Abstract
MicroRNAs (miRNAs) are ~22 nucleotide long, noncoding, endogenous RNA molecules which exert their functions by base pairing with messenger RNAs (mRNAs), thereby regulate protein-coding gene expression. In eukaryotic cells, miRNAs play important roles in regulating biological processes such as proliferation, differentiation, apoptosis, and stem cell self-renewal. The human genome may contain as many as 1,000 miRNAs, and more than 700 of them have been identified. miRNAs are predicted to target up to one third of mRNAs. Each miRNA can target hundreds of transcripts directly or indirectly, while more than one miRNA can converge on a single transcript target. Therefore, the potential regulatory circuitry afforded by miRNA is enormous. Recently, mounting evidence implicates miRNAs as a new class of modulator for human tumor initiation and progression. Therefore, it has been proposed that manipulating miRNA activity and miRNA biogenesis may be a novel avenue for developing efficient therapies against cancer.
Key words: cancer, microRNA, noncoding RNA, therapy
INTRODUCTION
Twenty years ago, most people believed that the only functional product from any given gene is protein, and noncoding sequences in the genome were nothing but redundancy generated during evolution. This view was shattered by the discovery of lin-4, a gene that controls the timing of Caenorhabditis elegans larval development by repressing LIN-14 protein expression (1,2). Lin-4 gene does not encode any protein; instead, it produces a pair of small RNAs, later known as microRNA (miRNA). It was then found that the small lin-4 RNAs suppressed lin-14 translation by binding to the complementary sites in the lin-14 3′ untranslated region (UTR) (1,2). The repression of Lin-14 translation by lin-4 represents the first example of gene expression regulation mediated by miRNAs.
Since the discovery of lin-4, miRNAs have been identified in many organisms including plants, zebrafish, worms, flies, mice, and human. To date, 706 Homo sapiens, 547 Mus musculus, 152 Drosophila melanogaster, 155 C. elegans, 184 Xenopus tropicalis, and 336 Danio rerio miRNAs are reported in mirBase (http://microrna.sanger.ac.uk) (3). About 3% of human genes encode for miRNAs, and up to 30% of human protein-coding genes may be regulated by miRNAs (4). Most miRNA genes are transcribed by RNA polymerase II, generating primary transcripts with local hairpin structures and flanking sequences. Subsequently, two RNase III ribonucleases, Drosha and Dicer, trim off the flanking sequence to produce a ~22-bp duplex, which in turn unwind and incorporate into the RNA-induced silencing protein complex (RISC) (5,6). Mature miRNAs guide RISC complex to target mRNAs by complementary base pairing with miRNA binding sites within the target transcripts and exert their regulatory effects by either messenger silencing or translation inhibition (7). Numerous efforts have been invested to elucidate the biological importance of miRNAs. Recently discovered functions range from control of timing of development, neuronal patterning, and hematopoietic lineage differentiation to modulation of metabolism. Computational approaches for identifying miRNA targets indicate that the aforementioned examples may only represent a small fraction of all the biological processes miRNAs involve. Intriguingly, studies on miRNA expression in samples from various types of cancer demonstrate that tumor cells often have a distinctive pattern of miRNA expression (8,9). In many cases, miRNA expression can serve as prognostic markers. Accordingly, many functionally validated miRNA targets are oncogenes and tumor suppressors. Consistent with these observations, gain or loss of function of individual miRNA has been reported to affect tumor cell proliferation, apoptosis, and invasion (8–10). Taken together, these evidences strongly suggest that miRNAs represent an important class of regulators for tumorigenesis as well as a new class of therapeutic targets for curing cancer.
BIOGENESIS OF miRNA
The majorities of miRNA genes exist in clusters in genome and are expressed polycistronically from their own promoter; while some other miRNA genes are found in intronic regions and as a result, are transcribed as part of the annotated genes. The transcriptions of miRNA genes are typically performed by RNA polymerase II (Pol II), generating primary miRNA transcripts (pri-miRNAs) that are capped, polyadenylated, and often contain introns. In some cases, Pol II-transcribed miRNAs can also be derived from intronic sequence of a protein-coding transcript following RNA splicing process. As a result of being transcribed by Pol II, miRNA gene expression can be elaborately regulated in specific conditions and cell types by various Pol II-associated transcription factors (5,6). In addition, RNA polymerase III has also been reported to transcribe miRNAs with upstream Alu, transfer RNA- or mammalian-wide interspersed repeat (MWIR)-based promoter elements (11).
The pri-miRNAs typically contain imperfect duplex structure of 33 base pairs, a terminal loop, and flanking sequences. Two steps of ribonuclease processing reactions are generally required to trim a pri-miRNA transcript into mature miRNA. The first processing event occurs in the nucleus and involves in releasing a ~70 nt hairpin structure (pre-miRNA) from the RNA duplex in the pri-miRNA transcript by nuclear RNase III-type protein, Drosha (12). Pri-miRNA processing is a cotranscriptional event as Drosha is often found to localize to the transcription sites, and pri-miRNAs are most abundant in chromatin-associated nuclear fractions (13). Many studies suggested that Drosha-mediated pri-miRNA cleavage is extensively regulated; however, the exact biochemical mechanism of Drosha regulation is unknown. Drosha-mediated pri-miRNA processing requires a cofactor, called the DiGeorge syndrome critical region gene 8 (DGCR8) protein, a double-stranded RNA-binding protein which interacts with pri-miRNAs and assists Drosha to cleave the substrate ~11 bp away from the single-stranded/double-stranded RNA junction of pri-miRNA transcript. The importance of DGCR8 in miRNA biogenesis is manifested by the finding that Dgcr8-deficient mouse embryonic stem (ES) cells fail to produce miRNAs and have proliferation and differentiation defects (5,6). Once cleaved by Drosha, pre-miRNAs are exported to the cytoplasm via exportin 5 which transports pre-miRNA by hydrolyzing of GTP from Ran proteins in the cytoplasm (5,6). miRNA maturation process in cytoplasm is carried out by Dicer, a highly conserved RNase III type endoribonuclease, present in almost all eukaryotic organisms. Dicer cleaves the pre-miRNA at ~22 nt from the pre-existing terminus generated by Drosha and releases ~22 nt miRNA duplexes with 2-nucleotide 3′ overhang, similar to small interfering RNAs (siRNAs). The mature miRNA duplex is short-lived. While one strand (the guide strand or miRNA) is associated with Argonaute proteins, the other strand (the passenger strand, miRNA*) is degraded (5,6). Dicer-associated proteins have been identified. However, current studies suggest that they are not required for miRNA processing but more likely to contribute to the formation of the RISC (7). The expression of many miRNA genes can be regulated by their own targets. For example, Drosophila mir-7 transcription can be repressed by Yan, an ETS domain transcription factor and mir-7 target (14). Let-7 transcription in C. elegans is inhibited by Lin 28 protein, which is a Let-7 target (15). These observations suggest the presence of a double-negative feedback loop during miRNA biogenesis. Such a regulatory mechanism can prevent adverse consequence from overexpression of miRNAs and underlines the importance of tight control of miRNA homeostasis.
MECHANISM OF SILENCING
Mature miRNAs directly bind to Argonaute proteins which are the central components of the RISCs. Argonaute proteins are composed of four domains: the PAZ domain, which can bind the guide strand at 3′ terminus; the PIWI domain, which harbors RNase H like activity and can catalyze the initial cleavage of a miRNA base-paired target, eventually leading to messenger degradation; MID-domain, which is responsible for binding the 5′ end of the guide strand, and the N-domain (6). The miRNA guides RISC to specifically recognize and repress target mRNAs. The specificity in choosing target transcripts is mainly decided by sequence complementarity between mRNA target sites and the nucleotide sequence from position 2 to 8 at the 5′ end of miRNAs (the seed). Although base pairing to the 3′ end of miRNA is thought to be less important in target recognition, it may contribute in target selection especially when sites have weaker miRNA seed matches. In most cases, miRNA binding sites are located in 3′ UTRs of target transcripts and often present in multiple copies (7). However, recent studies suggest that miRNA can repression target gene expression with miRNA binding site located in 5′ UTRs as well as within the coding region (16). Binding of a miRNA to the target mRNA typically leads to either translational repression or exonucleolytic mRNA decay. The degree of miRNA–mRNA complementarity is critical in deciding the types of regulatory mechanism. Perfect complementarity, which is rare in animal miRNA:mRNA base pairing, allows Ago-catalyzed cleavage of the mRNA strand; whereas central mismatches exclude cleavage and promote repression of mRNA translation. The mechanism by which miRISC regulates translation is a matter of controversy. Two models have emerged based available data. While one model proposes that miRNA-mediated repression occurs at translation initiation, the second model proposes postinitiational repression by miRISC. Moreover, recent studies suggested miRISC may repress elongation as well. The inconsistencies among different reports indicate that miRNA-mediated repression may be exerted at multiple stages depending on the various experimental systems used in different studies (7).
FUNCTIONS OF miRNAs
Computational analysis has been developed to predict miRNA targets based primarily on conserved seed pairing, local sequence, and structural features. Results from such studies suggest that, in mammals, individual miRNAs can have more than 100 targets, and at least 20–30% of animal transcripts bear one or more conserved miRNA binding sites in their 3′ UTR (4,17). The predicted regulatory targets of mammalian miRNAs were enriched for genes involved in transcriptional regulation but also encompassed an unexpectedly broad range of other functions (18).
miRNAs involvement in development was manifested by the functions of the very first two miRNA discovered, lin-4 and let-7, both of which control the timing of larva development in C. elegans. Further studies on worms, flies, fish, and Xenopus with defect in miRNA biogenesis pathway provided more evidence on the importance of miRNAs in neuronal, muscle, and germline development. In C. elegans, dcr-1 mutants display defects in germline development and embryonic morphogenesis; in Drosophila, dcr-1 mutant germline stem cell clones divide slowly; in zebrafish, lacking both maternal and zygotic Dicer resulted in embryogenesis defect (19). These discoveries underline the importance of normal miRNA homeostasis on proper development. Individual miRNAs have also been found to function in specific tissues, at specific times during development, and other specific processes. Covering this vast body of work is beyond the scope of this review; the cited reviews provide valuable insight (reviewed by (20,21)).
Studies on mice bearing deletions of genes in miRNA biogenesis pathway provided early evidence on importance of miRNAs on stem cell regulation. Dicer deficiency causes early embryonic deficiency due to devoid of stem cells. Loss of Dgcr8, the cofactor of Drosha, resulted in embryonic lethality and differentiation deficiency in ES cells; and loss of Argonaute proteins resulted in lethality during early embryonic stages. Consistent with the overall function of miRNA in stem cell function, ES cells have distinct miRNA signatures. For example, mir-290 ~ 295 cluster and mir-296 levels decrease during stem cell differentiation, while mir-21 and mir-22 increase in this process. And functional analysis indicated that individual miRNA may have specific roles in controlling stem cell differentiation and renewal. miRNA functions in somatic tissue stem cells include regulating multiple steps of hematopoiesis, modulating myogenesis, and cardiogenesis, identifying cell fate during neural development as well as preventing osteogenic differentiation and skin differentiation (reviewed by (22)). In addition, let-7 is found to regulate multiple stem cell-like properties in breast tumor-initiating cells (BT-IC) by silencing H-RAS and HMGA2, suggesting that miRNAs are also involved in modulating cancer stem cells (23).
miRNA control has emerged as a critical regulatory principle in the mammalian immune system. Genetic ablation of the miRNA machinery, as well as loss or deregulation of certain individual miRNAs, severely compromises immune development and response and can lead to immune disorders like autoimmunity and cancer (reviewed by (24)).
It is known for many years that some clinically important viruses encode abundant amounts of noncoding RNAs (ncRNAs) during infection. Although the understanding is still in its infancy, several recent reports have identified new functions for viral miRNAs and larger ncRNAs (25). There are over 120 known viral miRNAs, mostly from the large DNA genome herpesvirus family and additional few from the small DNA genome tumor viruses (http://microrna.sanger.ac.uk) (3). Human cytomegalovirus (HCMV) encodes a miRNA, miR-UL112, to evade host innate immune responses. Cells infected with HCMV with mutant miR-UL112 are more susceptible to killing by cocultured NK cells (26). Kaposi's sarcoma-associated herpesvirus miR-k12-11 and host mir-155 regulate a common set of mRNA targets (27), and both miRNAs share an identical seed sequence (28). Simian virus 40 expresses a precursor miRNA (pre-miRNA) late in infection, which matures into two miRNAs that bind to the transcripts encoding immunogenic early genes, thereby inducing their cleavage to evade the adaptive immune response in vivo (29). Other viruses like HCMV and Epstein–Barr virus encode miRNAs that downregulate expressions of viral genes that are involved in signaling and replication (30,31). Thus, it is possible that viral miRNAs evade the host innate immune response, regulate host gene expression, and contribute to viral-mediated tumorigenesis. These studies not only advanced our understanding on host–viral interactions, but also provide key insights into the diversity, regulation, and evolution of miRNA pathways.
miRNA EXPRESSION IS DEREGULATED IN HUMAN CANCER
Since miRNA functions are involved in regulating crucial biological processes, such as development, differentiation, apoptosis, and proliferation, it has long been suspected that miRNA expression can be deregulated in cancer, and abnormal miRNA activity may lead to tumorigenesis. Altered miRNA expression in cancer was first documented in chronic lymphocytic leukemia (CLL). For years, it is known that deletion at locus 13q14 occurs in more than 65% of CLL cases and in 50% mantle cell lymphomas, 16–40% of multiple myelomas, and 60% of prostate cancers (32); however, search for a tumor suppressor within this region failed despite numerous attempts. In 2002, Calin et al. identified two miR genes, mir-15a and mir-16-1, within 13q14 locus, and demonstrated that both miRNAs are deleted or downregulated in the majority of CLL cases (32). Later, Cimmino et al. showed that mir-15a and mir-16-1 can target BCL2, suggesting a possible molecular mechanism by which losing mir-15a and mir-16-1 can cause CLL (33). Since the report of miRNA alteration in CLL, many efforts have been invested into characterizing the expression profile of miRNA in different tumors. To date, high-throughput miRNA expression profile analysis has been conducted in various human cancers, including CLL, breast cancer, glioblastoma, thyroid papillary carcinoma, hepatocellular carcinoma, lung cancer, colon cancer, and pancreatic cancer (8,10). While the most commonly used technique is oligonucleotide miRNA microarray, other methods, such as bead-based flow cytometry technique, quantitative real-time polymerase chain reaction genome-wide miRNA analysis with serial analysis of gene expression, and high-throughput array-based Klenow enzyme assay, have also been applied for the assessment of cancer-specific miRNA expression levels (8). Due to space limitation, we will not discuss specific deregulation in each cancer. Instead, we will summarize some common characteristics of miRNA deregulation in different tumors. First of all, in every type of tumor analyzed, miRNA profiles of tumor cells are significantly different from normal cells from the same tissue (34,35), underlining the biological significance of miRNA function during the cancer progression. Moreover, miRNA expression profiles in tumors from similar developmental origins appear to have similar alterations, providing a tool for cancer diagnosis and prognosis (36–38). On the other hand, a 21 miRNA signature was identified to be differentially expressed in tumor types that are from different embryological origins (35), suggesting that these miRNAs may participate in fundamental signaling pathways altered in many types of malignancies.
Aberrant miRNA expression in cancer can be the result of at least four different mechanisms, including chromosomal abnormalities, genomic mutations and polymorphism, epigenetic changes, and alterations in miRNA biogenesis. Almost 50% of annotated human miRNAs are located in fragile sites (39). Consistent with this observation, high frequency of genomic alterations in miRNA loci was revealed in human melanomas, ovarian, and breast cancer using high-resolution array-based genomic hybridization (40,41). Chromosomal alterations in tumor samples can result in altered miRNA expression, which in turn could participate to the process of tumorigenesis. Mutations and polymorphisms in miRNA transcripts can also affect miRNA expression by altering miRNA processing or sequence complementarity. For example, mutations in the pri-miRNA of mir-15a and mir-16-1 that cause lower expression of corresponding miRNAs have been reported in breast cancer and CLL patients as well as in a mouse model of CLL (36,42). On the other hand, single-nucleotide polymorphisms within miRNA binding site of target mRNA have been also been reported, suggesting an alternative mean to modulate miRNA function by disrupting the base pairing complementarity between miRNA and target mRNAs (reviewed by (43)). Epigenetic modification can also affect miRNA expression. Several studies on breast and bladder cancer cells have demonstrated that inhibiting histone deacetylase (HDAC) activity has significant impact on miRNA expression (41,44,45). However, the effect of HDAC inhibitors seems to be tissue-specific as similar treatment on nonsmall cell lung cancer cell has little effect on miRNA expression (37). Lastly, the miRNA biogenesis activity in tumor samples can be different from normal tissue. Downregulation of Dicer and Drosha expressions has been observed in various types of cancer (46,47). Deletion of Ago locus in human chromosome 1 are often associated with Wilms tumors of kidney and neuroectodermal tumors (48).
Another potential regulatory mechanism for miRNA processing and activity is by adenosine-to-inosine (A-to-I) RNA editing, a modification process converting adenosine residue to inosine, therefore changing an A–U base pair into an I–G base pair. The enzymes catalyzing A-to-I editing, ADAR1 and ADAR2, specifically recognize imperfect RNA duplex structures, the main feature of pre-miRNA transcripts. Therefore, it has recently been proposed that miRNA precursors can be RNA-editing substrates. Indeed, several studies have identified A-to-I RNA-editing events in pri-miRNAs (49). While, in some cases, RNA editing suppresses the processing of pri- to pre-miRNA by Drosha; it can also redirect the miRNA-targeting specificity (50). The activity of ADARs is critical for embryonic development and is tightly regulated under normal circumstances (51–53). Intriguingly, many types of tumors were found to have altered level of RNA editing as well as ADAR expressions (54). Therefore, further investigations are needed to explore the impact of RNA editing on miRNA functions and the exact role of miRNA editing in tumor development and progression.
miRNAs AS ONCOGENES AND TUMOR SUPPRESSORS
Given miRNAs' roles in regulating cell proliferation, differentiation, and apoptosis, and the fact that tumor cells have distinct miRNA expression profiles, it has been proposed that some miRNAs may be functionally involved in tumor initiation and progression. These miRNAs that possess oncogenic or tumor-suppressive activities are termed as “oncomir” (9). A more complete list of miRNA functions in cancer can be found in a comprehensive summary recently published by Spizzo et al. (10). So far, expression profiling studies have implicated many miRNAs in the process of tumor development. The search for relevant targets of these miRNAs suggests that some may promote tumor cell growth by inhibiting tumor suppressor genes or genes that control cell cycle progression, differentiation, or apoptosis; while others can induce metastasis by targeting genes involved in epithelial–mesenchymal transition process and cell invasion. Although many oncogenic miRNAs can enhance the malignant phenotype of cancer cells, only one study showed the sufficiency of a single miRNA to promote de novo transformation. It may be consistent with the model in which miRNAs tune, rather than silence, expression of target genes.
So far, miR-155 represents the only example that a single miRNA is sufficient to induce tumorigenesis. Transgenic mice with mir-155 overexpression exhibit a preleukemic pre-B cell proliferation which progresses to B cell leukemia and high-grade lymphoma. miRNA expression profiling study between malignant B cells from transgenic mice and normal B cells from nontransgenic animals identified several genes, including AGTR1, AID, IKBKE, and TB53INP1, as potential targets of mir-155 (55). Consistent with its oncogenic activity, high expression of mir-155 has been reported in several types of cancer and was found to indicate poor prognosis in lung cancer, diffuse large B cell lymphoma, and aggressive CLL (56). Another example of oncomir, mir-17~92, is a cluster of miRNAs whose genomic locus is often amplified in tumors (57). The oncogenic activity of mir-17~92 cluster was revealed in the Eu–Myc transgenic mouse model of B cell lymphoma. Expressing this miRNA cluster significantly accelerated lymphomagenesis and decreased Myc-induced apoptosis (57). As mir-17~92 cluster targets multiple genes that are involved in apoptosis pathway, it is speculated that the combination of suppressing many target mRNAs is responsible for the antiapoptotic effect (57,58). Additional examples of oncogenic miRNAs include mir-21, which promotes both proliferation and metastasis and is overexpressed in glioblastoma, cholangiocarcinoma, pancreatic, breast as well as hepatocellular cancer (59,60); and mir-372 and mir-373 that can neutralize p53-mediated CDK inhibition and cooperate with Ras in cellular transformation (61).
In contrast to oncogenic miRNAs, miRNAs whose expression is decreased in malignant cells may function as tumor suppressor by negatively inhibiting oncogenes and/or genes that inhibit cell differentiation or apoptosis. miRNAs functioning as tumor suppressor genes include the let-7 (62), who negatively regulates Ras and HMGA2; mir-15a and mir-16-1 (32), which negatively regulate BCL2; as well as the mir-34 (63), that is induced by DNA damage and oncogenic stress in a p53-dependent manner which leads to apoptosis or cellular senescence. The role of let-7 in cancer was first demonstrated by the Slack group when they found that the let-7 family negatively regulates let-60/RAS in C. elegans by binding to the multiple let-7 complementary sites in let-60/RAS 3′ UTR (62). Moreover, let-7 expression is found lower in lung tumors than in normal lung tissue, whereas RAS protein is significantly higher in lung tumor. Therefore, it was proposed that let-7 acts as a tumor suppressor. Supporting this hypothesis, forced expression of let-7 family members is able to suppress cancer cell growth both in vitro (64,65) and in vivo (64,66). In addition, reduced expression of let-7 has been associated with shortened postoperative survival in various types of cancer (67–70). Let-7 probably performs those functions by targeting various genes, including oncogenic proteins, cell cycle-associated proteins, oncofetal genes, and Toll-like receptor-4 (62,69–72). Given the multiplexity in miRNA targeting strategy, the potential regulatory circuitry in cancer afforded by let-7 might be enormous.
miRNAs AND CANCER STEM CELLS
Recent research in cancer has provided strong support for the cancer stem cell hypothesis which proposes that a rare subpopulation of tumor cells have the unique ability to initiate and perpetuate tumor growth. These cells are called cancer stem cells or tumor-initiating cells which share various characteristics with embryonic and somatic stem cells including self-renewal and multipotent differentiation. Cancer stem cells may be highly resistant to radiation and chemotherapy; therefore, the development of more effective therapies for cancer requires effective targeting of this cell population (73). Accumulating evidence indicates that miRNAs play functional roles in normal and cancer stem cell maintenance and differentiation. First, embryonic stem cells with mutation of the key proteins in miRNA biogenesis pathway fail to maintain the self-renewal and differentiation capacities. Second, both embryonic and somatic stem cells exhibit distinct miRNA expression signatures comparing to differentiated cells. Third, certain miRNAs such as let-7 have been found to play critical roles in regulation of self-renewal and/or differentiation in both normal and cancer stem cells (reviewed by (22)).
Let-7 is one of the examples that miRNAs play a functional role in normal and cancer stem cell differentiation. In the C. elegans, let-7 times the differentiation of seam cells, the stem cells that divide asymmetrically during each larval stage, possibly by acting as a regulator of multiple genes required for cell cycle and proliferation (69,70). Second, in mammalian embryonic and somatic stem cells, let-7 interacts with two induced pluripotent stem cell genes, MYC and LIN28; and these autoregulatory loops, i.e., MYC-let-7 and LIN28-let-7, may control stem cell self-renewal and differentiation (70). Third, Nishino et al. show that during aging, elevated let-7b blocks HMGA2 and contributes to declining neural stem cell function; in contrast, HMGA2 maintains neural stem cell function in young mice through repression of the Ink4a/Arf locus (74). Forth, Ibarra et al. found that let-7 is depleted in a population of self-renewing mammary epithelial progenitor cells that can reconstitute the mammary gland; and enforced let-7 expression induces loss of these self-renewing cells from mixed cultures, suggesting its role in the regulation of progenitor maintenance (75). Fifth, by comparing miRNA expression in self-renewing and differentiated breast tumor cells, Yu et al. found that the let-7 was markedly reduced in breast cancer stem cells and increased with differentiation. It is also shown that the let-7 regulates multiple cancer stem cell properties and tumorigenicity of breast cancer cells by silencing multiple targets, including H-RAS and HMGA2 (23). Since miRNAs seem to be involved in controlling renewal and differentiation of cancer stem cells, targeting miRNA may be a novel strategy to treat cancer by modulating cancer stem cells.
TARGETING ONCOGENIC miRNAs
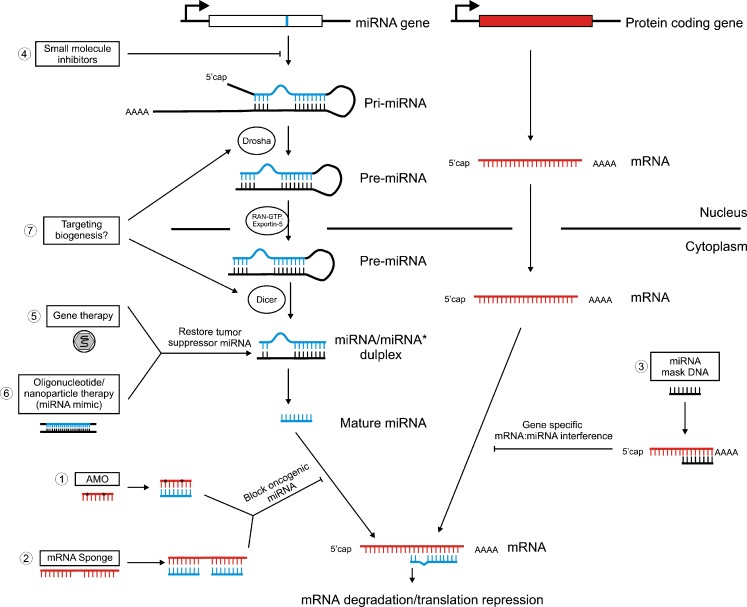
Many studies reviewed above have indicated that miRNAs can serve as novel therapeutic targets for cancer. For miRNAs with oncogenic capabilities, potential therapies include anti-miRNA oligonucleotides, microRNA sponges, miRNA masking, and small molecule inhibitor. For tumor-suppressor miRNAs, restoring suppressor miRNAs by forced expression of those miRNAs may be a useful strategy. In several tumor types with global decreasing miRNA biogenesis, approach to enhance miRNA biogenesis processing is discussed (Fig. 1).
Fig. 1.
Schematic diagram of miRNA biogenesis and the therapeutic strategies. 1 Anti-miRNA oligonucleotides (AMO) base pair with miRNA, therefore inhibit miRNA binding to target mRNAs; 2 mRNA sponges contain multiple binding sites for a specific miRNA which in turn prevent the binding of this miRNA with its endogenous targets; 3 miRNA mask DNA is complementary to miRNA binding site, resulting in gene-specific interference of miRNA:mRNA interaction; 4 small molecule inhibitor for mir-21 has been reported to inhibit the level of mature miRNA as well as pri-miRNA; 5, 6 gene therapy using virus delivery system and nanoparticle can force the expression of specific tumor suppressive miRNA to achieve therapeutic effect; 7 targeting miRNA biogenesis has been proposed, however, the feasibility of this approach need future evaluation
Anti-miRNA Oligonucleotides
The binding of miRNAs to their binding targets are simply and elegantly governed by the rules of Watson–Crick base pairing. Therefore, an obvious inhibitory molecule of miRNA is anti-miRNA oligonucleotides (AMOs) which blocks the interactions between miRNA and its target mRNAs by competition (76). AMOs are chemically modified in a variety of ways to improve the stability. One example is locked nucleic acid (LNA), often referred to as inaccessible RNAs, which is bicyclic high-affinity RNA analogs where the ribose moiety is chemically locked in a RNA-mimicking N-type (C3′-endo) conformation by the introduction of an extra 2′-O, 4′-C methylene bridge (77). The locked ribose conformation enhances base stacking and backbone preorganization and significantly increases the thermal stability upon hybridization with complementary single-stranded RNA target molecules. In addition, LNA is compatible with RNase H cleavage and display high aqueous solubility, low toxicity in vivo (77). Other oligonucleotide analogs, such as 2′-O-methyl- (78), and 2′-O-methoxyethyl-modified (2′-MOE) oligonucleotides (79) have also been proven to be efficient in functional inhibition of miRNAs. Besides chemical modifications, some improvement in inhibitor potency was observed by increasing the length of the AMOs (80). Optimized secondary structural elements that flanked the antisense core were highly potent and specifically block RISC activity in vitro for extended periods of time, thus suggesting structures surrounding or adjacent to the antisense core sequence are major determinants of inhibitor potency (80). In summary, a potent AMO may need a combination of optimization of sequences, structures, and/or chemical modifications.
Studies targeting mir-21 represent one of the first examples of inhibiting cancer development by downregulating oncogenic miRNA expression. mir-21 is overexpressed in many different cancer types and has been suggested to play a critical role in cell proliferation by downregulating the tumor suppressor genes Tpm1 and PTEN (60). Using a xenograft carcinoma model, Si et al. injected MCF-7 cells transiently transfected with 2-O-methyl oligonucleotides complementary to mir-21 and found that tumors derived from MCF-7 cells transfected with anti-mir-21 were 50% smaller in size than control MCF-7 tumor (81). In glioblastoma cell lines, knockdown mir-21 in vitro resulted in increased apoptosis (59). Complete eradication of mir-21 was observed in LNA-antimir-21–treated gliomas with the presence of neural precursor cells expressing a secretable variant of tumor necrosis factor-related apoptosis inducing ligand (S-TRAIL) in the murine brain (82). These studies suggest that AMOs may be promising reagents in treating cancer by suppressing oncogenic miRNAs.
MicroRNA Sponges
A microRNA sponge is defined as a synthetic mRNA containing multiple binding sites for an endogenous miRNA, therefore preventing the interaction between miRNA and its endogenous targets. Ebert et al. engineered such molecules by inserting a bulge between the microRNA binding sites at the position normally cleaved by Argonaute 2, therefore enabling stable association of microRNA sponges with microribonucleoprotein complexes loaded with the corresponding microRNA. In addition, they specifically designed sponges with complementary heptameric seed, so that a single sponge can be used to effectively repress an entire miRNA seed family (83). In in vitro experiments, these “sponges” derepressed miRNA targets as strongly as chemically modified AMOs (83). However, the efficacy of these stably expressed sponges in applications in vivo need to be evaluated.
miRNA Masking
Each miRNA may regulate hundreds of genes, and each gene can be regulated by multiple miRNAs. Similar to endogenous miRNA, the action of AMOs is sequence-specific but not gene-specific. Thus, AMOs may elicit off-target side effects and unwanted toxicity. Xiao et al. designed alternative strategy called “miR-Mask” which refers a sequence with perfect complementarity to the binding site for an endogenous miRNA in the target gene, which can form duplex with the target mRNA with higher affinity, therefore blocking the access of endogenous miRNA to its binding site without the potential side effects of mRNA degradation by AMOs (84). miR-Masks complementary to cardiac pacemaker channel-encoding genes HCN2 and HCN4 prevented the repressive actions of mir-1 and mir-133 on protein expression of these genes and cause acceleration of heart rate in rat model. This gene-specific, miRNA-interfering strategy was also validated in a study of zebrafish mir-430 in regulating TGF-β nodal agonist squint and antagonist lefty. The masking morpholinos complementary to mir-430 binding sites in target mRNAs elicited disruption of the specific mir-430–mRNA binding and resulted in enhancing or reducing the nodal signaling pathway, respectively (85). One should note that the efficacy of the miR-Mask strategy partially depends on the target gene selection, in applications such as cancer therapy, choosing key tumor suppressive, or oncogenic genes that is critical.
Small Molecule Inhibitor
Small molecule inhibitors against specific miRNAs have also been investigated. Gumireddy et al. identified azobenzene as a specific and efficient inhibitor of biogenesis of mir-21 from a screening. Such specific inhibitors of the miRNA pathway provide not only unique tools for the investigation of miRNA function, but also promising reagents to boost patient response to existing chemotherapies or stand-alone cancer drugs (86). The in vivo efficacy of such small-molecule inhibitors needs to be explored.
RESTORING SUPPRESSOR miRNAs
Since expressing protein-coding tumor suppressors can often inhibit tumor growth, it has been proposed that restoring tumor suppressive miRNAs may also have antitumor effect. Studies on several tumor suppressor miRNAs supported this hypothesis. In an in vitro culturing system, overexpressing Let-7 in lung cancer cell lines inhibited cell growth (64–67). Furthermore, stably expressing Let-7 from engineered lentiviral vector in BT-IC inhibited tumor formation in xenograft mouse model, suggesting that restoring Let-7 expression by viral vectors may serve as a potential cancer gene therapy (23). Since Lin28 has been found to block Let-7 processing and eventually cause pre-let-7 degradation (69,70), therefore, it will be worth of investigating whether inhibiting Lin28 will restore let-7 expression and inhibit tumorigenesis. A second example of miRNA replacement therapy is mir-15 and mir-16, which target BCL2 (33) and are often deleted in CLL patients (32). It has been reported that transfecting mir-15/16 expressing construct resulted in reduction of BCL2 protein levels and increased apoptosis in cancer cell lines (33,87). This study highlighted the possibility of treat tumors with BCL2 overexpression by restoring miR-15a and mir-16-1 expression. Most recently, miR-26a has been demonstrated as another example of tumor suppressive miRNA in hepatocellular carcinoma (HCC) and systemic administration of this miRNA using adeno-associated virus (AAV) in an animal model of HCC results in inhibition of cancer cell proliferation, induction of tumor-specific apoptosis, and significant protection from disease progression without toxicity (88). Since AAV vectors do not integrate into the host genome and eventually will be eliminated, this delivery approach minimizes the risk of vector-related toxicities. These findings also suggest that selection of miRNAs that are highly expressed and therefore, tolerated in normal tissues but lost in cancer cells can be a general strategy for restoring tumor suppressor miRNAs as therapy. Restoring miR-26a levels in hepatic cancer cells using AAV vectors is potentially a novel avenue to the clinic given the high prevalence of liver cancer worldwide. One should note, while the liver is well suited for AAV-mediated targeted therapy, the in vivo efficacy of such AAV vector-based therapy against solid tumors originated from other tissues needs thorough evaluation.
Besides viral vector-based gene restoration, miRNA mimics has also been used to in the gain-of-function experiments. These miRNA mimics are small, chemically modified, double-stranded RNA molecules that mimic endogenous mature miRNA molecules. miRNA mimics for many genes, such as pre-miR™ miRNA precursors (Ambion) and miRIDIAN™ microRNA mimics (Thermo Scientific Dharmacon), are now commercially available. To achieve strong therapeutic effect with these oligonucleotides in vivo, lipid- and polymer-based nanoparticles for systemic delivery in vivo have been developed, and promising results are reported (89–91). Since miRNA mimics do not have any vector-based toxicity, if their delivery agents do not cause side effect over long-term use, it can be a very promising therapeutic approach to treat tumors.
ENHANCING miRNA BIOGENESIS PROCESSING
Decreased miRNA biogenesis has been associated with tumor progression. For example, reduced Dicer1 expression in a subset of nonsmall cell lung cancers is found to correlate with poor prognosis (47); and high Dicer and Drosha expression were associated with increased median survival in ovarian cancer patients (46). However, increased Dicer expression was observed in early stage of lung adenocarcinoma relative to normal alveolar epithelium (92); and in a study conducted by Flavin et al., high Dicer expression is significantly correlated with the absence of lymph node metastases as well as greater prevalence of Ki-67 proliferation index, but not survival (93). These observations suggest that the effects of miRNA biogenesis may vary among different types of cancer, and other genomic and epigenetic events may also be important in determining the level of miRNAs. Supporting an overall antitumorigenic role of miRNAs, miRNA processing-impaired cells have enhanced tumorigenic activity and form tumors with accelerated invasiveness than control tumors. Though, downregulation of miRNA biogenesis is not sufficient to promote de novo transformation of wild-type mouse embryo fibroblasts (MEFs), conditional knockout of Dicer1 enhanced tumor development in a K-Ras–induced mouse model of lung cancer (94). In aggregate, much more studies are needed to evaluate modulating miRNA biogenesis as therapeutic approaches in treating cancer.
COMPARISON OF miRNA- AND siRNA-BASED THERAPIES
The potential clinical benefits of modulating miRNAs can be deduced from parallel studies of siRNA in cancer therapies. The two types of small RNAs differ in their origins as well as gene-targeting strategies. While siRNAs are often originated from synthetic/exogenous long dsRNAs and require perfect complementarity to a particular target mRNA to cleave that target, miRNAs are derived from genome-encoded hairpin-shaped precursors and only need partial sequence match to repress target gene expression. However, they do share the same gene-silencing machinery to silence target gene expression. (5). Therefore, therapeutic approaches based on miRNA and siRNA have intrinsic similarities and differences. The potential clinical benefits and limitations of miRNA therapy can be gleaned by comparing to siRNA therapies. Recently, it is shown that sustained, high-level short-hairpin expression produced lethal, dose-dependent liver injury. The displacement of endogenous miRNA precursor processing leads to the downregulation of liver-derived miRNAs and consequently morbidity (95). The risk of oversaturating endogenous small RNA pathways may be minimized by better understanding the feedback mechanisms in the miRNA cellular processing and effector mechanism which may provide an extra safety measure inapplicable to exogenous, artificial siRNAs.
The global upregulation of interferon-related genes and inflammatory cytokines is a major component of the “off-target activity” following introducing siRNA in vivo. Generally, the interferon response was primarily attributable to the activation of innate immunity by the delivery vehicle (96). The presence or absence of intrinsic immunostimulatory motifs in the siRNAs may also play a role in inducing interferon response (97). Recent studies suggest that exogenous administration of miRNAs may not cause interferon responses as siRNAs. For example, expression of the relevant miRNA by an inducible episomal vector effectively knocked down p53 expression without elicited an interferon response in vitro (98). However, potential toxicity from off-target effects and immune activation appear to be relevant to the small RNA concentration (99). Therefore, when design miRNA approaches, we need to use the most potent miRNA candidate at lowest concentration possible to interfere with tumor growth.
In comparison to siRNA approaches, the nature that one miRNA regulates multiple genes adds a unique layer of complexity to miRNA therapy. For example, while miR-155 alone is sufficient to induce tumorigenesis, systematic knockdown of miR-155 as antitumor therapy may collaterally affect immune proficiency because miR-155 modulates innate immune responses as well as lymphocytes and dendritic cell functions. Therefore, using tumor-specific delivery agents, such as tumor-specific nanoparticles or viral vectors, may obviate the concern of specific delivery.
FUTURE CHALLENGES
With all the efforts and advances made in developing miRNA-mediated therapy, two major hurdles still remain. First is to maintain target specificity. miRNA targeting is known to be sequence-specific instead of gene-specific. It is especially challenging since off-target gene silencing only requires partial complementary binding between miRNA and protein-coding transcripts. Therefore, it is important to evaluate the effect of a specific miRNA-mediated therapy on a proteome-wide scale to prevent unwanted gene alteration. The second hurdle is to achieve high therapeutic efficiency. Two factors that can limit miRNA therapeutic efficiency is the amplitude of target gene modulation and the number of cells that can be targeted. To address the first limitation, one should optimize target gene selection as well as therapeutic molecule design. A partial effect by miRNA knockdown-mediated therapy has been found to be therapeutic in neurodegenerative diseases, such as Alzheimer's disease; however, further evaluation is needed in cancer therapies. The second limitation comes from delivery efficiency. Lipid- and polymer-based nanoparticles for systemic delivery of siRNAs have been developed and tested. While lipid-based delivery of miRNA is efficient, it tends to induce an inflammatory response. On the other hand, biodegradable polymers induce less inflammatory responses but deliver less efficiently and have shorter effects. Another approach of targeted delivery takes advantage of viral vectors such as adeno-associated virus and lentivirus-based vectors. Different approaches may be suitable to different types of tumors; further investigations are needed to specifically evaluate these approaches in various tumors.
CONCLUSIONS
Studies from recent years have placed miRNAs as a critical class of regulator to protein-coding gene expression. As more and more evidence point out the importance of miRNA function during tumor development and progression, it is exciting to apply our knowledge and technology on miRNA into developing therapeutic reagents for treating cancer. Current antitumor treatments include surgery, radio/chemotherapy, hormonal treatment, and oncogene-targeted therapy. Many clinical studies have demonstrated that combined therapy based on patients' molecular profiles can deliver better response. With better understanding on miRNA's function in tumor progression and more sophisticated design of miRNA-modulating molecules, miRNA-mediated therapy will give a new impetus to cure cancer.
Acknowledgments
This work was supported by research grants from the Breast Cancer Alliance, the Ovarian Cancer Research Fund, National Cancer Institute and Department of Defense. Many studies greatly contributed to our knowledge on miRNAs and their therapeutic value in cancer treatment, due to the space limitation of this review, we could not cite all these papers. We apologize for that.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in Cancer. Cell. 2009;137(3):586–e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 11.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15(9):902–9. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123(7):1267–77. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243(2):215–25. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 16.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 17.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 18.Smalheiser NR, Torvik VI. Complications in mammalian microRNA target prediction. Methods Mol Biol. 2006;342:115–27. doi: 10.1385/1-59745-123-1:115. [DOI] [PubMed] [Google Scholar]
- 19.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579(26):5911–22. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 20.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 21.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9(10):789–96. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10(2):116–25. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 24.Petrocca F, Lieberman J. Micromanagers of immune cell fate and function. Adv Immunol. 2009;102:227–44. doi: 10.1016/S0065-2776(09)01204-8. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 26.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450(7172):1096–9. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair V, Zavolan M. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 2006;14(4):169–75. doi: 10.1016/j.tim.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–6. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 30.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3(11):e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 35.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 37.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103(24):9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105(19):7004–9. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109(12):5079–86. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24(10):489–97. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66(3):1277–81. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 45.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 46.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359(25):2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16(21):2733–42. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 49.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10(8):1174–7. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8(8):763–9. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279(6):4894–902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 52.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290(5497):1765–8. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 53.Feng Y, Sansam CL, Singh M, Emeson RB. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol Cell Biol. 2006;26(2):480–8. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17(11):1586–95. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103(18):7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102(10):3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 59.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 60.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124(6):1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 62.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 63.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008 Feb 28. [DOI] [PMC free article] [PubMed]
- 65.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 66.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008 Mar 3;7(6). [DOI] [PubMed]
- 67.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 68.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68(24):10307–14. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 70.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008 Jul 30. [DOI] [PubMed]
- 71.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 74.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135(2):227–39. doi: 10.1016/j.cell.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21(24):3238–43. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13(6):496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 77.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 78.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 79.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 80.Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, et al. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13(5):723–30. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 82.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67(19):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 83.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiao J, Yang B, Lin H, Lu Y, Luo X, Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J Cell Physiol. 2007;212(2):285–92. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]
- 85.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318(5848):271–4. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 86.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47(39):7482–4. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 88.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang H-W, et al. Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model. Cell. 2009;137(6):1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 90.Merritt WM, Lin YG, Spannuth WA, Fletcher MS, Kamat AA, Han LY, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100(5):359–72. doi: 10.1093/jnci/djn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–9. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67(5):2345–50. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 93.Flavin RJ, Smyth PC, Finn SP, Laios A, O'Toole SA, Barrett C, et al. Altered eIF6 and Dicer expression is associated with clinicopathological features in ovarian serous carcinoma patients. Mod Pathol. 2008;21(6):676–84. doi: 10.1038/modpathol.2008.33. [DOI] [PubMed] [Google Scholar]
- 94.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 95.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 96.Zhou R, Norton JE, Zhang N, Dean DA. Electroporation-mediated transfer of plasmids to the lung results in reduced TLR9 signaling and inflammation. Gene Ther. 2007;14(9):775–80. doi: 10.1038/sj.gt.3302936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457–62. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 98.Epanchintsev A, Jung P, Menssen A, Hermeking H. Inducible microRNA expression by an all-in-one episomal vector system. Nucleic Acids Res. 2006;34(18):e119. doi: 10.1093/nar/gkl624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Snove O, Jr, Rossi JJ. Toxicity in mice expressing short hairpin RNAs gives new insight into RNAi. Genome Biol. 2006;7(8):231. doi: 10.1186/gb-2006-7-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]