Abstract
The unique cytokine interleukin-18 (IL-18) acts synergistically with IL-12 to regulate T-helper 1 and 2 lymphocytes and, as such, seems to underlie the pathogenesis of various autoimmune and allergic diseases. Several anti-IL-18 agents are in clinical development, including the recombinant human antibody ABT-325, which is entering trials for autoimmune diseases. Given competing cytokine/receptor and cytokine/receptor decoy interactions, understanding the structural basis for recognition is critical for effective development of anti-cytokine therapies. Here we report three crystal structures: the murine antibody 125-2H Fab fragment bound to human IL-18, at 1.5 Å resolution; the 125-2H Fab (2.3 Å); and the ABT-325 Fab (1.5 Å). These structures, along with human/mouse IL-18 chimera binding data, allow us to make three key observations relevant to the biology and antigenic recognition of IL-18 and related cytokines. First, several IL-18 residues shift dramatically (>10 Å) upon binding 125-2H, compared with unbound IL-18 (Kato, Z., Jee, J., Shikano, H., Mishima, M., Ohki, I., Ohnishi, H., Li, A., Hashimoto, K., Matsukuma, E., Omoya, K., Yamamoto, Y., Yoneda, T., Hara, T., Kondo, N., and Shirakawa, M. (2003) Nat. Struct. Biol. 10, 966–971). IL-18 thus exhibits plasticity that may be common to its interactions with other receptors. Related cytokines may exhibit similar plasticity. Second, ABT-325 and 125-2H differ significantly in combining site character and architecture, thus explaining their ability to bind IL-18 simultaneously at distinct epitopes. These data allow us to define the likely ABT-325 epitope and thereby explain the distinct neutralizing mechanisms of both antibodies. Third, given the high 125-2H potency, 10 well ordered water molecules are trapped upon complex formation in a cavity between two IL-18 loops and all six 125-2H complementarity-determining regions. Thus, counterintuitively, tight and specific antibody binding may in some cases be water-mediated.
Interleukin (IL)3 -18 is a proinflammatory cytokine that participates in the regulation of innate and acquired immunity (2, 3). IL-18 acts alone or in concert with IL-12 to amplify the induction of proinflammatory and cytotoxic mediators, such as interferon-γ. For example, in IL-18 knock-out mice, levels of interferon-γ and cytotoxic T cells decrease despite the presence of IL-12. Inhibition of IL-18 activity has been found to be beneficial in several autoimmune disease animal models (e.g. collagen-induced arthritis (4) and colitis (5)). Furthermore, IL-18 expression is dramatically increased by the chronic inflammatory state extant in human autoimmune diseases, such as rheumatoid arthritis (6), multiple sclerosis (7, 8), and Crohn's disease (9). These observations suggest that blockade of IL-18 may be a useful human therapeutic modality (10).
Despite functional divergence from the IL-1 cytokine family, IL-18 shares many similarities with IL-1. First, human IL-18 is synthesized as a biologically inactive 24-kDa precursor. Like IL-1β, IL-18 is activated and secreted following cleavage by caspase-1 (and possibly other proteases) that generates the mature 18-kDa polypeptide. Despite low sequence homology to IL-1β (17%), the three-dimensional structure of IL-18 closely resembles the IL-1β β-trefoil fold, as shown by a recent IL-18 NMR structure determination (1). The IL-1 and IL-18 receptors are also homologous; IL-18 binds either to the IL-18Rα chain alone or to the heterodimeric IL-18Rα/IL-18Rβ receptor complex. IL-18 binds to IL-18Rα with ∼20 nm affinity, but signaling occurs only upon formation of the high affinity (0.2 nm) IL-18Rα·IL-18·IL-18Rβ ternary complex (11, 12). Surface mutational analysis has identified two sites for IL-18 binding to IL-18Rα that are similar to those observed in the IL-1β·IL-1Rα binary complex (13) as well as one site important for binding to IL-18Rβ (1).
In a recent study, a potent (0.2 nm) IL-18-neutralizing murine monoclonal antibody (mAb), 125-2H, inhibited binding of IL-18 to IL-18Rα alone but not the heterodimeric IL-18Rα/IL-18Rβ receptor complex, despite rendering the ternary complex with IL-18 non-functional (14). The structural basis for the unusual properties of 125-2H are unclear; the authors suggested that conformational changes in IL-18Rα occur upon formation of the IL-18Rα/IL-18Rβ receptor, thereby altering the interactions with 125-2H (14).
To understand the intricate interactions between IL-18 and this antibody, we have determined the co-crystal structure of human IL-18 and the 125-2H antigen-binding fragment (Fab) at 1.5 Å resolution. This structure rationalizes epitope mapping data, based on human/murine IL-18 chimeras (14), in which the primary antigenic recognition loop is located near the COOH terminus. A secondary loop bolsters the interactions between IL-18 and several 125-2H complementarity-determining regions (CDRs). Comparison of this complex structure with that of the unbound 125-2H Fab (2.3 Å resolution) shows that 125-2H is preorganized for antigen binding. Last, we also have determined the 1.5 Å resolution crystal structure of the Fab fragment of a fully human mAb, ABT-325, that binds a distinct IL-18 epitope, as confirmed by biochemical studies. ABT-325 is entering clinical trials for a variety of autoimmune disease indications.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human IL-18
Recombinant human pro-IL-18, in which the five cysteine residues at positions 10, 74, 104, 112, and 163 were mutated to alanine (“pro-IL-18–5C→A”; hereafter simply pro-IL-18; following UniProt entry Q14116, mature IL-18 comprises residues 37–193), was expressed with an amino-terminal His6 affinity purification tag followed by a tobacco etch virus protease cleavage peptide in Escherichia coli BL21 cells. The following procedure was carried out at 4 °C unless specified otherwise. Cells from a 1-liter culture (stored frozen at −80 °C) were thawed, resuspended in 25 ml of Buffer A (1× PBS (150 mm NaCl, 10 mm NaH2PO4, pH 7.2 (NaH2PO4 solution in which the pH was adjusted to 7.2 using NaOH)), 1 “protease tab” (EDTA-free complete protease inhibitor; Roche Applied Science, Part 1-873-580), and 10% glycerol), sonicated on ice (six 30-s iterations, 40% duty cycle, medium output), and centrifuged (GSA rotor, 17,000 rpm, 25 min). A 5-ml Ni2+-nitrilotriacetic acid affinity column (Qiagen) was prepared by washing sequentially with H2O (25 ml), 100 mm NiCl2 (50 ml), H2O (25 ml), and Buffer B (1× PBS, 10% glycerol, 10 ml). After applying the cell lysate supernatant to the column (2 ml/min flow rate), the column was washed with Buffer B plus 25 mm imidazole until nonspecifically bound proteins were eluted (monitored by absorbance at 280 nm). Pro-IL-18 was eluted with Buffer B plus 100 mm imidazole. Fractions containing a protein concentration greater than 0.3 mg/ml (Coomassie protein assay; Bio-Rad) were pooled. The pooled sample was diluted 1:2 with 50 mm Tris, pH 7.5. Caspase-1 (1 ml of caspase-1 per 36 mg of pro-IL-18; in a spectrophotometric enzymatic assay, 10 μl of this preparation gave a signal of 5.0 milliabsorbance units/min at 405 nm in a 10-min assay with 100 μm Ac-YVAD-p-nitroanilide (15)) was added to the pro-IL-18, and the mixture was incubated for 40 min at 30 °C. The sample was dialyzed against Buffer C (50 mm Tris, pH 8.0, 10% glycerol, 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride) overnight at 4 °C. The mixture was centrifuged to remove precipitated protein, filtered (0.2 μm), and loaded onto a MonoQ 10/10 anion exchange column (GE Healthcare; previously washed with Buffer C (40 ml); 2 ml/min). The column was washed with 5–7 column volumes (∼50 ml) of Buffer C until the A280 returned to base line. Mature IL-18 was eluted with a linear gradient of 0–0.5 m NaCl in Buffer B (50 column volumes (∼400-ml total volume)). A major peak eluted at ∼120 mm NaCl. The sample was concentrated to ∼20 mg/ml (Ultrafree-15 Biomax 10,000 molecular weight cut-off; Millipore) and frozen at −80 °C. Sample purity and identity were assessed with SDS-PAGE and mass spectrometry.
125-2H Fab Fragment
Murine IgG 125-2H was prepared from the hybridoma cell line (16) by the ascites method at Maine Biotechnology Services (Portland, ME). Papain gel slurry (Pierce) was activated with 3 volumes of Buffer D (20 mm Na2HPO4, 10 mm EDTA, 20 mm cysteine). The mAb was concentrated from 2.1 to 20 mg/ml in 1× PBS (Ultrafree-15 Biomax, 10 kDa), mixed with 50% papain gel slurry, and incubated at 37 °C for 24 h with gentle shaking. After overnight dialysis at 4 °C against Buffer E (50 mm Tris, pH 7.0) to remove cysteine, the sample was applied to a Protein A-Sepharose 4 Fast Flow affinity column (GE Healthcare; 25 ml; prepared by washing with Buffer E (100 ml)) at 2 ml/min. 125-2H Fab fractions (monitored by A280) were collected in the flow-through. Fractions containing the 125-2H Fab at >0.3 mg/ml were pooled, dialyzed overnight against Buffer F (50 mm Tris, pH 8.25), and then applied to a MonoQ 10/10 column (pre-equilibrated with Buffer F) at 2 ml/min. The column was washed with 3 column volumes of Buffer F, followed by elution with a 0–50% gradient of Buffer F/Buffer F + 500 mm NaCl. Four peaks, which corresponded to four species of the 125-2H Fab with distinct pI values, eluted. The major first peak was collected, concentrated (Ultrafree-15 Biomax, 10 kDa) to ∼20 mg/ml, and frozen at −80 °C.
ABT-325 Fab Fragment
ABT-325 IgG was expressed in Chinese hamster ovary cells in SR-286 medium. The supernatant after cell lysis was filtered through a 0.5-μm filter and loaded onto a Protein A affinity column (pre-equilibrated with 1× PBS). After washing, the IgG was eluted with Buffer G (150 mm NaCl, 0.1 m NaOAc, pH 3.5). The pooled IgG was concentrated to 20 mg/ml; papain digestion and Protein A purification was performed as described for 125-2H. Fractions containing the ABT-325 Fab at >0.3 mg/ml were pooled, concentrated to ∼20 mg/ml, and frozen at −80 °C.
IL-18·125-2H Fab Fragment Complex
IL-18 and the 125-2H Fab fragment were mixed in a 1:3 mass ratio and incubated for 1 h at 4 °C. After overnight dialysis against Buffer H (50 mm Tris, pH 8.0, 10% glycerol, 2.5 mm EDTA), the sample was applied to a MonoQ 10/10 column (pre-equilibrated with Buffer H) at 2 ml/min. The column was washed with 3 column volumes of Buffer H, and the IL-18·125-2H Fab complex was eluted with a 0–40% gradient of Buffer H/Buffer H + 500 mm NaCl. The complex was concentrated to ∼10 mg/ml and frozen at −80 °C.
Crystallization
Frozen 125-2H Fab stock (∼13 mg/ml) was thawed on ice. The Fab (2 μl) was mixed with 2 μl of a reservoir solution consisting of 10% polyethylene glycol 6000, 100 mm HEPES, pH 7.5, 5% 2,4-methylpentanediol and suspended over the reservoir (siliconized glass coverslip) at 4 °C. Rodlike crystals appeared within 1 day.
Frozen ABT-325 Fab stock (∼20 mg/ml) was thawed on ice. The Fab (2 μl) was mixed with 2 μl of a reservoir solution consisting of 25–30% polyethylene glycol 400, 100 mm CAPS, pH 10.5, and suspended over the reservoir at 4 °C. Rodlike crystals appeared within 1 day.
Frozen IL-18·125-2H Fab complex stock (∼10 mg/ml) was thawed on ice. The complex (1.5 μl) was mixed with 1.8 μl of reservoir solution (30% polyethylene glycol 4000, 100 mm Tris, pH 8.5, 0.2 m MgCl2), and 0.3 μl of 300 mm Sulfo-Betaine 201. The mixture was suspended over the reservoir (siliconized glass coverslip) at 18 °C. Rodlike crystals appeared within 1 week.
Structure Determination
Crystals of the ABT-325 Fab were harvested directly from their mother liquor using a fiber loop. Crystals of the IL-18·125-2H Fab complex and the 125-2H Fab were harvested in mother liquor plus 20% propylene glycol or 25% glycerol, respectively. All crystals were then flash-cooled by plunging into liquid nitrogen and stored in a liquid nitrogen refrigerator until x-ray diffraction data were collected.
X-ray diffraction data were collected by the rotation method at a temperature of 100 K either at the COM-CAT/32-ID (ABT-325 Fab and IL-18·125-2H Fab complex) or IMCA-CAT/17-ID (125-2H Fab) beam lines at the Advanced Photon Source (Argonne, IL). Data were indexed, integrated, and scaled and merged with HKL-2000 (17) (ABT-325 Fab and IL-18·125-2H Fab complex) or MOSFLM (18) and SCALA (19) (125-2H Fab) and then placed on an absolute scale with TRUNCATE (20). Five percent of the unique reflections for each crystal were randomly assigned to the “free” set, for calculation of the free R-factor (21). Further data manipulation was performed with the CCP4 Program Suite (22). Diffraction data statistics are summarized in Table 1.
TABLE 1.
X-ray data collection and refinement statistics
| Structure | ABT-325 Fab | 125-2H Fab | IL-18·125-2H Fab complex |
|---|---|---|---|
| Protein Data Bank entry | 2vxv | 2vxu | 2vxt |
| Data collection | |||
| Resolution (Å) | 50.0-1.49 | 17.5-2.33 | 50.0-1.50 |
| Space group | P212121 | P212121 | P21 |
| Unit cell lengths (Å) | a = 64.4 | a = 79.1 | a = 44.4 |
| b = 74.1 | b = 92.5 | b = 62.4 | |
| c = 107.3 | c = 137.7 | c = 104.7 | |
| Unit cell β angle (degrees) | 90 | 90 | 100.4 |
| Unique reflections | 83,126 | 41,229 | 86,865 |
| Mosaicity (degrees) | 0.3 | 0.4 | 1.0 |
| Overall statistics (highest shell) (Å) | 1.54-1.49 | 2.39-2.33 | 1.54-1.49 |
| Rsym (%) | 0.085 (0.414) | 0.046 (0.238) | 0.056 (0.354) |
| I/σI | 9.8 (3.1) | 18.9 (4.9) | 10.4 (2.3) |
| Data completeness (%) | 99.0 (92.3) | 94.0 (25.2) | 94.8 (56.6) |
| Mean multiplicity | 6.4 (4.1) | 5.2 (2.4) | 3.5 (2.1) |
| Wilson plot B-factor (Å2) | 16 | 41 | 21 |
| Refinement | |||
| Reflections used in refinement | 78,910 | 39,136 | 82,477 |
| Rcryst (%) | 15.3 | 18.4 | 16.2 |
| Rfree (%) | 17.9 | 24.2 | 19.6 |
| r.m.s. deviations, bond lengths (Å) | 0.015 | 0.014 | 0.012 |
| r.m.s. deviations, bond angles (degrees) | 1.6 | 1.5 | 1.7 |
| Ramachandran plot (% most favored and additionally allowed residues) | 99.2 | 99.4 | 99.0 |
| Atoms (including alternate conformations; protein, ligands, water molecules) | 3554, 20, 576 | 6702, NA,a 488 | 4779, 3, 593 |
| Average B-factors (protein, ligands, water molecules) (Å2) | 16.0, 26.8, 33.1 | 49.1, NA, 50.8 | 21.7, 25.1, 35.9 |
a NA, not applicable.
The ABT-325 Fab and IL-18·125-2H Fab complex structures were solved by molecular replacement using AMORE (23) in space groups P212121 and P21, respectively, using the NMC-4 Fab (Protein Data Bank entry 1FNS (24)) as the search model. For each, there was one obvious cross-rotation function and translation function solution. The structure of the 125-2H Fab was solved using PHASER (25, 26) in space group P212121, using the (refined) 125-2H Fab coordinates (from the complex) as the search model. The structure determination was complicated by non-crystallographic translational pseudosymmetry between the two Fab molecules in the asymmetric unit (177° rotation axis, offset 2.2° from the a axis; self-Patterson function peak at [0, ½, 0.38].
Refinement of the ABT-325 Fab and IL-18·125-2H Fab complex structures began with rigid body refinement using CNS (commercial version CNX-2002; Accelrys) (27). De novo IL-18 electron density was calculated with ARP/WARP (28). Slowcool simulated annealing molecular dynamics refinement (29) (CNS; 3000 to 300 K) was followed by cycles of Powell positional refinement at increasingly higher resolutions (up to 1.5 Å), including bulk solvent correction and individual temperature factor refinement; refinement alternated with manual rebuilding in the molecular graphics program O (30) into σA-weighted 2Fo − Fc and Fo − Fc electron density maps (31). Refinement concluded in REFMAC (32) with the addition of water molecules, residues in alternate conformations, refinement of overall anisotropic temperature factors, and translation-libration-screw modeling (33). Refinement of the 125-2H Fab structure was carried out similarly, using exclusively REFMAC. In addition to the proteins and water molecules, the ABT-325 Fab structure includes one buffer molecule (CAPS) and one glycerol molecule; the IL-18·125-2H complex structure includes one Mg2+ and two Cl− ions. Refinement statistics are summarized in Table 1.
The quality of all models was evaluated using PROCHECK (34) and WHATCHECK (35). Geometrical parameters and surface areas were calculated with SC, AREAIMOL, and CONTACT (22). The surface shape complementarity statistic, Sc, of the IL-18/125-2H Fab interface is 0.73, which is typical for antigen/antibody interactions (36). Structural alignments were performed with LSQMAN (37), and cavity volumes were defined with VOIDOO (38). Water molecules were ignored in all surface and cavity calculations. Analysis of pockets and cavities present in Protein Data Bank structures made use of the CASTp (39) (available on the World Wide Web) and GPSS (40) (available on the World Wide Web) servers; additional details are provided in the supplemental material. Figures were prepared with PyMOL (60) and VMD (41); surfaces and electrostatic potentials were calculated with MSMS (42) and APBS (43). Atomic coordinates and observed structure factors have been deposited at the Protein Data Bank (Table 1).
Epitope Mapping
Human/mouse and mouse/human IL-18 chimeric proteins were produced by in vitro transcription and translation in the pro-IL-18 form, with COOH-terminal V5 and His tags. Caspase-1 cleavage generated the mature, tagged IL-18 chimeras. Binding assays in a sandwich enzyme-linked immunosorbent assay format were carried out by capturing the IL-18 chimeras with the test antibody, followed by detection with an anti-tag antibody. Full experimental details are provided in Ref. 14.
RESULTS
We present here crystal structures of the Fab fragments of the human IL-18-specific monoclonal antibodies (mAbs) ABT-325 (fully human; also known as A-794722.0 or 2.5(E)mg1) and 125-2H (murine), as well as that of the human IL-18·125-2H Fab complex, at 1.5-, 2.3-, and 1.5-Å resolution.
Throughout, we used a form of human IL-18 in which all cysteine residues were mutated to alanine. An analogous Cys/Ser mutant has been reported (44). Expression and purification of this mutant IL-18 was greatly simplified, compared with the wild type protein, probably due to inhibition of polymerizing oxidation of surface-exposed residues Cys74 and Cys104.
Wild type and mutant IL-18 exhibited comparable antibody binding characteristics and biological activities. Mutant IL-18 binds both 125-2H and ABT-325 (45) with a KD of ∼0.2 nm. Both antibodies neutralize recombinant (human myelomonocytic cell line KG-1 bioassay; IL-18Rα/IL-18Rβ-driven interferon-γ production) and natural (whole blood assay; lipopolysaccharide + IL-12-driven interferon-γ production) human IL-18 with IC50 values of ∼0.2 nm (14, 45).
Importantly, both mature IL-18 surface cysteines, which are mutated to alanine in the construct used here (i.e. Ala74 and Ala104), are located >30 Å from the center of the 125-2H epitope and >15 Å from the nearest 125-2H residue at the combining site periphery. The two internal cysteines (here, Ala112 and Ala163) are similarly distant (center: >15 Å; nearest: >10 Å). Given these structural observations and the data demonstrating the biological equivalence of wild type and mutant IL-18s, we consider it very likely that the observations and conclusions we present herein will carry over without exception to natural, wild type IL-18.
Major IL-18 Loop Rearrangements Accompany 125-2H Binding
In the complex with 125-2H, mature human IL-18 (residues 37–193) adopts the compact, single domain “β-trefoil” fold characteristic of IL-1β (Fig. 1). Residues 38–192 were well ordered; the surface-exposed loops from 90 to 94 and from 166 to 169 were more mobile, and the terminal residues Tyr-37 and Asp-193 were not observed. The “top” of IL-18 is formed by a six-stranded antiparallel β-barrel, and the “bottom” is formed by three groups of antiparallel β-hairpins. These regular secondary structural elements are connected by irregular loops and a short α-helix (residues 70–77), which cover much of the surface of the molecule. Alternatively, in keeping with the “β-trefoil” name, IL-18 consists of a three-sided barrel, the sides of which are composed of three centrally located, highly curved antiparallel β-sheets surrounded by surface loops (Fig. 2a).
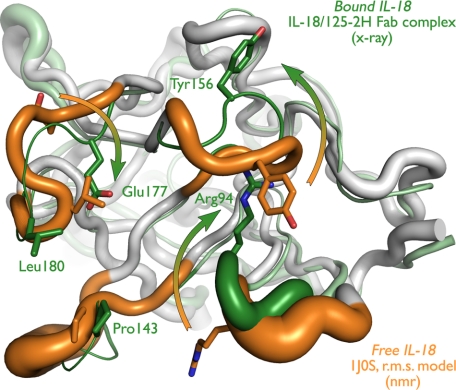
FIGURE 1.
Free and 125-2H-bound IL-18 exhibit large conformational differences. Free (white and orange; NMR (Protein Data Bank entry 1J0S)) and 125-2H-bound (gray and green; x-ray (this work)) IL-18 are overlaid. In these schematic diagrams, the tube width represents the (equivalent) temperature factor of the corresponding Cα atom (equivalent temperature factor Bequiv = 8π2〈u2〉/3, where 〈u2〉 is the r.m.s. deviation of the 20 NMR models). The four IL-18 loops that bind to 125-2H are colored orange (free) or green (bound). Note the significant movement (arrows) of Arg94, Tyr156, Glu177, and Leu180, and associated surface loops, which far exceeds the observed positional variability (tube diameter) in the structures. See also supplemental Fig. 1, a and b.
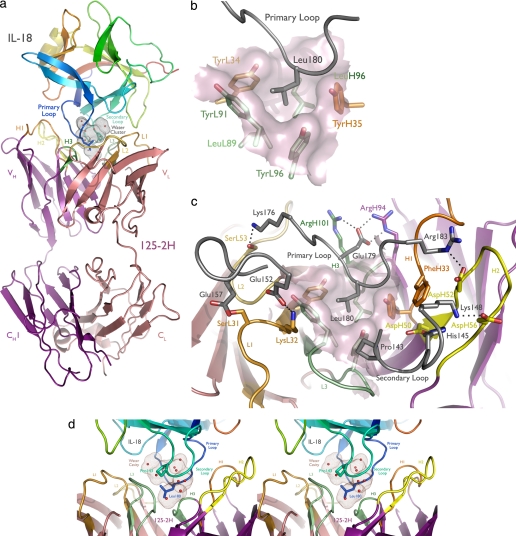
FIGURE 2.
125-2H binds human IL-18 residue Leu180 in a deep pocket, trapping 10 water molecules. a, overview of the complex. The antibody engages the primary (Leu180) and secondary (Pro143) IL-18 loops. IL-18 is colored as a rainbow, from the NH2 to the COOH terminus; CDRs 1, 2, and 3 of the 125-2H Fab fragment (purple, heavy chain; pink, light chain) are colored orange, yellow, and green, with the heavy chain in darker tones. The solvent-inaccessible, water-filled cavity trapped between 125-2H Fab and IL-18 is shown (brown dots). b, the center of the combining site, viewed from the perspective of IL-18 (gray). Note the deep hydrophobic pocket, formed by heavy and light chain Tyr and Leu residues, that binds Leu180. c, the periphery of the combining site is ringed by charge-charge and hydrogen bonding interactions involving all six 125-2H CDRs. d, stereoview illustrating the large cavity (brown dots) formed between the IL-18 primary and secondary loops and the 125-2H CDRs, trapping 10 well ordered water molecules. The detailed hydrogen bond interactions are shown in supplemental Fig. 2.
The core of the IL-18 crystal structure reported here is quite similar to the structure determined in solution using NMR techniques (Protein Data Bank entry 1J0S) (1). The r.m.s. deviation between the two structures is 1.5 Å (141 Cα atoms). It is apparent that mutation of the four mature cysteine residues to alanine caused no substantive change to the core IL-18 structure.
The short α-helix from Thr-70 to Asn-77 does pull inward by ∼7.5 Å, toward the IL-18 core, compared with the NMR structure. This inward motion, which is much larger than the void left by mutation of Cys74 to alanine, occurs on the IL-18 surface away from the 125-2H binding site. Although weak crystal contacts in this region may contribute, most of the altered positioning of this helix seems due to the high positional variability of the helix in the NMR structure (1).
In contrast to this stable core structure, the four IL-18 surface loops that interact with 125-2H undergo significant rearrangement upon binding to the antibody (Fig. 1). (Some rearrangement may result from the different pH values used in the NMR (pH 6.0) and crystallographic (pH 8.5) experiments; we believe a pH effect is likely to be minor, however, as the shift of the single histidine residue in IL-18, His-145, located at the periphery of the binding site, is spatially well separated from the other, much larger shifts discussed below.) These loops comprise residues Ser91–Gly95, Arg140–Lys148, Glu152–Gly158, and Lys176–Arg183. In each case, the 20 reported NMR structural models compare much more closely with each other than they do with the crystal structure reported here (Fig. 1). Among the NMR structures, the r.m.s. deviation within each of these loops is 0.8, 0.9, 0.3, and 0.2 Å, respectively. Between the antibody-bound IL-18 crystal structure and the free NMR structure (the “central” chain for each loop), however, the r.m.s. deviations increase to 2.1, 1.8, 1.6, and 1.4 Å.
These averages belie the true magnitude of the conformational changes. In the first loop, the Cα atoms of Pro93 and Arg94 move 4.9 and 5.2 Å into the binding site; the arginine side chain completely reorients, displacing the Cζ atom by 14.9 Å. Asp146 in the second loop moves 5.9 Å inward (Cα atom), dragging the rest of the loop with it. Tyr156 in the third loop shifts most dramatically, pulling its Cα atom 6.0 Å away from a steric clash with CDR L1 of 125-2H. The tyrosine side chain flips 180°, moving Tyr156 Oη by 17.6 Å. Finally, at the center of the combining site, Glu177 and Leu180 move 3.4 and 2.8 Å toward 125-2H, and their side chains move even more (Glu177 Cδ 8.8 Å; Leu180 Cγ, 4.5 Å) to make intimate contact with the antibody. Thus, significant alteration in the positions of these four IL-18 surface loops occurs upon binding to 125-2H.
All 125-2H Complementarity-determining Regions Contact IL-18
All six 125-2H CDRs contact IL-18, with VH forming the bulk of the interactions. Two IL-18 loops, Lys176–Arg183 (primary) and Arg140–Lys148 (secondary), mediate most of the antigenic contacts (Fig. 2a). Leu180, in the middle of the primary loop, plunges deep into the mAb combining site (Fig. 2b). Its side chain nestles into a pocket formed by aromatic and hydrophobic residues contributed by CDRs L1, L3, H1, and H3 (counterclockwise as viewed from IL-18 toward 125-2H: TyrH35, LeuH96, TyrL34, TyrL91, and TyrL96; LeuL89 forms the pocket base). The rather short CDR H3 (4 residues) helps to accommodate Leu180. The primary loop also makes numerous polar or charged interactions with 125-2H (Table 2).
TABLE 2.
Polar or charged interactions between IL-18 and 125-2H
| IL-18 residue/atom | 125-2H residue/atom | Distance |
|---|---|---|
| Å | ||
| Primary Loop | ||
| Lys176 Nζ | SerL53 Oγ | 2.9 |
| Asp178 O | ArgL46 Nη1/2 | 2.8/2.8 |
| Glu179 Oϵ1 | ArgH101 Nη1 | 2.8 |
| Glu179 Oϵ2 | ArgH94 Nϵ | 2.8 |
| Leu180 N | GlyH95 O | 3.2 |
| Gly181 N | TyrH35 Oη | 2.9 |
| Gly181 O | PheH33 N | 2.9 |
| Arg183 N | AspH31 O | 3.0 |
| Arg183 Nη1 | ThrH30 O | 2.9 |
| Secondary loop | ||
| Arg140 Nη2 | AlaL92 O | 2.9 |
| His145 Nϵ2 | AspH50 Oδ1 | 2.8 |
| Lys148 Nζ | AspH52 Oδ2 | 2.7 |
| Other loops | ||
| Glu152 Oϵ1 | LysL32 Nζ | 2.8 |
| Glu157 Oϵ2 | SerL31 Oγ | 2.6 |
The binding site is completed by the three other IL-18 loops. The secondary loop, Arg140–Lys148, is sandwiched between CDRs H2 and L3. It provides additional polar and van der Waals interactions that buttress the primary interaction. The other two loops, Ser91–Gly95 and Glu152–Gly158, make mostly van der Waals contacts with CDRs L1 and L2 (Table 2). Many charge-charge and hydrogen bonding interactions tie together IL-18 and 125-2H at the periphery of the binding site (Fig. 2c).
Unusually, TyrH35 Oη very closely approaches the IL-18 Leu180-Gly181 peptide bond π-system from the side (3.3 Å to Leu180 C; 2.9 and 3.1 Å to Gly181 N and Cα). These residues are very well ordered (mean B-factors ∼15 Å2) and lie in unambiguous electron density. The approach geometry is poor for a hydrogen bond between Oη and either Cα or N (Gly181 N is hydrogen-bonded to a water). Interestingly, only a small change in the TyrH35 χ1 torsion angle, from 60 to 40°, would bring Oη within 2.3 Å of the glycine carbonyl carbon, approximating nucleophilic addition into the Leu180-Gly181 peptide bond. The energetic favorability of this close interaction between IL-18 and 125-2H is unclear.
The IL-18·125-2H Interface Is Large and Filled with Water
Formation of the IL-18·125-2H complex buries an unusually large amount of solvent-accessible surface area at the interface. IL-18 loses ∼1,250 Å2 of surface area, and the antibody loses ∼1,100 Å2 (∼600 Å2 from VH, ∼500 Å2 from VL). The total buried area, ∼2,350 Å2, is at the high end for antigen-antibody complexes (46). IL-18 residue Leu180 contributes ∼150 Å2 to this total, much more than any other residue. As such, it qualifies as the initial “anchor” residue around which the rest of the complex then adapts (47). Glu179 and Arg183, which each bury ∼110 Å2, serve as “latch” residues that lock the proteins together (47).
Surprisingly, a cluster of 10 well ordered water molecules is trapped within a cavity formed by binding of the primary and secondary IL-18 loops to 125-2H (Fig. 2d). Leu180 and Pro143 define two ends of the cavity, which has a volume of ∼225 Å3 (38). Another water molecule, hydrogen-bonded to Leu180 O, PheH33 O, and GlyH95 N, is trapped in its own small void. There are seven hydrogen-bonding interactions among these water molecules themselves, 10 additional hydrogen bonds between the water molecules and 125-2H, and 11 more between the water molecules and IL-18. Thus, these water molecules form a well ordered (average temperature factor of 19 Å2), icelike network (35 of 40 possible hydrogen bonds made). This network fills a substantial void between the two proteins, thereby stabilizing the antigen/antibody interface through both van der Waals interactions and the nearly maximal number of hydrogen bonds that knit the two proteins together.
Have we intercepted an intermediate in the binding process? Could these water molecules be squeezed out of the binding interface in some (hypothetical) “final” bound state? Three points argue against this possibility. First, the intermolecular interface comprises a realistic and geometrically reasonable mix of central hydrophobic and peripheral charge-charge or hydrogen bond interactions. There are no obvious rearrangements of either antibody or antigen that one might postulate to convert the observed (“intermediate”?) structure to the “final” bound state. Second, it is not clear what residues should or could move, either on 125-2H or IL-18, to “squeeze” the water molecules out of the interface. Third, the crystallization was carried out at nearly neutral pH (pH 8.5) in a low ionic strength solution (i.e. approximately “physiological”), yet the crystals took a week to appear at room temperature. Thus, there is no a priori reason to suggest that what we have crystallized and observe here is a kinetically trapped intermediate rather than the final, thermodynamically stable bound conformation.
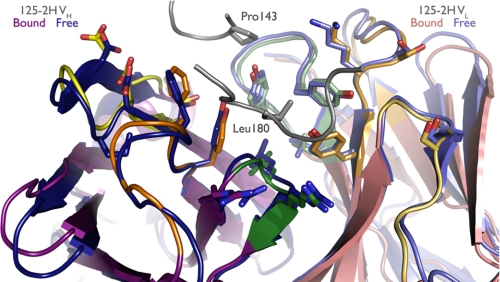
125-2H Is Preorganized to Bind IL-18
Comparison of the free and IL-18-bound 125-2H structures reveals that 125-2H is preorganized for binding (Fig. 3). The VH domains superimpose with an r.m.s. deviation of 0.4 Å (111 Cα), and the VL domains align equally well (0.3 Å, 107 Cα). With prealigned free and bound VH domains, the free Fab VL domain rotates by ∼1.7°, about an axis roughly perpendicular to the FV pseudo-2-fold axis, upon binding, thereby bringing the CDRs slightly closer to IL-18.
FIGURE 3.
125-2H is preorganized to bind IL-18. Bound (purple) and free (blue) 125-2H Fab are overlaid. The structure is essentially invariant. Side chains of residues shown in Fig. 2c are shown here as well.
The CDRs themselves adopt nearly identical conformations in the free and bound 125-2H structures. Aligning each CDR individually, r.m.s. deviations range only from 0.1 (L2) to 0.3 (H3) Å. In the context of prealigned free and bound VH or VL domains, all CDR r.m.s. deviations are less than 0.3 Å, except for L3 (0.4 Å) and H3 (0.9 Å). CDR H3 is slightly less well ordered in the free Fab structure. GlyH95 Cα is displaced by 1.5 Å, but the rest of the loop is shifted by less than 0.7 Å. Beyond the observation that the GlyH95-LeuH96 peptide bond adopts two alternate conformations, one of which matches that found in the complex, all of the CDR side chains that interact with IL-18 are found in the same conformation in the free and bound 125-2H structures.
The two crystallographically unique molecules in the free 125-2H Fab structure are essentially identical (r.m.s. deviation 0.4 Å, 426 Cα atoms). In both combining sites, which are mostly unhindered by crystal lattice contacts, there is unexplained spherical positive electron difference density (∼5σ) at the same position as IL-18 Leu180 Cα in the IL-18·125-2H Fab complex, midway between TyrL34 Oη and TyrH35 Oη. It is unclear what this density represents, but it could be a very loosely bound chloride ion. Overall, the 125-2H Fab itself shows no remarkable structural characteristics compared with other Fab structures.
Thus, crystallization of the 125-2H Fab either free or bound to IL-18, in two very different crystal lattices, under distinctly different crystallization conditions (see “Experimental Procedures”), yields the same detailed three-dimensional structure for the antigen combining site. It appears, therefore, that 125-2H is extensively preorganized for binding IL-18, explaining in part its high affinity and capture rate (kon ∼1.1 × 105 m−1 s−1).
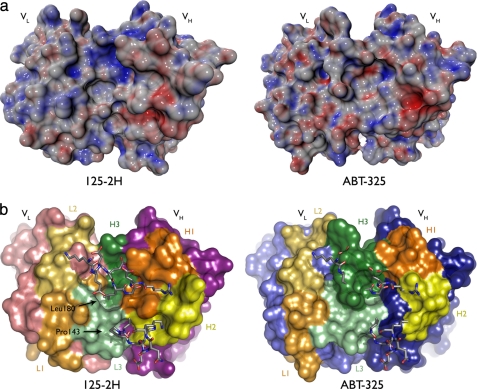
The ABT-325 and 125-2H Combining Sites Are Distinct
ABT-325 and 125-2H present very different binding surfaces to IL-18. They differ in CDR length, conformation, composition, and surface shape (Fig. 4). Despite the absence of the antigen, all ABT-325 CDRs were well ordered except for a small part of the center of H3, particularly the side chain of TrpH99. The combining site was mostly free of crystal lattice contacts, with some interactions with CDR H2 at the edge of the site.
FIGURE 4.
ABT-325 is incapable of binding IL-18 at the 125-2H epitope. ABT-325 and 125-2H differ in CDR composition and thus electrostatic and steric character. Molecular surface representations of both antibodies, oriented the same, are shown. The combining sites are viewed from the perspective of IL-18. a, surfaces colored by electrostatic potential (±25 kT/e). The concentration of positive potential (blue; top middle) on 125-2H is the Glu177 binding site. The highly negative (red; right), narrow pocket on ABT-325 is presumably where Lys165 binds. b, same view, colored by CDRs (as in Fig. 2). The IL-18 primary and secondary loops are overlaid, showing the incompatibility of the much larger CDR H3 in ABT-325 with binding Leu180 and the rest of the primary loop. Other differences, notably L3, provide the combining site with a very different steric character from 125-2H.
A key characteristic that distinguishes ABT-325 from 125-2H is the length of H3 (13 residues in ABT-325, compared with just 4 in 125-2H). The sinuous conformation of H3, first out of the center of the combining site and then back in, is incompatible with binding to the 125-2H-defined epitope of IL-18; the primary loop, especially Leu180, would clash with ABT-325 H3 (Fig. 4b). H3 residue TyrH100 defines the center of the combining site, thrusting its side chain up toward the antigen. H3 is rich in aromatic and aliphatic residues: two tyrosines and one each of tryptophan, valine, and proline. This hydrophobic character defines the rest of the combining site; there are two other tryptophan residues (TrpL94, L3; TrpH33, H1) and a relative lack of charged residues. An extra L3 residue in ABT-325 compared with 125-2H (10 versus 9 residues) promotes a distinct conformation as well. Here, TrpL94 swings back into the center of site, packing against TyrH100 and TrpH33. Thus, compared with 125-2H, ABT-325 presents a more uniform combining site to IL-18, in terms of both shape and electronic character.
In summary, it appears that ABT-325 binds to a somewhat more hydrophobic region of IL-18 that is distinct from the 125-2H epitope, consistent with Biacore experiments that show simultaneous binding of both antibodies to IL-18.
DISCUSSION
Three main conclusions may be drawn from our work. First, despite its small size, IL-18 is an extremely flexible molecule that adjusts among several conformations, including some apparently only rarely sampled in solution, to bind to its receptors (here, the murine antibody 125-2H). Second, the murine 125-2H and human ABT-325 antibodies present strikingly different binding surfaces to IL-18 and thereby bind the cytokine at distinct epitopes simultaneously.
Third, surprisingly, 10 water molecules are sequestered within a large cavity that is formed between 125-2H and IL-18 upon binding. These water molecules, which are trapped between the two key IL-18 antigenic surface loops (Arg140–Lys148 and Lys176–Arg183) and the antibody CDRs, form an icelike network (see Fig. 2d and supplemental Fig. 2) that knits the two proteins together. Thus, a very tight interaction between cytokine and antibody does not appear to require the complete (or even substantial) expulsion of surface-bound water molecules.
How unusual are such large, buried water clusters at antigen/antibody interfaces? To address this question, we examined the nearly 300 antibody (Fab, scFV, etc.)-protein complex structures in the Protein Data Bank (details are provided in the supplemental material). Compared with these reference structures, the water-filled cavity observed in the IL-18·125-2H Fab complex appears to be unique.
In terms of both inaccessible cavity volume at the combining site and especially the number of well ordered, buried water molecules, no extant antigen-antibody complexes are comparable with the IL-18·125-2H Fab complex. For example, several such complexes possess highly hydrated combining sites, but these generally involve one or more water-filled crevices rather than inaccessible cavities. The hen egg white lysozyme·HyHEL-5 complex (Protein Data Bank entry 1YQV), Kd ∼ 25 pm, does have six buried water molecules at the interface (48); notably, however, these water molecules are present in two discrete cavities (the largest has a volume of 136 Å3) rather than as a single block of “ice.” Several other lysozyme-Fab (or scFV) complexes (e.g. D1.3, Protein Data Bank entry 1VFB) (49) exhibit similar small cavities, usually filled with five or fewer water molecules (see supplemental Table 1).
Perhaps most similar is the complex between the scFV fragment of the KB5-C20 T-cell receptor and a specific, anti-clonotypic antibody Désiré-1 (Protein Data Bank entry 1KB5) (50). With a Kd of ∼4 nm, this complex is ∼20-fold weaker than the IL-18/125-2H interaction. About 2,700 Å2 of solvent-accessible surface area are buried at the KB5-C20/Désiré-1 interface. Seven water molecules are bound loosely in a very large, hydrophobic cavity (718 Å3) that is adjacent to a deep, water-filled pocket open to bulk solvent.
Beyond antibody/antigen interfaces, similar and even (much) larger water-filled cavities have been observed in other protein-protein complexes. Representative examples are discussed in a recent review (51). Buried water-filled cavities as large as that observed at the IL-18/125-2H Fab binding interface are, nonetheless, rather unusual.
In addition to the interfacial water molecules, the interaction between human IL-18 and the murine antibody 125-2H is dominated by the interaction of the Arg140–Lys148 and Lys176–Arg183 loops with all of the 125-2H CDRs (Fig. 2). The most important IL-18 anchor residue, Leu180, binds deeply into a pocket between the VL and VH domains of 125-2H. This hydrophobic anchoring event is presumably entropically favored, due to burial of significant hydrophobic surfaces on both IL-18 and 125-2H, and it then enables the peripheral, more hydrophilic, interactions to latch the two proteins together. Our structural and biochemical data allow us to make a number of additional observations, discussed below.
125-2H Is Specific for Human IL-18
The human IL-18·125-2H Fab structure clearly explains the inability of this antibody to bind murine IL-18. Although human and murine IL-18 exhibit 65% identity and 83% homology, one key amino acid difference would be expected to ablate binding to the murine protein; the anchor residue Leu180 is changed to asparagine in murine IL-18. This single difference probably dramatically alters the binding energetics, given the complete burial of the hydrophobic leucine when bound to 125-2H (Fig. 2b). Other changes may contribute to loss of binding as well. The cis Gln92-Pro93 peptide bond in human IL-18 makes a tight turn that contacts CDR L1 closely. In murine IL-18, the Glu-Val peptide bond would presumably adopt the trans configuration, extending the loop so that it would clash with 125-2H. Three other changes, namely deletion of Asp146 and the E177K and R183K mutations, would diminish other specific interactions between IL-18 and 125-2H by alteration of loop conformation, introduction of charge-charge repulsion, and loss of multiple hydrogen bonds, respectively.
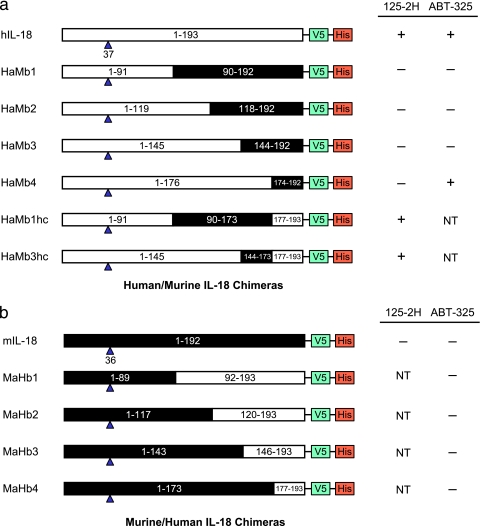
The crystallographically defined 125-2H epitope is consistent with and provides a structural explanation for binding and neutralization data obtained with murine IL-18 and human/murine IL-18 chimeras (14). In that work, 125-2H was unable to bind either to murine IL-18 or to four chimeras in which the COOH-terminal human IL-18 residues 92–193, 120–193, 146–193, or 177–193 were replaced by the corresponding murine sequence (Fig. 5a). Each of these chimeras changes the key Leu180 residue to asparagine. Two other chimeras, in which residues 1–91 and 177–193 or residues 1–145 and 177–193 were human, with the internal segment taking on the murine sequence, were tested. 125-2H was able to neutralize both, both of which restored the critical Leu180 residue.
FIGURE 5.
IL-18 chimeras help define binding epitopes. a, human IL-18 and six human (NH2 terminus)/murine (COOH terminus) chimeras (14). b, murine IL-18 and four murine (NH2 terminus)/human (COOH terminus) chimeras. Human sequences are shown as white boxes; murine sequences are black. Residue ranges, the first mature IL-18 residue following caspase-1 cleavage (blue triangle), and epitope tags are noted for each chimera. NT, not tested.
Structural Basis for the Ability of IL-18 to Bind Diverse Partners
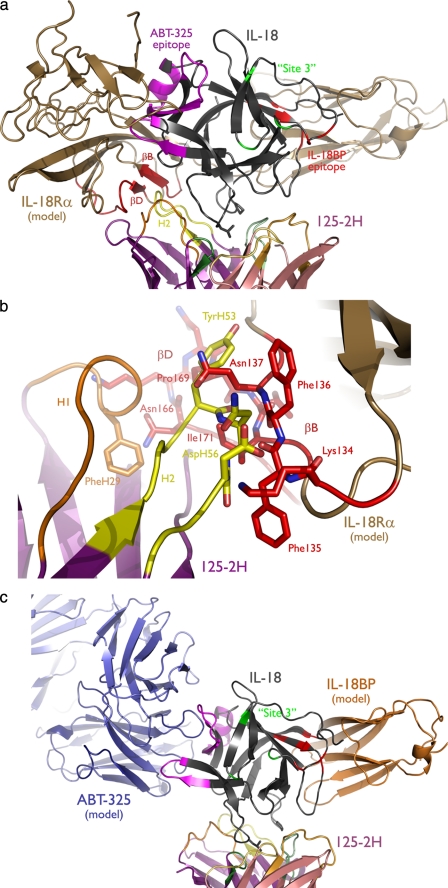
Other antibodies and proteins can bind IL-18 simultaneously with 125-2H. For example, Biacore and enzyme-linked immunosorbent assay binding data demonstrate that ABT-325 (52) and other antibodies (16), as well as the naturally occurring IL-18-binding protein (IL-18BP) (14), can do so, indicating that their epitopes do not overlap each other or the 125-2H epitope. Modeling allows access to the structural basis for how IL-18 is able to bind so many partners and consequently the impact of these bound partners upon the ability of IL-18 to engage its receptor (Fig. 6a).
FIGURE 6.
Multiple IL-18 epitopes mediate binding to multiple receptors. IL-18Rα and IL-18BP were modeled based on the IL-1R1 crystal structure (13) (Protein Data Bank entry 1ITB) and are shown bound to IL-18. a, engagement of IL-18 (gray) by IL-18Rα completely blocks the proposed ABT-325 epitope (magenta), and β-strands B and D (red) in domain 2 collide with 125-2H CDR H2. The IL-18BP (red) and IL-18Rβ (green; “Site 3” (see Ref. 1)) epitopes are also shown. b, close up view of the collision between 125-2H and IL-18Rα, viewed from behind relative to a. c, 125-2H, ABT-325, and IL-18BP can all bind IL-18 simultaneously, since their epitopes do not overlap.
First, can the 125-2H Fab·IL-18 crystal structure explain the intricate way in which 125-2H interferes with IL-18 receptor binding? Recall that although 125-2H inhibits binding of IL-18 to the low affinity IL-18Rα receptor, it allows IL-18 binding to the high affinity, heterodimeric IL-18Rα/IL-18Rβ receptor complex but renders the ternary complex nonfunctional (14). Modeling of IL-18 binding to IL-18Rα on the basis of the IL-1β·IL-1R1 crystal structure (13), as was done previously with the IL-18 NMR structure (1) but now in the context of bound 125-2H, indicates that the 125-2H heavy chain collides with domain 2 of IL-18Rα (Fig. 6a). Several residues primarily in CDR H2, but also H1, clash with IL-18Rα residues Lys134–Thr139 (β-strand B) and Glu161–Lys173 (loop and β-strand D) (Fig. 6b). Simultaneous binding of 125-2H and IL-18Rα would appear to be sterically incompatible, thus explaining the binding (and hence functional) blockade by the antibody. The IL-18 site on which the accessory chain IL-18Rβ binds, namely Lys115, Lys120, and Asp134 (“Site 3”) (1) is accessible in the 125-2H Fab·IL-18 complex (Fig. 6a). In analogy with an earlier suggestion (14), it appears that IL-18 and/or IL-18Rα conformational changes occur upon binding of IL-18Rβ. Those changes presumably alter the position of β-strands B and D, which need to shift only a few Å to allow binding of 125-2H. However, these conformational changes presumably also alter the relative positioning of 125-2H, IL-18, IL-18Rα, and IL-18Rβ, such that the antibody-cytokine-heterodimeric receptor quaternary complex cannot achieve the signaling-competent configuration accessible by the normal cytokine-heterodimeric receptor ternary complex.
A model for how IL-18 binds to IL-18BP has been proposed (53), again based on homology with IL-1β and IL-1R1 and the IL-1β·IL-1R1 complex crystal structure (13). When two proposed IL-18/IL-18BP interface residues, Glu42 and Lys89, were mutated to alanine, the mutant IL-18 could no longer be neutralized by IL-18BP (53, 54). Thus defined, the IL-18BP epitope is rotated ∼90° from the 125-2H epitope observed here (Fig. 6c), explaining how 125-2H and IL-18BP are able to bind simultaneously. Our modeling, which was completed 2 years ago, is completely in accord with the recent IL-18·poxvirus IL-18BP crystal structure (55).
In the absence of an ABT-325·IL-18 complex structure, what can we say about the ABT-325 binding site on IL-18? Epitope mapping of ABT-325 was performed using the aforementioned human/murine IL-18 chimeras (14). Murine IL-18 does not bind to ABT-325, and neither do chimeras in which the COOH-terminal human IL-18 residues 92–193, 120–193, or 146–193 were replaced by the corresponding murine sequence (Fig. 5a). But, unlike the case with 125-2H, the human (residues 37–176)/murine (residues 174–192) IL-18 chimera did bind to ABT-325, approximately equivalently to human IL-18. Thus, there is an important contribution to the ABT-325 epitope between residues 146 and 176, since only restoration of this portion restored binding. No significant binding contribution comes from residues 177–193, or binding there is due only to residues conserved between human and murine IL-18. We also tested four reversed IL-18 chimeras (NH2 terminus murine, COOH terminus human) (Fig. 5b), which comprise human IL-18 residues 92–193, 120–193, 146–193, or 177–193 (14). ABT-325 was unable to bind to any of these chimeras, indicating that an additional critical epitope lies within residues 37–91 of human IL-18.
Excluding regions that either overlap the 125-2H epitope or that are internal, IL-18 residues 146–176 contain a prominent, highly charged surface loop, Glu164–Leu169, which is rotated ∼90° from the crystallographically determined 125-2H epitope (Fig. 6c). Furthermore, only residues 59–76 are adjacent to this loop, surface-exposed, and within the extreme NH2-terminal (residues 37–91) segment. Thus, the chimera binding data suggest that ABT-325 binds to a conformational epitope consisting of residues 59–76 and 164–169. Engagement of this bipartite epitope by ABT-325 is consistent with simultaneous binding of ABT-325 and both 125-2H and IL-18BP to human IL-18 (Fig. 6c).
Modeling was attempted to further refine the ABT-325 epitope. Docking was performed with the ZDOCK (56), ClusPro (DOT) (57, 58), and GRAMM-X (59) Web servers, using the IL-18·125-2H Fab complex as a positive control. Neither ZDOCK nor ClusPro could reproduce the known complex structure, whereas the top GRAMM-X model matched crystallographic IL-18 perfectly, with an impressive r.m.s. deviation of 0.3 Å (155 Cα atoms; 1.2° rotation, <0.3 Å translation; 125-2H Fabs superimposed first). Unfortunately, although GRAMM-X provided many plausible models for the ABT-325·IL-18 complex, none clearly distinguished itself based on visual examination of putative binding interactions combined with the known epitope and simultaneous co-binding data. One model, on which the positioning of ABT-325 in Fig. 6c is based, scored ninth; slight manual adjustment allowed Phe57, Leu65, Met69, and Leu169 to make hydrophobic interactions with several ABT-325 CDRs and Lys165 to engage the negatively charged pocket in CDR H2 (Fig. 4), among other plausible antigen/antibody interactions.
Our inability to provide a more detailed model of how IL-18 and ABT-325 interact is probably due to two factors. First, CDR H3 is slightly disordered in the ABT-325 Fab structure. TrpH99 especially was poorly resolved, yet the well resolved main chain trace around this residue indicates that it projects in toward IL-18. Alteration of the tryptophan side chain conformation upon binding IL-18 or all or part of CDR H3 could preclude successful docking.
Second, more significantly, as shown in Fig. 1, the surface loops of IL-18 exhibit large positional variability between the crystal structure reported here and the earlier NMR structure (1). Such mobility is probably an attribute of IL-18 that allows it to bind to many different receptors (IL-18BP (e.g. see Ref. 55), the low affinity receptor IL-18Rα, the high affinity receptor IL-18Rα/IL-18Rβ, and of course many different antibodies) in distinct fashions. Given that many IL-18 residues moved >5 Å between solution structure and 125-2H complex, any similar mobility accompanying ABT-325 binding would prevent successful docking with the protocols we used. The loop mobility exhibited by IL-18 is probably a feature common to related cytokines, which also bind a panoply of partners.
Supplementary Material
Acknowledgments
We thank Richard Dixon for the insightful design of the human IL-18 cysteine mutant and Deborah Davis, John Mankovich, Ramu Sadhukhan, Serry Sousa, Sue Welsh, Lisa Quinn, and Jean Yang for the cloning and large scale expression of this protein. David Banach prepared the large quantities of caspase-1 needed for IL-18 production. Paul Sakorafas determined mAb kinetic constants (Biacore), and Renee Miller performed the enzyme-linked immunosorbent assay binding and KG-1 assays. We also thank an anonymous reviewer for prompting us to examine water cavities at antigen/antibody interfaces in detail, Joseph Dundas and Andrew Binkowski for help with the CASTp and GPSS servers, and Warren DeLano for help with PyMOL scripting. Last, we are grateful to Winnie Wong, Bernd Janssen, and Jochen Salfeld for support of this work.

The on-line version of this article (available at http://www.jbc.org) contains supplemental material, Figs. 1 and 2, and Table 1.
The atomic coordinates and structure factors (codes 2VXV, 2VXU, and 2VXT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IL
- interleukin
- mAb
- monoclonal antibody
- CDR
- complementarity-determining region
- PBS
- phosphate-buffered saline
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- r.m.s.
- root mean square
- IL-18BP
- IL-18-binding protein.
REFERENCES
- 1.Kato Z., Jee J., Shikano H., Mishima M., Ohki I., Ohnishi H., Li A., Hashimoto K., Matsukuma E., Omoya K., Yamamoto Y., Yoneda T., Hara T., Kondo N., Shirakawa M. (2003) Nat. Struct. Biol. 10, 966–971 [DOI] [PubMed] [Google Scholar]
- 2.Okamura H., Tsutsi H., Komatsu T., Yutsudo M., Hakura A., Tanimoto T., Torigoe K., Okura T., Nukada Y., Hattori K. (1995) Nature 378, 88–91 [DOI] [PubMed] [Google Scholar]
- 3.Nakanishi K., Yoshimoto T., Tsutsui H., Okamura H. (2001) Annu. Rev. Immunol. 19, 423–474 [DOI] [PubMed] [Google Scholar]
- 4.Plater-Zyberk C., Joosten L. A., Helsen M. M., Sattonnet-Roche P., Siegfried C., Alouani S., van De Loo F. A., Graber P., Aloni S., Cirillo R., Lubberts E., Dinarello C. A., van Den Berg W. B., Chvatchko Y. (2001) J. Clin. Invest. 108, 1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegmund B., Fantuzzi G., Rieder F., Gamboni-Robertson F., Lehr H. A., Hartmann G., Dinarello C. A., Endres S., Eigler A. (2001) Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R1264–R1273 [DOI] [PubMed] [Google Scholar]
- 6.Yamamura M., Kawashima M., Taniai M., Yamauchi H., Tanimoto T., Kurimoto M., Morita Y., Ohmoto Y., Makino H. (2001) Arthritis Rheum. 44, 275–285 [DOI] [PubMed] [Google Scholar]
- 7.Losy J., Niezgoda A. (2001) Acta Neurol. Scand. 104, 171–173 [DOI] [PubMed] [Google Scholar]
- 8.Karni A., Koldzic D. N., Bharanidharan P., Khoury S. J., Weiner H. L. (2002) J. Neuroimmunol. 125, 134–140 [DOI] [PubMed] [Google Scholar]
- 9.Ludwiczek O., Kaser A., Novick D., Dinarello C. A., Rubinstein M., Tilg H. (2005) Eur. Cytokine Netw. 16, 27–33 [PubMed] [Google Scholar]
- 10.Bombardieri M., McInnes I. B., Pitzalis C. (2007) Expert Opin. Biol. Ther. 7, 31–40 [DOI] [PubMed] [Google Scholar]
- 11.Yoshimoto T., Takeda K., Tanaka T., Ohkusu K., Kashiwamura S., Okamura H., Akira S., Nakanishi K. (1998) J. Immunol. 161, 3400–3407 [PubMed] [Google Scholar]
- 12.Azam T., Novick D., Bufler P., Yoon D. Y., Rubinstein M., Dinarello C. A., Kim S. H. (2003) J. Immunol. 171, 6574–6580 [DOI] [PubMed] [Google Scholar]
- 13.Vigers G. P., Anderson L. J., Caffes P., Brandhuber B. J. (1997) Nature 386, 190–194 [DOI] [PubMed] [Google Scholar]
- 14.Wu C., Sakorafas P., Miller R., McCarthy D., Scesney S., Dixon R., Ghayur T. (2003) J. Immunol. 170, 5571–5577 [DOI] [PubMed] [Google Scholar]
- 15.Brady K. D. (1998) Biochemistry 37, 8508–8515 [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi M., Nagaoka K., Kunikata T., Kayano T., Yamauchi H., Nakamura S., Ikeda M., Orita K., Kurimoto M. (1997) J. Immunol. Methods 206, 107–113 [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18.Leslie A. G. W. (1992) CCP4 and ESF-EACMB Newsletter on Protein Crystallography 26 [Google Scholar]
- 19.Evans P. R. (1997) Joint CCP4 and ESF-EACBM Newsletter 33 [Google Scholar]
- 20.French S., Wilson K. (1978) Acta Crystallogr. Sect. A 34, 517–525 [Google Scholar]
- 21.Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 22. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 23.Navaza J. (1994) Acta Crystallogr. Sect. A 50, 157–163 [Google Scholar]
- 24.Celikel R., Ruggeri Z. M., Varughese K. I. (2000) Nat. Struct. Biol. 7, 881–884 [DOI] [PubMed] [Google Scholar]
- 25.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 26.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 27.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 28.Lamzin V. S., Wilson K. S. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 29.Brünger A. T., Kuriyan J., Karplus M. (1987) Science 235, 458–460 [DOI] [PubMed] [Google Scholar]
- 30.Jones T. A., Zou J. Y., Cowan S. W., Kjelgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 31.Read R. J. (1986) Acta Crystallogr. D Biol. Crystallogr. A42, 140–149 [Google Scholar]
- 32.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1983) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 35.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence M. C., Colman P. M. (1993) J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 37.Kleywegt G. J. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 [DOI] [PubMed] [Google Scholar]
- 38.Kleywegt G. J., Jones T. A. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 39.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binkowski T. A., Joachimiak A. (2008) BMC Struct. Biol. 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 42.Sanner M. F., Olson A. J., Spehner J. C. (1996) Biopolymers 38, 305–320 [DOI] [PubMed] [Google Scholar]
- 43.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto Y., Kato Z., Matsukuma E., Li A., Omoya K., Hashimoto K., Ohnishi H., Kondo N. (2004) Biochem. Biophys. Res. Commun. 317, 181–186 [DOI] [PubMed] [Google Scholar]
- 45.Wu C., Ying H., Grinnell C., Bryant S., Miller R., Clabbers A., Bose S., McCarthy D., Zhu R. R., Santora L., Davis-Taber R., Kunes Y., Fung E., Schwartz A., Sakorafas P., Gu J., Tarcsa E., Murtaza A., Ghayur T. (2007) Nat. Biotechnol. 25, 1290–1297 [DOI] [PubMed] [Google Scholar]
- 46.Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 47.Rajamani D., Thiel S., Vajda S., Camacho C. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11287–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen G. H., Silverton E. W., Padlan E. A., Dyda F., Wibbenmeyer J. A., Willson R. C., Davies D. R. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 628–633 [DOI] [PubMed] [Google Scholar]
- 49.Bhat T. N., Bentley G. A., Boulot G., Greene M. I., Tello D., Dall'Acqua W., Souchon H., Schwarz F. P., Mariuzza R. A., Poljak R. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazza G., Housset D., Piras C., Grégoire C., Fontecilla-Camps J. C., Malissen B. (1999) J. Mol. Biol. 287, 773–780 [DOI] [PubMed] [Google Scholar]
- 51.Sonavane S., Chakrabarti P. (2008) PLoS Comput. Biol. 4, e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghayur T., Labkovsky B., Voss J. W., Green L., Babcock J., Jia X.-C., Wieler J., Kang J. S., Hedberg B. (2005) World Patent WO May 26, 2005/047307 [Google Scholar]
- 53.Kim S. H., Eisenstein M., Reznikov L., Fantuzzi G., Novick D., Rubinstein M., Dinarello C. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S. H., Azam T., Novick D., Yoon D. Y., Reznikov L. L., Bufler P., Rubinstein M., Dinarello C. A. (2002) J. Biol. Chem. 277, 10998–11003 [DOI] [PubMed] [Google Scholar]
- 55.Krumm B., Meng X., Li Y., Xiang Y., Deng J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20711–20715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen R., Li L., Weng Z. (2003) Proteins 52, 80–87 [DOI] [PubMed] [Google Scholar]
- 57.Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]
- 58.Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) Nucleic Acids Res. 32, W96–W99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tovchigrechko A., Vakser I. A. (2006) Nucleic Acids Res. 34, W310–W314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.