Abstract
The x-ray diffraction structure of the non-illuminated D96A bacteriorhodopsin mutant reveals structural changes as far away as 15 Å from residue 96, at the retinal, Trp-182, Ala-215, and waters 501, 402, and 401. The Asp-to-Ala side-chain replacement breaks its hydrogen-bond with Thr-46, and the resulting separation of the cytoplasmic ends of helices B and C is communicated to the retinal region through a chain of covalent and hydrogen-bonds. The unexpected long-range consequences of the D96A mutation include breaking the hydrogen bond between O of Ala-215 and water 501, and formation of a new hydrogen-bond between water molecules 401 and 402 in the extracellular region. Because in the T46V mutant a new water molecule appears at Asp-96 and its hydrogen-bond to Ile-45 replaces Thr-46 as its link to helix B, the separation of helices B and C is smaller than in D96A and there are no atomic displacements elsewhere in the protein. Propagation of conformational changes along the chain between the retinal and Thr-46 had been observed earlier in the crystal structures of the D96N and E204Q mutants, but in the trapped M state. Consistent with perturbation of the retinal region in D96A, little change of the Thr-46 region occurs between the non-illuminated and M states of this mutant. It appears that a local perturbation can propagate along a “track” in both directions between the retinal and the Asp-96/Thr-46 pair, either from photoisomerization of the retinal in the wild-type protein in one case or from the D96A mutation in the other.
Light-driven proton transport in bacteriorhodopsin is dependent on a few key residues, such as Lys-216, Asp-85, and Asp-96, and they have been identified in part from the distinctive phenotypes of mutants in which the replaced amino acids are unable, or much less able, to perform their normal roles (1–3). Other residues, such as Tyr-185, Glu-194, Glu-204, and Arg-82, play more dispensable roles, as suggested by the altered but less defective phenotypes of their site-specific mutations (4–9). As usual, such assignments make the assumption that the effects of mutations are local and interpretable in terms of differences in side-chain volume, hydrogen-bonding, ability to protonate and deprotonate, etc. Of necessity, they ignore the possibility of changes distant from the site of mutation. As often acknowledged, this assumption might not be justified in all cases, and for an unambiguous interpretation of the effects of mutations structural information about the non-illuminated state, as well as the intermediates of the altered reaction cycles, would be needed.
How good is the assumption of purely local perturbation in site-specific bacteriorhodopsin mutants? Crystal structures are available for several mutants, and they show that the extent of perturbation depends greatly on the residue that is exchanged, on the residue that replaced it, and on the changing local environment during the photocycle. The V49A mutation causes no structural alteration of the non-illuminated state other than the shorter side-chain (10), but the decay of the N state in the photocycle is considerably lengthened, suggesting that there may be greater structural consequences in this intermediate (11). The V219L mutation results in a cavity filled with two additional water molecules (10), but otherwise little change in the structure, although the N state has a longer lifetime also. The E204Q mutation causes local changes in the positions of water molecules and the position of the nearby Glu-194, but these displacements do not extend far into the protein (12). In the D96N mutant a new water molecule appears (13) as far as 7 Å from residue 96. The lifetime of the M and O states of the photocycle are strongly altered in the latter two mutants. Introducing a proline (14) causes little change in the structure when near the surface (in K41P), but distorts the main-chain when in the middle of a transmembrane helix (in A51P). The greatest global effect is by the D85S mutation, which radically alters the entire structure and results in changed crystal contacts, changed helical tilts, and redistribution of bound water (15).
There is reason to believe that at least some mutations should have more than purely local effects. Point mutations and the photoisomerization of retinal are both structural perturbations at a single, well-defined location. The thermal reactions in the photocycle are the consequences of the rotation of the C13=C14 retinal double-bond, but eventually they include atomic displacements throughout the protein. If the conformational possibilities available to the protein include only a few stable (or quasi-stable) states, the global structural changes might be assumed not only transiently in the functional cycle but also when induced by an appropriate side-chain replacement. In some mutants this seems to be indeed the case. The outward tilt of helix F that characterizes the late M and N intermediates (16–22) was observed in the non-illuminated states of D85N at alkaline pH, D85N/D96N at neutral pH (20, 23–25) and the D96G/F171C/F219L triple mutant (17, 18). Coupling between the protonation state of Asp-96 and the isomeric state of the retinal was observed not only in the N photointermediate but also in non-illuminated mutants of Asp-85 (26).
One of the long-range effects in the photocycle is the destabilization of the protonated Asp-96 in the cytoplasmic region, so it will become the proton donor to the Schiff base. This report explores, therefore, the stable structural changes caused by mutation of Asp-96 and its hydrogen-bonding partner Thr-46. The role of this region in proton transport has been described in crystallographic studies (27). Creation of a hydrogen-bonded chain of four water molecules in the photochemical cycle begins with formation of a water cluster at Asp-96. A water molecule intercalates between the side-chains of Asp-96 and Thr-46, and as the cluster grows it reaches the retinal during the M to N reaction (10) allowing reprotonation of the Schiff base by Asp-96. The redistribution and entry of water into this region is very likely the consequence of side-chain repacking (12) in response to the movements of Lys-216 and Trp-182 upon relaxation of the isomerized retinal into its bent shape. The observation that small cavities are formed before the N state already had suggested (10) that the chain of water molecules between Asp-96 and the retinal is created by gradual opening of the cavities and recruitment of water by wat502 to fill them.
Understanding of how the Schiff base is reprotonated in the M to N reaction has been aided considerably by studies (28–31) of the photocycles of mutants of Asp-96. Replacement of Asp-96 with a non-ionizable residue, such as Gly, Ala or Asn, removes the internal proton donor. Reprotonation of the Schiff base becomes pH dependent, suggesting that the proton is taken up from the bulk. The change of the volume of the side-chain may be expected to alter the distribution of water nearby, and thereby the conduction of the proton to the Schiff base. The possibility cannot be excluded that the phenotypes depend not only on the removal of Asp-96, but also on the kind of side-chain that replaced it. Indeed, the observations that the decay of M is much slower (20, 31) in D96N than in D96A and D96G, and that azide, a weak acid, strongly accelerates the reprotonation of the Schiff base in the D96N mutant but not in the wild-type or even in D96A and D96G, are hints (20) that the proton conduction path in D96N is unique and depends on the introduced asparagine. This idea is supported by the crystallographic structure of this mutant (13). In D96N, the hydrogen-bond between the side-chains of Asp-96 and Thr-46 is removed and the amide nitrogen of Asn-96 is connected to the Thr via a new water molecule, wat504.
These observations had suggested that the nature of the side-chain of residue 96 might have influence on the distribution of water molecules in the cytoplasmic region. Mutations of Asp-96 affect the photocycle not only because this residue is the proton donor to the Schiff base, but also because the Asp-96/Thr-46 pair will organize the pathway for proton conduction. We report here on the crystallographic structure of the non-illuminated and M states in the D96A mutant, and compare them to the non-illuminated states of the T46V and the wild type protein, and the M state of D96N. The results indicate that changes of the side-chain of residue 96 have consequences that are not only local but communicated to other locations, including the extracellular region, through atomic displacement along a “track” comprised of Thr-46, wat502, and Lys-216, and result in redistribution of bound water throughout the protein. This track appears to function also in the photocycle. In contrast to D96A, no such cascade of changes is observed in the less perturbed T46V mutant.
EXPERIMENTAL PROCEDURES
The bacteriorhodopsin crystals, grown in cubic lipid phase as described (32), were thin hexagonal plates about 120 µm × 120 µm ×10–15 µm. Pieces of the cubic phase with encased crystals were soaked overnight in the mother liqueur plus 0.5% octylglucoside1, and the crystals were removed and mounted by mechanical manipulation with a nylon loop.
Diffraction data were collected at 100K from either non-illuminated crystals, or after illumination for 3 sec at room temperature and rapid cooling with a cold nitrogen stream to 100K. Illumination was with a 5 mW He-Ne laser at 628 nm. The diffraction measurements were at beamlines BL 9-1 and 11-1 of SSRL (Stanford, California), using a 3 × 3 array CCD detector (ADSC, San Diego). For each data set 90 images with 1° oscillation angle were collected, integrated and scaled with HKL2000 (33), in the P63 space group, with minor variations from a = 61 Å, b = 61 Å, c= 108 Å, and α = 90°, β = 90°, γ = 120°.
Refinement of models was with SHELXL-97, as before (34), but with single conformations because occupancy of the desired state (non-illuminated or M) was essentially 100%. The input model was first 1C3W to produce omit maps that contained evidence for the replaced side-chains, and then with Asp-96 replaced with Ala or Thr-46 replaced with Val. Superimposition of coordinate sets for calculating displacements of selected atoms was with the program package CCP4i. Cavities were calculated with Molsoft ICM version 3.4–5 (Molsoft L.L.C. La Jolla, CA, U S A). The coordinates and the diffraction data for the non-illuminated states of D96A and T46V and the M state of D96A are deposited at the Protein Data Bank with accession codes XXXX, XXXX, and XXXX, respectively.
RESULTS AND DISCUSSION
In the following, we examine specific structural features of wild-type bacteriorhodopsin and the D96A and T46V mutants. In such comparisons, if the differences in the atomic positions or the cavity sizes are real they must exceed the magnitude of positional error between different crystals of the same kind. For the wild-type, this variability was estimated from the averages and standard deviations in independent models refined from diffraction data from five crystals, with resolutions ranging from 1.43 to 1.60 Å. Two of these are published, alone or as part of determination of a photocycle intermediate (35, 36), but three are from newly collected data. For the T46V mutant the positional errors are calculated from models for four crystals, with resolutions between 1.84 and 1.92 Å. The crystallographic resolution for D96A is 2.0 Å (two crystals). There was no detectable twinning in the crystals of the two mutants, i.e. the quality of the data is more or less equivalent to the wild-type. The statistics for the data and the refinement of models for the D96A and T46V mutants are given in Table 1. The standard deviations for inter-atomic distances in the five wild type models will be given in the comparisons below, and are ± 0.06 – 0.20 Å depending on the location, in line with earlier attempts to determine crystal-to-crystal reproducibility in our bacteriorhodopsin crystals (37). When we conclude that the structural feature is different it is only when the change of inter-atomic distance or cavity size is well above the corresponding standard deviation (or in the case of D96A the range from two independent crystals).
Table 1.
X-ray data collection and refinement statistics for data sets from a non-illuminated and an illuminated crystal of the D96A bacteriorhodopsin mutant. The illumination at 295K with red laser, as described in Methods, converted bacteriorhodopsin in the crystal to the M state with virtually full occupancy.
| Non-illuminated D96A |
Illuminated D96A |
Non-illuminated T46V |
|
|---|---|---|---|
| Data resolution range, Å | 2.0 – 25.0 | 2.08 – 25.0 | 1.84 – 25.0 |
| Total observations | 306,070 | 207,357 | 356,040 |
| Unique reflections | 15,618 | 13,966 | 19,393 |
| Rmerge a, b , % | 6.2 (75.6) | 6.6 (62.5) | 4.1 (53.7) |
| Average I/σ (I)b, c | 23.4 (2.9) | 23.4 (3.8) | 28.9 (2.1) |
| Completenessb, % | 99.9 (99.9) | 100.0 (100.0) | 95.5 (90.5) |
| Mosaicity, o | 0.57 | 0.61 | 0.84 |
| Refinement range, Å | 2.0 – 25.0 | 2.08 – 25.0 | 1.84 – 25.0 |
| Structure factors | 14,807 | 13,226 | 18,360 |
| Restraints | 8,206 | 8,240 | 8,223 |
| Parameters | 8,284 | 8,272 | 8,300 |
| Twin ratio | 97:3 | 58:42 | 99:1 |
| Protein atoms | 1717 | 1717 | 1720 |
| Retinal atoms | 20 | 20 | 20 |
| Water molecules | 23 | 23 | 24 |
| Lipid atoms | 310 | 310 | 310 |
| R-factord, e, % | 23.9 (19.8) | 15.6 (15.3) | 19.5 (17.2) |
| Rfree e, f, % | 29.9 (25.9) | 26.0 (23.7) | 26.0 (23.2) |
| Average protein B, Å2 | 32.0 | 29.2 | 30.9 |
| Average retinal B, Å2 | 25.0 | 22.7 | 20.5 |
| Average water B, Å2 | 38.3 | 39.5 | 34.0 |
| Average lipid B, Å2 | 68.5 | 71.1 | 72.6 |
| Deviation from ideal bond lengths, Å |
0.015 | 0.015 | 0.015 |
| Deviation from ideal bond angle distances, Å |
0.054 | 0.055 | 0.054 |
Rmerge (I) = Σhkl Σi | Ihkl,i − 〈Ihkl〉 | /Σhkl Σi | Ihkl,i | , where 〈Ihkl〉 is the average intensity of the multiple Ihkl,i observations for symmetry-related reflections.
I/σ(I), average of the diffraction intensities, divided by their standard deviations.
values in parentheses are for the 2.00 to 2.09 Å and the 2.08 to 2.17 Å shell resolution shells for the non-illuminated and the illuminated D96A crystal, respectively, and the 1.84 to 1.92 Å shell resolution shell for the T46V crystal.
R-factor = Σhkl | Fobs − Fcalc | / Σhkl | Fobs | , where Fobs and Fcalc are observed and calculated structure factors, respectively.
values are for all data, those in parentheses for F > 4σ(F).
Rfree = Σhkl ε T | Fobs − Fcalc | / Σhkl ε T | Fobs | , where a test set (5% of the data) is omitted from the refinement in such a way that all structure factors in each of several thin resolution-shells were selected to avoid bias from merohedral twinning.
Our intent was to compare also the structural changes in the M (last M substate, or M2’) states produced by illumination of the D96A and D96N mutants under the same conditions, i.e., at ambient temperature. Although it would be more relevant to transport function to use the M state of the wild-type as reference, this is not possible. In the wild-type protein, M decays too rapidly at room temperature to accumulate in amounts more than a few percent under usual illumination conditions. To populate this state, in the two studies of wild-type M produced in cubic phase grown crystals at ambient temperature (10, 38), the illumination was continued for a short time (1 sec) as the crystals were rapidly cooled, and thus the M was produced at some undefined temperature below ambient. Such a regime produces either a claimed mixture (38) of early and late M, or in our experience (10) an earlier M state than M2’. In both cases, the retinal assumed different configurations than in M of D96N. In two other studies, the illumination of the crystals was at 210 K (36) and 230 K (40), and the M produced was also earlier M, i.e., M1, with lesser changes than the later M states. In yet another study of the wild-type M intermediate, with crystals of a different kind where the crystal contacts were suggested to slow the decay of M and allow its accumulation at ambient temperature (41), the structural changes do not seem to be comparable to those in cubic phase crystals. In this case, there was virtually no change from the BR state near residue 96, where our attention is focused, but a sliding movement of helix G not seen in the other M structures was reported. The M state of the E204Q mutant (12), on the other hand, shows similarities to the structure of the M intermediate of D96N. Although the M produced is an earlier sub-state because the E204Q mutation blocks proton release to the extracellular surface, i.e., the M2 → M2’ reaction (6), the illumination conditions are most similar to those used for the D96A and D96N mutants.
Because the illuminated crystals contained little of the non-illuminated state of the protein, both models for M, from D96A in this study and from D96N earlier (13), are the results of refinements from data with full occupancy. The resolution of the data for M of D96N was 2.0 Å, and for D96A it is 2.08 Å (the latter from a crystal with merohedral twinning, see Table 1, as in the earlier report).
Structural Changes in the Non-illuminated State from Replacement of Asp-96
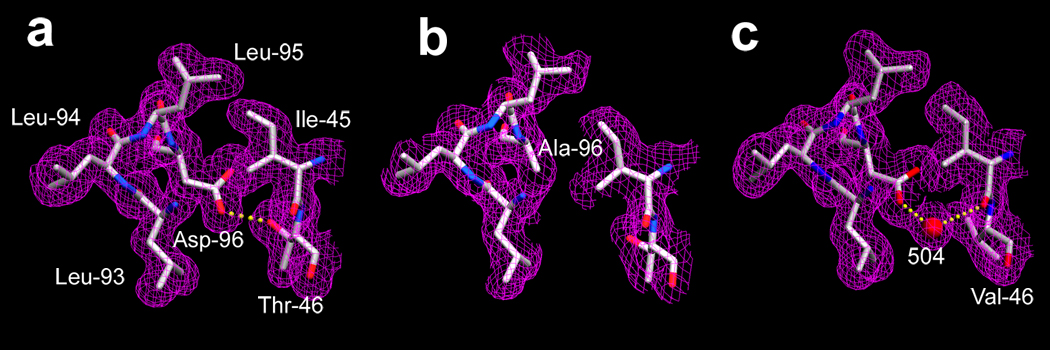
The crystal structure of the non-illuminated F219L mutant had revealed (10) that the cavity created by the side-chain replacement is filled by two water molecules hydrogen-bonded to each other and to an existing water (wat501). Comparison of the 2Fobs − Fcalc electron density maps of the immediate neighborhood of residue 96 in the D96A mutant and the wild type (Figures 1a and b) shows, however, that this is not always the case. The cavity between Ala-96 and Thr-46 from the smaller Ala side-chain, with a volume of 23–25 Å3, contains no density. As in the wild-type, the side-chain of Thr-46 donates a hydrogen-bond to the peptide C=O of Phe-42 (not shown in Figure 1), but the energy penalty of inserting a water molecule into this low-dielectric matrix is evidently not compensated by the hydrogen-bond this water would donate to OG1 of Thr-46. In contrast, replacement of the protonated carboxyl of residue 96 with an amide in D96N causes (13) insertion of a water molecule to bridge the amide ND2 and OG1 of Thr-46, and creates no cavity.
FIGURE 1.
Electron density (2Fobs − Fcalc) maps of the Asp-96 region in the non-illuminated state, in wild-type bacteriorhodopsin (a), the D96A mutant (b), and in T46V (c). Contour level at 1 σ. Wild-type model and map from 1C3W (32).
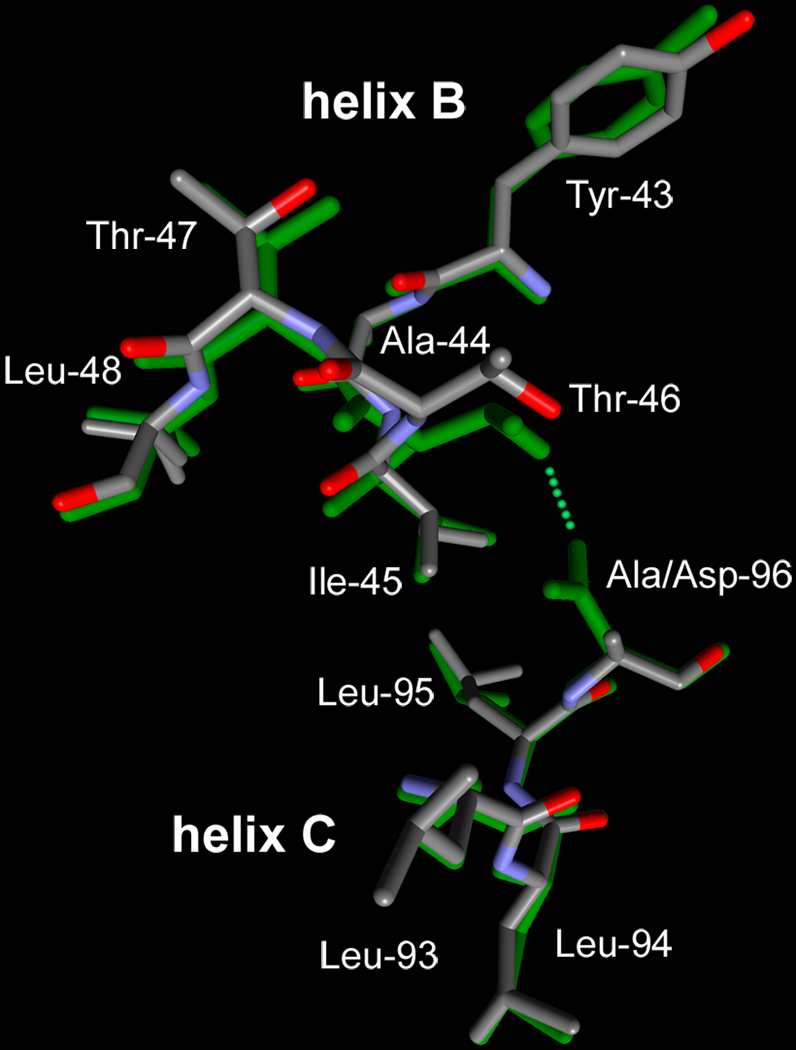
The Asp to Ala residue replacement in D96A, and the consequent loss of the hydrogen-bond between Asp-96 and Thr-46 that connects helices B and C, will perturb this region. As shown in Figure 2, where the model of D96A is overlaid on the wild type structure (latter in green), the cytoplasmic ends of helices B and C move apart. As a result, the distance between CA atoms of residue 96 and Thr-46 increases by nearly 1 Å. Another indication of perturbation is the increase of a small cavity in the protein, bounded by helices B, C, and G, that extends from the Thr-46 region toward the retinal (not shown). In the wild-type this cavity has a volume of 75 ± 4 Å3, but in D96A it nearly doubles in size, to 136 – 143 Å3. Otherwise, the main-chain and side-chain displacements from the D96A mutation are local, however, and do not extend significantly toward the center of the protein beyond Leu-48 on helix B and Leu-93 on helix C (Figure 2).
FIGURE 2.
Comparison of the Asp-96/Thr-46 region that connects helices B and C, in the non-illuminated states in D96A and the wild-type. The model for the mutant is shown with atomic colors, the wild-type model (32) in green.
Although the primary main-chain and side-chain perturbations from the D96A mutation in Figure 2 do not reach the retinal region, water molecules are affected as far away as the Schiff base and beyond, i.e. at a distance as far as 15 Å. Because these water molecules form hydrogen-bonds with functionally important residues and play prominent roles in the proton transport mechanism (42, 43), their displacements may be meaningful.
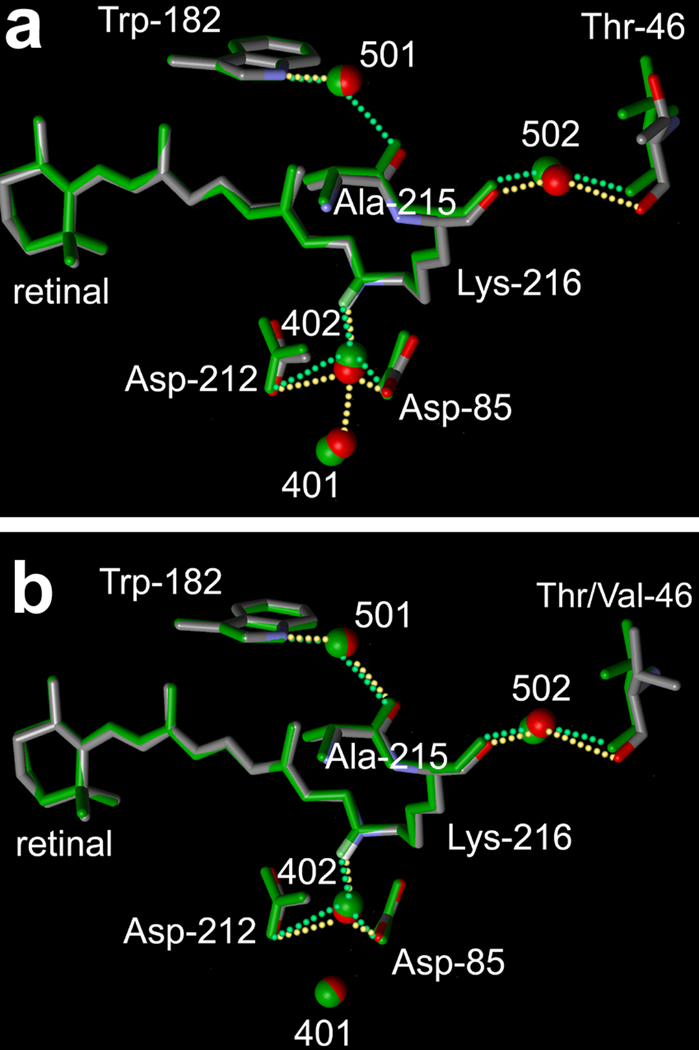
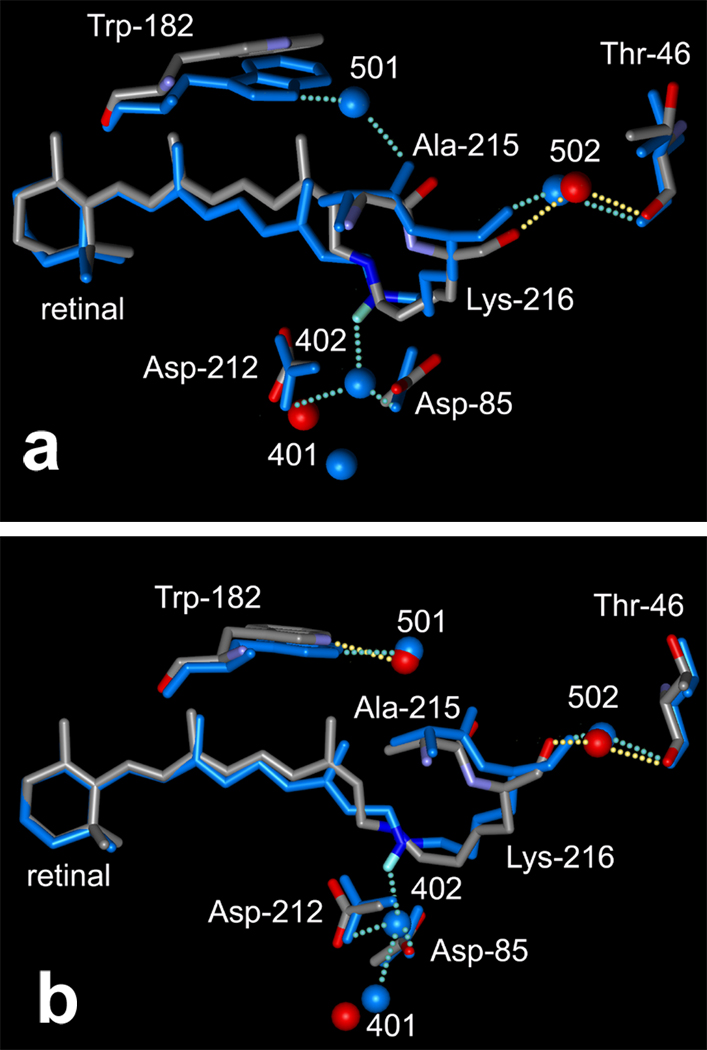
We had suggested earlier that the retinal region is connected to the Asp-96/Thr-46 region through a continuous chain of covalent and hydrogen-bonds that appear to have a functional role during the photocycle (12). The structure of the D96A mutant (atomic color model in Figure 3a) reveals that the retinal Schiff base region is influenced by the displacement of Thr-46 via the same chain. In the wild type, wat501 bridges helices G and F through hydrogen-bonds to the peptide O of Ala-215 and NH1 of Trp-182 (green model in Figure 3a). The hydrogen-bond of wat501 to Trp-182 is essentially unchanged in the D96A mutant (its length is 2.91 – 3.05 Å vs. 2.80 ± 0.06 Å in the wild-type). However, the hydrogen-bond between wat501 and the O of Ala-215 is broken as the inter-atomic distance increases from 2.97 ± 0.07 Å in the wild-type to 3.30–3.52 Å. This appears to be an indirect consequence of the displacement of helix B at Thr-46, as follows. Wat502 moves with the movement of Thr-46 described above because its hydrogen-bonds with the peptide O of Thr-46 (2.87 – 3.00 Å vs. 2.95 ± 0.09 Å in the wild type) and with the peptide O of Lys-216 also (2.94 – 2.95 Å vs. 3.02 ± 0.12 Å in the wild-type) are maintained (Figure 3a). Ala-215 is linked to the cytoplasmic region via the hydrogen-bond of the peptide O of Lys-216 to wat502. Displacement of wat502 therefore causes the main-chain of helix G to move at Lys-216 and Ala-215, and the peptide O of Ala-215 moves away from wat501, its hydrogen-bonding partner in the wild type protein.
FIGURE 3.
Comparison of the retinal region of the non-illuminated states of the D96A (a) and T46V (b) mutants (shown with atomic colors) with the wild-type (32) (in green).
Unexpectedly, as shown in Figure 3a, water molecules are displaced in the D96A mutant in the extracellular region also. Wat402 is coordinated by the Schiff base nitrogen (NZ of Lys-216) and OD2 of two anionic residues, Asp-85 and Asp-212. Wat402 is displaced strongly toward Asp-85 (the length of its hydrogen-bond with OD2 of Asp-85 is 2.30–2.46 Å vs. 2.67 ± 0.13 Å in the wild type), while its distance to Asp-212 is nearly unaffected (2.99–3.30 Å vs. 3.04 ± 0.20 Å in the wild type). Surprisingly, in the D96A mutant, wat401, the link of Asp-85 to the extracellular aqueous network, moves close enough to wat402 to form a new hydrogen-bond (inter-water distance is decreased from 3.68 ± 0.17 Å in the wild type to 2.87 – 3.06 Å). The approach of wat401 and wat402 is partly from the displacement of wat402, but mostly from the movement of wat401 (Figure 3a). In spite of this movement, the hydrogen-bonds of wat401 to its partners in the wild-type (Asp-85 and wat406) are relatively unaffected (not shown in Figure 3a). Likewise, although there is minor redistribution of side-chains in the extracellular region, no hydrogen-bonds are broken or formed as a result of the movement of wat401.
The observed movements of protein atoms, and particularly water far from the site of the D96A mutation, urge caution in interpreting the phenotypes of all mutations in terms of local effects, and in assigning O-H bands to specific water molecules solely on the basis of the location of mutations that affect them.
Little of this cascade of displacements between Thr-46 and the retinal region is evident in the earlier determined structure (13) of the more conservative mutant, D96N. The chain of hydrogen-bonds through O of Thr-46, wat502, and O of Lys-216 is unaffected by the smaller movement of helix C in this mutant, and the hydrogen-bond between O of Ala-215 and wat501 is not broken (not shown). The cytoplasmic cavity between Thr-46 and the retinal is only slightly increased, if at all, relative to the wild-type (to 82 Å3 vs. 75 ± 4 Å). The changes in the extracellular region are also less in D96N. Thus, although wat402 moves closer to OD2 of Asp-85 in D96N (2.30 Å vs. 2.67 ± 0.13 Å in the wild type) as in D96A, the inter-atomic distance between wat401 and wat402 is decreased (to 3.36 Å vs. 3.68 ± 0.17 Å in the wild type) but not sufficiently to form a hydrogen-bond.
Structural Changes in the Non-illuminated State from Replacement of Thr-46
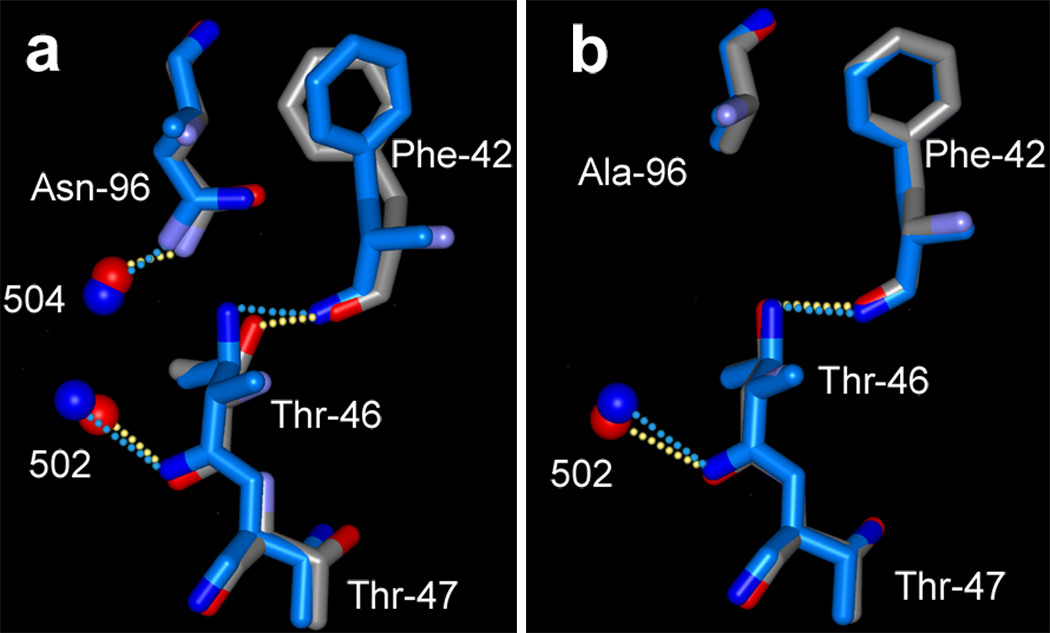
The Thr to Val change for residue 46 breaks its hydrogen-bond with Asp-96 as does the D96A mutation, but there are two important differences. First, the side-chain is replaced without change of volume. Second, the energy penalty of burying the highly polar COOH of residue 96 without a hydrogen-bonding partner in T46V must be greater than burying the OH of the Thr in D96A. Indeed, the 2Fobs − Fcalc electron density map in this region is different in the T46V and the D96A mutants (compare Figures 1b and 1c). There is no new cavity at residue 96 as in D96A, because a new water molecule, wat504, intercalates and forms a hydrogen-bond with OD2 of Asp-96 and O of Ile-45.
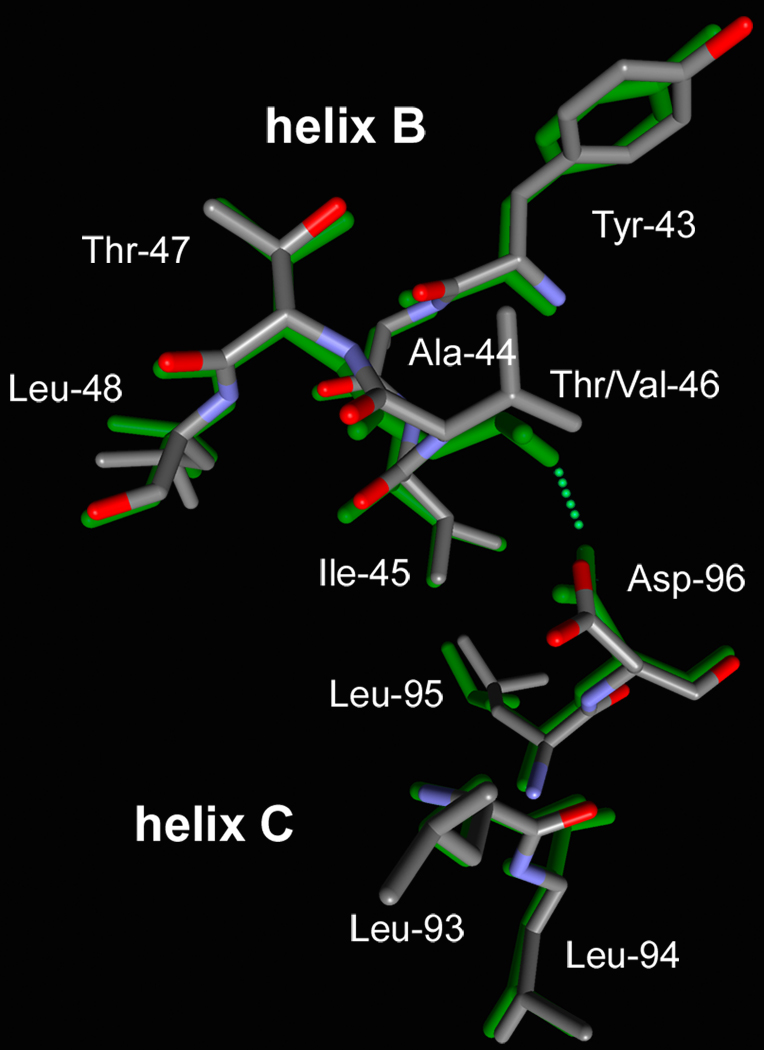
Consistent with the absence of a cavity at residue 96 and the formation of a new hydrogen-bond that retains the link between helices B and C, perturbation of the Asp-96 region in the T46V mutant is less than in D96A (Figure 4). The distance between the CA atoms of Asp-96 and residue 46 is increased by less than 0.5 Å relative to the wild-type, and this change is from movement of both residues rather than mainly of Thr-46 as in D96A (Figure 2). The cavity that extends toward retinal is increased in size, but less than in D96A, from 75 ± 4 Å3 in the wild-type to 94 ± 5 Å3. As in the D96A mutant, the perturbation is local, and little change is observed a few residues away on either helix B or C (Figure 4).
FIGURE 4.
Comparison of the retinal region of the non-illuminated states of the D96A (a) and T46V (b) mutants (shown with atomic colors) with the wild-type (32) (in green).
Consistent with the smaller perturbation at residue 46, the retinal region is less affected. As shown in Figure 3b, the hydrogen-bond of Ala-215 with wat501 is maintained (inter-atomic distance 2.97 ± 0.06 Å). Wat401 does not move significantly, and a new hydrogen-bond between wat402 and wat401 is not formed (inter-atomic distance 3.41 ± 0.09 Å).
In an earlier study of 2-dimensional crystals of the T46V mutant (17), a density feature at helix C was observed in the non-illuminated T46V vs. non-illuminated wild type difference projection map. Its origin may well be the separation of helices B and C in Figure 4.
Structural Changes in the M State from Replacement of Asp-96
Given the structural changes in the non-illuminated D96A mutant described above, it would be informative to know how they affect the structure of the M state. The decay of the M state is greatly slowed by replacement of Asp-96 with a non-protonatable residue, particularly at higher pH (31–34), and this allows accumulation of the last M state (M2’) in a photostationary state. Indeed, as with D96N crystals at neutral pH (13), a full color-change from purple to yellow upon illumination of D96A crystals at pH 8.5 at ambient temperature indicated the virtually complete conversion to the M state. This state was then trapped by rapidly dropping the temperature to 100K, as before (13).
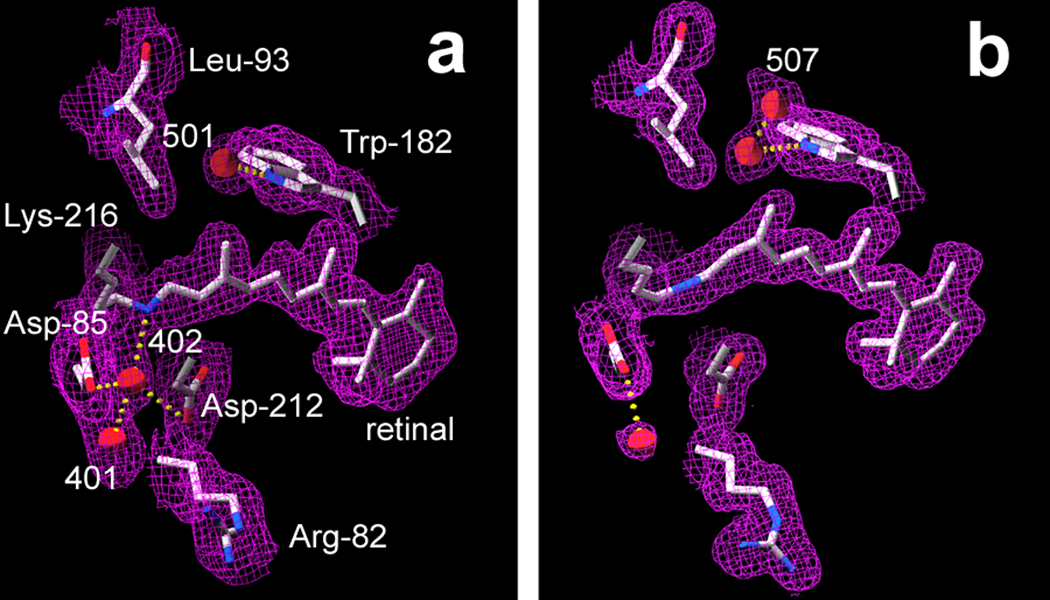
Figures 5a and 5b compare 2Fobs − Fcalc electron density maps of the non-illuminated and M states of D96A, respectively. As expected, the trapped M state of this mutant bears considerable similarity to the earlier reported (13) M of D96N: the retinal is 13-cis,15-anti, wat402 and wat406 are absent in the map (latter not shown), wat401 forms a hydrogen-bond with OD2 of Asp-85, and the Arg-82 side-chain is rotated toward the extracellular surface. An unexpected feature in M is a new water molecule, wat507, hydrogen-bonded to wat501. A water molecule had been reported (13) at this location in the non-illuminated, although not in the M state, of D96N.
FIGURE 5.
Electron density (2Fobs − Fcalc) maps of the retinal region in the non-illuminated state (a) and the trapped M state (b), in the D96A mutant. Contour level at 1 σ.
Elsewhere in the protein, however, the M states of D96A and D96N are different. Figures 6a and 6b show models of the retinal region in the M states (atomic colors) of D96N and D96A, respectively, each superimposed on the non-illuminated state of the same mutant (blue color). In D96N (Figure 6a) the rotation of the retinal C15=NZ-CE segment with the deprotonated Schiff base from the extracellular to the cytoplasmic direction and the accompanying changes of the geometry of the retinal polyene chain and the Lys-216 side-chain move the main-chain of helix G at Lys-216 and therefore at Ala-215 (for a detailed discussion of this, see refs. 12, 13). Displacement of O of Ala-215 breaks its hydrogen-bond with wat501, and there is no electron density for this water in M. The connection of helix G to helix F through this water and Trp-182 is thereby eliminated, and the Trp ring is free to tip upward from the approach of the 13-methyl group of the retinal (Figure 6a). Movement of Lys-216 moves the connected wat502 and the O of Thr-46, moving OG1 of Thr-46 away from Asn-96. Inasmuch as the same kinds of changes occur at these locations in the trapped M state of the E204Q mutant (12), this structural shift may occur in the wild-type M as well. It should have the rationale to lower the pKa of Asp-96 so it will become the proton donor to the Schiff base.
FIGURE 6.
Comparison of the retinal region of the trapped M states of the D96N (13) (a) and D96A (b) mutants (shown with atomic colors) with their respective non-illuminated states (in blue).
The displacements of Ala-215, Lys-216, wat502 and wat401 (but not Trp-182, and obviously not the retinal) in the M of D96N (Figure 6a) are similar to those in the non-illuminated state of the D96A mutant (Figure 3a), although they are greater in magnitude. Thus, in both models the O of Ala-215 moves away from wat501, wat502 is moved by displacement of Lys-216, and wat401 approaches the two aspartate carboxyls. In the M state the perturbation originates at the retinal and spreads to Asn-96 via Thr-46 (Figure 3a), while in D96A the perturbation originates at residue 96 and spreads to Thr-46 and then the retinal region (Figure 6a). It appears that the same “track” of covalent and hydrogen-bonds can be the means for propagating structural perturbation in both directions.
The changes in the position of Ala-215, wat502, and wat401 in M of D96N will have occurred partly in the non-illuminated D96A already. Consistent with perturbation of this region, while the movements of the retinal in the M state of D96A (Figure 6b) are similar to those of the M state of D96N (Figure 6a), the ensuing displacements of Lys-216, Ala-215, Lys-216, and wat501 are less extensive. For example, the movement of O of Ala-215 in the M state of D96N relative to the non-illuminated state is 0.97 Å, but in the M state of D96A it is only 0.55 Å. Water 502, connected to O of Lys-216, moves by 1.00 Å in M of D96N but 0.51 Å in the M state of D96A. As a result, in the M state of D96A communication of the retinal region with the Thr-46 region will have been impaired and Thr-46 should be less displaced.
Figures 7a and 7b compare the region of Thr-46 in the non-illuminated and M states, in D96N and D96A, respectively. In M of D96N the displacement of wat502 toward O of Thr-46 allows movement of the peptide segment, and the ensuing torsion of the main-chain moves OG1 of Thr-46 away from wat504 (by 0.88 Å). The inter-atomic distance between these atoms increases from 3.29 Å to 4.02 Å and the weak hydrogen-bond breaks. Because the other hydrogen-bonds in this region are maintained, the bulky side-chain of Phe-42, a participant of the cytoplasmic hydrophobic barrier (10) like Phe-219, moves, presumably in anticipation of the increased hydration that allows conducting a proton to the retinal Schiff base in the next step of the photocycle.
FIGURE 7.
Comparison of the Asp-96/Thr-46 region in the trapped M states of the D96N (13) (a) and the D96A (b) mutants. The models for M are shown with atomic colors, the models for their respective non-illuminated states in blue.
In M of D96A the lesser, and more importantly lateral, displacement of wat502 relative to O of Thr-46 results in lesser changes at Thr-46 (Figure 7b). In D96A OG1 of Thr-46 undergoes virtually no movement in M (its displacement is only 0.14 Å).
In the earlier described trapped M state of D96N, the cytoplasmic ends of helices F and G, as well as part of the E-F interhelical loop, were too disordered to model (13). The disorder along helix F begins at Val-177, which is displaced in a manner consistent with the outward tilt of the cytoplasmic end of helix F detected in many other studies (16–22). Thus, it may be the result of the tilt that disrupts the crystal locally, and as such constitutes evidence for this large-scale conformational shift. However, we detect neither disorder nor observable tilt of helix F in the trapped M of D96A, consistent with the lack of structural changes at the Ala-96/Thr-46 pair (Figure 7b). In this respect also, the M states of D96A and D96N seem to differ. Such a difference was reported before. In the projection map of the M state of a 2-dimensional crystal of D96G mutant (17), density changes were detected at helices F and G, but the difference map had considerably less magnitude at helix F than in the M state of the wild-type and D96N. The lesser tilt of helix F was attributed to an alteration of conformational flexibility in the D96G mutant, but not in the more conservative D96N mutant. This observation and its interpretation are consistent with the 3-dimensional structure we report here for the D96A mutant.
CONCLUSIONS
The D96A and the T46V mutations in the cytoplasmic region of bacteriorhodopsin both break the functionally important hydrogen-bond between Asp-96 and Thr-46, but in the former mutant the structural perturbation is greater and propagates to the retinal and the extracellular region by a “track” of covalent and hydrogen-bonds that includes wat502 that links helix B to helix G. The same track is utilized in the functional cycle of this proton pump to propagate structural changes from the photoisomerized retinal to Asp-96. If one can generalize from these findings, it seems possible that the structure of well-chosen mutants of bacteriorhodopsin-like heptahelical membrane proteins will reveal the details of functionally relevant conformational changes otherwise difficult to measure.
ACKNOWLEDGEMENTS
We thank the beamline staff at SSRL for their essential assistance, and F. Jurnak and S. Heffron for help with cavity calculations.
The work as supported in part by grants to J.K.L. from NIH (R01-GM29498) and from DOE (DEFG03-86ER13525).
Footnotes
Abbreviation: octylglucoside, n-octyl-β-D-glucopyranoside.
REFERENCES
- 1.Brown LS. Proton transport mechanism of bacteriorhodopsin as revealed by site- specific mutagenesis and protein sequence variability. Biochemistry (Mosc. ) 2001;66:1249–1255. doi: 10.1023/a:1013131318620. [DOI] [PubMed] [Google Scholar]
- 2.Butt HJ, Fendler K, Bamberg E, Tittor J, Oesterhelt D. Aspartic acids 96 and 85 play a central role in the function of bacteriorhodopsin as a proton pump. EMBO J. 1989;8:1657–1663. doi: 10.1002/j.1460-2075.1989.tb03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mogi T, Stern LJ, Marti T, Chao BH, Khorana HG. Structure-function studies on bacteriorhodopsin. VII. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rath P, Krebs MP, He Y, Khorana HG, Rothschild KJ. Fourier transform Raman spectroscopy of the bacteriorhodopsin mutant Tyr-185→Phe: Formation of a stable O-like species during light adaptation and detection of its transient N-like photoproduct. Biochemistry. 1993;32:2272–2281. doi: 10.1021/bi00060a020. [DOI] [PubMed] [Google Scholar]
- 5.Balashov SP, Imasheva ES, Ebrey TG, Chen N, Menick DR, Crouch RK. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. doi: 10.1021/bi970744y. [DOI] [PubMed] [Google Scholar]
- 6.Brown LS, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi JK. Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J. Biol. Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 7.Richter HT, Needleman R, Lanyi JK. Perturbed interaction between residues 85 and 204 in Tyr-185 Phe and Asp-85→Glu bacteriorhodopsins. Biophys. J. 1996;71:3392–3398. doi: 10.1016/S0006-3495(96)79532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dioumaev AK, Richter HT, Brown LS, Tanio M, Tuzi S, Saito H, Kimura Y, Needleman R, Lanyi JK. Existence of a proton transfer chain in bacteriorhodopsin: participation of Glu-194 in the release of protons to the extracellular surface. Biochemistry. 1998;37:2496–2506. doi: 10.1021/bi971842m. [DOI] [PubMed] [Google Scholar]
- 9.Govindjee R, Misra S, Balashov SP, Ebrey TG, Crouch RK, Menick DR. Arginine-82 regulates the pKa of the group responsible for the light- driven proton release in bacteriorhodopsin. Biophys. J. 1996;71:1011–1023. doi: 10.1016/S0006-3495(96)79302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schobert B, Brown LS, Lanyi JK. Crystallographic structures of the M and N intermediates of bacteriorhodopsin: assembly of a hydrogen-bonded chain of water molecules between Asp96 and the retinal Schiff base. J. Mol. Biol. 2003;330:553–570. doi: 10.1016/s0022-2836(03)00576-x. [DOI] [PubMed] [Google Scholar]
- 11.Dioumaev AK, Brown LS, Needleman R, Lanyi JK. Coupling of the reisomerization of the retinal, proton uptake, and reprotonation of asp-96 in the N photointermediate of bacteriorhodopsin. Biochemistry. 2001;40:11308–11317. doi: 10.1021/bi011027d. [DOI] [PubMed] [Google Scholar]
- 12.Luecke H, Schobert B, Richter HT, Cartailler J-P, Rosengarth A, Needleman R, Lanyi JK. Coupling photoisomerization of the retinal in bacteriorhodopsin to directional transport. J. Mol. Biol. 2000;300:1237–1255. doi: 10.1006/jmbi.2000.3884. [DOI] [PubMed] [Google Scholar]
- 13.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999;286:255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 14.Yohannan S, Yang D, Faham S, Boulting G, Whitelegge J, Bowie JU. Proline substitutions are not easily accommodated in a membrane protein. J. Mol. Biol. 2004;341:1–6. doi: 10.1016/j.jmb.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 15.Rouhani S, Cartailler JP, Facciotti MT, Walian P, Needleman R, Lanyi JK, Glaeser RM, Luecke H. Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate. J. Mol. Biol. 2001;313:615–628. doi: 10.1006/jmbi.2001.5066. [DOI] [PubMed] [Google Scholar]
- 16.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramaniam S, Lindahl M, Bullough P, Faruqi AR, Tittor J, Oesterhelt D, Brown L, Lanyi J, Henderson R. Protein conformational changes in the bacteriorhodopsin photocycle. J. Mol. Biol. 1999;287:145–161. doi: 10.1006/jmbi.1999.2589. [DOI] [PubMed] [Google Scholar]
- 18.Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 19.Dencher NA, Dresselhaus D, Zaccai G, Bueldt G. Structural changes in bacteriorhodopsin during proton translocation revealed by neutron diffraction. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7876–7879. doi: 10.1073/pnas.86.20.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kataoka M, Kamikubo H, Tokunaga F, Brown LS, Yamazaki Y, Maeda A, Sheves M, Needleman R, Lanyi JK. Energy coupling in an ion pump. The reprotonation switch of bacteriorhodopsin. J. Mol. Biol. 1994;243:621–638. doi: 10.1016/0022-2836(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 21.Kamikubo H, Kataoka M, Varo G, Oka T, Tokunaga F, Needleman R, Lanyi JK. Structure of the N intermediate of bacteriorhodopsin revealed by x-ray diffraction. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1386–1390. doi: 10.1073/pnas.93.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamikubo H, Oka T, Imamoto Y, Tokunaga F, Lanyi JK, d Kataoka M. The last phase of the reprotonation switch in bacteriorhodopsin: the transition between the M-type and the N-type protein conformation depends on hydration. Biochemistry. 1997;36:12282–12287. doi: 10.1021/bi9712302. [DOI] [PubMed] [Google Scholar]
- 23.Brown LS, Kamikubo H, Zimanyi L, Kataoka M, Tokunaga F, Verdegem P, Lugtenburg J, Lanyi JK. A local electrostatic change is the cause of the large-scale protein conformation shift in bacteriorhodopsin. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5040–5044. doi: 10.1073/pnas.94.10.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorgeirsson TE, Xiao W, Brown LS, Needleman R, Lanyi JK, Shin YK. Transient channel-opening in bacteriorhodopsin: an EPR study. J. Mol. Biol. 1997;273:951–957. doi: 10.1006/jmbi.1997.1362. [DOI] [PubMed] [Google Scholar]
- 25.Wegener AA, Chizhov I, Engelhard M, Steinhoff HJ. Time-resolved detection of transient movement of helix F in spin- labelled pharaonis sensory rhodopsin II. J. Mol. Biol. 2000;301:881–891. doi: 10.1006/jmbi.2000.4008. [DOI] [PubMed] [Google Scholar]
- 26.Dioumaev AK, Brown LS, Needleman R, Lanyi JK. Partitioning of free energy gain between the photoisomerized retinal and the protein in bacteriorhodopsin. Biochemistry. 1998;37:9889–9893. doi: 10.1021/bi980934o. [DOI] [PubMed] [Google Scholar]
- 27.Lanyi JK, Schobert B. Local-global conformational coupling in a heptahelical membrane protein: transport mechanism from crystal structures of the nine states in the bacteriorhodopsin photocycle. Biochemistry. 2004;43:3–8. doi: 10.1021/bi035843s. [DOI] [PubMed] [Google Scholar]
- 28.Tittor J, Soell C, Oesterhelt D, Butt HJ, Bamberg E. A defective proton pump, point-mutated bacteriorhodopsin Asp96 Asn is fully reactivated by azide. EMBO J. 1989;8:3477–3482. doi: 10.1002/j.1460-2075.1989.tb08512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto H, Marti T, Holz M, Mogi T, Lindau M, Khorana HG, Heyn MP. Aspartic acid-96 is the internal proton donor in the reprotonation of the Schiff base of bacteriorhodopsin. Proc. Natl. Acad. Sci. U. S. A. 1989;86:9228–9232. doi: 10.1073/pnas.86.23.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y, Varo G, Chang M, Ni B, Needleman R, Lanyi JK. Water is required for proton transfer from aspartate-96 to the bacteriorhodopsin Schiff base. Biochemistry. 1991;30:10972–10979. doi: 10.1021/bi00109a023. [DOI] [PubMed] [Google Scholar]
- 31.Miller A, Oesterhelt D. Kinetic optimization of bacteriorhodopsin by aspartic acid 96 as an internal proton donor. Biochim. Biophys. Acta. 1990;1020:57–64. [Google Scholar]
- 32.Rummel G, Hardmeyer A, Widmer C, Chiu ML, Nollert P, Locher KP, Pedruzzi I, Landau EM, Rosenbusch JP. Lipidic cubic phases: new matrices for the three-dimensional crystallization of membrane proteins. J. Struct. Biol. 1998;121:82–91. doi: 10.1006/jsbi.1997.3952. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Sheldrick GMST. High resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 35.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 36.Schobert B, Cupp-Vickery J, Hornak V, Smith S, Lanyi J. Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal. J. Mol. Biol. 2002;321:715–726. doi: 10.1016/s0022-2836(02)00681-2. [DOI] [PubMed] [Google Scholar]
- 37.Lanyi JK, Schobert B. Mechanism of proton transport in bacteriorhodopsin from crystallographic structures of the K, L, M-1, M-2, and M-2 ' intermediates of the photocycle. J. Mol. Biol. 2003;328:439–450. doi: 10.1016/s0022-2836(03)00263-8. [DOI] [PubMed] [Google Scholar]
- 38.Sass HJ, Buldt G, Gessenich R, Hehn D, Neff D, Schlesinger R, Berendzen J, Ormos P. Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin. Nature. 2000;406:649–653. doi: 10.1038/35020607. [DOI] [PubMed] [Google Scholar]
- 39.Lanyi JK, Schobert B. Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: the switch in the bacteriorhodopsin photocycle. J. Mol. Biol. 2002;321:727–737. doi: 10.1016/s0022-2836(02)00682-4. [DOI] [PubMed] [Google Scholar]
- 40.Facciotti MT, Rouhani S, Burkard FT, Betancourt FM, Downing KH, Rose RB, McDermott G, Glaeser RM. Structure of an early intermediate in the M-state phase of the bacteriorhodopsin photocycle. Biophys. J. 2001;81:3442–3455. doi: 10.1016/S0006-3495(01)75976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda K, Matsui Y, Kamiya N, Adachi S, Okumura H, Kouyama T. Crystal structure of the M intermediate of bacteriorhodopsin: allosteric structural changes mediated by sliding movement of a transmembrane helix. J. Mol. Biol. 2004;341:1023–1037. doi: 10.1016/j.jmb.2004.06.080. [DOI] [PubMed] [Google Scholar]
- 42.Maeda A. Internal water molecules as mobile polar groups for light-induced proton translocation in bacteriorhodopsin and rhodopsin as studied by difference FTIR spectroscopy. Biochemistry (Mosc ) 2001;66:1256–1268. doi: 10.1023/a:1013183302690. [DOI] [PubMed] [Google Scholar]
- 43.Kandori H. Hydration switch model for the proton transfer in the Schiff base region of bacteriorhodopsin. Biochim. Biophys. Acta. 2004;1658:72–79. doi: 10.1016/j.bbabio.2004.03.015. [DOI] [PubMed] [Google Scholar]