Abstract
Large-scale comparison of sequence polymorphism and divergence at numerous genomic loci within and between closely related species can reveal signatures of natural selection. Here, we present a population genomics study based on direct sequencing of 61 mitotic cell cycle genes from 30 Arabidopsis thaliana accessions and comparison of the resulting data to the close relative Arabidopsis lyrata. We found that the Arabidopsis core cell cycle (CCC) machinery is not highly constrained but is subject to different modes of selection. We found patterns of purifying selection for the cyclin-dependent kinase (CDK), CDK subunit, retinoblastoma, and WEE1 gene families. Other CCC gene families often showed a mix of one or two constrained genes and relaxed purifying selection on the other genes. We found several large effect mutations in CDKB1;2 that segregate in the species. We found a strong signature of adaptive protein evolution in the Kip-related protein KRP6 and departures from equilibrium at CDKD;1 and CYCA3;3 consistent with the operation of selection in these gene regions. Our data suggest that within Arabidopsis, the genetic robustness of cell cycle–related processes is more due to functional redundancy than high selective constraint.
INTRODUCTION
The molecular components underlying the mitotic cell cycle show a high degree of functional and structural evolutionary conservation across eukaryotes. However, the transcriptional and posttranslational regulation of the cell cycle can be dramatically different among species (Jensen et al., 2006). This regulatory variation reflects a degree of evolutionary flexibility in the cell cycle that can help explain how plants have evolved several specific cell cycle features, such as lifelong cell divisions leading to new organs or the intrinsic cell totipotency found in most plant species (Gutierrez, 2005; Inzé and De Veylder, 2006). In addition, natural phenotypic variation has been recorded in Arabidopsis thaliana for several plant traits related to the cell cycle, such as root growth rate, developmental speed, endoreduplication rate, and shoot size (Beemster et al., 2002; Koornneef et al., 2004; Lisec et al., 2008). However, given the two opposing evolutionary faces of the cell cycle observed between species (conserved in its components yet highly diverged in its regulation), it is hard to predict a priori if genetic variation present in the cell cycle genes could contribute to natural phenotypic variation within species.
A promising way to evaluate the molecular evolution of cell cycle genes in plants is to incorporate population genetics methods to assess the contribution of positive and negative selection in shaping genetic variation and to determine the overall levels of constraint (Maynard Smith and Haigh, 1974; Nei, 1987; Charlesworth et al., 1993). An added benefit of a population genetics approach is that where evidence of selection is found, it implicitly means that mutations in that locus can result in a change in phenotype that affects the fitness of the individual. Hence, a population genetics approach can equally assist in selecting candidate genes from a molecular system for follow-up studies (Shimizu and Purugganan, 2005; Weigel and Nordborg, 2005; Wright and Gaut, 2005).
A number of population genetic test statistics have been developed to identify single nucleotide polymorphism (SNP) patterns that deviate from expected neutral patterns, both for within-species studies (denoted as polymorphisms) and/or between species (denoted as divergence). A number of neutrality tests contrast polymorphism and divergence patterns in nonsynonymous (NS) and synonymous (S) codon sites (e.g., McDonald and Kreitman, 1991). Other tests make use of the increase (under positive selection) or decrease (under negative selection) in frequency of SNPs or patterns of polymorphism in all sites of a given locus to detect deviations from patterns expected under neutrality (Bamshad and Wooding, 2003; Nielsen, 2005; Biswas and Akey, 2006; Sabeti et al., 2006). Nevertheless, a well-recognized problem of these population genetic tests is that non-neutral signatures can also be generated by demographic forces, such as rapid population expansion or population bottlenecks (Tajima, 1989; McDonald and Kreitman, 1991; Eyre-Walker, 2002; Wright and Gaut, 2005). As a consequence, the standard neutral model may not apply for species with complex demographic histories, which can lead to erroneous conclusions about the significance of neutrality test statistics results. However, demographic forces are expected to affect the pattern of polymorphism genome-wide, as opposed to selection, which is expected to act locally on a targeted mutation (Cavalli-Sforza, 1966). One approach to address this issue is to generate an empirical null distribution of test statistic values from a large set of random loci (under the assumption that the vast majority of mutations are essentially neutral; Kimura, 1983). Then, if a test statistic value calculated for a given locus falls in the tails of this null, there is an increased probability that the signature of the locus is a result of selection rather than demography. An alternative approach to differentiate patterns of selection from demography is to model the demographic history of the species and as such estimate the null distribution of the test statistics (Tenaillon et al., 2004; Tenaillon and Tiffin, 2008). However, when the demographic history of a species is complex, a population model that explains all aspects of the data can be elusive. For A. thaliana, the demographic models developed so far can only explain parts of the observed data (Nordborg et al., 2005; Schmid et al., 2005; François et al., 2008). Therefore, the empirical approach has been advocated for population genetic analyses in A. thaliana (Tenaillon and Tiffin, 2008) and has been successfully applied to reveal potential targets of selection (Kelley et al., 2006; Toomajian et al., 2006; Borevitz et al., 2007).
In this article, we present a study of the mitotic cell cycle in Arabidopsis using a population genomics approach. Vandepoele et al. (2002) defined a core set of 61 A. thaliana cell cycle genes containing 12 kinases, 30 cyclins, two CDK subunit (CKS) genes, seven Kip-related proteins (KRPs), eight E2F/DP genes, and the Retinoblastoma (Rb) and WEE1 genes. Progression through the four phases of the mitotic cell cycle (first gap, DNA synthesis and replication; second gap, mitosis or abbreviated G1-S-G2-M) is determined by the activity of cyclin-dependent kinases (CDKs) in complex with cyclins (CYCs). The activity of CDK/CYC complexes is induced by CDK-activating kinases and is inhibited in A. thaliana by regulators such as KRPs, CKS docking proteins, and WEE1. Other cell cycle components are E2F, DP, and Rb that form transcription complexes that activate S-phase genes. The complex interplay of these 61 core cell cycle (CCC) and other genes not only leads a cell through mitosis but also initiates the endocycle (when a cell replicates its DNA content without a subsequent mitotic phase) and induces a cell to differentiate (for reviews, see Inzé and De Veylder, 2006; De Veylder et al., 2007).
Here, we describe patterns of sequence variation in the 61 CCC genes in a collection of 30 A. thaliana accessions, selected to capture a large fraction of the allelic diversity present in A. thaliana. We estimated a series of commonly used population genetics and found evidence of non-neutral processes operating in CCC gene regions using the empirical background distribution approach. For brevity, we refer in this manuscript to the CCC genes as the CCC data set and the empirical background data set published by Nordborg et al. (2005) as the 2010 data set (Clark et al. 2007). Additional tests of polymorphism and divergence were conducted for the CCC genes using a single Arabidopsis lyrata outgroup sequence. The results suggest that only a subset of the CCC genes are subject to strong evolutionary constraints, while others appear to be less constrained or, in some cases, even subject to positive selection.
RESULTS
For all 61 CCC genes we resequenced three or four ∼600-bp long fragments in 30 A. thaliana accessions to identify SNPs. One fragment was located within 1000-bp upstream of the translation start, one or two fragments were intragenic and covered both intronic and exonic sequence, and one fragment was located within 1000 bp downstream of the translation end. Although we have only partially resequenced the CCC genes, our approach allowed us to assess patterns of variation in the CCC genes. In total, the CCC data set consists of 238 gene fragments from 61 genes in a sample of 30 A. thaliana accessions. A complete list of sequences in the CCC data set can be found in Supplemental Data Set 1 online. Of a total of 238 gene fragments resequenced, 99.4% were successful (>300 bp). The average fragment length is 593 bp, and the minimal sample size is 26 accessions. We identified 3329 SNPs in the CCC data set, or one SNP per 42 bp.
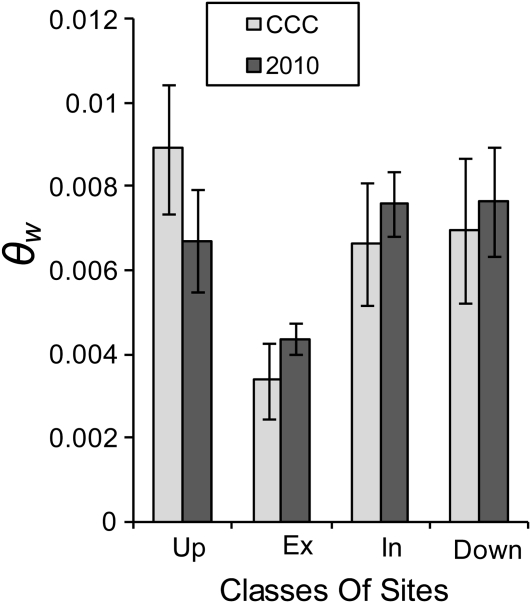
Similar to previous findings (Nordborg et al., 2005), we found a strong negative correlation (R2 = 0.118, P < 0.001; Pearson correlation) between polymorphism per site (θw) in the CCC genes and local gene density (see Supplemental Figure 1 online). No significant differences in exonic (P = 0.075), intronic (P = 0.372), and downstream (P = 0.501) θw values were observed between the CCC and the 2010 data sets (Figure 1; see Supplemental Table 2 online). Comparison of θw in upstream, exonic, intronic, and downstream site classes showed intronic and downstream θw to be similar both within the CCC genes (P = 0.596) as within the genes of the 2010 data set (P = 0.721). As expected, a significantly lower mean θw (P < 0.001) in the exonic than in the intronic CCC regions was found, indicating a greater constraint on coding sites. Unexpectedly, we observed a significantly (P = 0.018) higher θw in the upstream region of the CCC genes compared with the upstream regions in the 2010 data set. The CCC upstream θw value was also significantly higher than the CCC intronic and downstream θw (P = 0.002 and 0.017, respectively). These differences between upstream and intragenic/downstream θw values were absent for the 2010 data set. Mutation frequencies positively correlate with GC content (Hurst and Williams, 2000), so a higher GC% could explain θw differences. However, we found GC% in all four regions to be higher in the 2010 data set than in the CCC data set. Within the CCC data set, GC% in the upstream region (31.6%) was similar to GC% in intronic and downstream regions (32.4 and 31.5%, respectively). This is an unusually high level of nucleotide polymorphism in upstream gene regions. The difference may be explained by an unequal presence of functional promoter motifs in the upstream fragments of both data sets. It is also possible that what we observe is an excess of slightly deleterious mutations in the CCC upstream regions as a consequence of reduced selective constraint (Bustamante et al., 2002; Foxe et al., 2008).
Figure 1.
Histogram of the Average θw Values in the Upstream, Exonic, Intronic, and Downstream Sites Analyzed in the CCC Data Set and the 2010 Data Set.
Bars represent the 95% confidence interval. See Supplemental Table 2 online.
Patterns of SNP Diversity in CCC Coding Sequence
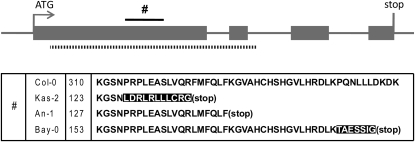
When mutations occur in the coding sequence of a gene, this can lead to amino acid or gene integrity changes, such as altered splice sites or premature stop codons. Such mutations can result in important alterations of protein function and may therefore be subject to natural selection. For CYCD7;1, CYCA3;3, and CDKB1;2, we found polymorphisms in the coding sequence that introduced premature stop codons in some accessions relative to the Col-0 reference (see Supplemental Table 3 online). The stop codons introduced in CYCD7;1 and CYCA3;3 were located at the C-terminal end of the amino acid sequence. In CDKB1;2, we identified three haplotypes in which a single base deletion resulted in early stop codons that truncated >50% of the protein sequence (see Supplemental Table 3 online). This is surprising because plant CDK sequences consist largely of functional domains (Joubès et al., 2000). In addition, we found that the A. lyrata ortholog of CDKB1;2 lacked most of the first two exons of the A. thaliana copy, equal to 70% of this protein (Figure 2), which we reconfirmed by PCR amplification and sequencing. However, we did not observe large-effect SNPs in the paralogous CDKB1;1 sequence. These observations suggest that CDKB1;2 is not essential for the fitness of the plant and may be on its way out of the Arabidopsis gene pool. Because of its disrupted integrity, CDKB1;2 was excluded from further coding sequence diversity analysis.
Figure 2.
Schematic Representation of CDKB1;2 in Arabidopsis and the Large-Effect Changes Observed.
Blocks in the figure represent exons annotated in A. thaliana. The dashed line delimits the gene region that was deleted in the A. lyrata orthologous CDKB1;2 gene. The solid line (#) delimits the amino acid region where early stop codons were observed in A. thaliana. Changes in the amino acid sequence within A. thaliana are summarized. The numbers are the position of the stop codon in the amino acid sequence. Each amino acid sequence represents a haplotype, symbolized by one accession. The Columbia-0 sequence is the reference. Changes in amino acid sequence due to a frame shift are marked in black.
Patterns of molecular evolution in CCC genes can be assessed with comparisons of the ratio of NS changes per NS site to the ratio of S changes per S site. We abbreviate these ratios as pN/pS for polymorphism and Ka/Ks for divergence. Because amino acid polymorphisms in the coding regions were not abundant enough to evaluate the pN/pS ratio at individual genes, the mean pN/pS for groups of CCC genes was used to evaluate the level of purifying selection within A. thaliana (Table 1). Overall, we found an average pN/pS of 0.304 in the 61 CCC genes, which was significantly higher (P < 0.05) than the genome-wide average in the 2010 data set (pN/pS = 0.252). The CDK family had a pN/pS value of 0.119, which is less than half of any other CCC gene family (P < 0.05). The high pN/pS value for cyclins is partly due to two outliers; CYCA3;3 has 24 NS to one S and CYCB1;3 has eight NS and no synonymous polymorphisms. Excluding these outliers lowered pN/pS for cyclins to 0.308, which is still high compared with the genome-wide average of 0.252 (P < 0.05).
Table 1.
Estimates of the Mean pN/pS and Ka/Ks Ratios
| CCC Group | Mean pN/pS | Mean Ka/Ks |
|---|---|---|
| CCC Gene Family | ||
| CDKs | 0.119a | 0.073a |
| Cyclins | 0.357 (0.308b) | 0.241 |
| E2f/DP | 0.324 | 0.239 |
| Interactors | 0.294 | 0.300 |
| CCC PAM cluster | ||
| Cluster 1 | 0.132 | 0.065 |
| Cluster 2 | 0.239 | 0.156 |
| Cluster 3 | 0.327 | 0.223 |
| Cluster 4 | 0.410 | 0.290 |
| Cluster 5 | 1.230 | 0.413 |
| Cluster 6 |
0.270 |
0.597 |
Without CDKB1;2.
Without CYCA3;3 and CYCB1;3.
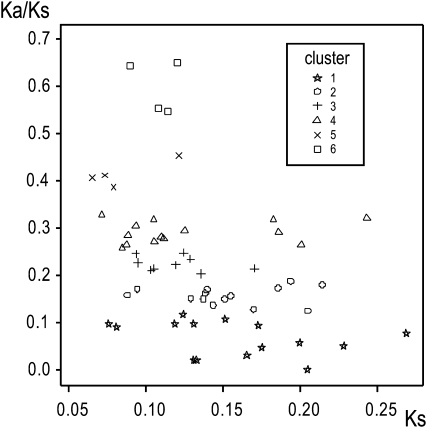
The Ka/Ks ratio of each CCC gene (see Supplemental Table 4 online) was estimated on the full reference coding sequence and clustered with the partitioning around medoids (PAM) algorithm (Kaufman and Rousseeuw, 1990). Clustering of Ka/Ks was done to illustrate different degrees of constraint across the cell cycle machinery. Wilks' Lambda test statistic suggested six clusters as optimal partitioning (Figure 3). We noticed that the cluster with the lowest mean Ka/Ks (cluster 1) contained all but one CDK (CDKF;1; cluster 2). Interestingly, the tandem duplicated genes CKS1 and CKS2 were also contained in cluster 1. The reported differential expression pattern of the CKS genes (Menges et al., 2005) and the high selective constraint suggests that neo- or subfunctionalization happened after the duplication event, which was prior to the divergence of the Arabidopsis lineage. Also, cluster 2, which contained the single-copy genes Rb and WEE1, showed high levels of selective constraint. Because the rate of evolution has been found to negatively correlate with gene expression (Wright et al., 2004; Foxe et al., 2008), we examined the effects of CCC expression levels (Menges et al., 2005) on the evolutionary rate by analysis of variance. We found that cluster 1 shows significant (P < 0.05) higher gene expression levels across all reported plant tissues compared with the other clusters. For all gene families other than the CDKs and CKSs, we saw different rates of evolution across the genes within the family. Some clear examples were the DEL and KRP families. The Ka/Ks ratios of the three DEL family genes were 0.123, 0.268, and 0.643. KRP2 and KRP3 were found in cluster 2, while KRP1, KRP5, KRP6, and KRP7 were all found in cluster 5 or 6. The variation in evolutionary rate might indicate which members of the family play a more evolutionary conserved role in the overall cell cycle machinery and which genes are more likely to be subject to adaptive change.
Figure 3.
Scatterplot of the Ka/Ks versus Ks Values per Gene.
Ka/Ks values are PAM clustered into six clusters and made visually distinguishable. Dotted lines were added to clarify the six clusters boundaries. Each marker represents one CCC gene. The gene content of each cluster is summarized in Supplemental Table 4 online.
We compared Ka/Ks and pN/pS values calculated for the six PAM clusters and for the four major functional groups of the CCC genes (Table 1). PAM cluster 1 and the CDK family had both the lowest pN/pS and Ka/Ks value. This suggests that purifying selection against NS polymorphisms within species mirrored the observed slow rate of evolution in between-species comparisons for these functional groups. Less concordance between pN/pS and Ka/Ks values was found in the other groups.
The MK test is based on the neutral expectation that the ratio of NS to S mutations will be equivalent for intraspecific polymorphism and between-species divergence (McDonald and Kreitman, 1991). The direction and average strength of selection on AA changes was estimated by the MKPRF selection parameter γ. Selection against NS replacements results in γ < 0, selection in favor of NS replacement in γ > 0. The MK test after Bonferroni correction was significant in CYCA3;3 and CYCB1;3 (P < 0.05) in the direction of an excess of NS polymorphisms. This excess of replacement polymorphism is consistent with selective processes, including a recent relaxation of functional constraints, segregation of slightly deleterious mutations, local adaptation, or some form of balancing selection. The results for MKPRF similarly suggest that purifying selection may be a primary form of selection on amino acid changes in this set of genes. Estimates of the selection parameter (γ) were significantly negative for CYCB2;1, CYCA3;1, CYCD7;1, CYCA3;3, and CDKD;1 but not for CYCB1;3 (see Supplemental Table 4 online). Many similar observations have been made for a variety of genes in Arabidopsis and have consistently been interpreted as evidence for an excess of slightly deleterious variation segregating in the Arabidopsis genome (Bustamante et al., 2002; Foxe et al., 2008). However, for CYCA3;3, CDKD;1, and CYCB2;1, additional signatures for departures from neutrality were observed in the genomic region of these genes (see below).
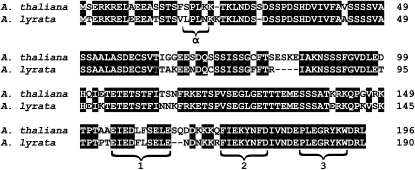
For one gene, KRP6, the MK test was significant (P < 0.05) due to an excess of replacement divergence relative to polymorphism, although this was not significant after Bonferroni correction. KRP6 also had the highest estimated selection parameter (γ) of all CCC genes, although again not significant. Because MK and MKPRF tested for selection from the resequenced KRP6 fragments only (for KRP6 this equals 80% of the total reference coding sequence), we estimated Ka/Ks again on the resequenced part of KRP6. For this section of the gene, we found Ka/Ks to be 1.10, which also is consistent with adaptive substitution. We estimated the 95% Ka/Ks confidence interval for KRP6 from 0.3955 to 3.8685 (Comeron 1995), implying that we cannot strictly reject that Ka/Ks equals one. However, the only reported genes with similarly high Ka/Ks ratios are a very few cases of R genes (Xiao et al., 2004; Bakker et al., 2006). Closer inspection of the KRP6 residues that have fixed since divergence with A. lyrata indicated that two replacement fixations were located in a conserved C-terminal CDK and cyclin binding motif region (Figure 4). Also, the N-terminal CDK phosphorylation site, which has been suggested to determine the stability and temporal degradation of the protein (De Veylder et al., 2001), showed two replacement fixations. These results are consistent with a scenario of adaptive evolution of KRP6 since the divergence of the two Arabidopsis species.
Figure 4.
Protein Sequence Alignment of KRP6.
The position of the CDK consensus phosphorylation site is indicated by α, and the positions of the conserved amino acid motifs in plant KRPs are indicated by 1, 2, and 3.
Patterns of SNP Diversity across the CCC Sequence Loci
Positive selection on a locus can result in patterns of polymorphism and divergence that depart from neutral patterns driven by mutation and genetic drift. Frequency spectrum-based statistics provide one mean of quantifying these patterns and can detect departures from equilibrium generated by two major types of positive selection. A selective sweep (when a beneficial allele becomes fixed in a population) is hallmarked by an excess of rare SNPs within a region of overall low nucleotide diversity. Balancing selection (when more than one allele of the gene is maintained in a population) is characterized by an excess of intermediate frequency SNPs in a region of high nucleotide diversity (Hudson and Kaplan, 1988; Bamshad and Wooding, 2003; Nielsen, 2005; Biswas and Akey, 2006; Walsh, 2008).
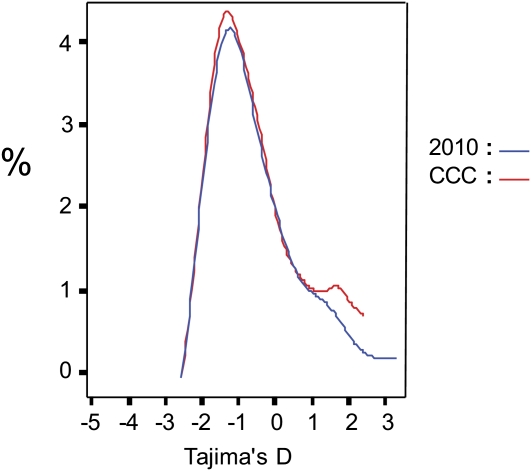
Departures from neutrality at the CCC loci can be tested with the frequency-spectrum based statistics Tajima's D (Tajima, 1989) and Fu and Li's D* and F* (Fu and Li, 1993) or with comparable tests Fu and Li's D and F (Fu and Li, 1993) and Fay and Wu's H (Fay and Wu, 2000) that require an outgroup (see Supplemental Data Set 2 online). In this article, we will only discuss Tajima's D statistics and refer the reader to Supplemental Data Set 2 online for the Fu and Li statistics. Significant positive values for Tajima's D statistics are consistent with the pattern of a locus under balancing selection. Significantly negative values suggest that the locus has been subjected to a selective sweep. A significantly positive Fay and Wu's H indicates a deficit of moderate- and high-frequency derived SNPs relative to equilibrium expectations, whereas a significant negative Fay and Wu's H indicates an excess of high-frequency derived SNPs. We assessed potential significance of Tajima's D using the empirical approach and weighed Tajima's D values against the genome-wide distribution of Tajima's D for the selected 30 accessions, as inferred from the 2010 data. A potential problem for the empirical approach is that the fragments for each CCC gene in our data sets are linked within a 5-kb window. The distribution of Tajima's D calculated on the CCC data set was similar to the distribution of the 2010 data set (two-sample smooth test: P = 0.990; Figure 5). We observed an unexpected small increase in positive values for Tajima's D (Figure 5), but this was not significant (skewness component of two sample smooth test: P = 0.626, F-test on distribution variance: P = 0.76). We did not have genome-wide outgroup sequence data at hand to calculate the genome-wide distribution of Fay and Wu's H. To avoid confusion, we report significance for Tajima's D values that exceed the negative or positive 2.5% thresholds of the 2010 genome wide distribution but report percentiles of the CCC distribution for the Fay and Wu's H values.
Figure 5.
Estimated Genome-Wide Distribution Plots for Tajima's D Neutrality Test Statistics.
Red lines represent the test statistic values calculated for 234 CCC gene fragments and blue lines the empirical genome-wide distribution calculated based on 973 fragments from the 2010 data set. The y axis indicates the frequency of the test statistic value in the data set.
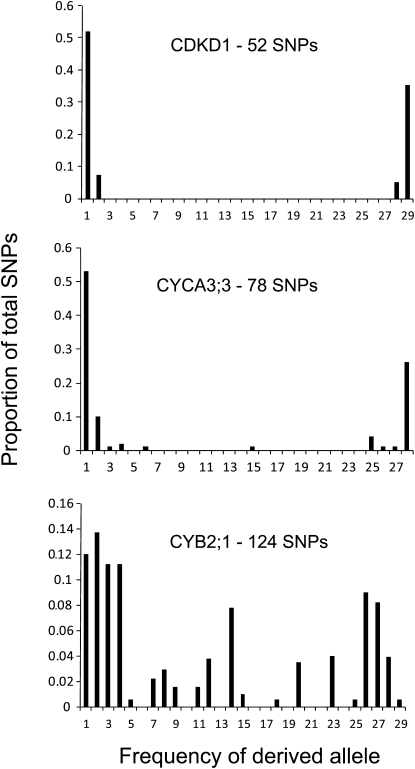
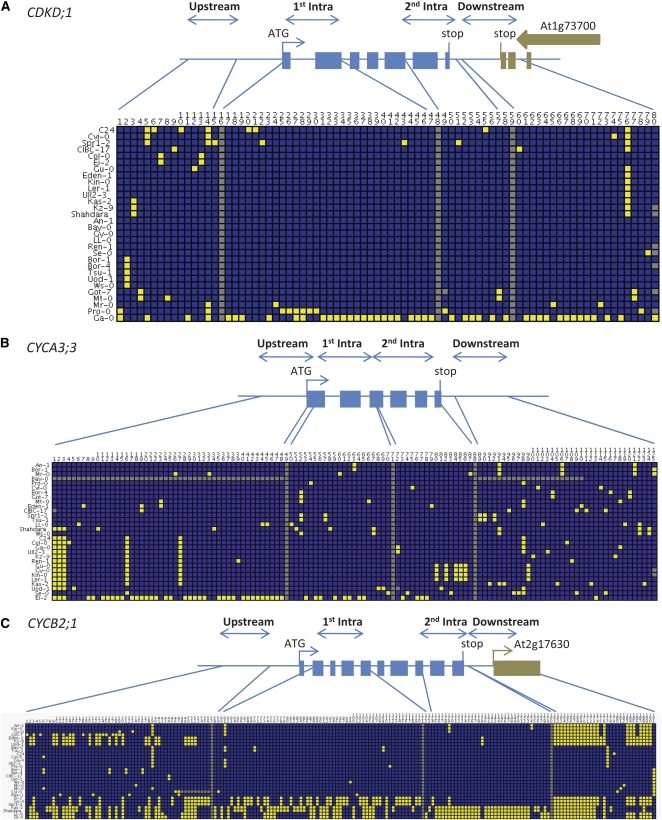
Many CCC genes showed outlier test statistic values, yet we observed high variation in nucleotide diversity levels and frequency patterns between the resequenced fragments even within the same CCC gene. For example, only 32 CCC genes have similar Tajima's D values (coefficient of variation range from −1 to 1) across all the resequenced fragments in the particular gene region. Such high variation has been reported before in A. thaliana (Tian et al., 2002; Shepard and Purugganan, 2003; Ding et al., 2007; Moore and Stevens, 2008), necessitating a conservative approach when interpreting population genetics statistics. The unusual patterns of variation in CDKD;1, CYCA3;3, and CYCB2;1 seemed most suggestive of a departure from neutrality due to selection on these loci or closely linked loci. CDKD;1 showed high negative Tajima's D values in all four fragments and was significant in two. Fay and Wu's H was also negative in all four fragments in the region, with the most extreme value of −19.5 in the first intragenic fragment (98th percentile). CYCA3;3 showed a very similar pattern with negative test statistics throughout the region. Examination of the derived site frequency spectrum showed a U-shaped pattern with SNPs limited to the lowest and highest frequency classes for both genes (Figure 6). The high frequency–derived polymorphism patterns were driven by the presence of a single haplotype that harbored mostly ancestral variation (represented by Ga-0 for CDKD;1 [Figure 7A] and Ei-2 for CYCA3;3 [Figure 7B]).
Figure 6.
Derived Site Frequency Spectra for CDKD;1, CYCA3;3, and CYCB2;1.
The x axis represents the count frequency of the derived SNP allele within 30 accessions. The y axis represents the frequency of SNP sites that have the given derived SNP frequency of the x axis. Bay-0 was removed from the CYCA3;3 analysis because of missing data.
Figure 7.
Visual Representation of Haplotypes of the CDKD;1, CYCA3;3, and CYCB2;1 Loci.
Each row represents an accession and each column a polymorphic site. For each polymorphic site, minor alleles are in yellow, major alleles in blue, and missing data in gray. Accessions are clustered by haplotype. Gray columns separate the resequenced fragments on the locus. On the schematic gene representation, blocks represent exons.
CYCB2;1 showed a very complex pattern of variation, with significant positive Tajima's D values in the upstream region and a highly negative Fay and Wu's H value (99th percentile) for the first intragenic fragment. This signature suggests balancing selection; however, the frequencies of the different haplotype blocks varied between the resequenced regions, most probably as a result of intragenic recombination (Figure 7C). We also observed many derived polymorphisms both at high and at intermediate frequencies (Figure 6). Despite being an interesting pattern (especially given the significantly negative selection parameter γ for CYCB2;1; see Supplemental Table 4 online), which has been observed and linked to balancing selection before (Innan et al., 1996; McDowell et al., 1998), it is hard to draw any clear conclusions on the history of selection for this locus.
DISCUSSION
In summary, we have found that the patterns of DNA sequence variation in the 61 CCC genes studied here suggest that within Arabidopsis the molecular cell cycle machinery is not a rigorously conserved system but that it is subject to different types of selection. When comparing divergence between A. thaliana and A. lyrata, all CCC genes showed Ka/Ks < 1, yet there is a high degree of variation in the rate of protein evolution across this system of genes. As expected, we found that many CCC genes showed evidence of strong selection against amino acid divergence (e.g., across the CDK family). Highly constrained genes are believed to occupy the vital positions in the molecular network, as has been demonstrated for some CCC genes: loss-of-function mutations in Rb or CDKA;1 lead to gametophytic lethality (Ebel et al., 2004; Iwakawa et al., 2006; Nowack et al., 2006), and overexpressing WEE1 arrests the cell cycle progression (De Schutter et al., 2007). However, we also found high rates of amino acid substitution, suggesting fundamental differences in levels of selective constraint across different CCC gene families. Most unexpected was the observation that the mean pN/pS ratio in the CCC genes is higher than in genes selected randomly, again suggesting lower selective constraints on many cell cycle genes despite their posited functional importance.
This variation in selective constraint of the CCC genes has several potential origins. One explanation is that gene expression negatively correlates with the rate of protein evolution in Arabidopsis (Wright et al., 2004; Foxe et al., 2008), and differences in expression level could therefore account for this pattern. Consistent with this hypothesis, many of the CCC genes where Ka/Ks was low have the highest expression among CCC genes across different plant tissues (Menges et al., 2005). Less-constrained CCC genes have low expression or only high temporal expression levels in specific tissues (Menges et al., 2005). Another possible contributing factor to the observed differences in selective constraints might be related to differences in the ratio of functional domains relative to the size of the protein. Kinases have more functional domains (Joubès et al., 2000) relative to their size than KRPs (De Veylder et al., 2001) or cyclins (Wang et al., 2004). Third, differences in the evolutionary rates may also reflect a difference in the degree of functional redundancy within the different families. Functional redundancy is usually, but not exclusively, a consequence of gene duplication (Pickett and Meeks-Wagner, 1995). If two genes are redundant, this can lead to a decrease in selective constraint on one or both genes (Ohno, 1970). However, selective constraint to maintain this redundancy has also been reported (Moore and Purugganan, 2005). The core cell cycle machinery contains many duplicated genes (Vandepoele et al., 2002), and functional redundancy has been reported for CYCA2-type genes (Imai et al., 2006) and knockout studies of the CDKB1 type (Boudolf et al., 2004). In our data set, we see examples of increased, as well as decreased, rates of evolution for duplicated and functionally redundant genes. The tandem duplicated CKS genes showed Ka/Ks values indicative for strong constraint, while for the CDKB1;2 gene, integrity changes lead to molecular nonfunctionalization in both A. thaliana and A. lyrata. This apparent nonfunctionalization of CDKB1;2 in both Arabidopsis species could be a consequence of functional redundancy with CDKB1;1 (Boudolf et al., 2004). However, CDKB1;2 seems to be the only CCC gene where nonfunctionalization is as clear despite the high level of duplication and redundancy in the rest of the cell cycle.
The model that emerges from these observations suggests that the robustness of plant development and cell cycle progression is secured by functional redundancy rather than by selective constraint against amino acid substitution. Under this scenario, changes in the timing and mode of expression are more likely to have an impact on plant physiology and therefore drive functional diversification in the cell cycle (Blanc and Wolfe, 2004; Menges et al., 2005; Jensen et al., 2006). Interesting in that regard, yet speculative, is the observed significantly increased θw in the upstream fragments of the CCC genes relative to the genome-wide upstream θw. This may be the result of differences in the density of functional promoter sites between both data sets. Alternatively, variation in purifying selection between the CCC genes and the 2010 data sets could explain our observation. The decrease in selective constraint on the CCC promoters would allow an increase in CCC regulatory variation across A. thaliana populations and drive the evolution of the cell cycle, as demonstrated by Jensen et al. (2006).
In addition to the observation that many CCC genes are not subject to strong selective constraints, we compiled evidence to suggest that some CCC genes harbor molecular patterns consistent with the operation of positive selection. KRP6 showed a Ka/Ks ratio of 1.1 and had fixed mutations in known functional domains. Furthermore, KRP6 showed a significant MK test (P < 0.05) due to an excess in replacement divergence and had the highest positive MKPRF selection parameter (γ) of all CCC genes. This excludes the possibility of relaxed purifying selection for this gene in A. thaliana. The high Ka/Ks value together with the MK test is consistent with historical positive selection operating on this gene. It is therefore interesting that KRP6 is the only KRP to have transcriptional activation at the S-to-G2 transition in the cell cycle (Menges et al., 2005) and so far the only KRP identified to be degraded at the protein level at the spindle assembly checkpoint during mitosis (G. De Jaeger, personal communication).
Two other CCC genes, CDKD;1 and CYCA3;3, showed negative values for the frequency spectrum-based statistics and a U-shaped derived site frequency spectrum. Demographic scenarios are capable of generating a U-shaped frequency distribution (Caicedo et al., 2007). However, we find demography alone to be an unlikely explanation because for Ga-0 and Ei-2, we observed only one CCC locus with an ancestral haplotype outlier in these accessions. Under a demographic scenario, we would see haplotype outliers in Ga-0 and Ei-0 over multiple loci across the genome, as observed in the geographical isolates Cvi-0 and Mr-0 (Nordborg et al., 2005; François et al., 2008). However, if high variability of unusual coalescents in the genome of this predominantly selfing species exists, almost any pattern, including the pattern we observed, could be generated at any locus in the genome, and a demographic scenario cannot be excluded. Both genes also had a significantly negative selection parameter γ in MKPRF because of an excess of amino acid polymorphism. A negative γ is consistent with purifying selection on slightly deleterious mutations, but for CDKD;1, we found five out of seven amino acid mutations in Ga-0 and for CYCA3;3 13 out of 24 amino acid mutations in Ei-2. Possible selective explanations for the negative γ and frequency statistic values are balancing selection (with the selective signature being masked by the sample strategy), local adaptation, haplotype introgression, or that the genes are located in a region that underwent a partial selective sweep.
A few interesting biological features for CDKD;1 and CYCA3;3 provide a working hypothesis on how natural variation in these genes would affect fitness. Increased expression of the Oryza sativa CDKD;1 accelerated S-phase progression and cell growth rates (Fabian-Marwedel et al., 2002). From this result it is thought that cyclin activating kinases, like CDKD;1, can play an important role in determining the growth rate and differentiation status of cells by controlling CDK activity. However, knockout studies of CDKD;1 in A. thaliana showed no change in phenotype under normal growth conditions (Shimotohno et al., 2006), suggesting functional redundancy with other CDKs. CYCA3;3 is the only A-type cyclin without a Destruction-box protein domain (needed for rapid degradation of cyclins to terminate the activity of the CDK/CYC complex; Wang et al., 2004). The gene also shows a constant expression profile during the cell cycle, which is atypical for A3-type cyclins (Menges et al., 2005), but higher expression specifically in anthers (Wang et al., 2004). This could suggest that neofunctionalization of this cyclin after duplication shifted it out of the mitotic cell cycle into the pathway for floral organ differentiation.
The plant cell cycle system coordinates many essential cell fate decisions, such as cell differentiation, cell proliferation, mitotic arrest under stress, and endoreduplication. It was therefore somewhat surprising that the cell cycle machinery, where conservation was expected, displayed a variety in signatures of natural selection. This evolutionary flexibility in the genes driving the cell cycle might contribute to a plant's ability to adapt to environmental changes. The population genomic approach demonstrated in this article contributes to a better understanding of how the cell cycle evolves and has the additional value to assist in nominating candidate genes for extended genetic studies. The CCC genes that display interesting allelic variation, in particular in combination with signals of positive selection, are prime candidate genes for follow-up studies, such as linkage and association studies, ascertaining the effect of cloned CCC alleles across different accessions or cloning A. lyrata paralogous genes in an A. thaliana background.
METHODS
Selected Accessions
The sample of 30 Arabidopsis thaliana accessions was selected from the collection of 96 accessions described by Nordborg et al. (2005) (Nottingham Arabidopsis Stock Centre stock number N22660). A set of 11 accessions was chosen based on their use in generating mapping populations (An-1, C24, Col-0, Cvi-0, Gu-0, Gy-0, Kas-2, Ler-1, Shahdara, Tsu-1, and Ws-0). This initial core of accessions was further enriched for allelic variation through the M strategy of Schoen and Brown (1993) as implemented in the software algorithm Mstrat v1.0 (Gouesnard et al., 2001). The M strategy selects a set of 30 accessions that captures almost all of the allelic diversity present in the set of 96 accessions (McKhann et al., 2004). We ran Mstrat on 25 polymorphic fragments from the 2010 data set, evenly dispersed over the genome and in linkage equilibrium. Using 100 iterations per Mstrat run (with Nei's diversity index [Nei, 1987] as second criterion of maximization and the 11 predefined accessions were set as kernel core), we constructed 300 independent core sets of 30 accessions and retained the 30 most selected ones. The final core collection (see Supplemental Table 1 online) successfully picked up accessions from every geographic subpopulation identified by Nordborg et al. (2005).
DNA Samples and Sequencing
The genes studied were the 61 core cell cycle genes of A. thaliana as defined by Vandepoele et al. (2002). For A. thaliana, we resequenced four ∼600-bp fragments per CCC gene positioned as follows: one fragment in the 1000-bp upstream sequence from the translation start, one intragenic fragment covering both exonic and intronic DNA close to the translation start, one intragenic fragment located near the 3′ end of the gene, and one fragment in the 1000-bp sequence downstream of the translation end. For the 2010 data set, we used the same 1000-bp window criterion to classify sites as up- or downstream. For the CCC data set, we used the most recent gene models and 1000-bp up- and downstream sequences from the Arabidopsis Genome Initiative reference sequence available on The Arabidopsis Information Resource at the moment of analysis (TAIR6; http://www.Arabidopsis.org). The choice to resequence well-positioned fragments instead of the whole gene region was to keep resequencing costs and efforts at a minimum, while still generating a complete overview of the nucleotide diversity in each cell cycle gene and its direct surroundings. Only one intragenic fragment was designed in CKS1, CKS2, KRP5, and KRP7 because of their small gene size. All primers were designed with the PRIMER3 software (Rozen and Skaletsky, 2000). Primers were chosen without regard to predicted functional regions. Genomic DNA was extracted from the shoot tissues with the CTAB method (Doyle and Doyle, 1990). Amplification was done using PCR protocols for direct sequencing with SilverStar DNA polymerase from Eurogentec. DNA fragments were purified with ExoSAP-IT (USB) and sequenced directly in forward orientation using cycle sequencing with the BigDye Terminator v3.1 kit (Applied Biosystems) on an ABI-3000 automated sequencer. The resequencing of the CDKF;1 upstream region recurrently failed for unclear reasons. We found the upstream region of the unrelated gene At4g09390 to be very similar to the CDKF;1 upstream region and suggest the problem to be coamplification with this region. From the downstream sequence of KRP2, only 52 bp was resequenced because of poly-T regions in the fragment. A complete list of sequences in the CCC data set and the primers used can be found in Supplemental Data Set 1 online. An 8x coverage genome assembly of Arabidopsis lyrata subsp lyrata was provided to us by the Department of Energy Joint Genome Initiative A. lyrata genome sequencing project. The A. lyrata sequence trace archives are accessible at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Orthologous A. lyrata CCC genes were localized with BLAST software (Altschul et al., 1990) and locally aligned with A. thaliana with the Smith-Waterman algorithm (Smith and Waterman, 1981).
SNP Detection
Base calling, sequence assembly, sequence alignment, and polymorphism identification were performed with the Phred/Phrap/Consed package (Ewing et al., 1998; Gordon et al., 1998). All sequences and polymorphisms were validated by visual inspection of the chromatograms and edited where appropriate. In case of low-quality sequences (Phred quality score <20) or ambiguous SNP identification, the whole fragment was again resequenced in both forward and reverse orientations. Heterozygous SNPs and sequences whose quality remained low despite multiple attempts were removed from further analysis. Processing and archiving in multi-fasta files was done with the MEGA3 version 3.1 software package (Kumar et al., 2004). Haplotype visualization was done with the online VH1 software package (http://pga.gs.washington.edu/VH1.html).
Population Genetic Analysis
All analyses within or between the CCC and 2010 data sets were done on the 30 selected A. thaliana accessions only. Sites with insertions, deletions, or with more than two SNP alleles were discarded from all population genetic analyses because of the complex mutational patterns. All estimates of local nucleotide diversity θw are based on the number SNPs per site (Watterson, 1975) and were calculated with Nei's θw estimation (normalized θw for analyzed sites per locus; Nei, 1987; equation 10.3, n = 30). θw calculations in up- and downstream fragments were done on noncoding sites only. Local gene densities surrounding the CCC genes were retrieved from Nordborg et al. (2005), and correlations with θw were calculated with Pearsons correlation. θw of a fragment or of a class of sites (e.g., synonymous) was only calculated when the number of resequenced sites was at least 100 bp. For calculations of θw, θN, and θS in exons, a minimum of 240-bp exonic sites per fragment was applied to retain sufficient amounts of synonymous sites per fragment. Assessment of the significance of differences in mean θw values between classes of sites within or between the CCC and 2010 data sets was done with one-way analysis of variance on log-normalized θw values.
Estimation of exonic S and NS sites, pN/pS and Ka/Ks values, and calculating MK tests were conducted with polydNdS, Gestimator, and MKtest in the libsequence software package (Thornton, 2003). pN/pS comparisons were tested for significance using χ2 on the pooled NS and S counts per tested proportion. Ka/Ks was estimated using the method of Comeron (1995) and tested for significance using the K-estimator software package (Comeron, 1999). Ka/Ks estimates are based on the full coding sequence in the Columbia-0 and A. lyrata comparison. Ka/Ks was clustered using the PAM algorithm (Kaufman and Rousseeuw, 1990) using the R environment for statistical computing (http://www.r-project.org/). PAM searches for k representative objects or medoids for the values in the data set. Then, k clusters are constructed by assigning each observation to the nearest medoid. Optimization of k was done with Wilks' Lambda test. MK tests and MKPRF analysis were done on coding sites from the resequenced CCC fragments only. Significance of the MK test was Bonferroni corrected for multiple testing. The population selection parameter γ and its 95% credibility interval were estimated using Markov chain Monte Carlo algorithms and the default settings of the priors on the MKPRF web server (http://cbsuapps.tx.cornell.edu/mkprf.aspx), which is partially funded by Microsoft. The MKPRF algorithm scales the intensity of γ for every individual gene to the selection intensity distribution observed for all genes using a Bayesian implementation of Poisson random field (Sawyer and Hartl, 1992; Bustamante et al. ,2002), and estimates of γ represent the mean of the posterior distribution. Because MKPRF tests NS/S counts against the expected model derived from all genes tested, it is considered more powerful to detect selection than MK (Bustamante et al., 2002) but has shown to have a linear dependency between the number of detected genes with γ > 0 and the priors used for the model (Li et al., 2008).
Neutrality test statistics Tajima's D (Tajima, 1989), Fu and Li's D/D* and F/F* (Fu and Li, 1993), and Fay and Wu's H (Fay and Wu, 2000) were calculated using the software VariScan v2.0 (Hutter et al., 2006). The neutrality test statistic formulas in Variscan use Nei's θw estimator and a modification of the pairwise nucleotide diversity statistic θπ (Tajima, 1989), πm, which can handle missing data at sites without the need of discarding them (Hutter et al., 2006). All test statistics calculations were done with the Variscan parameters set to only analyze sites with <10% missing data (referred to as informative sites). In both the CCC and the 2010 data set, neutrality test statistics were calculated only for fragments with at least 300 analyzed nucleotide sites.
The distribution of polymorphism neutrality test statistic values for the 238 analyzed CCC fragments was compared with the genome-wide distribution of the same statistics estimated from 990 fragments from the 2010 data set. The graphs of the distributions are normal Kernel smoothing plots for the two-point average interval frequency scatter. We used the two-sample smooth test of Janic-Wróblewska and Ledwina (2000) to test the similarity between the two distributions. This statistic models a distribution in k dimensions and has been shown to be a powerful test to compare distributions (Ducharme and Ledwina, 2003). Moreover, if the test is significant, the k different components in the decomposition of the statistic give additional information about how the distributions differ. We analyzed the smooth plots to the order k = 4. The first component, which is exactly the Mann-Witney statistic, is related to a difference in location, the second component to difference in scale, the third to difference in skewness, and the fourth to difference in kurtosis.
For Tajima's D and Fu and Li's D and F, we identified outliers in the empirical distributions of the statistics. Under the assumption of neutrality for the vast bulk of the genome-wide fragments, the ranked distribution of neutrality test statistic values calculated for the data set of 2010 fragments can be considered as the null. The test statistic values at the negative and positive 2.5 percentiles of the genome-wide distribution are set as thresholds. Neutrality test statistics values for the 238 analyzed CCC fragments were considered significant if they exceeded the threshold value, referred to as a rejection of neutrality at the 5% significance level. This nonparametric empirical approach is not a formal test of neutrality, and values in the 2.5% tails are interpreted as outliers in the empirical distribution, where we must accept a reasonable chance for false positives (Kelley et al., 2006). However, for A. thaliana, it is considered as the preferred approach to follow (Schmid et al., 2005; Tenaillon and Tiffin, 2008) and has been used in a number of studies (Carlson et al., 2005; Toomajian et al., 2006; Borevitz et al., 2007).
Accession Numbers
A complete list of accession numbers for the sequences in the CCC data set can be found in Supplemental Data Set 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Correlation between θw and Gene Density.
Supplemental Figure 2. Visual Representation of CYCB1;4, CDKD;3, CYCB1;2, and E2Fa Haplotypes.
Supplemental Table 1. List of Resequenced A. thaliana Accessions.
Supplemental Table 2. Summary of Nucleotide Diversity.
Supplemental Table 3. Data for the CCC Frame Shifts.
Supplemental Table 4. Summary of Codon Site Statistics for the CCC Genes.
Supplemental Data Set 1. Summary of Sequence Data and Primer Sequences.
Supplemental Data Set 2. Summary of SNP Frequency Pattern Statistics for the CCC Genes.
Supplementary Material
Acknowledgments
We thank Emelie Bovyn, Caroline Buysschaert, Wilson Ardiles Diaz, Jan Gielen, Debbie Rombaut, and Raimundo Villarroel for excellent technical assistance in sequencing and data processing, Lieven Sterck for bioinformatics assistance, Heidi Wouters for statistical assistance, Martine De Cock for help in preparing the manuscript, and Juliette de Meaux (Max-Planck-Institut für Züchtungsforschung) for critical reading of the manuscript and advice. We thank the Department of Energy Joint Genome Initiative A. lyrata genome sequencing project for sharing selected sequences prior to publication. Part of this work was supported by a grant from the MORPH research coordination network for R.S. and conducted in the lab of Michael Purugganan of the New York University Department of Biology, who we thank for financial support and stimulating discussions. R.S. and R.K. are indebted to the Institute for the Promotion of Innovation by Science and Technology in Flanders for a predoctoral fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Marnik Vuylsteke (marnik.vuylsteke@psb.vib-ugent.be).
Online version contains Web-only data.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Bakker, E.G., Toomajian, C., Kreitman, M., and Bergelson, J. (2006). A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad, M., and Wooding, S.P. (2003). Signatures of natural selection in the human genome. Nat. Rev. Genet. 4 99–111. [DOI] [PubMed] [Google Scholar]
- Beemster, G.T.S., De Vusser, K., De Tavernier, E., De Bock, K., and Inze, D. (2002). Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin dependent kinase activity. Plant Physiol. 129 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S., and Akey, J.M. (2006). Genomic insights into positive selection. Trends Genet. 22 437–446. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz, J.O., et al. (2007). Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104 12057–12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf, V., Vlieghe, K., Beemster, G.T.S., Magyar, Z., Torres Acosta, J.A., Maes, S., Van Der Schueren, E., Inzé, D., and De Veylder, L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C.D., Nielsen, R., Sawyer, S.A., Olsen, K.M., Purugganan, M.D., and Hartl, D.L. (2002). The cost of inbreeding in Arabidopsis. Nature 416 531–534. [DOI] [PubMed] [Google Scholar]
- Caicedo, A.L., Williamson, S.H., Hernandez, R.D., Boyko, A., Fledel-Alon, A., York, T.L., Polato, N.R., Olsen, K.M., Nielsen, R., McCouch, S.R., Bustamante, C.D., and Purugganan, M.D. (2007). Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C.S., Thomas, D.J., Eberle, M.A., Swanson, J.E., Livingston, R.J., Rieder, M.J., and Nickerson, D.A. (2005). Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 15 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza, L.L. (1966). Population structure and human evolution. Proc. R. Soc. Lond. B. Biol. Sci. 164 362–379. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., Morgan, M.T., and Charlesworth, D. (1993). The effect of deleterious mutations on neutral molecular variation. Genetics 134 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.M., et al. (2007). Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342. [DOI] [PubMed] [Google Scholar]
- Comeron, J.M. (1995). A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J. Mol. Evol. 41 1152–1159. [DOI] [PubMed] [Google Scholar]
- Comeron, J.M. (1999). K-Estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15 763–764. [DOI] [PubMed] [Google Scholar]
- De Schutter, K., Joubès, J., Cools, T., Verkest, A., Corellou, F., Babiychuk, E., Van Der Schueren, E., Beeckman, T., Kushnir, S., Inzé, D., and De Veylder, L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., Krols, L., Terras, F., Landrieu, I., Van Der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., and Inzé, D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8 655–665. [DOI] [PubMed] [Google Scholar]
- Ding, J., Cheng, H., Jin, X., Araki, H., Yang, Y., and Tian, D. (2007). Contrasting patterns of evolution between allelic groups at a single locus in Arabidopsis. Genetica 129 235–242. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J., and Doyle, J.L. (1990). A rapid total DNA preparation procedure for fresh plant tissue. Focus 12 13–15. [Google Scholar]
- Ducharme, G.R., and Ledwina, T. (2003). Efficient and adaptive nonparametric test for the two-sample problem. Ann. Stat. 31 2036–2058. [Google Scholar]
- Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429 776–780. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A. (2002). Changing effective population size and the McDonald-Kreitman test. Genetics 162 2017–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian-Marwedel, T., Umeda, M., and Sauter, M. (2002). The rice cyclin-dependent kinase-activating kinase R2 regulates S-phase progression. Plant Cell 14 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J.C., and Wu, C.I. (2000). Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxe, J.P., Dar, V.U., Zheng, H., Nordborg, M., Gaut, B.S., and Wright, S.I. (2008). Selection on amino acid substitutions in Arabidopsis. Mol. Biol. Evol. 25 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François, O., Blum, M.G.B., Jakobsson, M., and Rosenberg, N.A. (2008). Demographic history of european populations of Arabidopsis thaliana. PLoS Genet. 4 e1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y.X., and Li, W.H. (1993). Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. (1998). Consed: A graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Gouesnard, B., Bataillon, T.M., Decoux, G., Rozale, C., Schoen, D.J., and David, J.L. (2001). MSTRAT: An algorithm for building germ plasm core collections by maximizing allelic or phenotypic richness. J. Hered. 92 93–94. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (2005). Coupling cell proliferation and development in plants. Nat. Cell Biol. 7 535–541. [DOI] [PubMed] [Google Scholar]
- Hudson, R.R., and Kaplan, N.L. (1988). The coalescent process in models with selection and recombination. Genetics 120 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, L.D., and Williams, E.J. (2000). Covariation of GC content and the silent site substitution rate in rodents: Implications for methodology and for the evolution of isochores. Gene 261 107–114. [DOI] [PubMed] [Google Scholar]
- Hutter, S., Vilella, A.J., and Rozas, J. (2006). Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinformatics 7 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, K.K., Ohashi, Y., Tsuge, T., Yoshizumi, T., Matsui, M., Oka, A., and Aoyama, T. (2006). The A-Type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., Tajima, F., Terauchi, R., and Miyashita, N.T. (1996). Intragenic recombination in the Adh locus of the wild plant Arabidopsis thaliana. Genetics 143 1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40 77–105. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Shinmyo, A., and Sekine, M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45 819–831. [DOI] [PubMed] [Google Scholar]
- Janic-Wróblewska, A., and Ledwina, T. (2000). Data driven rank test for two-sample problem. Scand. J. Stat. 27 281–297. [Google Scholar]
- Jensen, L.J., Jensen, T.S., de Lichtenberg, U., Brunak, S., and Bork, P. (2006). Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature 443 594–597. [DOI] [PubMed] [Google Scholar]
- Joubès, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inzé, D., Umeda, M., and Renaudin, J.P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43 607–620. [DOI] [PubMed] [Google Scholar]
- Kaufman, L., and Rousseeuw, P.J. (1990). Finding Groups in Data: An Introduction to Cluster Analysis. (New York: Wiley).
- Kelley, J.L., Madeoy, J., Calhoun, J.C., Swanson, W., and Akey, J.M. (2006). Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 16 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. (1983). The Neutral Theory of Molecular Evolution. (Cambridge, UK: Cambridge University Press.)
- Koornneef, M., Alonso-Blanco, C., and Vreugdenhil, D. (2004). Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Li, Y.F., Costello, J.C., Holloway, A.K., and Hahn, M.W. (2008). “Reverse Ecology” and the power of population genomics. Evolution 62 2984–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec, J., Meyer, R.C., Steinfath, M., Redestig, H., Becher, M., Witucka-Wall, H., Fiehn, O., Torjek, O., Selbig, J., Altmann, T., and Willmitzer, L. (2008). Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J. 53 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J.H., and Kreitman, M. (1991). Adaptive protein evolution at the Adh locus in Drosophila. Nature 351 652–654. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G., Goff, S., Holub, E.B., and Dangl, J.L. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann, H.I., Camilleri, C., Bérard, A., Bataillon, T., David, J.L., Reboud, X., Le Corre, V., Caloustian, C., Gut, I.G., and Brunel, D. (2004). Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J. 38 193–202. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., and Haigh, J. (1974). The hitch-hiking effect of a favourable gene. Genet. Res. 23 23–25. [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Moore, R.C., and Purugganan, M.D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8 122–128. [DOI] [PubMed] [Google Scholar]
- Moore, R.C., and Stevens, M.H. (2008). Local patterns of nucleotide polymorphism are highly variable in the selfing species Arabidopsis thaliana. J. Mol. Evol. 66 116–129. [DOI] [PubMed] [Google Scholar]
- Nei, M. (1987). Molecular Evolutionary Genetics. (New York: Columbia University Press).
- Nielsen, R. (2005). Molecular signatures of natural selection. Annu. Rev. Genet. 39 197–218. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack, M.K., Grini, P.E., Jakoby, M.J., Lafos, M., Koncz, C., and Schnittger, A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38 63–67. [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (New York: Springer Verlag).
- Pickett, F.B., and Meeks-Wagner, D.R. (1995). Seeing double: Appreciating genetic redundancy. Plant Cell 7 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Sabeti, P.C., Schaffner, S.F., Fry, B., Lohmueller, J., Varilly, P., Shamovsky, O., Palma, A., Mikkelsen, T.S., Altshuler, D., and Lander, E.S. (2006). Positive natural selection in the human lineage. Science 312 1614–1620. [DOI] [PubMed] [Google Scholar]
- Sawyer, S.A., and Hartl, D.L. (1992). Population genetics of polymorphism and divergence. Genetics 132 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K.J., Ramos-Onsins, S., Ringys-Beckstein, H., Weisshaar, B., and Mitchell-Olds, T. (2005). A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen, D.J., and Brown, A.H.D. (1993). Conservation of allelic richness in wild crop relatives is aided by assessment of genetic-markers. Proc. Natl. Acad. Sci. USA 90 10623–10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard, K.A., and Purugganan, M.D. (2003). Molecular population genetics of the Arabidopsis CLAVATA2 region. The genomic scale of variation and selection in a selfing species. Genetics 163 1083–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K.K., and Purugganan, M.D. (2005). Evolutionary and ecological genomics of Arabidopsis. Plant Physiol. 138 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno, A., Ohno, R., Bisova, K., Sakaguchi, N., Huang, J., Koncz, C., Uchimiya, H., and Umeda, M. (2006). Diverse phosphoregulatory mechanisms controlling cyclin-dependent kinase-activating kinases in Arabidopsis. Plant J. 47 701–710. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., and Waterman, M.S. (1981). Identification of common molecular subsequences. J. Mol. Biol. 147 195–197. [DOI] [PubMed] [Google Scholar]
- Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M.I., and Tiffin, P.L. (2008). The quest for adaptive evolution: a theoretical challenge in a maze of data. Curr. Opin. Plant Biol. 11 110–115. [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, U'Ren J, Tenaillon O, Gaut BS. (2004). Selection versus demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 21 1214–1225. [DOI] [PubMed] [Google Scholar]
- Thornton, K. (2003). Libsequence: A C++ class library for evolutionary genetic analysis. Bioinformatics 19 2325–2327. [DOI] [PubMed] [Google Scholar]
- Tian, D., Araki, H., Stahl, E., Bergelson, J., and Kreitman, M. (2002). Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian, C., Hu, T.T., Aranzana, M.J., Lister, C., Tang, C., Zheng, H., Zhao, K., Calabrese, P., Dean, C., and Nordborg, M. (2006). A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4 e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouzé, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, B. (2008). Using molecular markers for detecting domestication, improvement, and adaptation genes. Euphytica 161 1–17. [Google Scholar]
- Wang, G., Kong, H., Sun, Y., Zhang, X., Zhang, W., Altman, N., DePamphilis, C.W., and Ma, H. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G.A. (1975). On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nordborg, M. (2005). Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiol. 138 567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S.I., and Gaut, B.S. (2005). Molecular population genetics and the search for adaptive evolution in plants. Mol. Biol. Evol. 22 506–519. [DOI] [PubMed] [Google Scholar]
- Wright, S.I., Yau, C.B., Looseley, M., and Meyers, B.C. (2004). Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol. Biol. Evol. 21 1719–1726. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Emerson, B., Ratanasut, K., Patrick, E., O'Neill, C., Bancroft, I., and Turner, J.G. (2004). Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol. Biol. Evol. 21 1661–1672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.