Abstract
Target of rapamycin (TOR) is a highly conserved serine/threonine kinase that controls cell growth, primarily via regulation of protein synthesis. In Saccharomyces cerevisiae, TOR can also suppress the transcription of stress response genes by a mechanism involving Tap42, a serine/threonine phosphatase subunit, and the transcription factor Msn2. A physical association between mammalian TOR (mTOR) and the transcription factor signal transducer and activator of transcription-1 (STAT1) was recently identified in human cells, suggesting a similar role for mTOR in the transcription of interferon-γ-stimulated genes. In the current study, we identified a macromolecular protein complex composed of mTOR, STAT1, the Tap42 homologue α4, and the protein phosphatase 2A catalytic subunit (PP2Ac). Inactivation of mTOR enhanced its association with STAT1 and increased STAT1 nuclear content in PP2Ac-dependent fashion. Depletion of α4, PP2A, or mTOR enhanced the induction of early (i.e. IRF-1) and late (i.e. caspase-1, hiNOS, and Fas) STAT1-dependent genes. The regulation of IRF-1 or caspase-1 by mTOR was independent of other known mTOR effectors p70 S6 kinase and Akt. These results describe a new role for mTOR and α4/PP2A in the control of STAT1 nuclear content, and the expression of interferon-γ-sensitive genes involved in immunity and apoptosis.
The macrocyclic lactone rapamycin (Sirolimus, RapamuneTM), as well as its analogues temsirolimus (CCI-779, ToriselTM) and everolimus (RAD-001, CerticanTM), are approved for immunosuppression after organ transplantation, treatment of renal cell carcinoma, and the prevention of coronary artery in-stent restenosis (1). Their only known target is mammalian target of rapamycin (mTOR),3 a highly conserved protein that controls cell growth in response to mitogens and changes in cellular metabolism. The effects of mTOR on cell growth involve the phosphorylation of p70 S6 kinase (S6K) and the translation inhibitor 4E-BP1, key regulators of ribosomal biogenesis and the initiation of protein synthesis (2). In contrast to its role in the initiation of translation, the current study focuses on mTOR as a regulator of mammalian gene transcription.
Studies in Saccharomyces cerevisiae have revealed possible mechanisms by which mTOR might control mammalian transcription factors. TOR-regulated transcriptional control pathways include ribosomal biogenesis, the nutrient deprivation response, and the stress response (3). TOR stimulates 35 S ribosomal RNA expression and ribosomal biogenesis in nutrient-dependent fashion. Inhibition of TOR (e.g. rapamycin or amino acid depletion) reproduces a catabolic response in part by inhibiting rRNA synthesis. In the nutrient deprivation and stress responses, TOR controls the nuclear localization of key transcription factors by mechanisms that require its associated serine/threonine phosphatases (i.e. Pph21, Pph22, or Sit4) and their adaptor, Tap42 (3). Rapamycin blocks phosphorylation of the transcription factor Gln3, as well as its cytosolic scaffolding protein Ure2, permitting Gln3 translocation to the nucleus and the induction of nutrient discrimination pathway genes (4). During heat shock or osmotic stress, inactivation of TOR or Tap42 leads to sustained transcription of stress response genes by augmenting the nuclear content of the transcription factor Msn2 (5). TOR does not appear to affect the phosphorylation of Msn2. Interactions between the mammalian homologues mTOR, α4, and the protein phosphatase 2A catalytic subunit (PP2Ac) have been described, but little is known regarding the mechanism by which TOR controls the induction of stress transcriptional responses in higher eukaryotes.
We recently reported a physical association between mTOR, the transcription factor signal transducer and activator of transcription-1 (STAT1), and the STAT1 kinase protein kinase C-δ (PKCδ) (6). In human lung epithelial carcinoma (A549) cells, mTOR·STAT1 interactions were enhanced by Escherichia coli lipopolysaccharide and the cytokine interferon-γ (IFN-γ), a key inducer of genes involved in microbial killing, inflammation, and apoptosis. Although mTOR is a protein kinase, STAT1 did not appear to be its substrate. We subsequently proposed that, like Msn2 in yeast, mTOR might control the nuclear trafficking of STAT1 independent of its phosphorylation.
IFN-γ transcriptional programs are determined, in large part, by the control of STAT1 nuclear content, and binding of STAT1 to target gene promoters. Phosphorylation of its Tyr-701 residue is required for IFN-γ-induced STAT1 dimerization, accelerated translocation to the nucleus, and physical interaction with DNA-binding elements. STAT1 is also phosphorylated at its Ser-727 residue by novel protein kinase C isoforms (i.e. PKCδ and PKCϵ) (7); this modification controls STAT1 transcriptional activity (8), as well as its nuclear export (9). The induction of early IFN-γ-stimulated genes (e.g. IRF-1 and STAT1) is required for, and amplifies, the maximal expression of late IFN-γ-stimulated genes (10, 11).
Because mechanisms by which TOR controls gene transcription in yeast might be conserved, we hypothesized that inactivation of mTOR augments STAT1 nuclear localization by an α4- and PP2Ac-dependent mechanism. Here we demonstrate a dynamic association between mTOR, STAT1, PP2Ac, and α4 that is regulated by mTOR kinase activity. Inactivation of mTOR, by a mechanism that requires α4 and PP2Ac, increases the nuclear content of STAT1 and amplifies the induction of IFN-γ-stimulated genes involved in inflammation and apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Cytokine Induction
A549, 2fTGH, and U3A cells were cultured as previously described (6, 12). HEK 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, 100 units/ml, and streptomycin, 100 μg/ml. Cells were incubated without or with rapamycin (Biomol), 50 ng/ml, in serum-free or serum-containing media for 1 h before addition of IFN-γ (Roche Applied Science), 100 units/ml, for the indicated times.
Transfection of siRNAs and Plasmids
A549 cells were transfected with 10–30 nm siRNA (siGENOME SMARTpool, Dharmacon) directed against the indicated mRNAs (supplemental Table S1) using Dharmafect I, according to the manufacturer's protocol. A non-targeting siRNA pool (siCONTROL) was used as a negative control. Specificity and potential off-target effects of the siRNA pool were tested using each of two individual siRNA duplexes per mRNA target (supplemental Table S1 and Fig. S3). For the expression of recombinant proteins, sub-confluent A549 or HEK 293T cells were incubated with serum-free medium and mammalian expression vectors, 0.5–1.0 μg of plasmid DNA per 9.6-cm2 culture surface area, mixed with Lipofectamine 2000 or LTX (Invitrogen) in a 2:3 ratio for 5 h before replacement with fresh culture medium. Mammalian expression vectors pRK5 engineered to express Myc-tagged wild-type or dominant-negative (kinase-dead) mTOR (Dr. D. Sabatini, Massachusetts Institute of Technology), were obtained from Addgene. After 48–72 h, experimental protocols were initiated, and lysates were prepared for detection of protein or mRNA.
Preparation of Cell Lysates for Detection of Proteins or Protein Complexes
Endogenous or recombinant proteins in whole cell or purified lysates were detected by Western blot analysis using antibodies listed in Table S2. Whole cell A549 or HEK 293T lysates were generated after washing once with cold phosphate-buffered saline, and incubating for 15 min on ice in lysis buffer A (20 mm Tris, pH 8.0, 0.3% CHAPS, 1 mm EDTA, 10 mm β-glycerophosphate, aprotinin, 10 μg/ml, leupeptin, 10 μg/ml, 1 mm phenylmethylsulfonyl fluoride, 50 mm NaF, 100 μm sodium orthovanadate). After freezing and thawing, cells were homogenized on ice and cleared (1,000 × g for 5 min). Supernatants were further separated (16,000 × g for 30 min) to generate particulate-free lysates. For immunoprecipitation experiments, proteins (1 mg) from whole cell HEK 293T cells lysate were incubated with control IgG, anti-STAT1, or anti-Myc antibody, each 4 μg, overnight at 4 °C before addition of 20 μl of protein G-Sepharose for 1 h. Pellets were washed three times with phosphate-buffered saline containing 0.3% CHAPS before solubilization of bound proteins in SDS sample buffer for 5 min at 95 °C, separation by SDS-PAGE, and detection of bound proteins by Western blot.
Preparation of Nuclear Lysates
A549 cell nuclear lysates were prepared as described previously (13). Briefly, cell pellets were resuspended in nuclear lysis buffer 1 (20 mm Tris, pH 7.5, 10 mm KCl, 1 mm dithiothreitol, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 0.5 mm phenylmethylsulfonyl fluoride, 100 μm sodium orthovanadate) before homogenization (Dounce, Pestle B) and centrifugation (1,100 × g for 3 min). Supernatants were then centrifuged (16,000 × g for 30 min) and used as cytosolic fractions, whereas pelleted nuclei were resuspended in 2 volumes of nuclear lysis buffer 2 (20 mm Tris, pH 7.5, 500 mm KCl, 1 mm dithiothreitol, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 0.5 mm phenylmethylsulfonyl fluoride, 100 μm sodium orthovanadate) before freezing, thawing, and centrifugation for 30 min at 16,000 × g.
GST Pulldown Assay
The cDNA encoding wild-type STAT1 (amino acids 1–750) was cloned by PCR into the Gateway pDONR 221 Entry vector (Invitrogen) using human lung cDNA template and oligonucleotide primers listed in Table S3, and verified by automated sequencing. cDNAs were transferred to Gateway destination vector pDEST15 by recombination, before transformation of E. coli BL21 cells, and induction of GST-STAT1 protein synthesis with 2% arabinose as per the manufacturer's protocol (Invitrogen). Purification of GST-STAT1 from crude bacterial lysates was performed by immobilization on glutathione-Sepharose (Amersham Biosciences), before recovery in elution buffer (50 mm Tris-HCl, pH 8, 40 mm glutathione). Purity was verified by Coomassie blue staining, and confirmed by Western blot analysis with antibodies against STAT1 or GST. Recombinant proteins, 10 μg each, were added to 30 μl of glutathione-Sepharose 4B (Amersham Biosciences), and mixed for 1 h at 4 °C, before addition of equal amounts of whole cell lysate from HEK 293T cells. Beads were washed with phosphate-buffered saline containing 1 mm phenylmethylsulfonyl fluoride and 0.3% CHAPS. Proteins were eluted in SDS sample buffer, separated by SDS-PAGE, and detected by Western blot analysis.
Purification of Endogenous Protein Complexes by Chromatography
Whole cell lysates for ion-exchange and gel-filtration chromatography were prepared using lysis buffer A without detergent. HEK 293T whole cell homogenates were applied to a SP Sepharose column (Amersham Biosciences) before washing with 5 column volumes of buffer B (20 mm Tris-HCl, pH 8.0), and collection of unbound proteins (i.e. flow-through). Protein content was monitored by continuous UV absorption spectrometry. Bound proteins were eluted with a continuous NaCl gradient formed by automated mixing with buffer C (20 mm Tris-HCl, pH 8.0, 1 m NaCl), and collected in 1-ml fractions. Proteins in fractions from chromatographic peaks were detected by Western blot analysis or used for subsequent gel-filtration chromatography.
For gel-filtration chromatography, 500-μl aliquots of the partially purified mTOR·STAT1·PP2Ac-containing fractions generated by cation-exchange chromatography were applied to a Superose 6 column (Amersham Biosciences, 10-mm diameter/24-cm bed height), before perfusion with gel-filtration elution buffer (Buffer A without detergent). Proteins or protein complexes were collected in 0.5-ml fractions and detected by Western blot analysis. The column was calibrated using Dextran blue (2000 kDa), aldolase (158 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa) molecular mass standards dissolved in Buffer A.
Quantification of Protein Levels by Band Densitometry
Images of Western blot films were acquired using an Alpha Imager (Alpha Inotech Corp.) and analyzed with Alpha Ease FC software (version 4.1.0). Band density was obtained using the spot density and autobackground functions. The means of band density values from multiple experiments were compared by Student's t test.
Real-time PCR
RNA was extracted (Illustra RNAspin kit, Amersham Biosciences), and cDNA was generated by reverse transcription from 1 μg of RNA (Superscript II, Invitrogen). TaqMan- or SYBR green-based real-time PCR was performed using 2 μl of cDNA and Power SYBR green or TaqMan Universal PCR master mix (ABI), as per the manufacturers' instructions. Primers are listed in supplemental Tables S4 and S5. PCR reactions were carried out for 45 cycles (ABI 7500 Real Time PCR System). Results are expressed as -fold induction in mRNA levels as calculated by the ΔΔCt method (14).
Fluorescence Imaging of STAT1 or STAT1·PKCδ Heterodimers
For detection of endogenous STAT1, A549 cells were fixed with 4% paraformaldehyde for 15 min at room temperature before permeabilization with 0.2% Triton X-100 and incubation with mouse αSTAT1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and then Alexa Fluor 568 conjugated to anti-mouse antibody (Invitrogen). For expression of enhanced cyan fluorescent protein (ECFP)-tagged STAT1, gateway destination vector pcDNA3.1/nV5/ECFP-DEST was constructed by PCR-amplifying ECFP cDNA using pECFP-C1 (Clontech) as a template, and the following primers, each containing an EcoO109I restriction site: forward, 5′-acaaagggcccaatggtgagcaagggcga-3′; reverse, 5′-acaaagggcccttgtacagctcgtccat-3′. EcoO109I digestion products of pcDNA3.1/nV5-DEST (Invitrogen) and the PCR product were ligated according to the manufacturer's protocol (Invitrogen); the resulting construct was propagated in DB3.1 E. coli-competent cells (Invitrogen), and verified by automated sequencing. The cDNA encoding wild-type STAT1was transferred from pDONR 221-STAT1 WT to pcDNA3.1/nV5/ECFP-DEST by recombination. For experiments employing co-expression of PP2Ac isoforms, wild-type PP2Acα, or its constitutively active Y307F form, were cloned by PCR into Gateway entry vector pDONR 221 using plasmid templates PCDNA3.1- HA-PP2Acα WT and PCDNA3.1-HA-PP2Acα Y307F (Dr. P. Branton, McGill University), and primers listed in supplemental Table S3. The dominant-negative PP2Acα form (L199P) inhibits endogenous PP2Acα function, and cannot bind or hydrolyze its substrates (15); this mutant was engineered by site-directed mutagenesis (Stratagene) using pDONR 221-PP2Acα WT as template and the following primers: forward, 5′-ccaatgtgtgacttgccgtggtcagatccagatgac-3′; reverse, 5′-gtcatctggatctgaccacggcaagtcacacattgg-3′. The cDNAs encoding PP2Ac isoforms forms were transferred from their respective pDONR 221 plasmids to pcDNA3.1/nV5/DEST by recombination. A549 cells were transfected with pcDNA3.1/nV5/ECFP-STAT1 alone (1 μg/9.6-cm2 dish surface area), or with empty vector or pcDNA3.1/nV5-PP2Acα isoforms. For co-expression experiments, STAT1 (0.7 μg/9.6-cm2 dish surface area) and PP2Ac (0.3 μg/9.6-cm2 dish surface area) mammalian expression plasmids were co-transfected.
Plasmid vectors for protein fragment complementation assays (PCAs) were adapted for the Gateway cloning system and provided by Dr. S. Michnick (University of Montreal). These pcDNA3.1 destination vectors contain Gateway recombination sites inserted downstream from cDNAs for yellow fluorescent protein fragments YF1 (amino acids 1–158) or YF2 (amino acids 159–239) (16) (supplemental Fig. S2). Wild-type PKCδ was cloned by PCR into the Gateway entry vector pDONR 221 using the primers and template listed in supplemental Table S3. cDNAs encoding wild-type STAT1 or PKCδ were transferred to the pcDNA3.1-based PCA vectors by recombination. Plasmids, 1 μg/9.6-cm2 dish surface area, were co-transfected in a 1:1 ratio (VF1-STAT1:VF2-PKCδ) by liposomal transfection (Lipofectamine LTX Reagent, Invitrogen) according to the manufacturer's recommendation, before initiation of the indicated experimental protocols. Cells were fixed in 4% paraformaldehyde for 15 min at room temperature, rinsed with phosphate-buffered saline, and mounted in Vectashield Mounting Medium containing the DNA-binding dye 4′,6-diamidino-2-phenylindole (Vector Laboratories).
Fluorescence was detected by multitrack image acquisition (ECFP: excitation 458 nm, emission 475 nm; YFP: excitation 512 nm, emission 529 nm; Alexa Fluor 568: excitation 578 nm, emission 603 nm; 4′,6-diamidino-2-phenylindole: excitation 405 nm, emission 475 nm) using a Plan-Neofluar 40×/1.3× oil differential interference contrast objective and Zeiss LSM 510 META scanning confocal microscope. Images were acquired at room temperature using the integrated Zeiss AxioCam HR digital sensor and LSM 510 software. Fluorescence intensity was assessed by measuring relative pixel density with ImageJ software (Version 1.38x, NIH), and images were displayed using a Zeiss LSM Image Browser (version 4.2.0.121, Carl Zeiss MicroImaging). Results are expressed as the ratio of nuclear to cytoplasmic pixel density.
RESULTS
The Phosphatase Subunits α4 and PP2Ac Associate with mTOR and STAT1 in Mammalian Cells
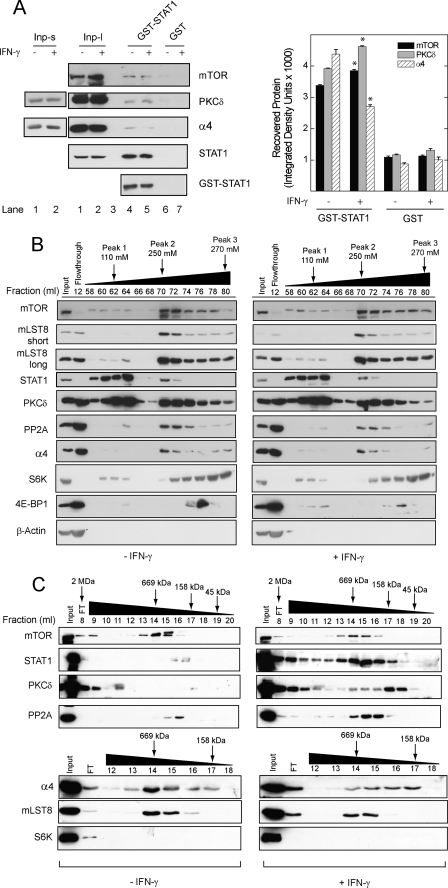
We identified additional components of the mTOR·STAT1 complex by affinity purification of proteins from HEK 293T whole cell lysates, a commonly used system for studying mTOR protein interactions. In GST pulldown assays, recombinant GST-STAT1, but not GST alone, associated with mTOR and PKCδ in HEK 293T lysates, as well as the mTOR-associated phosphatase subunit α4 (Fig. 1A). As expected, GST-STAT1 was strongly associated with endogenous STAT1. Consistent with our previous work (6), IFN-γ led to a small but significant increase in the amount of mTOR or PKCδ that bound recombinant STAT1 (Fig. 1A, right panel). The association between α4 and STAT1 was reduced in lysates from cells exposed to IFN-γ. These results indicate that mTOR, PKCδ, and α4 can associate with recombinant STAT1; however, the levels of mTOR, PKCδ, or α4 bound to GST-STAT1 were a small fraction of the total cellular input (Fig. 1A, lanes 4 and 5 versus lanes 1 and 2), suggesting that the complex may not be abundant, or its assembly might require STAT1 post-translational modification. We therefore proceeded to characterize the complex using endogenous proteins from intact cells.
FIGURE 1.
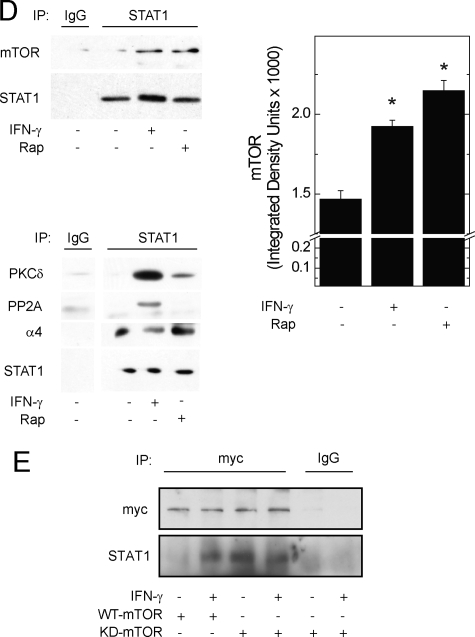
STAT1 associates with mTOR, α4, and PP2Ac in a macromolecular complex. A, HEK 293T cells were exposed to serum-free medium for 1 h before incubation without or with IFN-γ, 100 units/ml for 30 min. Proteins in whole cell homogenates, 2.5 mg, were incubated with recombinant GST alone or GST-STAT1, each 10 μg, bound to glutathione-Sepharose. Bound proteins and sample inputs (Inp) 0.5% of the cell lysate) were separated by SDS-PAGE and detected by Western blot analysis. Long (Inp-l) and short (Inp-s) exposures of the same gel are shown. Average band density for recovered mTOR ± S.E. from three independent experiments is shown (right panel, *, p < 0.05 versus control). B, proteins in whole cell homogenates, 40 mg, from cells serum-deprived cells (1 h) incubated without or with IFN-γ were separated by cation-exchange chromatography. Beginning at the first chromatographic peak (flow-through), 1-ml fractions were collected. A NaCl gradient was applied, beginning at 54 ml (0 mm NaCl). 30-μl aliquots of fractions from the flow-through and the indicated three peaks were analyzed by Western blot analysis. C, samples, 500 μl, from an mTOR·STAT1-containing fraction (Peak 2, 70 ml) were resolved by gel-filtration chromatography, and the indicated proteins were detected by Western blot analysis. The gel-filtration column was calibrated with molecular mass markers (shown above). D, HEK 293T cells, or E, HEK 293T cells expressing Myc-tagged wild-type (WT) or kinase-dead (KD) mTOR, were serum-deprived for 1 h in the absence or presence of rapamycin before incubation without or with IFN-γ for 30 min. Proteins from whole cell lysates, 1 mg, were immunoprecipitated with αSTAT1 (D), α-Myc (E), or normal IgG antibody before detection of protein G-Sepharose-purified proteins by Western blot analysis. αSTAT1 immunoprecipitates were run on separate gels for the detection of mTOR (left) and other mTOR·STAT1 complex proteins (right), respectively. Average band density for recovered mTOR (mean ± S.E.) from three independent experiments is shown (right panel; *, p < 0.05 versus control). All gels are representative of 3–5 independent experiments.
Endogenous proteins in HEK 293T whole cell lysates were separated by ion-exchange and gel-filtration chromatography. HEK 293T cells were serum-deprived for 1 h before incubation without or with IFN-γ for 30 min. After cation-exchange chromatography (Fig. 1B), STAT1, and PKCδ were detected in two chromatographic peaks at elution volumes of 60–64 ml (Peak 1, 110 mm NaCl) and 70–72 ml (Peak 2, 250 mm NaCl), respectively. mTOR, and its associated phosphatase subunit α4, was detected with PKCδ, STAT1, and PP2Ac, but only in fractions from Peak 2 (Fig. 1B). mLST8, a known mTOR-interacting adaptor protein, was also present in Peak 2. The peak elution of mTOR-associated translational control proteins 4E-BP1 and S6K occurred at higher concentration of NaCl. Therefore, the transcription factor STAT1 co-purifies with mTOR, mLST8, PKCδ, PP2Ac, and α4, but not with the translational control factors 4E-BP1 or S6K.
Fractions containing mTOR, STAT1, PP2Ac, and α4 (Peak 2) were next subjected to gel-filtration chromatography (Fig. 1C). In unstimulated cells, mTOR and mLST8 were detected at a molecular mass similar to that of thyroglobulin (∼669 kDa). In cells exposed to IFN-γ, STAT1, PKCδ, and PP2Ac were detected in high molecular weight fractions with mTOR; with the exception of PKCδ, monomeric forms of proteins in the mTOR·STAT1 complex were absent in this cation exchange-purified fraction (i.e. Peak 2). In contrast to PP2Ac, mTOR, and PKCδ, α4 no longer co-eluted with STAT1 in cells exposed to IFN-γ. Although a small amount of S6K was recovered in Peak 2 (Fig. 1B), it did not co-fractionate with STAT1 after gel-filtration chromatography (Fig. 1C). Therefore, the subset of mTOR that binds PKCδ and STAT1 co-elutes with PP2Ac and α4 and is distinct from that which regulates translation initiation or ribosomal biogenesis. The amount of STAT1 that co-elutes with mTOR and α4 represents a fraction of the input, because STAT1 is present in multiple complexes (Fig. 1, B and C). These data are consistent with variation in the amounts of mTOR-associated proteins that bound GST-STAT1 (Fig. 1A).
To determine whether mTOR kinase activity regulates the association between STAT1 and mTOR, we exposed HEK 293T cells to serum-free media, rapamycin, or IFN-γ. Blockade of mTOR activity with rapamycin strengthened the association between STAT1 and mTOR as compared with baseline (Fig. 1D), but had no discernable effect on the association between STAT1 and PP2A or α4. As previously shown (6), IFN-γ increased the association between STAT1 and mTOR. Similar to findings in gel-filtration experiments (Fig. 1C), IFN-γ strengthened the association between STAT1 and PKCδ or PP2Ac.
These results suggest that STAT1 is preferentially associated with inactivated mTOR. Consistent with the effect of rapamycin, the recombinant kinase-dead form of Myc-mTOR exhibited a stronger association with STAT1 than that with the wild type (Fig. 1E, lane 3 versus 1). IFN-γ enhanced the association between wild-type Myc-mTOR and STAT1 (Fig. 1E, lane 2 versus 1) but failed to further increase that between kinase-dead Myc-mTOR and STAT1 (Fig. 1E, lane 4 versus 3). Despite an increased association with kinase-dead mTOR, the phosphorylation of STAT1, as determined by phospho-specific antibodies or gel retardation, did not appear to be altered by rapamycin or expression of the kinase-dead mTOR mutant (supplemental Fig. S1). In vitro kinase assays using mTOR immunoprecipitates were not employed, because at least one other STAT1 kinase is present in the complex (i.e. PKCδ). Taken together, these data indicate that mTOR can be found in a dynamic macromolecular complex with STAT1, PKCδ, and the mTOR-associated phosphatase subunits α4 and PP2Ac. Moreover, mTOR kinase activity regulates its association with STAT1.
Rapamycin Increases the Nuclear Content of mTOR·STAT1 Component Proteins
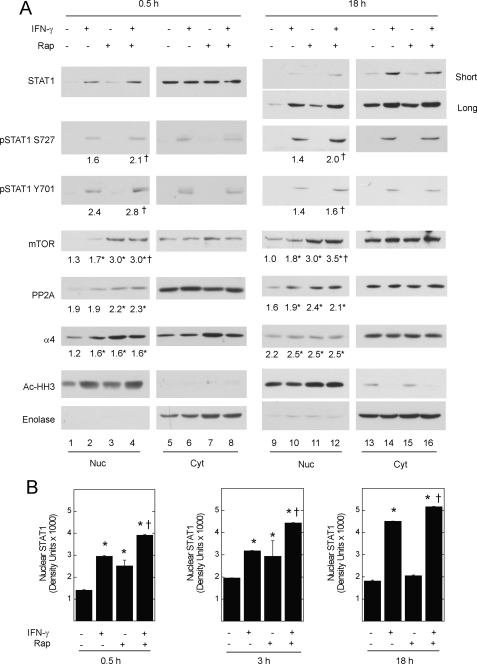
STAT1 did not appear to be a kinase substrate for mTOR (6), and we reasoned that, like the yeast TOR/Msn2 pathway (3), mTOR might control the nuclear content of STAT1. To test for mTOR effects, we used transient exposure to its inhibitor rapamycin, because prolonged genetic or molecular manipulation of mTOR, or its endogenous suppressor TSC2, can alter STAT1 levels and activity (6, 18). Subsequent experiments were performed in lung epithelial adenocarcinoma (A549) cells, because HEK 293T cells do not express many IFN-γ- and STAT1-dependent genes (data not shown) that regulate apoptosis or inflammation (6, 19, 20). A549 cells were serum-deprived for 1 h in the absence or presence of rapamycin, before incubation with serum-free medium or IFN-γ for 0.5, 3, or 18 h, and preparation of nuclear and cytosolic lysates. In cells exposed to IFN-γ, rapamycin further enhanced nuclear STAT1 levels (Fig. 2A, lanes 4 versus 2 and lanes 12 versus 10; Fig. 2B). The enhancing effect of rapamycin was observed for the Tyr-701- and Ser-727-phosphorylated forms of STAT, indicating the accumulation of STAT1 in its transcriptionally active form. Separation of nuclear and cytoplasmic proteins was confirmed by co-localization of acetylated histone H3 and enolase, respectively.
FIGURE 2.
The effect of rapamycin on nuclear STAT1 content. A549 cells were serum deprived for 1 h without or with rapamycin (50 ng/ml) before incubation without or with IFN-γ, 100 units/ml, for 0.5, 3, or 18 h. A, proteins were detected by Western blot analysis after generation of nuclear and cytosolic fractions. Gels shown are representative of six experiments. Average integrated band density × 1000 (S.E. < 5% of the mean for each condition, †, p < 0.05 rapamycin with IFN-γ versus IFN-γ alone; *, p < 0.05 versus control) is indicated below Western blots for nuclear phospho-STAT1, mTOR, PP2Ac, or α4. B, average integrated band density for nuclear STAT1 ± S.E. (*, p < 0.05 versus untreated control; †, p < 0.05 rapamycin with IFN-γ versus IFN-γ alone).
Incubation of cells with rapamycin alone for 0.5, 3, or 18 h was sufficient to increase the nuclear content of STAT1 in its unphosphorylated form (Fig. 2, A (lanes 3 and 11) and B). The nuclear accumulation of STAT1 was accompanied by elevated nuclear levels of mTOR, α4, and PP2Ac seen at all time points (Fig. 2A, lanes 3 and 11). We did not consistently observe an inverse correlation between cytoplasmic and nuclear levels of mTOR·STAT1-associated proteins. These results suggest that a fraction of total cellular STAT1, mTOR, α4, and PP2Ac undergoes nuclear trafficking, and that inactivation of mTOR can increase the nuclear content of mTOR·STAT1-associated proteins.
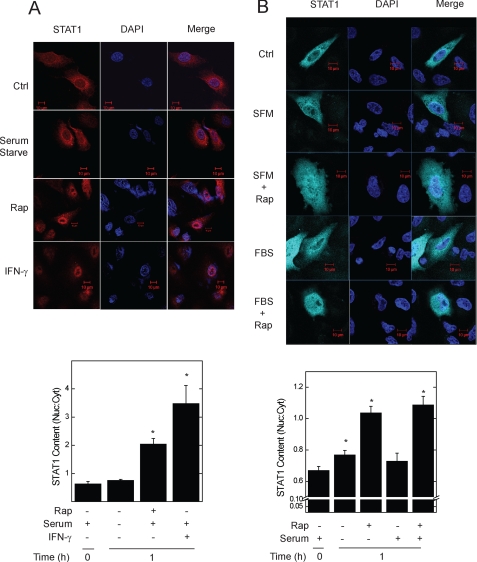
Although rapamycin significantly enhanced the nuclear content of STAT1 (Fig. 2B), changes in nuclear STAT1 content observed by Western blot analysis were of small magnitude, and low levels of the endoplasmic reticulum marker protein Erp57 were detected in nuclear fractions (data not shown). To better determine the effect of mTOR inactivation on constitutive STAT1 nuclear trafficking, we performed fluorescence confocal imaging of untransfected A549 cells, or those expressing wild-type cyan fluorescent protein (CFP)-tagged STAT1 (Fig. 3). As expected, IFN-γ led to a significant increase in nuclear STAT1 as measured by its nuclear to cytoplasmic ratio (Fig. 3A). Incubation of cells with rapamycin for 1 h also augmented endogenous (Fig. 3A) or CFP-STAT1 (Fig. 3B) levels in the nucleus. Serum deprivation minimally increased nuclear levels of recombinant CFP-STAT1, but not those of endogenous STAT1. In cells exposed to IFN-γ, rapamycin-induced changes in nuclear to cytoplasmic STAT1 ratio were difficult to quantify by fluorescence microscopy due to saturation of the nuclear signal (data not shown). Together, results from biochemical and imaging experiments indicate that the increased association between inactivated mTOR and STAT1 (Fig. 1, D and E) is accompanied by enhanced STAT1 nuclear content.
FIGURE 3.
Rapamycin induces STAT1 translocation to the nucleus. A, untransfected A549 cells, or B, those expressing recombinant CFP-STAT1 alone, were incubated without or with serum in the absence or presence of rapamycin, 50 ng/ml, for 0 (Ctrl) or 1 h. Endogenous STAT1 (red) and CFP-STAT1 (cyan blue) were detected by indirect immunofluorescence, and direct fluorescence, confocal microscopy, respectively. Slides were mounted with solution containing the nuclear marker 4′,6-diamidino-2-phenylindole (DAPI, navy blue). Summarized data (mean nuclear to cytoplasmic pixel density ratio ± S.E., n = 3–5 cells per experiment) are shown below the images, and are representative of three independent experiments. *, p < 0.05 versus control.
PP2Ac Mediates the Effect of Rapamycin on STAT1 Nuclear Content
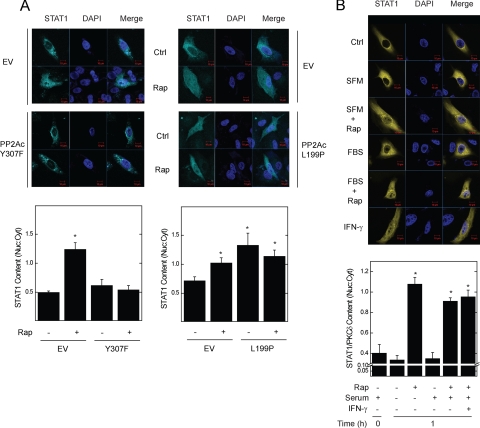
In mammalian cells, mTOR and α4 associated with PP2Ac (21). To determine whether rapamycin effects on STAT1 nuclear content requires PP2Ac activity, we co-expressed its constitutively-active (Y307F) or dominant-negative (L199A) form with CFP-STAT1 (Fig. 4A). Expression of the constitutively active form blocked the rapamycin-dependent increase in nuclear CFP-STAT1 levels. Expression of dominant-negative PP2Ac caused a 2-fold increase in basal CFP-STAT1 levels. In contrast to untransfected (Fig. 3A) or empty vector-transfected cells (Figs. 3B, 4A, and 4B), rapamycin failed to further augment nuclear STAT1 levels in cells expressing dominant-negative PP2Ac. These results demonstrate that PP2Ac activity mediates the effects of mTOR inactivation on STAT1 nuclear trafficking.
FIGURE 4.
mTOR and PP2Ac activity regulate STAT1 nuclear content. A, A549 cells co-expressing CFP-STAT1 and PP2Ac isoforms (Y307F, constitutively active; L199P, dominant-negative) were incubated without or with serum in the absence or presence of rapamycin, 50 ng/ml, for 0 (Ctrl) or 1 h, before detection of CFP-STAT1 (cyan blue) by confocal fluorescence microscopy. B, STAT1 and PKCδ, each linked to complementary fragments of YFP were expressed in A549 cells. Physical interaction of the recombinant proteins leads to reconstitution of active YFP permitting detection by fluorescence microscopy. A549 cells expressing STAT1 and PKCδ PCA constructs were incubated without or with serum in the absence or presence of rapamycin, 50 ng/ml, or IFN-γ, 100 units/ml, for 1 h. Summarized data (mean nuclear to cytoplasmic pixel density ratio ± S.E., n = 3–5 cells per experiment) are shown below representative images of CFP-STAT1 (cyan blue) or STAT1·PKCδ heterodimers (yellow) and nuclei (navy blue), and represent four independent experiments. *, p < 0.05 versus control.
Because rapamycin also enhanced the association between STAT1 and PKCδ (Fig. 1D), we used PCA to test whether mTOR can regulate the nuclear levels of STAT1 complexes. Wild-type recombinant STAT1 and PKCδ, each linked to a complementary fragment of YFP, were expressed in A549 cells (Fig. 4B and supplemental Fig. S2). After 48 h, physical association between STAT1 and PKCδ resulted in the detection of YFP fluorescence in the cytoplasm. Consistent with measurements of endogenous or CFP-tagged STAT1 (Fig. 3), incubation of cells with rapamycin alone for 1 h led to a 2.5-fold increase in STAT1·PKCδ nuclear content. Serum deprivation alone did not alter the nuclear content of the STAT1·PKCδ heterodimer. STAT1·PKCδ translocation to the nucleus elicited by rapamycin was similar to that induced by IFN-γ, and slightly attenuated by the presence of serum. YFP fluorescence was not detected in cells co-transfected with empty vector controls (data not shown).
Taken together, complementary biochemical and imaging studies demonstrate that STAT1 nuclear levels are regulated by mTOR in PP2A-dependent fashion. Rapamycin can increase the nuclear content of unphosphorylated STAT1 in the absence of IFN-γ, or enhance the nuclear content of activated STAT1 in cells exposed to IFN-γ. We next determined whether inactivation of mTOR, or the depletion of α4/PP2Ac phosphatase subunits, leads to enhanced induction of STAT1-dependent genes.
Inactivation of mTOR Enhances the Induction of Early IFN-γ-stimulated Genes
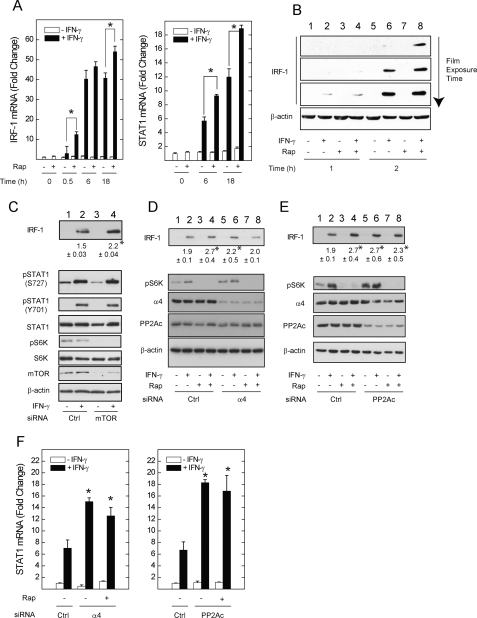
Interferon regulatory factor-1 (IRF-1) is an early STAT1-dependent IFN response gene, the product of which, in a feed-forward positive feedback loop, directly activates the STAT1 gene promoter (10). Newly synthesized STAT1 and IRF-1 then contribute to the induction of other late IFN-γ-stimulated genes (22). Because rapamycin enhanced nuclear STAT1 protein levels (Figs. 2–4), we reasoned that inactivation of mTOR would augment IRF-1 expression in cells exposed to IFN-γ. A549 cells were serum-deprived in the absence or presence of rapamycin before incubation with IFN-γ for up to 18 h. Cells exposed to rapamycin and IFN-γ exhibited greater induction of IRF-1 mRNA (Fig. 5A, left panel; 13- versus 3-fold) and protein (Fig. 5B, lane 8 versus 6) than those exposed to IFN-γ alone (Fig. 5A; 13- versus 3-fold). Subsequent induction of STAT1 mRNA at 6 h, which is primarily regulated by IRF-1, was also significantly enhanced by rapamycin (Fig. 5A, right panel). Interestingly, the effect of rapamycin was less pronounced at 6 h, and then resumed at 18 h, suggesting a secondary signaling event due to new protein synthesis or autocrine signaling. Consistent with increased mRNA levels, the enhancing effect of rapamycin on IRF-1 protein induction was pronounced 2 h after exposure to IFN-γ (Fig. 5B). Incubation of cells with rapamycin alone (i.e. in the absence of IFN-γ) did not appreciably increase IRF-1 mRNA or protein levels (Fig. 5, A and B).
FIGURE 5.
Inhibition of mTOR, or depletion of α4/PP2Ac, augments STAT1 and IRF-1 expression in response to IFN-γ. A549 cells were incubated with serum-free medium in the absence or presence of rapamycin, 50 ng/ml, for 1 h before addition of IFN-γ, 100 units/ml, for 0.5, 6, or 18 h before detection of IRF-1 or STAT1 mRNA (A) or 0.5, 1, or 2 h before detection of IRF-1 protein (B). In panels C–E, cells were transfected with control or siRNA targeting mTOR, α4, or PP2Acα for 72 h before serum withdrawal for 1 h in the absence or presence of rapamycin, incubation without or with IFN-γ for 2 h, and detection of the indicated proteins by Western blot. Average integrated band density for IRF-1 ± S.E. (*, p < 0.05 rapamycin with IFN-γ versus IFN-γ) is shown below each Western blot. F, similarly siRNA-transfected cells were exposed to IFN-γ in the absence or presence of rapamycin for 18 h before detection of STAT1 mRNA levels by real-time PCR. Changes in mRNA levels are expressed as -fold change relative to control mRNA levels = 1 (ΔΔCt method, means of triplicate samples ± S.E.), and are representative of three experiments (*, p < 0.05 versus IFN-γ alone in control-transfected cells). Gels in panels B–E represent data from 3–4 independent experiments.
To further demonstrate an inhibitory role for mTOR, α4, or PP2Ac during the induction of IFN-γ-stimulated genes, we transfected cells with pooled siRNA duplexes to maximize target mRNA and protein depletion while limiting potential off-target effects. In agreement, pooled siRNA targeting α4 or PP2Ac reduced mRNA levels more effectively than each of two individual siRNA duplexes (supplemental Fig. S3, A and B). Moreover, neither pooled nor single siRNA duplexes significantly depleted the levels of other mRNAs encoding proteins in the mTOR pathways (supplemental Fig. S3, C and D). siRNA-mediated depletion of mTOR reproduced the effect of rapamycin on IRF-1 levels (Fig. 5C, lanes 4 versus 2). Phosphorylation of S6K at Thr-389 in mTOR-depleted cells was reduced (lanes 3 and 4), confirming inhibition of mTOR activity.
We next determined whether the enhancing effect of mTOR blockade on IRF-1 or STAT1 expression requires α4 or PP2Ac (Fig. 5, D–F). Like rapamycin (Fig. 5, D and E, lanes 4 versus 2), siRNA-mediated depletion of α4 (Fig. 5D, lanes 6 versus 2) or PP2Ac (Fig. 5E, lanes 6 versus 2) increased IRF-1 protein levels in cells exposed to IFN-γ. Rapamycin failed to further increase IRF-1 protein induction in α4- or PP2Ac-deficient cells (lanes 8 versus 6). Phosphorylation of S6K at the PP2Ac/mTOR-sensitive T389 residue (23) was increased upon depletion of PP2Ac, indicating a reduction in PP2Ac activity (Fig. 5E, lanes 5 versus 1). Depletion of α4 also increased phosphorylation of S6K, indicating that, as previously shown (24, 25), α4 can be a positive regulator of PP2Ac activity (Fig. 5D, lane 5 versus 1). IRF-1 induction was enhanced whether phosphorylation of S6K was reduced (mTOR depletion (Fig. 5C)) or augmented (PP2Ac depletion (Fig. 5E)), suggesting that phosphorylation of S6K is not involved in mTOR regulation of STAT1. Depletion of mTOR, α4, or PP2Ac using each of two single siRNA duplexes reproduced the effects of pooled siRNAs on IRF-1 induction (supplemental Fig. S4). Consistent with elevated IRF-1 levels, depletion of α4 or PP2Ac enhanced STAT1 mRNA induction by 2- and 3-fold, respectively (Fig. 5F). Rapamycin failed to potentiate STAT1 mRNA induction in α4- or PP2Ac-deficient cells. These data demonstrate a requirement for α4 and PP2Ac in the enhancing effect of rapamycin on IRF-1 and STAT1 induction. The effect of rapamycin on STAT1 expression is sustained, and follows the de novo synthesis of IRF-1.
Inhibition of mTOR Enhances the Induction of Late IFN-γ-stimulated Genes
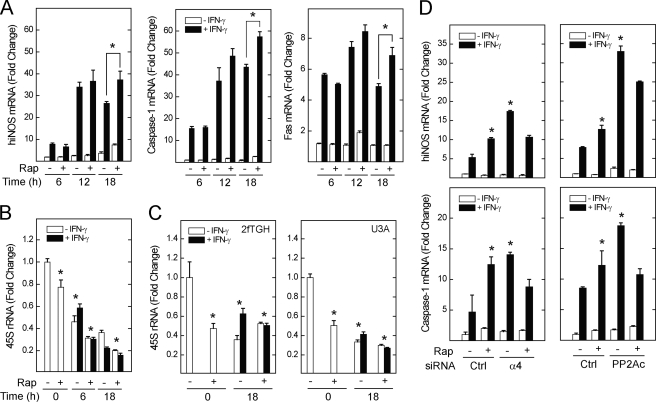
Given the sustained increase in STAT1 and IRF-1 levels upon inactivation of mTOR or its associated phosphatases, we hypothesized that mTOR, PP2Ac, and α4 also control the induction of late IFN-γ-stimulated STAT1-dependent genes. We further proposed that this effect of mTOR should be independent of ribosomal biogenesis, which in yeast is Tap42 (α4)-independent. The human inducible nitric oxide synthase (hiNOS), Fas (CD95), and caspase-1 genes encode pro-inflammatory and pro-apoptotic proteins that play essential roles in the innate immune response and the control of cell death. All three are induced by IFN-γ in STAT1- and IRF-1-dependent fashion (22, 26, 27). Consistent with its enhancing effect on IRF-1 and STAT1 (Fig. 5), rapamycin augmented subsequent hiNOS, Fas, and caspase-1 mRNA levels by 25–38% at 18 h after exposure to IFN-γ (Fig. 6A). There was no significant effect of rapamycin on late IFN-γ-stimulated gene expression at 6 or 12 h. Although this is consistent with a requirement for de novo IRF-1 and STAT1 synthesis, a STAT1-independent effect of rapamycin, or an autocrine mechanism, could not be ruled out.
FIGURE 6.
Inhibition of mTOR, α4, or PP2A augments the induction of STAT1- and IRF-1-dependent IFN-γ-stimulated genes. A549 cells were incubated with serum-free medium in the absence or presence of rapamycin, 50 ng/ml, before addition of IFN-γ, 100 units/ml, for 6, 12, or 18 h. Changes in caspase-1, Fas, and hiNOS mRNA (*, p < 0.05 rapamycin with IFN-γ versus IFN-γ alone) (A), or 45 S rRNA levels (*, p < 0.05 versus control) (B) are expressed as -fold increase relative to control = 1 (ΔΔCt method, means of triplicate samples ± S.E.), and represent data from three experiments. C, STAT1-deficient (U3A) or control (2fTGH) transformed human fibroblasts were exposed to serum-free medium in the absence or presence of rapamycin before addition of IFN-γ for 0 or 18 h and measurement of 45 S rRNA levels (*, p < 0.05 versus control). D, A549 cells were transfected with siRNAs targeting α4 or PP2Acα for 72 h before serum deprivation in the absence or presence of rapamycin and incubation with IFN-γ for 18 h. Changes in hiNOS or caspase-1 mRNA levels are representative of three individual experiments. *, p < 0.05 versus IFN-γ alone in control-transfected cells.
Unlike STAT1-dependent genes, inhibition of mTOR did not augment levels of precursor ribosomal RNA (i.e. 45 S rRNA). In contrast, and as observed in previous studies (28), serum deprivation without or with rapamycin blocked the synthesis of 45 S rRNA; this response was not significantly altered by IFN-γ (Fig. 6B). The inhibitory effect of rapamycin on 45 S rRNA levels was preserved in STAT1-deficient human U3A cells (Fig. 6C), whereas Fas, hiNOS, or caspase-1 mRNA was undetectable (data not shown). As was the case for STAT1 and IRF-1, depletion of α4 or PP2Ac significantly increased hiNOS and caspase-1 mRNA levels in cells exposed to IFN-γ (Fig. 6D). Depletion of α4 or PP2Ac blocked the enhancing effect of rapamycin, indicating that both are required for the effect of mTOR on caspase-1 or hiNOS mRNA levels.
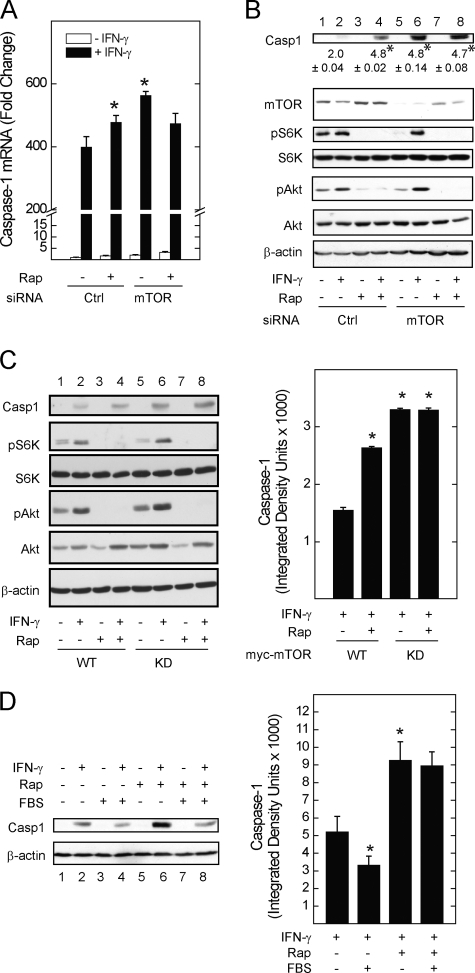
To confirm that the effect of rapamycin reflected a requirement for mTOR activity, we used molecular mTOR inhibitors and measured caspase-1 induction. siRNA-mediated depletion of mTOR (Fig. 7, A and B), as well as the expression of kinase-dead recombinant mTOR (Fig. 7C), significantly enhanced the induction of caspase-1 mRNA and protein levels at 18 h. As expected, phosphorylation of S6K or Akt was reduced in unstimulated mTOR-deficient cells (Fig. 7, B and C, lane 5 versus 1). In contrast to the 2-h time point (Fig. 5C), phosphorylation of S6K or Akt in IFN-γ-stimulated cells was intact, perhaps due to preserved endogenous mTOR activity, or the contribution of other signaling pathways. As was the case for IRF-1 (Fig. 5), caspase-1 levels were enhanced by rapamycin regardless of Akt or S6K phosphorylation state (Fig. 7, B and C, lane 6 versus lanes 4 and 8). Depletion of mTOR or expression of kinase-dead mTOR (Fig. 7, A–C) blocked the enhancing effect of rapamycin on caspase-1 induction, indicating that the effect of rapamycin occurred via mTOR.
FIGURE 7.
mTOR kinase activity is required for the suppression of Caspase-1 induction. A549 cells were transfected with control or siRNA targeting mTOR for 72 h before serum withdrawal for 1 h in the absence or presence of rapamycin, incubation without or with IFN-γ for 18 h, and detection of caspase-1 mRNA levels by real-time PCR (A) or caspase-1 protein levels by Western blot (B). C, Myc-tagged wild-type (WT) or kinase-dead (KD) mTOR were expressed in A549 cells, before incubation without or with rapamycin, 50 ng/ml, in serum-free medium for 1 h followed by the addition of IFN-γ for 18 h. D, A549 cells were exposed to IFN-γ in the absence or presence of rapamycin or serum for 18 h, before preparation of whole cell lysates and detection of caspase-1 by Western blot analysis. Changes in mRNA levels (A) are expressed as -fold change relative to control mRNA levels = 1 (ΔΔCt method, means of triplicate samples ± S.E.), and are representative of three experiments (*, p < 0.05 versus IFN-γ alone in control-transfected cells). Protein levels (mean integrated band density for nuclear STAT1 ± S.E.) were measured by densitometry and are shown to the right of each Western blot. Results from Western blots (B–D) are representative of 3–4 individual experiments (*, p < 0.05 versus IFN-γ alone).
To determine whether caspase-1 induction by IFN-γ is regulated by growth factors, and whether mTOR is required, A549 cells were incubated without or with serum in the absence or presence of rapamycin. The induction of caspase-1 protein levels by IFN-γ was enhanced by rapamycin (Fig. 7D, lane 6 versus 2). Incubation of cells with serum suppressed the induction of caspase-1 by IFN-γ (lane 4 versus 2), and this effect was reversed by the addition of rapamycin (lane 8 versus 4), indicating a role for mTOR in dampening IFN-γ-stimulated gene and protein induction by growth factors.
DISCUSSION
In this study, we identified a novel mechanism by which mTOR controls the transcription factor STAT1. mTOR and the phosphatase subunits α4 and PP2Ac associate with STAT1 in a dynamic protein complex. Inactivation of mTOR favors its association with STAT1 and promotes both constitutive and IFN-γ-induced STAT1 nuclear content. Inactivation of mTOR, α4, or PP2Ac enhances the expression of STAT1-dependent genes and proteins involved in inflammation and apoptosis. The effect of mTOR on STAT1 does not correlate with the phosphorylation status of other known mTOR cell growth effectors (e.g. S6K and Akt) (Figs. 5–7). The regulation of STAT1 by mTOR and α4 is reminiscent of TOR/Tap42-dependent control of Msn2 in yeast (5) and suggests an evolutionarily conserved mechanism by which mTOR controls cellular stress responses.
We show that inactivation of mTOR amplifies the induction of early (i.e. IRF-1) and late (i.e. hiNOS, caspase-1, and Fas) IFN-γ-stimulated genes. Others have shown that early induction of IRF-1 requires rapid translocation of activated STAT1 to the nucleus (29). Newly synthesized IRF-1 permits additional synthesis of STAT1 (10). The increased IRF-1 and STAT1 contribute to the induction of late IFN-γ-stimulated genes, which are regulated by multiple transcription factors, and by autocrine feedback (22). Our data are in agreement with this feed-forward control mechanism and its regulation by mTOR. That is, rapamycin enhanced IRF-1 mRNA at 0.5 but not at 6 h. Similarly, the effect of rapamycin on late IFN-γ-stimulated genes (i.e. 12 and 18 h) followed the new synthesis of IRF-1 and STAT1. Although we cannot exclude a STAT1-independent effect of rapamycin on late IFN-γ-stimulated genes, the enhancement of their mRNA and protein levels coincided with elevated nuclear STAT1 levels (Fig. 2, A and B) and required mTOR, α4, and PP2Ac (Figs. 6 and 7).
The enhancing effect of mTOR inactivation on STAT1 exhibits several similarities to the TOR/Msn2-mediated “stress response” in S. cerevisiae (5). As was the case for TOR and Msn2 (30), we could not detect a role for mTOR in the phosphorylation of STAT1 (6) (Figs. 2 and 5C, and supplemental Fig. S1). Furthermore, rapamycin increased the physical association between mTOR and STAT1 (Fig. 1D, rapamycin alone), as well as STAT1 nuclear content (Figs. 2 (lane 3), 3, and 4), in the absence if IFN-γ (i.e. under conditions where STAT1 was not phosphorylated at Ser-727 or Tyr-701). Therefore, mTOR regulates the constitutive trafficking of STAT1 via a rapamycin-sensitive mechanism that involves an enhanced association between inactive mTOR and STAT1. As is the case for yeast TOR/Msn2, the precise molecular target for regulation of STAT1 nuclear transport by mTOR kinase or PP2Ac phosphatase activity is unknown and awaits further phospho-proteomic characterization of the complex and its components.
In contrast to its stimulation of protein synthesis and cell growth, mTOR attenuated the induction of STAT1-dependent genes. Yeast TOR and Tap42 similarly modify stress response genes by reducing the amount of Msn2 in the nucleus. In contrast to Msn2- or STAT1-dependent stress-induced genes, TOR can also control constitutive levels of nutrient-sensitive genes (5, 31). In yeast, “nutrient discrimination pathway” genes are regulated by a different transcription factor (i.e. Gln3) and its scaffolding protein Ure2. Unlike Msn2 or STAT1, however, rapamycin blocked the phosphorylation of Ure2 and Gln3 (32). As was the case for yeast Msn2 or Gln3, mTOR control of STAT1 was functionally and genetically distinct from its effect on the synthesis of ribosomal RNA precursors (Fig. 6, B and C). Our experiments in α4-depleted cells, or those expressing constitutively active or dominant-negative PP2Ac mutants, place α4 and PP2Ac downstream of mTOR in its control of STAT1, and STAT1-dependent genes (Figs. 4–7). These results implicate mTOR, its associated phosphatases, and STAT1 in a partially conserved mechanism that regulates the levels of stress-induced genes.
Enhancement of IFN-γ/STAT1 signaling is one of several mechanisms by which inactivation of mTOR might control gene transcription in mammalian cells. Increased mTOR activity coincided with inhibition of NF-κB-dependent inflammatory genes (33), and enhanced activity of the transcription factors p53 and c-Jun (34, 35). Sequestration of mTOR in the nucleus prevented its ability to stimulate the synthesis of hypoxia inducible factor-1α protein in the cytoplasm (36). With respect to direct physical interactions with transcription factor complexes, TOR can bind and stimulate control elements in the pre-rRNA gene of S. cerevisiae (37) or those in mammalian mitochondrial biogenesis genes (38). The current study outlines an additional mechanism involving a physical association between mTOR and STAT1 and its effect on STAT1 nuclear content and STAT1-dependent gene transcription.
Intact mTOR kinase activity prevented its association with unphosphorylated STAT1, resulting in reduced constitutive STAT1 nuclear content. In contrast, other described mechanisms that can limit STAT1 activity target phosphorylated STAT1. STAT1-specific cytosolic and nuclear phosphatases and protein inhibitors (e.g. TCPTP and SOCS) can rapidly suppress IFN-γ signaling effects (39). In addition, the nuclear import and export of STAT1 in cells exposed to IFNs is highly regulated (40). The co-repressor protein inhibitor of activated STAT-1 blocks STAT1 DNA-binding activity and leads to its sequestration in nuclear bodies (41). Studies in cells exposed to prolactin demonstrated an effect of α4 on protein inhibitor of activated STAT-1 and STAT1, suggesting an additional mechanism by which mTOR, and its associated phosphatase subunits, might control the expression of STAT1-dependent genes (42).
The role of mTOR in cell growth, proliferation, or death, is a principal preoccupation in research on clinical proliferative disorders, such as tuberous sclerosis complex and lymphangioleiomyomatosis, which involve germ line or mosaic loss of the endogenous mTOR suppressor tuberin (43). However, in animal models and clinical trials, rapamycin, which is intended to block cell growth and proliferation, only partially inhibits tumor or disease progression (44, 45), suggesting that alternative signaling mechanisms might be targeted. In fact, tuberous sclerosis complex 2-deficient tumors are particularly sensitive to the growth inhibitory effects of IFN-γ and exhibit dysregulated STAT1 signaling (46). Consistent with these in vivo observations, we show that, when mTOR activity is reduced, assembly of the mTOR·STAT1 complex can enhance STAT1 nuclear content and the induction of genes involved in apoptosis independently of other known mTOR effectors, such as S6K, Akt, or 4E-BP1. Targeting the physical association between mTOR and STAT1 might therefore represent an additional opportunity to amplify tumor suppressor activity (e.g. IRF-1 and STAT1) and curb the progression of clinical conditions that exhibit excessive mTOR activity. In addition, the involvement of α4, an essential mammalian inhibitor of apoptosis (47), is consistent with a central role for the mTOR·STAT1 complex in the control of cell growth, proliferation, and death.
Supplementary Material
Acknowledgments
We thank Drs. J. Moss and S. Levine (NHLBI, National Institutes of Health) for helpful comments on the manuscript, Drs. G. Stark (Cleveland Clinic, Ohio) for providing STAT1-deficient cells, P. Branton for PP2Ac constructs, S. Michnick for PCA assays, and T. Moss (Laval University, Quebec) for rDNA plasmids and advice regarding rRNA expression experiments. We also thank Dr. D. Sabatini (Massachusetts Institute of Technology) for providing Myc-mTOR mammalian expression vectors via Addgene.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01CA125436. This work was also facilitated by the McGill University Health Centre Confocal Core Facility (S. Laporte) and funded by the Canadian Institutes for Health Research and by an American Thoracic Society/Lymphangioleiomyomatosis Foundation Award in partnership with the Tuberous Sclerosis Alliance and Tuberous Sclerosis Canada.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S5.
- mTOR
- mammalian target of rapamycin
- STAT1
- signal transducer and activator of transcription-1
- IRF-1
- interferon regulatory factor-1
- hiNOS
- human inducible nitric-oxide synthase
- IFN-γ
- interferon-γ
- PKCδ
- protein kinase C-δ
- PP2Ac
- protein phosphatase 2A catalytic subunit
- S6K
- p70 S6 kinase
- siRNA
- small interference RNA
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- GST
- glutathione S-transferase
- PCA
- protein fragment complementation assay
- WT
- wild type
- CFP
- cyan fluorescent protein
- ECFP
- enhanced CFP
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Huang S., Bjornsti M. A., Houghton P. J. (2003) Cancer Biol. Ther. 2, 222–232 [DOI] [PubMed] [Google Scholar]
- 2.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 3.De Virgilio C., Loewith R. (2006) Oncogene 25, 6392–6415 [DOI] [PubMed] [Google Scholar]
- 4.Cooper T. G. (2002) FEMS Microbiol. Rev 26, 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Düvel K., Santhanam A., Garrett S., Schneper L., Broach J. R. (2003) Mol. Cell 11, 1467–1478 [DOI] [PubMed] [Google Scholar]
- 6.Kristof A. S., Marks-Konczalik J., Billings E., Moss J. (2003) J. Biol. Chem. 278, 33637–33644 [DOI] [PubMed] [Google Scholar]
- 7.Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 8.Decker T., Kovarik P. (2000) Oncogene 19, 2628–2637 [DOI] [PubMed] [Google Scholar]
- 9.Lödige I., Marg A., Wiesner B., Malecová B., Oelgeschläger T., Vinkemeier U. (2005) J. Biol. Chem. 280, 43087–43099 [DOI] [PubMed] [Google Scholar]
- 10.Wong L. H., Sim H., Chatterjee-Kishore M., Hatzinisiriou I., Devenish R. J., Stark G., Ralph S. J. (2002) J. Biol. Chem. 277, 19408–19417 [DOI] [PubMed] [Google Scholar]
- 11.Hu X., Herrero C., Li W. P., Antoniv T. T., Falck-Pedersen E., Koch A. E., Woods J. M., Haines G. K., Ivashkiv L. B. (2002) Nat. Immunol. 3, 859–866 [DOI] [PubMed] [Google Scholar]
- 12.McKendry R., John J., Flavell D., Müller M., Kerr I. M., Stark G. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J. E., Chen J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14340–14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 15.Evans D. R., Myles T., Hofsteenge J., Hemmings B. A. (1999) J. Biol. Chem. 274, 24038–24046 [DOI] [PubMed] [Google Scholar]
- 16.Remy I., Montmarquette A., Michnick S. W. (2004) Nat. Cell Biol. 6, 358–365 [DOI] [PubMed] [Google Scholar]
- 17.Deleted in proof
- 18.El Hashemite N., Zhang H., Henske E. P., Kwiatkowski D. J. (2003) Lancet 361, 1348–1349 [DOI] [PubMed] [Google Scholar]
- 19.Kristof A. S., Marks-Konczalik J., Moss J. (2001) J. Biol. Chem. 276, 8445–8452 [DOI] [PubMed] [Google Scholar]
- 20.Kristof A. S., Fielhaber J., Triantafillopoulos A., Nemoto S., Moss J. (2006) J. Biol. Chem. 281, 23958–23968 [DOI] [PubMed] [Google Scholar]
- 21.Nanahoshi M., Nishiuma T., Tsujishita Y., Hara K., Inui S., Sakaguchi N., Yonezawa K. (1998) Biochem. Biophys. Res. Commun. 251, 520–526 [DOI] [PubMed] [Google Scholar]
- 22.Kim H. S., Lee M. S. (2007) Cell Signal. 19, 454–465 [DOI] [PubMed] [Google Scholar]
- 23.Peterson R. T., Desai B. N., Hardwick J. S., Schreiber S. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4438–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inui S., Sanjo H., Maeda K., Yamamoto H., Miyamoto E., Sakaguchi N. (1998) Blood 92, 539–546 [PubMed] [Google Scholar]
- 25.Murata K., Wu J., Brautigan D. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura T., Ishihara M., Lamphier M. S., Tanaka N., Oishi I., Aizawa S., Matsuyama T., Mak T. W., Taki S., Taniguchi T. (1995) Nature 376, 596–599 [DOI] [PubMed] [Google Scholar]
- 27.Kirchhoff S., Sebens T., Baumann S., Krueger A., Zawatzky R., Li-Weber M., Meinl E., Neipel F., Fleckenstein B., Krammer P. H. (2002) J. Immunol. 168, 1226–1234 [DOI] [PubMed] [Google Scholar]
- 28.Moss T. (2004) Curr. Opin. Genet. Dev. 14, 210–217 [DOI] [PubMed] [Google Scholar]
- 29.Pine R., Canova A., Schindler C. (1994) EMBO J. 13, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck T., Hall M. N. (1999) Nature 402, 689–692 [DOI] [PubMed] [Google Scholar]
- 31.Peng T., Golub T. R., Sabatini D. M. (2002) Mol. Cell Biol. 22, 5575–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertram P. G., Choi J. H., Carvalho J., Ai W., Zeng C., Chan T. F., Zheng X. F. (2000) J. Biol. Chem. 275, 35727–35733 [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S., Tergaonkar V., Rothlin C. V., Correa R. G., Bottero V., Bist P., Verma I. M., Hunter T. (2006) Cancer Cell 10, 215–226 [DOI] [PubMed] [Google Scholar]
- 34.Huang S., Shu L., Dilling M. B., Easton J., Harwood F. C., Ichijo H., Houghton P. J. (2003) Mol. Cell 11, 1491–1501 [DOI] [PubMed] [Google Scholar]
- 35.Huang S., Liu L. N., Hosoi H., Dilling M. B., Shikata T., Houghton P. J. (2001) Cancer Res. 61, 3373–3381 [PubMed] [Google Scholar]
- 36.Bernardi R., Guernah I., Jin D., Grisendi S., Alimonti A., Teruya-Feldstein J., Cordon-Cardo C., Simon M. C., Rafii S., Pandolfi P. P. (2006) Nature 442, 779–785 [DOI] [PubMed] [Google Scholar]
- 37.Li H., Tsang C. K., Watkins M., Bertram P. G., Zheng X. F. (2006) Nature 442, 1058–1061 [DOI] [PubMed] [Google Scholar]
- 38.Cunningham J. T., Rodgers J. T., Arlow D. H., Vazquez F., Mootha V. K., Puigserver P. (2007) Nature 450, 736–740 [DOI] [PubMed] [Google Scholar]
- 39.Shuai K., Liu B. (2003) Nat. Rev. Immunol. 3, 900–911 [DOI] [PubMed] [Google Scholar]
- 40.Reich N. C. (2007) Cytokine Growth Factor Rev. 18, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shuai K., Liu B. (2005) Nat. Rev. Immunol. 5, 593–605 [DOI] [PubMed] [Google Scholar]
- 42.Nien W. L., Dauphinee S. M., Moffat L. D., Too C. K. (2007) Mol. Cell Endocrinol. 263, 10–17 [DOI] [PubMed] [Google Scholar]
- 43.Kristof A. S., Moss J. (2003) in Interstitial Lung Disease ( Schwarz M. I., King T. E. eds) pp. 851–864, B. C. Decker, Hamilton, Ontario [Google Scholar]
- 44.Bissler J. J., McCormack F. X., Young L. R., Elwing J. M., Chuck G., Leonard J. M., Schmithorst V. J., Laor T., Brody A. S., Bean J., Salisbury S., Franz D. N. (2008) N. Engl. J. Med. 358, 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee L., Sudentas P., Donohue B., Asrican K., Worku A., Walker V., Sun Y., Schmidt K., Albert M. S., El Hashemite N., Lader A. S., Onda H., Zhang H., Kwiatkowski D. J., Dabora S. L. (2005) Genes Chromosomes Cancer 42, 213–227 [DOI] [PubMed] [Google Scholar]
- 46.El Hashemite N., Kwiatkowski D. J. (2005) Am. J. Respir. Cell Mol. Biol. 33, 227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong M., Fox C. J., Mu J., Solt L., Xu A., Cinalli R. M., Birnbaum M. J., Lindsten T., Thompson C. B. (2004) Science 306, 695–698 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.