Abstract
We previously found that a plasmid bearing a replication initiation region efficiently initiates gene amplification in mammalian cells and that it generates extrachromosomal double minutes and/or chromosomal homogeneously staining regions. During analysis of the underlying mechanism, we serendipitously found that hairpin-capped linear DNA was stably maintained as numerous extrachromosomal tiny episomes for more than a few months in a human cancer cell line. Generation of such episomes depended on the presence of the replication initiation region in the original plasmid. Despite extrachromosomal maintenance, episomal gene expression was epigenetically suppressed. The Southern blot analysis of the DNA of cloned cells revealed that the region around the hairpin end was diversified between the clones. Furthermore, the bisulfite-modified PCR and the sequencing analyses revealed that the palindrome sequence that derived from the original hairpin end or its end-resected structure were well preserved during clonal long term growth. From these data, we propose a model that explains the formation and maintenance of these episomes, in which replication of the hairpin-capped DNA and cruciform formation and its resolution play central roles. Our findings may be relevant for the dissection of mammalian replicator sequences.
Gene amplification plays a pivotal role in mammalian malignant transformation through acquisition of growth advantages or drug resistances (for recent reviews, see Refs. 1–4). Amplified genes are cytogenetically detected at extrachromosomal double minutes (DMs)2 or in chromosomal homogeneously staining region (HSRs). DMs are acentric, atelomeric chromatin bodies composed of amplified DNA of genomic origin. We previously found that a plasmid with a mammalian replication initiation region (IR) and a matrix attachment region (MAR) is efficiently amplified to high copy number in mammalian cells and that it generates DMs and/or HSRs composed of plasmid sequences (5, 6). Because IRs and MARs are scattered throughout the mammalian genome in a frequency of once per several tens of kilobase pairs and because it was proposed that circular molecules excised from the genome may mediate gene amplification in cancer cells (7, 8), we considered the plasmid system as a novel one that efficiently reproduces gene amplification in cultured cells. Thus, we examined the mechanism by which the IR/MAR plasmid mimics gene amplification (5, 9). We suggested that the IR/MAR plasmid undergoes multiplication to produce a large extrachromosomal circle consisting of tandem repeats of plasmid sequences. If multiplication is extensive, the circle forms DMs. If the large circle is integrated into the chromosome, it efficiently initiates a breakage-fusion-bridge cycle, which generates an HSR. Plasmid-borne genes are expressed more actively when they are amplified in DMs than in HSRs (10). Indeed, most of an HSR is heterochromatic, and transcription is detected only at few discrete spots inside the HSR (11). If an inducible promoter in the HSR is activated, the entire HSR is loosened, and active transcription is detected throughout this region (11). Such transcriptional activation, in collaboration with DNA demethylation induced by 5-azacytidine treatment, results in fragmentation of the HSR and the generation of extrachromosomal DMs (10). In addition, gene amplification generated by the IR/MAR plasmid has been used for chromosome studies (12–16), because it creates large HSRs or numerous DMs composed of a defined plasmid sequence.
In previous experiments on IR/MAR plasmid-mediated gene amplification, we always transfected supercoiled covalently closed plasmid DNA into cells. Thus, the initial aim of this study was to examine the effect of the physical structure of the plasmid molecule at the time of transfection to further understand the gene amplification process. Thus, in addition to the supercoiled and the linear IR/MAR plasmid, we examined the effect of a hairpin cap at the end of a linear molecule, because it was suggested that inverted repeats, cruciform formation, and hairpin-capped chromosome ends have implications for gene amplification in yeast (17–20). As a result, we found that the plasmid generated unanticipated structures, numerous extrachromosomal tiny episomes (ETEs). ETEs are tiny dots revealed by fluorescence in situ hybridization (FISH) using a plasmid probe, and they are distinct from the far larger DMs. The appearance of ETEs depends on the presence of the IR sequence, and they are stably maintained in cells for more than 3 months. We propose a model that explains how the hairpin-capped IR/MAR plasmid is maintained extrachromosomally, and the persistence of this structure resembles a recently reported phenomenon in yeast cells (21). Our finding may benefit the analysis of replication initiation in the extrachromosomal context.
EXPERIMENTAL PROCEDURES
Plasmids and DNA
The plasmids pSFVdhfr and pSFV-V (see Fig. 1A) were previously described (5). pSFVdhfr bears a 4.6-kilobase pair fragment of a replication IR from the 3′ region downstream of the DHFR gene, which contains a sequence showing MAR activity; the plasmid also has blasticidine and hygromycin resistance genes. pSFV-V is a control plasmid that lacks the DHFR IR/MAR segment. pΔBM-d2EGFP was constructed by digesting pSFV-dhfr with BamHI/MluI to remove the hygromycin expression cassette, and the remaining portion was ligated to a synthetic 62-bp double-stranded oligonucleotide that bears a multiple cloning site (MCS; 5′-BamHI*-KpnI-AscI-SalI-SwaI-AsiSI-SbfI-BamHI-MluI*-3′, where the asterisk indicates a cohesive end to the denoted enzyme) to produce pΔBM-MCS. pΔBM-d2EGFP (see Fig. 1B) was constructed by digesting pΔBM-MCS with AscI and AsiSI at unique sites in the MCS and ligating it to a PvuI/BssHII fragment derived from pCMV-d2EGFP (10) that contains the d2EGFP gene driven by a cytomegalovirus promoter and terminated by a SV40 poly(A) signal.
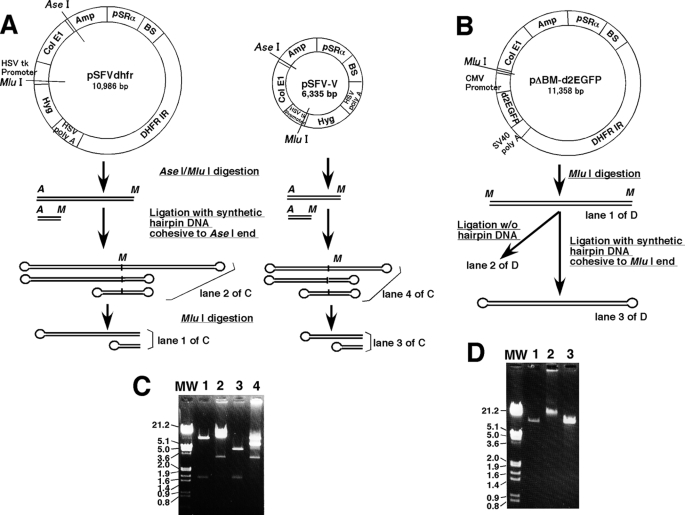
FIGURE 1.
Plasmid DNA used for transfection. The structures of pSFVdhfr, pSFV-V, and pΔBM-d2EGFP are depicted. The plasmids were linearized at the indicated restriction enzyme sites and ligated to synthetic oligonucleotides to generate hairpin-capped ends, as described under “Experimental Procedures.” For pSFVdhfr and pSFV-V, the linear dimer DNA with hairpin ends at both sides was digested with MluI to produce the monomer DNA with hairpin end at one side (A). In C and D, the images of analytical gel for the transfected DNA preparation that appear in A and B are shown. The sizes of all bands were as anticipated from the diagrams in A and B. MW, molecular weight.
Supercoiled plasmid DNA was isolated from bacteria with a plasmid purification kit (Qiagen) and linearized by digesting with the indicated restriction enzyme. Hairpin-capped linear plasmid DNA was prepared as illustrated in Fig. 1. For the experiment in Fig. 3B, pSFVdhfr or pSFV-V was digested by AseI/MluI, and the product was ligated with an excess molar amount of synthetic AseI hairpin DNA (see Fig. 1A). The sequence was 5′-TAA TAT GCT GCA CTG ACG TCA GTG CAG CAT AT, which generates an AseI cohesive end after fold-back. The reaction product should be a linear inverted plasmid dimer in which both ends are hairpin-capped; electrophoretic analysis revealed a predominant band of a size corresponding to the plasmid dimer (see Fig. 1C). Digestion of this preparation with MluI resulted in a linear plasmid monomer, of which one end was hairpin-capped. In other experiments, pΔBM-d2EGFP was digested with MluI, which cuts in the MCS, and the digest was ligated with an excess molar amount of synthetic MluI hairpin DNA (see Fig. 1B), whose sequence 5′-CGC GAT ATG CTG CAC TGA CGT CAG TGC AGC ATA T generates an MluI cohesive end after fold-back. The ligation product was about the same length with the original DNA, whereas the ligation without hairpin DNA produced much larger DNA, as anticipated (see Fig. 1D). Such a preparation was treated with phenol/chloroform, precipitated by ethanol, and used for the transfection.
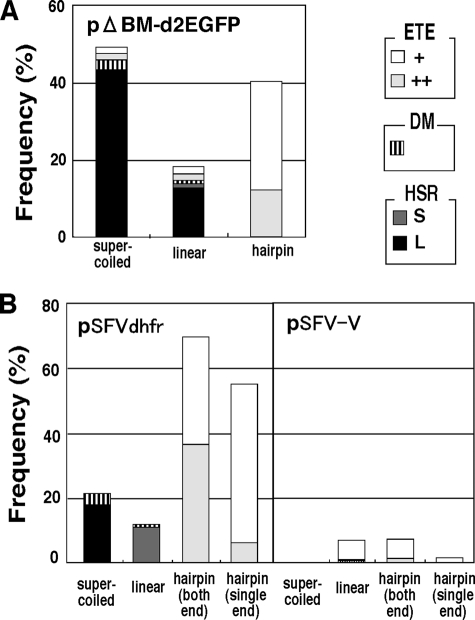
FIGURE 3.
ETEs are generated by hairpin-capped DNA in an IR-dependent fashion. Supercoiled, linear, or hairpin-capped pΔBM-d2EGFP (A) and pSFVdhfr or pSFV-V (B) were prepared as shown in Fig. 1. For the latter plasmids, the DNA was hairpin-capped at both ends or a single end. DNA was transfected into COLO 320DM cells, and after 1 month, the plasmid sequences in stable transformants were analyzed by FISH. Plasmid signals in interphase nuclei are classified as large (L) or small (S) HSRs, DMs, and few (+) or many (++) ETEs, as shown in Fig. 2 (A–F). The frequency of cells bearing each structure was determined by viewing more than 500 interphase nuclei.
Cells, Culture, and Cytogenetic Procedures
Human COLO 320DM (CCL 220) neuroendocrine tumor cells were maintained as previously described (22). There are on average ∼64 copies of the c-myc oncogene in these cells, which localize to DMs. The cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum. All transfections were performed using the Gene Porter 2 lipofection kit (Genlantis Co.). Stable transformants were selected by 5 μg/ml blasticidine (Funakoshi, Co., Tokyo, Japan) and continuously grown in this concentration of blasticidine. More than 1 month after the transfection, the polyclonal transformant was analyzed by FISH to determine the location of plasmid sequence or by genomic Southern blot hybridization. From the same polyclonal transformant, clonal cell populations were obtained by limiting dilution and further grown for ∼2 months.
Metaphase chromosome spreads were prepared from these cells, and the location of plasmid sequences was determined by FISH using a digoxigenin-labeled probe prepared from the plasmid. Probe preparation and FISH procedures were as described (23). The slide was counterstained with propidium iodide and examined with an epifluorescence microscopy (ECLIPSE TE2000-U, Nikon) equipped with a 100× objective lens (Nikon Plan Fluor, NA 1.30 oil) and an appropriate filter set. Digital images were acquired with a Fuji FinePix S1Pro digital camera (Fuji Film Co.) and processed with Adobe Photoshop CS version 8.0.1 (Adobe Systems, Inc.). Sodium butyrate (Sigma), trichostatin A (Sigma), MS-275 (Alexis Biochemical, San Diego, CA), oxamflatin (Calbiochem Biochemicals, Darmstadt, Germany), dimethyl sulfoxide (Nakarai Tesque, Kyoto, Japan), and 5-azacytidine (Sigma) were added to the cultures at the indicated concentrations.
Molecular Biological Procedures
Extraction of total genomic DNA and Southern blot hybridization were as described (5). To amplify the palindrome sequence, we used bisulfite-modified PCR, as described by others (24, 25). Namely, we treated the DNA with bisulfite to convert unmethylated cytosine to uracil by using the EpiTect bisulfite kit (Qiagen) according to the manufacturer's recommendations. Using the bisulfite-modified DNA, PCR was performed as the following condition. The 20-μl reaction mixture contained 1× Blend Taq Buffer (Toyobo Co., Osaka, Japan), 200 μm dNTP (Toyobo), 0.25 unit of Blend Taq Plus (Toyobo), 1 μm primer, and the bisulfite-modified DNA. The PCR condition was 35 cycles or the number of cycles in Fig. 1 at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 60 s. The 20–24-mer primer sequences were designed from the plasmid sequence at the region where no CpG sequence appeared, and all of the cytosine residues in the original sequence were substituted by thymine in the primer sequence. The sequence of the primer will be provided upon request.
To sequence the PCR product, it was cloned by using pGEM-T Easy Vector Systems (Promega) and Escherichia coli DH5α (TOYOBO, Co., Osaka). DNA for sequencing was directly amplified form the E. coli colony by PCR using Blend Taq Plus (TOYOBO). The product was purified by Dye Terminator Removal gel filtration cartridge (Edge BioSystems) and analyzed by the ABI PRISM dGTP BigDye terminator ready reaction kit (Applied Biosystems) and the ABI PRISM 310 genetic analyzer (Applied Biosystems).
RESULTS
A Hairpin-capped Linear IR/MAR Plasmid Persists as ETEs in Transformants
We prepared supercoiled, linear, and hairpin-capped plasmid DNA as described in the legend to Fig. 1 and under “Experimental Procedures,” introduced them into human colorectal carcinoma COLO 320DM cells, and selected stable transformants by blasticidine for 1 month. We prepared metaphase chromosome spreads from transformants and detected plasmid sequences by FISH. As we previously reported (5, 6, 9–12, 14, 15, 23, 26), the supercoiled IR/MAR plasmid efficiently generated HSRs and DMs. Small (Fig. 2A) and large HSRs (Fig. 2B) or DMs (Fig. 2, C and D) could be identified not only in metaphase cells but also in interphase nuclei. We determined their frequencies as shown in Fig. 3. Supercoiled pΔBM-d2EGFP (Fig. 3A) or supercoiled pSFVdhfr (Fig. 3B) generated large HSRs in 40 or 20% of the transformants, respectively, as well as DMs at lower frequencies. As reported, such gene amplification depended on the presence of the IR/MAR sequence in the plasmid, because supercoiled pSFV-V did not generate HSRs or DMs. We also found that the linearized IR/MAR plasmid generated HSRs or DMs. The data in Fig. 3 show that the frequency for linear DNA appeared lower than that for supercoiled DNA. However, the efficiency strongly depended on the site where the plasmid was linearized.3
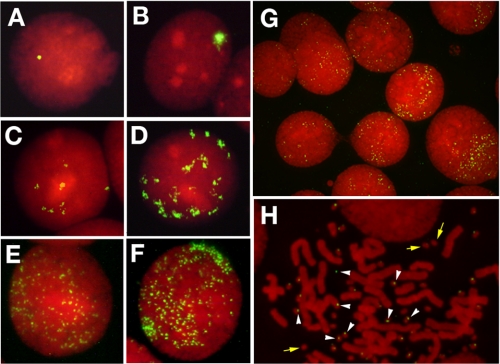
FIGURE 2.
FISH detects DMs, HSRs, or ETEs in cells stably transformed by the IR/MAR plasmid. COLO 320DM cells were transfected with the supercoiled, linear, or hairpin-capped IR/MAR plasmid. One month after selection of stable transformants, metaphase spreads were prepared and hybridized with the plasmid probe. The hybridized probe was detected with FITC (green), and DNA was counterstained with propidium iodide (red). Small (A) or large (B) HSRs, as well as few (C) or many (D) DMs are identified in the interphase nuclei of cells transformed by supercoiled or linear plasmid DNA. In contrast, ETEs are formed in cells transformed by the hairpin-capped IR/MAR plasmid (E–H). The number/nucleus varies from few (E) to many (F). Cells with ETEs are very frequent among transformants (G). In H, a metaphase chromosome spread is shown, where ETEs appear as tiny extrachromosomal entities (white arrowheads) that frequently adhere to DMs (yellow arrows) (6, 23) that form during oncogenesis of this cell line. These small dots are not simple background signal, because they did not appear in the control slide.
We expected that the addition of a hairpin cap to the linear IR/MAR plasmid might affect the frequencies of DM or HSR formation. However, we were surprised to see that hairpin-capped DNA scarcely generated HSRs and conventional DMs, but instead the plasmid sequence appeared as many tiny FISH signals in interphase (Fig. 2G) or metaphase (Fig. 2H) cells. In our previous papers (5, 6, 10, 14, 15, 22, 23, 26), we had detected DMs in COLO 320DM cells by FISH using DM-painting probe or c-myc cosmid probe. However, the chromatin of DMs in these cells is large enough; thus we could easily detect them by DNA staining (yellow arrows in Fig. 2H). In the figure, the tiny FISH spots (white arrowheads) that were invisible by DNA staining stuck to such DMs or apart from them. It is consistent that the extrachromosomal elements frequently stick to the DMs, which was shown for the autonomous replicating plasmid in the same COLO 320DM cells (23). The frequencies of the cells bearing such tiny FISH signals are shown in Fig. 3. Such tiny episomal sequences appeared in initial polyclonal transformants that were grown for 1 month after transfection (Figs. 2 and 3) and cloned cells (see below) that were grown for a few months after transfection. Therefore, these episomal plasmid sequences were stably maintained. We named these extrachromosomal tiny episomes ETEs. The appearance of ETEs depended not only on the presence of the hairpin cap of the transfected DNA (Fig. 3A) but also on the presence of the IR/MAR sequence. Namely, hairpin-capped linear pSFVdhfr generated ETEs, whereas pSFV-V yielded far fewer ETEs (Fig. 3B). On the other hand, linear DNA that was hairpin-capped at one or both ends generated ETEs. This result suggests that the end without the hairpin cap may be ligated intracellularly to generate a molecule with two hairpin-capped ends.
Expression from ETEs Is Epigenetically Silenced
We previously showed that expression from DMs is always much higher than that from HSRs (10). Now, we analyzed expression from ETEs. Because the plasmid pΔBM-d2EGFP contains the d2EGFP expression cassette, which produces an enhanced GFP with a very short cellular half-life, we measured green fluorescence by flow cytometry as previously described (10). We found that d2EGFP expression was lower in cells with the linear plasmid than in cells with the supercoiled plasmid (Fig. 4A). This difference may be explained by the frequency of HSRs (Fig. 3A), although most of the HSRs were silenced (10). Another explanation is that exonucleolytic digestion of the linear molecule in transfected cells might destroy the promoter, because the cytomegalovirus promoter that drives the d2EGFP gene is adjacent to the end of the linear DNA. Southern analysis showed that such digestion actively took place (see below). In contrast, if the hairpin cap was added to the same linear DNA, it generated cells with much higher levels of expression (Fig. 4A). Because the HSR was not detected between the cell population (Fig. 3A), the expression seen in Fig. 4 mostly reflects the expression from ETEs. We show below that the plasmid with a hairpin end was not digested as a linear molecule but that the structure diverged during clonal growth, which might also destroy the flanking cytomegalovirus promoter. Therefore, gene expression from hairpin-derived ETEs might be higher than that from supercoil-derived HSRs. Therefore, application of the hairpin-capped IR/MAR plasmid to protein production may be a future task.
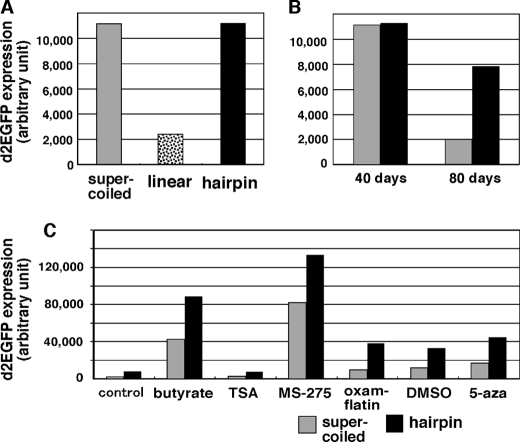
FIGURE 4.
Expression from ETEs is epigenetically silenced. Supercoiled (gray bars), linear (dotted bars in A), or hairpin-capped (black bars) pΔBM-d2EGFP was transfected into COLO 320 cells, and transformants were selected on blasticidine for 40 days (A and C) or 40 or 80 days (B). The expression of d2EGFP was measured with a cell sorter as described (10) and is expressed as arbitrary units. For C, the cells were treated with 2 mm butyrate, 100 nm trichostatin A, 3 μm MS-275, 0.1 μg/ml oxamflatin, 1.6% dimethyl sulfoxide (DMSO), or 3 μm 5-azacytidine for 3 days before measuring d2EGFP expression.
Gene expression in supercoil-derived transformants decreased significantly during cellular passage, which suggests the progression of epigenetic gene silencing in HSR, as we previously showed (10, 11). In contrast, we found that expression in hairpin molecule-derived transformants did not significantly decrease (Fig. 4B). This suggested that epigenetic gene silencing might be weak in ETEs. Because epigenetic silencing is usually mediated by DNA methylation and/or histone deacetylation, we examined the effect of drugs that inhibit such processes (Fig. 4C). Indeed, many of the histone deacetylase inhibitors examined (butylate, trichostatin A, MS-275, oxamflatin, and dimethyl sulfoxide) as well as the DNA methylation inhibitor 5-azacytidine greatly increased gene expression from ETEs, showing that the epigenetic mechanism suppressed gene expression not only in HSRs but also in ETEs. The extrachromosomal environment is generally supposed to be refractory to epigenetic silencing. However, the result reported here suggests that this is not always the case.
Southern Blot Analysis Suggests the Structural Heterogeneity at the Region That Was Hairpin Ends
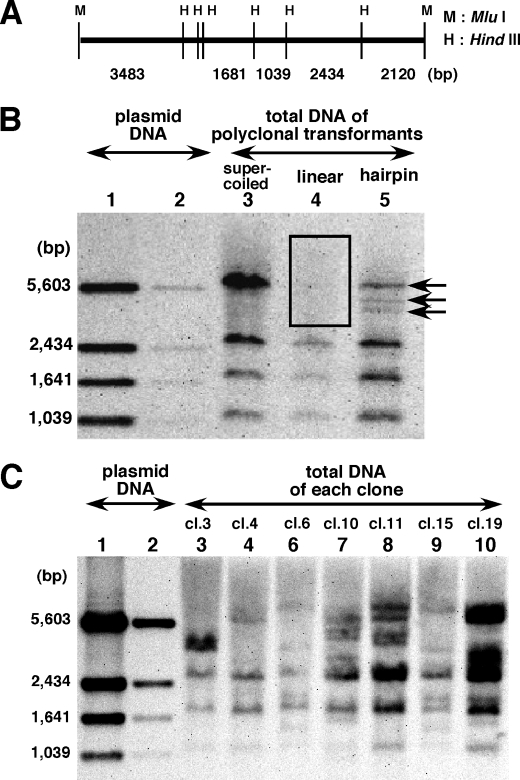
We extracted total DNA from polyclonal transformants that were generated with supercoiled, linear, or hairpin-capped pΔBM-d2EGFP plasmid DNA. We digested the DNA with HindIII, which cuts the original plasmid at several sites (Fig. 5A), and analyzed these restriction products by Southern hybridization using the entire plasmid DNA as a probe (Fig. 5B). DNA from the supercoiled molecule-derived transformant produced hybridization bands that were identical to those of the original supercoiled pΔBM-d2EGFP DNA (compare lane 2 with lane 1). This confirmed our previous result, and it showed that the covalently closed circular IR/MAR plasmid was amplified and arranged as a direct repeat in DMs or HSRs (5). In contrast, the 5,603-bp fragment was not observed in the linear molecule-derived transformant (open box in lane 3), whereas another three bands (1,039, 1,631, and 2,434 bp) were present. Because the original 5,603-bp band contained the MluI site where the plasmid was linearized, this result strongly suggested that linear plasmids were exonucleolytically digested from the ends before ligating with each other in transfected cells, so that the 5,603-bp band became a faint smear. Strikingly, the result was rather different for hairpin-capped molecule-derived transformants. Namely, in addition to the conserved three bands from the internal sequence, there were many hybridization bands instead of the 5,603-bp band. These bands were likely derived from hairpin-capped MluI ends.
FIGURE 5.
Genomic Southern blot analysis suggests both dynamic and static natures of the hairpin end. The MluI (M) and HindIII (H) sites of pΔBM-d2EGFP are shown in A. The plasmid was linearized and hairpin-capped at the MluI site. Supercoiled, linear, or hairpin-capped plasmid DNA was transfected into COLO 320 cells, and polyclonal transformants were selected on blasticidine for 40 days. From cells transfected with hairpin-capped DNA, several clones were isolated after an additional 30 days of culture. Clones bearing ETEs were identified by FISH. The genomic DNA from polyclonal (B) or monoclonal (C) cells was isolated and digested with HindIII, and 2 μg of each was applied to the gel. After electrophoresis, it was subjected to Southern blot hybridization using the entire plasmid DNA as a probe. As a control, purified plasmid DNA was used in lanes 1 (10 ng) and 2 (1 ng) of B and C. The 5,603-bp band disappeared because of smearing in lane 4 of B (indicated by box) or appeared as multiple bands in lane 5 of B (indicated by arrows). Several fragment profiles are seen for different clones, instead of the 5,603-bp band (C).
To understand these extra bands, we isolated several clones from hairpin-capped plasmid transformants. We analyzed 20 clones by FISH and selected eight clones that bear ETEs as the sole location of plasmid sequences. Because the images were very close to the representative ETE images (Fig. 2, E–H), they are not shown. Another 12 clones had no significant FISH signals, which was consistent with the result on polyclonal transformant (Fig. 3). For such ETE-bearing clones, we performed Southern analysis as above (Fig. 5C). As a result, a few discrete bands appeared for each clone in addition to the conserved three bands. These extra bands were rather different between the clones and even different from the bands appearing for the polyclonal cells, from which the clones derived. Because the cloned cells were cultured for more than 2 months after the analysis on the polyclone, the structure of the hairpin-capped end may intermittently diverge during cellular propagation.
Palindrome Sequences around Hairpin-capped Ends Are Specifically Detected in ETE-bearing Cells
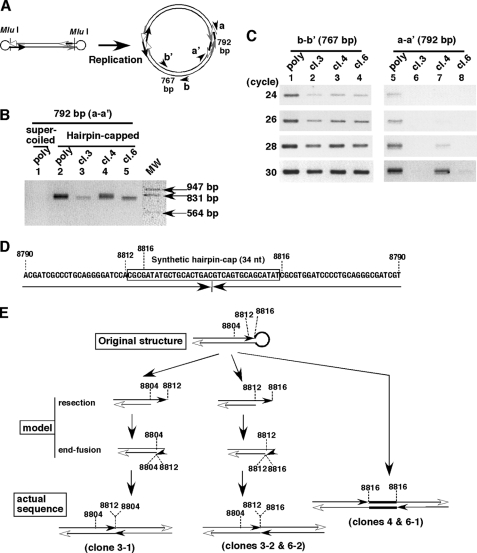
We showed that the generation of ETEs from the hairpin-capped plasmid required the IR sequence (Fig. 3), suggesting a requirement for DNA replication. If a hairpin-capped linear molecule is replicated, it necessarily produces a palindrome sequence, as shown in Fig. 6A. Therefore, we wanted to detect this palindrome in transformants. However, amplification of the palindrome by conventional PCR was difficult, because such a sequence might form a cruciform extrusion. Therefore, we treated total DNA from transformants with sodium bisulfite to convert nonmethylated cytosine to uracil. After that, we performed PCR using a specific primer set (Fig. 6B, a-a′) that would amplify a product if the palindrome were present. If total DNA from a supercoiled plasmid-derived transformant was used as a template, PCR did not amplify any product (Fig. 6B, lane 1). In contrast, a product with the expected size (792 bp; Fig. 6A) was amplified from a hairpin-derived transformant (Fig. 6B, lane 2). On the other hand, a product of the same size was amplified from the original hairpin-capped linear DNA, as predicted (data not shown).
FIGURE 6.
The palindrome sequence that corresponds to the original hairpin end or its end-resected structure was well preserved during clonal long term growth. A, if the hairpin-capped IR/MAR plasmid is replicated, it should contain palindromes. The primer sets a-a′ and b-b′ were designed to amplify such palindromes and the internal region, respectively. B, total DNA was isolated from a polyclonal supercoiled plasmid-derived transformant (lane 1), polyclonal (lane 2), or clones 3, 4, and 6 (lanes 3–5) of hairpin-capped plasmid transformants. The DNA was treated with sodium bisulfite to prevent formation of secondary structures. PCR was performed using primer set a-a′. After 30 cycles, the products were separated by agarose electrophoresis and stained with ethidium bromide. The molecular weight (MW) positions are shown at right. C, the same amount of bisulfite-treated DNA was used for PCR using primer set b-b′ (lanes 1–4) or a-a′ (lanes 5–8). At the cycle number indicated, a portion was removed and analyzed by electrophoresis as in B. D and E, from the bisulfite-modified PCR shown in B, the products were cloned using plasmid vector, and they were subjected to the base sequencing. The sequences were aligned to the theoretical palindrome from the hairpin-capped plasmid sequence (D), and the structures are shown in the bottom of E. By comparing those structures with the one of the introduced molecule, a model that explains how the former structure was formed from the latter is depicted in the middle part of this panel.
We next ligated the PCR product with the vector, cloned it in E. coli, and determined the base sequences of the inserts. For clones 3 and 6, there were two kinds of inserts that differ their length, and we sequenced both of them. By comparing the sequence with the original hairpin-capped plasmid sequence, we found that the sequences from clone 4 and clone 6-1 were identical, and they perfectly matched to the complete palindrome sequence from the original hairpin end (Fig. 6, D and E). On the other hand, the sequences from clones 3-2 and 6-2 were identical. This sequence and the one from clone 3-1 also contained almost the complete palindrome deduced from the MluI end of the plasmid, but they lacked the loop region of the synthetic hairpin (Fig. 6E). Generation of such a structure was most plausibly explained by the fact that the loop region of the hairpin was resected by a single-strand specific nuclease and an exonuclease, followed by the intrastrand annealing using microhomology and repair, which generate another hairpin end. Such a mechanism was suggested to be involved in the initiation of gene amplification (27). It was very surprising that the structure identical to the hairpin-capped end or its replicated form, i.e. palindrome, was preserved for more than 3 months after the transfection.
We next addressed how frequently plasmid copy number was associated with the palindrome structure. Thus, we simultaneously performed a conventional PCR by using another set of primers (Fig. 6A, b–b′) that amplified a portion of the plasmid apart from the hairpin. The intensity of the b-b′ product should reflect the amount of plasmid sequence, whereas the intensity of the a-a′ product should reflect the amount of palindrome. It is uncertain whether the efficiency of amplification might differ between two sets of primer, but we considered the comparison would provide a rough estimate. The result (Fig. 6C) showed that the polyclonal transformant contained a significant amount of the palindrome (compare lanes 1 and 5 at 24 and 26 cycles). However, the amount of palindrome varied significantly among individual clones (compare lanes 2–4 and lanes 6–8, respectively). This result was consistent with the Southern blot data, which suggested the presence of heterogeneity among clones near the hairpin cap-derived sequence (Fig. 5).
DISCUSSION
How Is the Hairpin-capped IR/MAR Plasmid Maintained Long Term in Transfected Cells?
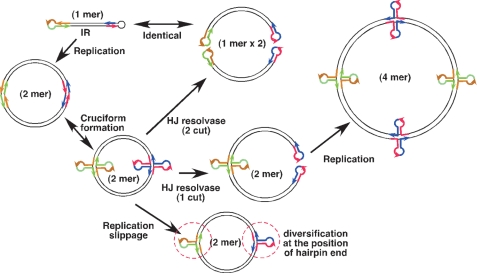
From the above evidence, we propose a model that explains how the hairpin-capped plasmid is maintained in cells in an IR/MAR-dependent fashion (Fig. 7). Our results suggest that the hairpin-capped molecule is replicated in cells to form a circular inverted dimer. Such a molecule has two palindromes, which may be converted to a cruciform. This structure is recognized by the Holliday junction resolvase and cleaved to form an additional hairpin-capped end. If the replication machinery reaches the cruciform, then so-called “replication slippage” might occur. This may explain why the structure around the originally hairpin-capped end becomes diverged among clones. On the other hand, the intensity of bands observed by Southern hybridization applied on cloned cells (Fig. 5C) suggested that between 200 and 2,000 copies of the plasmid sequences are present in each cloned cell, whereas FISH images indicated that each cell contained ∼50–200 copies of ETEs. Therefore, a rough calculation suggests that each ETE consists of 5–10 copies of plasmid sequences on average. On the other hand, the data in Fig. 6 showed that the ETE contained a palindrome. Therefore, transcription of the palindrome would produce a double-stranded RNA that efficiently induces RNA interference and silences plasmid sequences. This proposal is consistent with the finding that ETEs were epigenetically silenced (Fig. 4).
FIGURE 7.
A model that explains how the hairpin-capped IR/MAR plasmid is stably maintained in the cells. See the text for details.
The conversion of a palindrome to a hairpin-capped end has been suggested for yeast. Such hairpin-capped ends may trigger a breakage-fusion-bridge cycle and gene amplification, if they are chromosomally located (17, 19). This process preferentially occurs under conditions where hairpin processing activity is compromised (18). Furthermore, a recent report showed that episomal palindromes launch a self-perpetuating breakage-fusion-bridge-independent increase in copy number termed “escape,” if the hairpin is not processed by Mre11 nuclease activity (21). This phenomenon appears very similar to what happens in our experimental system. Here, ETE formation from the hairpin-capped plasmids was observed in human COLO 320DM cells, but we could not detect this in HeLa or CHO-K1 cells,4 suggesting that a hairpin processing activity might be missing in COLO 320 cells. However, microarray analysis revealed that Mre11 as well as the mammalian Holliday junction resolvase (containing Eme1 and Mus81) (28) is expressed normally in COLO 320 cells.5 Therefore, determining the genetic background that supports the stable maintenance of hairpin-capped IR/MAR plasmids is an important future task.
Implication of This Study
We have made a serendipitous finding that a hairpin-capped linear molecule with an IR/MAR sequence may be maintained as numerous episomes in mammalian cells, and we have proposed a mechanism underlying this process. This study provides a novel method to develop an episomal vector in mammalian cells. Until now, several papers have reported the development of episomal vectors for mammalian cells (29, 30); however, such vectors still pose a difficult challenge except for those using viral elements for replication/segregation. However, sequences that support autonomous replication (the replicator sequence) have been identified in bacteria and yeast cells by isolating sequences that support episomal maintenance. Because of the lack of an episomal system in mammalian cells, studies of replicator activity have completely depended on the analysis of replication initiation at ectopic chromosomal loci, which in turn depends on chromosomal context (31). The findings reported here will provide an ideal system for analyzing replicator activity in mammalian cells.
This work was supported in part by Grant-in-Aid for Scientific Research (B) 17370002 from the Japan Society for the Promotion of Science (to N. S.) and Grant-in-Aid for Scientific Research on Priority Areas, Nuclear Dynamics 19038016 from the Ministry of Education, Science, Sports and Culture of Japan (to N. S.).
M. Uchida, unpublished results.
M. Uchida and N. Shimizu, unpublished observation.
S. Harada, unpublished data.
- DM
- double minute
- ETE
- extrachromosomal tiny episome
- FISH
- fluorescence in situ hybridization
- HSR
- homogeneously staining region
- IR
- initiation region
- MAR
- matrix attachment region
- MCS
- multiple cloning site.
REFERENCES
- 1.Albertson D. G. (2006) Trends Genet. 22, 447–455 [DOI] [PubMed] [Google Scholar]
- 2.Haber J. E., Debatisse M. (2006) Cell. 125, 1237–1240 [DOI] [PubMed] [Google Scholar]
- 3.Bailey S. M., Murnane J. P. (2006) Nucleic Acids Res. 34, 2408–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myllykangas S., Knuutila S. (2006) Cancer Lett. 232, 79–89 [DOI] [PubMed] [Google Scholar]
- 5.Shimizu N., Hashizume T., Shingaki K., Kawamoto J. K. (2003) Cancer Res. 63, 5281–5290 [PubMed] [Google Scholar]
- 6.Shimizu N., Miura Y., Sakamoto Y., Tsutsui K. (2001) Cancer Res. 61, 6987–6990 [PubMed] [Google Scholar]
- 7.Carroll S. M., DeRose M. L., Gaudray P., Moore C. M., Needham-Vandevanter D. R., Von Hoff D. D., Wahl G. M. (1988) Mol. Cell. Biol. 8, 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Von Hoff D. D., Needham-VanDevanter D. R., Yucel J., Windle B. E., Wahl G. M. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4804–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu N., Shingaki K., Kaneko-Sasaguri Y., Hashizume T., Kanda T. (2005) Exp. Cell Res. 302, 233–243 [DOI] [PubMed] [Google Scholar]
- 10.Shimizu N., Hanada N., Utani K., Sekiguchi N. (2007) J. Cell. Biochem. 102, 515–529 [DOI] [PubMed] [Google Scholar]
- 11.Utani K., Shimizu N. (2009) Nucleic Acids Res. 37, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu N., Shingaki K. (2004) J. Cell Sci. 117, 5303–5312 [DOI] [PubMed] [Google Scholar]
- 13.Bosisio D., Marazzi I., Agresti A., Shimizu N., Bianchi M. E., Natoli G. (2006) EMBO J. 25, 798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu N., Misaka N., Utani K. (2007) Genes Chrom. Cancer 46, 865–874 [DOI] [PubMed] [Google Scholar]
- 15.Utani K., Kawamoto J. K., Shimizu N. (2007) Mol. Cancer Res. 5, 695–704 [DOI] [PubMed] [Google Scholar]
- 16.Diefenbacher M., Sekula S., Heilbock C., Maier J. V., Litfin M., van Dam H., Castellazzi M., Herrlich P., Kassel O. (2008) Mol. Endocrinol. 22, 1767–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanan V., Mieczkowski P. A., Kim H. M., Petes T. D., Lobachev K. S. (2006) Cell 125, 1283–1296 [DOI] [PubMed] [Google Scholar]
- 18.Lobachev K. S., Gordenin D. A., Resnick M. A. (2002) Cell 108, 183–193 [DOI] [PubMed] [Google Scholar]
- 19.Lobachev K. S., Rattray A., Narayanan V. (2007) Front. Biosci. 12, 4208–4220 [DOI] [PubMed] [Google Scholar]
- 20.Voineagu I., Narayanan V., Lobachev K. S., Mirkin S. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9936–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coté A. G., Lewis S. M. (2008) Mol. Cell 31, 800–812 [DOI] [PubMed] [Google Scholar]
- 22.Shimizu N., Kanda T., Wahl G. M. (1996) Nat. Genet. 12, 65–71 [DOI] [PubMed] [Google Scholar]
- 23.Hashizume T., Shimizu N. (2007) J. Cell. Biochem. 101, 552–565 [DOI] [PubMed] [Google Scholar]
- 24.Tanaka H., Tapscott S. J., Trask B. J., Yao M. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8772–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rattray A. J. (2004) Nucleic Acids Res. 32, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu N., Ochi T., Itonaga K. (2001) Exp. Cell Res. 268, 201–210 [DOI] [PubMed] [Google Scholar]
- 27.Tanaka H., Cao Y., Bergstrom D. A., Kooperberg C., Tapscott S. J., Yao M. C. (2007) Mol. Cell. Biol. 27, 1993–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiyama T., Katsura M., Yoshihara T., Ishida M., Kinomura A., Tonda T., Asahara T., Miyagawa K. (2006) Nucleic Acids Res. 34, 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenke A. C., Stehle I. M., Herrmann F., Eisenberger T., Baiker A., Bode J., Fackelmayer F. O., Lipps H. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11322–11327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaarschmidt D., Baltin J., Stehle I. M., Lipps H. J., Knippers R. (2004) EMBO J. 23, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aladjem M. I., Fanning E. (2004) EMBO Rep. 5, 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]