Abstract
Caspase-3, 6 and 7 cleave many proteins at specific sites to induce apoptosis. Their recognition of the P5 position in substrates has been investigated by kinetics, modeling and crystallography. Caspase-3 and -6 recognize P5 in pentapeptides as shown by enzyme activity data and interactions observed in the crystal structure of caspase-3/LDESD and in a model for caspase-6. In caspase-3 the P5 main-chain was anchored by interactions with Ser209 in loop-3 and the P5 Leu side-chain interacted with Phe250 and Phe252 in loop-4 consistent with 50% increased hydrolysis of LDEVD relative to DEVD. Caspase-6 formed similar interactions and showed a preference for polar P5 in QDEVD likely due to interactions with polar Lys265 and hydrophobic Phe263 in loop-4. Caspase-7 exhibited no preference for P5 residue in agreement with the absence of P5 interactions in the caspase-7/LDESD crystal structure. Initiator caspase-8, with Pro in the P5-anchoring position and no loop-4, had only 20% activity on tested pentapeptides relative to DEVD. Therefore, caspases-3 and -6 bind P5 using critical loop-3 anchoring Ser/Thr and loop-4 side-chain interactions, while caspase-7 and -8 lack P5-binding residues.
Keywords: Enzyme catalysis, Cysteine protease, Protein recognition, Apoptosis, Induced fit
Introduction
Caspase family members induce apoptosis by cleavage of diverse protein substrates, although many of the signaling pathways and potential substrates are poorly characterized. Altered caspase-mediated apoptosis is associated with many diseases. Increased caspase activity and apoptosis are observed in stroke and neurodegenerative diseases like Alzheimer’s, Parkinson’s and Huntington’s disease [1–4]. Apoptosis has been implicated in the death of cardiomyocytes during acute myocardial infarction, as well as for the progressive loss of surviving cells in failing hearts [5,6]. In contrast, reduced caspase activity is associated with cancer, autoimmune diseases, and viral infections [7–10]. Pan-caspase inhibitors are in clinical trials for acute myocardial infarction and liver transplants [11,12]. Knowledge of the substrate specificity of caspases is vital for identifying signaling pathways leading to apoptosis in normal cells and disease states and for the rational design of selective therapeutic agents to control cell death.
The caspase family comprises cysteine proteases that cleave the peptide bond after an aspartic acid in their protein substrates. They are subdivided into inflammatory and apoptotic caspases based on their function and prodomain structure. The mammalian apoptotic caspases include initiators and effectors of apoptosis. The initiator caspases-2, -8, -9, -10 and -12 activate the effector, or executioner, caspases-3, -6, and -7 by cleaving the procaspase to form the active enzyme [13,14]. The active caspases-3, -6, and -7 hydrolyze many protein substrates in the signaling pathways leading to apoptosis [15]. The crystal structures have been solved for the apoptotic caspase-2, -3, -7, -8, -9, in their complexes with protein inhibitors, peptide analogs and non-peptide inhibitors [16–18]. The enzymatically active caspase is a heterotetramer of two large and two small subunits formed by cleavage of a procaspase dimer. Peptide substrates or inhibitors bind in two similar active sites formed between the large and small subunits of the two heterodimers.
The substrate specificity of apoptotic caspases has been defined by studies of peptides. Most studies have focused on tetrapeptide substrates of residues P4-P1, where cleavage occurs after the P1 residue. Caspase-2, -3 and -7 prefer the tetrapeptide DEXD, while caspase-6, -8 and -9 recognize L/VEXD [13,19]. The effector caspases-3 and -7 are very similar; they share 56% sequence identity and preferentially recognize DEVD. Recently, the substrate recognition sequence was extended to P5 for caspase-2 and -3. The P5 position was determined to be essential for recognition by caspase-2 [17]. Similarly, caspase-3 was shown to preferentially recognize pentapeptides with hydrophobic amino acids at P5 over tetrapeptides [20]. Caspase-6 shares 37 and 41% sequence identity with caspase-7 and -3, respectively. However, caspase-6 differs from the other two caspases in preferring the substrate sequence LEHD rather than DEVD. The possibility of recognizing the P5 residue in substrates has not been explored for caspase-6. Moreover, many cellular protein substrates are cleaved at non-canonical sequences and it is often not known which caspase acts on a particular protein [15,21].
In order to provide additional specificity data for identification of the protein substrates and signaling pathways of each executioner caspase we have further investigated caspase recognition of the P5 position in peptide substrates. The specificity of the executioner caspases was investigated for polar or hydrophobic P5 residues using the colorimetric substrates Ac-QDEVD-pNA and Ac-LDEVD-pNA, and tetrapeptide Ac-DEVD-pNA as a control, where Ac is the acetyl group and pNA is p-nitroanilide. Crystal structures were solved of caspase-3 and -7 in their complexes with the pentapeptide Ac-LDESD-CHO, and molecular models were constructed for the caspase-6 complex. The relative activity on the substrates agrees with the caspase residues forming the S5 site and their interactions with the P5 position.
Materials and methods
Reagents
Restriction enzymes were purchased from both New England BioLabs, Inc. and Stratagene. PCR SuperMix was purchased from Stratagene. All media were obtained from Difco or Gibco BRL. SDS–PAGE standards were purchased from Pharmacia. Other chemicals were obtained from Sigma. The colorimetric substrates were Ac-DEVD-pNA from Biomol International, PA, Ac-QDEVD-pNA and Ac-LDEVD-pNA from EZBiolab, IN, where Ac is the acetyl group and pNA is p-nitroanilide. The inhibitor Ac-LDESD-CHO was obtained from EMD Chemicals, Inc., NJ.
Vector construction for caspase-6
Human caspase-6 (EC 3.4.22.59) was engineered as a truncated, prodomain-less, construct MetSerPhe25-Asn293 (similar to [22]) of the caspase-6 isoform alpha. The pOTB7 vector (Invitrogen), containing full-length cDNA of caspase-6 (NCBI database ID: NM_001226, NCBI accession BC000305) was used as a template and amplified by PCR using the primers: 5′-GAATTCATGTCGTTCTATAAAAGAGAA-3′ and 3′-CTCGAGATTAGATTTTGGAAAGAAATG-5′ to introduce EcoR1 and XhoI restriction sites, respectively. The PCR transcript was subcloned into the TOPO XL vector (Invitrogen) through the blunt ends. The amplified DNA was excised with EcoRI/XhoI endonucleases and the caspase-6 construct was recloned into the bacterial expression vector pET23b as an EcoRI-to-XhoI fragment by ligation reaction and expressed in Escherichia coli BL21DE3 (Stratagene Inc) cells with a C-terminal (His)6 tag. The sequence of the construct was confirmed by the dideoxynucleotide chain-termination method using primers to T7 promoter and terminator.
Expression and purification of caspase-6
The expression construct was transformed into E. coli BL21DE3 cells (Stratagene Inc) according to the manufacture’s protocol and the cells were grown on an LB/ampicillin plate overnight. Next day, one colony was transferred into 10 ml LB medium containing ampicillin (100 mg/ml) and the culture was grown overnight at 37°C with shaking. The 1000 ml of LB medium containing ampicillin was inoculated with the overnight culture and incubated at 37°C with shaking until OD600 = 0.6 – 0.8 (about 3–4 h) followed by induction with 2mM isopropyl-β, D-thiogalactopyranoside (final concentration) for four hours at 30°C. Cells were collected by centrifugation at 5,000g for 20 min, resuspended in 40 ml working buffer (50 mM Tris–HCl, pH 7.4, 250 mM NaCl) and lysed by ultra-sonication without either protease inhibitors or lysozyme. The cellular debris was removed by centrifugation (20,000g, 25 min). The supernatant was examined for the presence of the caspase-6 by SDS–PAGE and stored at −20°C. The protein was purified from the soluble cell fraction using Ni-NTA chromatography (GE-Healthcare) according to the manufacturer’s standard protocol. The protein fractions eluted in 0.35–0.5 M imidazole in working buffer were collected, dialyzed against working buffer and concentrated to approximately 3 mg/ml using Centricon-10 ultrafiltration (Amicon Inc). During the concentration step the caspase-6 processed (cleaved) itself into two subunits to become an active protease. The final purification step was done on Sephacryl S-100HR column (Pharmacia). The purity of caspase-6 was above 95% as estimated from Coomassie blue stained 10–20% (w/v) polyacrylamide gels, which were done in denaturing, reducing conditions. The purified protein was equilibrated with 100 mM HEPES, pH 7.25, containing 100 mM NaCl, 0.1% CHAPS, 10% sucrose, using Centricon-5 ultrafiltration, and stored at −20°C. The final yield of catalytically active caspase-6 was about 1.5 mg per liter of bacterial culture.
Expression and purification of caspase-3, -7 and -8
Recombinant human caspase-3, -7 and -8 were expressed in E. coli and purified as described previously [20,23].
Determination of activity
Measurements of the caspase activity were performed with the colorimetric assay as described previously [20]. Caspase-3, -7, and -8 were preincubated in assay buffer (50 mM HEPES, 100 mM NaCl, 0.1% CHAPS, 10% glycerol, 1 mM EDTA and 10 mM DTT, pH7.5) at room temperature for 5 mins prior to the addition of substrate at different concentrations. The assay buffer for caspase-6 was 50 mM HEPES, pH 7.25, containing 100 mM NaCl, 10% sucrose, 0.1% CHAPS. The p-nitroanilide released by the substrate hydrolysis was measured at a wavelength of 405 nm using a Polarstar Optima microplate reader (BMG Labtechnologies, NC). All assays were performed in triplicate and the mean values were plotted. Kinetic constants were calculated by direct fits of the data, obtained at less than 20% substrate proteolysis, to the Michaelis–Menten equation using non-linear regression analysis in SigmaPlot 9.0 (SPSS Inc). The catalytic constant kcat of caspase-3 substrates Ac-DEVD-pNA, Ac-LDEVD-pNA and Ac-QDEVD-pNA were determined by using the equation kcat = Vmax/[E], where [E] values were measured by active site titration during Ki determination as described below. The same methods were used for other caspases tested in this study. Caspase-3 substrate analog inhibitors Ac-DEVD-Cho, Ac-QDEVD-Cho, and Ac- LDEVD-Cho form covalent bonds between the aldehyde (-CHO) group of the inhibitor and the mercapto (-SH) group of Cys163 on the protein. According to the vendor’s instructions, the binding of these inhibitors is reversible, although it is strong. Therefore, they were treated as reversible tight-binding inhibitors. For the measurement of inhibition constant Ki, caspase-3 was incubated with its substrate analog inhibitors in reaction buffer at room temperature for 30 min. Then, substrate was added and the reaction velocity was calculated according to substrate cleavage. The inhibition constants of each inhibitor were determined by a dose-response curve described by the equation:
where [E], [S] and IC50 correspond to active enzyme concentration, substrate concentration and the inhibitor concentration needed to suppress half of the enzyme activity, respectively.
Crystallization, X-ray data collection and refinement of caspase-3 and -7 with LDESD
The crystals of caspase-7 complexed with inhibitor Ac-LDESD-CHO were grown by hanging drop vapor diffusion method at room temperature using 1:15 molar ratio of the enzyme (6 mg/ml) to inhibitor. The crystals were obtained with a well solution of 0.1M sodium citrate buffer (pH 5.0–5.5) and 2.1 M sodium formate. Caspase-3 was incubated at room temperature with Ac-LDESD-CHO at 10-fold molar excess. Crystals were grown by the hanging-drop vapor-diffusion method using a well solution of 100 mM sodium citrate (pH 7.0), 5% glycerol, 10 mM dithiothreitol, and 14–16% (w/v) PEG 6000. Crystals grew at room temperature within 24 h. The crystals were soaked in the mother liquor with 22% (caspase-7/LDESD) and 20% (caspase-3/LDESD) glycerol as cryoprotectant for ~1 min and immediately frozen in liquid nitrogen. Diffraction data were collected at 100°K on beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory. The data were integrated and scaled with HKL2000 [24].
The crystal structures were solved by molecular replacement using PHASER [25] in the CCP4i suite of programs [26] using the starting models of caspase-7 complexed with Ac-DEVD-CHO (PDB code 1F1J) [16] and caspase-3 complexed with Ac-VDVAD-CHO (PDB code 2H65). The structures were subjected to several rounds of refinement in CNS or SHELX97 [27] for caspase-7 and -3, respectively. The molecular graphics programs O 9.0 [28] and Coot 0.33 [29] were used in model rebuilding. The inhibitor was fitted into unambiguous electron density. The solvent molecules were inserted at stereochemically reasonable positions based on the peak height of the 2Fo-Fc and Fo-Fc electron density maps, hydrogen bond interactions and interatomic distances. The geometry of the refined structures was validated according to the Ramachandran plot criteria of [30]. Sequence alignment was performed by using the ClustalW server [31] and GeneDoc (http://www.cris.com/2Ketchup/genedoc.shtml) used to generate the figure. Molecular figures were prepared with Molscript, Raster3D [32] and PyMol (http://www.pymol.org).
The crystal structures have been deposited in the Protein Data Bank with accession codes 3EDQ for caspase-3/LDESD, and 3EDR for caspase-7/LDESD.
Construction of molecular model of caspase-6
The model for caspase-6 in complex with the peptide aldehyde Ac-LDESD-CHO was constructed by homology modeling from the new high resolution crystal structure of caspase-3/LDESD. Sequence alignment for caspase-6 was performed with a profile-profile method using full dynamics programming and the Kullback entropy [33] with frequency profiles obtained from Psi-BLAST [34]. The program AMMP [35,36] was used with the latest version of the potential set, which was reparameterized to minimize RMS deviation when applied to high resolution crystal structures (parameter set atoms.tuna). All hydrogen atoms were included in this potential. The adduct of the catalytic cysteine 163 with P1 Asp, and the N-acetyl Leu at P5 were explicitly modeled with charges derived with the method of moments approximation [37]. Side chains were built using the analytic atom building routines in AMMP [38] followed by annealing over local dominating sets, minimization and conjugate gradients.
Results and Discussion
Predictions for caspase recognition of P5 in substrates
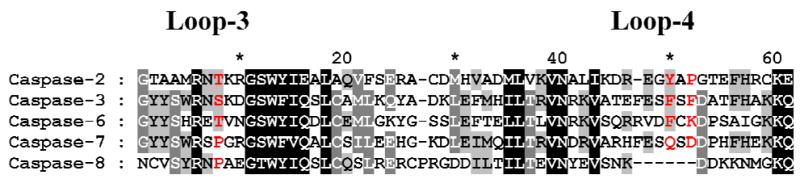
The P5 specificity of caspase-6 was predicted from comparison of the sequences and structures of caspases. Molecular models were generated for human caspase-6 due to the absence of diffraction quality crystals. Alignment of the amino acid sequences (Fig. 1) suggested that caspase-6 shared the P5 binding loop observed in the crystal structures of caspase-3 [20,23] and caspase-2 with pentapeptides [17]. The hydrophobic P5 residue interacts with Phe250 and Phe252 in the S5 site of caspase-3 and with Tyr273 and Pro275 at the structurally equivalent positions in caspase-2. Caspase-6 with Phe263 and Lys265 at the equivalent positions was predicted to recognize both hydrophobic and polar P5 residues in substrates. In contrast, caspase-7 has the polar residues Gln276 and Asp278 in the corresponding positions of S5, and was predicted to recognize polar P5 residues. Caspase-8 was used as a control since there was no evidence for an S5 binding pocket in the sequence alignment or structure. Therefore, caspase-8 was expected to prefer tetrapeptides, or else to bind P5 of pentapeptides in a different manner to that observed for caspase-2 and -3. These predictions were evaluated by solving the crystal structures of caspase-3 and -7 with the peptide aldehyde inhibitor Ac-LDESD-CHO, and determining the activity of caspase-3, -6, -7 and -8 for the pentapeptides containing either hydrophobic Leu or polar Gln at P5 at the N-terminus.
Fig. 1.
Sequence homology among five caspase family members. Only the regions of loop-3 and loop-4 in small subunits are compared. The residues involved in recognition and binding of P5 in substrates are colored in red.
Kinetic data for P5-containing substrates
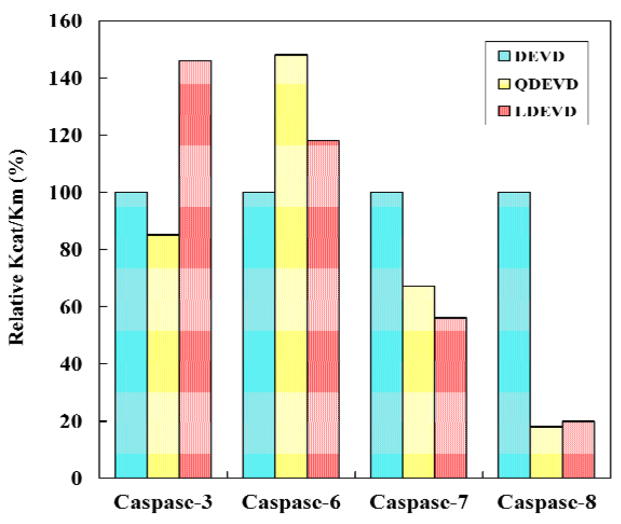
The substrate hydrolysis was analyzed for the effector caspases-3, -6, and -7 and the initiator caspase-8. The effect of P5 in substrates was investigated using the pentapeptides Ac-LDEVD-pNA containing a hydrophobic P5 residue and Ac-QDEVD-pNA with a polar P5 residue. The tetrapeptide substrate Ac-DEVD-pNA, which is hydrolyzed efficiently by all four caspases, was used as a reference. The kinetic parameters for activity of caspase-3, -6, -7 and -8 are listed in Table 1 and the relative activities are compared in Figure 2. Caspase-3 showed increased relative activity of 146% for the pentapeptide with P5 Leu, in agreement with the previous study [20], while the relative activity was lower (85%) for the peptide with the polar P5 Gln, due primarily to changes in Km. Caspase-6 showed increased activity for both pentapeptides; a small increase to 118% for LDEVD and a substantial increase to 148% for QDEVD, due to changes in both kcat and Km values relative to the activity on DEVD. LEHD, however, is a better substrate than DEVD for caspase-6 with 3-fold higher kcat/Km (0.19 min−1μM−1 and 0.058 min−1μM−1, respectively), in agreement with the known tetrapeptide specificity [19]. Caspase-7 showed decreases in relative activity to 56% and 67% for the substrates with P5 Leu and Gln, respectively, with changes in both kcat and Km. Dramatic changes were observed for caspase-8 where the relative activity dropped sharply to about 20% for both pentapeptides compared to DEVD with 3 to 4-fold increased values of Km.
Table 1.
Kinetic parameters of caspases for peptide substrates.
| kcat (min−1) | Km (μM) | kcat/Km (min−1μM−1) | Relative kcat/Km (%) | |
|---|---|---|---|---|
| Caspase-3 | ||||
| DEVD | 55±3 | 44±2 | 1.3±0.1 | 100 |
| QDEVD | 57±3 | 54±3 | 1.1±0.1 | 85 |
| LDEVD | 53±3 | 28±1 | 1.9±0.1 | 146 |
| Caspase-6 | ||||
| DEVD | 28±3 | 490±60 | 0.058±0.01 | 100 |
| QDEVD | 92±10 | 1070±110 | 0.086±0.01 | 148 |
| LDEVD | 30±3 | 440±40 | 0.068±0.01 | 118 |
| Caspase-7 | ||||
| DEVD | 77±4 | 44±2 | 1.8±0.1 | 100 |
| QDEVD | 63±3 | 51±3 | 1.2±0.1 | 67 |
| LDEVD | 58±3 | 61±3 | 1.0±0.1 | 56 |
| Caspase-8 | ||||
| DEVD | 14±1 | 34±3 | 0.40±0.02 | 100 |
| QDEVD | 11±1 | 151±8 | 0.07±0.01 | 18 |
| LDEVD | 9±1 | 107±5 | 0.08±0.01 | 20 |
Fig. 2.
Relative activity on substrates. The relative kcat/Km values are shown for caspase hydrolysis of Ac-DEVD-pNA (cyan), Ac-QDEVD-pNA (yellow) and Ac-LDEVD-pNA (pink).
Crystal structures of caspase-3/LDESD and caspase-7/LDESD
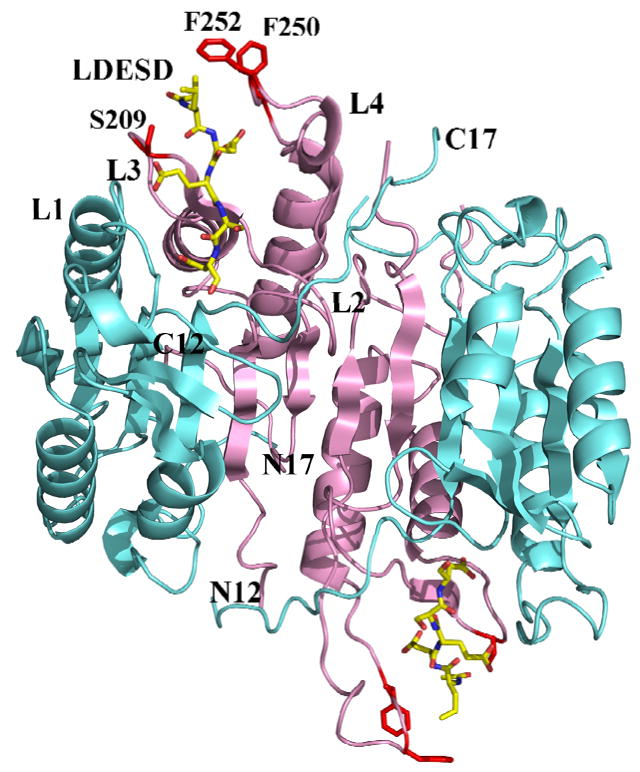
Caspase-3 and caspase-7 were crystallized in with the peptide aldehyde inhibitor Ac-LDESD-CHO. The caspase-3 complex was crystallized in the orthorhombic space group of P212121 and caspase-7 complex was crystallized in the trigonal space group of P3221. The structures were refined to the resolutions of 1.61 Å and 2.45 Å, respectively. The data collection and refinement statistics are listed in Table 2. The caspase-7 structure is at lower resolution with higher B-factor likely due to the higher solvent content of 65% compared to 52% for the caspase-3 crystals. The mature caspase-3 contains two heterodimers (p17/p12)2, which are derived from processing of procaspase-3 (Fig. 3). The residues 35–174 and 34–174 are visible in the electron density for the two p17 subunits, respectively, and 186–278 in the two p12 subunits in the heterotetramer. The overall structure of caspase-3/LDESD is very similar to the previously reported structures of caspase-3 with the canonical tetrapeptide DEVD and pentapeptide VDVAD [20], with RMS deviations of 0.38 Å and 0.17 Å for Cα atoms, respectively. The asymmetric unit of caspase-7/LDESD contains a complete catalytic unit of two p20-p10 heterodimers. Residues 58–196 and 52–196 are visible in the two p20 subunits, respectively, and residues 211–303 are visible in both p10 subunits in the heterotetramer. The refined overall structure of the caspase-7/LDESD is very similar to the previously reported structure of caspase-7 in complex with the canonical tetrapeptide DEVD [16]. The entire catalytic domain with the two heterodimers can be superposed with that of caspase-7/DEVD with RMS differences of 0.27 for 460 topologically equivalent Cα atoms.
Table 2.
Crystallographic data collection and refinement statistics.
| Caspase3/Ac-LDESD-CHO | Caspase7/Ac-LDESD-CHO | |
|---|---|---|
| Space group | P212121 | P3221 |
| a (Å) | 67.37 | 88.64 |
| b (Å) | 93.56 | 88.64 |
| c (Å) | 97.62 | 187.71 |
| β (°) | 90 | 120 |
| Resolution range | 50–1.61 | 50–2.45 |
| Total observations | 358,844 | 175,449 |
| Unique reflections | 72,895 | 28,712 |
| Completeness | 90.1 (63.9)a | 90.3 (57.3)a |
| <I/σ(I)> | 15.3 (2.4) | 18.5 (2.7) |
| Rsym (%)b | 7.5 (41.6) | 9.5 (38.9) |
| Resolution range | 10–1.61 | 10–2.45 |
| Rcryst (%)c | 17.4 | 20.1 |
| Rfree (%)d | 22.3 | 24.8 |
| Mean B-factor (Å2) | 32.4 | 64.2 |
| Number of atoms | ||
| Protein | 3803 | 3777 |
| Inhibitor | 84 | 84 |
| Water | 276 | 82 |
| r.m.s. deviations | ||
| Bond length (Å) | 0.009 | 0.006 |
| Angles | 0.029(Å)e | 1.35(°)f |
Values in parentheses are given for the highest resolution shell
Rsym = Σhkl|Ihkl − 〈Ihkl〉|/ΣhklIhkl.
R = Σ|Fobs − Fcal|/ΣFobs.
Rfree = Σtest(|Fobs| − |Fcal|)2/Σtest|Fobs|2.
The angle rmsd in SHELX97 is indicated by distance in Å.
The angle rmsd in CNS is indicated by angle in degree.
Fig. 3.
Overall structure of caspase-3/LDESD. Two heterodimers (p17/p12)2 of caspase-3/LDESD are shown in a ribbon representation with the large and small subunits colored cyan and pink, respectively. The inhibitor Ac-LDESD-CHO is colored by element type. The side chains of caspase-3 residues that interact with P5 Leu are shown in red. The N and C termini are indicated for the 12 kDa and 17 kDa chains. L1 to L4 indicate loops 1 to 4 that form the substrate binding site.
Interactions of caspase-3 with LDESD
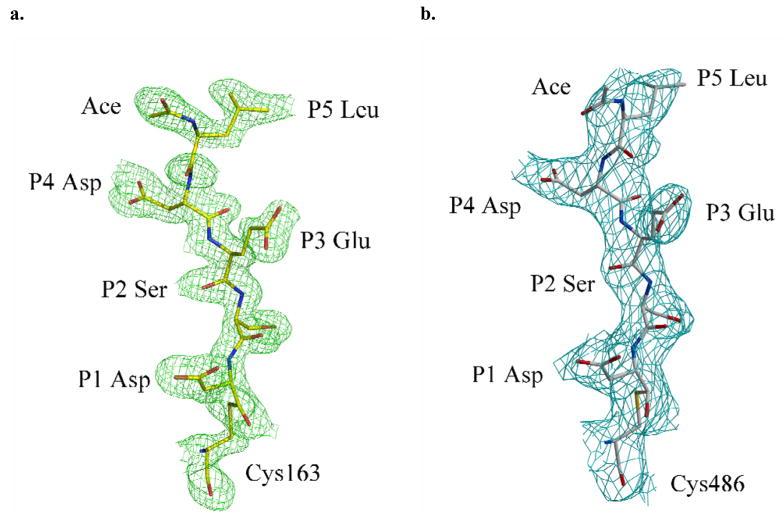
Most of the residues in the active site of caspase-3/LDESD are in similar positions and orientations when compared to those of the caspase-3/VDVAD complex [20]. The reported alternate conformations of His121 were also observed in the current structure, supporting the proposed Cys-His catalytic dyad hydrolysis mechanism [39]. All the residues in the peptide substrate are clearly visible in the electron density as shown in Figure 4. The pentapeptide of caspase-3/LDESD can be superimposed with those of caspase-2/LDESD and caspase-3/VDVAD structures with RMS deviations of 0.33 (0.17) Å and 0.09 (0.18) Å for the 5 topologically equivalent Cα atoms, respectively. (The values in the parenthesis show the RMS deviations of the inhibitor in the second catalytic site.) The interactions between caspase-3 and the peptide analog Ac-LDESD-CHO are illustrated in Figure 5a. Compared to the caspase-3/VDVAD structure, caspase-3/LDESD has substitutions at three positions, P2 (Ala/Ser), P3 (Val/Glu) and P5 (Val/Leu). The interactions at the P2 position are similar in both structures. Both Ala and Ser have van der Waals interactions with caspase residues Tyr204 and Trp206. However, the main chain amide, carbonyl oxygen and side chain oxygen of P2 Ser form hydrogen bond interactions with three water molecules since the P2 side chain is exposed to solvent. The side chain of P3 Val in VDVAD forms van der Waals interactions with caspase-3 atoms. In contrast, the negatively charged side chain of P3 Glu in LDESD forms a strong ionic interaction with the side chain of Arg207 and their main chains are connected by two hydrogen bond interactions between the carbonyl oxygen atoms and amide groups.
Fig. 4.
Structure of peptide analog inhibitor Ac-LDESD-CHO. 2Fo – Fc electron density map for Ac-LDESD-CHO in one heterodimer of the caspase-3/LDESD complex (a) and caspase-7/LDESD complex (b). The active site cysteine of caspase-3/-7 forms a hemithioacetal bond with the aldehyde group of the inhibitor.
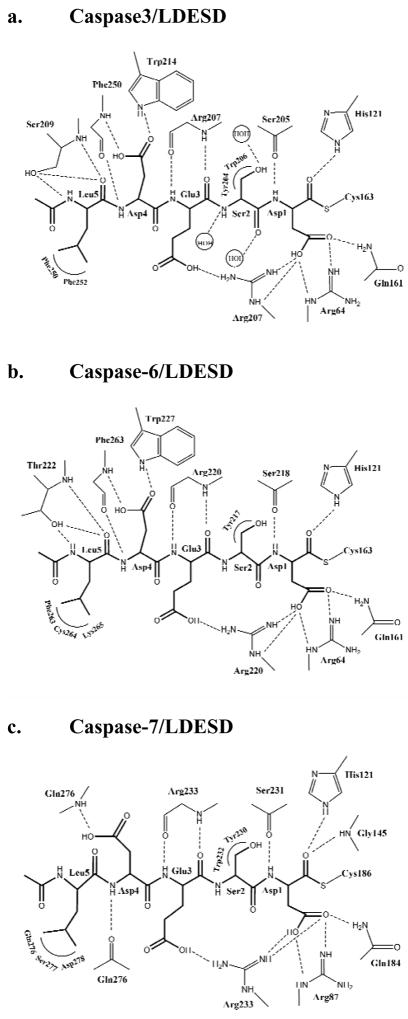
Fig. 5.
Interactions of caspase-3, 6 and 7 with Ac-LDESD-CHO. (a) Caspase-3/LDESD. (b) Caspase-6/LDESD. (c) Caspase-7/LDESD. Thicker lines represent the peptide analog inhibitor. The inhibitor is covalently bonded to the catalytic cysteine. Dashed lines represent hydrogen bonds and ion pairs, while curved lines indicate van der Waals interactions. Similar interactions were observed in the two binding sites in the heterotetramers.
At the P5 position, the main chain amide of Leu forms a hydrogen bond interaction with the side chain hydroxyl of Ser209, and the main chain P5 carbonyl oxygen forms hydrogen bond interactions with both the hydroxyl side chain and the main chain amide of Ser209. The hydrophobic Leu side chain is accommodated in the hydrophobic pocket formed by the side chains of Phe250 and Phe252, which is similar to the P5 interactions in the caspase-3/VDVAD structure. The CD1 of P5 Leu forms van der Waals interactions with the aromatic side chain of Phe252. In addition, CD2 of P5 forms van der Waals interactions with the side chains of both Phe250 and Phe252. The acetyl group of Ac-LDESD-CHO protrudes into the solvent region. It has been suggested that the P5 residue is unnecessary for the efficient cleavage of caspase-3 and -7 substrates [40]. However, the current investigation confirmed that caspase-3 has a hydrophobic S5 pocket, which recognizes hydrophobic P5 residues.
Predicted interactions of caspase-6 with LDESD
The modeled structure of caspase-6/LDESD showed very similar overall structure and interactions with the pentapeptide as observed in the crystal structure of caspase-3/LDESD. These two caspases are predicted to have conserved hydrogen bond interactions with the pentapeptide, with the exception of the interactions with P5 (Fig. 5b). The caspase-6 has different S5 amino acids in loop-3 and -4, forming similar interactions with P5 Leu to those in caspase-3/LDESD. The main chain carbonyl oxygen and amide of P5 are anchored by hydrogen bonds with the amide and side chain hydroxyl of Thr222 in caspase-6 rather than the structurally equivalent Ser209 in caspase-3. The side chain of P5 Leu forms hydrophobic interactions with loop-4 residues Phe263 and Lys265 in caspase-6, which correspond to Phe250 and Phe252 in caspase-3.
Interactions of caspase-7 with LDESD
The pentapeptide inhibitor in the caspase-7/LDESD complex adopts an extended conformation as seen for the canonical tetrapeptide DEVD. The P5 Leu in the pentapeptide buries an additional ~ 208 Å2 of surface area when compared to DEVD in the caspase-7/DEVD complex. The pentapeptide of caspase-7/LDESD can be superposed with those of caspase-2/LDESD, caspase-3/LDESD and caspase-3/VDVAD structures with RMS deviation of 0.54(0.48) Å, 0.22(0.35) Å and 0.31(0.43) Å for the 5 topologically equivalent Cα atoms, respectively (The values within the parenthesis are those of the inhibitor in the second catalytic site.) The orientation and position of the P1-P4 residues of caspase-7/LDESD are very similar to those in the caspase-7/DEVD complex. The only difference is the Val/Ser substitution at the P2 position, and both side chains form similar hydrophobic interactions with Tyr230 of caspase-7 (Fig. 5c). The P5 Leu side chain of LDESD interacts with the residues of the loop-4 in the active site cleft. However, the main chain amide and carbonyl oxygen of P5 Leu do not form hydrogen bonds with caspase-7, unlike the interactions observed in caspase-3/LDESD (Fig. 5a). The CD1 of P5 Leu has van der Waals interactions with the side chain of Asp278. The P5 CD2 forms van der Waals interactions with Cα atom of Ser277, and the main chain carbonyl groups of both Gln276 and Ser277. The hydrogen bond interaction between the main chain carbonyl of Gln276 and the P4 amide in the caspase-7/DEVD structure is conserved in the caspase-7/LDESD complex, while the acetyl group of Ac-LDESD-CHO is exposed to the solvent.
Comparison of executioner caspase/LDESD structures
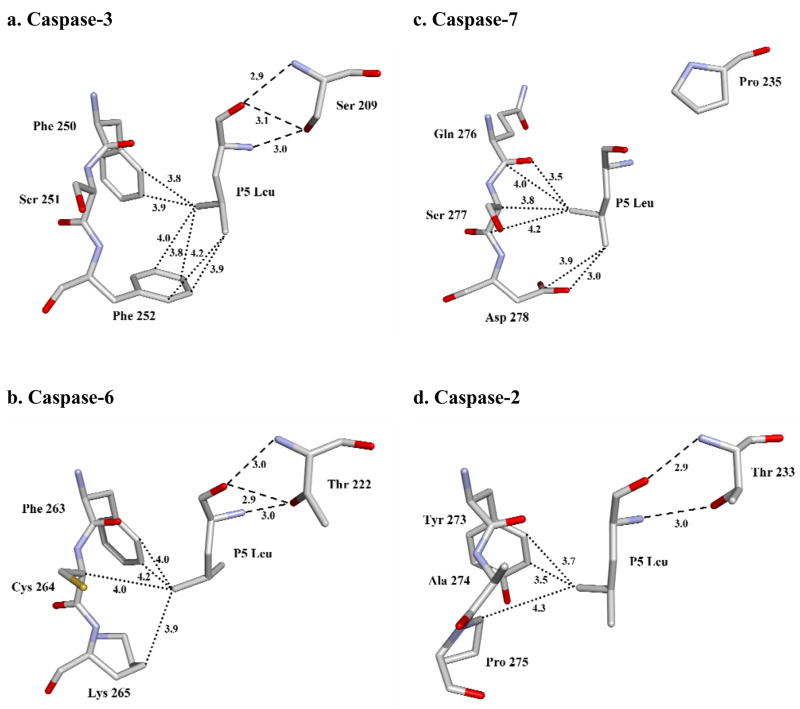
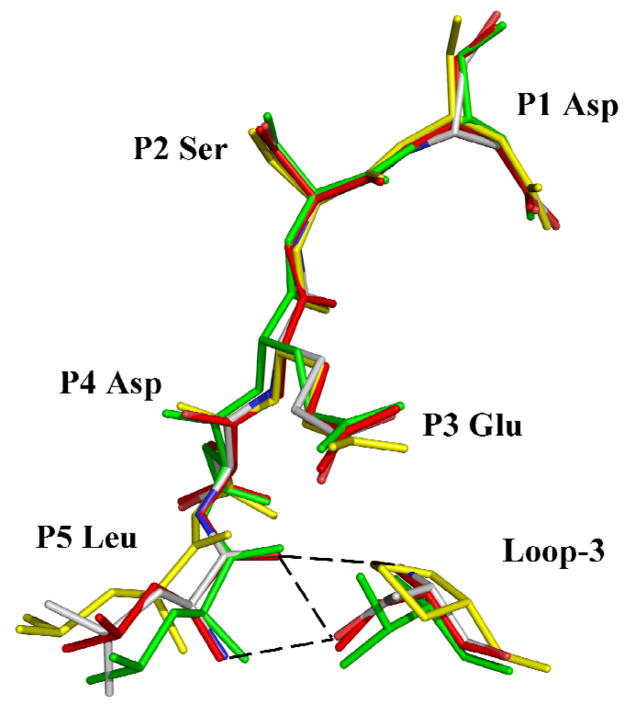
The P5 interactions were compared for the pentapeptide complexes with caspase-2, -3, -6 and -7 (Fig. 6). The P5 Leu side chain forms similar hydrophobic interactions with the side chains of equivalent residues Phe250 and Phe252 in caspase-3, Tyr273 and Pro275 in caspase-2, Phe263 and Lys265 in caspase-6. However, in caspase-7 the P5 Leu side chain forms van der Waals interactions with the main chain atoms of residues 276–277 and the side chain of Asp278. In the other caspases, however, the P5 main chain interacts with Thr243 in the loop-3 of caspase-2, Ser209 in caspase-3, and Thr222 in caspase-6. In caspase-3, the P5 amino acid is secured by the interaction of its main chain amide with side chain hydroxyl of Ser209, and by the two hydrogen bonds formed by its carbonyl with main chain amide and side chain hydroxyl of Ser209. Similar interactions are predicted for Thr222 in the caspase-6 complex. In caspase-2, the main chain amide of Thr243 forms a hydrogen bond interaction with the carbonyl of P5 Leu and the side chain hydroxyl group of Thr243 interacts with the backbone amide of P5 [17]. Therefore, the hydroxyl of Ser or Thr of loop-3 in caspase-2, -3 and -6 forms stabilizing hydrogen bonds with the main chain of the P5 residue. In contrast, Pro235 is the corresponding loop-3 residue of caspase-7, which cannot form the hydrogen bonds that anchor P5 in the other caspases. When caspase-2/LDESD, caspase-3/LDESD and caspase-7/LDESD structures are superposed, the residues P1-P4 of the peptides have almost identical position and orientation (Fig. 7). However, the Cα of the P5 residue of caspase7/LDESD is ~2 Å further away from Pro235 in loop-3 when compared to the corresponding positions in the pentapeptide complexes of caspase-2 and caspase-3. This change is due to loss of the critical hydrogen bonds in the caspase-7/LDESD complex. Instead, the P5 Leu side chain forms closer interactions with the loop-4 residues Ser277 and Asp278. Therefore, unlike caspase-2, -3 and -6, caspase-7 is not structurally suited to accommodate the hydrophobic P5 residue in peptides.
Fig. 6.
Interactions of P5 Leu in S5 subsite of caspases. (a) caspase-3/LDESD. (b) caspase-6/LDESD. (c) caspase-7/LDESD. (d) caspase-2/LDESD. The atoms are colored by element type. Hydrogen bond interactions are represented by broken lines and the van der Waals contacts are shown in dotted lines. Similar interactions were observed in the two binding sites in the heterotetramers.
Fig. 7.
Superposition of pentapeptide substrate analogs in the caspase active site. The inhibitor and the critical loop-3 residue that fixes the P5 position in structures of caspase-2, -3, - 6 and -7 complexes with LDESD are shown in green, red, element type and yellow, respectively. Hydrogen bonds between P5 and Thr/Ser of loop-3 in caspase-2, -3 and -6 are indicated by broken lines. The substitution of Pro in loop-3 of caspase-7 eliminates these crucial hydrogen bonds.
Induced-fit conformational change in caspase-3 and not in caspase-7
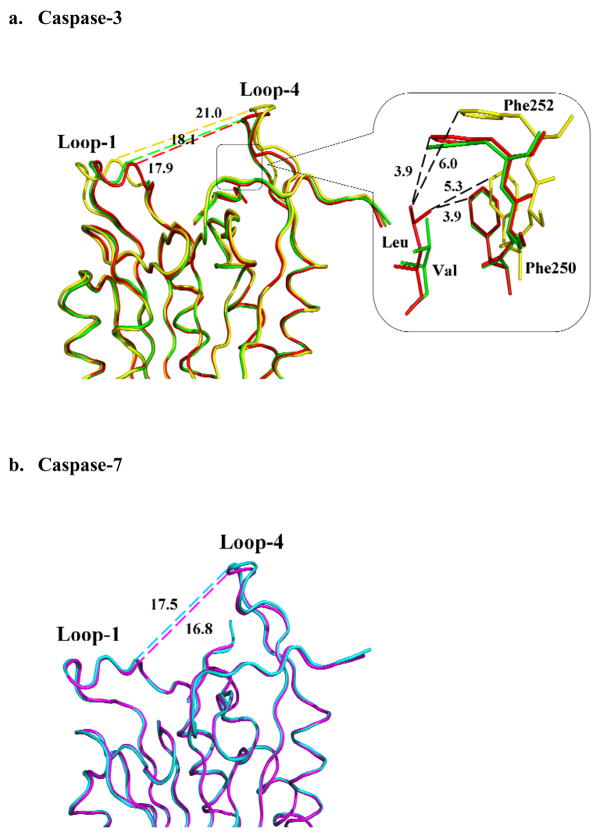
Interactions between P5 residues and caspase-3 residues forming the S5 subsite were associated with conformational changes in loop-1 and loop-4 regions when compared with the complexes with tetrapeptides [20]. Structures of caspase-3 with tetrapeptide DEVD, pentapeptide LDESD and VDVAD were superimposed, and the distance between the Cα atoms of Gly60 (in loop-1) and Asp253 (in loop-4) was measured to assess the conformational differences (Fig. 8). Although these three caspase-3 crystal structures are not isomorphic, loop-1 has no crystal contacts, while loop-4 has similar hydrophobic interactions with a symmetry-related molecule. Therefore, the conformational change at S5 subsite is mainly due to the interactions of P5 residue with the loop residues. The two loops have moved closer by about 3 Å in the structures of caspase-3/LDESD and caspase-3/VDVAD compared with that of caspase-3/DEVD. This conformational change partly closes the binding pocket formed by the four loops and enables the P5 side chain to form favorable van der Waals interactions with caspase-3 residues. In the caspase-3/VDVAD structure, only one of the P5 Val CD atoms has hydrophobic interactions within the S5 pocket formed by Phe250 and Phe252 (Fang et al., 2006). In the caspase-3/LDESD structure, however, both CD atoms of the P5 Leu have van der Waals contacts with Phe250 and Phe252 (Fig. 6a and Fig. 8a). This observation confirms that loop-1 and loop-4 regions of caspase-3 are flexible and accommodate different P5 residues by an induced-fit mechanism. However, it is not possible to determine from these static crystal structures whether the observed conformational differences result from entropic or enthalpic contributions.
Fig. 8.
Comparison of caspase complexes with pentapeptides and tetrapeptide. (a) Superposition of Cα backbone of caspase-3 with LDESD (red), VDVAD (green) and DEVD (yellow). Interatomic distances in Å are shown by broken lines. Conformational change is indicated by the altered separation of loop-1 and loop-4 in different complexes. The S5 binding site (boxed region) is shown in detail, where Phe250 and Phe252 form hydrophobic interactions with the P5 residues. (b) Superposition of Cα backbone of caspase-7 with LDESD (cyan) and DEVD (magenta). No significant structural change is observed in the two complexes of caspase-7.
Similar analysis was performed for the structures of caspase-7 in complex with tetrapeptide and pentapeptide (Fig. 8b). Loop-1 and -4 of caspase-7 do not show any crystal contacts. The separation of loop-1 and -4 is very similar in caspase-7/DEVD and caspase-7/LDESD (19.9 and 20.3 Å, respectively), indicating that no conformational change was induced in agreement with the kinetic data. The existence of a conformational change in caspase-6 upon binding of P5 residue cannot be verified in the absence of crystal structures. However, its preference for a P5 residue in the kinetic analysis and structural similarity with caspase-3 suggested that caspase-6 also has a flexible S5 binding site formed by the four loops.
Correlation of kinetic and structural data
Knowledge of the molecular structures of caspases has been critical for understanding their recognition of substrates and inhibitors. The kinetic data can be interpreted in light of the caspase residues in the loops of the S5 subsite interacting with the P5 residue. The caspase-3/LDESD structure showed hydrophobic interactions between the side chains of P5 Leu and Phe250 and Phe252 in the S5 pocket, similar to those reported for P5 Val in caspase-3/VDVAD and consistent with the increased activity (150%) for pentapeptides with hydrophobic P5 Leu compared to tetrapeptides of the same P1-P4 sequence (Fig. 2 and [20]). However, the activity was similar for the tetrapeptide DEVD and the pentapeptide QDEVD, suggesting little effect of polar P5 residues. The putative S5 pocket of caspase-6 shares similarities with those of caspase-3 and -2 since Phe and Lys are present at equivalent positions in loop-4, and Thr is at the P5 anchoring position in loop-3 (Fig. 6b). Moreover, the Phe263 in S5 can form hydrophobic contacts with P5 Leu, while the Lys265 may interact with polar P5 residues like Gln. Hence, caspase-6 was predicted to form favorable interactions with both hydrophobic and polar P5 residues in agreement with the relative activity data, although there is a preference for polar P5 (Fig. 2).
Caspase-7 has the polar residues Gln and Asp at structurally equivalent positions in the S5 pocket (Fig. 6c) and was predicted to prefer polar residues at P5. However, caspase-7 showed lower activity for P5 Gln and P5 Leu in pentapeptides relative to DEVD suggesting no preference for polar P5 residues (Fig. 2). This specificity difference of caspase-7 compared to the caspase-2, -3 and -6 correlated with the observed interactions of P5 with the residue at the loop-3 anchoring position. Structural analysis suggests the loop-3 residue Thr243/Ser209 in caspase-2, -3 and -6 is crucial for the recognition and anchoring of P5 residues. In caspase-7, the substitution of an imino acid, proline, at the structurally equivalent position in the loop-3 prevents the formation of the hydrogen bond with the carbonyl of P5, while the nonpolar side chain of proline abolishes the other hydrogen bond with the amide of P5 that anchors this residue in the S5 binding pocket (Fig. 7). The presence of Pro rather than Ser or Thr explains the lower activity of caspase-7 with P5-containing substrates.
Caspase-8 lacks the long surface loop-4 that forms the S5 pocket in the executioner caspases, while the P5 anchoring residue in loop-3 is proline as in caspase-7 (Fig. 1). Therefore, caspase-8 strongly prefers the tetrapeptide substrate DEVD compared to the two tested pentapeptides. Comparison of the primary structure of other human caspases reveals that inflammatory caspase-1 and apoptotic initiator caspase-9 also have proline as the S5 residue in loop-3 and hence it is likely that these caspases have no preference for the P5 residue in their substrates.
Our structural and kinetic analysis of the preference of executioner caspases for the P5 position of substrates will help identify the specific protein substrates and apoptotic pathways activated by each caspase. The P1 to P4 substrate recognition sequences of the executioner caspases are similar and hard to differentiate, especially between caspase-3 and -7. The P5 residue will contribute less than the P1 to P4 residues to the overall affinity for caspases and therefore, the differences in the relative kcat/Km are also small. Our studies have shown that the hydrophobicity of the S5 subsite in caspase-3 and -6 is consistent with the relative kcat/Km for cleavage of peptides with hydrophobic or polar P5 residues, while caspase-7 does not strongly interact with P5 of peptides. These findings are valuable in understanding the presence of redundant executioner caspases. Importantly, differences were identified in P5 recognition of caspase-3 and –7, which otherwise recognize very similar substrates, and these differences were correlated with the residue present in the newly defined loop-3 anchoring position and the caspase conformation with pentapeptides rather than tetrapeptides. Such differences in substrate recognition can be exploited in the rational design of selective inhibitors, which has proved a challenge, especially for caspase-3 and –7 [21,41]. Selective pharmacological inhibitors of specific caspases are desirable to reduce cell death for treatment of diseases characterized by increased apoptosis, such as stroke, heart disease and neurodegenerative diseases.
Acknowledgments
G.F. and B.F. were supported in part by the Georgia State University Research Program Enhancement award. B.F. was supported by the Georgia State University Molecular Basis of Disease Fellowship. I.T.W. and R.W.H. are Georgia Cancer Coalition Distinguished Cancer Scholars. This research was supported in part by the Georgia Research Alliance, the Georgia Cancer Coalition and the National Institutes of Health award GM065762. We thank the staff at the SER-CAT beamline at the Advanced Photon Source, Argonne National Laboratory, for assistance during X-ray data collection. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Abbreviations
- Ac
acetyl
- CHO
aldehyde
- pNA
p-nitroanilide
- RMS
root mean square
Contributor Information
Guoxing Fu, Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA.
Alexander A. Chumanevich, Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA
Johnson Agniswamy, Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA.
Bin Fang, Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA.
Robert W. Harrison, Department of Computer Science, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA
Irene T. Weber, Department of Biology, Molecular Basis of Disease Program, Georgia State University, Atlanta, GA 30303, USA.
References
- 1.Tacconi S, Perri R, Balestrieri E, et al. Increased caspase activation in peripheral blood mononuclear cells of patients with Alzheimer’s disease. Exp Neurol. 2004;190(1):254–262. doi: 10.1016/j.expneurol.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Hartmann A, Troadec JD, Hunot S, et al. Caspase-8 is an effector in apoptotic death of dopaminergic neurons in Parkinson’s disease, but pathway inhibition results in neuronal necrosis. J Neurosci. 2001;21(7):2247–2255. doi: 10.1523/JNEUROSCI.21-07-02247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermel E, Gafni J, Propp SS, et al. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell Death Differ. 2004;11(4):424–438. doi: 10.1038/sj.cdd.4401358. [DOI] [PubMed] [Google Scholar]
- 4.LeBlanc AC. The role of apoptotic pathways in Alzheimer’s disease neurodegeneration and cell death. Curr Alzheimer Res. 2005;2(4):389–402. doi: 10.2174/156720505774330573. [DOI] [PubMed] [Google Scholar]
- 5.Zidar N, Jera J, Maja J, Dusan S. Caspases in myocardial infarction. Adv Clin Chem. 2007;44:1–33. doi: 10.1016/s0065-2423(07)44001-x. [DOI] [PubMed] [Google Scholar]
- 6.Takemura G, Fujiwara H. Morphological aspects of apoptosis in heart diseases. J Cell Mol Med. 2006;10(1):56–75. doi: 10.1111/j.1582-4934.2006.tb00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HS, Lee JW, Soung YH, et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology. 2003;125(3):708–715. doi: 10.1016/s0016-5085(03)01059-x. [DOI] [PubMed] [Google Scholar]
- 8.Soung YH, Lee JW, Kim SY, et al. CASPASE-8 gene is inactivated by somatic mutations in gastric carcinomas. Cancer Res. 2005;65(3):815–821. [PubMed] [Google Scholar]
- 9.Volkmann X, Cornberg M, Wedemeyer H, et al. Caspase activation is required for antiviral treatment response in chronic hepatitis C virus infection. Hepatology. 2006;43(6):1311–1316. doi: 10.1002/hep.21186. [DOI] [PubMed] [Google Scholar]
- 10.Lakhani SA, Masud A, Kuida K, et al. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311(5762):847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer U, Schulze-Osthoff K. Apoptosis-based therapies and drug targets. Cell Death Differ. 2005;12(Suppl 1):942–961. doi: 10.1038/sj.cdd.4401556. [DOI] [PubMed] [Google Scholar]
- 12.Baskin-Bey ES, Washburn K, Feng S, et al. Clinical Trial of the Pan-Caspase Inhibitor, IDN-6556, in Human Liver Preservation Injury. Am J Transplant. 2007;7(1):218–225. doi: 10.1111/j.1600-6143.2006.01595.x. [DOI] [PubMed] [Google Scholar]
- 13.Lavrik IN, Golks A, Krammer PH. Caspases: pharmacological manipulation of cell death. J Clin Invest. 2005;115(10):2665–2672. doi: 10.1172/JCI26252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384(Pt 2):201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10(1):76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Y, Fox T, Chambers SP, et al. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000;7(6):423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 17.Schweizer A, Briand C, Grutter MG. Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. J Biol Chem. 2003;278(43):42441–42447. doi: 10.1074/jbc.M304895200. [DOI] [PubMed] [Google Scholar]
- 18.Shiozaki EN, Chai J, Rigotti DJ, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11(2):519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 19.Thornberry NA, Rano TA, Peterson EP, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272(29):17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 20.Fang B, Boross PI, Tozser J, Weber IT. Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition. J Mol Biol. 2006;360(3):654–666. doi: 10.1016/j.jmb.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 21.Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14(1):66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Calvo M, Peterson EP, Rasper DM, et al. Purification and catalytic properties of human caspase family members. Cell Death Differ. 1999;6(4):362–369. doi: 10.1038/sj.cdd.4400497. [DOI] [PubMed] [Google Scholar]
- 23.Agniswamy J, Fang B, Weber IT. Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis. FEBS J. 2007;274(18):4752–4765. doi: 10.1111/j.1742-4658.2007.05994.x. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. Ref Type: Generic. [DOI] [PubMed] [Google Scholar]
- 25.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 26.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59(7):1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 27.Sheldrick GM, Schneider TR. SHELXL: High-resolution refinement. Macromolecular Crystallography B. 1997;277:319–343. [PubMed] [Google Scholar]
- 28.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(121):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Lovell SC, Davis IW, Arendall WB, III, et al. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merritt EA, Murphy ME. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994;50(6):869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 33.Kullback S. Information Theory and Statistics. John Wiley and Sons, Inc; New York; N.Y: 1959. [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison RW. Stiffness and Energy Conservation in Molecular Dynamics: an Improved Integrator. J Comp Chem. 1993;14:1112–1122. [Google Scholar]
- 36.Harrison RW, Chatterjee D, Weber IT. Analysis of six protein structures predicted by comparative modeling techniques. Proteins. 1995;23(4):463–471. doi: 10.1002/prot.340230402. [DOI] [PubMed] [Google Scholar]
- 37.Bagossi P, Tozser J, Weber IT, Harrison RW. Modification of parameters of the charge equilibrium scheme to achieve better correlation with experimental dipole moments. J Mol Model. 1999;5:143–152. [Google Scholar]
- 38.Harrison RW. A Self-Assembling Neural Network for Modeling Polymers. J Math Chem. 1999;26:125–137. [Google Scholar]
- 39.Wilson KP, Black JA, Thomson JA, et al. Structure and mechanism of interleukin-1 beta converting enzyme. Nature. 1994;370(6487):270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 40.Talanian RV, Quinlan C, Trautz S, et al. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272(15):9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 41.Onteniente B. Natural and synthetic inhibitors of caspases: targets for novel drugs. Curr Drug Targets CNS Neurol Disord. 2004;3(4):333–340. doi: 10.2174/1568007043337210. [DOI] [PubMed] [Google Scholar]