Abstract
Phototropins (phot1 and phot2) are plasma membrane–associated receptor kinases that respond specifically to blue and UV wavelengths. In addition to a C-terminal Ser/Thr kinase domain, phototropins contain two N-terminal chromophore binding LOV domains that function as photoswitches to regulate a wide range of enzymatic activities in prokaryotes and eukaryotes. Through domain swapping, we show that the photochemical properties of Arabidopsis thaliana phot1 rely on interactions between LOV1 and LOV2, which are facilitated by their intervening linker sequence. Functional analysis of domain-swap proteins supports a mechanism whereby LOV2 acts as a dark-state repressor of phot1 activity both in vitro and in vivo. Moreover, we find a photoactive role for LOV1 in arresting chloroplast accumulation at high light intensities. Unlike LOV2, LOV1 cannot operate as a dark-state repressor, resulting in constitutive receptor autophosphorylation and accelerated internalization from the plasma membrane. Coexpression of active and inactive forms of phot1 demonstrates that autophosphorylation can occur intermolecularly, independent of LOV1, via light-dependent receptor dimerization in vivo. Indeed, transphosphorylation is sufficient to promote phot1 internalization through a clathrin-dependent endocytic pathway triggered primarily by phosphorylation of Ser-851 within the kinase activation loop. The mechanistic implications of these findings in regard to light-driven receptor activation and trafficking are discussed.
INTRODUCTION
Higher plants possess at least four different classes of photoreceptor proteins: the phytochromes (Bae and Choi, 2008), cryptochromes (Li and Yang, 2007), phototropins (Christie, 2007), and members of the Zeitlupe (ZTL) family (Demarsy and Fankhauser, 2009). Phytochromes are photoreversible red/far-red photoreceptors, whereas cryptochromes, phototropins, and members of the ZTL family specifically absorb blue/UV wavelengths. Plants also respond to UV-B wavelengths (Jenkins, 2009) and green light (Folta and Maruhnich, 2007); however, the photosensors responsible for their detection remain elusive.
Phototropins regulate a variety of responses that serve to optimize photosynthetic efficiency and promote plant growth under weak light conditions (Christie, 2007). These processes include phototropism (Liscum and Briggs, 1995), light-induced stomatal opening (Kinoshita et al., 2001), chloroplast relocation movements (Kagawa et al., 2001), as well as leaf expansion (Sakamoto and Briggs, 2002; Takemiya et al., 2005) and positioning (Inoue et al., 2008b). Arabidopsis thaliana contains two phototropins (phot1 and phot2) that exhibit overlapping and distinct functions. phot1 and phot2 act redundantly to regulate phototropism, stomatal opening, leaf expansion, and chloroplast accumulation movement at low light intensities but exhibit different photosensitivities (Christie, 2007). While phot1 mediates the rapid inhibition of hypocotyl growth upon transfer of dark-grown seedlings to light (Folta and Spalding, 2001), phot2 is responsible for chloroplast avoidance movement at high light intensities (Kagawa et al., 2001) and mediates the chloroplast accumulation at the cell base in darkness (Tsuboi et al., 2007). Phototropins are also found in lower plants, including the unicellular green alga Chlamydomonas reinhardtii where it regulates the algal sexual lifecycle in response to blue light (Huang and Beck, 2003).
Phototropins comprise two distinct functional units: an N-terminal photosensory input region coupled to a C-terminal Ser/Thr kinase effector domain (Christie, 2007; Matsuoka et al., 2007) that belongs to the AGC family of protein kinases (Christie, 2007). The N-terminal region contains two LOV (light, oxygen, or voltage sensing) domains designated LOV1 and LOV2. Each binds one molecule of flavin mononucleotide noncovalently (Christie et al., 1999) and shares a common tertiary fold, which creates a hydrophobic binding pocket for the flavin chromophore (Crosson and Moffat, 2001; Nakasako et al., 2008). Blue light excitation invokes a reversible photocycle that involves the formation of a covalent adduct between the flavin and a conserved Cys residue within the LOV domain (Salomon et al., 2000; Swartz et al., 2001). Flavin-cysteinyl adduct formation induces subsequent protein conformational changes (Corchnoy et al., 2003; Harper et al., 2003) that, in turn, trigger activation of the C-terminal kinase domain resulting in receptor autophosphorylation (Christie, 2007), a prerequisite for phototropin signaling (Inoue et al., 2008a). Although hydrophilic proteins, phototropins are associated with the plasma membrane (Briggs et al., 2001). Upon blue light irradiation, a fraction of phot1 is rapidly released from the plasma membrane in Arabidopsis (Sakamoto and Briggs, 2002), whereas phot2 associates with the Golgi apparatus (Kong et al., 2006). The mechanistic basis and biological significance of this partial light-induced internalization is not fully understood but may represent a mode of phototropin signaling (Kong and Nagatani, 2008; Wan et al., 2008) or receptor desensitization (Christie, 2007).
LOV1 and LOV2 domains of higher plant phototropins typically share 40% amino acid identity (Christie et al., 1999) but exhibit different photochemical properties (Salomon et al., 2000) and distinct functional roles in regulating phototropin activity (Christie et al., 2002; Cho et al., 2007). LOV2 plays a major role in light-activated receptor autophosphorylation owing to specific protein changes that occur in response to photoexcitation. This involves displacement of a conserved α-helix, designated Jα, that likely functions as a helical connector coupling LOV2 photoexcitation to kinase activation (Harper et al., 2003). Indeed, disruption of the Jα-helix through site-directed mutagenesis results in phot1 kinase activation in the absence of light (Harper et al., 2004; Jones et al., 2007). LOV2 is also reported to bind in trans to the kinase domain of phot2 and inhibit in vitro phosphorylation of the artificial substrate casein in darkness (Matsuoka and Tokutomi, 2005), leading to the proposal that LOV2 functions as repressor of phototropin kinase activity in the dark (Christie, 2007; Matsuoka et al., 2007). However, it has not been established whether LOV2 regulates autophosphorylation by a similar mode of action or whether this can occur in trans.
In contrast with LOV2, the functional role(s) of LOV1 remains poorly understood. Hence, the distinct roles of LOV1 and LOV2 are particularly intriguing given their high degree of sequence identity (Christie et al., 1999) combined with their almost identical tertiary structures (Crosson and Moffat, 2001; Nakasako et al., 2008). These factors prompted us to investigate whether these domains were functionally interchangeable. In this study, we employed a domain-swapping strategy to investigate the mechanistic basis underlying phot1 kinase activation and endocytosis by light. Our findings uncover new insights into the photochemical and functional roles of LOV1 and LOV2, in addition to the mode receptor activation, and demonstrate that phot1 endocytic recruitment is promoted by an intermolecular phosphorylation mechanism that is clathrin dependent in plant cells.
RESULTS
Photochemical Properties of Phot1 are Dependent on LOV1 and LOV2
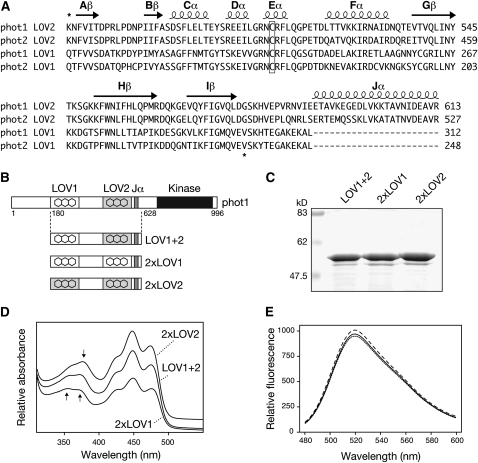
Based on the predicted secondary structures of LOV1 and LOV2, a region of 104 amino acids was selected to engineer novel variants of Arabidopsis phot1 containing altered combinations of LOV domains (Figure 1A). Our initial aim was to create domain-swap proteins to assess the relative contributions of LOV1 and LOV2 on phot1 photochemistry. Fragments of phot1, designated LOV1+2 (Figure 1B), expressed in Escherichia coli offer a convenient system to study phot1 reactivity (Christie et al., 2002), as their photochemical properties reflect those of full-length photoreceptor (Kasahara et al., 2002). We therefore generated truncated domain-swap proteins whereby LOV1 was replaced by LOV2 and vice versa. The resulting proteins, designated 2xLOV1 and 2xLOV2 (Figure 1B), were purified by affinity chromatography (Figure 1C) and their spectral properties assessed by UV-visible absorbance and circular dichroism (CD) spectroscopy.
Figure 1.
Domain Swapping of Arabidopsis phot1 LOV Domains.
(A) Sequence alignment of LOV1 and LOV2 domains of Arabidopsis phot1 and phot2. Predicted secondary structure is indicated on top of the sequences. Regions used for domain swapping of phot1 LOV domains are indicated by asterisks. The conserved Cys required for photochemical reactivity is boxed.
(B) Schematic illustration showing regions of wild-type phot1 (LOV1+2) and 2xLOV1 and 2xLOV2 proteins generated. Relative positions of phot1 domain structures are indicated.
(C) Coomassie-stained SDS-PAGE gel showing purified preparations of wild-type phot1 (LOV1+2), 2xLOV1, and 2xLOV2. Sizes of molecular weight marker proteins are indicated on the left in kilodaltons.
(D) Absorption spectra of wild-type (LOV1+2), 2xLOV1, and 2xLOV2 proteins. Spectra are offset for clarity. Arrows indicate absorbance differences between 2xLOV1 and 2xLOV2 in the UV-A region of the spectrum.
(E) Fluorescence emission spectra of flavin fluorescence released upon denaturation of equal concentrations (0.4 mg mL−1) of wild-type LOV1+2 (solid line), 2xLOV1 (dotted line), and 2xLOV2 (dashed line).
Absorption spectra for the domain-swap proteins were similar to that of the wild type (LOV1+2) but showed key differences in the near-UV region (Figure 1D) The spectral properties of 2xLOV1 are typical for LOV1, showing two absorbance peaks of equal intensity around 360 and 370 nm (Kasahara et al., 2002). 2xLOV2 showed a more prominent peak in this region that is red shifted to around 380 nm, consistent with the spectral properties of LOV2 (Kasahara et al., 2002). The spectral differences between 2xLOV1 and 2xLOV2 were also evident from their respective fluorescence excitation spectra (see Supplemental Figure 1 online), whereas the wild type showed intermediate absorbance (Figure 1D) and fluorescence (see Supplemental Figure 1 online) properties. Importantly, flavin binding for 2xLOV1 and 2xLOV2 matched that of the wild type (Figure 1E), implying that domain swapping had not comprised protein tertiary structure. Minor differences in secondary structure were detected by CD spectroscopy for 2xLOV1 and 2xLOV2 relative to the wild type (see Supplemental Figure 2 online). However, as found for UV-visible absorbance spectroscopy (Figure 1D), the CD properties for the wild type resembled an average of the two and may simply reflect secondary structure differences between LOV1 and LOV2 that are maintained in the domain-swap proteins.
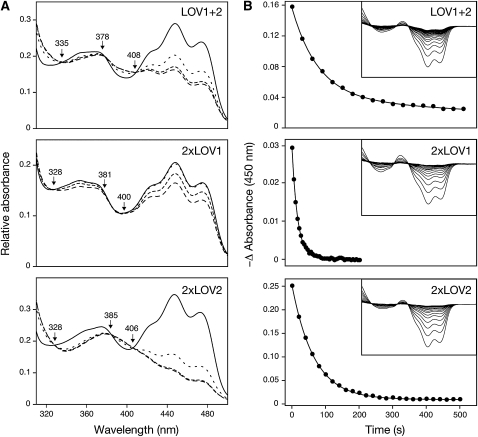
Irradiation of the domain-swap proteins resulted in a loss of absorbance in the blue region of the spectrum and the appearance of three isosbestic points (Figure 2A) corresponding to flavin-cysteinyl adduct formation within the LOV domains (Salomon et al., 2000; Swartz et al., 2001). Irradiation with a 1-ms camera strobe flash produced difference spectra that were fully reversible in the dark (Figure 2B). 2xLOV1 recovered with a half-life of 12 s, coinciding with the recovery kinetics for the single LOV1 domain under identical conditions (see Supplemental Figure 3A online). Likewise, 2xLOV2 recovered with a half-life of 46 s, which corresponded to the recovery properties of LOV2 (see Supplemental Figure 3B online). We therefore conclude that the spectral and photochemical properties of 2xLOV1 and 2xLOV2 reflect those of the respective individual domains.
Figure 2.
Photochemical Properties of 2xLOV1 and 2xLOV2.
(A) Light-induced absorption changes recorded for the wild type (LOV1+2), 2xLOV1, and 2xLOV2 in response to blue light irradiation (300 μmol m−2 s−1, 470 ± 10 nm). Consecutive spectra were recorded after irradiations of 0, 10, 30, and 60 s at a protein concentration of 2.5 mg mL−1. In each case, isosbestic points are indicated.
(B) Dark-recovery kinetics for wild-type (LOV1+2), 2xLOV1, and 2xLOV2 proteins. Dark-minus-light absorption difference spectra are shown monitoring recovery back to the initial dark state following photoexcitation (inset). Spectra were recorded every 30 s (LOV1+2), 5 s (2xLOV1), and 20 s (2xLOV2). Absorption changes after light excitation measured at 450 nm is shown in the main panel. Wild-type (LOV1+2) decay fits to two exponentials with half-lives of 60 and 735 s. Decay for 2xLOV1 and 2xLOV2 fit to single exponentials with half-lives of 12 and 46 s, respectively.
Presence of the LOV Linker Region Promotes Interactions between LOV1 and LOV2
In contrast with the domain-swap proteins, dark-recovery kinetics for the wild type could be resolved into two components, with half-lives of 60 s and 735 s, respectively (Figure 2B). These values vary somewhat from previous measurements of Arabidopsis phot1 LOV1+2 fragments (Christie et al., 2002; Kasahara et al., 2002) and most likely arise from differences in experimental conditions.
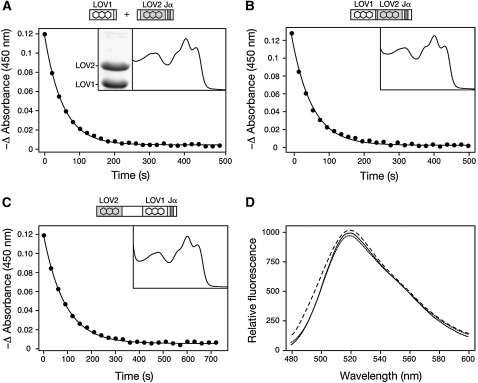
Neither of the domain-swap proteins displayed the slow phase of dark recovery characteristic of the wild type, suggesting that this property is dependent on the presence of LOV1 and LOV2. This property was also not detected for a mixed population of LOV1 and LOV2 expressed separately in E. coli (Figure 3A) or when the linker region between LOV1 and LOV2 was removed from the protein (Figure 3B). Hence, the presence of the linker region appears to influence phot1 photoreactivity by promoting some heterointeraction between LOV1 and LOV2 that is lost in the domain-swap proteins. Consequently, LOV1 and LOV2 were inverted to examine whether their position contributes to this photochemical behavior. Dark-recovery kinetics for the resulting domain swap designated LOV2+1 fit to a single exponential with a half-life of 60 s (Figure 3C) matching with the initial recovery component detected for the wild type (Figure 2B). Absence of a slow recovery phase suggests that this property of phot1 is not only dependent on LOV1 and LOV2, but also on their position within the receptor protein. Once again, flavin binding relative to the wild type was used to confirm the structural integrity of these additional variants (Figure 3D).
Figure 3.
Impact of the LOV Linker Region and LOV Domain Positioning on phot1 Photoreactivity.
(A) Dark recovery after light excitation measured at 450 nm for a mixed population of LOV1 and LOV2 domains. Decay fits to a single exponential with a half-life of 37 s. The absorption spectrum is shown (inset) alongside a Coomassie blue–stained SDS-PAGE gel showing equal protein levels.
(B) Dark recovery after light excitation measured at 450 nm for LOV1+2 lacking the intervening linker region. Decay fits to a single exponential with a half-life of 60 s. Absorption spectrum is shown (inset).
(C) Dark recovery after light excitation measured at 450 nm for the LOV2+1 domain-swap protein. Decay fits to a single exponential with a half-life of 60 s. Absorption spectrum is shown (inset). In each case, protein concentrations were 2 mg mL−1.
(D) Fluorescence emission spectra of flavin fluorescence released upon denaturation of equal concentrations (0.4 mg mL−1) of wild-type LOV1+2 (solid line), LOV2+1 (dotted line), and LOV1+2 lacking the intervening linker region (dashed line).
Replacing LOV1 with LOV2 Reduces Phot1 Activity in Vitro
Previous studies have shown that LOV2 photoexcitation plays a major role in regulating phot1 kinase activation by blue light (Christie et al., 2002). Given the structural integrity of 2xLOV1 and 2xLOV2 as monitored by flavin binding (Figure 1E), full-length domain-swap proteins (designated phot12xLOV1 and phot12xLOV2, respectively) were created to assess the functional roles of LOV1 and LOV2 in regulating phot1 activity. The rationale behind this approach was twofold: to determine whether LOV1 could functionally replace LOV2 and to investigate whether replacement of LOV1 with LOV2 could provide further insights into the function of LOV1.
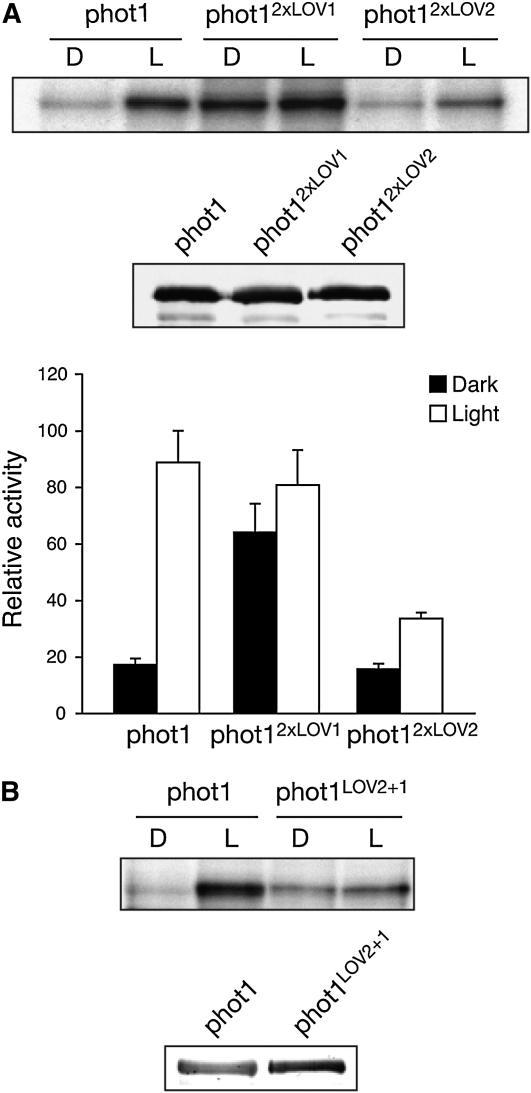
As reported previously (Christie et al., 2002), wild-type phot1 typically exhibits a fivefold increase in receptor autophosphorylation upon irradiation when expressed in insect cells (Figure 4A). By contrast, a substantial increase in dark-level phosphorylation was observed for phot12xLOV1. This constitutive mode of activity is similar to that described for phot1 carrying a single point mutation I608E within the Jα-helix (Harper et al., 2004; Jones et al., 2007) that resides C-terminal to LOV2 (Figure 1A). LOV1, unlike LOV2, is therefore unable to operate as an effective photoswitch to control phot1 kinase activity. Given the constitutive activity of phot12xLOV1, these findings also suggest that LOV2 functions as a dark-state repressor. Consistent with this conclusion, autophosphorylation for phot12xLOV2 was attenuated under saturating light conditions (Figure 4A). Moreover, the reduced activity of phot12xLOV2 did not result from an alteration in phosphorylation kinetics compared with the wild type (see Supplemental Figure 4 online). Thus, introduction of a second LOV2 domain in phot12xLOV2 appears to cause additional repression of phot1 kinase activity. To further investigate this possibility, we engineered a full-length version of phot1 in which LOV1 and LOV2 were interchanged. We rationalized that if insertion of LOV2 into the position of LOV1 was able to repress phot1 autophosphorylation, the resulting protein designated phot1LOV2+1 should exhibit constitutive kinase activity similar to that of phot12xLOV1, but at a reduced level. Indeed, in vitro phosphorylation analysis demonstrated this to be the case (Figure 4B). Thus, coupling of LOV2 to the kinase domain via the Jα-helix appears to be necessary to create an effective repression mechanism that can be deactivated by light.
Figure 4.
Autophosphorylation Activity of Full-Length phot1 Domain-Swap Proteins Expressed in Insect Cells.
(A) Autoradiograph showing light-dependent autophosphorylation of wild-type phot1, phot12xLOV1, and phot12xLOV2 in protein extracts isolated from insects cells. Samples were given a mock irradiation (D) or irradiated with white light (L) at a total fluence of 10,000 μmol m−2 prior to the addition of radiolabeled ATP. Immunoblot analysis of phot1 protein levels is shown below. Kinase activity was quantified by phosphor imaging and expressed as a percentage of maximal phosphorylation activity. Standard errors are shown (n = 3).
(B) Autophosphorylation activity of the phot1LOV2+1 domain-swap protein relative to wild-type phot1. Immunoblot analysis of phot1 protein levels is shown below.
Phot12xLOV2 Exhibits Reduced Activity in Planta
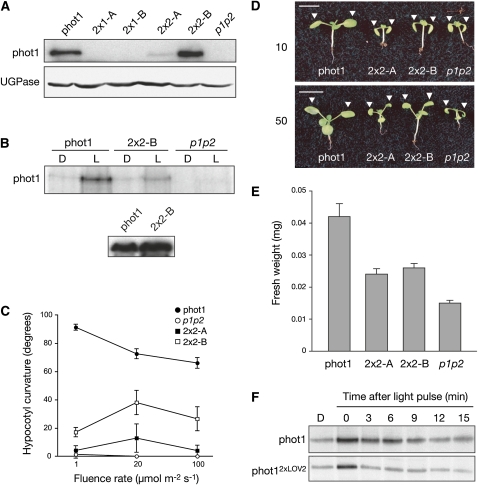
To assess their functionality in Arabidopsis, phot12xLOV1 and phot12xLOV2 were expressed in the phot1 phot2 double mutant. The cauliflower mosaic virus 35S promoter was used to drive expression of all constructs, including wild-type phot1, which was included as a control. Unfortunately, we were unable to retrieve transgenic lines expressing sufficient levels of phot12xLOV1 to assess its functionality (Figure 5A) despite the presence of detectable transcript (see Supplemental Figure 5 online). Similar issues were also encountered in our attempts to generate lines expressing phot1I608E, although transcripts were present (see Supplemental Figure 5 online). Nevertheless, independent lines expressing low and moderate levels of phot12xLOV2 were obtained (2x2-A and 2x2-B, respectively), the latter being comparable to wild-type phot1 protein levels driven by the 35S promoter (Figure 5A).
Figure 5.
Functional Analysis of phot12xLOV2.
(A) Immunoblot analysis of total protein extracts (30 μg) from 3-d-old etiolated seedlings expressing wild-type phot1 or phot12xLOV1 (2x1-A and 2x1-B) or phot12xLOV2 (2x2-A and 2x2-B) in the phot1 phot2 double mutant (p1p2). Protein extracts were probed with anti-phot1 antibody (top panel) and antibody raised to against UDP-glucose pyrophosphorylase (UGPase; bottom panel) as a loading control.
(B) In vitro autophosphorylation activity in membrane extracts isolated from 3-d-old etiolated transgenic seedlings expressing wild-type phot1 and phot12xLOV2 (2x2-B). Protein extracts were given a mock irradiation (D) or irradiated with 10,000 μmol m−2 white light (L) prior to the addition of radiolabeled ATP. Protein extracts were probed with anti-phot1 antibody (bottom panel).
(C) Phototropism fluence-rate response of 3-d-old etiolated transgenic seedlings expressing wild-type phot1 and phot12xLOV2 (2x2-A and 2x2-B). Phototropic curvatures were measured after exposure to unilateral blue light for 24 h. Each value is the mean of at least 20 seedlings. Standard errors are shown.
(D) Leaf positioning in transgenic seedlings expressing wild-type phot1 and phot12xLOV2 (2x2-A and 2x2-B) in response to 10 or 50 μmol m−2 s−1 continuous white light. White arrows indicate the positioning of the first true leaves. Bars = 1 cm.
(E) Fresh weight measurements of 15-d-old transgenic plants expressing wild-type phot1 and phot12xLOV2 (2x2-A and 2x2-B).
(F) Effect of dark incubation on the autophosphorylation activity of wild-type phot1 and phot12xLOV2 expressed in insect cells. Samples were irradiated with white light (10,000 μmol m−2) and incubated in the dark at room temperature for the times indicated prior to the addition of radiolabeled ATP.
Microsomal membranes were prepared from dark-grown seedlings for in vitro phosphorylation analysis. phot12xLOV2 autophosphorylation was attenuated compared with the wild type (Figure 5B), correlating with the diminished activity of this protein in insect cells (Figure 4A). 35S-driven phot1 restored phototropic responsiveness in etiolated seedlings over a broad range of fluence rates of blue light (1 to 100 μmol m−2 s−1), whereas no curvature was detected for the phot1 phot2 double mutant (Figure 5C). Weaker curvatures were observed at higher blue light intensities even in wild-type (gl-1) seedlings (see Supplemental Figure 6 online) that may result from a decrease in phot1 protein levels (Sakamoto and Briggs, 2002). Seedlings expressing phot12xLOV2 showed reduced phototropic curvature that correlated with protein expression. Notably, reduced phototropism in line 2x2-B did not result from lower protein levels compared with the wild type (Figure 5A), indicating that phot12xLOV2 exhibits reduced functionality.
35S-driven phot1 also restored leaf positioning in the phot1 phot2 double mutant at 10 and 50 μmol m−2 s−1 white light (Figure 5D), while phot-deficient seedlings lacked this response. Surprisingly, leaf positioning was restored at 50 μmol m−2 s−1 in both 2x2-A and 2x2-B despite their differences in protein levels. A similar trend was observed for their growth under equivalent light conditions (Figure 5E). The comparable functionality of lines 2x2-A and 2x2-B (expressing low and moderate levels of phot12xLOV2, respectively) under these circumstances may depend on the photochemical properties of phot12xLOV2 rather than protein levels. The dark recovery of 2xLOV2 is faster than that of the wild type (Figure 2B) and would be expected to yield lower steady state levels of photoproduct. Similarly, in vitro autophosphorylation of phot12xLOV2 has a shorter memory for a light pulse relative to the wild type when subsequently transferred to darkness (Figure 5F). Thus, a shorter-lived signaling state for phot12xLOV2 may partly account for its reduced photosensitivity in planta.
Domain Swapping Uncovers a Role for LOV1 in Arresting Chloroplast Accumulation at High Light Intensities
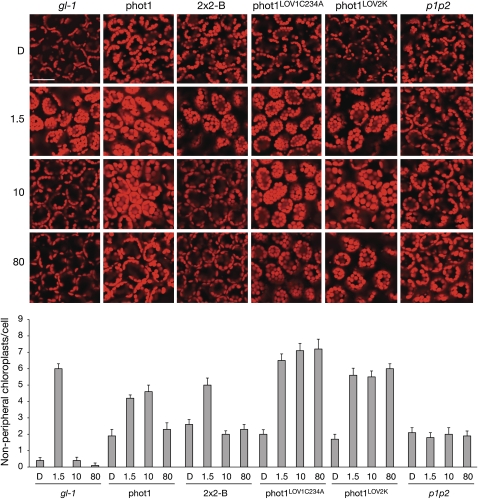
phot12xLOV2-expressing lines were also investigated for their ability to restore chloroplast accumulation movement. 35S-driven phot1 restored chloroplast accumulation at 1.5 μmol m−2 s−1 blue light similar to that observed in wild-type (gl-1) plants (Figure 6). Chloroplast accumulation was evident in 2x2-B under these conditions, indicating that phot12xLOV2 is functional for this response. Chloroplasts of wild-type plants (gl-1) underwent avoidance movement under higher light intensities (10 and 80 μmol m−2 s−1) owing to the presence of phot2 (Kagawa et al., 2001). Likewise, chloroplasts were less randomly localized in the absence of light relative to the phot1 phot2 double mutant since phot2 is known to mediate their positioning to the anticlinal cell wall in darkness (Tsuboi et al., 2007).
Figure 6.
Light-Dependent Chloroplast Positioning in Mesophyll Cells of Transgenic Arabidopsis.
Rosette leaves were detached from 4-week-old plants and illuminated with 1.5, 10, and 80 μmol m−2 s−1 of blue light or kept in darkness for 3 h. Chloroplast autofluorescence was monitored (red) by a confocal laser scanning microscope. Bar = 20 mm. Each panel represents cells viewed from above. Chloroplast relocation was monitored in the wild type (gl-1) and the phot1 phot2 double mutant (p1p2) as a control. In the wild type, chloroplasts accumulate to the cell surface when irradiated with 1.5 μmol m−2 s−1 blue light and undergo avoidance movement to the cell sidewalls at higher light intensities. Chloroplasts in the phot1 phot2 double mutant remain randomly localized by comparison. Transgenic Arabidopsis expressing wild-type phot1, phot12xLOV2 (2x2-B), phot1 with LOV1 inactivated (phot1LOV1C234A), and the LOV2 kinase region of phot1 (phot1LOV2K) were selected for analysis. The number of chloroplasts at the front face per mesophyll cell after light treatment is quantified below. Each value represents the mean from 25 cells. Standard errors are shown.
Interestingly, functional differences between the lines expressing comparable levels of phot1 and phot12xLOV2 were detected under high light conditions. At 10 μmol m−2 s−1 blue light, chloroplasts continued to accumulate in plants expressing phot1 (Figure 6), while chloroplasts in plants expressing phot12xLOV2 appeared randomly localized as in the phot1 phot2 double mutant. Random chloroplast positioning was also observed at 80 μmol m−2 s−1 blue light in plants expressing phot1, suggesting that phot1 responsiveness for chloroplast accumulation is ameliorated at high blue light intensities and that phot12xLOV2 is more photosensitive for this response. Since LOV2 exhibits greater quantum efficiency than LOV1 (Salomon et al., 2000; Kasahara et al., 2002), we reasoned that the difference in photosensitivity observed could be attributed to substitution of LOV1 by LOV2. To investigate this in more detail, we monitored chloroplast accumulation in two additional transgenic lines in the phot1 phot2 double mutant. The first expresses a mutated form of phot1 in which the reactivity of LOV1 has been abolished by mutation of the photoactive Cys residue to Ala (Cho et al., 2007). The second expresses a truncated form of phot1 comprising only the LOV2-kinase region of the protein (Sullivan et al., 2008). As shown in Figure 6, chloroplasts continued to accumulate at 80 μmol m−2 s−1 blue light when LOV1 was either inactivated (phot1LOV1C234A) or absent (phot1LOV2K), providing additional support for its role in arresting chloroplast accumulation in excess light.
Phosphorylation Status Impacts the Subcellular Localization of phot1
The kinase activity of phot2 is reported to enhance its association with the Golgi apparatus upon blue light excitation (Kong et al., 2006). Determining the subcellular localization of phot12xLOV1 was therefore of particular interest given its constitutive kinase activity. Owing to the difficulties encountered in generating stable transgenic lines expressing phot12xLOV1, an alternative expression strategy was employed.
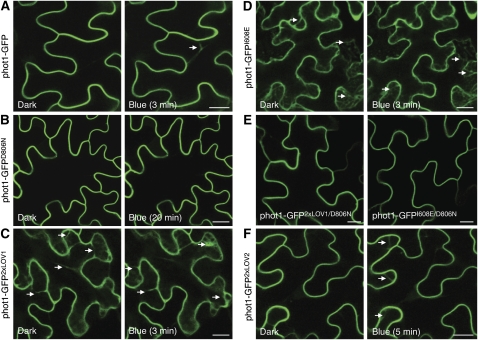
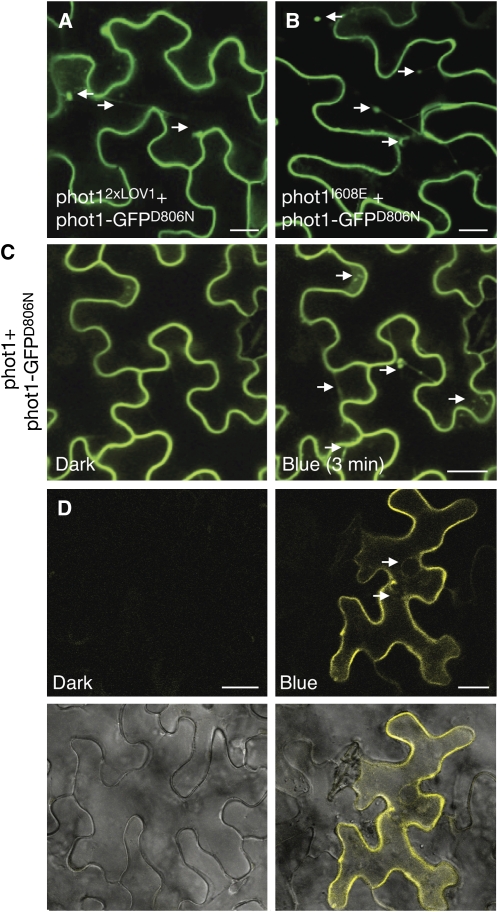
Agrobacterium tumefaciens–mediated expression of phot1 fused to green fluorescent protein (phot1-GFP) in tobacco (Nicotiana benthamiana) epidermal cells produced plasma membrane localization in the dark (Figure 7A). As reported for functionally active phot1-GFP in Arabidopsis (Sakamoto and Briggs, 2002; Wan et al., 2008), the high-intensity blue light laser used to excite GFP induced phot1 internalization from the plasma membrane in tobacco epidermal cells. Internalization was observed in >90% of transformed cells (n = 25) and was rapid, occurring within several minutes as visualized by the appearance of dynamic cytosolic structures (Figure 7A), which intensified over time (see Supplemental Movie 1 online). Incorporation of the D806N mutation that inhibits phot1 kinase activity (Christie et al., 2002) abolished phot1-GFP relocalization, even after prolonged irradiation (Figure 7B). Conversely, phot1-GFP2xLOV1 was internalized even under noninductive conditions (Figure 7C). Enhanced internalization of phot1-GFP2xLOV1 was not due to increased expression, as protein levels were comparable to that of phot1-GFP (see Supplemental Figure 7 online). Moreover, an identical subcellular localization pattern was observed for phot1 harboring the I608E mutation (phot1-GFPI608E) that also triggers constitutive kinase activation (Figure 7D), implying that receptor relocalization is directly linked to phosphorylation status. Consistent with this conclusion, internalization of phot1-GFP2xLOV1 and phot1-GFPI608E was negated by kinase inactivation (Figure 7E). phot1-GFP2xLOV2 showed light-induced internalization similar to that of the wild type but was somewhat slower in appearance (Figure 7F), which might be expected given its shorter-lived photoproduct (Figure 5F).
Figure 7.
Subcellular Localization of phot1-GFP and Domain-Swap Proteins in N. benthamiana.
(A) Fluorescence images showing the rate of phot1-GFP internalization in tobacco leaf epidermal cells at 3 min after a 30-s irradiation with blue light. The white arrow indicates internalization.
(B) Inactivation of phot1 kinase activity (phot1-GFPD806N) inhibits blue light–dependent internalization.
(C) phot1-GFP2xLOV1 shows internalization in the absence of a blue light stimulus. The white arrows indicate internalization.
(D) phot1-GFPI608E also shows constitutive internalization as indicated by the white arrows.
(E) Inactivation of phot1 kinase activity by incorporation of the D806N mutation blocks internalization of phot1-GFP2xLOV1 and phot1-GFPI608E in the absence of light.
(F) phot1-GFP2xLOV2 undergoes blue light–dependent internalization, similar to phot1-GFP.
Bars = 20 μm.
[See online article for color version of this figure.]
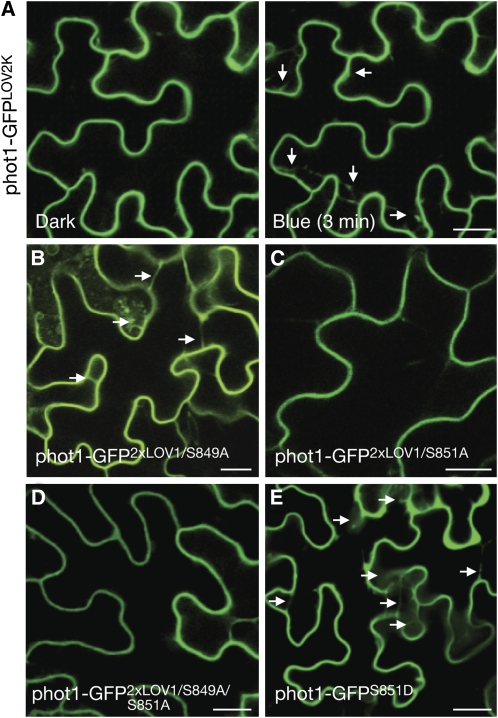
The subcellular localization properties of phot1-GFP2xLOV1 and phot1-GFPI608E are unprecedented and were therefore exploited to map the phosphorylation sites responsible for instigating phot1 movement from the plasma membrane. Arabidopsis phot1 is phosphorylated predominately on Ser residues situated within three portions of the protein: N-terminal to LOV1, the LOV linker region, and the C terminus of the protein, including the catalytic kinase domain (Inoue et al., 2008a, 2008b; Sullivan et al., 2008). A truncated version of phot1 comprising the LOV2-kinase region was still found to exhibit blue light–induced internalization (Figure 8A), implying that phosphorylation sites located in this region are involved in this process. Autophosphorylation of Ser-851 within the kinase activation loop is required for phot1 action, whereas Ser-849 plays an accessory role (Inoue et al., 2008a). Substitution of Ser-849 to Ala within phot1-GFP2xLOV1 had little impact on receptor localization under noninductive conditions (Figure 8B). However, mutation of Ser-851 alone (Figure 8C) or both Ser-849 and Ser-851 (Figure 8D) severely attenuated phot1-GFP2xLOV1 internalization. Furthermore, replacement of Ser-851 with Asp in wild-type phot1, which is expected to mimic phosphorylation (Inoue et al., 2008a), caused internalization of phot1-GFP under noninductive conditions (Figure 8E), confirming that autophosphorylation of Ser-851 is primarily responsible for instigating phot1 movement from the plasma membrane.
Figure 8.
Identification of Phosphorylation Sites Regulating phot1 Internalization.
(A) The LOV2-kinase region of phot1 (phot1-GFPLOV2K) undergoes blue light–dependent relocalization (indicated by the white arrows).
(B) Mutation of Ser-849 to Ala in phot12xLOV1 does not impair internalization in darkness (indicated by white arrows).
(C) Mutation of Ser-851 to Ala in phot12xLOV1 severely impacts the formation of cytosolic structures in darkness.
(D) Mutation of both Ser-849 and Ser-851 to Ala in phot12xLOV1 impairs internalization in darkness.
(E) Mutation of Ser-851 to Asp in phot1-GFP induces the formation of cytosolic structures in darkness (indicated by white arrows).
Bars = 20 μm.
[See online article for color version of this figure.]
Phot1 Internalization Is Promoted by Intermolecular Phosphorylation
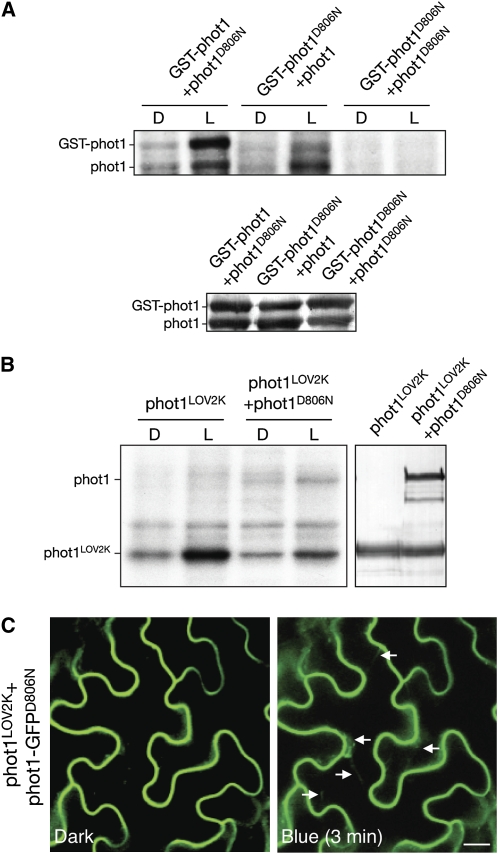
Both cis- and trans-interactions are implicated in autophosphorylation-dependent activation of protein kinases. We therefore used the constitutive nature of phot12xLOV1 to examine whether autophosphorylation could occur in trans. Coexpression of phot1-GFPD806N with nonfluorescently labeled phot12xLOV1 resulted in receptor internalization under noninductive conditions (Figure 9A), suggesting that receptor autophosphorylation can occur in trans to stimulate trafficking from the plasma membrane. Coexpression of phot1-GFPD806N with phot1I608E produced similar results (Figure 9B). In addition, coexpression of phot1-GFPD806N with wild-type phot1 caused it to be internalized in a light-dependent manner (Figure 9C). A direct interaction between distinct phot1 molecules was confirmed using bimolecular fluorescence complementation (BiFC) (Walter et al., 2004). Reconstitution of yellow fluorescent protein (YFP) fluorescence both at the plasma membrane and in cytosolic structures was evident only after irradiation of tobacco epidermal cells with blue light (Figure 9D). No fluorescence was detected in cells under noninductive conditions (Figure 9D) or when phot1 was coexpressed with empty vector controls (data not shown). In vitro transphosphorylation between glutathione S-transferase (GST)-tagged and non-GST-tagged versions of phot1 was also observed in insect cells using native (Figure 10A) or constitutively active (see Supplemental Figure 8 online) forms of the receptor, further demonstrating that phot1 is capable of intermolecular phosphorylation.
Figure 9.
Transphosphorylation between phot1 Molecules Promotes Receptor Internalization.
(A) and (B) Coexpression of a kinase-inactive version of phot1-GFP (phot1-GFPD806N) with nonfluorescently labeled phot12xLOV1 (A) or phot1I608E (B) shows that both these constitutively active forms of phot1 can induce internalization of phot1-GFPD806N in darkness (as indicated by the white arrows).
(C) Fluorescence images of a kinase inactive version of phot1-GFP (phot1-GFPD806N) coexpressed with nonfluorescently labeled phot1 in tobacco leaf epidermis show that phot1 can induce blue light–dependent internalization of phot1-GFPD806N (as indicated by the white arrows).
(D) Fluorescence images of phot1-YN and phot1-YC coexpressed in tobacco leaf epidermis. Reconstitution of YFP fluorescence was only detected following blue light irradiation. Bright-field images are shown below. Internalization of phot1 is indicated (white arrows).
Bars = 20 μm.
[See online article for color version of this figure.]
Figure 10.
Transphosphorylation between phot1 Molecules in the Presence or Absence of LOV1.
(A) Autoradiograph showing light-dependent transphosphorylation of a kinase-inactive version of phot1 (phot1D806N) by GST-phot1 and vice versa in protein extracts from insect cells. Immunoblot analysis of phot1 protein levels is shown below.
(B) Autoradiograph showing light-dependent transphosphorylation of a kinase-inactive version of phot1 (phot1D806N) by the phot1LOV2K in protein extracts from insect cells. Immunoblot analysis of phot1 protein levels is shown on the right.
(C) Fluorescence images of a kinase inactive version of phot1-GFP (phot1-GFPD806N) coexpressed with nonfluorescently labeled phot1LOV2K in tobacco leaf epidermis show that phot1LOV2K can induce light-dependent internalization of phot1-GFPD806N (indicated by white arrows). Bar = 20 μm.
[See online article for color version of this figure.]
Isolated LOV1 domains of Arabidopsis phot1 form dimers and are proposed to function as sites of receptor dimerization (Nakasako et al., 2004, 2008; Katsura et al., 2009). However, we found that the LOV2K region of phot1 retains light-induced kinase activity when expressed in insect cells and is able to transphosphorylate phot1D806N (Figure 10B). Moreover, coexpression of phot1-GFPD806N with phot1LOV2K resulted in its light-induced internalization from the plasma membrane (Figure 10C), indicating that LOV1 is not essential to stimulate phot1 endocytosis via intermolecular phosphorylation.
Phot1 Endocytosis in Response to Autophosphorylation Involves Clathrin
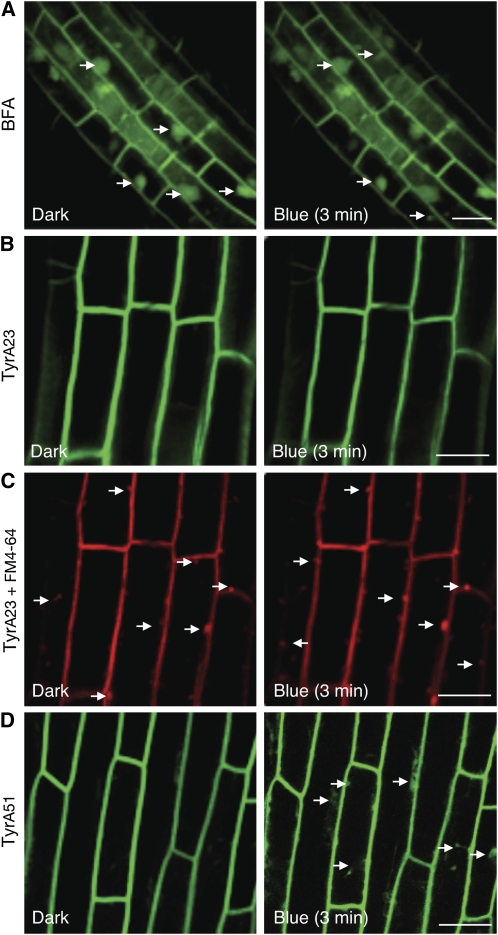
Clathrin plays a crucial role in endocytosis in animal cells as a scaffolding protein underlying membrane invagination and surrounding the primary endocytic vesicle (Mousavi et al., 2003). Recently, endocytosis via clathrin-mediated processes has been reported in plants (Dhonukshe et al., 2007; Leborgne-Castel et al., 2008). The Tyr analog Tyrphostin A23 has been shown to inhibit clathrin-mediated endocytosis in both animal (Banbury et al., 2003) and plant cells (Dhonukshe et al., 2007). Treatment with Tyrphositn A23 attenuated constitutive internalization of phot1-GFP2xLOV1 and phot1-GFPI608E in tobacco epidermal cells, while equivalent concentrations of Tyrphostin A51, an inactive structural analog, were ineffective (see Supplemental Figure 9 online). Brefeldin A (BFA), an inhibitor of endosomal trafficking, caused phot1-GFP2xLOV1 and phot1-GFPI608E to accumulate in intracellular endosomal aggregates, the so-called BFA compartments (see Supplemental Figure 9 online). Similar effects of BFA were observed in hypocotyl cells of dark-grown Arabidopsis seedlings expressing phot1-GFP driven by the native PHOT1 promoter (Figure 11A), demonstrating that phot1 is trafficked through the endosomal recycling pathway, as are other plasma membrane–associated proteins, including phot2 (Kong et al., 2006).
Figure 11.
Pharmacological Interference of phot1-GFP Localization in Arabidopsis.
(A) Confocal image of phot1-GFP in 3-d-old etiolated Arabidopsis hypcotyl cells treated with 50 μM BFA. BFA induces internalization and aggregation of phot1-GFP from the plasma membrane into BFA compartments. The white arrows indicate BFA aggregation. Bar = 20 μm.
(B) and (C) Treatment with 30 μM Tyrphostin A23 (Tyr23) inhibits blue light–induced phot1-GFP internalization in the hypocotyls of 3-d-old etiolated Arabidopsis seedlings (B) but does not abolish internalization of the endocytic marker FM4-64 (C) as indicated by the white arrows.
(D) Treatment with 30 μM Tyrphostin A51 (TyrA51), an inactive analog of Tyrphostin A23, has no effect on the blue light–dependent internalization of phot1-GFP from the plasma membrane of hypocotyl cells (as indicated by the white arrows).
[See online article for color version of this figure.]
Diffusion of phot1-GFP was also apparent at the plasma membrane of hypocotyl cells following fluorescence recovery after photobleaching (FRAP). Recovery was rapid, occurring within several minutes (see Supplemental Figure 10 online). Dynamic internalization of phot1-GFP coincided with FM4-64 localization in cells neighboring the area of FRAP, indicating that a fraction of phot1 enters the endocytic pathway (see Supplemental Figure 10 online). Again, Tyrphostin A23 attenuated the stimulatory effect of blue light on phot1 internalization (Figure 11B). Uptake of the fluorescent endocytic tracer FM4-64 was still detected (Figure 11C), in accordance with a significant fraction of vesicle trafficking in plant cells that is not dependent on clathrin (Dhonukshe et al., 2007) As observed in tobacco, Tyrphostin A51 did not interfere with phot1-GFP internalization in Arabidopsis seedlings (Figure 11D). More importantly, equivalent concentrations of Tyrphostin A23 did not inhibit phot1 kinase activity in insect cells (see Supplemental Figure 11 online), indicating that its effect on phot1 endocytic recruitment was not due to impaired autophosphorylation.
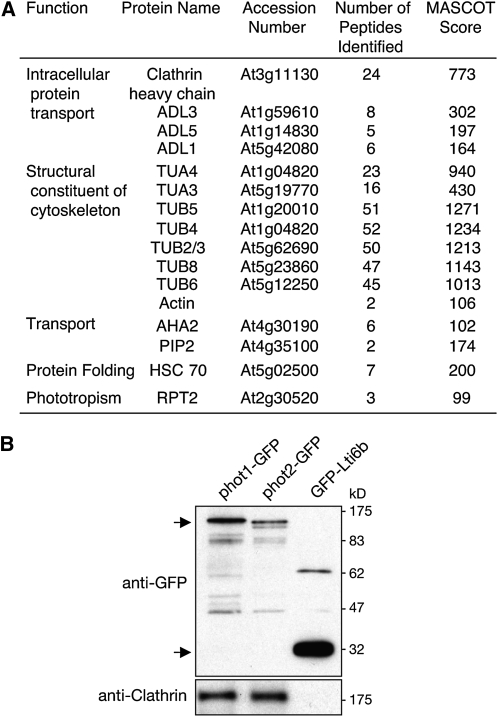
To further investigate the molecular mechanisms governing phot1 endocytosis, phot1-GFP was immunoprecipitated from Arabidopsis under irradiated conditions in an attempt to identify additional components of the photoreceptor complex by liquid chromatography–tandem mass spectrometry (LC-MS/MS; see Supplemental Data Set 1 online). The plasma membrane marker fusion GFP-Lti6b was used as control to distinguish proteins that specifically coimmunoprecipitate with phot1 (see Supplemental Data Set 2 online). Proteins known to interact directly with phot1, such as Root Phototropism 2 (Inada et al., 2004), were identified in our analysis demonstrating the veracity of the approach (Figure 12A). The phot1-interacting protein Nonphototropic Hypocotyl 3 (Motchoulski and Liscum, 1999) was not identified in our LC-MS/MS analysis of phot1-GFP immunoprecipitates, indicating that this approach, although extremely informative, carries with it some limitations. In addition to cytoskeleton components and proteins involved in ion/solute transport, clathrin was identified in phot1-GFP immunoprecipitations, the presence of which was verified using an antibody raised to plant clathrin heavy chain (Figure 12B). Clathrin was also detected in immunoprecipitations of phot2-GFP but not with GFP-Lti6b (Figure 12B). While further proteins associated with vesicle trafficking were prevalent in phot1-GFP immunoprecipitations, including tubulin (Rappoport et al., 2003), heat shock cognate protein 70 kD (HSC70; Lu et al., 2007), and Arabidopsis dynamin-like proteins, high molecular weight GTPases that compose a major component of the clathrin-mediated endocytic pathway in animal cells (Mousavi et al., 2003), coimmunoprecipitation of clathrin provided further support for its role in phot1 endocytic trafficking.
Figure 12.
Immunopurification of phot1-GFP from Arabidopsis.
(A) Identification of putative phot1-interacting proteins by LC-MS/MS of phot1-GFP immunoprecipitations. ADL, Arabidopsis dynamin-like; TUA, tubulin α-chain; TUB, tubulin β-chain; AHA2, Arabidopsis H+-ATPase 2; PIP3, plasma membrane intrinsic protein 3; HSC70-1, heat shock cognate 70 kD protein 1; and RPT2, root phototropism 2. TAIR accession numbers are given where MS data match a single accession.
(B) phot1-GFP and phot2-GFP associates with clathrin in vivo. Four-day-old light grown seedlings were dark adapted overnight and treated with 100 μmol m−2 s−1 of blue light for 30 min. phot1-GFP and phot2-GFP were immunoprecipitated from the microsomal membrane fraction with anti-GFP antibody linked to magnetic microbeads and subjected to immunoblot analysis with anti-GFP and anti-clathrin antibodies. The plasma membrane marker line, GFP-Lti6b, was used as a control for nonspecific interactions. Sizes of molecular weight marker proteins are indicated on the left in kilodaltons.
DISCUSSION
Autophosphorylation within the Kinase Activation Loop Stimulates phot1 Endocytic Recruitment
The findings gained from the domain-swapping strategy employed in this study provide numerous insights into the mode action of the blue light receptor phot1. A major discovery was constitutive nature of phot12xLOV1 that was used to gain new information regarding the mechanistic basis of phot1 endocytosis by light. Retrieval of receptor proteins from the plasma membrane typically involves their sorting either for recycling back to the plasma membrane or targeting to late endosomes and eventual degradation in the vacuole (Geldner and Robatzek, 2008). Prime examples include the Leu-rich repeat receptor-like kinases Brassinosteroid Insensitive 1 (BRI1) (Kinoshita et al., 2005) and Flagellin Sensing 2 (Robatzek et al., 2006). By comparison, phototropins offer a useful system to study receptor endocytosis as their retrieval from the plasma membrane stimulated by blue light (Sakamoto and Briggs, 2002). Both pharmacological interference (Figure 11) and coimmunoprecipitation (Figure 12) demonstrate a role for clathrin in phot1 endocytic recruitment. To date, links between clathrin and endocytosis in plants have been limited, despite the presence of homologs to key proteins of the mammalian endocytic machinery (Holstein, 2002). However, a role for clathrin in mediating constitutive endocytosis of PIN auxin efflux carriers has been demonstrated (Dhonukshe et al., 2007). Members of the PIN family are reported to play important roles in the redistribution of auxin underlying phototropic curvature (Friml et al., 2002; Blakeslee et al., 2004). phot1 may therefore influence clathrin-dependent trafficking of PIN family members to mediate changes in auxin transport. This possibility is particularly intriguing given that PIN proteins are known substrates for AGC kinase activity (Zourelidou et al., 2009).
Clathrin was also identified in immunoprecipitations of phot2-GFP (Figure 12), suggesting that light-driven endocytic recruitment of phot1 and phot2 is initiated by similar mechanisms. Indeed, phot1 localization, like phot2 (Kong et al., 2006), is sensitive to BFA (Figure 11A), demonstrating that their export to the plasma membrane is likely to pass though the trans-Golgi network. Recently, a novel function for clathrin in regulating protein kinase activity has been reported in animal cells. Mutation of a potential clathrin binding motif within G Protein–coupled Receptor Kinase 2 affects its ability to phosphorylate and promote internalization of ligand-stimulated receptors (Mangmool et al., 2006). Investigation of whether such a mode of action also occurs in Arabidopsis is certainly warranted since this motif is present in a wide range of kinases, including phot1, phot2, and the related AGC kinase PINOID (928LLQRD932, 840LLNRD844, and 370LLNKD374, respectively).
Subcellular localization analysis of constitutively active forms of phot1 showed that receptor autophosphorylation of Ser-851 triggers phot1 endocytic recruitment (Figures 7 and 8). Localization of FLG2 (Robatzek et al., 2006) and Somatic Embryogenesis Receptor Kinase 1 (Shah et al., 2002) are also strongly influenced by their phosphorylation status, but their endocytic recruitment is promoted by receptor dephosphorylation (Shah et al., 2002; Serrano et al., 2007). Phosphorylation of Ser-851 may, however, trigger phot1 internalization by stabilizing its association with the endocytic machinery in a manner similar to animal Tyr-kinase paradigms, such as epidermal growth factor receptor (Sorkin and Goh, 2009).
To date, the functional consequences of phot1 relocalization still remain poorly understood. Retention of phot1 at the plasma membrane within the elongating regions of Arabidopsis hypocotyls is proposed to account for the red light enhancement of phototropic sensitivity to low intensities of blue light (Han et al., 2008). Therefore, rapid internalization of phot1 from the plasma membrane may function to attenuate receptor signaling. Prolonged exposure to blue light results in phot1 degradation (Sakamoto and Briggs, 2002) and most likely reflects a long-term adaptation process. Although there is currently no evidence linking phot1 autophosphorylation to receptor turnover, signal attenuation as such may account for the problems encountered in generating lines expressing phot12xLOV1 and phot1I608E. Since autophosphorylation of Ser-851 has been shown to be a primary step in phot1 signaling (Inoue et al., 2008a), we cannot exclude the possibility that endocytic recruitment represents a mode of receptor signaling in addition to receptor desensitization. Endosomal signaling is common in animal cells and has recently been established in plants enabling efficient signal transfer to specific subcellular targets (Geldner and Robatzek, 2008). The phosphomimic mutation S851D increased phot1 internalization in tobacco epidermal cells (Figure 8E). phot1 carrying this mutation mediates sustained signaling for blue light–induced proton pumping and stomatal opening (Inoue et al., 2008a). Whether this increased activity correlates with an enhancement of phot1 endosomal trafficking now requires further investigation.
Intermolecular Autophosphorylation of Phot1 Is Not Dependent on LOV1
Although phot1 autophosphorylation has been monitored in vitro and in vivo for a number of plant species (Briggs et al., 2001), little effort has been made to address whether this occurs intra and/or intermolecularly. The use of epitope- and non-epitope-tagged forms of phot1 showed that intermolecular autophosphorylation occurs both in vitro and in vivo (Figures 9 and 10). Reciprocal transphosphorylation may function to quantitatively enhance phot1 signaling by maximizing receptor kinase affinity for target substrates in a manner similar to that proposed for BRI1 and BRI1-associated Receptor Kinase 1 (Wang et al., 2008). phot2 has also been shown to transphosphorylate phot1 at least in vitro (Cho et al., 2007), but the functional consequences of this interaction are not known. Transphosphorylation between phot1 molecules has been reported upon mixing membrane preparations from different plants species (Reymond et al., 1992). However, transphosphorylation from endogenous phot1 was not evident in our analysis with tobacco epidermal cells, as phot1-GFPD806N was retained at the plasma membrane even after prolonged light exposure (Figure 7B).
BiFC analysis demonstrated that full-length Arabidopsis phot1 undergoes light-dependent dimerization in vivo (Figure 9D), coinciding with a mechanism of light-driven intermolecular autophosphorylation. While LOV1 domains of phot1 form homodimers in vitro (Nakasako et al., 2004, 2008; Katsura et al., 2009), the ability of phot1LOV2K to mediate intermolecular phosphorylation in vitro and in vivo indicates that this mechanism can occur in the absence of LOV1 (Figure 10). The LOV2 domain of phot1 is known to dimerize in solution depending on its concentration (Nakasako et al., 2004; Katsura et al., 2009). Studies are now aimed at defining the site(s) of receptor dimerization and to assess whether intermolecular phosphorylation is essential for phototropin signaling.
Functional Conservation between LOV1 and LOV2
Unlike other LOV-photosensory proteins in plants and bacteria, phototropins are the only candidates identified that contain two LOV domains (Christie, 2007). LOV2 is the principle light-sensing domain (Christie et al., 2002; Cho et al., 2007; Sullivan et al., 2008) and is proposed to act as a repressor of phototropin kinase activity by interacting with the C-terminal kinase domain. The domain-swapping studies presented here concur with such a mode of receptor activation but highlight the necessity of the Jα-helix to create an efficient repression mechanism that is deactivated by light. phot12xLOV2 did not show a complete loss of activity, suggesting that an additional LOV2 domain is unable to fully counteract kinase regulation by the LOV2-Jα-helix photoswitch.
LOV domains from plant blue light receptors, including Arabidopsis phot1, have been shown to replace the activity of the LOV domain from the fungal photoreceptor White Collar 1 (Cheng et al., 2003), indicating that these photosensory modules are functionally conserved. Yet, our findings demonstrate that LOV1 is unable to replace the function of LOV2 as a dark-state repressor in the regulation of phot1 kinase activity (Figure 4A). The increased basal kinase activity observed for phot12xLOV1 could arise indirectly from disruption of the Jα-helix analogous to that described for the I608E mutation of phot1 (Harper et al., 2004). Although minor differences in secondary structure were observed for 2xLOV1 relative to the wild type by CD spectroscopy, these were not indicative of a loss in α-helicity (see Supplemental Figure 2 online). Alternatively, protein surface differences could account for the inability of LOV1 to replace LOV2 function. Recent structural modeling highlights the presence of a potential kinase-docking site within LOV2 that is absent from LOV1 (Tokutomi et al., 2007). This region conforms to the phosphorylation site consensus sequence of Arabidopsis phot1 (Sullivan et al., 2008) except that it contains an Ala residue in place of the phosphorylatable Ser. Phototropin kinase may therefore be regulated in a manner similar to cAMP-dependent protein kinase A and protein kinase C, whereby activation is mediated by displacement of a pseudosubstrate inhibitor peptide sequence from the catalytic kinase subunit.
Since their discovery over a decade ago, a major role for LOV2 in regulating phototropin action is well established (Christie, 2007). However, a biological function for LOV1 has been unclear. Functional analysis of the domain-swap proteins generated in this study was performed using the phot1 phot2 double mutant, which enabled us to monitor phot1 responsiveness for chloroplast accumulation over a range of light intensities. Together, these findings identify a photoactive function for LOV1 in arresting chloroplast accumulation at high light intensities (Figure 6). Such a role would serve to promote an efficient transition from accumulation to avoidance movement in excess light and consequently prevent a tug of war scenario arising from these two modes of relocation movement. Substitution of LOV1 by LOV2 not only complemented this inhibitory action but also increased its photosensitivity (Figure 6). We interpret these findings to indicate that LOV2 can replace LOV1 function given its higher quantum efficiency (Salomon et al., 2000; Kasahara et al., 2002). Given that inactivation of LOV1 does not affect phot1 responsiveness for phototropism and leaf expansion (Cho et al., 2007), the inhibitory action of LOV1 identified here may be limited to chloroplast accumulation movement. It is interesting to note that LOV1 domains of phot2 exhibit higher quantum efficiency compared with those of phot1 (Kasahara et al., 2002). This greater photosensitivity could enable phot2 to effectively change its mode of signaling to promote chloroplast avoidance movement.
Implications of Domain Swapping on Phot1 Reactivity
So far, much attention has focused on the study of individual LOV domains outside the context of the full-length photoreceptor protein. One exception is the fungal photoreceptor VIVID, which consists mostly of a LOV domain and plays an important role in mediating photoadaptation responses (Schwerdtfeger and Linden, 2003) and facilitating circadian clock entrainment (Elvin et al., 2005). The domain-swapping studies presented here support the hypothesis that cysteinyl-adduct decay for phot1 is affected by interactions between the LOV domains (Kasahara et al., 2002). Inactivation of LOV1 does not appear to impact these photochemical properties (Christie et al., 2002). Rather, the position of LOV1 and LOV2 (Figure 3C) in combination with the intervening linker sequence (Figure 3B) largely affects the recovery of phot1 back to its ground state. Such properties are absent in fusion proteins carrying identical LOV domains (Figure 2B), suggesting that interactions between LOV1 and LOV2 are required to bring about the slow recovery that is unique to phot1 (Kasahara et al., 2002). Dark recovery of phot2 is typically 10-fold faster in comparison to phot1 (Kasahara et al., 2002). Thus, a longer lifetime of receptor activation would contribute to the greater photosensitivity of phot1 over phot2 observed in vivo (Aihara et al., 2008). Structural analysis of extended phototropin fragments, including both LOV domains, will help determine the molecular basis underlying these photochemical differences.
Dark recovery of phototropins might be rate limited by protein conformational changes that differ between phot1 and phot2. The linker region of phot1 contains phosphorylation sites important for 14-3-3 binding (Inoue et al., 2008a; Sullivan et al., 2009), but the biological significance of this interaction remains to be determined. Phosphorylation and subsequent 14-3-3 binding could further modulate receptor activity, given the importance of the linker region in mediating potential interactions between LOV1 and LOV2 (Figure 3B). Nonetheless, a functional requirement for LOV1 in influencing photoreceptor reactivity together with a potential photoactive role in arresting chloroplast accumulation movement would provide evolutionary selective pressure to retain this photosensory module within the phototropin molecule. Recent interest in LOV photosensors has expanded toward the engineering of novel regulatory light switches to control target entities with specific applications (Wu et al., 2009). Our findings not only shed light on the mechanisms underlying phot1 reactivity, but also provide further opportunities to fine-tune the sensitivity of such protein-based light switches.
METHODS
Plant Material and Growth
Wild type (gl-1, ecotype Columbia) and the phot1 phot2 double mutant have been described previously (Kagawa et al., 2001) as have transgenic Arabidopsis thaliana expressing phot1-GFP (Sakamoto and Briggs, 2002), phot2-GFP (Kong et al., 2006), GFP-Lti6b (Cutler et al., 2000), phot1LOV1C234A (Cho et al., 2007), and phot1LOV2K (Sullivan et al., 2008). Seeds were planted on soil or surface sterilized and planted on half-strength Murashige and Skoog medium with 0.8% agar (w/v). After cold treatment (4°C) for 3 d, seedlings were grown in a controlled environment room (Fitotron; Weiss-Gallenkamp) unless otherwise stated. Fluence rates for all light sources were measured with a Li-250A and quantum sensor (LI-COR).
Domain Swapping and Mutagenesis
The cloning vector pBluescript (Stratagene) carrying the PHOT1 cDNA of Arabidopsis was used as template for all mutagenesis. Primer sequences can be found in Supplemental Table 1 online. Domain swapping was performed first by removing the LOV domain of interest by inverse PCR (primers LOV1 DEL_F/R and LOV2 DEL_F/R, respectively). Concomitantly, a unique SphI site was introduced within the PHOT1 coding region in which an alternative LOV domain was introduced. PCR fragments containing LOV1 (104 amino acid residues between 197 and 300, primers LOV1 SphI_F/R) and LOV2 (104 amino acid residues between 475 and 578, primers LOV2 SphI_F/R) were used to create chimeric PHOT1 sequences by insertion into the appropriate SphI site. Additional residues encoded by the SphI sites flanking the newly inserted LOV domain were eliminated using the QuikChange multi site-directed mutagenesis system (Stratagene) to create exact domain replacements within the PHOT1 sequence. The PHOT12xLOV1 coding sequence was used as a starting template to create PHOT1LOV2+1 using the same procedure. All constructs were confirmed by sequencing the entire PHOT1 coding region before subcloning into the appropriate expression vector.
Protein Expression in Escherichia coli and Purification
The coding region of PHOT1 containing both LOV domains (amino acid residues 180 to 628) was amplified by PCR and cloned into pCALn-EK (Stratagene) via EcoRI and NcoI to create N-terminal calmodulin binding peptide fusions (primers pCAL LOV1_F and pCAL LOV2_R). Primer sequences can be found in Supplemental Table 1 online. An identical cloning strategy was used to create constructs expressing LOV1 (amino acid residues 180 to 325, primers pCAL LOV1_F/R) and LOV2 (amino acid residues 448 to 628, primers pCAL LOV2 F/R) separately. Using inverse PCR, the linker region of phot1 (amino acid residues 305 to 475, primers Link DEL_F/R) was replaced by an SphI site encoding Ala and Cys. Proteins were expressed using E. coli Rosetta BL21(DE3)pLysS (Novagen) and purified by calmodulin affinity chromatography (Christie et al., 1999).
Spectroscopic Analysis
Absorption spectra were collected with a Shimadzu MultiSpec-1501 diode array spectrophotometer at room temperature. The optical path length was 0.5 cm, and a white-light camera strobe flash provided the excitation pulse. Fluorescence emission and excitation spectra were recorded using a Perkin-Elmer LS-55 luminescence spectrometer. Fluorescence excitation spectra were recorded by monitoring the emission at 520 nm. Fluorescence emission spectra were obtained using an excitation wavelength of 450 nm. Protein concentrations were determined by the Bradford protein assay (Bio-Rad). CD spectroscopy was performed as described previously (Jones et al., 2007). Flavin was released from the chromoproteins by protein denaturation (Jones et al., 2007).
Insect Cell Expression
Coding sequences of PHOT1, PHOT12xLOV1, PHOT12xLOV2, and PHOT1LOV2+1 were subcloned from the relevant pBluescript templates into the baculovirus transfer vector pAcHLT-A (BD Biosciences) via EcoRI. PCR was used to clone PHOT1 into pAcG3X (BD Biosciences) via BamHI and EcoRI (primers pAcG3X PHOT1_F/R) to create an N-terminal GST fusion. Primer sequences can be found in Supplemental Table 1 online. Single amino acid changes were introduced by the QuikChange site-directed mutagenesis system (Stratagene). The coding region encoding the LOV2-kinase region of phot1 (amino acid residues 448 to 996, primers pAcHLT-A LOV2K_F/R) was amplified by PCR and cloned into pAcHLT-A via EcoRI and SmaI. Recombinant baculovirus was titered by end-point dilution and used to infect Spodoptera frugiperda insect cells. Expression of recombinant phot1 was performed in darkness as described previously (Christie et al., 1998). For coinfections, viral stocks were mixed prior to inoculation (Cho et al., 2007).
In Vitro Phosphorylation Analysis
In vitro phosphorylation analysis was performed under dim red light as described previously (Christie et al., 2002). Plant microsomal membrane proteins were assayed in the presence of 1% Triton X-100 (Sullivan et al., 2008).
Transformation of Arabidopsis
35S-driven wild-type PHOT1, PHOT12xLOV1, and PHOT12xLOV2 transformation vectors were constructed using the modified binary expression vector pEZR(K)-LC as described previously (Christie et al., 2002). In each case, coding regions were subcloned from the relevant pBluescript templates via EcoRI. Constructs were transformed into the phot1 phot2 double mutant with Agrobacterium tumefaciens as described previously (Christie et al., 2002). Based on the segregation of kanamycin resistance, T2 lines containing a single transgene were selected, and homozygous T3 seeds were harvested for further analysis.
Measurement of Phototropic Curvature
Measurement of hypocotyl curvatures (1 and 20 μmol m−2 s−1) and light sources used were performed as described previously (Sullivan et al., 2008). Blue light (100 μmol m−2 s−1) was obtained using a slide projector (TLP-T50; Toshiba) and passing the light through a water filter and one layer of blue Plexiglas (Liscum and Briggs, 1995). In each case, blue light treatments were 470 ± 10 nm.
Leaf Positioning Measurements
Plants were grown for 7 d on soil under a 16/8-h 22/18°C light-dark cycle (50 μmol m−2 s−1) and transferred to continuous light (10 or 50 μmol m−2 s−1) for 5 d before representative plants were photographed (Sullivan et al., 2008).
Chloroplast Relocation
Chloroplast accumulation and avoidance were examined as described previously (Sullivan et al., 2008). Plants were grown for 4 weeks on soil under a 16/8-h 22/18°C light-dark cycle (50 μmol m−2 s−1). Rosette leaves were detached and placed on half-strength Murashige and Skoog agar plates and illuminated with blue light (1.5, 10, or 80 μmol m−2 s−1) or kept in darkness for 3 h. Chloroplast fluorescence in mesophyll cells was monitored by confocal scanning microscopy.
RNA Extraction and RT-PCR
Total RNA was extracted from leaf tissue using the RNeasy plant mini kit (Qiagen) according to the instructions of the supplier. cDNA synthesis and RT-PCR of PHOT1 and control ACTIN2 transcripts (primers PHOT1 trans_F/R and ACTIN trans_F/R, respectively) were performed as described (Cho et al., 2007; Kaiserli and Jenkins, 2007). Primer sequences can be found in Supplemental Table 1 online. For each transcript, amplification was assayed over a range of cycle numbers to select optimal conditions for visualization of the PCR product and quantification. Transcript levels in different RNA samples were compared using cycle numbers within the linear range of amplification.
Transient Expression in Nicotiana benthamiana by Agrobacterium Infiltration
Coding sequences of PHOT1, PHOT12xLOV1, and PHOT12xLOV2 and the LOV2-kinase region of phot1 were amplified by PCR from the relevant pBluescript templates and cloned into the binary expression vector pEZR(K)-LN via EcoRI and BamHI (primers pEZR PHOT1_F/R and pEZR LOV2K_F/R, respectively) to create C-terminal GFP fusions. Primer sequences can be found in Supplemental Table 1 online. Modified pEZR(K)-LC constructs were used to express nonfluorescently labeled forms of phot1 and phot1LOV2K (Sullivan et al., 2008). Amino acid changes were introduced by the QuikChange site-directed mutagenesis system (Stratagene). Agrobacterium transformed with the plasmid DNA of interest was inoculated in Luria-Bertani broth at 28°C and continuously shaken (220 rpm) until an OD550 of 1.0 was reached. Agrobacterium was pelleted by centrifugation at 2000g for 5 min, diluted to an OD550 of 0.4 with a solution containing 10 mM MgCl2, 10 mM MES-KOH, pH 5.6, and 200 mM acetosyringone, and incubated at room temperature for 2 h (Waadt et al., 2008). For coinfections, bacterial stocks were mixed prior to inoculation. Agrobacterium was infiltrated into N. benthamiana with a syringe through small incisions made at the lower leaf side. Plants were incubated at 30°C in white light for ∼2 d and then kept in darkness (noninductive conditions) for 24 h prior to imaging. In each case, representative images are shown that are indicative of at least three independent experiments in which similar results obtained (>90% of the transformed cells, n = 25 to 50). Protein extraction was performed by grinding tissue from the infiltrated leaf area in a solution containing 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 10% glycerol, 5 mM DTT, and 2% polyvinylpolypyrrolidone in the presence of a protease inhibitor cocktail mixture (Roche). The crude soluble protein fraction was separated by centrifugation at 16,000g at 4°C for 10 min. Protein concentration was estimated using the Bradford assay (Bio-Rad) and 30 μg separated by 7.5% SDS-PAGE.
Confocal Microscopy
Subcellular localization of GFP-tagged proteins in Arabidopsis seedlings and in leaves of N. benthamiana were visualized by a confocal laser scanning microscope (Zeiss LSM 510) under water with a ×40 objective under dark conditions. GFP was excited using an argon laser at 488 nm, and its emission was collected between 505 and 530 nm. To induce blue light activation of phot1, plant samples mounted on a slide were irradiated with an argon laser (488 nm, 20% power) for a period of 30 s after the initial scan. Subsequent images were taken after a period of 3 to 20 min as indicated. FM4-64 labeling and visualization was performed as described previously (Sutter et al., 2007). For FRAP measurements, 100% laser power was used with the following settings: one prebleach scan, bleaching for 12 s, and three postbleach scans every 60 s.
BiFC
The coding sequence of Arabidopsis PHOT1 was cloned by PCR via BamHI and SmaI (primers pSPYCE/SPYNE PHOT1_F/R) into the binary vectors pSPYNE-35S and pSPYCE-35S (Walter et al., 2004) to create protein fusions with the N-terminal and C-terminal halves of YFP at the C terminus of phot1 (phot1-YN and phot1-YC, respectively). Primer sequences can be found in Supplemental Table 1 online. Coinfiltration of Agrobacterium containing both BiFC constructs was performed an OD600 of 0.4:0.4. Epidermal cell layers of tobacco leaves were assayed for fluorescence 2 to 3 d after infiltration. YFP was excited with an argon laser at 488 nm and emission detected between 525 and 540 nm.
Inhibitor Treatments
BFA (Sigma-Aldrich), Tyrphostin A23 (Novagen), and Tyrphostin A51 (Novagen) were used from DMSO stock solutions (50 mM for BFA and 30 mM for Tyrphostin). Etiolated seedlings were infiltrated under vacuum with BFA and Tyrphostin A23 or A51 and kept in water containing the inhibitors for 1 h (BFA) or 30 min (Tyr A23 and A51) prior to microscopy analysis and blue light irradiation.
Protein Extraction and Immunoprecipitation of GFP-Tagged Proteins
Plant proteins and microsomal membranes were extracted as described previously (Sullivan et al., 2008). GFP immunoprecipitations were performed using the μMACS GFP isolation kit (Miltenyi Biotech). Immunoprecipitated proteins were eluted in 0.1 M triethylamine, pH 11.8/0.1% Triton X-100 and neutralized with 3 μL of 1 M MES, pH 3.
Antibodies and Immunoblot Analysis
After separation by SDS-PAGE, proteins were transferred onto nitrocellulose membrane (Bio-Rad) by electroblotting. Proteins were detected by immunoblot analysis with anti-GFP monoclonal antibody (BD Biosciences), anti-clathrin heavy chain monoclonal antibody (Abcam), anti-phot1 antibody (Cho et al., 2007), or anti-UGPase antibody (Sullivan et al., 2008). Blots were developed with horseradish peroxidase–linked secondary antibodies and Immobilon Western chemiluminescence horseradish peroxidase substrate (Millipore).
Identification of Proteins by LC-MS/MS
Elutions from GFP immunoprecipitations were separated ∼1 cm by SDS-PAGE to remove Triton X-100 from the samples. The whole gel lane was excised and analyzed by mass spectrometry by the Fingerprints Proteomics Facility (University of Dundee, Dundee, UK). Proteins were identified using the National Center for Biotechnology Information database with the MASCOT search engine (www.matrixscience.com). Arabidopsis was used as the taxonomic restriction. The following search parameters were applied: trypsin as the cleaving enzyme, peptide mass tolerance 1.5 D, fragment mass tolerance 0.5 D, and one missed cleavage allowed. Carbamidomethyl of Cys was set as a fixed modification, and variable modifications were Met oxidation and phosphorylation of Ser, Threonine and tyrosine. The significance threshold was P < 0.01 and proteins required at least one bold red peptide. For protein identification at least two peptide matches were required.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number NM 114447 (PHOT1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fluorescence Properties of 2xLOV1 and 2xLOV2.
Supplemental Figure 2. Dark State CD Spectra of LOV1+2, 2xLOV1, and 2xLOV2, Measured in the Far-UV Region (190 to 260 nm).
Supplemental Figure 3. Dark-Recovery Kinetics for Individual LOV1 and LOV2 Domains of Arabidopsis phot1.
Supplemental Figure 4. Kinetics of Light-Dependent Autophosphorylation for Wild-Type phot1 and phot12xLOV2 in Extracts from Insect Cells.
Supplemental Figure 5. RT-PCR Analysis of PHOT1 Transcripts.
Supplemental Figure 6. Phototropism Fluence Rate Response of 3-d-Old Etiolated Wild-Type (gl-1) and phot1 phot2 Mutant Seedlings (p1p2).
Supplemental Figure 7. Immunoblot Analysis of Nicotiana benthamiana Expressing phot1-GFP Fusions.
Supplemental Figure 8. Transphosphorylation between phot1 Molecules.
Supplemental Figure 9. Pharmacological Interference of phot1-GFP Localization in N. benthamiana.
Supplemental Figure 10. FRAP Analysis of phot1-GFP in 3-d-Old Etiolated Arabidopsis.
Supplemental Figure 11. Effect of Tyrphostin A23 (Tyr A23) on phot1 Kinase Activity in Protein Extracts from Insect Cells.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. MASCOT Search Results of Proteins Identified in phot1-GFP Immunoprecipitations by LC-MS/MS.
Supplemental Data Set 2. MASCOT Search Results of Proteins Identified in GFP-Lti6b Immunoprecipitations by LC-MS/MS.
Supplemental Movie 1. Phot1-GFP Relocalization in Response to Blue Light.
Supplementary Material
Acknowledgments
We thank Winslow Briggs and Mike Blatt for critically reading the manuscript and for their continual encouragement. We also thank Gareth Jenkins for helpful discussions. In addition, we thank Margaret Ennis for excellent technical support, Sharon Kelly for help with CD spectroscopy, Tong-Seung Tseng and Winslow Briggs for providing the phot1LOV1C234A transgenic line, Akira Nagatani for providing the phot2-GFP transgenic line, and Annegret Honsbein for supplying and providing help with BiFC vectors. This work was supported by UK Biotechnology and Biological Sciences Research Council (BBSRC) Grants C17551 and BB/C000366/1 (to J.M.C.), by the Gatsby Charitable Foundation, and by a BBSRC Wain International Travel Fellowship (to S.S). J.M.C. is extremely grateful to the Royal Society for the award of a University Research Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: John M. Christie (j.christie@bio.gla.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Aihara, Y., Tabata, R., Suzuki, T., Shimazaki, K., and Nagatani, A. (2008). Molecular basis of the functional specificities of phototropin 1 and 2. Plant J. 56 364–375. [DOI] [PubMed] [Google Scholar]
- Bae, G., and Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59 281–311. [DOI] [PubMed] [Google Scholar]
- Banbury, D.N., Oakley, J.D., Sessions, R.B., and Banting, G. (2003). Tyrphostin A23 inhibits internalization of the transferring receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278 12022–12028. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., Bandyopadhyay, A., Peer, W.A., Makam, S.N., and Murphy, A.S. (2004). Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 134 28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., Christie, J.M., and Salomon, M. (2001). Phototropins: A new family of flavin-binding blue light receptors in plants. Antioxid. Redox Signal. 3 775–788. [DOI] [PubMed] [Google Scholar]
- Cheng, P., He, Q., Yang, Y., Wang, L., and Liu, Y. (2003). Functional conservation of light, oxygen, or voltage domains in light sensing. Proc. Natl. Acad. Sci. USA 100 5938–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.Y., Tseng, T.-S., Kaiserli, E., Sullivan, S., Christie, J.M., and Briggs, W.R. (2007). Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol. 143 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J.M. (2007). Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58 21–45. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282 1698–1701. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Salomon, M., Nozue, K., Wada, M., and Briggs, W.R. (1999). LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96 8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J.M., Swartz, T.E., Bogomolni, R.A., and Briggs, W.R. (2002). Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32 205–219. [DOI] [PubMed] [Google Scholar]
- Corchnoy, S.B., Swartz, T.E., Lewism, J.W., Szundi, I., Briggs, W.R., and Bogomolni, R.A. (2003). Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem. 278 724–731. [DOI] [PubMed] [Google Scholar]
- Crosson, S., and Moffat, K. (2001). Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98 2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffiths, J.S., and Somerville, C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy, E., and Fankhauser, C. (2009). Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 12 69–74. [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P., Aniento, F., Hwang, I., Robinson, D.G., Mravec, J., Stierhof, Y.D., and Friml, J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17 217–219. [DOI] [PubMed] [Google Scholar]
- Elvin, M., Loros, J.J., Dunlap, J.C., and Heintzen, C. (2005). The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes Dev. 19 2593–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta, K.M., and Maruhnich, S.A. (2007). Green light: A signal to slow down or stop. J. Exp. Bot. 58 3099–3111. [DOI] [PubMed] [Google Scholar]
- Folta, K.M., and Spalding, E.P. (2001). Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 26 471–478. [DOI] [PubMed] [Google Scholar]
- Friml, J., Wiśniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Geldner, N., and Robatzek, S. (2008). Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 147 1565–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, I.S., Tseng, T.-S., Eisinger, W., and Briggs, W.R. (2008). Phytochrome A regulates the intracellular distribution of phototropin 1-green fluorescent protein in Arabidopsis thaliana. Plant Cell 20 2835–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, S.M., Christie, J.M., and Gardner, K.H. (2004). Disruption of the LOV-J alpha helix interaction activates phototropin kinase activity. Biochemistry 43 16184–16192. [DOI] [PubMed] [Google Scholar]
- Harper, S.M., Neil, L.C., and Gardner, K.H. (2003). Structural basis of a phototropin light switch. Science 301 1541–1544. [DOI] [PubMed] [Google Scholar]
- Holstein, S.E. (2002). Clathrin and plant endocytosis. Traffic 3 614–620. [DOI] [PubMed] [Google Scholar]
- Huang, K., and Beck, C.F. (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 100 6269–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada, S., Ohgishi, M., Mayama, T., Okada, K., and Sakai, T. (2004). RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S., Kinoshita, T., Matsumoto, M., Nakayama, K.I., Doi, M., and Shimazaki, K. (2008. a). Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. USA 105 5626–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S., Kinoshita, T., Takemiya, A., Doi, M., and Shimazaki, K. (2008. b). Leaf positioning of Arabidopsis in response to blue light. Mol. Plant 1 1–12. [DOI] [PubMed] [Google Scholar]
- Jenkins, G.I. (2009). Signal transduction in response to UV-B radiation. Annu. Rev. Plant Biol. 60 407–431. [DOI] [PubMed] [Google Scholar]
- Jones, M.A., Feeney, K.A., Kelly, S.M., and Christie, J.M. (2007). Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J. Biol. Chem. 282 6405–6414. [DOI] [PubMed] [Google Scholar]
- Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., Tabata, S., Okada, K., and Wada, M. (2001). Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291 2138–2141. [DOI] [PubMed] [Google Scholar]
- Kaiserli, E., and Jenkins, G.I. (2007). UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19 2662–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, M., et al. (2002). Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 129 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura, H., Zikihara, K., Okajima, K., Yoshihara, S., and Tokutomi, S. (2009). Oligomeric structure of LOV domains in Arabidopsis phototropin. FEBS Lett. 583 526–530. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Caño-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., and Chory, J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433 167–171. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M., and Shimazaki, K. (2001). Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414 656–660. [DOI] [PubMed] [Google Scholar]
- Kong, S.G., and Nagatani, A. (2008). Where and how does phototropin transduce light signals within the cell? Plant Signal. Behav. 3 275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, S.G., Suzuki, T., Tamura, K., Mochizuki, N., Hara-Nishimura, I., and Nagatani, A. (2006). Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 45 994–1005. [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel, N., Lherminier, J., Der, C., Fromentin, J., Houot, V., and Simon-Plas, F. (2008). The plant defense elicitor cryptogein stimulates clathrin-mediated endocytosis correlated with reactive oxygen species production in bright yellow-2 tobacco cells. Plant Physiol. 146 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q.H., and Yang, H.Q. (2007). Cryptochrome signaling in plants. Photochem. Photobiol. 83 94–101. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, A.J.H., Sun, T.-X., Matsuzaki, T., Yi, H.-X., Eswara, J., Bouley, R., McKee, M., and Brown, D. (2007). Heat shock protein 70 interacts with Aquaporin-2 and regulates its trafficking. J. Biol. Chem. 282 28721–28732. [DOI] [PubMed] [Google Scholar]
- Matsuoka, D., Iwata, T., Zikihara, K., Kandori, H., and Tokutomi, S. (2007). Primary processes during the light-signal transduction of phototropin. Photochem. Photobiol. 83 122–130. [DOI] [PubMed] [Google Scholar]
- Matsuoka, D., and Tokutomi, S. (2005). Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc. Natl. Acad. Sci. USA 102 13337–13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangmool, S., Haga, T., Kobayashi, H., Kim, K.M., Nakata, H., Nishida, M., and Kurose, H. (2006). Clathrin required for phosphorylation and internalization of beta2-adrenergic receptor by G protein-coupled receptor kinase 2 (GRK2). J. Biol. Chem. 281 31940–31949. [DOI] [PubMed] [Google Scholar]
- Motchoulski, A., and Liscum, E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286 961–964. [DOI] [PubMed] [Google Scholar]
- Mousavi, S.A., Malerød, L., Berg, T., and Kjeken, R. (2003). Clathrin-dependent endocytosis. Biochem. J. 377 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasako, M., Iwata, T., Matsuoka, D., and Tokutomi, S. (2004). Light-induced structural changes of LOV domain-containing polypeptides from Arabidopsis phototropin 1 and 2 studied by small-angle X-ray scattering. Biochemistry 43 14881–14890. [DOI] [PubMed] [Google Scholar]
- Nakasako, M., Zikihara, K., Matsuoka, D., Katsura, H., and Tokutomi, S. (2008). Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J. Mol. Biol. 381 718–733. [DOI] [PubMed] [Google Scholar]
- Rappoport, J.Z., Taha, B.W., and Simon, S.M. (2003). Movement of plasma-membrane-associated clathrin spots along the microtubule cytoskeleton. Traffic 4 460–467. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Short, T.W., and Briggs, W.R. (1992). Blue light activates a specific protein kinase in higher plants. Plant Physiol. 100 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., and Briggs, W.R. (2002). Cellular and subcellular localization of phototropin 1. Plant Cell 14 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon, M., Christie, J.M., Knieb, E., Lempert, U., and Briggs, W.R. (2000). Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39 9401–9410. [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger, C., and Linden, H. (2003). VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, M., Robatzek, S., Torres, M., Kombrink, E., Somssich, I.E., Robinson, M., and Schulze-Lefert, P. (2007). Chemical interference of pathogen-associated molecular pattern-triggered immune responses in Arabidopsis reveals a potential role for fatty-acid synthase type II complex-derived lipid signals. J. Biol. Chem. 282 6803–6811. [DOI] [PubMed] [Google Scholar]
- Shah, K., Russinova, E., Gadella, T.W., Jr., Willemse, J., and De Vries, S.C. (2002). The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 16 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin, A., and Goh, L.K. (2009). Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 315 683–696. [DOI] [PubMed] [Google Scholar]
- Sullivan, S., Thomson, C.E., Kaiserli, E., and Christie, J.M. (2009). Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett. 583 2187–2193. [DOI] [PubMed] [Google Scholar]
- Sullivan, S., Thomson, C.E., Lamont, D.J., Jones, M.A., and Christie, J.M. (2008). In vivo phosphorylation site mapping and functional characterization of Arabidopsis phototopin 1. Mol. Plant 1 178–194. [DOI] [PubMed] [Google Scholar]
- Sutter, J.U., Sieben, C., Hartel, A., Eisenach, C., Thiel, G., and Blatt, M.R. (2007). Abscisic acid triggers the endocytosis of the arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17 1396–1402. [DOI] [PubMed] [Google Scholar]
- Swartz, T.E., Corchnoy, S.B., Christie, J.M., Lewis, J.W., Szundi, I., Briggs, W.R., and Bogomolni, R.A. (2001). The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276 36493–36500. [DOI] [PubMed] [Google Scholar]
- Takemiya, A., Inoue, S., Doi, M., Kinoshita, T., and Shimazaki, K. (2005). Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutomi, S., Matsuoka, D., and Zikihara, K. (2007). Molecular structure and regulation of phototropin kinase by blue light. Biochim. Biophys. Acta 1784 133–142. [DOI] [PubMed] [Google Scholar]
- Tsuboi, H., Suetsugu, N., Kawai-Toyooka, H., and Wada, M. (2007). Phototropins and Neochrome1 mediate nuclear movement in the fern Adiantum capillus-veneris. Plant Cell Physiol. 48 892–896. [DOI] [PubMed] [Google Scholar]
- Waadt, R., Schmidt, L.K., Lohse, M., Hashimoto, K., Bock, R., and Kudla, J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56 505–516. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wan, Y.-L., Eisinger, W., Ehrhardt, D., Kubitscheck, U., Baluska, F., and Briggs, W.R. (2008). The subcellular localization and blue-Light-induced movement of phototropin 1-GFP in etiolated seedlings of Arabidopsis thaliana. Mol. Plant 1 103–117. [DOI] [PubMed] [Google Scholar]
- Wang, X., Kota, U., He, K., Blackburn, K., Li, J., Goshe, M.B., Huber, S.C., and Clouse, S.D. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15 220–235. [DOI] [PubMed] [Google Scholar]
- Wu, Y.I., Frey, D., Lungu, O.I., Jaehrig, A., Schlichting, I., Kuhlman, B., and Hahn, K.M. (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou, M., Müller, I., Willige, B.C., Nill, C., Jikumaru, Y., Li, H., and Schwechheimer, C. (2009). The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136 627–636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.