Abstract
Overproduction of amyloid precursor protein (APP) and β-amyloid likely contribute to neurodegeneration seen in Alzheimer's Disease (AD). APP mRNA contains several, 3′-untranslated region (UTR), cis-acting regulatory elements. A 52 base element (52sce), immediately downstream from the stop codon, has been previously shown to complex with uncharacterized cytoplasmic proteins. In this study, we purify and identify six proteins that specifically bind to the 52sce, and show that these proteins interact with each other and with APP mRNA in intact human neuroblastoma cells. We also present evidence that at least one of these proteins, the DEAD-box helicase rck/p54, is involved in post-transcriptional regulation, as the its overexpression in cultured cells results in elevated levels of APP mRNA and protein. These findings suggest a novel mechanism for post-transcriptional regulation of APP mRNA.
Keywords: Alzheimer's disease, β-Amyloid Precursor Protein, 3′-UTR regulatory elements, rck/p54
1. Introduction
Alzheimer's Disease (AD) is characterized by the accumulation of neuritic plaques and neurofibrillary tangles in the neocortex, limbic system and hippocampus (Braak and Braak, 1995; Braak and Braak, 1996), resulting in synaptic dysfunction, neuronal loss and relentlessly progressive dementia (Mattson, 2004). A major component of senile plaques are various sized β-amyloid (Aβ) peptides, typically 39 – 42 amino acids long, with longer species more prone to aggregation (Balbach et al, 2000; Pike et al, 1995). Aβ is generated by β- and γ-secretase on Amyloid Precursor Protein (APP) (Greenfield et al, 1999).
Overexpression of APP mRNAs are associated with the development of AD and Down Syndrome (DS). Overexpression of wild-type APP mRNA and protein leads to increased production, secretion and intracellular accumulation of amyloidogenic fragments (Mattson, 1997). Patients with DS trisomy 21, on which the APP gene resides, ubiquitously overexpress APP mRNA by 1.5 – 2 fold and invariably develop the full histological lesions of Alzheimer's Disease by the age of 40 (Buckland et al, 1993; Neve et al, 1988; Oyama et al, 1994). Both in DS (Buckland et al, 1993; Rumble et al, 1989) and in early-onset AD (Higgins et al, 1988), overexpression of APP mRNA is closely and positively correlated with downstream APP protein expression and Aβ deposition. This suggests that dysregulation of transcription and/or APP mRNA degradation can promote development of AD pathology.
All isoforms of APP mRNA share a common 1.8 kb 3′ untranslated region (UTR), which is encoded by exon 18. This 3′-UTR contains several cis – acting regulatory elements which influence the stability of APP mRNA (Amara et al, 1999; Westmark et al, 2006; Zaidi and Malter, 1994). We have previously shown that one such element is located in the first 52 bases downstream of the stop codon. Deletion of this 52-base stop codon element (52sce) dramatically destabilized APP mRNA in intact cells (Westmark et al, 2006).
Cytoplasmic proteins expressed across a variety of human cell types specifically bind 52sce RNA in electrophoretic mobility shift assays (EMSA). The expression of eGFP-APP 3′-UTR mRNAs in SHSY5Y cells caused the destabilization of endogenous APP mRNA, presumably through titration of 52sce – binding proteins (Westmark et al, 2006). This suggests that the interaction between the 52sce and its' protein co-factors is important for APP mRNA stability.
In this study we purify and identify six cytoplasmic proteins that specifically interact with 52sce. All six are known mRNA-binding proteins. With the exception of nucleolin, which has been shown to interact with an additional regulatory element in the APP 3′ – UTR (Zaidi and Malter, 1995), these proteins were not known to bind to APP mRNA. The identified proteins are part of the complex with APP 3′-UTR sequences based on EMSA, and specifically co-immunoprecipitate with APP mRNA from cultured human neuroblastoma. We also show that one of these proteins, the DEAD-box helicase rck/p54 (Akao et al, 1995), is involved in the regulation of cellular APP mRNA and protein levels. Augmentation of intracellular rck/p54 protein levels, either through TAT-protein transduction or through rck/p54 cDNA transfection into live cells, resulted in APP mRNA and protein overexpression comparable to that seen in Down's Syndrome (Tanzi et al, 1987) and AD (Palmert et al, 1988) brain tissue. These data suggest a novel mechanism in post-transcriptional regulation of APP mRNA, which may be relevant to AD pathology.
2. Materials and methods
2.1. Antibodies
The anti-nucleolin (catalog #sc-8031), anti – β-tubulin (catalog #sc-8035) monoclonal antibodies, as well as anti-MeCP2 (catalog #sc-20700) and anti – La (catalog #sc-33593) rabbit polyclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-TatYB1 polyclonal antibody was raised as described previously (Capowski et al, 2001). Anti-eFF1α monoclonal antibody (catalog #05-235) was purchased from Upstate Cell Signaling Solutions (Lake Placid, NY). Human anti-La/SS-B polyclonal antibody (catalog #L1380) was purchased from United States Biological (Swampscott, MA). SW1, SW3 and SW5.8 anti-La/SS-B monoclonal antibodies (Pruijn et al, 1995) were a generous gift from Ger Pruijn.
Anti-rck/p54 polyclonal antibody was donated by Yukihiro Akao or purchased from MBL International (Woburn, MA; catalog #PD009). Anti-rck/p54 monoclonal antibody (DDX6 monoclonal antibody, clone 3D2) was purchased from Abnova Corporation (Taipei, Taiwan; catalog #H00001656-M01). Anti-PAI-RBP1 polyclonal antibody was a generous gift from Thomas Gelehrter. Anti-PAI-RBP1 monoclonal antibody was purchased from GeneTex (San Antonio, TX; catalog #GTX90457). Anti-APP polyclonal antibody was from Zymed Laboratories (South San Francisco, CA; catalog #51-2700). Anti-mouse β-actin antibody (catalog #A5441), mouse IgG (catalog #I5381) were purchased from Sigma Chemical Company (St. Louis, MO).
2.2. In vitro transcription
DNA oligonucleotides containing a T7 polymerase start site 5′ to APP specific sequences were generated by Integrated DNA Technologies. The APP specific sequences were as follows: 52sce FL: APP751 cDNA (accession #X06989) 2381 – 2432; Δ22: APP751 cDNA 2403 – 2432; sh52sce: APP751 cDNA 2403 – 2454. Oligonucleotides containing the reverse complement sequences were also generated. Forward and reverse sequences were annealed to generate double-stranded DNA templates for transcription. In vitro transcription was carried out with T7 RNA polymerase. Large quantities of RNA were generated with the MEGAscript T7 kit (Ambion). RNA was radiolabelled or biotinylated by including 32P-α-CTP (Amersham) or biotin-14-CTP (Invitrogen), respectively, in the transcription reaction. Transcription was carried out at 37°C for 2 hours. Transcripts were extracted with phenol-chloroform, precipitated with isopropanol, dissolved in diethyl pyrocarbonate-treated H2O, and quantified by absorbance at 260 nm.
2.3. Cell culture and lysates
SH-SY5Y cells were cultured on 12-well plates (Falcon) or on cover slips coated with Human Placental Collagen Type VI (Sigma) in 50/50 mix of Dulbeco's Modified Eagles Medium and Ham's F12 (50/50 DMEM/F12 mix; Gibco), supplemented with 1mM sodium pyruvate, 0.1mM L-glutamine, 50 μg/ml penicillin/streptomycin and 10% fetal bovine serum (FBS, Gibco) in a 5% CO2 atmosphere at 37°C. K562 cells were cultured in RPMI 1640 supplemented with 0.1mM L-glutamine, 50 μg/ml penicillin/streptomycin and 10% fetal bovine serum (FBS) in a 5% CO2 atmosphere at 37°C.
Cells were directly pelleted (K562 cells) or trypsinized and pelleted (SH-SY5Y cells), washed with DPBS and resuspended in IP buffer (20 mM HEPES, pH 7.9, 50 mM KCl, 2.5 mM MgCl2, 0.1% NP-40, protease inhibitor cocktail, 1 mM DTT). Cells were lysed by swelling on ice for 10 min and the nuclei were pelleted at 600g for 20 min at 4 °C. The supernate fraction was collected as the cytosolic lysate. Lysates were used directly or stored at −80 °C.
2.4. Northwestern and immunoblotting
Proteins were denatured by incubation at 95 °C for 5 – 10 min in SDS sample buffer (4% SDS, 12% glycerol, 50 mM Tris (pH 6.8), 2% β-mercaptoethanol, 0.06% bromophenol blue) and electrophoresed on a Tris-Glycine-SDS polyacrylamide gel. Proteins were transferred onto a nitrocellulose membrane using the Trans-Blot Cell (Bio-Rad), according to the manufacturer's protocol.
For northwestern blotting, the membrane was incubated sequentially at room temperature in buffer NW (25mM HEPES, pH 8.0, 40mM KCl, 5mM MgCl2, 0.75mM DTT) containing 6M, 3M, 1.5M, 0.75M, 0.37M, 0.182M and 0M Guanidinium HCl. The membrane was blocked in buffer NW, 5% nonfat dry milk (NFDM) at room temperature for 90 min, and incubated at room temperature overnight with 107 cpm of radiolabelled RNA probe in buffer NW, 40U/ml RNasin Plus. The membrane was washed extensively in buffer NW, air-dried for 20 – 30 min, and imaged on a Storm® 860 Phosphorimager (Molecular Dynamics).
For immunoblotting, the membrane was blocked at room temperature for 1 hr in TBS-T (15mM TrisHCl, pH 7.6, 137mM NaCl, 0.1% Tween®-20), 5% NFDM, washed in TBS-T and incubated at room temperature for 240 min with a dilution of the primary antibody in TBS-T. Anti-nucleolin and anti-La/SS-B antibodies were diluted as per manufacturer's recommendations. Anti-rck/p54, anti-PAI-RBP1, anti-TatYB1 and anti-EF1α antibodies were biotinylated with the FluoReporter® Mini-Biotin-XX Protein Labelling Kit (Molecular Probes), according to the manufacturer's protocol. The biotinylated antibodies were diluted as follows for use in immunoblotting: 1:500 anti-TatYB1; 1:1000 anti-rck/p54; 1:500 anti-PAI-RBP1 and 1:1000 anti-EF1α. After incubation with the primary antibody, the membrane was washed in TBS-T, and incubated with a 1:1000 dilution of the appropriate HRP-conjugated secondary antibody, or 1:10000 dilution of peroxidase-conjugated streptavidin in TBS-T. Blots were imaged with ECL+ substrate on a Storm® 860 Phosphorimager (Molecular Dynamics).
2.5. RNA mobility shift assays
Radiolabeled RNA (100,000 cpm or 2.5 ng) was mixed with 5 μg of total cell lysate, and gel shifts were performed as described previously (Zaidi et al, 1994; Zaidi and Malter, 1995).
2.6. Protein purification and identification
K562 S100 lysate (900 μg) was pre-cleared by incubation at 37 °C for 30 minutes with 20 μg biotinylated 52sce FL RNA tethered to Dynabeads M-280 Strepavidin (Invitrogen) in Buffer L (10 mM Tris-HCl, pH 7.5, 0.1M KCl, 1mM DTT), 1X protease inhibitor cocktail, 0.1% bovine serum albumin, 2850 Units/ml RNasin Plus, for 30 min at 37 °C. Precleared lysate was incubated at 37 °C for 1 hr with 30 μg biotinylated sh52sce RNA tethered to Dynabeads M-280 Strepavidin, under the same buffer conditions. Dynabeads were washed with Buffer L, and proteins were eluted for 30 min at 37 °C with 300 μg free sh52sce RNA. A second elution with one bed volume of 2x SDS sample buffer (8% SDS, 24% glycerol, 100 mM Tris (pH 6.8), 4% β-mercaptoethanol, 0.12% bromophenol blue) was carried out at 95 °C for 5 min. Eluates were subjected to the Northwestern Blot procedure with the sh52sce radiolabelled RNA as probe. Active bands were excised out of an identically ran SDS-PAGE gel stained with Coomassie Brilliant Blue and subjected to MALDI-MS Peptide Mass Mapping at the Columbia University Protein Chemistry Core Facility (Columbia University, New York, NY). Peptide mass values were queried against the Mascot protein sequence database (Matrix Science, www.matrixscience.com).
2.7. Immunoprecipitation, RNA isolation, cDNA synthesis and PCR analysis of APP mRNA
SH-SY5Y cell cytosol from 2X107 cells was pre-cleared with protein G Dynabeads in IP buffer containing phosphatase inhibitors ( 0.1mM Na3VO4, 10mM NaF, 20mM α-glycerophosphate) and 1000U/ml RNasin Plus. Pre-cleared lysate was incubated with 5 μg antibody and 50 μl Protein G Dynabeads (stock suspension volume) at 4°C for 5 hours. For La/SS-B immunoprecipitation, 50 μl Protein G Dynabeads (stock suspension volume) were pre-incubated for 1 hour at 4 °C with 50 μl SW1 and 50 μl SW5.8 hybridoma (Pruijn et al, 1995) supernates; pre-cleared SH-SY5Y cytosol was mixed with the antibody-bound beads and incubated as above. Beads were washed 5 times with IP buffer and a portion of the IP reaction was transferred to TRI-Reagent (MRC, Inc.), total RNA isolated and precipitated in the presence of 50 μg glycogen. The final pellet was solubilized in diethyl pyrocarbonate treated H2O and reverse-transcribed with Qiagen Omniscript and random nonamer primer (60 min at 37 °C, 5 min at 93 °C). The cDNA was PCR-amplified [15 min at 95 °C, (40 cycles: 30 sec at 95 °C, 1 min at 68 °C, 1 min at 72 °C), 10 min at 72 °C] with HotStarTaq DNA polymerase (Qiagen) and primers against the human APP gene (accession #X06989; forward: 899 – gacgaaccctacgaagaagcca – 920 and reverse: 1248 – gctttctggaaatgggcatgttc – 1226) which span the alternatively spliced site in the APP cDNA sequence. PCR reactions were analyzed on 1.5% agarose gels containing EtBr. Alternatively, cDNA was subjected to real time PCR with APP ORF FWD and APP ORF REV primers as described below. The amount of APP signal, n = 3, was quantified and normalized to that seen in the pre-immune mouse IgG immunoprecipitate, ± S.E.M.
2.8. Plasmid preparation
Preparation of pTAT-HA-GFP (Capowski et al, 2001), CMV-eGFP-APP 3′-UTR and CMV-eGFPAPPΔ52sce 3′-UTR (Westmark et al, 2006) has been described previously.
For pTAT-HA-RCK plasmid preparation, the open reading frame of rck/p54 (NM_004397 bases 367-1785) was amplified with primers designed to add a unique NcoI site to the 5′ end, and a unique XhoI site to the 3′ end. The PCR product was digested with NcoI and XhoI and ligated to NcoI/XhoI digested pTAT-HA vector (Vocero-Akbani et al, 2000), which upon translation produces rck/p54 with N-terminal TAT and HA tags.
For pIRES-RCK-hrGFPII plasmid preparation, the open reading frame of rck/p54, except for the stop codon (NM_004397 bases 367-1782) was amplified. The forward primer was designed to add a unique BamHI site, followed by a consensus Kozak sequence (GCC ACC) immediately 5′ to the ATG start codon (NM_004397 bases 367- 369). The reverse primer was designed to generate a unique NotI site 3′ to the rck/54 open reading frame. The PCR product was digested and ligated to BamHI/NotI digested pIRES-hrGFP II plasmid (Stratagene), to produce pIRES-RCK-hrGFP II, which upon transcription produces a bicistronic mRNA with rck/p54 ORF followed by an internal ribosome entry site and an hrGFP ORF.
Rck/p54 insert sequence and in-frame insertion were verified by DNA sequencing at the UW Biotech Center DNA Sequencing Facility (University of Wisconsin – Madison, Madison, WI). The sequencing primers used were as follows: Rck/p54 (accession #NM_004397): 367 – atgggtctgtccagtcaa - 384; 867 - acaggtcagtcaaatttg – 884; 1367 – agcgagttgaattgctag – 1384; 617 – taatcttcaaactcatttc – 600; 1117 – tagcgtgagaataatatc – 1000; 1617 – gctaagccaagatgacca – 1600.
2.9. His-tagged protein purification and transduction
For purification of His-tagged TAT-RCK and TAT-GFP, BL21 strain E. coli transformed with the appropriate plasmid were grown and induced with 1mM IPTG for 10 hours as in (Capowski et al, 2001). Proteins were purified from inclusion bodies as described in (Capowski et al, 2001). Eluted proteins were dialyzed against 1X PBS, 10% glycerol and concentrated with Centricon-10 (TAT-GFP) or Centricon-30 (TAT-RCK) centrifugal concentrators (Amicon) according to manufacturer's instructions.
2.10. TAT-fusion protein transduction into SH-SY5Y cells
SH-SY5Y cells were seeded at a density of 1.5×105 cells/well and used for TAT-fusion protein transduction when cell density reached approximately 80%. TAT-fusion protein was added into the media to a final concentration of 50 nM. Every 6 hours, media was aspirated and replaced with fresh media, to which fresh TAT-fusion protein was added.
2.11. cDNA transfection of SH-SY5Y cells
SH-SY5Y cells were seeded onto Human Placental Collagen Type VI coated coverslips in 12-well plates, at a density of 1.5×105 cells/well and cultured for 48 hours prior to transfection. Transfection was performed with 0.4 μg of pIRES-RCK-hrGFP II or pIRES-hrGFP II, or 1 μg of CMV-eGFP-APP 3′-UTR or CMV-eGFP-APP52sce 3′-UTR, and Lipofectamine® (Invitrogen) according to the manufacturer's instructions.
2.12. Real Time PCR
PCR primers were designed with Primer Express software from Applied Biosystems (Foster City, CA) and BLAST homology searches of the amplicons revealed the primers to be gene-specific. PCR reactions were optimized for primer and template concentrations and contained 200 nM APP ORF primers (accession #X06989; ORF FWD: 1479 – tggccctggagaactacatca – 1499; ORF REV: 1565 – cgcggacatacttctttagcatatt – 1541), 50 nM eGFP primers (accession #AJ890283 FWD: 625 - agcaaagaccccaacgagaa – 644; REV: 684 – ggcggcggtcacgaa – 660), 200nM α-tubulin primers (accession #NM_006000 FWD:666 – tggtggacaacgaagcaatc – 685; REV: 726 – gttgggcgctcgatgtct – 709) or 100 nM S26 primers (accession #NM_001029; forward: 406 – cgcagcagtcagggacattt – 425, reverse: 474 – ttcacatacagcttgggaagc – 454), 10.5 μl 1:5 diluted RT reaction and 12.5 SYBR green PCR mix in a 25 μl reaction volume. The cycle conditions were: 2 min at 50 °C, 10 min at 95 °C, (40 cycles: 15 sec at 95 °C, 1 min at 60 °C), followed by a dissociation stage for 15 sec at 95 °C, 1 min at 60 °C, 15 sec at 95 °C. The average PCR efficiencies for all three sets of primers were 100 ± 5%.
2.13. Confocal microscopy and APP/RCK analysis by immunohistochemistry (IHC)
Immunohistochemistry and confocal microscopy were performed essentially as described in (Westmark et al, 2006). Briefly, SH-SY5Y cells were fixed to Collagen VI-coated coverslips with 4% paraformaldehyde and permeabilized with 100% methanol. Cells were stained with 0.5 μg/ml anti-Amyloid Precursor Protein polyclonal antibody (Zymed) and 3 μg/ml anti-rck/p54 monoclonal antibody (Amaxa) in 1X DPBS/1% FBS, stained with Rhodamine Red™-x goat anti-mouse IgG (H+L) and AlexaFluor® 633-x goat anti-rabbit IgG (H+L) (Invitrogen) diluted 1:500 in 1X DPBS/1% FBS, and mounted on microscope slides. GFP and rhodamine were imaged as in (Westmark et al, 2006); AlexaFluor® 633 was imaged at 633nm (excitation) and 650nm (emission). Image acquisition and analysis of relative red/green/blue fluorescence intensities with ImageJ software were carried out as described in (Westmark et al, 2006). Approximately 50 cells were quantitated for each data set.
3. Results
3.1. Protein-binding region within the 52sce RNA
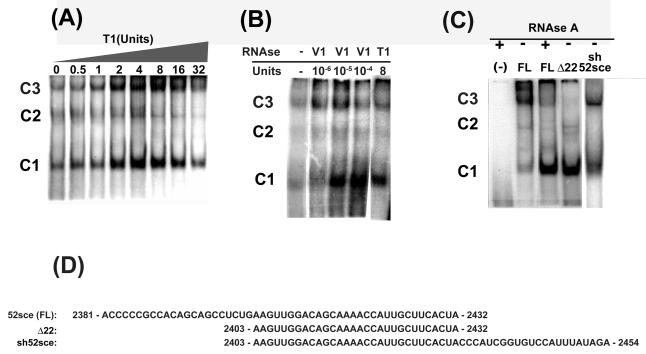
The 52sce element in the 3′UTR of APP mRNA specifically interacts with multiple cellular proteins based on electrophoretic mobility shift assay (EMSA; (Westmark et al, 2006)). EMSA procedure typically included treatment of the protein-RNA complex with RNAse T1 prior to electrophoresis. To test the suitability of the 52sce oligoribonucleotide for affinity purification of its' binding proteins, we tested whether the protein-RNA interactions would be preserved without T1 RNAse. We performed EMSA's with increasing amounts of RNAse T1, ranging from zero to 32 units RNAse T1 per reaction (Figure 1A). As shown, RNA-protein complexes 1, 2 and 3 (C1, C2, C3 in Figure 1, corresponding to bands 1, 2, 3, respectively, in (Westmark et al, 2006)) increased as the amount of T1 increased, reaching maximal levels at 4-8 units T1. These data suggested that only part of the 52sce was required for protein binding or that it adopted a secondary structure unfavorable to protein-RNA interactions. Treatment of 52sce RNA with RNAse V1, which recognizes and cuts double-stranded or helical RNA, with subsequent electrophoresis of RNA fragments on a denaturing sequencing gel showed that bases 1 – 22 of 52sce were predominantly double stranded (data not shown). When EMSAs were performed with RNAse V1, all three RNA-protein complexes increased as the amount of RNAse V1 increased (Figure 1B). RNAse V1 was much more potent than RNAse T1 in enhancing protein-RNA complex formation, requiring only 10−5 units to attain the levels seen in the control reaction performed with 8 units of RNAse T1. These data suggested that the 5′ 22 bases of 52sce contain a double stranded or helical region which interferes with protein binding. Deletion of the first 22 bases of 52sce markedly enhanced the formation of Complexes 1 and 2 as compared with the full length 52sce when no RNAse treatment was applied (Figure 1C). These data are consistent with earlier observations that the 5′ half of the 52sce has only 0.4% of the element's total protein-binding affinity. (Westmark et al, 2006). Complex 3, however, was decreased upon deletion of the first 22 bases of 52sce, as well as upon treatment of the full-length 52sce with RNAse A. Complex 3 was recovered when the 22 bases immediately 3′ to the 52sce in the APP 3′-UTR (APP751 cDNA bases 2433 -2454) were included in the RNA probe, restoring it to its' original length of 52 bases. This oligoribonucleotide (APP751 cDNA bases 2403 – 2454, hereafter “sh52sce”) displayed the same pattern of protein binding without exogenous RNAse treatment as the full length 52sce did when treated with RNAse, while retaining most, if not all, protein binding affinity (Figure 1C). Therefore, sh52sce oligoribonucleotide was used for affinity purification of 52sce binding proteins.
Figure 1.
The 3′ 30 bases of 52sce RNA are sufficient for protein-RNA complex formation, which is blocked by the 5′ 22 bases. (A) K562 whole cell lysate was incubated with radiolabelled 52sce, then treated with increasing amounts of RNAse T1. (B) K562 whole cell lysate was incubated with radiolabelled 52sce, then treated with indicated amounts of RNAse V1 or RNAse T1. (C) K562 whole cell lysate was incubated with radiolabelled full-length (FL) 52sce oligoribonucleotide, a truncated 52sce with 22 bases deleted from the 5′ end (Δ22), or an oligoribonucleotide containing the 3′ 30 bases of 52sce and the 22 bases immediately downstream of 52sce in the APP 3′ UTR (sh52sce). (D) FL, Δ 22 and sh52sce sequences are given. Numbering reflect APP751 cDNA sequence (accession #X06989).
3.2. Purification and identification of 52sce-binding proteins
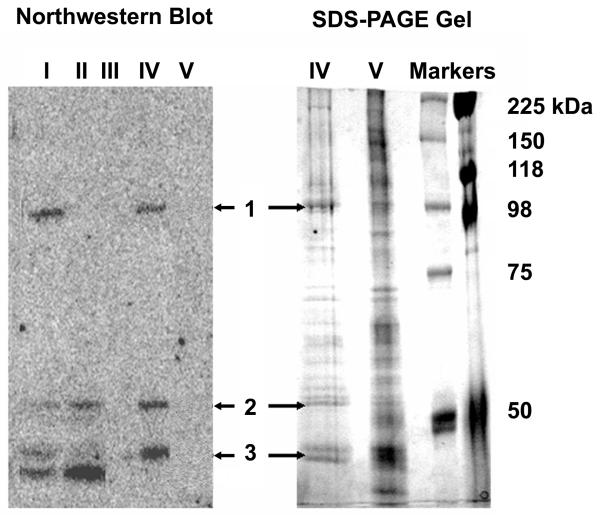
A variety of groups have used affinity chromatography for the rapid isolation of RNA binding proteins (Neupert et al, 1990; Pinol-Roma et al, 1990; Swanson and Dreyfuss, 1988). Therefore, we prepared biotinylated sh52sce oligoribonucleotide, and immobilized it on strepavidin-conjugated beads. Because 52sce binds the target proteins poorly without RNAse pre-treatment, we used 52sce-conjugated beads to pre-clear K562 whole cell lysate. The flow-through was subsequently incubated with sh52sce – conjugated beads to pull down the 52sce – binding proteins. After several washes, 52sce – binding proteins were eluted off the beads with a 10-fold excess of free sh52sce RNA. A second elution was performed by incubating the beads in 1% SDS at 95 °C. All fractions were separated by SDS-PAGE and analyzed for 52sce binding activity by northwestern blotting with radiolabeled sh52sce. As shown in Figure 2, crude K562 lysate showed 4 distinct bands, 3 of which were pulled down by the affinity purification and eluted with sh52sce RNA, but not with the denaturing elution. The 3 bands, one ~100 kD, and the other two ~50 kD, were excised and subjected to trypsin digestion and liquid chromatography/ mass spectroscopy of the resulting peptides. Since Band 3 corresponded to a closely running doublet on the SDS-PAGE gel, we could not readily determine which of the two bands represented the 52sce – binding activity and subjected both bands to mass spectroscopy.
Figure 2.
Purification and identification of 52sce-binding proteins. K562 whole cell lysate was incubated with biotinylated oligoribonucleotide (sh52sce; APP751 mRNA bases 2403 – 2454) tethered to a strepavidin affinity column matrix. The column was eluted with a 10-fold excess of free sh52sce, and column fractions were run out on a Northwestern blot probed with radiolabelled sh52sce. Active bands in the eluate (bands 1,2, and 3) were excised from an identically-ran SDS-PAGE gel and identified through Mass Spectroscopy. I: K562 total cell lysate; II: K562 total cell lysate pre-cleared with 52sce RNA; III: Affinity Column flow-through. IV: First eluate (with free sh52sce RNA); V: Second eluate (with SDS denaturing buffer).
A search of the mass spectroscopy data against the Mascot protein sequence database (www.matrixscience.com) produced 16 matches with a Mowse score above the threshold of significance (Supplementary Table 1). Of those 16, we chose 6 as likely to be bona-fide 52sce binding proteins (Table 1). All 6 of these proteins had the appropriate molecular weight, matched the mass spectroscopy data with very high significance and all 6 were known RNA-binding proteins. As these proteins were purified using the protein-binding region of 52sce as bait, eluted with free sh52sce RNA and shown to have 52sce-binding activity on the northwestern blot, they are likely bona-fide 52sce binding proteins. Therefore, these data strongly suggest that we have obtained positive identifications for many, if not all, 52sce – binding proteins.
Table 1. Putative 52sce – binding Proteins.
All peptide matches with a Mowse score above the threshold of significance (≥ 45) generated from peptide mass mapping for bands 1, 2 and 3. Predicted molecular weight in KD is included for each match, as well as the number of trypsin-digested peptides that were matched to each protein.
| Protein Name | NCBI Accession Numbers (H. sapiens) | ||
|---|---|---|---|
| Gene | mRNA(RefSeq) | Protein (RefSeq) | |
| Nucleolin | 4691 | NM_005381 | NP_005372 |
| Plasminogen Activator Inhibitor –RNA Binding Protein 1 (PAI-RBP1) |
26135 |
NM_015640 (Transcript Variant 4) |
NP_056455 (Transcript Variant 4) |
| Rck/p54 | 1656 | NM_004397 | NP_004388 |
| Autoantigen La/Sjogren Syndrome Antigen B (La/SS-B) |
6741 | NM_003142.2 | NP_003133 |
| Elongation Factor 1α (EF1α) | 1915 | NM_001402 | NP_001393 |
| Y Box Binding Protein 1 (YB-1). | 4904 | NM_004559.2 | NP_004550 |
3.2. RNA-dependent and RNA-independent interactions between 52sce-binding proteins
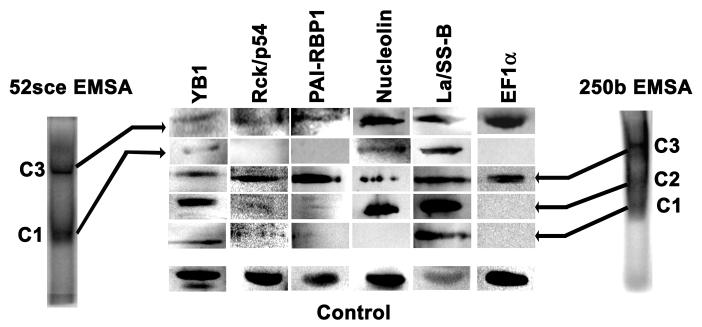
To confirm that the identified proteins were indeed present in the protein-RNA complexes seen by EMSA (Westmark et al, 2006), RNA-protein complexes obtained with K562 cell lysate and radiolabelled sh52sce or the first 250 bases of the APP 3′ UTR, were excised and subjected to SDS-PAGE and immunoblotting. As shown in Figure 3, several complexes of different apparent molecular weight were observed for each probe. These complexes were different with respect to their protein composition. All six putative 52sce binding proteins were found in the same complex with the first 250 bases of the 3′ UTR. Two other complexes with the same RNA did not contain rck/p54, PAI-RBP1 or EF1α, and one of those also did not contain nucleolin. All six proteins were found in Complex 3 with sh52sce, but only Nucleolin, La/SS-B and YB-1 were seen in Complex 1.
Figure 3.
Purified 52sce-binding proteins complex with APP mRNA sequences in electrophoretic mobility shift assay. K562 total cell lysate was incubated with radiolabelled sh52sce RNA or with RNA corresponding to the first 250 bases of APP751 3′UTR (bases 2381 – 2630). Protein-RNA complexes were excised from native gels, and gel fragments electrophoresed into SDS-PAG followed by transfer and and immunoblot against indicated targets. Control: K562 total cell lysate.
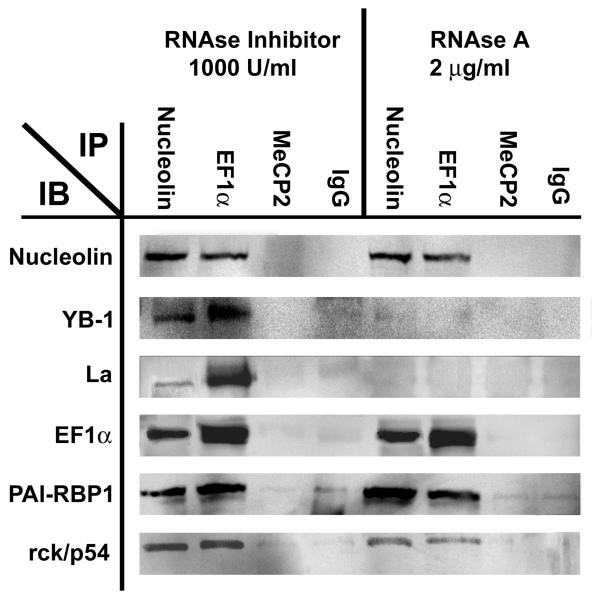
To determine whether the interactions between 52sce-binding proteins are RNA-dependent, we performed immunoprecipitations for nucleolin and EF1α either under RNA-protecting conditions (in the presence of RNAse inhibitors) or RNA-destroying conditions (RNAse inhibitors omitted and an excess of RNAse A added to the starting material). The immunoprecipitates were subjected to immunoblotting with antisera against the 52sce binding proteins. As shown in Figure 4, nucleolin and EF1α both coimmunoprecipitated with all other 5 proteins in question. However, the co-immunoprecipitations of nucleolin with La/SS-B or with YB-1 were RNA-dependent, whereas those with of nucleolin with rck/p54, PAI-RBP1, or EF1α were RNA-independent. Likewise, EF1α co-immunoprecipitations with nucleolin, PAI-RBP1 and rck/p54 were RNA independent, but those with La and YB-1 were RNA-dependent. This suggests the 52sce-protein complex contains both protein-protein interactions (nucleolin, EF1α, rck/p54 and PAI-RBP1) and protein-RNA interactions (YB-1, La/SS-B).
Figure 4.
RNA-dependent and RNA-independent interactions within the 52sce – binding complex. Immunoprecipitation with an antibody specific against the indicated target was carried out either in the presence or absence of RNAse A. Immunoprecipitates were immunobloted with antibody against the indicated target.
Control immunoprecipitations with an irrelevant antibody against methyl CpG binding protein 2 (MeCP2) and with pre-immune mouse IgG pulled down negligible amounts of 52sce-binding proteins in comparison to nucleolin and EF1α immunoprecipitations.
3.3. In-vivo association between 52sce-binding proteins and APP mRNA
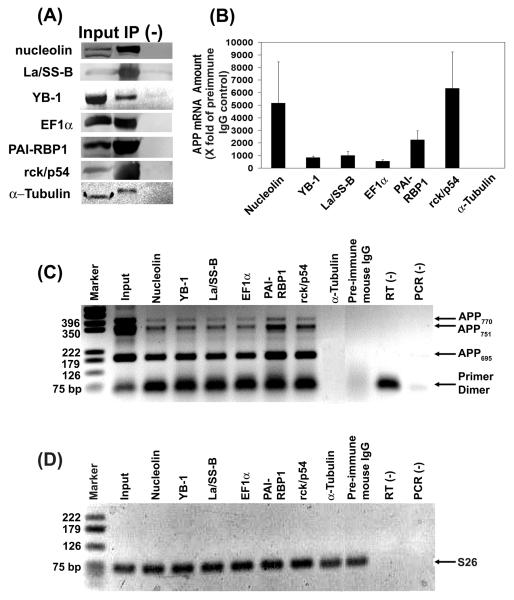
We have purified proteins that bind in vitro to the 52sce and the sequence immediately downstream of it. Interactions with this oligoribonucleotide may or may not mirror those seen in intact cells with full-length APP mRNA. Therefore, we immunoprecipitated all six 52sce-binding proteins from the cytosol fraction of SH-SY5Y human neuroblastoma cell line (Figure 5A). Based on RT/qPCR analysis of the control and specific antibody pellets (Figure 5B), all six proteins co-immunoprecipitated with APP mRNA. The APP mRNA signal in each precipitated fraction was at least 500-fold stronger than the one seen for the precipitated fraction from the negative control immunoprecipitation (with anti – α-tubulin antibody) or from the non-specific immunoprecipitation control (with pre-immune mouse IgG). Furthermore, the association between 52sce-binding proteins and APP mRNA held true for all three major isoforms of APP – APP695, APP751 and APP770, present in SH-SY5Y cells, whereas none of the three isoforms were detected in the precipitated fraction with anti – α-tubulin or with pre-immune mouse IgG (Figure 5C). This was to be expected, as the entire 3′ UTR of APP, including the 52sce, is conserved between the three isoforms (Sandbrink et al, 1993). Furthermore, since the APP695 and APP751 isoforms are obtained through alternative pre-mRNA splicing (Neve et al, 1988), their corresponding PCR products cannot be generated from genomic DNA with this set of primers. Likewise, the primers used in real-time PCR (Figure 5B) span a 19-kilobase intron in the APP gene, and thus are extremely unlikely to amplify APP from genomic DNA under the two-step protocol we used for real-time PCR. This, in combination with the absence of APP signal in the RT and PCR no template controls (Figure 5C, “RT (−)” and “PCR (−)” lanes, respectively) suggests that the amplified product is resulting from APP mRNA, and not from APP DNA contamination.
Figure 5.
52sce-binding proteins co-immunoprecipitate with APP mRNA from a human neuroblastoma cell line. (A) The indicated proteins were immunoprecipitated from SH-SY5Y cytosol fraction (“Input”) with a specific antibody (“IP”) or with pre-immune mouse IgG (“(−)”). (B) mRNA from immunoprecipitates was reverse-transcribed with random primer and subjected to qPCR with APP-specific primers. The amount of APP signal, n = 3, was quantified and normalized to that seen in the pre-immune mouse IgG immunoprecipitate, ± S.E.M. (C) cDNA obtained from immunoprecipitates in a representative experiment in (B) was amplified with primers specific for the APP alternative splice site, and the PCR products were electrophoresed on a 1.5% agarose gel (inverted color scheme). Marker: pGEM DNA size markers (Promega); Input: SH-SY5Y cytosol; RT(−): RT without template; PCR(−): PCR without template. (D) Same set of immunoprecipitates as in (C) was amplified with primers specific for S26, and the PCR products were electrophoresed on a 1.5% agarose gel (inverted color scheme). All lanes are the same as in (C).
The mRNA for S26, an abundant and constitutively expressed ribosomal component protein (Ivanov et al, 2005), was present in equal amounts in all the immunoprecipitates from the same set of reactions, presumably due to non-specific binding to the matrix (Figure 5D). This suggests that the presence of APP signal in the 52sce binding protein immunoprecipitates cannot be explained by non-specific mRNA binding, otherwise APP would also be detected in equal measure in the precipitated fraction with β-tubulin and pre-immune mouse IgG. These data confirm that the six 52sce-binding proteins specifically associate with APP mRNA in human neuroblastoma cells.
3.4. Overexpression of rck/p54 increases APP mRNA and protein levels in SH-SY5Y human neuroblastoma
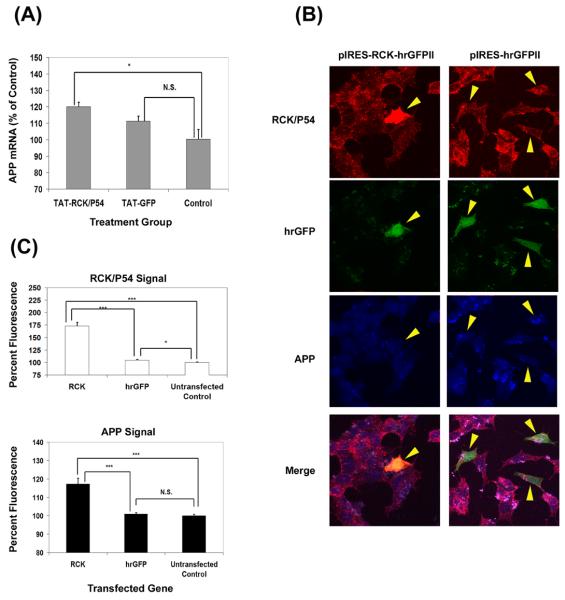
The expression of eGFP-APP 3′-UTR mRNAs in SH-SY5Y cells caused the destabilization of endogenous APP mRNA. This effect required only the first 250 bases of the APP 3′ UTR and was lost if the 52sce was deleted ((Westmark et al, 2006) and PR Westmark, manuscript in preparation). These data suggested that titration of binding proteins to the 52sce containing transgene reduced the stability of endogenous APP mRNA. Since rck/p54 and its homologues are members of a well-conserved family of RNA helicases, known to be involved in control of RNA degradation (Akao et al, 2003; Chu, and Rana, 2006; Coller et al, 2001; Tanner and Linder, 2001; Weston and Sommerville, 2006), we decided to investigate the effects of altering rck/p54 protein levels on APP mRNA and protein levels in human cells. Recombinant rck/p54 fused at the N-terminus to the HIV TAT protein membrane permeabilization domain (TAT-RCK) was transduced into SH-SY5Y cells. TAT-GFP transduction was used as a nonspecific control. Eighteen hours after TAT-fusion protein transduction, total RNA was isolated and APP mRNA levels quantified by qPCR. S26 mRNA was used as a loading control for the qPCR. APP mRNA levels were normalized to those in untreated control cells. As shown in Figure 6A, TAT-RCK transduction produced a statistically significant (p < 0.05) increase in APP mRNA steady state levels to 120 ± 2.6% of control. TAT-GFP transduction increased APP APP mRNA steady state levels to 111 ± 3% of control, but this increase was not statistically significant (p > 0.15).
Figure 6.
Rck/p54 overexpression increases APP mRNA and protein levels in SH-SY5Y cells. (A) 50nM TAT-RCK or TAT-GFP was transduced into SH-SY5Y cells. APP mRNA levels were quantified by real time PCR. The amount of signal, n = 3, was quantified and normalized to the amount present in untreated control cells, ± S.E.M. *: p<0.05; N.S. = not significant (p > 0.1) (two-tailed, homoscedasic T-test). (B) SH-SY5Y cells were transfected with various expression vectors as shown. Eighteen hours later, cells were fixed and stained with rhodamine-conjugated anti-rck/p54 and AlexaFluor® 633 conjugated anti-APP antibodies. Representative positive transfected cells are shown. (C) APP and rck/p54 staining was quantified for cells transfected with pIRES-RCK-hrGFP II (RCK) or pIRES-hrGFP II (hrGFP). The amount of signal from approximately 50 positively transfected cells was quantified and normalized to approximately the same number of untransfected cells. (*: p<0.05; ***: p <0.001; N.S. = not significant (p > 0.1) (two-tailed, heteroscedasic T-test).
To investigate the effect of rck/p54 overexpression on APP protein levels in cultured cells, we transfected rck/p54 cDNA into SH-SY5Y cells. The transfected vector coded for a bicistronic mRNA which coded for rck/p54 as well as GFP and transfected cells show green fluorescence. As a control, cells were transfected with GFP cDNA alone. At 18 hours after transfection, cells were fixed and stained with rhodamine-conjugated anti-rck/p54 and Alexa633 – conjugated anti-APP. Total APP expression was compared in transfected (green) to non-transfected cells. As shown in Figure 6B, approximately 5% of the cells were transfected. Analysis by confocal microscopy showed that rck/p54 was increased to 173 ± 8% over untransfected cells within the same field, which resulted in an increase of APP to 117 ± 3% compared to untransfected cells (Figure 6C). Both changes were highly significant statistically (p <10−5) when compared to either untransfected cells or cells transfected with GFP alone (Figure 6C). More cells were transfected with GFP cDNA and they displayed a slight, but statistically significant (p < 0.05) increase in endogenous levels of rck/p54 (104 ± 1.5%) compared to untransfected cells. However, APP levels were unchanged (100.9 ± 0.75%, p > 0.4; Figure 6C). These data strongly suggest that augmentation of rck/p54 levels in cultured cells increases total APP mRNA and protein levels and are consistent with the hypothesis that 52sce – binding proteins function as APP mRNA stabilizers.
3.5 Overexpression of rck/p54 increases eGFP-APP 3′-UTR mRNA levels in SH-SY5Y human neuroblastoma in a 52sce – dependent manner.
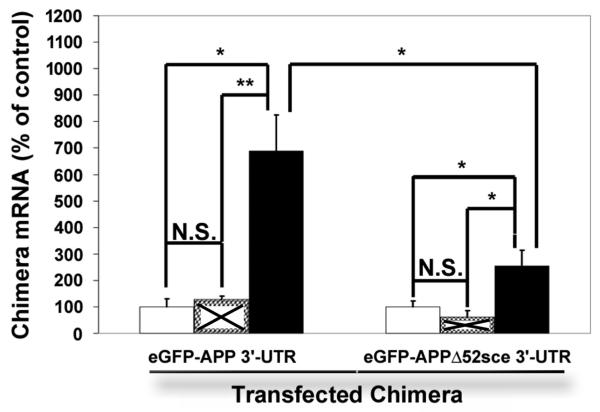
To determine whether the stabilizing effect of rck/p54 overexpression on APP mRNA was modulated by the 52sce, we transfected SH-SY5Y human neuroblastoma with CMV-eGFP–APP 3′-UTR or CMV-eGFP-APPΔ52sce 3′-UTR. Eighteen hours after transfection, cells were transduced with 50nM TAT-RCK or TAT-GFP. Twelve hours after transduction, total RNA was isolated and eGFP - APP chimera mRNA levels measured by qPCR. S26 mRNA was used as a loading control for the qPCR. eGFP-APP mRNA levels were normalized to those in untreated control cells. TAT-RCK transduction resulted in a 6.8 fold ± 19% increase in the eGFP – APP 3′-UTR mRNA levels (Figure 7). This increase was statistically significant as compared both to the untransduced control (p < 0.05) and to TAT-GFP transduced cells (p < 0.01). TAT-GFP transduction resulted in a 0.28 fold ± 12.5% increase in the eGFP – APP 3′UTR mRNA levels which was not statistically significant (p > 0.3). Likewise, TAT-GFP had no effect (p > 0.3) on the eGFP – APPΔ52sce 3′-UTR mRNA levels. TAT-RCK transduction increased APPΔ52sce 3′-UTR mRNA levels 2.53 fold ± 20% over untransduced control. This change was statistically significant as compared both to the untransduced control (p < 0.05) and the TAT-GFP transduced cells (p < 0.05), but was markedly and significantly (p<0.05) smaller than that seen for eGFP – APP 3′-UTR mRNA. Neither TAT-GFP nor TAT-RCK transduction resulted in an increase in S26 or α-tubulin mRNA levels (data not shown), demonstrating a specific effect on mRNAs containing the APP 3′ UTR rather than a nonspecific change in overall mRNA decay.
Figure 7.
Rck/p54 overexpression increases EGFP-APP chimera mRNA levels in SH-SY5Y cells in a 52sce – dependent manner. SH-SY5Y were transfected with EGFP-APPwt or EGFP-APPΔ52sce plasmids as shown. Eighteen hours after transfection, 50nM TAT-RCK (■) or TAT-GFP (☒) was transduced into the same cells. eGFP-APP chimera mRNA levels were quantified by qPCR. The amount of signal, n = 3, was quantified and normalized to the amount present in untransduced control cells (□), ± S.E.M. **:> p<0.01;*: p<0.05; N.S. = not significant (p > 0.1) (two-tailed, homoscedasic T-test).
4. Discussion
In this study, we describe the purification and identification of six proteins that bind to the 52sce in the 3′-UTR of APP mRNA. Our data do not support previous observations that the entire 52 bases of the element are necessary for protein binding (Westmark et al, 2006). The present results suggest the functional domain is located midway within the element (Westmark et al, 2006). Bases 22 -26 (from the 5′ end) appear to be essential and likely enable in vitro protein binding. Adding 22, 3′ additional bases also had a positive effect on protein binding, particularly for the high molecular weight complex (C3). This suggests that part of the loss of the protein-binding affinity of the 3′ half of the element was secondary to reduced length, which may have resulted in an unfavorable change in secondary structure.
Of the six purified 52sce – binding proteins, nucleolin is the only one previously known to bind APP mRNA and affect the rate of its decay (Westmark and Malter, 2001; Zaidi and Malter, 1995). It was not, however, previously known to bind to the 52sce. All six are established RNA-binding proteins. Nucleolin and YB-1 have been implicated in the post-transcriptional regulation of multiple mRNA species, including bcl-2 (Sengupta et al, 2004), CD154 (Singh et al, 2004), GM-CSF (Capowski et al, 2001), APP (Zaidi and Malter, 1995). There is little sequence similarity between the binding sites of these proteins on different mRNA targets, and their binding may be mediated by secondary or higher structure of the mRNA molecule. Consistent with that hypothesis, mutations that affect the secondary structure of 52sce also affect its affinity for 52sce-binding proteins (P.R. Westmark, manuscript in preparation).
PAI-RBP1 is recently described (Heaton et al, 2001) and appears to bind to an A-rich sequence in the 3′-UTR of Plasminogen Activator Inhibitor 1 (PAI1) mRNA and modulate cAMP-induced changes in PAI1 mRNA stability (Heaton et al, 2003). Neither the 52sce nor the 22 bases immediately downstream of it contains an A-rich stretch. Therefore, PAI-RBP1 likely participates in the APP mRNA – binding complex through a protein-protein interaction with nucleolin.
PAI-RBP1 has also been shown to associate with progesterone (P4) receptors and mediate anti-apoptotic effects of P4 on luteal and granulosa cells in rat ovaries (Engmann et al, 2006; Peluso et al, 2005). The effects of P4 include neuroprotection in several regions of the brain (reviewed in Wojtal et al, 2006). Since APP has been implicated in synaptogenesis and neuroprotection against excitotoxic, metabolic and oxidative insults, modulation of APP expression is a possible mechanism of progesterone-mediated neuroprotection. In fact, Chao et. al (1994) have shown that progesterone and estrogen treatment of ovariectomized female rats increased APP695 mRNA expression in the hippocampus, whereas treatment with estrogen alone did not have a significant effect. Whether PAI-RBP1 acts downstream of P4 in the nervous system, as it does in the ovaries, remains to be determined.
Autoantigen La controls the stability and initiates the translation of multiple viral RNA species (Chan and Tan, 1987; Pruijn et al, 1995), and binds to and stabilizes H4 histone mRNA (McLaren et al, 1997). It has also been found in complex with small nuclear RNAs, but the consequences of this association are unknown (Pruijn et al, 1995). EF1 regulates peptide chain elongation during translation. It uses energy from GTP hydrolysis to bind to aminoacyl-tRNA and facilitate tRNA binding to the ribosomal A site, followed by transfer of the amino acid onto the nascent peptide chain (Marco et al, 2004). EF1α contains the GTP-binding and GTP-hydrolyzing activities (Marco et al, 2004). It was not previously known to bind to mRNA or play a part in mRNA stabilization. In the case of the 52sce- binding protein complex, the role of EF1α may likewise involve using energy from GTP hydrolysis to facilitate an event such as the binding of cofactors or a change in the secondary structure of APP mRNA.
Rck/p54 is a member of the highly conserved DEAD-box family of RNA helicases, which are known to be involved in many post-transcriptional processes requiring a change of mRNA secondary structure, including RNA degradation and translational repression (Akao et al, 2003; Coller and Parker, 2005; Weston and Sommerville, 2006). Ski2p, a DEAD-box helicase is contained in a heterotrimeric complex which recruit the exosome to mRNA molecules for decay (Parker and Song, 2004). Ski2p may use energy from ATP hydrolysis to alter RNA structures or proteins that hinder exosome digestion (Parker and Song, 2004). Conversely, rck/p54 could participate in maintaining APP mRNA in a favorable conformation for interactions with stabilizing protein factors, attenuating exosomal digestion. In this study, modest increases in rck/p54 increased APP mRNA and protein levels by significant levels which was comparable to that seen in DS fetal brains (Tanzi et al, 1987). Palmert et. al. likewise report a 1.5 – 2 fold increase in APP mRNA expression in the nucleus basalis and locus coeruleus of AD patients as compared to age-matched healthy controls (Palmert et al, 1988).
Overexpression of rck/p54 had a much more pronounced effect upon the levels of eGFP-APP 3′-UTR chimera mRNAs than endogenous APP mRNA. This may be partially explained by eGFP-APP 3′-UTR expression under the control of a constitutive, highly active CMV promoter. Coupled to a higher rate of production, even small increases in mRNA stability would be expected to result in dramatic accumulation. Deletion of the 52sce reduced the TAT-RCK induction of the eGFP-APP chimera reporter mRNA steady-state levels by more than 2.5-fold, but did not completely abolish it. This suggests that although the 52sce is important for the stabilizing action of rck/p54, another domain in the APP 3′ UTR may also be involved.
Furthermore, since rck/p54 associates APP 3′-UTR as part of a multiprotein complex, it is likely that it is not solely responsible for the stabilizing effect of that complex. Consistent with that assumption, overexpression of 52sce in SH-SY5Y cells, a treatment that would be expected to decoy all the 52sce binding proteins, resulted in a ~3-fold reduction in APP protein levels and a ~4-fold reduction in APP mRNA levels (Westmark et al, 2006). 52sce-binding proteins other than rck/p54 may play a stabilizing role. Studies of the effects of overexpression and knock-down of other 52sce-binding proteins are under way in our laboratory.
Table 2. Identities of purified 52sce-binding proteins. NCBI database accession numbers for the human genes are included, as well as mRNA and amino acid reference sequences.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grants R01 AG10675 and P30 HD03352 (to J.S.M.), and by a private donation by William and Jean Roper (to the Waisman Center). We are indebted to Dr. Thomas Gelehrter (University of Michigan Medical School) for the generous gift of the anti-PAI-RBP1 polyclonal antibody, Dr. Ger J. M. Pruijn (Katholieke Universiteit Nijmegen, Nijmegen, Netherlands) for the gift of the SW1, SW3, and SW5.8 anti-La/SS-B monoclonal antiobides, and Dr. Yukihiro Akao (Gifu International Institute of Biotechnology, Gifu, Japan) for the gift of the anti-rck/p54 polyclonal antibody. We also wish to thank them for providing the respective immunoprecipitation and immunoblot protocols. We thank Dr. Mary Ann Gawinowicz (Columbia University Protein Core and DNA Sequencing Facility, Columbia University, New York, NY) for MALDI-MS peptide mass mapping, Dr. Cara Westmark for training in real-time PCR and the immunoprecipitation/RNA extraction protocol, and Drs. Janet Mertz and Jeffrey Ross for advice in experimental design.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
The authors hereby declare that they have no actual or potential conflicts of interest.
References
- Akao Y, Marukawa O, Morikawa H, Nakao K, Kamei M, Hachiya T, Tsujimoto Y. The rck/p54 candidate proto-oncogene product is a 54-kilodalton D-E-A-D box protein differentially expressed in human and mouse tissues. Cancer Res. 1995;55:3444–3449. [PubMed] [Google Scholar]
- Akao Y, Yoshida H, Matsumoto K, Matsui T, Hogetu K, Tanaka N, Usukura J. A tumour-associated DEAD-box protein, rck/p54 exhibits RNA unwinding activity toward c-myc RNAs in vitro. Genes Cells. 2003;8:671–676. doi: 10.1046/j.1365-2443.2003.00665.x. [DOI] [PubMed] [Google Scholar]
- Amara FM, Junaid A, Clough RR, Liang B. TGF-beta(1), regulation of alzheimer amyloid precursor protein mRNA expression in a normal human astrocyte cell line: mRNA stabilization. Brain Res. Mol. Brain Res. 1999;71:42–49. doi: 10.1016/s0169-328x(99)00158-8. [DOI] [PubMed] [Google Scholar]
- Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, Dyda F, Reed J, Tycko R. Amyloid fibril formation by A beta 16-22, a seven-residue fragment of the Alzheimer's beta-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–13759. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Evolution of the neuropathology of Alzheimer's disease. Acta Neurol. Scand. Suppl. 1996;165:3–12. doi: 10.1111/j.1600-0404.1996.tb05866.x. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol. Aging. 1995;16:271–8. doi: 10.1016/0197-4580(95)00021-6. discussion 278-84. [DOI] [PubMed] [Google Scholar]
- Buckland P, Tidmarsh S, Spurlock G, Kaiser F, Yates M, O'Mahony G, McGuffin P. Amyloid precursor protein mRNA levels in the mononuclear blood cells of Alzheimer's and Down's patients. Brain Res. Mol. Brain Res. 1993;18:316–320. doi: 10.1016/0169-328x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- Capowski EE, Esnault S, Bhattacharya S, Malter JS. Y box-binding factor promotes eosinophil survival by stabilizing granulocyte-macrophage colony-stimulating factor mRNA. J. Immunol. 2001;167:5970–5976. doi: 10.4049/jimmunol.167.10.5970. [DOI] [PubMed] [Google Scholar]
- Chan EK, Tan EM. Human autoantibody-reactive epitopes of SS-B/La are highly conserved in comparison with epitopes recognized by murine monoclonal antibodies. J. Exp. Med. 1987;166:1627–1640. doi: 10.1084/jem.166.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HM, Spencer RL, Frankfurt M, McEwen BS. The effects of aging and hormonal manipulation on amyloid precursor protein APP695 mRNA expression in the rat hippocampus. J. Neuroendocrinol. 1994;6:517–521. doi: 10.1111/j.1365-2826.1994.tb00614.x. [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation Repression in Human Cells by MicroRNA-Induced Gene Silencing Requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engmann L, Losel R, Wehling M, Peluso JJ. Progesterone regulation of human granulosa/luteal cell viability by an RU486-independent mechanism. J. Clin. Endocrinol. Metab. 2006;91:4962–4968. doi: 10.1210/jc.2006-1128. [DOI] [PubMed] [Google Scholar]
- Greenfield JP, Tsai J, Gouras GK, Hai B, Thinakaran G, Checler F, Sisodia SS, Greengard P, Xu H. Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer beta-amyloid peptides. Proc. Natl. Acad. Sci. U. S. A. 1999;96:742–747. doi: 10.1073/pnas.96.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton JH, Dlakic WM, Dlakic M, Gelehrter TD. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the Type-1 plasminogen activator inhibitor mRNA. J. Biol. Chem. 2001;276:3341–3347. doi: 10.1074/jbc.M006538200. [DOI] [PubMed] [Google Scholar]
- Heaton JH, Dlakic WM, Gelehrter TD. Posttranscriptional regulation of PAI-1 gene expression. Thromb. Haemost. 2003;89:959–966. [PubMed] [Google Scholar]
- Higgins GA, Lewis DA, Bahmanyar S, Goldgaber D, Gajdusek DC, Young WG, Morrison JH, Wilson MC. Differential regulation of amyloid-beta-protein mRNA expression within hippocampal neuronal subpopulations in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1297–1301. doi: 10.1073/pnas.85.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Malygin AA, Karpova GG. Human ribosomal protein S26 suppresses the splicing of its pre-mRNA. Biochim. Biophys. Acta. 2005;1727:134–140. doi: 10.1016/j.bbaexp.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Marco E, Martin-Santamaria S, Cuevas C, Gago F. Structural basis for the binding of didemnins to human elongation factor eEF1A and rationale for the potent antitumor activity of these marine natural products. J. Med. Chem. 2004;47:4439–4452. doi: 10.1021/jm0306428. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer's disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol. Rev. 1997;77:1081–1132. doi: 10.1152/physrev.1997.77.4.1081. [DOI] [PubMed] [Google Scholar]
- McLaren RS, Caruccio N, Ross J. Human La protein: a stabilizer of histone mRNA. Mol. Cell. Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert B, Thompson NA, Meyer C, Kuhn LC. A high yield affinity purification method for specific RNA-binding proteins: isolation of the iron regulatory factor from human placenta. Nucleic Acids Res. 1990;18:51–55. doi: 10.1093/nar/18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RL, Finch EA, Dawes LR. Expression of the Alzheimer amyloid precursor gene transcripts in the human brain. Neuron. 1988;1:669–677. doi: 10.1016/0896-6273(88)90166-3. [DOI] [PubMed] [Google Scholar]
- Oyama F, Cairns NJ, Shimada H, Oyama R, Titani K, Ihara Y. Down's syndrome: up-regulation of beta-amyloid protein precursor and tau mRNAs and their defective coordination. J. Neurochem. 1994;62:1062–1066. doi: 10.1046/j.1471-4159.1994.62031062.x. [DOI] [PubMed] [Google Scholar]
- Palmert MR, Golde TE, Cohen ML, Kovacs DM, Tanzi RE, Gusella JF, Usiak MF, Younkin LH, Younkin SG. Amyloid protein precursor messenger RNAs: differential expression in Alzheimer's disease. Science. 1988;241:1080–1084. doi: 10.1126/science.2457949. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A, Losel R, Wehling M. Expression and function of PAIRBP1 within gonadotropin-primed immature rat ovaries: PAIRBP1 regulation of granulosa and luteal cell viability. Biol. Reprod. 2005;73:261–270. doi: 10.1095/biolreprod.105.041061. [DOI] [PubMed] [Google Scholar]
- Pike CJ, Overman MJ, Cotman CW. Amino-terminal deletions enhance aggregation of beta-amyloid peptides in vitro. J. Biol. Chem. 1995;270:23895–23898. doi: 10.1074/jbc.270.41.23895. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Swanson MS, Matunis MJ, Dreyfuss G. Purification and characterization of proteins of heterogeneous nuclear ribonucleoprotein complexes by affinity chromatography. Methods Enzymol. 1990;181:326–331. doi: 10.1016/0076-6879(90)81133-f. [DOI] [PubMed] [Google Scholar]
- Pruijn GJ, Thijssen JP, Smith PR, Williams DG, Van Venrooij WJ. Anti-La monoclonal antibodies recognizing epitopes within the RNA-binding domain of the La protein show differential capacities to immunoprecipitate RNA-associated La protein. Eur. J. Biochem. 1995;232:611–619. doi: 10.1111/j.1432-1033.1995.611zz.x. [DOI] [PubMed] [Google Scholar]
- Rumble B, Retallack R, Hilbich C, Simms G, Multhaup G, Martins R, Hockey A, Montgomery P, Beyreuther K, Masters CL. Amyloid A4 protein and its precursor in Down's syndrome and Alzheimer's disease. N. Engl. J. Med. 1989;320:1446–1452. doi: 10.1056/NEJM198906013202203. [DOI] [PubMed] [Google Scholar]
- Sandbrink R, Banati R, Masters CL, Beyreuther K, Konig G. Expression of L-APP mRNA in brain cells. Ann. N. Y. Acad. Sci. 1993;695:183–189. doi: 10.1111/j.1749-6632.1993.tb23049.x. [DOI] [PubMed] [Google Scholar]
- Sengupta TK, Bandyopadhyay S, Fernandes DJ, Spicer EK. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- Singh K, Laughlin J, Kosinski PA, Covey LR. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J. Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Gusella JF, Watkins PC, Bruns GA, George-Hyslop P, Van Keuren ML, Patterson D, Pagan S, Kurnit DM, Neve RL. Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235:880–884. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- Vocero-Akbani A, Lissy NA, Dowdy SF. Transduction of full-length Tat fusion proteins directly into mammalian cells: analysis of T cell receptor activation-induced cell death. Methods Enzymol. 2000;322:508–521. doi: 10.1016/s0076-6879(00)22046-6. [DOI] [PubMed] [Google Scholar]
- Westmark CJ, Malter JS. Extracellular-regulated kinase controls beta-amyloid precursor protein mRNA decay. Brain Res. Mol. Brain Res. 2001;90:193–201. doi: 10.1016/s0169-328x(01)00112-7. [DOI] [PubMed] [Google Scholar]
- Westmark PR, Shin HC, Westmark CJ, Soltaninassab SR, Reinke EK, Malter JS. Decoy mRNAs reduce beta-amyloid precursor protein mRNA in neuronal cells. Neurobiol. Aging. 2006;27:787–796. doi: 10.1016/j.neurobiolaging.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtal K, Trojnar MK, Czuczwar SJ. Endogenous neuroprotective factors: neurosteroids. Pharmacol. Rep. 2006;58:335–340. [PubMed] [Google Scholar]
- Zaidi SH, Denman R, Malter JS. Multiple proteins interact at a unique cis-element in the 3′-untranslated region of amyloid precursor protein mRNA. J. Biol. Chem. 1994;269:24000–24006. [PubMed] [Google Scholar]
- Zaidi SH, Malter JS. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J. Biol. Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]
- Zaidi SH, Malter JS. Amyloid precursor protein mRNA stability is controlled by a 29-base element in the 3′-untranslated region. J. Biol. Chem. 1994;269:24007–24013. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.