Abstract
Ovarian serous carcinoma, the most common and lethal type of ovarian cancer, was thought to develop from two distinct molecular pathways. High-grade (HG) serous carcinomas contain frequent TP53 mutations while low-grade (LG) carcinomas arise from serous borderline tumors (SBT) and harbor mutations in KRAS/BRAF/ERBB2 pathway. However, the molecular alterations involved in the progression from SBT to LG carcinoma remain largely unknown. As well, the extent of deletion of tumor suppressors in ovarian serous carcinomas has not been well-studied. To further address these two issues, we assessed DNA copy number changes among affinity-purified tumor cells from 37 ovarian serous neoplasms including SBT, LG and HG tumors using high density 250K SNP arrays. Chromosomal instability index as measured by changes in DNA copy number was significantly higher in HG than in LG serous carcinomas. Hemizygous ch1p36 deletion was common in LG serous carcinomas but was rarely seen in SBT. This region contains several candidate tumor suppressors including miR-34a. In contrast, in HG serous carcinomas, significant numbers of amplifications and deletions including homozygous deletions were identified. Among homozygous deletions, loci containing Rb1, CDKN2A/B, CSMD1, and DOCK4 were most common, being present in 10.6%, 6.4%, 6.4% and 4.3%, respectively, in independent 47 affinity-purified HG serous carcinomas. Except the CDKN2A/B region, these homozygous deletions were not present in either SBT or LG tumors. Our study provides a genome-wide homozygous deletion profiles in HG serous carcinomas, serving as a molecular foundation to study tumor suppressors in ovarian cancer.
Introduction
In the United States, ovarian cancer is responsible for more cancer deaths than any other neoplasms of the female reproductive organs, with an estimated 15,520 deaths in 2008 (1). Ovarian cancer is a heterogeneous group of diseases, and among them, serous carcinoma is the most common type, representing more than half of ovarian cancers. The majority of serous carcinomas are high-grade and had been thought to arise in a progressive fashion from benign serous cystadenoma to serous borderline tumor (SBT, also known as serous tumor of low malignant potential or atypical proliferative serous tumor), to low-grade (LG) serous carcinoma, and then finally to high-grade (HG) serous carcinoma (2). However, clinicopathological observations and recent molecular genetic studies from several research groups have challenged this paradigm. Two distinct pathways are now thought to lead to the development of LG and HG serous carcinomas (3–10). LG carcinomas are now thought to develop from SBTs, and progress in a stepwise fashion because histological transitions can be found in SBT and LG serous carcinomas from the same specimen and furthermore, both SBT and LG lesions share similar molecular genetic changes (3). They are slow-growing, indolent tumors that have a relatively good prognosis as compared to HG carcinomas. Molecular genetic analysis has demonstrated that SBT/LG serous carcinomas typically display sequence mutations in KRAS/BRAF/ERBB2, but with infrequent mutations in TP53 (11–13). In contrast, HG serous carcinomas often present in advanced stages (stages III-IV) and rarely harbor mutations in KRAS/BRAF/ERBB2. HG serous carcinomas grow rapidly and are highly aggressive and over than 75% of these tumors harbor TP53 mutations (14–18).
DNA copy number alterations including chromosomal amplification, deletion and aneuploidy are the hallmarks of neoplasia (19). Amplification is one of the mechanisms that leads to an increase in activity of oncogenes and development of drug resistance (20, 21), while allelic deletion results in inactivation of tumor suppressors. Identification and characterization of genes within the amplified and deleted chromosomal loci not only provide new insights into the pathogenesis of cancer but may also lead to new approaches to diagnosis and therapy. Previous studies have applied different approaches to analyze DNA copy number changes on a genome-wide scale (22–28), but most combined different histological types of ovarian carcinomas such as serous, endometrioid, clear cell and mucinous tumors in the analysis, and very few included LG serous carcinomas and SBTs. A head-to-head comparison between HG and LG serous carcinomas and between LG serous carcinomas and their precursor lesions, SBTs, has not been published. Furthermore, although studies have identified several convincing amplification events in ovarian cancer, detection of deletions, especially homozygous ones, has been challenging because (1) it requires technologies with a sufficient resolution and (2) it requires samples that are highly enriched with tumor cells because the presence of normal stromal cells or endothelial cells can mask deletions. Thus, in this study we applied high density (250K) SNP arrays and used affinity-purified tumor cells from fresh specimens to address two biological questions that are related to the pathogenesis of ovarian cancer and have not yet been elucidated yet: (1) what is the cardinal molecular genetic alteration(s) during the transition from serous borderline tumor to low-grade serous carcinoma, and (2) what are the deleted tumor suppressor genes in high-grade serous tumors?
Material and Methods
Tissue Samples
Tissue samples from 12 SBTs, 12 LG ovarian serous carcinomas, and 13 HG ovarian serous carcinomas were collected from the Department of Pathology, Johns Hopkins Hospital (Baltimore, Maryland). The acquisition of the anonymous tissue specimens for this study was approved by the Institutional Review Board at the Johns Hopkins Medical Institutions. The status of primary/recurrence and the mutational status of TP53/KRAS/BRAF of each sample are listed in supplementary Table 1. Fresh tumor specimens were digested with collagenase (1 mg/ml), washed with RPMI medium, and affinity-purified using the anti-Ber-EP4-conjugated magnetic beads (Invitrogen, Carlsbad, CA) as previously described (29, 30). The purity of the isolated tumor cells was confirmed by immunostaining using an anti-cytokeratin 8 antibody (CAM5.2, Becton Dickinson, Franklin Lakes, NJ). The negatively sorted cellular fraction which represented the tumor adjacent normal stromal cells was separately cultured. The stromal fibroblast demonstrated distinct morphology and its purity were confirmed by positive vementin immunoreactivity and negative Ber-EP4 and CD31 immunoreactivities. Anti-vementin was purchase from Sigma (St. Louis, MO), anti-Ber-EP4 from Abcam (Cambridge, MA), and anti-CD31 from Invitrogen (Carlsbad, CA).
Single nucleotide polymorphism (SNP) array
SNPs were genotyped using 250K StyI arrays (Affymetrix, Santa Clara, CA) in the Microarray Core Facility at the Dana-Farber Cancer Institute (Boston, MA). A detailed protocol is available on the Core center webpage1. Briefly, genomic DNA was fragmented using the restriction enzyme, StyI, ligated with linkers, followed by PCR amplification. The PCR products were purified and then digested with DNaseI to a size ranging from 250 bp to 2,000 bp. Fragmented PCR products were labeled with biotin and hybridized to the arrays. Arrays were then washed with the Affymetrix fluidics stations. The bound DNA was fluorescently labeled using streptavidin-phycoerythrin conjugates and scanned using the Gene Chip Scanner 3000.
Analysis of SNP array data was performed using the dChip 2006 program (24, 31). Data was normalized to a baseline array with median signal intensity at the probe intensity level using the invariant set normalization method. A model-based (PM/MM) method was employed to obtain the signal values for each SNP in each array. Signal values for each SNP were compared with the average intensities from 15 normal samples, among which 13 samples are tumor adjacent stromal fibroblasts purified from the same patients whose tumor samples were analyzed by SNP array. The rest two samples are normal fallopian tube tissues. To infer the DNA copy number from the raw signal data, we used the Hidden Markov Model (31), based on the assumption of diploid for normal samples. Mapping information of SNP locations and cytogenetic bands were based on curation of Affymetrix and University of California Santa Cruz hg17. In this study, we used an arbitrary cutoff of >3 copies in more than 6 consecutive SNPs to define amplification, a cutoff of < 0.5 copy to define homozygous deletion, and a copy number between 0.5 and 1.5 to define hemizygous deletions. If there were intermittent, non-consecutive probes with inferred copy numbers that deviated less than +/- 0.1 copies from the cutoff criteria, this probe was considered within the cutoff range. The DNA copy number changes in all samples were analyzed using the same criteria.
Index of DNA copy number changes (Chromosome Instability Index)
To facilitate the quantification of sub-chromosomal copy number alterations, we applied the Circular Binary Segmentation algorithm (32) to count copy number changes and used a CIN index to determine the levels of DNA copy number changes. In this study, amplification was defined as a region with a copy number greater than 3 in at least 6 contiguous SNPs, while homozygous deletion was defined as a region with a copy number less than 0.5 in at least 6 contiguous SNPs. Hemizygous deletion was defined as a region where the copy number fell between 0.5 and 1.5 in at least 6 contiguous SNPs. We further computed the CIN index for each chromosome at a genome-wide scale based on the total amount of gain and loss. The chromosome-specific CIN index was defined as the sum of amplitudes of all gain/loss segments divided by the total number of SNPs in the chromosome, and the genome-wide CIN index was defined as log(C1+1) + … + log(Ci+1) + … + log(C23+1), where Ci is the CIN index of chromosome i. For a gain segment, the amplitude was the average intensity of SNP signals within the segment. For a loss segment, in order to match the effect of losses to the same scale of gains, the amplitude was calculated by 2.5+(A-2.5)(1.5-a)/1.5, where a was the average intensity of SNP signals within the loss segment and A was the maximum gain amplitude across all cases on the same chromosome.
Western blot analysis
Western blot analysis was performed on representative HG tumors to test if tumors with apparent homozygous deletions expressed the gene products. Similar amounts of protein were loaded in 10–12% Tris-acrylamide gels and after transblotting, the membranes were probed with either anti-pRb antibody (clone IF8, Santa Cruz, CA) or anti-p16 antibody (clone G175-1239, BD Biosciences, San Jose, CA). GAPDH was used as the loading control. After secondary antibody incubation, membranes were developed using chemiluminescence and detected using Chemidoc XRS (BioRad, Hercules, CA).
Detailed methods for quantitative real-time PCR, statistic analysis, and transfection and functional study of miR-34a are described in Supplementary Materials and Methods.
Results
Global DNA copy number alterations detected in ovarian serous tumors
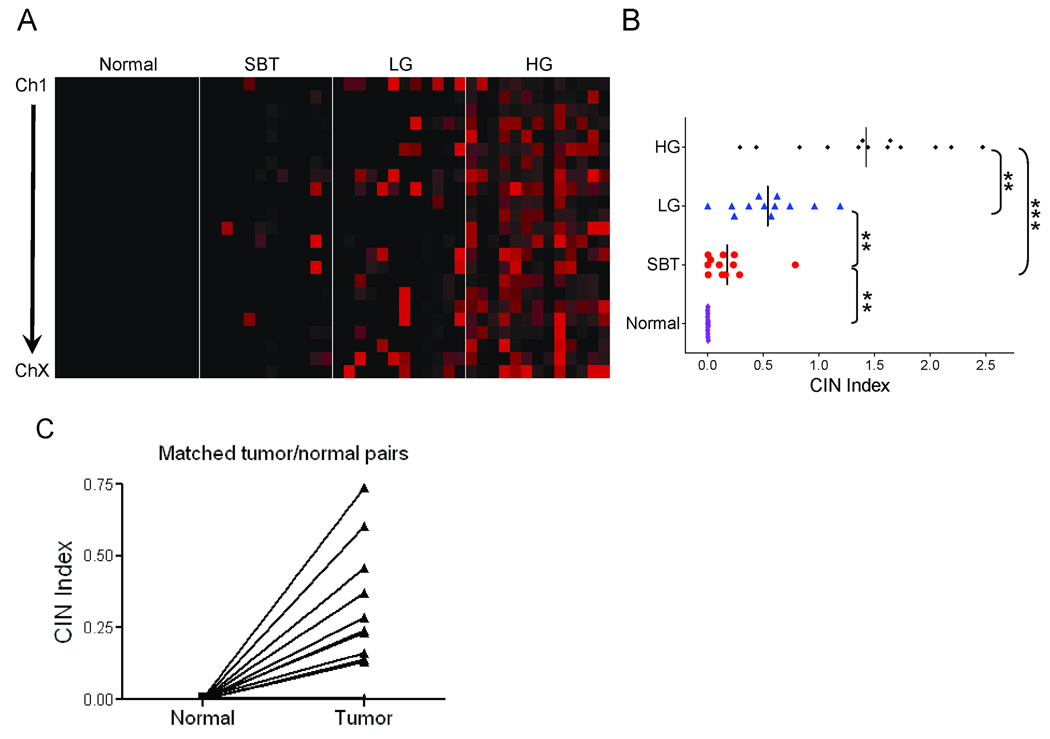
The genome-wide DNA copy number profiles in 12 SBTs, 12 LG, and 13 HG serous carcinomas are shown in supplementary Fig 1. As compared to SBTs and LG serous carcinomas, HG serous carcinomas demonstrated widespread DNA copy number gains and losses involving all chromosomes. To compare the overall levels of DNA copy number changes in different serous neoplasms, we used the Chromosome Instability (CIN) index based on counting the total numbers of discrete DNA fragments showing either gain or loss in each sample. First, we compare the CIN index between tumor adjacent stromal fibroblasts (isolated from 7 SBTs and 6 LGs) and their corresponding tumors. Fig 1A showed CIN index at each chromosome and Fig 1B showed genome-wide CIN index (the combined CIN index from all chromosomes) of each specimen. The results demonstrated that the CIN index of normal samples consistently approximated to zero in all chromosomes, while their tumor counterparts showed elevated CIN index. Paired t-test was performed to demonstrate a statistically significant difference in CIN index between normal samples and matched tumors (p<0.01). In contrast, serous neoplasms, especially LG and HG carcinomas demonstrated an elevated CIN index (Fig 1). In LG serous carcinomas, the chromosomes with the highest CIN index were chromosomes 1, 22, and X, while chromosomes 2, 3, 5, 7, 11, and 12 had the lowest CIN index (supplementary Fig 2). In HG tumors, all chromosomes had a high CIN index (supplementary Fig 2). As compared to SBTs and LG tumors, HG tumors demonstrated the highest level of DNA copy number alterations as their overall CIN index was significantly higher than the CIN index of either LG serous carcinomas or SBTs (Fig 1B). LG serous carcinomas also exhibited a significantly higher CIN index than SBTs, indicating that there were more chromosomal rearrangements in LG serous carcinomas than in SBT tumors (Fig 1B).
Figure 1. Genome-wide chromosome instability (CIN) index in ovarian serous tumors.
A. The CIN index in individual chromosome of each serous tumor is plotted using a pseudo-color gradient indicating the copy number alteration level (low to high: dark to red). Normal: stromal fibroblast from the tumor; SBT: serous borderline tumor (atypical proliferative serous tumor); LG: low-grade serous carcinoma; HG: high-grade serous carcinoma. B. Genome-wide CIN index for each tumor. C. CIN index in matched normal and tumor pairs from 7 SBT and 6 LG tumors. Tumor samples are found to harbor significantly higher CIN index than adjacent normal samples (p<0.01).
Frequent deletions in the ch1p36 and ch9p21 loci in LG tumors
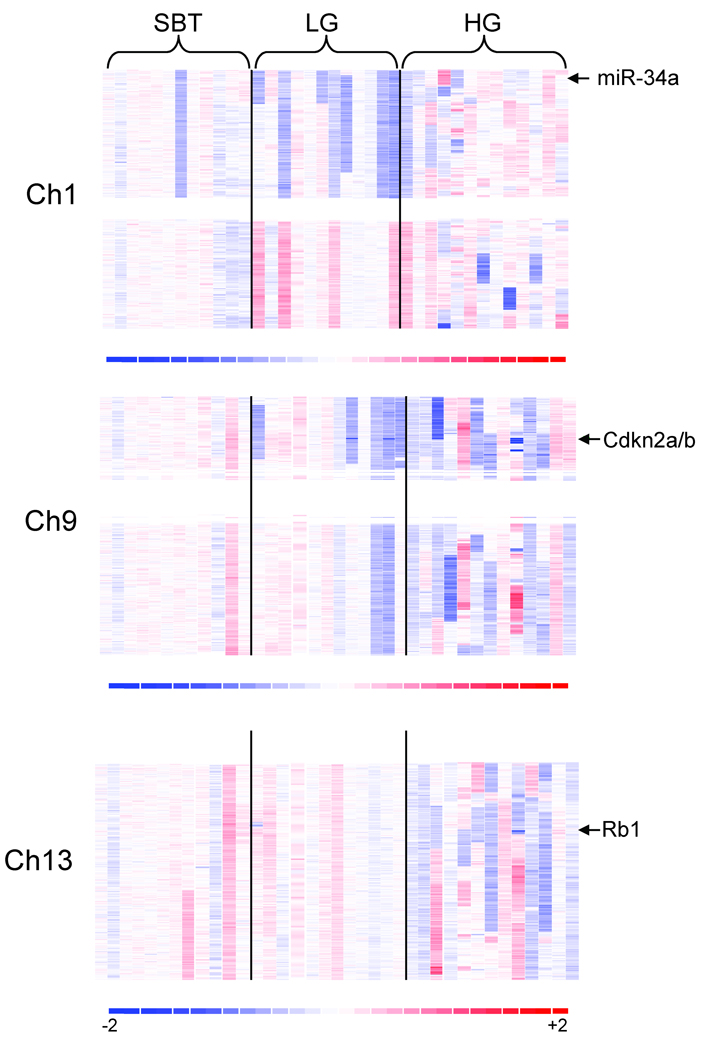
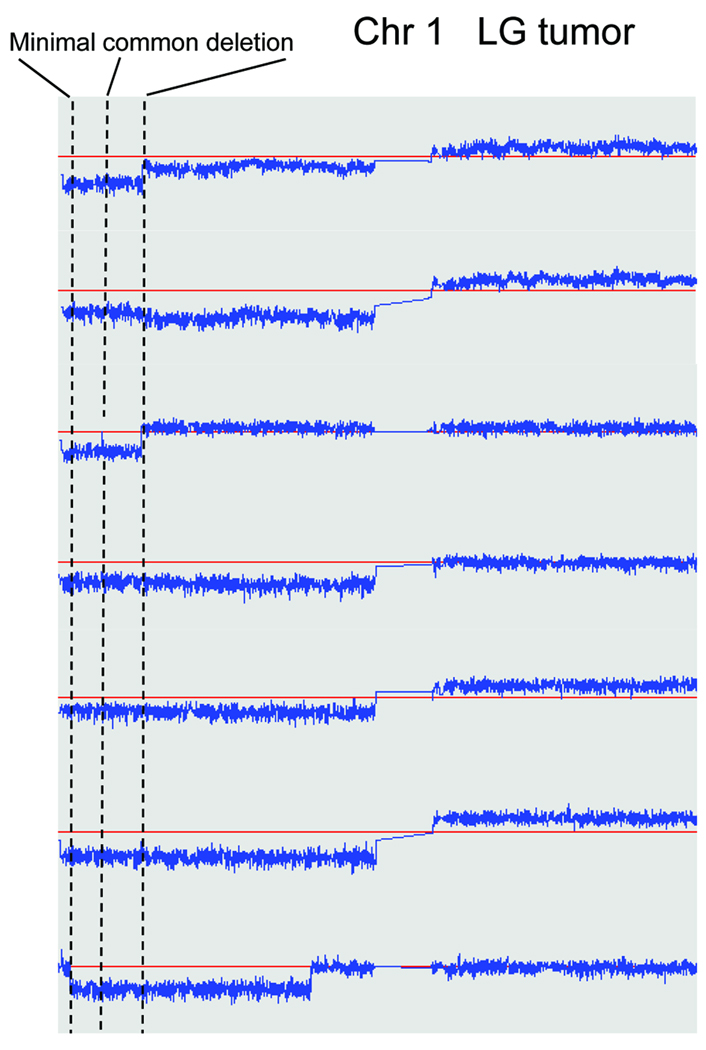
To identify sub-chromosomal regions that are potentially involved in the progression from SBT to LG serous carcinoma, we compared the profiles of DNA copy number changes between both types of ovarian tumors. Our results demonstrated that the most significant DNA copy number changes in LG carcinomas were deletions at ch1p36 and ch9p21.3 which occurred in 7/12 and 6/12 of LG tumors, respectively (Fig 2). Of note, ch1p deletions were all hemizygous whereas 2 specimens of ch9p21.3 deletions were homozygous and 4 were hemizygous. One of the SBTs also harbored ch1p36 hemizygous deletion but none of the SBTs contained detectable ch9p21.3 deletions. Minimal mapping of the ch1p36 deletion in these 7 LG serous carcinomas revealed that there were two immediately adjacent deleted regions; one spanning the interval from 5,579,980 to 16,540,200 bp, and the other spanning the interval from 17,035,400 to 31,700,900 bp (Fig 3). Notably, CHD5 and miRNA-34a, previously reported tumor suppressor genes, are located within the first common deleted region in LG carcinomas. To determine if CHD5 was inactivated by point mutations, we performed DNA sequence analysis on the 12 LG carcinomas used in the SNP array analysis and on the SBT that also harbored the ch1p36 hemizygous deletion. However, we did not identify somatic mutations in CHD5.
Figure 2. Copy number alteration in chromosome 1, 9, and 13.
DNA copy number changes are represented as pseudo-color gradients corresponding to the copy number increase (red boxes) and decrease (blue boxes) as compared to pooled normal samples. Each column represents an individual tumor sample. Arrow indicates candidate tumor suppressor gene at each region.
Figure 3. Minimal mapping of chromosome 1p hemizygous deletions in LG tumors.
Copy number of chromosome 1 of all seven LG tumors harboring ch1p36 deletion is plotted against chromosomal location. The minimally deleted region (delineated by dotted lines) contains two adjacent deletion fragments. Red horizontal lines indicate normal copy number: 2. Candidate tumor suppressor genes, CHD5 and miRNA-34a, are located within the first deletion fragment.
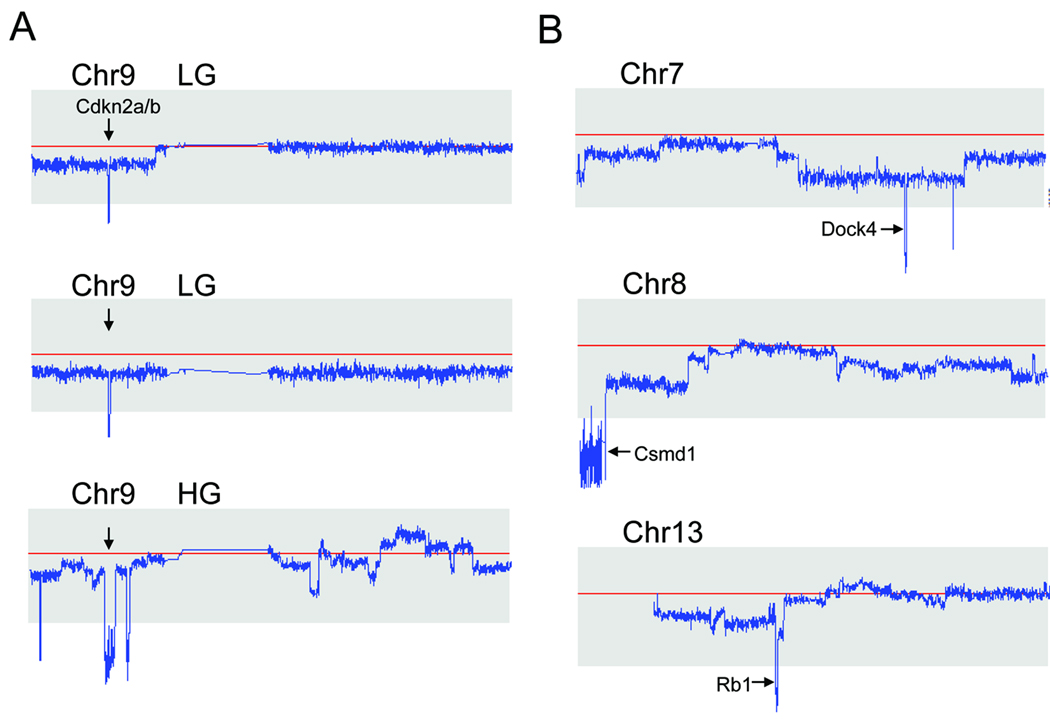
Minimal mapping of the two LG serous carcinomas with homozygous deletion in ch9p21.3 region demonstrated that the core deletion, spanning 21,945,000 to 22,009,700 bp and which harbors the CDKN2A/B locus, encodes p16, p15, and Arf (p14) (Fig 4A). To determine if additional mutations were present in both CDKN2A and CDKN2B, we performed DNA sequence analysis of these genes among 10 LG serious carcinomas and 9 SBTs, but did not detect somatic point mutations in any of these specimens.
Figure 4. Representative chromosomes containing homozygous deletions.
A. Two LG serous carcinomas and one HG serous carcinoma harbor the ch9p21.3 homozygous deletion. Red horizontal lines represent normal copy number: 2. The deletions in LG serous carcinomas are small and discrete, but the deletion in the HG tumor is larger and encompasses 2.92 Mb. B. Representative homozygous deletions in ch7, ch8, and ch13 in HG serous carcinomas. Candidate tumor suppressor genes residing within the deleted regions are indicated by arrows.
Additional genomic alterations present in LG serous carcinoma but not in SBTs included a 3-fold gain at ch1q and a hemizygous deletion of the entire X chromosome. These regions may contain genes important in the progression from SBT to LG serous carcinoma.
Profiles of DNA copy number changes in HG serous carcinomas
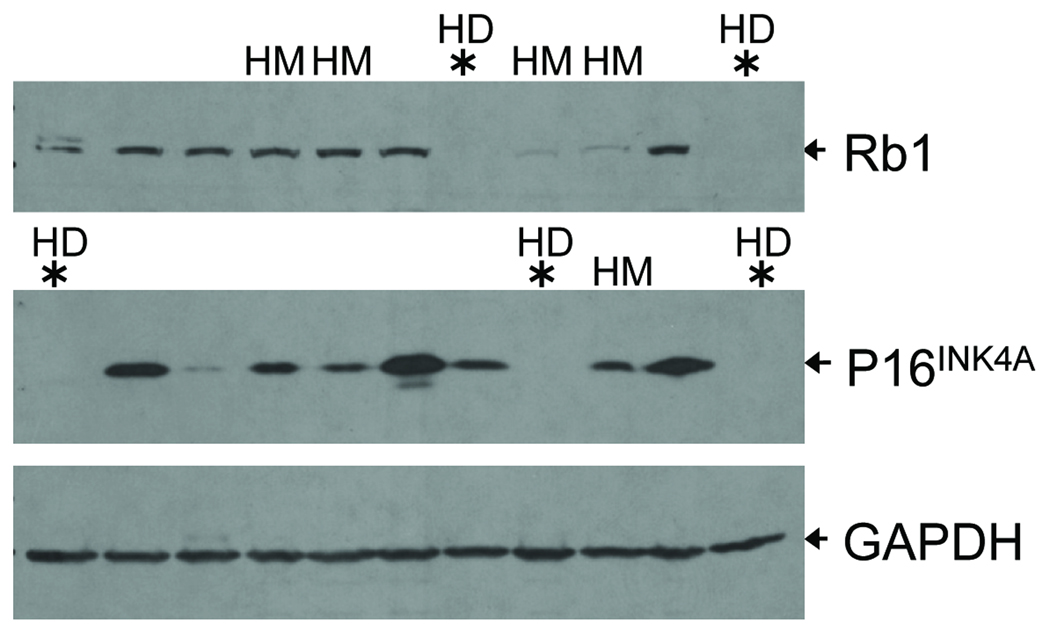
The current 250K SNP array analysis revealed that the most frequently amplified regions in HG serous carcinomas were the loci at ch3q, ch12p, and ch19p, all of which have been reported in our previous study using a 10K SNP array platform (27). Supplementary Table 2 summarizes the most common amplicons identified among HG serous carcinomas in this study. Because genomic amplifications in the ovarian cancer have been previously characterized in several studies including our own (9, 26, 27, 33–37), in this report, we focused on analyzing homozygous deletions which were difficult to be discovered by previous approaches. In this regard, we took advantage of affinity purification of tumor cells which would enhance the sensitivity in detecting homozygous deletions. Furthermore, we used the relatively high density 250K SNP platform because our pilot study has shown that it can reliably detect a previously reported 450 kb homozygous deletion in DiFi cells (32). Supplementary Table 3 summarizes the most common regions carrying deletions identified in this study. Among the loci that were deleted were Rb1 (ch13q14.2), CDKN2A/B (ch9p21.3), CSMD1 (ch8q23.1-23.3), and DOCK4 (ch7q31.1), all of which have been previously reported to be homozygously deleted in human cancers (Fig 4B). Ten of the homozygous deletion loci were randomly selected for validation by quantitative real-time PCR. We were able to confirm each of the selected deletions (data shown in Supplementary Table 3). None of the deletions were present in the matched normal tissues, indicating that these homozygous deletions in HG serous carcinomas were somatic changes acquired during tumor progression. To further assess the frequency of deletions of Rb1, CDKN2A, CSMD1 and DOCK4, we used genomic quantitative real-time PCR on an independent panel of 47 affinity-purified HG serous carcinomas. The frequency of homozygous deletion of Rb1, CDKN2A, CSMD1 and DOCK4 was 10.6%, 6.4%, 6.4% and 4.3%, respectively. Specific antibodies against two of the known tumor suppressors, pRb and p16 (encoded by CDKN2A), were available which permitted us to determine if the deletion affected protein expression. Eleven representative HG serous carcinomas with known DNA copy number status in both regions were included in Western blot analysis. As expected, the results were entirely consistent with the deletional status of each locus. As shown in Figure 5, two HG serous carcinomas with homozygous deletions in the Rb1 locus completely lost pRb protein expression. Two tumors of the four tumors harboring hemizygous deletions at the Rb1 locus exhibited low pRb expression. Other tumors with retained Rb1 alleles showed a robust pRb protein band. Three HG serous carcinomas harboring homozygous deletions of the CDKN2A locus did not express detectable p16 protein. The remaining 8 tumors which retained one or both CDKN2A alleles expressed a variable but detectable amount of p16 protein.
Figure 5. Loss of p16 and Rb1 protein expression in tumors with homozygous deletion at 9p21.3 and 13q14.2, respectively.
Western blot analysis was performed using HG serous tumors with known DNA copy number at 9p21.3 and 13q14.2 regions. Tumors with homozygous deletions (HD) at those loci were found to completely lose protein expression of p16 or Rb1, respectively. Hemizygous deletion (HM) was found to variably affect protein expression of p16 and Rb1. The remaining tumors contain normal copy number (2 copies). GAPDH was used as the sample loading control.
Growth inhibition by miR-34a in low-grade carcinomas
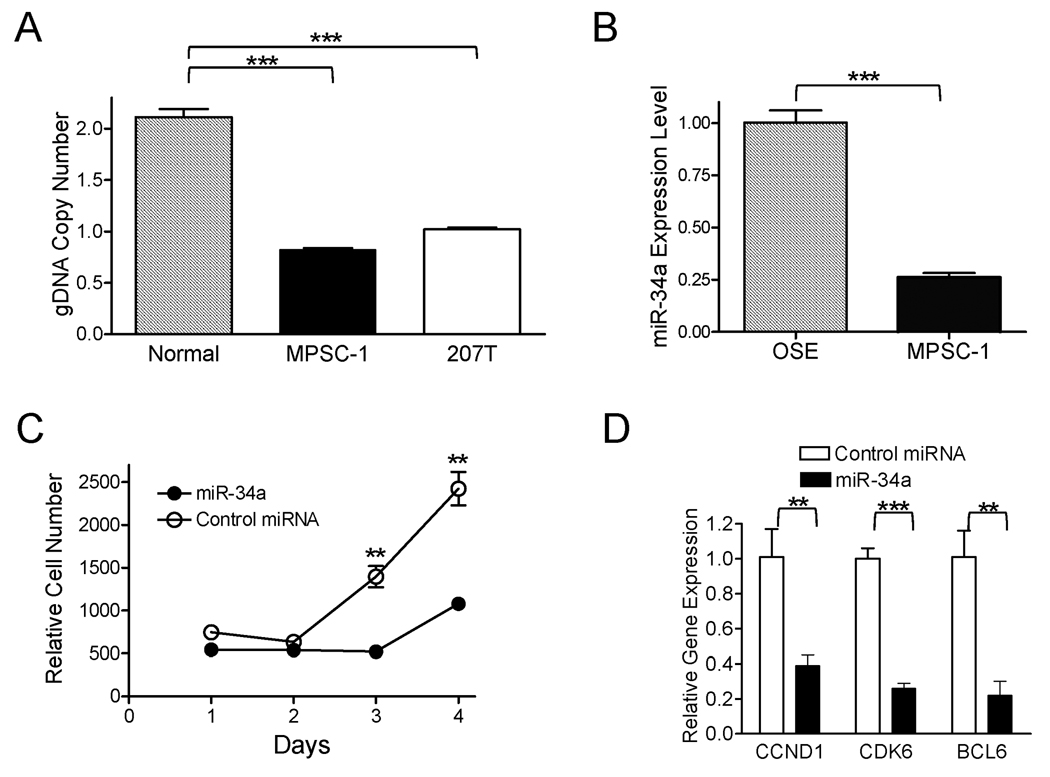
miR-34a has been demonstrated as a potential tumor suppressor in several tumors including neuroblastoma, colorectal cancer, pancreatic cancer, and non-small cell lung cnacer (38–43). Given the frequent hemizygous deletion at the miR-34a locus in LG carcinomas, we decided to examine if miR-34a participated in the pathogenesis of LG serous carcinomas. MPSC-1, a LG carcinoma cell line that harbored hemizygous deletion at the miR-34a locus and expressed a low level of endogenous miR-34a as compared to normal ovarian surface epithelial cells was selected for functional study (Fig 6 A & B). MPSC-1 cells were transfected with synthetic miR-34a mimic and cell growth was monitored. We found that miR-34a mimic significantly suppressed the cell growth of MPSC-1 when compared to the control miRNA treated cells (Fig 6C). Furthermore, miR-34a mimic treatment was associated with a higher percentage of apoptotic cells in MPSC-1 cells than the mimic control (t test, p<0.05, data not shown), a result consistent with previous reports (39, 41). To examine if miR-34a can regulate mRNA stability of its target genes, following miR-34a transfection we performed quantitative RT-PCR to determine mRNA levels of eleven reported miR-34a target genes (listed in supplementary Table 4). Our results demonstrated that CCDN1, CDK6, and BCL2 mRNAs were significantly down-regulated by miR-34a 24 hours after transfection as compared to the control miRNA (Fig 6D).
Figure 6. Biological effects of miR-34a in a LG carcinoma cell line, MPSC-1.
(A) Quantitative PCR demonstrates a reduced genomic DNA (gDNA) copy number at the miR-34a locus in a LG carcinoma cell line, MPSC-1. The calculated copy number in normal tissue, MPSC-1, and 207T is 2.1, 0.8 and 1.0, respectively. 207T is a LG carcinoma with hemizygous deletion at the miR-34a locus based on SNP array analysis. (B) miR-34a expression measured by the TagMan microRNA assay. OSE: normal ovarian surface epithelial cells. (C) miR-34a inhibits cell proliferation as measured by the cell-titer blue assay. (D) mRNA levels of candidate miR-34a target genes measured by quantitative RT-PCR.
Discussion
Ovarian carcinoma is a complex disease comprised of a variety of neoplasms that have been termed surface epithelial tumors. These tumors have vastly different clinical, histopathologic and molecular features and their pathogenesis differs significantly. A major challenge, in so far as their pathogenesis is concerned, has been to identify precursor lesions and shed light on the molecular pathways that lead to the development of invasive ovarian carcinoma. Among the various types of surface epithelial tumors the serous group is by far the most common, accounting for approximately half of all surface epithelial tumors. They are also arguably the most lethal. Accordingly, in this study we focused exclusively on the serous tumors. Although several previous reports have applied genome-wide molecular analyses to study ovarian carcinomas (9, 34–36), there are several unique approaches in this report that aim to elucidate these fundamental questions. First, we applied high resolution SNP arrays on affinity-purified tumor cells, an approach that provides high sensitivity and specificity in detecting subtle DNA copy number alterations, especially hemizygous and homozygous deletions. Second, the focus on a single histological subtype of ovarian cancer rather than grouping different tumor subtypes together permitted a comprehensive view of molecular genetic changes in serous ovarian tumors. Third, we performed quantitative PCR to validate the homozygous deletions detected by SNP arrays and demonstrated high specificity of the current approach in detecting homozygous deletions.
Several conclusions can be drawn from this study in terms of the molecular etiology of ovarian SBT/LG carcinoma. First, the observation that ch1p36 hemizygous deletions and ch9p21 homozygous or hemizygous deletions occurred far more frequently in LG serous carcinomas than in SBTs suggests that these regions may harbor potential tumor suppressors, the loss of which contribute to tumor progression from SBT to LG carcinoma. The ch1p36 region contains several potential tumor suppressor genes (44, 45). We chose miR-34a to investigate its function and found that transfected miR-34a can suppress cellular proliferation and induce apoptosis in LG carcinoma cells. The functional effects of miR-34a reported here are similar to previous studies (38–43). Furthermore, in LG carcinoma cells, miR-34a was found to down-regulated several of the miR-34a target genes including CCDN1, CDK6, and BCL2. The results support the role(s) of miR-34a in regulating cell proliferation and apoptosis in LG serous carcinoma. It should be noted that the 1p36 deletion encompasses a large chromosomal region containing more than 80 genes; therefore, further studies are required to delineate the culprit tumor suppressor gene(s) located at this region.
Second, frequent deletion at the ch9p21 region corresponding to the CDKN2A/B locus encodes three well-known tumor suppressor proteins, p14 (Arf), p16 and p15, was observed in LG serous carcinoma. Arf is a potent tumor suppressor that blocks cell-cycle progression by interfering with the p53 negative regulator, MDM2, thereby stabilizing p53 protein expression. CDKN2A and CDKN2B share similar if not redundant function in inhibiting cyclin-dependent kinase (CDK) which regulates the pRb pathway. Besides, the expression level of CDKN2A was enhanced in response to oncogene-induced stress such as by the activation of RAS-RAF-MEK signaling pathway. This observation is of considerable interest because SBTs and LG carcinomas frequently contain activating KRAS and BRAF mutations, but rarely contain TP53 mutations. Thus, retained CDKN2A/B in SBTs may prevent progression to LG carcinoma despite the presence of activating KRAS and BRAF mutations frequently observed in SBTs. When CDKN2A/B locus is deleted, the check point becomes defective and tumor cells may escape from cell cycle arrest and exploit the oncogenic functions of mutated KRAS/BRAF, leading to LG tumor progression.
The genome-wide analysis from this study uncovered a handful numbers of homozygously deleted genes that maybe implicated in tumorigenesis of HG serous carcinomas. These include relatively frequent homozygous deletions of Rb1 and CDKN2A/B. Homozygous deletions in either of these regions accounted for 17% (8/47) HG serous carcinomas. Since both genes are probably silenced by additional molecular mechanisms such as promoter hypermethylation, the inactivation in either gene in HG serous carcinomas is expected to be very frequent. In fact, we observed that more than 50% (6/11) of HG serous carcinomas had a significant reduction in either pRb or p16 protein expression based on Western blot analysis supporting this interpretation. pRb and p16 proteins both participate in regulating progression of the cell cycle from G1 to S phase. It is well-known that HG serous carcinomas harbor frequent TP53 mutation; however the molecular genetic changes in Rb1 and CDKN2A were previously not recognized because somatic mutations in these genes are rare in HG serous carcinomas. In addition, our results demonstrating that deletions and down-regulation of the Rb/p16 pathway often coexist with TP53 mutations, suggest that Rb1 and TP53 act independently and perhaps are equally important in the development of HG serous carcinomas. The genetic analysis reported here provides an explanation why mouse ovarian cancer model generated by inducible deletion of Rb1 and TP53 in ovarian epithelium results in tumors with morphology closely resembled its human counterpart (46).
In addition to the well-characterized tumor suppressors discussed above, in HG serous carcinoma we also identified candidate tumor suppressor genes that are less known to be implicated in human cancers. These include CUB and Sushi Multiple Domains 1 (CSMD1) which is located in ch8p23.1-23.3 (47) and Dictator of Cytokinesis 4 (DOCK4) which is located in ch7q31.1 (48). Deletions in ch8p23 have been detected in several other types of human cancer including squamous cell carcinoma. As well, comprehensive sequencing analysis has revealed somatic mutations of CSMD1 in colon and breast cancers (49, 50). DOCK4 encodes regulator of small GTPase and was originally identified as a homozygously deleted tumor suppressor (48). The function of both CSMD1 and DOCK4 are less clear and future studies are required to define their role(s) in the pathogenesis of HG serous carcinoma.
The current data using 250K SNP array has validated our previous study using 10K SNP array in profiling amplicons in 33 HG and 10 LG ovarian serous carcinomas (27). In addition, the much higher resolution of 250K SNP arrays permitted a reliable detection of small sub-chromosomal deletions. However, it needs to be noted that the sample sizes performed in the current study is relatively small and the observed mutation rate should be confirmed in future large-scale studies.
Our quantitative measurement of DNA copy number changes as defined by the CIN index, provided cogent evidence that HG serous carcinomas are characterized by a much higher level of DNA copy number changes as compared to LG serous carcinomas and SBTs. In addition, LG serous carcinomas harbor higher numbers of sub-chromosomal alterations than do SBTs. In 13 pairs of matched tumors and tumor stromal fibroblasts, the CIN index was found to be significantly higher in the tumors than the normal stromal fibroblasts, refuting the proposal of co-evolution of tumor stromal cells during tumorigenesis.
In summary, the results from this study shed new light on the possible tumor suppressor roles of miR-34a and CDKN2A/B in the progression of SBT to LG carcinoma. For HG tumors, deletions in the Rb-p16 pathway may contribute to its development, and CSMD1 and DOCK4 may represent new tumor suppressor genes. The reported unique molecular genetic landscape in different types of ovarian serous neoplasm can serve as a roadmap for future studies aiming at elucidating molecular pathogenesis, and developing new diagnostic test and target-based therapy.
Supplementary Material
Acknowledgements
This study was supported by grants from US Department of Defense Research Council OC0400600 (TLW), American Cancer Society RSG-08-174-01-GMC (TLW), Ovarian Cancer Research Fund (TLW), NIH RO1CA116184 (RJK), NIH RO1CA129080 (IMS), NIH RO1 R01CA103937 (IMS) and NIH R33CA109872 (YW).
Footnotes
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Link CJ, Jr, Kohn E, Reed E. The relationship between borderline ovarian tumors and epithelial ovarian carcinoma: epidemiologic, pathologic, and molecular aspects. Gynecol Oncol. 1996;60:347–354. doi: 10.1006/gyno.1996.0054. [DOI] [PubMed] [Google Scholar]
- 3.Singer G, Kurman RJ, Chang H-W, Cho SKR, Shih I-M. Diverse tumorigenic pathways in ovarian serous carcinoma. Am J Pathol. 2002;160:1223–1228. doi: 10.1016/s0002-9440(10)62549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shih Ie M, Kurman RJ. Molecular pathogenesis of ovarian borderline tumors: new insights and old challenges. Clin Cancer Res. 2005;11:7273–7279. doi: 10.1158/1078-0432.CCR-05-0755. [DOI] [PubMed] [Google Scholar]
- 5.Shih I-M, Kurman RJ. Ovarian tumorigenesis- a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/s0002-9440(10)63708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malpica A, Deavers MT, Lu K, et al. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol. 2004;28:496–504. doi: 10.1097/00000478-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Bonome T, Lee JY, Park DC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 8.Gilks CB, Vanderhyden BC, Zhu S, van de Rijn M, Longacre TA. Distinction between serous tumors of low malignant potential and serous carcinomas based on global mRNA expression profiling. Gynecol Oncol. 2005;96:684–694. doi: 10.1016/j.ygyno.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Meinhold-Heerlein I, Bauerschlag D, Hilpert F, et al. Molecular and prognostic distinction between serous ovarian carcinomas of varying grade and malignant potential. Oncogene. 2005;24:1053–1065. doi: 10.1038/sj.onc.1208298. [DOI] [PubMed] [Google Scholar]
- 10.Bell DA. Origins and molecular pathology of ovarian cancer. Mod Pathol. 2005;18 Suppl 2:S19–S32. doi: 10.1038/modpathol.3800306. [DOI] [PubMed] [Google Scholar]
- 11.Singer G, Oldt R, 3rd, Cohen Y, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 12.Nakayama K, Nakayama N, Kurman RJ, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5:779–785. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 13.Sieben NL, Macropoulos P, Roemen GM, et al. In ovarian neoplasms, BRAF, but not KRAS, mutations are restricted to low-grade serous tumours. J Pathol. 2004;202:336–340. doi: 10.1002/path.1521. [DOI] [PubMed] [Google Scholar]
- 14.Dansonka-Mieszkowska A, Ludwig AH, Kraszewska E, Kupryjanczyk J. Geographical variations in TP53 mutational spectrum in ovarian carcinomas. Ann Hum Genet. 2006;70:594–604. doi: 10.1111/j.1469-1809.2006.00257.x. [DOI] [PubMed] [Google Scholar]
- 15.Kohler MF, Marks JR, Wiseman RW, et al. Spectrum of mutation and frequency of allelic deletion of the p53 gene in ovarian cancer. J Natl Cancer Inst. 1993;85:1513–1519. doi: 10.1093/jnci/85.18.1513. [DOI] [PubMed] [Google Scholar]
- 16.Salani R, Kurman RJ, Giuntoli R, 2nd, et al. Assessment of TP53 mutation using purified tissue samples of ovarian serous carcinomas reveals a higher mutation rate than previously reported and does not correlate with drug resistance. Int J Gynecol Cancer. 2008;18:487–491. doi: 10.1111/j.1525-1438.2007.01039.x. [DOI] [PubMed] [Google Scholar]
- 17.Kupryjanczyk J, Thor AD, Beauchamp R, et al. p53 gene mutations and protein accumulation in human ovarian cancer. Proc Natl Acad Sci U S A. 1993;90:4961–4965. doi: 10.1073/pnas.90.11.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen WH, Reles A, Runnebaum IB, et al. p53 mutations and expression in ovarian cancers: correlation with overall survival. Int J Gynecol Pathol. 1999;18:29–41. doi: 10.1097/00004347-199901000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Kinzler KW, Vogelstein B. The Genetic Basis of Human Cancer. 2nd edition. Toronto: McGraw-Hill; 2002. [Google Scholar]
- 20.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 21.Wang TL, Diaz LA, Jr, Romans K, et al. Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc Natl Acad Sci U S A. 2004;101:3089–3094. doi: 10.1073/pnas.0308716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farley J, Smith LM, Darcy KM, et al. Cyclin E expression is a significant predictor of survival in advanced, suboptimally debulked ovarian epithelial cancers: a Gynecologic Oncology Group study. Cancer Res. 2003;63:1235–1241. [PubMed] [Google Scholar]
- 23.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 24.Cheng JQ, Godwin AK, Bellacosa A, et al. AKT2, a putative oncogene encoding a member of a subfamily of protein- serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci U S A. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu R, Lin L, Beer DG, et al. Amplification and overexpression of the L-MYC proto-oncogene in ovarian carcinomas. Am J Pathol. 2003;162:1603–1610. doi: 10.1016/S0002-9440(10)64294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih Ie M, Sheu JJ, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci U S A. 2005;102:14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama K, Nakayama N, Jinawath N, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–2617. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 28.Gorringe KL, Jacobs S, Thompson ER, et al. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res. 2007;13:4731–4739. doi: 10.1158/1078-0432.CCR-07-0502. [DOI] [PubMed] [Google Scholar]
- 29.Shih Ie M, Wang TL. Apply innovative technologies to explore cancer genome. Curr Opin Oncol. 2005;17:33–38. doi: 10.1097/01.cco.0000147382.97085.e4. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama K, Nakayama N, Davidson B, et al. Homozygous Deletion of MKK4 in Ovarian Serous Carcinoma. Cancer Biol Ther. 2006;5:630–634. doi: 10.4161/cbt.5.6.2675. [DOI] [PubMed] [Google Scholar]
- 31.Slamon DJGW, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 32.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 33.Park JT, Li M, Nakayama N, et al. Notch-3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 34.Schraml P, Schwerdtfeger G, Burkhalter F, et al. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. Am J Pathol. 2003;163:985–992. doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuda H, Birrer MJ, Ito YM, et al. Identification of DNA copy number changes in microdissected serous ovarian cancer tissue using a cDNA microarray platform. Cancer Genet Cytogenet. 2004;155:97–107. doi: 10.1016/j.cancergencyto.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Bernardini M, Lee CH, Beheshti B, et al. High-resolution mapping of genomic imbalance and identification of gene expression profiles associated with differential chemotherapy response in serous epithelial ovarian cancer. Neoplasia. 2005;7:603–613. doi: 10.1593/neo.04760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Israeli O, Gotlieb WH, Friedman E, et al. Genomic analyses of primary and metastatic serous epithelial ovarian cancer. Cancer Genet Cytogenet. 2004;154:16–21. doi: 10.1016/j.cancergencyto.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 39.Cole KA, Attiyeh EF, Mosse YP, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun F, Fu H, Liu Q, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 43.Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 44.Bagchi A, Papazoglu C, Wu Y, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 45.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flesken-Nikitin A, Choi KC, Eng JP, Shmidt EN, Nikitin AY. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 2003;63:3459–3463. [PubMed] [Google Scholar]
- 47.Sun PC, Uppaluri R, Schmidt AP, et al. Transcript map of the 8p23 putative tumor suppressor region. Genomics. 2001;75:17–25. doi: 10.1006/geno.2001.6587. [DOI] [PubMed] [Google Scholar]
- 48.Yajnik V, Paulding C, Sordella R, et al. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–684. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
- 49.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 50.Farrell C, Crimm H, Meeh P, et al. Somatic mutations to CSMD1 in colorectal adenocarcinomas. Cancer Biol Ther. 2008;7:609–613. doi: 10.4161/cbt.7.4.5623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.