Abstract
Allergic asthma is associated with persistent functional and structural changes in the airways and involves many different cell types. Many proteins involved in allergic asthma have been identified individually, but complete protein profiles (proteome) have not yet been reported. Here we have used a differential proteome mapping strategy to identify tissue proteins that are differentially expressed in mice with allergic asthma and in normal mice. Mouse lung tissue proteins were separated using two-dimensional gel electrophoresis over a pH range between 4 and 7, digested, and then analyzed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MS). The proteins were identified using automated MS data acquisition. The resulting data were searched against a protein database using an internal Mascot search routine. This approach identified 15 proteins that were differentially expressed in the lungs of mice with allergic asthma and normal mice. All 15 proteins were identified by MS, and 9 could be linked to asthma-related symptoms, oxidation, or tissue remodeling. Our data suggest that these proteins may prove useful as surrogate biomarkers for quantitatively monitoring disease state progression or response to therapy.
Keywords: Electrophoresis, Gel; Two-Dimensional; Disease Models, Animal; Asthma; Proteomics; MALDI-TOF/MS; Chi313 protein, mouse
INTRODUCTION
Asthma is a multifactorial disease characterized by chronic inflammation in the lungs. The signature of the disease is massive infiltration of the bronchial mucosa by several cell types including lymphocytes, eosinophils, and mast cells, which is accompanied by a pronounced elevation in serum IgE. Desquamation of the lung epithelium occurs, along with goblet cell hyperplasia, thickening of the mucosal layer, and mucus production within the bronchioles, with the overall result being impeded airflow and airway hyperreactivity (AHR) (1).
Asthma is also defined as a complex genetic disorder with a heterogeneous phenotype attributed to the interactions among many genes and the environment (2). To comprehend the pathogenetic mechanisms underlying the many variants of asthma, it is essential to identify factors that initiate, intensify, and modulate the inflammatory response of the airway and to determine how these immunological and biological processes produce the characteristic airway abnormalities (3).
The complex nature of the asthma phenotype, together with genetic heterogeneity and environmental influences, has made it difficult to uncover the aspects that underlie this common disease (4). Recently, a number of factors have been reported that might predispose or protect against asthma, but there is no single factor which can completely explain the development of asthma (5-10). Thus, large-scale, high-throughput, and whole-proteome studies are needed to understand the genetic contribution to asthma.
Two-dimensional polyacrylamide gel electrophoresis (2-DE), which separates polypeptides by their charge (pI) and relative molecular mass (Mr), has been shown to be a powerful technique for analyzing complex mixtures of proteins (11). The improvement of two-dimensional electrophoresis by the development of immobilized pH gradients (IPGs; 12), together with improved solubilization techniques (13, 14), now permits the reproducible separation of up to 2000 proteins on a single two-dimensional electrophoresis gel. Such gel-separated proteins can be identified rapidly by mass spectrometry (MS), and, if genomic information is also available, such analyses permit the systematic identification of the protein complement of a genome, known as the proteome (15-18). In this study, we have adopted a proteomics approach to better understand the protein basis of this pathology of asthma on the whole, and to development of marker for ovalbumin induces asthma.
MATERIALS AND METHODS
Animals
Male BALB/c mice (5 weeks old) were obtained from Charles River Laboratories (MA, U.S.A.) and were held under specific pathogen-free conditions. The experiments, approved by our institutional Committee on Animal Experimentation, were performed in compliance with governmental and international guidelines on animal experimentation.
Sensitization and provocation procedures
The experimental mice were sensitized to ovalbumin (OVA, Grade V; Sigma-Aldrich, St. Louis, MO, U.S.A.) on days 1 and 14 by administration of three intraperitoneal (i.p.) injections of 10 µg OVA emulsified in 200 µL phosphate-buffered saline (PBS) with 1.5 mg aluminum hydroxide (Sigma-Aldrich). The mice were subsequently challenged with an OVA aerosol (1% in PBS) for 20 min on days 28, 29, and 30 (Fig. 1A). The aerosol was delivered to a 9-L Perspex exposure chamber by a nebulizer (model 646; DeVilbiss, Somerset, PA, U.S.A.) driven by an airflow of 40 L/min, which provided an aerosol with an output of 0.33 mL/min. The control animals received i.p. injections of aluminum hydroxide in PBS and were subsequently challenged with PBS aerosol.
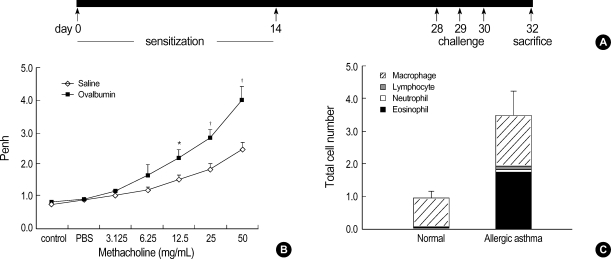
Fig. 1.
Construction of the mouse model for allergic asthma. (A) Schematic representation of ovalbumin (OVA) sensitization and challenge protocol. (B) Airway responsiveness to methacholine in OVA-sensitized mice at 24 hr after the last challenge with saline (diamonds) or OVA (black rectangles). The results are expressed as the arithmetic means±SEM (n=6 per group). Significant differences from saline-challenged animals: *p<0.05; †p<0.01. (C) Numbers of mononuclear cells recovered in bronchial alveolar (BAL) fluid after saline or OVA challenge. Differential cell counts were performed on a minimum of 500 cells per sample to identify eosinophils (black), neutrophils (open), macrophages (hatched), and lymphocytes (gray). Challenge with OVA significantly increased the total cell numbers. Eosinophils and macrophages recovered in BAL fluid were significantly increased, whereas the neutrophil and lymphocyte counts were unchanged in OVA-challenged mice.
Measurement of airway hyperreactivity
Responsiveness to methacholine was assessed in conscious, unrestrained mice by barometric plethysmography, using an apparatus and software from All Medicus (Seoul, Korea). The measurements were conducted using standard methods, similar to those described previously (19). Each mouse was placed in a plethysmograph chamber and exposed to an aerosol of PBS for baseline readings, followed by increasing concentrations of aerosolized methacholine (2.5-20 mg/mL; Sigma-Aldrich) for 3 min each. AHR was assessed using the enhanced pause parameter (Penh), which was calculated automatically by the software based on the mean pressure generated in the plethysmograph chambers during inspiration and expiration, combined with the time of each respiratory phase. Penh readings were recorded for 5 min after each aerosol exposure and averaged. The results are presented as the mean percent change in Penh at each methacholine concentration compared with the baseline readings, calculated from the data from groups of six mice.
Sample preparation
All reagents used in 2-DE were obtained from Amersham Biosciences (Seoul, Korea). Fresh lung tissue was placed in a clean mortar containing liquid nitrogen and finely ground; 1 mL of sample buffer (7 M urea, 2 M thiourea, 4% CHAPS, 40 mM Tris-HCl, 20 mM DTT, 2% IPGphore buffer) was added and mixed thoroughly. The suspensions were sonicated for approximately 1 hr at 4℃ and centrifuged at 15,000 g for 30 min at 4℃; the supernatants were stored at -80℃ until use.
Two-dimensional gel electrophoresis
2-DE was performed in a horizontal apparatus (IPGphor and Hoefer 600 SE; Amersham Biosciences, Uppsala, Sweden). For the analytical gels, 200 µg of protein was applied onto immobilized pH gradient (IPG) strips (24 cm, pH 4-7) containing Immobilines pH 4-7, thiourea, CHAPS, and urea, according to the method of Rabilloud et al. (13). After isoelectric focusing, the strips were applied to the top of SDS-PAGE gels (12.5%), and the proteins were separated according to their molecular mass. The SDS-PAGE gels were silver-stained to detect proteins (20). For the analysis of selected proteins after tryptic digestion, 2 mg of digested protein was applied to the IPG strip, and the final SDS-PAGE gel was stained using Coomassie Brilliant Blue G250 (Sigma-Aldrich).
Image analysis
Digitized images of the stained gels were analyzed using the 2-DE gel analysis program ImageMaster 2D ver. 4.0 (Amersham Pharmacia Biotech, Uppsala, Sweden). A comparison report of the qualitative and quantitative differences between the samples for each data set of was generated.
In-gel digestion and mass spectrometric analysis
Differentially expressed protein spots were excised from the gels, cut into smaller pieces, and digested with trypsin (Promega, Madison, WI, U.S.A.), as previously described (21). For MALDI-TOF MS analysis, the tryptic peptides were concentrated on POROS R2 columns (Applied Biosystems, Foster City, CA, U.S.A.). After successive column washings with 40% methanol, 100% acetonitrile, and 50 mM ammonium bicarbonate, the samples were applied onto the R2 column and eluted in 2 mL of a-cyano-4-hydroxycinnamic acid before MALDI-TOF analysis (22, 23). The spectra for the protein samples were obtained using a Voyager DE PROMALDI-TOF spectrometer (Applied Biosystems). The searches of protein databases were performed with the MSFit program (http://prospector.ucsf.edu/ucsfhtml3.4/msfit.htm) using monoisotopic peaks. A mass tolerance within 50 ppm was allowed for the first analysis; the system was subsequently recalibrated at 20 ppm using the lists of proteins obtained from the initial analysis. The spectra were also internally calibrated using trypsin autolysis products. The resulting peptide masses were used to search the databases managed by SWISSPROT (http://kr.expasy.org) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Semiquantitative RT-PCR
Single-stranded cDNA was synthesized from 3 µg of total RNA using an oligodeoxythymidylic acid primer and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, U.S.A.). Each cDNA was diluted for subsequent PCR amplification. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was amplified as a quantitative control. Each PCR was carried out in a 20-µL volume of 1× PCR buffer with a program of 10 min at 94℃ for initial denaturing, followed by 26 cycles of 94℃ for 30 sec, 56℃ for 30 sec, and 72℃ for 30 sec, in a GeneAmp PCR system 9600 (Perkin-Elmer Applied Biosystems, Foster City, CA). The following primers were used: YM1-forward, 5'-taccagttgggctaagga-3'; YM1-reverse, 5'-atgctggaaatcccaca-3'; YM2-forward, 5'-gccattggaggatggaa-3'; and YM2-reverse, 5'-tggaatgtggttcaaagt.
Statistical analysis
The proteins from the asthmatic and control groups were analyzed for differences in levels of expression using Student's t-test or the Mann-Whitney U-test. Values of p<0.05 were considered significant.
RESULTS
Construction of the asthmatic mouse model
The intraperitoneal sensitization and nebulized provocation of mice with ovalbumin resulted in a dose-dependent increase in airway responsiveness to inhaled methacholine (Fig. 1B). The treated mice had significantly increased numbers of total cells in bronchoalveolar lavage (BAL) fluid, as well as in the number and percentage of eosinophils in the BAL fluid (Fig. 1C). Also, the levels of ovalbumin-specific IgE were increased in the plasma of ovalbumin-challenged asthmatic mice (data not shown).
2-DE analysis of mouse lung tissues from control and OVA-challenged mice
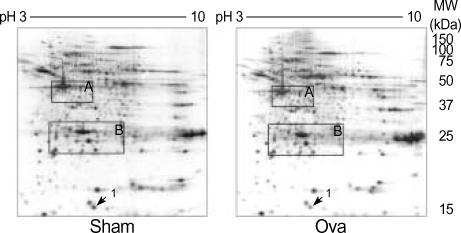
To examine the differential expression of proteins in lung tissues from asthmatic mice and normal controls, proteomic analysis was performed using high-resolution 2-DE. Fig. 2 is a representative master gel image showing the separation of proteins from normal mouse lung tissue. More than 1,000 protein spots with pIs between 3 and 10 and with relative molecular masses between 15 and 150 kDa were detected on the 2-DE gels. A computer-assisted comparative analysis of the respective silver-stained spot patterns of six paired samples (six normal controls and six OVA-challenged, asthmatic mice) showed that one protein spot appearing in the 2-DE gels of control tissues (Fig. 2, arrows) were slightly increased in the lung tissues of OVA-challenged mice. However, most of the variable spots were located in two limited areas of the gels (Fig. 2, boxed).
Fig. 2.
2-dimension electrophoresis (DE) of proteins from normal (Sham) and allergic asthma (OVA) mouse lung tissues. Proteins from the whole lung were extracted and separated on an IPG strip with a nonlinear gradient of pH 3-10, followed by separation in the second dimension on an 8-18% SDS polyacrylamide gel. The gel was stained with Coomassie Blue G-250. Spots 1 was determined to be transthyretin.
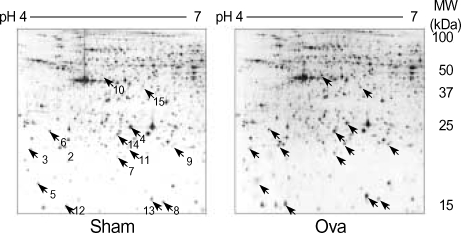
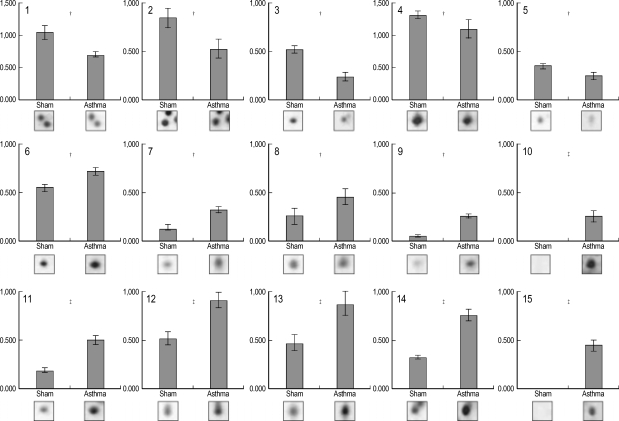
By conducting 2-DE using several types of IPG strips, we found that the 4-7 pH range covers a majority of the proteins of interest in mouse lung tissue, and thus was the best choice for initial survey investigations (data not shown). In order to preselect proteins exhibiting variations in expression levels, the protein patterns from asthmatic and control lung tissues were divided into two classes, and the quantities of all detected spots in both classes were compared by Student's t-test using ImageMaster 2-DE gel analysis software. After comparing the 2-DE protein patterns on duplicate gels of lung tissue from six asthmatic and six control mice, we found 14 protein spots that were significantly different in asthmatic and control tissues (Fig. 3). Among them, eight spots were meaningfully increased, and five spots were decreased in the lungs of OVA-challenged mice (Fig. 4). Two protein spots were exclusively expressed in asthmatic lung tissues (Fig. 3, spots 10 and 15).
Fig. 3.
2-DE of proteins from normal (Sham) and allergic asthma (OVA) mouse lung tissues. In the first dimension, proteins were loaded on a 24-cm IPG strip with a linear gradient of pH 4-7. A 12% SDS polyacrylamide gel was used for second dimension separation. Proteins were visualized by silver staining. Indicated spots represent proteins differentially expressed between normal and asthmatic lung tissues.
Fig. 4.
Protein expression levels were determined by relative intensity using image analysis. Normalized spot intensities of the asthma versus normal control (sham) group were compared. Mean intensity and spot intensities on individual gels are shown. *p<0.05; †p<0.01, ‡p<0.001 by Student's t-test.
Protein identification
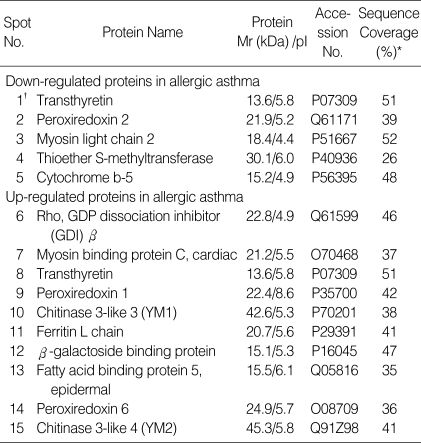
The protein spots that revealed statistically significant differences between asthmatic mice and normal controls were excised from the gels, trypsinized, and analyzed by MALDI-TOF MS. Of these, we have been able to identify 16 proteins (Table 1). The number of matching peptides, the percentage of sequence coverage, and the accuracy of mass estimates were used to evaluate the database search results. Trypsin autolysis peaks were regularly used for internal calibration of the mass spectra to achieve a 50 ppm mass accuracy. Also, in most cases the apparent Mr and pI determined from the 2-DE protein pattern were in agreement with the theoretical values of the identified proteins.
Table 1.
Differentially expressed proteins in the lungs of mice with OVA-induced allergic asthma
*Sequencing coverage is defined as the percentage of the whole length of the protein sequence which is covered by matched peptides identified by the MALDI-TOF MS analysis, †Spots shown in Fig. 2.
Differential expression of YM1 and YM2 genes
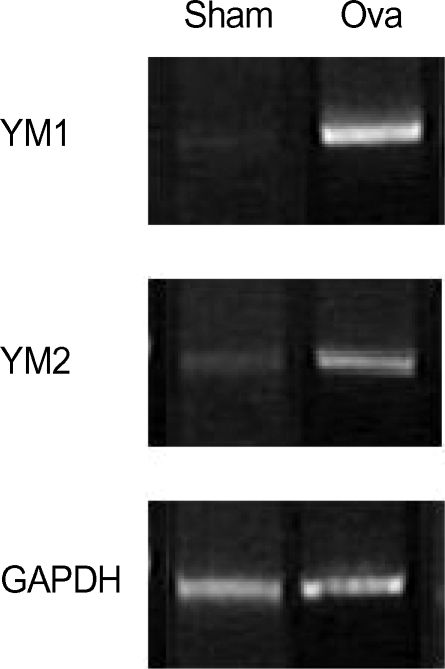
To determine whether the up-regulation of YM1 and YM2 expression occurred at the mRNA level, we performed semiquantitative RT-PCR analysis (Fig. 5) using YM1- and YM2-specific primers. The results confirmed that the mRNAs of YM1 and YM2 were expressed at very low levels in normal mouse lung tissue (Fig. 5, SHAM), and were present at considerably higher levels in the lungs of asthmatic mice (Fig. 5, OVA). These results substantiate the specific up-regulation of YM1 and YM2 expression in the lungs of asthmatic mice compared with those of normal mice.
Fig. 5.
RT-PCR analysis of the mRNA expression of differentially expressed YM proteins. Primers specific for YM1 and YM2 were used as indicated to amplify the transcripts from total RNA isolated from normal and asthmatic lung tissues. RT-PCR of the housekeeping gene GAPDH was used to control for RNA variation (bottom).
DISCUSSION
In this report, we present the comparative proteome analysis of normal and asthmatic lung tissues from mice. The pH ranges used for the first-dimension gel electrophoresis in this study (3-10 and 4-7) separated a proportion of the soluble proteins in the lung tissues, allowing a focus on the high-resolution, more unambiguous comparison of proteins with isoelectric points between pH 4 and 7.
To avoid individual and experimental variation, we produced a large number of replicate gels from the same sample or treatment group in order to obtain statistically significant changes in protein levels between the different groups, i.e., asthma versus normal. Using 2-DE, we found 15 proteins that were differentially expressed in the lung tissues of mice with OVA-induced allergic asthma. Five protein spots were down-regulated and eight protein spots were up-regulated in asthmatic lungs compared with normal lung. These protein spots were identified by in-gel digestion and MALDI-TOF MS.
The identified proteins could be classified into three different groups based on their cellular functions and participation in biochemical pathways. Four proteins (cytochrome b5, and peroxiredoxins 1, 2, and 6) are thought to be related to oxidation and reduction. As antioxidant enzymes in humans, peroxiredoxins appear to be involved in the redox regulation of cellular signaling and differentiation, displaying opposing effects (24). Our 2-DE data showed that peroxiredoxin 2 was down-regulated and peroxiredoxins 1 and 6 were up-regulated in asthmatic mouse lungs compared with normal lungs. This result suggests that oxidants play an important role in the development of asthma.
Hegesh et al. (25) reported that cytochrome b5 levels were very low in the red blood cells of a patient with congenital methemoglobinemia, but were normal in the healthy unrelated parents and in the patient's siblings. This finding confirmed the conclusion of Hultquist and Passon (26) that cytochrome b5 is required for methemoglobin reduction in vivo. The reductase converts ferric cytochrome b5 to ferrous cytochrome b5; the latter then converts methemoglobin (ferric hemoglobin) to normal hemoglobin (ferrous Hb). These reports together with our findings in the lungs of mice with experimental asthma suggest that the decreased levels of cytochrome b5 might be related to the increases in methemoglobin in the lung tissues of asthmatic patients.
Three other differentially expressed proteins (rho-GDP dissociation inhibitor-beta, myosin light chain 2, and myosin binding protein C) can be classified as structural proteins (27, 28). Airway remodeling is a term used to describe the dynamic processes that lead to structural changes in the airways of patients with asthma. These structural changes are thought to result in an irreversible component of the airway obstruction seen in asthma and perhaps also in the development of airway hyperresponsiveness. The dynamic processes underlying these structural changes are viewed as injury-repair processes driven by airway inflammation. Although the concept of airway remodeling and its role in chronic persistent airflow obstruction is widely accepted, it should be recognized that airway remodeling is still just a concept (27). Numerous histopathologic abnormalities have been described in asthmatic airways, and yet the functional consequences of these abnormalities and their role in the natural history of asthma remain unknown. In this context, we consider that cytoskeleton-related proteins such as myosin light chain 2 and myosin binding protein C might be directly involved with airway remodeling. Rho-GDP dissociation inhibitor-beta was also differentially expressed during the process of angiogenesis, which may indicate a link between its up-regulation in asthma and the cellular proliferation and migration events that are required for angiogenesis and vascular remodeling (28).
YM1 and YM2 were the most interesting among the identified proteins. According to our 2-DE, the protein spots for YM1 and YM2 were scarcely detectable in normal mice. In contrast, these two protein spots were dramatically up-regulated in asthmatic mouse lung tissues. The semi-quantitative RT-PCR results (Fig. 5) established that the 2-DE findings reflected increased gene transcription of YM1 and YM2. These findings were consistent with a previous report (29) demonstrating that both TH2 cytokines and allergic challenge both induced YM1 expression in macrophages by a STAT6-dependent mechanism. YM proteins are thought to be bi-functional with roles in eosinophil chemotaxis and tissue remodeling (30). More recently, Zhu et al. (31) reported that acidic mammalian chitinase (AMCase), a human homologue of YM1 and YM2, was induced via an interleukin-13 mediated pathway in epithelial cells and macrophages in an aeroallergen model and was expressed in very large quantities in human asthma. In addition, the neutralization of AMCase ameliorated Th2 inflammation and airway hyperresponsiveness (31).
In summary, we used 2-DE to identify proteins that are differentially expressed in normal and allergic asthmatic mouse lung tissues. Most of the identified proteins are related to oxidation-reduction and airway remodeling, which may provide an important connection to the thickening of airway walls observed in the allergic lung. Recently, many reports demonstrated associations between the expression of certain proteins and asthma. However, a comparative analysis of the total protein profiles for allergic asthmatic and normal lung tissues has not been reported. Our data suggest that the identified proteins may provide tools for gauging the success of therapeutic interventions designed to reduce the chronic inflammation underlying asthma.
Footnotes
This work was supported by a grant from the Korea Health 21 R&D Project, Ministry of Heath & Welfare, Republic of Korea (01-PJ3-PG6-01GN04-0003 and 03-PJ10-PG6-GP01-0002).
References
- 1.Jember AG, Zuberi R, Liu FT, Croft M. Development of allergic inflammation in a murine model of asthma is dependent on the costimulatory receptor OX40. J Exp Med. 2001;193:387–392. doi: 10.1084/jem.193.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hakonarson H, Halapi E. Genetic analyses in asthma: current concepts and future directions. Am J Pharmacogenomics. 2002;2:155–166. doi: 10.2165/00129785-200202030-00001. [DOI] [PubMed] [Google Scholar]
- 3.Busse WW, Lemanske RF., Jr Asthma. N Engl J Med. 2001;344:350–362. doi: 10.1056/NEJM200102013440507. [DOI] [PubMed] [Google Scholar]
- 4.Hakonarson H, Bjornsdottir US, Halapi E, Palsson S, Adalsteinsdottir E, Gislason D, Finnbogason G, Gislason T, Kristjansson K, Arnason T, Birkisson I, Frigge ML, Kong A, Gulcher JR, Stefansson K. A major susceptibility gene for asthma maps to chromosome 14q24. Am J Hum Genet. 2002;71:483–491. doi: 10.1086/342205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coyle AJ, Le Gros G, Bertrand C, Tsuyuki S, Heusser CH, Kopf M, Anderson GP. Interleukin-4 is required for the induction of lung Th2 mucosal immunity. Am J Respir Cell Mol Biol. 1995;13:54–59. doi: 10.1165/ajrcmb.13.1.7598937. [DOI] [PubMed] [Google Scholar]
- 6.Foster PS, Hogan SP, Ramsay AJ, Matthaei KI, Young IG. Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J Exp Med. 1996;183:195–201. doi: 10.1084/jem.183.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, Sheppard D, Mohrs M, Donaldson DD, Locksley RM, Corry DB. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn L, Homer RJ, MacLeod H, Mohrs M, Brombacher F, Bottomly K. Th2-induced airway mucus production is dependent on IL-4Ralpha, but not on eosinophils. J Immunol. 1999;162:6178–6183. [PubMed] [Google Scholar]
- 9.Townsend MJ, Fallon PG, Matthews DJ, Smith P, Jolin HE, McKenzie AN. IL-9-deficient mice establish fundamental roles for IL-9 in pulmonary mastocytosis and goblet cell hyperplasia but not T cell development. Immunity. 2000;13:573–583. doi: 10.1016/s1074-7613(00)00056-x. [DOI] [PubMed] [Google Scholar]
- 10.Gavett SH, Chen X, Finkelman F, Wills-Karp M. Depletion of murine CD4+ T lymphocytes prevents antigen-induced airway hyperreactivity and pulmonary eosinophilia. Am J Respir Cell Mol Biol. 1994;10:587–593. doi: 10.1165/ajrcmb.10.6.8003337. [DOI] [PubMed] [Google Scholar]
- 11.O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 12.Gorg A, Postel W, Gunther S. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1988;9:531–546. doi: 10.1002/elps.1150090913. [DOI] [PubMed] [Google Scholar]
- 13.Rabilloud T, Adessi C, Giraudel A, Lunardi J. Improvement of the solubilization of proteins in two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1997;18:307–316. doi: 10.1002/elps.1150180303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molloy MP, Herbert BR, Walsh BJ, Tyler MI, Traini M, Sanchez JC, Hochstrasser DF, Williams KL, Gooley AA. Extraction of membrane proteins by differential solubilization for separation using twodimensional gel electrophoresis. Electrophoresis. 1998;19:837–844. doi: 10.1002/elps.1150190539. [DOI] [PubMed] [Google Scholar]
- 15.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two-dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dainese P, Staudenmann W, Quadroni M, Korostensky C, Gonnet G, Kertesz M, James P. Probing protein function using a combination of gene knockout and proteome analysis by mass spectrometry. Electrophoresis. 1997;18:432–442. doi: 10.1002/elps.1150180318. [DOI] [PubMed] [Google Scholar]
- 17.Roepstorff P. Mass spectrometry in protein studies: From genome to function. Curr Opin Biotechnol. 1997;8:6–13. doi: 10.1016/s0958-1669(97)80151-6. [DOI] [PubMed] [Google Scholar]
- 18.Yates JR., III Mass spectrometry and the age of the proteome. J Mass Spectrom. 1998;33:1–19. doi: 10.1002/(SICI)1096-9888(199801)33:1<1::AID-JMS624>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 19.Hamelmann E, Schwarze J, Takeda K, Oshiba A, Larsen GL, Irvin CG, Gelfand EW. Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography. Am J Respir Crit Care Med. 1997;156:766–775. doi: 10.1164/ajrccm.156.3.9606031. [DOI] [PubMed] [Google Scholar]
- 20.Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, McNulty DE, Carr SA, Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 22.Beavis RC, Chait BT. a-Cyano-4-hydroxycinnamic Acid as a Matrix for Matrix assisted Laser Desorption mass Spectrometry. Org Mass Spectrom. 1992;27:156–158. [Google Scholar]
- 23.Vorm O, Roepstorff P, Mann M. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal Chem. 1994;66:3281–3287. [Google Scholar]
- 24.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 25.Hegesh E, Hegesh J, Kaftory A. Congenital methemoglobinemia with a deficiency of cytochrome b5. N Engl J Med. 1986;314:757–761. doi: 10.1056/NEJM198603203141206. [DOI] [PubMed] [Google Scholar]
- 26.Hultquist DE, Passon PG. Catalysis of methaemoglobin reduction by erythrocyte cytochrome b5 and cytochrome b5 reductase. Nature New Biol. 1971;229:252–254. doi: 10.1038/newbio229252a0. [DOI] [PubMed] [Google Scholar]
- 27.Fish JE, Peters SP. Airway remodeling and persistent airway obstruction in asthma J. Allergy Clin Immunol. 1999;104(3 Pt 1):509–516. doi: 10.1016/s0091-6749(99)70315-5. [DOI] [PubMed] [Google Scholar]
- 28.Adra CN, Ko J, Leonard D, Wirth LJ, Cerione RA, Lim B. Identification of a novel protein with GDP dissociation inhibitor activity for the ras-like proteins CDC42Hs and rac I. Genes Chromosomes Cancer. 1993;8:253–261. doi: 10.1002/gcc.2870080408. [DOI] [PubMed] [Google Scholar]
- 29.Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, Glass CK. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem. 2002;277:42821–42829. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 30.Webb DC, McKenzie AN, Foster PS. Expression of the Ym2 lectin-binding protein is dependent on interleukin (IL)-4 and IL-13 signal transduction: identification of a novel allergy-associated protein. J Biol Chem. 2001;276:41969–41976. doi: 10.1074/jbc.M106223200. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Z, Zheng T, Homer RJ, Kim Y, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]