Abstract
In contrast to Escherichia coli, initiation of mRNA decay in Gram-positive organisms is poorly understood. We studied the fate of the highly structured RNAs generated by premature transcription termination of S-adenosylmethionine (SAM)-dependent riboswitches in Bacillus subtilis. An essential protein of earlier unknown function, YmdA, was identified as a novel endoribonuclease (now called RNase Y) that was capable of preferential cleaving in vitro of the 5′ monophosphorylated yitJ riboswitch upstream of the SAM-binding aptamer domain. Antiterminated full-length yitJ mRNA was not a substrate for RNase Y in vivo and in vitro, transcripts capable of forming the antiterminator were only cleaved in the presence of SAM. Turnover of 10 other SAM-dependent riboswitches was also initiated by RNase Y. Depletion of this ribonuclease increased the half-life of bulk mRNA more than two-fold. This indicates that RNase Y might be not only important for riboswitch RNA turnover but also as a key player in the initiation of mRNA decay in B. subtilis. About 40% of the sequenced eubacterial species have an RNase Y orthologue.
Keywords: Bacillus subtilis, endoribonuclease, mRNA degradation, riboswitch, RNase Y
Introduction
The degradation of mRNA is a vital process in all organisms. By regulating this process, cells can control gene expression and react rapidly to changing growth conditions. Bacterial mRNA degradation has been best studied in the two-model organisms Escherichia coli and Bacillus subtilis and has revealed surprising differences in enzymes and pathways between Gram-negative and Gram-positive bacteria. In E. coli, mRNA decay is initiated by endonucleolytic cleavage followed by 3′–5′ exonucleolytic degradation. The major player in this process is RNase E, an essential endoribonuclease with a significant preference for 5′ monophosphorylated substrates (Carpousis et al, 2009). The removal of the 5′ triphosphate from primary transcripts by the pyrophosphatase RppH has been shown to be the rate limiting step in the 5′ end dependent action of RNase E on many mRNAs (Deana et al, 2008). Inactivation of RNase E increases bulk mRNA stability (Babitzke and Kushner, 1991) and the abundance of >40% of all E. coli mRNAs (Lee et al, 2002).
B. subtilis has no ortholog of RNase E but the essential RNase J1 (rnjA) and its paralog RNase J2 (rnjB) have endonucleolytic cleavage specificity similar to that of RNase E (Even et al, 2005). They also have a 5′–3′ exonucleolytic activity (Mathy et al, 2007) and are sensitive to the phosphorylation state of the 5′ end, with monophosphorylated or hydroxylated 5′ ends being the preferred substrates (Even et al, 2005; Mathy et al, 2007; Li de la Sierra-Gallay et al, 2008). The X-ray crystallographic structure of RNase J1 showed the presence of a mononucleotide-binding pocket at one nucleotide distance from the catalytic site that cannot accommodate a triphosphate group (Li de la Sierra-Gallay et al, 2008).
RNases J1 and J2 have overlapping substrate specificities but together affect the expression levels of hundreds of genes (Mäder et al, 2008). Individual case studies have shown that RNases J1/J2 are effectively involved in the degradation of specific structural mRNAs and in fine-tuning the expression of a polycistronic transcript through a processing event that generates matured transcripts with differential stabilities (Mäder et al, 2008). In theory, the polyvalence of RNases J1/J2 could explain all characteristics described for mRNA degradation in B. subtilis, including a decay-initiating cleavage followed by 5′ exonuclease degradation. However, in an rnjA/rnjB double mutant (RNase J1 was depleted) the half-life of global mRNA was only slightly increased (Even et al, 2005) an observation that does not support the idea of an important function for RNases J1/J2 in initiating global mRNA decay. More generally, direct evidence that endonucleolytic cleavage has a major function in mRNA decay in B. subtilis is still elusive.
Secondary structures in the untranslated leader are a common feature of many B. subtilis mRNAs. To gain more insight into the events initiating decay of structured mRNA we have studied the fate of S-adenosylmethionine (SAM)-dependent riboswitches. This conserved genetic system (S-box) controls the expression of 11 transcriptional units (about 26 genes) involved in biosynthesis and transport of methionine and SAM in B. subtilis (Grundy and Henkin, 1998, 2003; Winkler and Breaker, 2005; Henkin, 2008). Regulation occurs at the level of premature transcription termination (Grundy and Henkin, 1998). In the presence of methionine (when SAM levels are high) SAM binds directly to the leader RNA. This causes a structural rearrangement that prevents the formation of the antiterminator (which forms preferentially in the absence of the effector) by stabilizing an antiantiterminator structure, called the SAM aptamer (positions +58 to +177, Figure 1A). This favours the formation of the terminator structure and provokes premature termination of transcription (Epshtein et al, 2003; McDaniel et al, 2003, 2005; Winkler et al, 2003). Terminator read-through is induced in response to starvation for methionine when SAM levels are low (Figure 1B). Under noninducing conditions (SAM excess), this transcription antitermination system produces large amounts of prematurely terminated mRNA from a constitutive promoter. Efficient degradation of these highly structured RNAs with bound SAM (Tomsic et al, 2008) is not only important for nucleotide recovery but also for recycling of SAM, a costly metabolite.
Figure 1.
Model for regulation of S-box gene expression in response to SAM. (A) In the presence of methionine (high SAM level), binding of SAM (represented by an encircled S) stabilizes the antiantiterminator structure favouring the formation of the terminator structure, which causes premature termination of transcription. (B) When SAM levels are low, the default antiterminator structure forms by base pairing of sequences of the antiantiterminator (grey, named I) and the terminator (hatched, named II), allowing expression of the downstream coding region. The positions of oligonucleotides are shown. Relevant positions on the transcript are numbered from the transcription start point (determined in Figure 2C). The yitJ AUG start codon is indicated.
In this work, we examined the degradation of these complex structures. We have identified a novel endoribonuclease, encoded by an essential gene of earlier unknown function, ymdA that is responsible for initiating the decay of all SAM-dependent riboswitches in B. subtilis. We provide evidence that this enzyme, called RNase Y, is likely to be a key player in initiating mRNA decay in this organism and we discuss how RNases Y and J might act together.
Results
Turnover of the yitJ riboswitch is very efficient
Expression of the S-box gene yitJ was reported to be specifically induced by starvation for methionine (Grundy and Henkin, 1998). Northern blot analysis was carried out to monitor the presence of terminated and read-through transcripts in a methionine auxotroph grown in the presence or absence of methionine. Total RNA was gel separated and hybridized with an RNA probe complementary to the 3′ end of the yitJ leader and the beginning of the structural gene (positions 152–346, Figure 1, see Materials and methods). Care was taken to exclude yitJ leader sequences prone to cross-hybridize with other S-box gene leaders. As expected, the full-length transcript of 2.1 kb was only detected when the cells were starved for methionine (Figure 2A). The yitJ promoter is constitutively active and is not required for methionine regulation of yitJ expression (Grundy and Henkin, 1998). In the presence of methionine one would thus have expected to detect the prematurely terminated leader transcript of about 220 nt. However, we failed to detect any transcript of this size (Figure 2A and B, lane 1). This observation indicated that an efficient mechanism must operate to initiate the decay of this highly structured transcript.
Figure 2.
Analysis of yitJ leader RNA in vivo. (A) Northern blot analysis of yitJ mRNA. Total RNA from the methionine auxotrophic strain SSB441, isolated from cultures grown in the presence or absence of methionine were separated on a denaturing 1.2% agarose gel. The RNA was probed with a uniformly labelled antisense 195 nt RNA covering 3′ sequences of the leader transcript and extending into the yitJ coding region (positions 152–346 Figure 1B delimited by oligonucleotides HP1231 and HP1056). An RNA sequence ladder was run in parallel as a size marker. Migration of 23S and 16S ribosomal RNAs is indicated. (B) High resolution Northern analysis of the yitJ leader. Total RNA from an RNase J1/J2 double mutant (rnjA inducible), an rnc mutant and a ymdA mutant (inducible), was isolated in the presence or absence of inducer, separated on an 8% polyacrylamide gel and probed with the leader-specific oligonucleotide HP1400 (Figure 1A). A [32P]-labelled DNA ladder was transferred to the membrane together with the RNA. (C) Identification of the yitJ transcription start. Oligonucleotide HP1238 (Figure 1A) was hybridized to and extended on total RNA isolated from a wild type and Pspac-ymdA mutant strain (±IPTG). The same primer was used for generating the sequence ladder. +1 indicates the band corresponding to the yitJ transcription start; the first transcribed nucleotide of the yitJ mRNA is encircled. (D) Primer extension analysis of total RNA isolated from a methionine auxotrophic strain grown in the presence or absence of methionine. T indicates the region corresponding to the terminator stem. (E) Sequence of the yitJ sigma A type promoter preceding the new transcription start point identified in (C).
Initiation of riboswitch degradation requires an essential gene of unknown function
The most straight-forward hypothesis for the apparently efficient turnover of the yitJ riboswitch RNA was an initial cleavage preparing the structured RNA for exonucleolytic digestion. Obvious candidates for a decay-initiating cleavage were RNase III and RNases J1/J2. The former is a double strand-specific endoribonuclease that could cleave within the long structured 5′ noncoding region. RNases J1/J2 have been shown to have a function in the expression of various structured leader mRNAs (Even et al, 2005; Choonee et al, 2007; Deikus et al, 2008; Yao et al, 2008). We carried out a Northern blot analysis using an oligonucleotide probe (HP1400) hybridizing close to the putative 5′ end of the yitJ mRNA (positions 28–58, Figure 1) (Grundy and Henkin, 1998). In a rnjA, rnjB double mutant, which expressed no RNase J2 (gene deletion) and showed a 20-fold reduced level of RNase J1 from the Pxyl-rnjA gene in the absence of the inducer (Mäder et al, 2008), we were unable to detect any prematurely terminated transcripts (Figure 2B, lanes 2 and 3). A similar result was obtained in an rnc null mutant (Figure 2B, lane 6), suggesting that neither RNase III nor RNases J1 and J2 are involved in the initial event leading to the degradation of the riboswitch. We then turned our attention to the ymdA gene, one of the few remaining essential genes (Kobayashi et al, 2003) whose function remained unknown. We constructed a strain (SSB447) in which the ymdA chromosomal copy was under control of the IPTG-inducible Pspac promoter. Total RNA was extracted from this strain grown in the presence of 2 mM IPTG or after resuspension in the same medium without IPTG to deplete YmdA. The RNA was separated on a polyacrylamide gel and probed with the same oligonucleotide probe (HP1400) described above. In the absence of IPTG, we detected a strong signal of 220 nucleotides. This likely corresponded to the yitJ leader mRNA prematurely terminated at the leader terminator by the interaction of SAM with the riboswitch. In the presence of IPTG, the riboswitch RNA was undetectable, suggesting that the ymdA gene product could encode a ribonuclease initiating the decay of the riboswitch (Figure 2B, lanes 4 and 5).
We used the strain depleted for YmdA to determine experimentally the transcription start point of yitJ by primer extension using oligonucleotide HP1238 (Figure 1). Transcription starts with a G residue at position—265 with respect to the yitJ start codon and originates from a Sigma A type promoter located upstream (Figure 2C, lane 3 and Figure 2E). This transcription start site is 30 nucleotides upstream with respect to the earlier predicted 5′ end of the yitJ mRNA (Grundy and Henkin, 1998; Tomsic et al, 2008). The expected length of the leader transcript, originating at this position and terminating immediately downstream of the terminator is about 220 nucleotides. This corresponds to the prematurely terminated leader transcript stabilized in the YmdA depleted strain (Figure 2B, lane 5).
As the yitJ riboswitch is efficiently degraded in the presence of YmdA, we wanted to know whether read-through transcripts still contain the complete leader sequence. For that purpose, total RNA was extracted from a methionine auxotrophic strain (but wild type for ymdA) grown in the presence or after withdrawal of methionine. The RNA was used for primer extension with an oligonucleotide complementary to sequences within the yitJ ORF (HP1056, Figure 1B). We detected two major signals corresponding to the full-length transcript (346 nt) and a transcript of about 125 nt, which corresponds to a premature arrest of the reverse transcriptase at the terminator structure (Figure 2D, lane 2). An arrest at the leader terminator might seem at odds with the notion that the read-through transcript is not cleaved because it exists in the antiterminator conformation. However, the RNA used for extending the primer in vitro is denatured before the reaction and it is reasonable to assume that under these conditions some of the transcript will refold into the terminator conformation. No signal was detected at a position potentially corresponding to a cleavage site. This suggested that the riboswitch is in its antiterminator conformation, which folds first and is more stable than the terminator conformation (McDaniel et al, 2003; Winkler et al, 2003), and that in this form it is not an efficient substrate for YmdA. This in turn implies that YmdA should not have a significant impact on the expression level of yitJ (see Discussion).
YmdA is an endoribonuclease
A truncated form of YmdA lacking the 25 N-terminal amino acids comprising a membrane-binding domain was purified by affinity chromatography (see Materials and methods and Discussion). To characterize the enzymatic activity of YmdA, we performed in vitro assays on riboswitch RNAs synthesized with T7 RNA polymerase. The first substrate contained leader sequences from position 1 up to 196 (3′ end defined by primer HP1238, Figure 1A), which corresponds to the nucleotide immediately upstream of the terminator stem (Figure 3C). This RNA cannot fold into the antiterminator conformation (Figure 1B), thus increasing the potential to form the SAM-binding aptamer (Figure 1A). This configuration corresponds to that of the terminated leader transcript, which is the most likely substrate for YmdA in vivo. Incubation of the 5′-labelled monophosphorylated transcript with YmdA produced a major fragment resulting from endonucleolytic cleavage after an A residue at position 52 (Figure 3A and C). Two minor but not reproducible cleavages were also observed after positions 124 (in the apical loop of the aptamer) and 185 (between the aptamer and the terminator, Figure 3A).
Figure 3.
Endonucleolytic cleavage of yitJ leader transcripts by YmdA. (A) Cleavage of a 5′ end-labelled monophosphorylated transcript (3′ end defined by primer HP1238). The major cleavage site was mapped on a sequence ladder generated with a primer whose 5′ end corresponded to the transcription start point. (B) Effect of increasing SAM concentrations (25 and 80 μM) on the cleavage of a 5′ end-labelled monophosphorylated transcript (3′ end defined by primer HP1356). (C) Cartoon of the riboswitch leader. The vertical arrow indicates the cleavage site for YmdA in the leader mRNA as identified in panel A. The position of the primers delimiting the transcript 3′ ends on the PCR templates are shown. The encircled S stands for SAM.
Binding of SAM to the riboswitch aptamer precludes formation of the default antiterminator conformation (Epshtein et al, 2003; McDaniel et al, 2003; Winkler et al, 2003). We therefore tried to mimic the in vivo situation by using a substrate 12 nt longer, which contained the sequences required for the formation of the antiterminator (up to position 208, oligonucleotide HP1356, Figure 3D). Such a transcript should a priori not be a good substrate for RNase Y because, in the absence of SAM, it was shown to fold preferentially into the antiterminator conformation (Epshtein et al, 2003; McDaniel et al, 2003). However, the RNA should become accessible to cleavage on addition of SAM, which shifts the RNA structure towards the terminator conformation. As shown in Figure 3B, cleavage by YmdA at its major cleavage site (position +52) was barely detectable in the absence of SAM but was stimulated significantly by increasing over the physiological range the concentration of the metabolite, which in vivo reaches ∼300 μM in the presence of methionine (Tomsic et al, 2008). A very weak cleavage was also observed at position 54. This indicates that YmdA is sensitive to the structural context of the cleavage site and provides a rationale for the fact that the leader transcript, when part of the antiterminated full-length transcript, is not a good substrate for YmdA.
We therefore considered YmdA to be an endoribonuclease and renamed it RNase Y (rny). Cleavage by RNase Y required the presence of Mg+2 ions, which could be replaced by Mn+2 or Zn+2 (data not shown).
RNase Y activity is sensitive to the 5′ phosphorylation state of the substrate
The endoribonucleolytic activity of RNase Y had been detected on a 5′ monophosphorylated substrate (Figure 3). We wanted to determine whether cleavage by RNase Y was altered by the nature of the substrate 5′ end. The same transcript as described above (3′ end defined by primer 1238, Figure 3C) carrying either a tri- or monophosphorylated 5′ end was uniformly labelled with α-[32P]UTP and tested under our standard conditions. As expected, the 5′ monophosphorylated substrate was cleaved producing two fragments of 145 and 52 nt (Figure 4A, lanes 3 and 4) with identical yield, with respect to the number of U residues present in each fragment (data not shown), plus many additional products, some in significant yields.
Figure 4.
RNase Y cleavage of the yitJ riboswitch is sensitive to the 5′ phosphorylation state of the substrate. (A) Equal amounts of [32P] uniformly labelled tri- and monophosphorylated yitJ transcripts (3′ end defined by primer HP1238) were used as substrates in post-transcriptional cleavage assays with RNase Y as described in Material and methods. Reaction products were separated on an 8% polyacrylamide gel. The major cleavage site is indicated by scissors and the cleavage products are identified by schematic drawings on the right. A [32P]-labelled 50 bp DNA ladder was used as a size marker. (B) Kinetic analysis of RNase Y cleavage of 3′ end-labelled tri- and monophosphorylated yitJ transcripts (3′ end defined by primer HP1238). Samples from scaled-up cleavage assays were taken at the indicated time points. The reaction products were separated on a 5% polyacrylamide gel. The RNase Y-dependent cleavage product of 150 bases is indicated by an arrow. The band below corresponds to a spontaneous cleavage of the transcript after its purification and is also observed without addition of the enzyme. (C) Graph of kinetic analysis of RNase Y cleavage of yitJ transcripts bearing mono- and triphosphorylated 5′ ends. The reactions were carried out as in (B) but incubated up to 90 min to detect cleavage of 5′ tri-phosphorylated transcripts. The part of the gels containing the cleavage products are shown on the top (5′-P) and bottom of the graph (5′-PPP), respectively. The numbers on the Y axis represent arbitrary units generated by quantification with the Image J program.
In contrast, the transcript carrying a 5′ PPP group was resistant to cleavage under the conditions used (Figure 4A, lanes 1 and 2). This indicated that RNase Y is sensitive to the 5′ phosphorylation state of its substrate and requires a 5′ P for significant cleavage to occur. A kinetic analysis using 3′ end-labelled mono- and triphosphorylated substrates confirmed the preference of RNase Y for transcripts carrying a monophosphate group at the 5′ end (Figure 4B). The initial reaction rate observed for the 5′ P substrate was about 20-fold faster than that observed with the 5′ PPP substrate but after 90 min a similar amount of cleavage product was produced with both substrates (Figure 4C).
Degradation of the yitJ riboswitch in vivo requires RNase Y, RNase J1, PNPase and RNase R
We have shown that the turnover of the yitJ riboswitch depends on the presence of RNase Y in vivo (Figure 2B, lanes 4 and 5). The discovery that RNase Y is an endoribonuclease capable of cleaving in vitro the riboswitch leader in its terminator conformation provided a rationale for how this nuclease might initiate the decay of the riboswitch in vivo. Therefore, we performed a primer extension analysis using primer HP1134 (Figure 5D) on total RNA isolated from a number of endo- and exoribonuclease mutants. As expected, the full-length leader transcript was greatly stabilized when RNase Y expression was reduced (Figure 5A, lanes 6 and 7). In a strain depleted for RNase J1, we detected a weak signal at position +50 (indicated by a scissors symbol labelled 1 in Figure 5A, lane 3), which corresponds quite well to the RNase Y cleavage site identified in vitro (position 52, Figure 3C), and a number of smaller degradation intermediates (Figure 5A, lane 3), implying that the RNase Y cleaved mRNA is less efficiently degraded from its 5′ end under these conditions. However, we observed a more significant accumulation of degradation intermediates in strains devoid of the major 3′–5′ exoribonucleases, polynucleotide phosphorylase and ribonuclease R (Figure 5A, lanes 4 and 5). The two accumulating transcripts have 5′ ends corresponding either to the original transcription start or to position +58 (Figure 5D). The quite dramatic effect of the 3′–5′ exonuclease mutations on the stabilization of the riboswitch degradation intermediates suggested that the leader transcript must be cleaved somewhere else to create an entry site for these nucleases. Extension of primer HP1238 complementary to sequences immediately upstream of the terminator (Figure 1A) failed to detect any degradation intermediates on total RNA isolated from the pnp/rnr and rnjA mutants (data not shown). This indicated that the riboswitch RNA was cleaved at a second site somewhere between primers HP1134 and HP1238. We performed a 5′/3′ RACE experiment by first ligating total RNA from the wild type, rnjA/B and pnp/rnr double-mutant strains. RT–PCR across the junction of circularized mRNA using primers HP1394 (positions +146 to +121) and HP1395 (positions +148 to +166, Figure 5D) specifically produced fragments of ∼130 bp for the mutant strains (Figure 5B, lanes 1 and 2). This was the size expected for a transcript cleaved on either side of the aptamer domain (Figure 5D). The RT–PCR products were purified cloned and 20 recombinant plasmids were sequenced. This confirmed the site of RNase Y cleavage upstream of the aptamer (Figure 5D, cleavage 1) and is in agreement with the primer extension product observed in the RNase J1/J2 mutant (Figure 5A, lane 3, position +50). The second cleavage by RNase Y (see Discussion) occurred after the A residue +182, also in a single-stranded region between the aptamer and the terminator structures (Figure 5D). Further confirmation of the site 2 cleavage was obtained through an S1 mapping analysis of the downstream cleavage product containing the transcription terminator. A fragment of expected size (25 nucleotides, see legend to Figure 5C) accumulated in cells depleted for RNase J1 presumably because 5′–3′ exonucleolytic degradation of this fragment was inefficient under these conditions (Figure 5C, lane 3). On the basis of our data, turnover of the yitJ riboswitch in its terminator conformation likely proceeds as illustrated schematically in Figure 5D (see Discussion).
Figure 5.
Degradation of the yitJ riboswitch in vivo requires RNase Y, RNase J1, PNPase and RNase R. (A) Extension of primer HP1134 (see panel D) on total RNA isolated from wild-type and different RNase mutant strains (pnpA and rnr encode polynucleotide phosphorylase and RNase R, respectively). The scissors symbol ○1 indicates the RNase Y in vivo cleavage site upstream of the aptamer (position +50, panel D, see text). Extension products corresponding to the transcription start (+1) and a degradation intermediate (+58) stabilized in the pnpA and rnr mutants (lanes 4 and 5) are indicated. (B) Mapping of two RNase Y cleavage sites in vivo by 5′/3′ RACE. Total RNA isolated from the wild-type, pnpA/rnr and rnjB/rnjA double-mutant strains, was ligated to create circular RNA from potential degradation intermediates. RT–PCR was used to amplify the sequences across the 5′/3′ junction. The amplified products with an expected size of ∼130 bp (not present in the wild type) were gel purified, cloned and sequenced. The cleavage sites were deduced from sequences representing the majority of clones from the two mutants (20 independent clones). All sequences from the remaining clones identified ligated junctions, which, probably due to partial degradation, were located within the aptamer domain (data not shown). The products with a size of ∼100 bp present in all lanes were generated by PCR amplification from tandemly ligated aptamer RNAs not relevant here (data not shown). (C) Detection of the 3′ terminal fragment of the yitJ leader created by RNase Y cleavage 2. Total RNA from a wild-type and rnjB/rnjA double mutant was subjected to S1 mapping analysis using oligonucleotide HP1442 (complementary to positions +207 to +131 on the mRNA). The band corresponding in size to the expected 3′ terminal fragment is indicated. (D) Scheme of the yitJ riboswitch RNA on which the two RNase Y cleavage sites (as determined by 5′–3′ RACE) on either side of the aptamer domain are indicated. The degradation pathway of the riboswitch following initial cleavage by RNase Y is illustrated based on the data shown in panels A and C (see Discussion).
RNase Y acts on all riboswitch controlled S-box gene family members
We next investigated whether RNase Y initiated decay of the SAM-dependent riboswitches of the other S-box genes. For that purpose, we performed Northern blot analysis of the other 10 SAM riboswitches present in B. subtilis. mRNA was prepared from strain SSB447, which is wild type for methionine biosynthesis and contains the IPTG-inducible rny gene. Specific oligonucleotide probes (Supplementary Table I) for each riboswitch were used to avoid potential cross-hybridization. With the exception of cysH, a prematurely terminated riboswitch transcript was detected for all the genes in the culture depleted for RNase Y (Figure 6). The cysH operon has been shown earlier to be regulated primarily at the level of transcription initiation in response to O-acetyl-serine (Mansilla et al, 2000) and no prematurely terminated leader transcript was thus expected to be present. The 11 S-box genes were also analysed in the RNase J1/J2 double mutant. Using gene-specific probes, we observed no effect of the absence/depletion of RNases J1/J2 on the expected full-length leader RNA levels of any of the S-box genes (data not shown). Taken together, this indicates that RNase Y is the key enzyme for the turnover of SAM-dependent riboswitches in B. subtilis.
Figure 6.
Turnover of all functional S-box riboswitches in B. subtilis is initiated by RNase Y. Northern blot analysis of the leader transcripts of 10 S-box genes. Total RNA isolated from the rny (ymdA) depletion strain SSB447 grown in the presence and absence of IPTG was separated on 8% acrylamide gels and probed with leader-specific oligonucleotides (Supplementary Table I). The putative full-length leader transcripts are marked by an arrow head. The shorter transcripts are likely degradation intermediates. A [32P]-labelled DNA ladder was transferred to the membrane along with the RNA.
Bulk mRNA stability is increased in an RNase Y depleted strain
We next investigated whether RNase Y has an impact on global mRNA stability. The chemical decay rate of pulse-labelled RNA was determined in strain SSB447 (containing the IPTG-inducible rny copy) grown in the presence or absence of IPTG inducer. The measured mRNA half-lives were 6.1±1.3 min (−IPTG) and 3.5±0.4 min (+IPTG), respectively, compared with 2.8±0.2 min for the wild-type strain. In the RNase Y depletion strain, global mRNA decay is thus slowed more than two-fold compared with the wild type.
RNase Y is a widely conserved HD domain protein
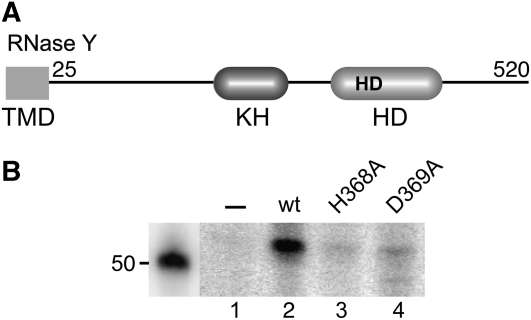
On the basis of the presence of the conserved HD domain, the YmdA protein had been assigned to the superfamily of metal-dependent phosphohydrolases (Aravind and Koonin, 1998). The presence of an RNA-binding KH domain (Grishin, 2001) together with the HD domain (Figure 7A) suggested that the primary function of this protein is in nucleic acid metabolism. As a putative metalloenzyme RNase Y possesses a distinctive combination of metal-chelating residues, essentially histidines and aspartates (Aravind and Koonin, 1998). To assess the importance of the HD domain for RNase Y activity, we mutated His368 and Asp369 that constitute the highly conserved HD motif. As shown in Figure 7B, the mutated proteins were severely impaired in cleavage of the yitJ riboswitch, indicating that the HD domain is important for the endonucleolytic activity of RNase Y.
Figure 7.
RNase Y domain structure and effect of point mutations on RNase Y activity. (A) The KH and HD domains are indicated together with the HisAsp doublet conserved in HD domain proteins. TMD indicates the N-terminal transmembrane domain. (B) Cleavage of the yitJ leader transcript by wild-type RNase Y and H368A and D369A mutant proteins. Cleavage products were separated on an 8% polyacrylamide gel (similar to as shown in Figure 3A and B). Only the portion of the gel corresponding to the major cleavage product (52 nt) is shown.
Discussion
The expression of many genes in B. subtilis and Gram-positive organisms in general is controlled by transcription antitermination mechanisms (Merino and Yanofsky, 2005; Grundy and Henkin, 2006), which produce large amounts of prematurely terminated transcripts from strong constitutive promoters. The SAM-dependent riboswitch controlling the yitJ gene belongs to this class (Tomsic et al, 2008) and forms a highly structured mRNA, especially in its terminator conformation, that is when complexed with SAM. It was surprising to find that this presumably very stable transcript was virtually undetectable in a wild-type strain (Figure 2B, lane 1). This meant that there must be a very efficient mechanism to initiate its decay, providing, a priori, a perfect role for RNases J1/J2. These enzymes, particularly RNase J1, with their dual endo- and 5′–3′ exoribonucleolytic activities are thought to constitute the heart of the RNA degradation machinery in Gram-positive organisms. However, they were not found to be involved in the rate-determining initiating events of SAM riboswitch decay (Figure 2B, lane 3). Instead, as we show here, the turnover of this transcript is dependent on the product of the ymdA gene (Figure 2B). In vitro experiments characterized the YmdA protein as an endoribonuclease (named RNase Y) capable of cleaving the yitJ riboswitch six nucleotides upstream of the aptamer structure (Figure 3A and C). Interestingly, RNase Y appeared to be very sensitive to the RNA conformation downstream of its cleavage site. Consequently, an in vitro transcript including 3′ sequences, allowing the formation of the antiterminator but not the terminator (Figure 3C, primer HP1356), was a poor substrate for RNase Y. However, more efficient cleavage was obtained by adding increasing amounts of SAM (Figure 3B), which is known to favour the terminator conformation by binding and stabilizing the mutually exclusive aptamer domain (McDaniel et al, 2005). This suggested that the principal substrate for RNase Y is the riboswitch complexed with SAM. Corroborating this hypothesis, extension of a primer specific for read-through (antiterminated) transcripts present in RNA isolated under methionine starvation conditions, detected essentially transcripts, which contained the complete leader sequences (Figure 2D). In this case, the more stable antiterminator conformation probably remains predominant and inhibits cleavage of the read-through transcripts. Such a cleavage would actually be counterproductive as this should destabilize the structural mRNA encoding the YitJ protein and reduce the effect of transcriptional antitermination. In support of this notion, depletion of RNase Y had no significant impact on the induced expression of a yitJ-lacZ fusion after methionine starvation (data not shown).
Initiation of the yitJ riboswitch decay in vivo turned out to be more complex than anticipated from the in vitro data. We could confirm that RNase Y cleaves the riboswitch upstream of the aptamer domain in vivo, two nucleotides upstream of the cleavage site identified in vitro (Figures 3C and 5D). Small differences in cleavage site specificity when comparing in vivo and in vitro conditions are not uncommon and probably reflect to some extent, conformational differences of the structured RNA when presented to the nuclease. More surprising was the fact that in vivo a second very efficient cleavage occurred downstream of the aptamer domain (14 nt upstream of the terminator structure, Figure 5D). Although cleavage 1 (also observed in vitro) was about 50% efficient (as estimated by the primer extension signals obtained in the pnpA-rnr mutants, Figure 5A, lanes 4 and 5), cleavage between the aptamer and the terminator was virtually 100% efficient on the prematurely terminated transcripts. The degradation intermediates observed in the pnpA-rnr mutant strains accumulated to even higher levels than the intact leader transcript in the RNase Y depleted strain (Figure 5A, lanes 4, 5 and 7). This difference is probably due to the fact that PNPase and RNase R are completely absent in the pnpA-rnr knock-out mutants, whereas RNase Y has only been depleted, a procedure in which some residual enzyme remains in the cell. The aptamer domain is thus predominantly degraded by the 3′–5′ exoribonucleases PNPase and RNase R (Figure 5A, lanes 4 and 5). However, the presence of degradation intermediates in the RNase J1 depleted strain (Figure 5A, lane 3) suggests that some degradation of the aptamer domain also occurs through the RNase J1 5′–3′ exonucleolytic activity attacking the riboswitch RNA cleaved at site 1. One of the major degradation intermediates identified in the pnpA-rnr mutants had a 5′ end at position +58 (Figure 5A, lanes 4 and 5). This corresponds to the nucleotide immediately upstream of the highly structured aptamer domain (Figure 5D) and presumably constitutes a stall site for the 5′–3′ exoribonucleolytic progression of RNase J1.
The riboswitch leader in its terminator conformation strongly accumulated in an RNase Y depleted strain (Figure 2B, lane 5), indicating that the terminator efficiently protects the leader riboswitch from 3′ to 5′ exonucleolytic attack. Degradation of the fragment containing the terminator thus had to occur principally by the 5′–3′ exonucleolytic activity of RNase J1 using the cleavage at site 2 as an entry site. This could be confirmed by S1 nuclease mapping, which showed an accumulation of the 25 nt terminal fragment in an RNase J1 depleted strain (Figure 5C, lane 3). Taken together, our data suggest that SAM riboswitch turnover initiates with rate determining endonucleolytic cleavages by RNase Y, which are likely dependent on the intracellular SAM concentration determining RNA folding. The cleavage products then become substrates for the 3′–5′ exonucleases PNPase and RNase R (the aptamer domain and probably the 50 nt 5′ terminal leader fragment) and the 5′–3′ exoribonucleolytic activity of RNase J1. The latter is required for the degradation of the 3′ terminal fragment and accessory for that of the aptamer domain.
We have not yet been able to show reproducibly in vitro that RNase Y is responsible for the site 2 cleavage on the riboswitch RNA. This might be due to the unfavourable structural conformation obtained with the in vitro transcripts chosen to optimize cleavage at site 1. As the full-length terminated riboswitch leader can only be detected in an RNase Y mutant (Figure 2B, lane 5), it is nevertheless very likely that RNase Y cleaves at both sites. As an alternative, cleavage at site 1 could be obligatory to create a structural context recognized by another RNase. However, if this was the case then cleavage at site 2 would be expected to be less or as efficient as cleavage at site 1. We have observed the contrary (see above).
RNase Y initiated the turnover of all SAM-dependent riboswitches in B. subtilis (Figure 6) with the exception of cysH, which was shown earlier to be regulated at the level of transcription initiation (Mansilla et al, 2000). The sizes of the stabilized full-length leader transcripts detected in the RNase Y depleted strain correlated quite well with the transcription initiation sites inferred from a sequence-based search for promoters (Grundy and Henkin, 1998 #1760). However, it remains to be analysed whether the degradation pathway of the yitJ riboswitch elucidated here is also applicable to the other S-box riboswitches. As the RNA–SAM complexes are very stable with half-lives ranging from 4 to 11 min (Tomsic et al, 2008), an efficient turnover of the large amounts of riboswitch RNA produced is not only important for nucleotide recycling but also for liberating bound SAM, a costly metabolite.
The chemical decay rate of bulk mRNA was slowed more than two-fold in the RNase Y depletion strain compared with the wild-type strain (6.1 versus 2.8 min, respectively). The effect of RNase Y on global mRNA stability is thus significant and distinctly greater than the 30% increase in mRNA stability observed in a RNase J1/J2 double-mutant strain (Even et al, 2005). This suggests that mRNA cleavage by RNase Y might have a major function in the initiation of mRNA degradation in B. subtilis.
RNase Y appeared to cleave in single-stranded A or AU rich sequences upstream of a secondary structure, a context resembling the cleavage specificities of RNases J1/J2 (Deikus et al, 2008; Even et al, 2005; Yao et al, 2007) and also E. coli RNase E (Callaghan et al, 2005). The requirement for a downstream secondary structure would explain why cleavage at site 1 only occurs in the presence of SAM: the lower stem of the aptamer domain (Figure 3C) only forms on binding of the metabolite (McDaniel et al, 2005). This suggested that cleavage site selection by RNase Y is extremely specific. However, SAM might only be needed to form a (unspecific) secondary structure 3′ to the cleavage site, a role played by the terminator structure downstream of cleavage site 2. In this case, the cleavage specificity of RNase Y would be compatible with a more general function in mRNA metabolism.
RNase Y preferred a 5′ monophosphorylated substrate and its depletion significantly increased bulk mRNA stability, two characteristics it shares with RNase E. Reduced cleavage of 5′ triphosphorylated RNA avoids attack of nascent RNA transcripts and is a priori a useful property for an enzyme responsible for setting the decay rate of many mRNAs. However, RNase Y seems to be able to discriminate also between distinct RNA conformations located downstream of its cleavage site. The rather poor cleavage efficiency observed in vitro might reflect this sensitivity. It thus remains to be seen to what extent the nature of the 5′ end and RNA structure, respectively, contribute to substrate specificity and enzyme activity.
A recent study using a bacterial two hybrid system showed that PNPase, RNase J1 and the glycolytic enzymes 6-phosphofructokinase and enolase can interact with the ymdA gene product and potentially form a degradosome-like complex in vivo (Commichau et al, 2009). It will be interesting to see if such a complex can be purified and how the addition of glycolytic enzymes might improve or alter RNase Y activity in vitro. The same authors also showed that an earlier reported processing of the B. subtilis gapA operon (Meinken et al, 2003) is diminished in vivo when ymdA expression is reduced (Commichau et al, 2009). Testing already known RNA cleavages in an RNase Y depleted strain might thus rapidly lead to the discovery of new potential substrates for RNase Y. An earlier systematic inactivation analysis of unknown but essential genes had shown that depletion of YmdA resulted in several effects on cell and chromosome morphology (Hunt et al, 2006). Interestingly, a H368A mutation (that caused a strong decrease in endonucleolytic activity, Figure 7B) showed a phenotype similar to the depleted strain, suggesting that it is the impaired endonucleolytic activity of RNase Y that is responsible for the observed effects.
Gfp-fusion experiments localized the YmdA protein to the cell periphery (Hunt et al, 2006), consistent with the presence of a predicted N-terminal transmembrane domain (Figure 7A). In our hands, the affinity purified, full-length His-tagged protein was not suitable for in vitro assays due to considerable contamination by a large number of proteins presumably associated with membrane fragments. We resolved this problem by deleting the 25 N-terminal amino acids comprising the transmembrane domain and using intein technology for purification.
Localization of RNase Y (alone or in complex with other proteins) at the membrane, further extends the analogy to E. coli RNase E. In fact, RNase E contains a short domain with the propensity to form an amphipathic α-helix that is necessary and sufficient to bind to membranes (Khemici et al, 2008), providing a rationale for the cytoskeletal-like structure of the degradosomal proteins in the cell (Taghbalout and Rothfield, 2007). However, the consequence of this localization on RNA processing and degradation is not clear, neither for RNase E nor for RNase Y. It was suggested that the membrane might serve as a storehouse for RNase E and that the active form of the enzyme is the small proportion of molecules that are cytoplasmic (Khemici et al, 2008). The conserved pseudo-compartmentalization of RNase Y and RNase E in two very different organisms (B. subtilis and E. coli, respectively) clearly suggests an evolutionary advantage for such a configuration. The membrane bound nucleases might provide an anchor for assembling potentially transient degradosome-type complexes thereby fine tuning the degradation machinery under various physiological conditions.
RNase Y is the first member of a new class of ribonucleases that constitute a subgroup within the superfamily of HD proteins by combining the metal-chelating HD domain with the RNA-binding KH domain (Aravind and Koonin, 1998). Single amino acid substitutions of the highly conserved HD doublet greatly reduced the ribonucleolytic activity of RNase Y, indicating that the catalytic activity resides within the HD domain. Some cleavage by the D369A mutant also occurred at a second site 2–3 nucleotides upstream corresponding closely to the in vivo cleavage site (Figure 7, lane 4).
Orthologs of RNase Y occur in about 40% of the currently sequenced eubacteria (Table I), with similarities ranging from 56 to 99%. RNase Y is absent from the archea and eukaryotic organisms with a single exception, Drosophila willistoni. Organisms containing RNase Y also have either an RNase E/G like enzyme or RNase J. A few bacteria actually have all three enzymes, like the Frankia and Salinispora species of Actinobacteria and 25–30% of the Bacillales (e.g. Listeria) and Clostridia (Table I). This raises interesting questions as to the sharing of functions for all three enzymes and whether one of these usually essential RNases becomes dispensable in this context. On the other hand, we only found a single eubacterial species (Sulcia muelleri in the Bacteroidetes) that has none of the three enzymes. This is also the case for the Crenarcheota and the majority of the Euryarcheota in which novel key ribonucleases probably remain to be identified.
Table 1.
Occurrence of RNase Y in procaryotes
 |
How does RNase Y fit into our current understanding of mRNA processing and degradation in bacteria? It is intriguing that two completely unrelated enzymes like RNase Y and RNase E, from a Gram-positive (B. subtilis) and Gram-negative organism (E. coli) that are thought to follow very different strategies concerning mRNA metabolism, share so many features including large effects on global mRNA stability and a 5′ end preference for mono phosphorylated substrates. This raises the possibility that mRNA processing and degradation might be more similar between B. subtilis and E. coli than presently assumed, with an endonucleolytic cleavage being the crucial step in initiating mRNA decay in both organisms. RNases J1/J2 have smaller effects on bulk mRNA than RNase Y (Even et al, 2005), suggesting that these enzymes are not initiating decay on a large scale (be it endo- or exonucleolytically) but are mostly involved in degrading cleavage products exonucleolytically in a 5′–3′ direction. This property is especially useful for degrading transcripts whose 3′ ends are protected by a secondary structure. With this in mind, the 5′–3′ exonucleolytic activity of RNase J may have evolved in organisms like B. subtilis that, in contrast to E. coli, does not appear to have an efficient polyadenylation pathway to allow 3′–5′ degradation of structured RNAs. In B. subtilis, 5′ road blocks (structural or formed by bound ribosomes, etc.) can substantially stabilize even nontranslated downstream sequences, implying that an endoribonuclease like RNase Y should not efficiently cleave the body of an mRNA. We are still ignorant of the exact parameters that determine substrate specificity for RNase Y and, for that matter, also for the endonucleolytic activity of RNase J. Further characterisation of RNase Y should include an analysis of the relative importance of the preference of RNase Y for the 5′ phosphorylation state and conformation, respectively, of the RNA substrate. Interestingly, each of these two characteristics has the potential to control access of RNase Y to nascent transcripts. Together, they could easily provide the specificity necessary to reconcile the activity of RNase Y with our current understanding of mRNA metabolism in Gram-positive bacteria.
Materials and methods
Bacterial strains and growth conditions
The prototrophic B. subtilis 168 strain 1A2 (BGSC) was the parent strain used for the construction of mutants. The RNase J1/J2 double-mutant strain SSB357 (rnjB∷spc, Pxyl∷rnjA) was described earlier (Even et al, 2005) and the RNase III mutant strain BG322 (Herskowitz and Bechhofer, 2000) was provided by D Bechhofer. For strain SSB447, the ymdA gene was put under control of the Pspac promoter by Campbell-type integration of plasmid pHMD1. Induction of yitJ expression by methionine starvation was carried out in strain SSB441 (metC2, lys1) provided by T Leighton. E. coli strain SSC91, used for the overexpression of YmdA, was constructed by lysogenisation of strain IBPC852 (Δrnb201∷tet, rph1, thyA715, rne1) kindly provided by P Regnier with λDE3. B. subtilis was grown in SMS minimal medium (Spizizen, 1958) supplemented with 0.1% L-glutamine. Methionine was added to a final concentration of 2 mM when required. Antibiotics were added, at the following concentrations: ampicillin (200 μg/ml), kanamycin (5 μg/ml), chloramphenicol (4 μg/ml). For depletion of the essential RNases J1 and Y, strains bearing inducible copies of these genes (Pxyl and Pspac, respectively) were grown overnight in the presence of inducer. The cells were centrifuged and resuspended in medium without inducer. Growth was monitored every 30 min and the first slowing of growth in the absence of inducer, compared with a culture grown with inducer, was detectable at an OD600 of ∼0.4. Cells were harvested immediately after the first detectable change in growth rate.
Construction of the YmdA depletion strain
Plasmid pHMD1 was used to make the ymdA depletion strain: a 500 bp fragment was PCR amplified using oligonucleotides HP1113/HP1114, digested SalI and SphI and ligated into the respective sites of plasmid pDG648 (kindly provided by P Stragier) downstream of Pspac promoter. Campbell-type integration of pHMD1 on the B. subtilis chromosome rendered ymdA expression IPTG dependent.
Purification of YmdA
Plasmid pHMD3 was used to overexpress YmdA: the coding sequence of the ymdA gene excluding the first 25 residues comprising a transmembrane domain was PCR amplified using oligonucleotides HP1182/HP754 and cloned in phase with the intein domain in plasmid pKYB1 (New England Biolabs) to give plasmid pHMD3. YmdA was isolated as intein fusion protein using the IMPACT system (New England Biolabs). Overexpression of the recombinant proteins was carried out in strain SSC91. Purified YmdA was heated 10 min at 45°C before use to inactivate the thermosensitive RNase E that might potentially contaminate the preparation.
Site-directed mutagenesis
The H368A and D369A mutations in the YmdA HD domain were introduced on plasmid pHMD3 using the Quikchange (Stratagene) strategy (oligonucleotides HP1298, HP1299, HP1300, HP1301) and KOD DNA polymerase (Novagen).
Northern blot analysis
Northern blot analysis was carried out as described (Putzer et al, 1992) using 6 μg of total RNA separated either on a 1.2% agarose or 8% acrylamide gel. A continuously α-[32P] UTP-labelled riboprobe was generated by T7 in vitro transcription of a PCR fragment made with the primers HP1231 (5′ end corresponds to position +152) and HP1058 (corresponds to HP1056 plus the upstream T7 promoter sequence) was used to detect both the terminated yitJ leader and full-length transcript. 5′ [32P]-labelled oligonucleotides (listed in Supplementary Table I) were used as probes to specifically detect the leader transcripts of all 11 S-box genes.
Primer extension analysis
A measure of 5 μg of total RNA was subjected to reverse transcription analysis by extending the 5′ [32P]-labelled primers with Superscript III reverse transcriptase (Invitrogen).
S1 nuclease mapping
A measure of 30 μg of total RNA was mixed with 5′ end-labelled oligonucleotide HP1442 (105 cpm, ∼10 ng), precipitated, and resuspended in 40 mM Pipes pH 6.4, 1 mM EDTA pH8, 0.4 M NaCl and 80% deionized formamide. Hybridization was carried out by incubating the mixture at 48°C overnight and S1 nuclease (Fermentas) digestion (200 U) was carried out according to the manufacturer's instructions at 37°C for 30 min. The reaction products were resolved on a 5% polyacrylamide gel.
5′/3′ RACE
Total RNA (5 μg) was circularized with RNA ligase (Biolabs) and subjected to RT–PCR across the 5′/3′ junction, using oligonucleotides HP1134 (RT) and HP1394 and HP1395 (PCR). The PCR fragments of ∼130 bp were isolated from agarose gels, digested with EcoRI and BamHI and cloned into plasmid pJRD184 (Heusterspreute and Thi, 1985) for sequencing. Ten clones of each mutant were sequenced.
Assay of RNase Y activity
Reactions were carried out using about 10 nM RNA substrate and 0.25 μM RNase Y in a reaction volume of 10 μl. The buffer contained 20 mM Hepes pH 8, 8 mM MgCl2, 0.48 U/μl RNAsin (Promega), 100 mM NaCl and the reactions were incubated for 10 min at 30°C. For time course experiments, samples of scaled-up reactions (50 μl) were taken at the indicated times. Reactions were stopped by the addition of 5 μl stop buffer (87.5% formamide, 0.05% xylene cyanol, 0.05% bromophenol blue, 5 mM EDTA) and samples were loaded on 5 or 8% denaturing polyacrylmide gels. The RNA substrates were synthesized with T7 RNA polymerase from PCR products (primer pairs HP1198/HP1238 and HP1198/HP1356). In both cases, the mRNA transcripts contained an additional 5′ G residue when compared with the wild-type mRNA. The in vitro transcripts were either 5′ end-labelled with γ-[32P] ATP using T4 polynucleotide kinase following dephosphorylation, continuously labelled with α-[32P] UTP or 3′ end-labelled with [32P]-pCp using T4 RNA ligase. 5′ monophosphorylated transcripts were synthesized by including a 50-fold excess of GMP over GTP in the reaction.
To test the effect of SAM on mRNA cleavage, SAM (25 and 80 μM) was added to the RNA substrate and the mixture was incubated at 70°C for 10 min and slowly cooled to 37°C, before the addition of RNase Y.
A 5′ end-labelled 50 bp DNA ladder was used as a size marker in most experiments.
Chemical decay of bulk mRNA
Total mRNA decay was measured as described earlier (Wang and Bechhofer, 1996; Even et al, 2005).
Supplementary Material
Supplementary Table I
Review Process File
Acknowledgments
We are grateful to Jackie Plumbridge for useful discussions and critical reading of the manuscript. We thank J Stülke and F Commichau for helpful discussions and sharing preliminary data. We thank Barry Holland for kindly commenting the manuscript. We are grateful to S Even and D Brechemier-Baey for their contributions in the very early stages of this work. This work was supported by funds from the CNRS (UPR 9073) and Université Paris VII-Denis Diderot.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravind L, Koonin EV (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci 23: 469–472 [DOI] [PubMed] [Google Scholar]
- Babitzke P, Kushner SR (1991) The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci USA 88: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF (2005) Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437: 1187–1191 [DOI] [PubMed] [Google Scholar]
- Carpousis AJ, Luisi BF, McDowall KJ (2009) Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Nucleic Acid Res Mol Biol 85: 91–135 [DOI] [PubMed] [Google Scholar]
- Choonee N, Even S, Zig L, Putzer H (2007) Ribosomal protein L20 controls expression of the Bacillus subtilis infC operon via a transcription attenuation mechanism. Nucleic Acids Res 35: 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J (2009) Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics 8: 1350–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG (2008) The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451: 355–358 [DOI] [PubMed] [Google Scholar]
- Deikus G, Condon C, Bechhofer DH (2008) Role of Bacillus subtilis RNase J1 endonuclease and 5′-exonuclease activities in trp leader RNA turnover. J Biol Chem 283: 17158–17167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E (2003) The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA 100: 5052–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H (2005) Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res 33: 2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV (2001) KH domain: one motif, two folds. Nucleic Acids Res 29: 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM (1998) The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol 30: 737–749 [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM (2003) The T box and S box transcription termination control systems. Front Biosci 8: d20–d31 [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM (2006) From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol 41: 329–338 [DOI] [PubMed] [Google Scholar]
- Henkin TM (2008) Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev 22: 3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz MA, Bechhofer DH (2000) Endoribonuclease RNase III is essential in Bacillus subtilis. Mol Microbiol 38: 1027–1033 [DOI] [PubMed] [Google Scholar]
- Heusterspreute M, Thi VH (1985) Vectors with restriction site banks. IV. pJRD184, a 3793-bp plasmid vector having 43 unique cloning sites. Gene 39: 299–304 [DOI] [PubMed] [Google Scholar]
- Hunt A, Rawlins JP, Thomaides HB, Errington J (2006) Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology 152(Part 10): 2895–2907 [DOI] [PubMed] [Google Scholar]
- Khemici V, Poljak L, Luisi BF, Carpousis AJ (2008) The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol 70: 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E et al. (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Bernstein JA, Cohen SN (2002) RNase G complementation of rne null mutation identifies functional interrelationships with RNase E in Escherichia coli. Mol Microbiol 43: 1445–1456 [DOI] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H (2008) Structural insights into the dual activity of RNase J. Nat Struct Mol Biol 15: 206–212 [DOI] [PubMed] [Google Scholar]
- Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H (2008) mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol 70: 183–196 [DOI] [PubMed] [Google Scholar]
- Mansilla MC, Albanesi D, de Mendoza D (2000) Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in L-cysteine biosynthesis in Bacillus subtilis. J Bacteriol 182: 5885–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C (2007) 5′-to-3′ Exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129: 681–692 [DOI] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM (2003) Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA 100: 3083–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Henkin TM (2005) A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol Microbiol 57: 1008–1021 [DOI] [PubMed] [Google Scholar]
- Meinken C, Blencke HM, Ludwig H, Stulke J (2003) Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology 149 (Part 3): 751–761 [DOI] [PubMed] [Google Scholar]
- Merino E, Yanofsky C (2005) Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet 21: 260–264 [DOI] [PubMed] [Google Scholar]
- Putzer H, Gendron N, Grunberg-Manago M (1992) Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J 11: 3117–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J (1958) Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA 44: 407–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghbalout A, Rothfield L (2007) RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc Natl Acad Sci USA 104: 1667–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsic J, McDaniel BA, Grundy FJ, Henkin TM (2008) Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol 190: 823–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Bechhofer DH (1996) Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol 178: 2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR (2005) Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59: 487–517 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol 10: 701–707 [DOI] [PubMed] [Google Scholar]
- Yao S, Blaustein JB, Bechhofer DH (2007) Processing of Bacillus subtilis small cytoplasmic RNA: evidence for an additional endonuclease cleavage site. Nucleic Acids Res 35: 4464–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Blaustein JB, Bechhofer DH (2008) Erythromycin-induced ribosome stalling and RNase J1-mediated mRNA processing in Bacillus subtilis. Mol Microbiol 69: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I
Review Process File