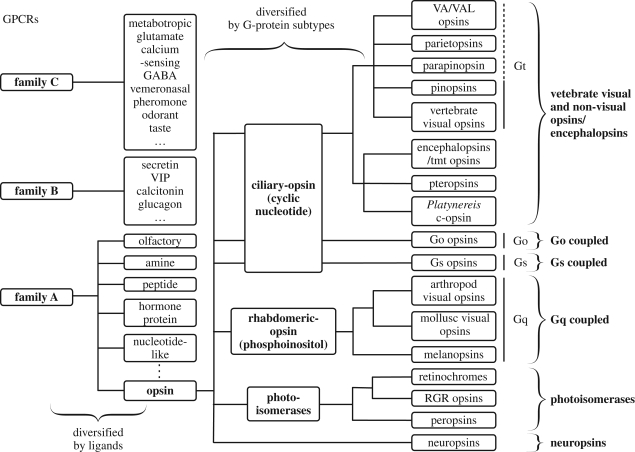
The phylogenetic tree of GPCRs indicates that GPCR initially diversified by responding to different ligands, following a diversification based on their response through different G proteins (Fryxell & Meyerowitz 1991; Fryxell 1995) (figure 4). Because all opsins use retinal as a ligand, it is reasonable to assume that rhodopsin evolved from a retinoid receptor that acquired the ability to covalently bind to its ligand, allowing it to evolve as a photoreceptor molecule. Eventually, the ancestral opsin or opsins went through a diversification process by coupling with different G proteins. This is reflected in the phylogenetic tree of opsins today, where there is a reliable correlation between different subfamilies and their functional characteristics.
Figure 4.
Schematic representation of the phylogenic relationship of opsins. Opsins belong to the family-A GPCRs, and they can be roughly subdivided into ciliary opsins, rhadbomeric opsins and photoisomerases. The ciliary opsins are characterized by their expression in ciliary photoreceptor cells and cyclic nucleotide signalling cascade. On the other hand, rhabdomeric opsins are expressed in rhabdomeric photoreceptor cells and have phosphoinositol signalling cascade. Finally, photoisomerases comprises proven and putative stereospecific photoisomerases.
(b). Phylogeny
Since the first opsin was sequenced in 1982, researchers have continuously unearthed more and more sequences, and today there are more than 1000 sequences of opsins available, from animals ranging from jellyfish to humans. Many of the opsins have a clear function; some opsins function as light sensors for visual systems, whereas others play a role in non-visual tasks such as circadian regulation. However, there are numerous opsins whose functions remain unknown.
Based on sequence homology, the opsin family can be categorized into six subfamilies, namely the vertebrate opsin/encephalopsin subfamily, the Go opsin subfamily, the recently characterized Gs opsin subfamily, the invertebrate Gq opsin subfamily, the photoisomerase subfamily and the neuropsin subfamily. These opsins share less than 20 per cent identity between subfamilies.
Although sequence comparison reveals that opsins can be clearly classified into six subfamilies, occasionally phylogeny alone is not enough to discern their relationships. Some opsins are far too diverged so that very often their differences do not explicitly manifest residue properties subject to natural selection. However, genomic structures such as synteny and intron positions are conserved across large evolutionary distances, consisting of billions of years, and they provide additional information about opsin relationships when phylogeny alone fails. Vertebrate opsins and the encephalopsins share three intron positions not observed in other opsin families (Velarde et al. 2005). Moreover, vertebrate opsins, encephalopsins, Go opsins and Gs opsins are all thought to be expressed in ciliary photoreceptor cells, characterized by an extended cilium, as opposed to Gq opsins, which are expressed in rhabdomeric photoreceptor cells, a morphologically different photoreceptor cell type. Melanopsin is thought to be expressed in cells derived from an ancestral rhabdomeric photoreceptor cell (Arendt 2003). On the basis of phylogeny and intron positions, retinochrome, retinal GPCR (RGR) opsins and peropsins clearly comprise a distinctive subclade. Because retinochrome and RGR opsins function as photoisomerases, we will tentatively call this group photoisomerases. Finally, neuropsins have intron positions conserved in the photoisomerases; however, this group has not been functionally characterized and we will consider it as a separate group. Therefore, opsins can be divided into three rough groups: ciliary opsins, rhabdomeric opsins and photoisomerases.
Ciliary and rhabdomeric photoreceptor cells have a characteristic cytoarchitecture that distinguishes them. It appears that these cells have increased the membrane-bound rhodopsin, allowing them to improve the probability of capturing a photon. Ciliary and rhabdomeric photoreceptor cells are present in both vertebrates and invertebrates, strongly suggesting that our common ancestor already used these two types of photoreceptor cells.
(i). Ciliary opsins
In addition to being expressed in ciliary photoreceptor cells characterized by an extended cilium, ciliary opsins also share some similarities in their phototransduction mechanism. Although they can produce different responses, all opsins in this group seem to function through signalling cascades that alter the concentration of cyclic nucleotides.
Vertebrate opsin/encephalopsin. The vertebrate opsin/encephalopsin subfamily consists of vertebrate opsins and encephalopsins.
Vertebrate opsins. The vertebrate opsin group comprises visual and non-visual opsins in vertebrates. Phylogenic analysis of vertebrate visual opsins reveals that they can be further subdivided into five subgroups consisting of four cone opsins and one rod opsin group, which are distinguished by their spectral sensitivity (Okano et al. 1992b). The S group consists of cone opsins that absorb UV or violet light, the M1 group absorbs blue light, the M2 group absorbs green light and the L group absorbs red or green light. The rod opsin group, denoted Rh, absorbs green/blue light and is a sister group to the M2 group. Cone and rod opsins function through the well-characterized Gt signalling pathway (figure 1). In addition to their role as ocular photoreceptor molecules, vertebrate visual opsins are also expressed in non-visual photoreceptor cells, such as the pineal photoreceptor cells; however, their role and/or contribution in non-visual photoreception remains unknown (Wada et al. 1998; Mano et al. 1999).
The non-visual opsins in the vertebrate opsin subfamily consist of pinopsins, parapinopsins, VA (vertebrate ancient) opsins and parietopsins. They are opsins that closely resemble vertebrate visual opsins, with more than 40 per cent identity. Non-visual opsins are presumed to be involved in light-dependent physiological phenomena, such as photic regulation of circadian rhythms, photoperiodicity and body colour change. Pinopsins are found in the pineal organ of avian species, reptiles and amphibians, where they may play a role in its regulation, but they seem to be absent from teleosts and mammals (Okano et al. 1994; Max et al. 1995; Taniguchi et al. 2001). Parapinopsins have been found in the photosensitive pineal and parapineal organs of jawless fish, teleost fish and amphibians (Blackshaw & Snyder 1997; Koyanagi et al. 2004). VA opsins were initially identified in salmon and they were named VA opsins because they seem to have diverged early in vertebrate opsin evolution (Soni & Foster 1997). They are localized in the inner retina and the brain in teleosts. Isoforms of VA opsins called VAL opsins have been reported, characterized by the extension of their carbonyl terminus (Kojima et al. 2000; Moutsaki et al. 2000; Minamoto & Shimizu 2002). A recent study reported that VAL opsins are duplicated in the teleost lineage and that the two copies of VAL opsin have a differential expression, suggesting that they have different physiological roles (Kojima et al. 2008).
Lizards and other non-mammalian vertebrates have been known to possess a photoreceptive organ on the top of their head, called the parietal eye, complete with a cornea, a lens and a retina. The opsin expressed in the ‘third eye’, named parietopsin, is closely related to the vertebrate visual opsins, with approximately 40 per cent identity to parapinopsins and VA opsins (Su et al. 2006). Interestingly, the photoreceptor cells of the parietal eye possess two signalling pathways, which are activated in response to different light. Blue light causes hyperpolarization through the pinopsin–gustducin signalling pathway, and green light causes depolarization through the parietopsin–Go signalling pathway.
Encephalopsins: There are two main groups in this opsin group: encephalopsins (or panopsins)/tmt opsins and Platyneresis c-opsin/pteropsins. Encephalopsins were initially identified in mammals, and their name was derived from their strong expression in the brain and testes (Blackshaw & Snyder 1999). However, subsequent studies showed that this opsin is also widely expressed, although weakly, in non-photoreceptive tissue, such as the heart, lung, liver, kidney, skeletal muscle and pancreas, as well as in the retina (Halford et al. 2001). Because of its wide distribution, it has been suggested that encephalopsin would be better named panopsin. Teleost multiple tissue (tmt) opsin is a homologue of mammalian encephalopsin, found in teleost fish, widely distributed in neuronal and non-neuronal tissue, where it has been proposed as the photopigment that regulates photic entrainment of peripheral clocks (Moutsaki et al. 2003).
A homologue of encephalopsin and tmt opsin was identified in ciliary photoreceptor cells in the brain of the marine rag worm, Platyneresis (Arendt et al. 2004). This finding surprised researchers because it was the first ciliary opsin to be identified in protostomes. Moreover, an orthologue (orthologues are genes in different species that originated from a common ancestral gene through speciation) called pteropsin was reported in insects (Hill et al. 2002; Velarde et al. 2005). Pteropsins and Platyneresis c-opsin have great similarity to vertebrate opsins, and they also have three common introns with vertebrate opsins, indicating a close relationship with vertebrate opsins. They are expressed in insect brains and not in their eyes. Interestingly, this lineage of opsins was lost in Drosophila, a phenomenon that delayed its discovery.
Go opsins. Go opsins have been identified in molluscs and in the chordate amphioxus, but they are not present in humans, mice, zebrafish or fruitfly (Kojima et al. 1997; Koyanagi et al. 2002). Like vertebrate opsins/encephalopsins, they are expressed in ciliary photoreceptor cells. Light stimulation of these ciliary photoreceptor cells results in an increase in cGMP, probably through the activation of membrane GC (guanylyl cyclase), that subsequently opens K+ selective channels and thus causes a hyperpolarization response (Gomez & Nasi 2000).
Gs opsins. The most recently described opsin subfamily is that of opsins found in cnidarians, including the sea anemone, hydra and jellyfish (Plachetzki et al. 2007; Suga et al. 2008). More recently, the box jellyfish opsins, which also cluster in this subfamily, have been shown to signal through the Gs signalling pathway, involving an increase in cAMP (Koyanagi et al. 2008b). These opsins are also expressed in ciliary photoreceptor cells.
(ii). Rhabdomeric opsins
Rhabdomeric opsins are localized in the microvilli of rhabdomeric photoreceptor cells, which are morphologically different from ciliary photoreceptor cells. These opsins transmit light signals through the G protein Gq subgroup, involving phospholipase C (PLC) (Terakita et al. 1993; Lee et al. 1994). Dissociated Gqα binds to its effector enzyme, PLCβ, and activates it. The substrate of PLC is a membrane phospholipid, PIP2, which is separated into two potential messengers: DAG (diacylglycerol) and IP3 (inositol 1,4,5-tris-phosphate). Exactly how these messengers act is still unknown, but similar to the vertebrate visual signalling cascade, they act on a membrane channel. However, contrary to the vertebrate visual CNG (cyclic nucleotide gated) channel, the invertebrate visual TRP (transient receptor potential) channel opens in response to light stimuli, creating a depolarization response. In contrast to the vertebrate visual opsins, most of the invertebrate Gq opsins produce a thermally stable active state and the photo-activated molecule can be reconverted to the ground state by absorption of a second photon. The biochemical study of these opsins has been delayed because they are difficult to express in cultured cells, and only recently did the exogenous expression of these opsins became possible (Terakita et al. 2008). Also recently, the crystal structure of squid rhodopsin was solved (Murakami & Kouyama 2008). These recent breakthroughs should catalyse our understanding of these opsins.
Invertebrate Gq opsins. The invertebrate Gq opsin family contains the arthropod and mollusc visual opsins, as well as melanopsins. Like the vertebrate visual opsins, arthropods possess a well-documented colour vision, supported by Gq opsins tuned to a specific colour (Briscoe & Chittka 2001; Koyanagi et al. 2008a). Melanopsins are found in vertebrates, but they closely resemble invertebrate visual opsins (Provencio et al. 1998, 2000). Initially identified in amphibians, they have been subsequently reported in all vertebrate classes. Melanopsins are the primary photoreceptor molecules for non-image forming function such as the photo-entrainment of the circadian clock and pupillary constriction in mammals (Hattar et al. 2003; Lucas et al. 2003; Panda et al. 2003). It has recently been shown that melanopsin also participates in visual tasks, by regulating optic inputs from photoreceptor cells, according to the circadian phase, i.e. the time of the day (Dacey et al. 2005; Barnard et al. 2006).
(iii). Photoisomerases
Photoisomerases are stereospecific isomerases that bind all-trans-retinal and generate 11-cis-retinal. As their biological role is not to initiate a signalling cascade, they are not coupled to a G protein and thus they do not generate a signalling cascade. The photoisomerase opsin family consists of retinochrome, RGR opsins and peropsins.
Retinochrome/RGR opsins. Retinochrome is the retinal isomerase that supplies 11-cis-retinal to the visual cycle in molluscs (Hara & Hara 1967). RGR opsin expression is confined to the retinal pigment epithelium (RPE) and Müller cells of vertebrates, and to the brain of a chordate ascidian (Jiang et al. 1993; Nakashima et al. 2003). Its endogenous chromophore is the all-trans form of retinal and it can photoisomerize it to 11-cis in a stereospecific manner, although the resulting 11-cis-retinal decays thermally back to all-trans-retinal (Hao & Fong 1999). It would appear that rather than supplying 11-cis-retinal to other opsins, RGR opsins regulate retinoid traffic in RPE in a light-dependent manner (Radu et al. 2008).
Peropsins. Peropsins have been reported in vertebrates and the chordate amphioxus (Sun et al. 1997; Koyanagi et al. 2002). Phylogenetically, they are closely related to photoisomerases, and their localization in the RPE suggests that they may also function as photoisomerases. In fact, they can bind all-trans-retinal and photoisomerize it to 11-cis-retinal (Koyanagi et al. 2002). However, contrary to other photoisomerases, peropsin possesses sequence motifs conserved among family-A GPCRs. The highly conserved D/ERY triad at the cytoplasmic surface and the NPXXY motif at H7 are important for the activation of the G protein. The presence of these structural features suggests that peropsin may bind and activate a G protein, although its physiological significance is unclear.
(iv). Neuropsins
Neuropsins are thought to be expressed predominantly in mammalian neural tissue, eye and brain, although weaker expression in testes and spinal cord is also reported (Tarttelin et al. 2003). Like peropsins they also possess the highly conserved D/ERY triad at the cytoplasmic surface and the NPXXY motif at H7. Although these opsins share intron positions with peropsins, they have not yet been functionally characterized and their function remains unknown.
(c). Genomes
Genomes provide us with valuable information, as they contain the complete set of opsins for a particular organism. For example, humans possess nine different opsins. Three opsins are expressed in cone photoreceptor cells, which determine the three colours in our vision: red, green and blue. A rhodopsin, which functions under dim light conditions, is expressed in rod photoreceptor cells. Melanopsin is the opsin that functions in the circadian regulatory system and pupil constriction of the eyes. In addition to these, we have encephalopsin, neuropsin, RGR opsin and peropsin. Furthermore, genomes tell us the location of these genes in the genome, which is generally conserved, and allow us to compare between species (Nordström et al. 2004; Kuraku et al. 2009). As gene locations are usually conserved among species, we can track opsin's evolutionary events such as gene losses, gene duplications and function-altering amino acid substitutions, which can contribute to our understanding of the diversification of biological functions.
3. Exploring Evolution Experimentally
In this section, we will focus our attention on the best characterized photoreceptor molecule, the rod opsin, or rhodopsin. As mentioned earlier, advances in molecular biology have allowed scientists to work with artificially manipulated proteins. This allows us to recreate opsins that may have once existed. We can now directly test the properties that were favoured by natural selection over the course of opsin evolution.
We will give two examples of molecular adaptations in rhodopsin: the evolution of vertebrate opsins and the evolution of rod opsins. Vertebrate opsin evolution involves the reduction of agonist binding, and the counterion displacement, whereas rod opsin evolution deals with the differences observed in rods and cones. These are adaptations that underlie rhodopsin's exceptionally high sensitivity to light.
(a). Evolution of vertebrate opsins
Sequence comparison of different opsins reveals sites that are clearly conserved within subfamilies and sites that are highly variable. Highly conserved sites are expected to be functionally important, and differences between subfamilies in these sites often reflect adaptive changes.
(i). Evolution of the counterion
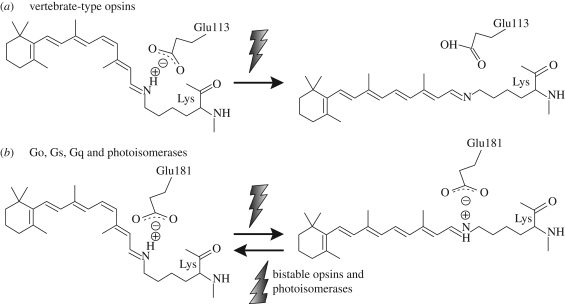
As mentioned earlier, the retinal attaches to the opsin in H7 in the transmembrane domain through a protonated Schiff base linkage. Although protonation of the Schiff base is necessary for visible light reception, a positive charge in the hydrophobic transmembrane environment is highly unstable. Opsins counter this problem by providing a negatively charged amino acid, a counterion that stabilizes the positive charge of the protonated Schiff base. There are two tentative sites that can serve as a counterion in opsins: 113 at H3 in the transmembrane domain, and 181 at the ECL2 (extracellular loop 2) between H4 and H5 (figures 2 and 5). The residue at 181 is a conserved glutamic (aspartic) acid among all opsin subfamilies, whereas 113 vary between them.
Figure 5.
The counterion of opsins. (a) Vertebrate visual and non-visual opsins have a negatively charged glutamic acid at 113 in H3 that functions as the counterion for the positive charge of the protonated retinal Schiff base. (b) The counterion of Go/Gs/Gq opsins and photoisomerases is E181.
Vertebrate opsins and encephalopsins generally have a glutamic acid on both 113 and 181. Mutations at E113 in vertebrate visual opsins result in a big blue shift in the absorbance maxima of the pigment, making it UV light sensitive, an indication that the Schiff base is deprotonated (Sakmar et al. 1989). On the other hand, mutations at E181 do not disturb the absorbance maxima. These experiments indicate that E113, and not E181, is the one that serves as the counterion in the vertebrate opsins group.
The counterion of other opsins was first identified in the mollusc photoisomerase retinochrome (Terakita et al. 2000). In this opsin, 113 is occupied by residues that do not possess a negative charge, methionine or histidine, while 181 is occupied by the conserved glutamic acid. Site-directed mutagenesis showed that mutations at 113 do not disturb the absorbance spectra, whereas mutations induced at E181 result in a deprotonated Schiff base, indicating that E181 serves as the counterion in photoisomerases.
In contrast to easily expressed vertebrate opsins and retinochrome, invertebrate Gq opsins are not readily expressed in cultured cells, and their functional characterization has been delayed.
Although there is no direct experimental evidence for the Gq opsin counterion, Gq, Go, peropsins and neuropsins all have a tyrosine or a phenylalanine residue at 113. Furthermore, mutational experiments on Go opsins and peropsins have shown that they behave similar to retinochrome (Terakita et al. 2004). Mutations at 113 do not cause deprotonation of the Schiff base, whereas mutations at E181 cause spectral changes characteristic of Schiff base deprotonation.
Taken together, these experiments show that only vertebrate opsins have a counterion at 113, whereas all other opsins seem to have a counterion at the conserved glutamic acid at 181 (figure 5). Interestingly, the counterion of Go opsins can be artificially moved from 181 to 113. As mentioned earlier, mutations at 181 in Go opsins cause spectral changes owing to the deprotonation of the Schiff base. However, a second mutation introducing a glutamic acid in 113 reverses the deprotonation, showing that the glutamic acid at 113 can act as a counterion (Terakita et al. 2004). This experiment proved that 113 can serve as a counterion in addition to its original counterion at 181, which is in contrast with the vertebrate opsins, where only 113 can serve as the counterion. Moreover, these experiments also revealed that the glutamic acid at 113 in Go opsins could serve as a counterion in the resting state but not in the signalling state. It seems that an ancestral pigment had a counterion at 181, and vertebrate opsins recruited E113 as a novel counterion. Acquisition of the novel counterion at 113 relaxed the function of 181 as a counterion, probably causing rearrangements in ECL2 that further prevented it from functioning as a counterion.
Detailed analyses of E113 in vertebrate visual opsins have revealed some interesting functions of this residue, in addition to its function as the counterion for visible light sensing. E113 has been shown to serve as a ‘molecular switch’ which suppresses rhodopsin activation in the absence of light (Robinson et al. 1992). The attraction of the positive charge of the protonated K296 and the negative charge of the deprotonated E113 maintain the receptor in an inactive resting state (figure 5). Light absorption diminishes this interaction and the proton of the Schiff base is transferred to E113, triggering the formation of a hydrogen network that activates the receptor (Jäger et al. 1994). Therefore, vertebrate visual pigments have a photoproduct with a deprotonated Schiff base, which contributes to the efficient activation of the G protein.
However, the newly acquired counterion in vertebrate opsins did not immediately act as a molecular switch. The analysis of the photoproducts of vertebrate opsins shows that only vertebrate visual opsins have a deprotonated photoproduct, suggesting that parapinopsins and encephalopsins behave more like invertebrate opsins, despite their sequence similarity to vertebrate visual opsins. It is believed that during the course of vertebrate visual opsin evolution, E113 acquired the capacity to act as a molecular switch. Moreover, it seems that E113 may have an additional role in enhancing light sensitivity, by increasing the isomerization efficiency upon light absorption (Tsutsui et al. 2008).
After E113 took over the role of counterion, the selective pressure on E181 must have decreased, allowing different amino acids at this position. An interesting mutation is observed in cone opsins, which are sensitive to red light, i.e. they absorb light at longer wavelengths. The position 181 is occupied by histidine instead of the highly conserved glutamic acid (Nathans et al. 1986; Kuwata et al. 1990). These unusual substitutions are considered to be special adaptations for the absorption of red light. It turns out that H181 is part of a chloride binding site, which causes the chromophore to maximally absorb light of longer wavelength (Wang et al. 1993). This adaptation was possible because of the counterion displacement from 181 to 113, which freed 181 from its structural constraints as the counterion, allowing it to acquire a new function.
(ii). Rhodopsin ligands
Although it is clear from sequence homology and structural similarities that rhodopsin is a GPCR, its behaviour is quite different from that of the typical GPCR. Most notably, rhodopsin has a covalent bond to its ligand, the retinal. In rhodopsin the inverse agonist is photoisomerized to the agonist, whereas other GPCRs employ diffusible ligands to regulate their activity. Vertebrate visual opsins have a peculiar adaptation, as they seem to have lost the ability to bind an exogenous agonist and form the signalling state. Although some studies report that the addition of all-trans-retinal enhances the intrinsic activation of the receptor, the spectroscopic formation of an active state is not observed and these are considered to be allosteric effects (Jäger et al. 1996; Surya & Knox 1998). If a photoreceptor molecule were to be activated by an exogenous agonist, it would produce false signals that do not originate from light. Therefore, avoiding the binding and activation by an exogenous agonist, i.e. the formation of a signalling state, reduces the ‘dark noise’, the activation detected in the dark.
An intriguing opsin was found recently in the amphioxus, which belongs to the Go opsin subfamily (Koyanagi et al. 2002). The amphioxus Go opsin retains agonist binding, and the complex of the opsin and the agonist is indistinguishable from its photoproduct (Tsukamoto et al. 2005). The opsin has 50 times higher affinity to its inverse agonist 11-cis-retinal than to its agonist all-trans-retinal. Although it still retains agonist binding, it is clearly reduced, indicating that amphioxus Go is an opsin with intermediate properties between a general GPCR that exhibits high affinity to agonist, and the vertebrate visual opsins that completely suppressed agonist binding to decrease dark noise. Mutational experiments on the amphioxus Go opsin indicate that W265 located in H6 suppresses the agonist binding (figure 2). Introducing mutations at W265 shifts the affinity of the opsin to favour the binding of all-trans-retinal, the agonist, indicating that W265 is responsible for the high affinity to 11-cis-retinal, the inverse agonist. Moreover, the shift in affinity correlates well with the volume of the amino acid introduced; smaller amino acids favour binding of all-tarns-retinal, whereas bulkier amino acids favour binding of 11-cis-retinal. This residue is well conserved in many opsin subfamilies, including vertebrate visual opsins, and mutation-induced reduction in inverse agonist affinity was also observed in vertebrate visual opsins (Reeves et al. 1999). The agonist binding is also reported for invertebrate Gq opsins (Koutalos et al. 1989). It seems that a common denominator of agonist-binding opsins is the ability to form a protonated photoproduct. As mentioned earlier, vertebrate visual opsins, which have a deprotonated photoproduct, have a molecular switch, consisting of a hydrogen bond network that suppresses the intrinsic activity of the receptor in the dark. Disruption of these interactions results in the formation of the signalling state. It would appear that in addition to suppressing the intrinsic activity of the opsin, these hydrogen bond interactions also prevent opsin from binding an exogenous agonist.
(b). Evolution of vertebrate vision: rods and cones
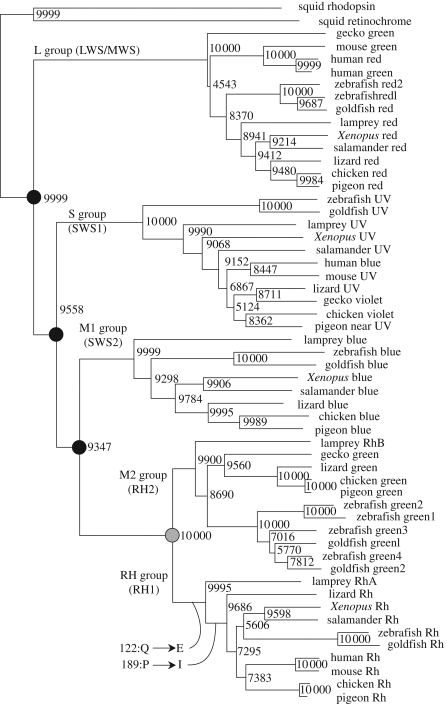
Sequence analysis revealed the phylogenetic relationship of vertebrate type opsins and it is now evident that vertebrates first acquired four kinds of cone opsins, the molecular basis of colour vision, after which a new opsin specialized for dim light reception emerged (figure 6). These opsins are expressed in morphologically different cells: rods and cones (figure 1). Rod cells contain rod opsin, rhodopsin, which functions in dim light and thus underlies our scotopic vision. Most animals have only one scotopic photoreceptor molecule, and so our vision is monochromatic in dim light. On the other hand, cone cells contain cone opsins, which function in well-lit conditions, allowing the so-called photopic vision, and also colour perception. It is surprising that colour vision, which would require more complex signal processing, evolved before the simpler chromatic vision.
Figure 6.
Phylogenetic tree of vertebrate visual opsins constructed by NJ (numbers at the nodes indicate the bootstrap values of 10 000 replicates). Five distinctive groups, which correspond well with their spectral sensitivities, can be identified: the UV or violet light-sensitive S (or SWS1) group, the blue light-sensitive M1 (or SWS2) group, the green light-sensitive M2 (or RH2) group, the red or green light-sensitive L (or LWS/MWS) group and the scotopic vision RH (or RH1) group. Note that the tree does not necessarily reflect the phylogenic relationship of organisms. Black circle, colour vision; grey circle, twilight vision.
(i). Properties of rods and cones
Rods and cones have well-defined response properties. Rods are characterized by high sensitivity, slow response and slow dark adaptation, whereas cones have low sensitivity, fast response and fast dark adaptation (figure 7). It may seem that rods compromised their fast response and fast dark adaptation for higher sensitivity. Such properties originate from the molecular properties of the functional proteins in these photoreceptor cells. In the 1980s, we successfully isolated and purified cone visual pigments from chicken photoreceptor cells. Moreover, we were able to obtain sufficient amounts of purified protein to carry out spectroscopic and biochemical analyses in order to elucidate the molecular properties of cone opsins. We compared the molecular properties of cone opsins and rod opsins, to test whether their molecular properties could account for the physiological response profiles of rod and cone photoreceptor cells.
Figure 7.
Response profiles of rods and cones. Rods are characterized by a large and slow response, high sensitivity and a slow dark adaptation. On the other hand, cones are characterized by a small and fast response, low sensitivity and a fast dark adaptation. These response profiles originate from the functional proteins in them.
One would think that the best way to create a dim-light photoreceptor molecule is to improve the sensitivity of the photoreceptor molecule itself, i.e. to increase the probability that the receptor would capture light and form a signalling state. Although increasing the sensitivity of the receptor would increase the sensitivity of the photoreceptor cell, this is not the mechanism that visual pigments employ. In fact, we discovered that rod opsins and cone opsins have the same sensitivity to light (Okano et al. 1992a; Shichida et al. 1994). How do opsins then cause high sensitivity in rods and low sensitivity in cones? Our results suggest that the answer is the lifetime of the signalling state. Rod opsins have a prolonged active state in comparison with cone opsins, which allows them to activate more G proteins (Imai et al. 1997b). Having a longer signalling state allows rods to amplify their response, and thus increases the sensitivity of the photoreceptor cell. The fast response of cones is also related to the signalling state. Cone opsins can form the signalling state much faster than rod opsins, and therefore their photoreceptor cells can respond faster (Shichida et al. 1994). Rod opsin has evolved dramatically prolonged signalling, allowing it to amplify light responses better than cone opsins, in exchange for slower responsiveness. Thus, under dim light, where only rods function, we cannot distinguish colours or see movement very well, but we can nevertheless see.
The slower dark adaptation in rods when compared with cones can be explained by the difference in reconstitution rates between the respective opsins. As previously stated, the signalling state of a vertebrate visual opsin is thermally unstable, and it dissociates into the apoprotein, opsin, and the chromophore, i.e. the retinal. The opsin must reincorporate the chromophore in order to function again as a photoreceptor molecule, and the uptake of the chromophore by the opsin is called reconstitution. Cone opsins can reincorporate retinal faster than rod opsins, explaining their differences in dark adaptation speed.
When we started investigating the response profiles of rods and cones by focusing on the opsins, there was no report of differences in the molecular properties of other functional proteins involved in phototransduction. However, owing in part to recent advances in techniques that allow researchers to separate cone cells from rod cells, differences in the molecular properties of other functional proteins are now being examined (Kawamura & Tachibanaki 2008; Nikonov et al. 2008). These studies show that, just like the opsins show properties that distinguish rod and cone photoreceptor cells, other functional proteins present in the photoreceptor cells also exhibit properties that correlate with the response profiles of the respective photoreceptor cell. In other words, it is likely that each one of the functional proteins involved in phototransduction has evolved properties that have given rods and cones their different roles. Therefore, future research should focus on identifying the amino acid substitutions that give rise to the molecular properties of these functional proteins and to identify what property of which functional proteins contributes to the physiological properties of the photoreceptor cells. In the following section, we will summarize our studies of the molecular properties of rod and cone opsins, and their possible contribution to the response profiles of the photoreceptor cells.
(ii). Molecular evolution of rod opsins from cone opsins
The molecular properties of opsins derive from their primary amino acid sequence. As rod opsins evolved from cone opsins, we can compare their sequences and identify the amino acid differences. Amino acid sites that are conserved in cone opsins but not in rhodopsin are likely to account for their differences. Sequence comparison with chicken vertebrate visual opsins revealed several such sites. Using site-directed mutagenesis, these putative amino acids were introduced to rod opsin, to test whether they affect the functional properties of rhodopsin. As a result, the amino acids located at sites 122 and 189 were determined as the main contributors to the functional properties of rod and cone opsins (Imai et al. 1997a; Kuwayama et al. 2002) (figure 2). Rod opsins have a glutamic acid at 122, whereas homologous positions are taken by glutamine in the chicken green cone opsin and isoleucine in the chicken red cone opsin. Introducing these residues (E122I and E122Q) in rod opsin dramatically increases its reconstitution rate, and it also causes an increase in the decay rate of the signalling state. More recent works show that E122 forms part of a hydrogen bond network that stabilizes the photoreceptor molecule (Beck et al. 1998; Patel et al. 2005). On the other hand, the position 189 is a conserved proline residue in cone opsins, whereas the same position is occupied by an isoleucine in rod opsin. The mutation P189I in cone opsins causes a reduction in the decay rate of the signalling state, whereas I189P in rod opsin causes an increase in the decay rate (Kuwayama et al. 2002). Together, these two sites can account for the differences in the molecular properties of rods and cones. In fact, the lifetime of the signalling state of cone opsins with Q/I122E and P189I is almost identical to that of rod opsin.
As shown above, 122 and 189 are the functional determinants of rod and cone visual pigments. Phylogenetic analysis of rod opsins shows that all rod opsins have a glutamic acid at 122. However, not all rod opsins have an isoleucine at 189. Lamprey rod opsin has a proline residue at 189, suggesting that rod opsins first acquired E122 and gained rod opsin like properties, and later I189 was introduced to further enhance these properties (see figure 6).
(iii). The ultimate proof
We showed that the molecular properties of the opsins expressed in the photoreceptor cells could in part explain the differences between rods and cones. In fact, rod photoreceptor cells from genetically engineered mice, carrying rod opsins with the mutation E122Q, showed that the molecular properties of opsin clearly affect the response profile of the photoreceptor cells (Imai et al. 2007). But how much do the molecular properties of opsins affect the response profiles of photoreceptor cells? The best way to elucidate this question is to exchange the opsins alone in the photoreceptor cells and test how their properties change the physiological responses of the photoreceptor cells. A genetically engineered, knock-in mouse whose rod opsin was replaced with the green cone opsin was generated (Sakurai et al. 2007). The homologous knock-in mouse expresses the green cone opsin instead of the rod opsin. Unfortunately, deletion of rod opsin causes degeneration of the photoreceptor cells, which complicates the analysis, because the response of these photoreceptor cells cannot be directly compared with normal mice and differences in the response profile may arise from the degeneration rather than from the properties of opsins. However, it is possible to analyse the response profile of photoreceptor cells under identical conditions, by generating a heterozygous knock-in mouse, which expresses both rod opsin and green cone opsin in its rods. Because they express in the same cell, all the phototransduction components are identical, and any difference in the physiological response of these cells arises from the opsins. In order to allow selective activation of the opsins, E122Q rod opsin was employed. This mutant exhibits similar properties to the wild-type rod opsin but its absorbance maxima is shifted, so that it can be preferentially light-activated without activating the green cone opsins in the same cell. Analyses of this knock-in mouse clearly showed that the photoresponse of green cone opsin was threefold lower than that of rod opsin.
An additional finding in these knock-in mice was that the thermal activation rate of green cone opsin is 860-fold higher than that of rod opsin. A thermal activation means that the receptor forms a signalling state without light stimuli, and it manifests itself as dark noise in the signal. In order to produce a true response to light, the photoreceptor cell must produce a signal that is higher than that of the dark noise. This essentially means that our eyes can effectively sense light in environments with 860 times less light, due to the exceptional thermal stability of rod opsins. These results show that the evolution of the scotopic light sensor, rod opsin, required the receptor to acquire a large response and to suppress the dark noise.
4. Conclusion
One of the differences between physical sciences and life sciences is the element of time. In life sciences, we use many words that implicitly contain the concept of time. Take the word ‘adaptation’, for example, which can be used in the context of physiology to denote a phenomenon that happens in a short time span, and it is a phenomenon in a time span where we can easily conduct experiments. However, ‘adaptation’ can also be used to describe evolutionary processes, which generally happen far beyond the time scales that we can manipulate. The topic of this article ‘opsin and phototransduction evolution’ is also a concept that implies time spans far beyond the grasp of our experimental reach. The photoisomerization of the retinal is an event that takes place in the order of 10−15 s. On the other hand, evolution of the functional proteins in our eyes is an ongoing process of billions of years, which is in the order of 1016 s.
Biological phenomena that manifest themselves in a time span that allow us to conduct experiments can be approached effectively at the molecular level, because they allow researchers to observe their reproducibility, a principle which is at the base of empirical sciences. If one can set up the experimental conditions so that the same result is reproduced, these experimental conditions can lead to the elucidation of the mechanism behind a particular phenomenon. We have conducted comparative studies of opsin molecules from various organisms to investigate the mechanism of functional diversification.
Exploring the diverse functions of diverse organisms can sometimes lead to universal principles. This is undoubtedly the reflection of the fact that living organisms evolved and diversified from a common ancestor over unfathomable periods of time. Moreover, it can also be speculated that an ancestral gene that performed a particular function has diversified over the course of evolution and has brought new functionality to that organism. A close look at the phylogenetic tree of opsins reveals the acquisition of diverse functions over time, even in the small domain of signal transduction. The functional diversity of opsins is a clear testimony of adaptation, and their diversity provides a window of opportunity to extract information about the time course of their evolution. As mentioned before, manipulation and analyses of protein functions using genetic engineering techniques can be achieved in feasible time spans. We hope that the use of site-directed mutagenesis based on the phylogenetic relationship of the molecules, combined with their functional characterization, will lay a new path in the field of evolutionary research.
Acknowledgements
We thank Dr M. Koyanagi of Osaka University for advice and discussion regarding the phylogenic relationship of opsins. We also thank the anonymous reviewers for their invaluable comments and critical reading of the manuscript. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (Grants-in-Aid for Scientific Research; the Global Center of Excellence Program ‘Formation of a Strategic Base for Biodiversity and Evolutionary Research: from Genome to Ecosystem’ (ProgramA06)) to Y.S.
Footnotes
One contribution of 13 to a Theme Issue ‘The evolution of phototransduction and eyes’.
References
- Arendt D.2003Evolution of eyes and photoreceptor cell types. Int. J. Dev. Biol. 47, 563–571 [PubMed] [Google Scholar]
- Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A. W., Wittbrodt J.2004Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871 (doi:10.1126/science.1099955) [DOI] [PubMed] [Google Scholar]
- Barnard A. R., Hattar S., Hankins M. W., Lucas R. J.2006Melanopsin regulates visual processing in the mouse retina. Curr. Biol. 16, 389–395 (doi:10.1016/j.cub.2005.12.045) [DOI] [PubMed] [Google Scholar]
- Beck M., Sakmar T. P., Siebert F.1998Spectroscopic evidence for interaction between transmembrane helices 3 and 5 in rhodopsin. Biochemistry 37, 7630–7639 (doi:10.1021/bi9801560) [DOI] [PubMed] [Google Scholar]
- Blackshaw S., Snyder S. H.1997Parapinopsin, a novel catfish opsin localised to the parapineal organ, defines a new gene family. J. Neurosci. 17, 8083–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S., Snyder S. H.1999Encephalopsin: a novel mammalian extraretinal opsin discretely localised in the brain. J. Neurosci. 19, 3681–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe A. D., Chittka L.2001The evolution of color vision in insects. Annu. Rev. Entomol. 46, 471–510 (doi:10.1146/annurev.ento.46.1.471) [DOI] [PubMed] [Google Scholar]
- Dacey D. M., Liao H.-W., Peterson B. B., Robinson F. R., Smith V. C., Pokorny J., Yau K.-W., Gamlin P. D.2005Melanopsin-expressing ganglion cells in primate retina signal colour and irradiance and project to the LGN. Nature 433, 749–754 (doi:10.1038/nature03387) [DOI] [PubMed] [Google Scholar]
- Dartnall H. J. A.1968The photosensitivities of visual pigments in the presence of hydroxylamine. Vision Res. 8, 339–358 (doi:10.1016/0042-6989(68)90104-1) [DOI] [PubMed] [Google Scholar]
- Emeis D., Kühn H., Reichert J., Hofmann K. P.1982Complex formation between metarhodopsin ii and gtp-binding protein in bovine photoreceptor membranes leads to a shift of the photoproduct equilibrium. FEBS Lett. 143, 29–34 (doi:10.1016/0014-5793(82)80266-4) [DOI] [PubMed] [Google Scholar]
- Fryxell K. J.1995The evolutionary divergence of neurotransmitter receptors and second-messenger pathways. J. Mol. Evol. 41, 85–97 (doi:10.1007/BF00174044) [DOI] [PubMed] [Google Scholar]
- Fryxell K. J., Meyerowitz E. M.1991The evolution of rhodopsins and neurotransmitter receptors. J. Mol. Evol. 33, 367–378 (doi:10.1007/BF02102867) [DOI] [PubMed] [Google Scholar]
- Gomez M. P., Nasi E.2000Light transduction in invertebrate hyperpolarizing photoreceptors: possible involvement of a Go-regulated guanylate cyclase. J. Neurosci. 20, 5254–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S., Freedman M. S., Bellingham J., Inglis S. L., Poopalasundaram S., Soni B. G., Foster R. G., Hunt D. M.2001Characterization of a novel human opsin gene with wide tissue expression and identification of embedded and flanking genes on chromosome 1q43. Genomics 72, 203–208 (doi:10.1006/geno.2001.6469) [DOI] [PubMed] [Google Scholar]
- Hao W., Fong H. K.1999The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium. J. Biol. Chem. 274, 6085–6090 (doi:10.1074/jbc.274.10.6085) [DOI] [PubMed] [Google Scholar]
- Hara T., Hara R.1967Rhodopsin and retinochrome in the squid retina. Nature 214, 573–575 (doi:10.1038/214573a0) [DOI] [PubMed] [Google Scholar]
- Hattar S., et al. 2003Melanopsin and rod–cone photoreceptive systems account for all major accessory visual functions in mice. Nature 424, 76–81 (doi:10.1038/nature01761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. A., et al. 2002G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178 (doi:10.1126/science.1076196) [DOI] [PubMed] [Google Scholar]
- Imai H., Kojima D., Oura T., Tachibanaki S., Terakita A., Shichida Y.1997aSingle amino acid residue as a functional determinant of rod and cone visual pigments. Proc. Natl Acad. Sci. USA 94, 2322–2326 (doi:10.1073/pnas.94.6.2322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Terakita A., Tachibanaki S., Imamoto Y., Yoshizawa T., Shichida Y.1997bPhotochemical and biochemical properties of chicken blue-sensitive cone visual pigment. Biochemistry 36, 12 773–12 779 (doi:10.1021/bi970809x) [DOI] [PubMed] [Google Scholar]
- Imai H., et al. 2007Molecular properties of rhodopsin and rod function. J. Biol. Chem. 282, 6677–6684 (doi:10.1074/jbc.M610086200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger F., Fahmy K., Sakmar T. P., Siebert F.1994Identification of glutamic acid 113 as the Schiff base proton acceptor in the metarhodopsin II photointermediate of rhodopsin. Biochemistry 33, 10 878–10 882 [DOI] [PubMed] [Google Scholar]
- Jäger S., Palczewski K., Hofmann K. P.1996Opsin/all-trans-retinal complex activates transducin by different mechanisms than photolyzed rhodopsin. Biochemistry 35, 2901–2908 (doi:10.1021/bi9524068) [DOI] [PubMed] [Google Scholar]
- Jiang M., Pandey S., Fong H. K.1993An opsin homologue in the retina and pigment epithelium. Invest. Ophthalmol. Vis. Sci. 34, 3669–3678 [PubMed] [Google Scholar]
- Kawamura S., Tachibanaki S.2008Rod and cone photoreceptors: molecular basis of the difference in their physiology. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 150, 369–377 (doi:10.1016/j.cbpa.2008.04.600) [DOI] [PubMed] [Google Scholar]
- Kojima D., Terakita A., Ishikawa T., Tsukahara Y., Maeda A., Shichida Y.1997A novel Go-mediated phototransduction cascade in scallop visual cells. J. Biol. Chem. 272, 22 979–22 982 (doi:10.1074/jbc.272.37.22979) [DOI] [PubMed] [Google Scholar]
- Kojima D., Mano H., Fukada Y.2000Vertebrate ancient-long opsin: a green-sensitive photoreceptive molecule present in zebrafish deep brain and retinal horizontal cells. J. Neurosci. 20, 2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima D., Torii M., Fukada Y., Dowling J. E.2008Differential expression of duplicated VAL-opsin genes in the developing zebrafish. J. Neurochem. 104, 1364–1371 (doi:10.1111/j.1471-4159.2007.05093.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutalos Y., et al. 1989Regeneration of bovine and octopus opsins in situ with natural and artificial retinals. Biochemistry 28, 2732–2739 (doi:10.1021/bi00432a055) [DOI] [PubMed] [Google Scholar]
- Koyanagi M., Terakita A., Kubokawa K., Shichida Y.2002Amphioxus homologs of Go-coupled rhodopsin and peropsin having 11-cis- and all-trans-retinals as their chromophores. FEBS Lett. 531, 525–528 (doi:10.1016/S0014-5793(02)03616-5) [DOI] [PubMed] [Google Scholar]
- Koyanagi M., Kawano E., Kinugawa Y., Oishi T., Shichida Y., Tamotsu S., Terakita A.2004Bistable UV pigment in the lamprey pineal. Proc. Natl Acad. Sci. USA 101, 6687–6691 (doi:10.1073/pnas.0400819101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M., Nagata T., Katoh K., Yamashita S., Tokunaga F.2008aMolecular evolution of arthropod color vision deduced from multiple opsin genes of jumping spiders. J. Mol. Evol. 66, 130–137 (doi:10.1007/s00239-008-9065-9) [DOI] [PubMed] [Google Scholar]
- Koyanagi M., Takano K., Tsukamoto H., Ohtsu K., Tokunaga F., Terakita A.2008bJellyfish vision starts with cAMP signaling mediated by opsin-Gs cascade. Proc. Natl Acad. Sci. USA 105, 15 576–15 580 (doi:10.1073/pnas.0806215105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S., Meyer A., Kuratani S.2009Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol. Biol. Evol. 26, 47–59 (doi:10.1093/molbev/msn222) [DOI] [PubMed] [Google Scholar]
- Kuwata O., et al. 1990The primary structure of iodopsin, a chicken red-sensitive cone pigment. FEBS Lett. 272, 128–132 (doi:10.1016/0014-5793(90)80465-U) [DOI] [PubMed] [Google Scholar]
- Kuwayama S., Imai H., Hirano T., Terakita A., Shichida Y.2002Conserved proline residue at position 189 in cone visual pigments as a determinant of molecular properties different from rhodopsins. Biochemistry 41, 15 245–15 252 (doi:10.1021/bi026444k) [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Shah S., Suzuki E., Zars T., O'Day P. M., Hyde D. R.1994The Drosophila dgq gene encodes a G alpha protein that mediates phototransduction. Neuron 13, 1143–1157 (doi:10.1016/0896-6273(94)90052-3) [DOI] [PubMed] [Google Scholar]
- Lucas R. J., Hattar S., Takao M., Berson D. M., Foster R. G., Yau W.2003Diminished pupillary light reflex at high irradiances in melanopsin-knockout mice. Science 299, 245–247 (doi:10.1126/science.1077293) [DOI] [PubMed] [Google Scholar]
- Mano H., Kojima D., Fukada Y.1999Exo-rhodopsin: a novel rhodopsin expressed in the zebrafish pineal gland. Brain Res. Dev. Brain Res. 73, 110–118 [DOI] [PubMed] [Google Scholar]
- Max M., McKinnon P. J., Seidenman K. J., Barrett R. K., Applebury M. L., Takahashi J. S., Margolskee R. F.1995Pineal opsin: a nonvisual opsin expressed in chick pineal. Science 267, 1502–1506 (doi:10.1126/science.7878470) [DOI] [PubMed] [Google Scholar]
- Minamoto T., Shimizu I.2002A novel isoform of vertebrate ancient opsin in a smelt fish, Plecoglossus altivelis. Biochem. Biophys. Res. Commun. 290, 280–286 (doi:10.1006/bbrc.2001.6186) [DOI] [PubMed] [Google Scholar]
- Morizumi T., Imai H., Shichida Y.2003Two-step mechanism of interaction of rhodopsin intermediates with the C-terminal region of the transducin alpha-subunit. J. Biochem. 134, 259–267 (doi:10.1093/jb/mvg139) [DOI] [PubMed] [Google Scholar]
- Moutsaki P., Bellingham J., Soni B. G., David-Gray Z. K., Foster R. G.2000Sequence, genomic structure and tissue expression of carp (Cyprinus carpio L.) vertebrate ancient (VA) opsin. FEBS Lett. 473, 316–322 (doi:10.1016/S0014-5793(00)01550-7) [DOI] [PubMed] [Google Scholar]
- Moutsaki P., Whitmore D., Bellingham J., Sakamoto K., David-Gray Z. K., Foster R. G.2003Teleost multiple tissue (tmt) opsin: a candidate photopigment regulating the peripheral clocks of zebrafish? Brain Res. Mol. Brain Res. 112, 135–145 (doi:10.1016/S0169-328X(03)00059-7) [DOI] [PubMed] [Google Scholar]
- Murakami M., Kouyama T.2008Crystal structure of squid rhodopsin. Nature 453, 363–367 (doi:10.1038/nature06925) [DOI] [PubMed] [Google Scholar]
- Nakashima Y., Kusakabe T., Kusakabe R., Terakita A., Shichida Y., Tsuda M.2003Origin of the vertebrate visual cycle: genes encoding retinal photoisomerase and two putative visual cycle proteins are expressed in whole brain of a primitive chordate. J. Comp. Neurol. 460, 180–190 (doi:10.1002/cne.10645) [DOI] [PubMed] [Google Scholar]
- Nathans J., Piantanida T. P., Eddy R. L., Shows T. B., Hogness D. S.1986Molecular genetics of inherited variation in human color vision. Science 232, 203–210 (doi:10.1126/science.3485310) [DOI] [PubMed] [Google Scholar]
- Nikonov S. S., Brown B. M., Davis J. A., Zuniga F. I., Bragin A., Pugh E. N., Craft C. M.2008Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron 59, 462–474 (doi:10.1016/j.neuron.2008.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Larsson T. A., Larhammar D.2004Extensive duplications of phototransduction genes in early vertebrate evolution correlate with block (chromosome) duplications. Genomics 83, 852–872 (doi:10.1016/j.ygeno.2003.11.008) [DOI] [PubMed] [Google Scholar]
- Okano T., Fukada Y., Shichida Y., Yoshizawa T.1992aPhotosensitivities of iodopsin and rhodopsins. Photochem. Photobiol. 56, 995–1001 (doi:10.1111/j.1751-1097.1992.tb09722.x) [DOI] [PubMed] [Google Scholar]
- Okano T., Kojima D., Fukada Y., Shichida Y., Yoshizawa T.1992bPrimary structures of chicken cone visual pigments: vertebrate rhodopsins have evolved out of cone visual pigments. Proc. Natl Acad. Sci. USA 89, 5932–5936 (doi:10.1073/pnas.89.13.5932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano T., Yoshizawa T., Fukada Y.1994Pinopsin is a chicken pineal photoreceptive molecule. Nature 372, 94–97 (doi:10.1038/372094a0) [DOI] [PubMed] [Google Scholar]
- Palczewski K., et al. 2000Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 (doi:10.1126/science.289.5480.739) [DOI] [PubMed] [Google Scholar]
- Panda S., et al. 2003Melanopsin is required for non-image-forming photic responses in blind mice. Science 301, 525–527 (doi:10.1126/science.1086179) [DOI] [PubMed] [Google Scholar]
- Patel A. B., Crocker E., Reeves P. J., Getmanova E. V., Eilers M., Khorana H. G., Smith S. O.2005Changes in interhelical hydrogen bonding upon rhodopsin activation. J. Mol. Biol. 347, 803–812 (doi:10.1016/j.jmb.2005.01.069) [DOI] [PubMed] [Google Scholar]
- Plachetzki D. C., Degnan B. M., Oakley T. H.2007The origins of novel protein interactions during animal opsin evolution. PLoS ONE 2, e1054 (doi:10.1371/journal.pone.0001054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I., Jiang G., de Grip W. J., Hayes W. P., Rollag M. D.1998Melanopsin: an opsin in melanophores, brain, and eye. Proc. Natl Acad. Sci. USA 95, 340–345 (doi:10.1073/pnas.95.1.340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I., Rodriguez I. R., Jiang G., Hayes W. P., Moreira E. F., Rollag M. D.2000A novel human opsin in the inner retina. J. Neurosci. 20, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu R. A., Hu J., Peng J., Bok D., Mata N. L., Travis G. H.2008Retinal pigment epithelium-retinal G protein receptor-opsin mediates light-dependent translocation of all-trans-retinyl esters for synthesis of visual chromophore in retinal pigment epithelial cells. J. Biol. Chem. 283, 19 730–19 738 (doi:10.1074/jbc.M801288200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P. J., Hwa J., Khorana H. G.1999Structure and function in rhodopsin: kinetic studies of retinal binding to purified opsin mutants in defined phospholipid-detergent mixtures serve as probes of the retinal binding pocket. Proc. Natl Acad. Sci. USA 96, 1927–1931 (doi:10.1073/pnas.96.5.1927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P. R., Cohen G. B., Zhukovsky E. A., Oprian D. D.1992Constitutively active mutants of rhodopsin. Neuron 9, 719–725 (doi:10.1016/0896-6273(92)90034-B) [DOI] [PubMed] [Google Scholar]
- Sakmar T. P., Franke R. R., Khorana H. G.1989Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc. Natl Acad. Sci. USA 86, 8309–8313 (doi:10.1073/pnas.86.21.8309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K., Onishi A., Imai H., Chisaka O., Ueda Y., Usukura J., Nakatani K., Shichida Y.2007Physiological properties of rod photoreceptor cells in green-sensitive cone pigment knock-in mice. J. Gen. Physiol. 130, 21–40 (doi:10.1085/jgp.200609729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidou M., Sugahara M., Uchiyama H., Hiraki K.1990On the three visual pigments in the retina of the firefly squid, watasenia scintillans. J. Comp. Physiol. A: Sensory 166, 769–773 [Google Scholar]
- Shichida Y., Imai H., Imamoto Y., Fukada Y., Yoshizawa T.1994Is chicken green-sensitive cone visual pigment a rhodopsin-like pigment? A comparative study of the molecular properties between chicken green and rhodopsin. Biochemistry 33, 9040–9044 (doi:10.1021/bi00197a002) [DOI] [PubMed] [Google Scholar]
- Soni B. G., Foster R. G.1997A novel and ancient vertebrate opsin. FEBS Lett. 406, 279–283 (doi:10.1016/S0014-5793(97)00287-1) [DOI] [PubMed] [Google Scholar]
- Su C.-Y., Luo D.-G., Terakita A., Shichida Y., Liao H.-W., Kazmi M. A., Sakmar T. P., Yau W.2006Parietal-eye phototransduction components and their potential evolutionary implications. Science 311, 1617–1621 (doi:10.1126/science.1123802) [DOI] [PubMed] [Google Scholar]
- Suga H., Schmid V., Gehring W. J.2008Evolution and functional diversity of jellyfish opsins. Curr. Biol. 18, 51–55 (doi:10.1016/j.cub.2007.11.059) [DOI] [PubMed] [Google Scholar]
- Sun H., Gilbert D. J., Copeland N. G., Jenkins N. A., Nathans J.1997Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc. Natl Acad. Sci. USA 94, 9893–9898 (doi:10.1073/pnas.94.18.9893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya A., Knox B. E.1998Enhancement of opsin activity by all-trans-retinal. Exp. Eye Res. 66, 599–603 (doi:10.1006/exer.1997.0453) [DOI] [PubMed] [Google Scholar]
- Suzuki T., Eguchi E.1987A survey of 3-dehydroretinal as a visual pigment chromophore in various species of crayfish and other freshwater crustaceans. Cell. Mol. Life Sci. 43, 1111–1113 (doi:10.1007/BF01956053) [Google Scholar]
- Taniguchi Y., Hisatomi O., Yoshida M., Tokunaga F.2001Pinopsin expressed in the retinal photoreceptors of a diurnal gecko. FEBS Lett. 496, 69–74 (doi:10.1016/S0014-5793(01)02395-X) [DOI] [PubMed] [Google Scholar]
- Tarttelin E. E., Bellingham J., Hankins M. W., Foster R. G., Lucas R. J.2003Neuropsin (Opn5): a novel opsin identified in mammalian neural tissue. FEBS Lett. 554, 410–416 (doi:10.1016/S0014-5793(03)01212-2) [DOI] [PubMed] [Google Scholar]
- Terakita A., Hariyama T., Tsukahara Y., Katsukura Y., Tashiro H.1993Interaction of GTP-binding protein Gq with photoactivated rhodopsin in the photoreceptor membranes of crayfish. FEBS Lett. 330, 197–200 (doi:10.1016/0014-5793(93)80272-V) [DOI] [PubMed] [Google Scholar]
- Terakita A., Yamashita T., Shichida Y.2000Highly conserved glutamic acid in the extracellular IV–V loop in rhodopsins acts as the counterion in retinochrome, a member of the rhodopsin family. Proc. Natl Acad. Sci. USA 97, 14 263–14 267 (doi:10.1073/pnas.260349597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A., Koyanagi M., Tsukamoto H., Yamashita T., Miyata T., Shichida Y.2004Counterion displacement in the molecular evolution of the rhodopsin family. Nat. Struct. Mol. Biol. 11, 284–289 (doi:10.1038/nsmb731) [DOI] [PubMed] [Google Scholar]
- Terakita A., Tsukamoto H., Koyanagi M., Sugahara M., Yamashita T., Shichida Y.2008Expression and comparative characterization of Gq-coupled invertebrate visual pigments and melanopsin. J. Neurochem. 105, 883–890 (doi:10.1111/j.1471-4159.2007.05184.x) [DOI] [PubMed] [Google Scholar]
- Tsukamoto H., Terakita A., Shichida Y.2005A rhodopsin exhibiting binding ability to agonist all-trans-retinal. Proc. Natl Acad. Sci. USA 102, 6303–6308 (doi:10.1073/pnas.0500378102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui K., Imai H., Shichida Y.2008E113 is required for the efficient photoisomerization of the unprotonated chromophore in a UV-absorbing visual pigment. Biochemistry 47, 10 829–10 833 (doi:10.1021/bi801377v) [DOI] [PubMed] [Google Scholar]
- Velarde R. A., Sauer C. D., Walden K. K. O., Fahrbach S. E., Robertson H. M.2005Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem. Mol. Biol. 35, 1367–1377 (doi:10.1016/j.ibmb.2005.09.001) [DOI] [PubMed] [Google Scholar]
- Wada Y., Okano T., Adachi A., Ebihara S., Fukada Y.1998Identification of rhodopsin in the pigeon deep brain. FEBS Lett. 424, 53–56 (doi:10.1016/S0014-5793(98)00138-0) [DOI] [PubMed] [Google Scholar]
- Wandell B. A., Dumoulin S. O., Brewer A. A.2007Visual field maps in human cortex. Neuron 56, 366–383 (doi:10.1016/j.neuron.2007.10.012) [DOI] [PubMed] [Google Scholar]
- Wang Z., Asenjo A. B., Oprian D. D.1993Identification of the Cl(−)-binding site in the human red and green color vision pigments. Biochemistry 32, 2125–2130 (doi:10.1021/bi00060a001) [DOI] [PubMed] [Google Scholar]