Abstract
c‐Src non‐receptor tyrosine kinase is an important component of the platelet‐derived growth factor (PDGF) receptor signaling pathway. c‐Src has been shown to mediate the mitogenic response to PDGF in fibroblasts. However, the exact components of PDGF receptor signaling pathway mediated by c‐Src remain unclear. Here, we used stable isotope labeling with amino acids in cell culture (SILAC) coupled with mass spectrometry to identify Src‐family kinase substrates involved in PDGF signaling. Using SILAC, we were able to detect changes in tyrosine phosphorylation patterns of 43 potential c‐Src kinase substrates in PDGF receptor signaling. This included 23 known c‐Src kinase substrates, of which 16 proteins have known roles in PDGF signaling while the remaining 7 proteins have not previously been implicated in PDGF receptor signaling. Importantly, our analysis also led to identification of 20 novel Src‐family kinase substrates, of which 5 proteins were previously reported as PDGF receptor signaling pathway intermediates while the remaining 15 proteins represent novel signaling intermediates in PDGF receptor signaling. In validation experiments, we demonstrated that PDGF indeed induced the phosphorylation of a subset of candidate Src‐family kinase substrates – Calpain 2, Eps15 and Trim28 – in a c‐Src‐dependent fashion.
Keywords: Kinase, Mass spectrometry, Phosphoproteomics, Phosphorylation, Signal transduction, SILAC, c-Src, PDGF
1. Introduction
The protein tyrosine kinase v‐Src is the transforming product of Rous sarcoma virus, the first oncogenic retrovirus to be identified. c‐Src, the cellular counterpart of v‐Src, is a membrane‐associated non‐receptor tyrosine kinase. Src‐family kinases (SFKs) regulate mammalian cell growth and mitogenesis. c‐Src tyrosine kinase plays versatile roles in cell responses induced by platelet‐derived growth factor (PDGF), that include cell growth, cell cycle progression, cell survival, cell migration, actin cytoskeleton rearrangement, DNA synthesis and receptor endocytosis (DeMali et al., 1999; Thomas and Brugge, 1997). The role of c‐Src downstream of PDGF receptor stimulation has been demonstrated in murine fibroblasts that express the PDGF receptor and respond to PDGF. Upon PDGF stimulation, an increase in the catalytic activity of Src‐family tyrosine kinases Src, Fyn and Yes, is observed in these cells (Ralston and Bishop, 1985). Further downstream signaling events following SFKs activation are mainly effected by the induction of cascade of phosphorylation events and protein–protein interactions. It has also been noted that PDGF may activate spatially distinct pools of SFKs leading to different biological outcomes (Veracini et al., 2006).
The activity and structural conformation of the Src‐family protein kinases are mainly regulated by phosphorylation events (Roskoski, 2005). Src kinase has been shown to be phosphorylated on serine, threonine and tyrosine residues. For example, Src kinase is phosphorylated on Thr24, Thr46 and Ser72 by CDK1/cdc2 kinase, which is important for the cell cycle transition (Shenoy et al., 1992). PDGF stimulation also induces a PKC mediated phosphorylation of Ser12 in c‐Src (Gould and Hunter, 1988). Phosphorylation of human c‐Src kinase on Tyr530, mediated by c‐terminal Src kinase (CSK), leads to Src kinase inactivation and locks it in a closed conformation under basal conditions (Cooper et al., 1986; Okada et al., 1991) and the activation of Src kinase requires dephosphorylation of Tyr530 (Bagrodia et al., 1991), a residue that is not present in its transforming homolog, v‐Src. Other members of the Src family also undergo similar regulation by such phosphorylation events at the C‐terminal tyrosine residue by CSK or by its homolog Chk (Hamaguchi et al., 1996). Src undergoes an autophosphorylation at Tyr419 located in the activation loop, which promotes its kinase activity (Boggon and Eck, 2004).
c‐Src kinase plays an important role in PDGF receptor signaling pathway and is actively regulated by a series of tyrosine phosphorylation events. Ligand‐induced activation of PDGF receptor beta subunit leads to the stepwise activation of c‐Src. First, dephosphorylation of Tyr530 allows a conformational change in c‐Src that leads to its autophosphorylation at Tyr419 (Kmiecik et al., 1988). The Tyr419‐phosphorylated Src kinase subsequently associates with the PDGF receptor which phosphorylates Src at Tyr216 rendering it active (Roskoski, 2005). Finally, the active c‐Src kinase phosphorylates PDGF receptor on Tyr934, which is located in its kinase domain (Hansen et al., 1996).
The use of small molecule pharmacological inhibitors as a complementary approach to current proteomic techniques like mass spectrometry provides additional selectivity to analyze signal transduction pathways. Src‐family inhibitors PP1 and PP2 have been extensively used in probing signal transduction pathways though both inhibit a broad range of receptor and non‐receptor tyrosine kinases, including PDGF receptor. Recently, SU6656 (2‐oxo‐3‐(4, 5, 6, 7‐tetrahydro‐1 H‐indol‐2‐ylmethylene)‐2, 3‐dihydro‐1H‐indole‐5‐sulfonic acid dimethylamide) was described as a selective inhibitor of Src‐family tyrosine kinases that can be used to probe PDGF receptor signaling. These studies have demonstrated that concentrations that inhibit Src, SU6656 do not inhibit the PDGF receptor activity (Blake et al., 2000); however at a higher concentration (10μM), SU6656 has also been shown to inhibit RSK1, AMPK, phosphorylase kinase and DYRK1A (Bain et al., 2003). Therefore, we used SU6656 to examine c‐Src signaling following activation of the PDGF receptor at a lower concentration (2μM) at which Src‐family kinases are inhibited. Further, in our studies, we carried out anti‐phosphotyrosine immunoprecipitation to specifically look at tyrosine‐phosphorylated substrates because Src is a tyrosine kinase. The non‐Src‐family kinases that could be inhibited by SU6656 are serine/threonine kinases whose substrates would not be identified in our strategy.
Stable isotope labeling with amino acids in cell culture (SILAC), an in vivo incorporation of isotopically labeled amino acids into the whole cell proteome for relative quantitation by mass spectrometry (Amanchy et al., 2005b), has been extensively used for the study of protein complexes (Wang and Huang, 2008), protein–protein interactions (Dobreva et al., 2008; Trinkle‐Mulcahy et al., 2008), dynamics of protein abundance in signaling pathways (Stokes et al., 2007) and post‐translational modifications (Blagoev et al., 2004; Kratchmarova et al., 2005; Kruger et al., 2008). SILAC‐based strategies have been successfully employed to study Src tyrosine kinase signaling in cells expressing active and inactive forms of c‐Src (Amanchy et al., 2008), in Src‐transformed cells (Luo et al., 2008) and in cells where inactive Src has been chemically activated (Qiao et al., 2006). However, only a small set of downstream signaling molecules activated by c‐Src in PDGF signaling are currently known.
Here, we employed a SILAC‐based quantitative proteomic approach to identify the downstream tyrosine kinase substrates of c‐Src in PDGF signaling. We enriched tyrosine‐phosphorylated proteins by immunoaffinity purification and quantitated the tyrosine phosphorylation status of proteins in NIH/3T3fibroblasts upon PDGF stimulation in the presence or absence of a Src‐family kinase inhibitor SU6656 (Blake et al., 2000). We observed that 43 proteins were phosphorylated on tyrosine residues upon PDGF stimulation and their phosphorylation were inhibited by pretreatment of SU6656. Among these proteins, 23 proteins are known c‐Src substrates, of which 16 proteins were already implicated in PDGF signaling while the remaining 7 proteins have not been shown to be involved in PDGF signaling from previous reports. The other 20 proteins were novel c‐Src substrates, of which 5 proteins have evidence of involvement in PDGF signaling as reported in literature while the remaining 15 proteins are novel PDGF signaling intermediates. We have experimentally showed that a subset of proteins (Calpain 2, Cortactin, cPLA2, Eps15, Ezrin, Fyb, Shp2 and Trim28) are Src‐family tyrosine kinase substrates downstream of PDGF signaling. Thus, by using SILAC‐based quantitative proteomic approaches, specific kinase inhibitors can be employed to dissect downstream of specific kinases in various signaling pathways.
2. Results and discussion
2.1. SILAC for in vivo identification of endogenous c‐Src substrates in PDGF receptor signaling pathway
SILAC allows complete labeling of cellular proteomes in vivo and simultaneous identification and quantitation of relative abundance of the peptides from cells under different conditions. This method also allows us to distinguish contaminating proteins that arise due to non‐specific binding to antibodies or the agarose matrix used in such experiments. SILAC has also been used previously for the dissection of Src kinase phosphoproteome (Amanchy et al., 2008; Luo et al., 2008; Qiao et al., 2006). The aim of our experiments was in vivo identification of endogenous Src kinase substrates in PDGF signaling.
We performed SILAC labeling of cellular proteins in a murine embryo fibroblast cell line, NIH/3T3, by growing the cells in DMEM containing different stable isotopes labeled arginine – 12C6‐arginine (light), 13C6‐arginine (medium), or 13C6, 15N4‐arginine (heavy) (Figure 1). The cells were adapted to the SILAC media for 5 passages as described earlier before they were probed for PDGF signaling. Thus, the whole proteome of NIH/3T3 cells was fully labeled with the stable isotope‐labeled arginine residues (Amanchy et al., 2005b). The NIH/3T3 cells grown in 12C6‐arginine‐containing medium were left unstimulated in basal condition as a control. The cells grown in 13C6‐arginine‐containing medium were stimulated by PDGF for 5min. The cells grown in 13C6,15N4‐arginine‐containing medium were pre‐treated with a potent inhibitor of c‐Src, SU6656, for 1h prior to stimulation with PDGF. Cells lysis was followed by the enrichment of tyrosine‐phosphorylated proteins using anti‐phosphotyrosine antibodies as described before (Amanchy et al., 2008). Src kinase activity in fibroblasts has been shown earlier (Gould and Hunter, 1988) to be maximal at 5–10min of PDGF stimulation shown by its multisite phosphorylation. Once Src kinase is activated, it initiates a cascade of downstream phosphorylation events. We chose a 5min time point of stimulation of NIH/3T3 cells with PDGF in our 3‐state SILAC experiment. NIH/3T3 cells demonstrated an increase in tyrosine phosphorylation upon stimulation of the PDGF receptor as evidenced by immunoblotting with anti‐phosphotyrosine antibodies (Figure 2) in both the whole cell extracts and anti‐phosphotyrosine immunoprecipitates. In cells pre‐treated with SU6656, an inhibition in tyrosine phosphorylation induced by PDGF was observed reflecting the role of c‐Src kinase downstream of PDGF receptor signaling. Western blotting also indicated that the levels of a sub‐population of Src molecules phosphorylated on tyrosine 419 was also increased upon stimulation but came back to basal levels upon treatment with SU6656 although the total amount of c‐Src remained similar in all three conditions (Figure 2). Phosphorylation of Src kinase on tyrosine 419 is a measure of its catalytic efficiency. This demonstrates that those proteins that are regulated by c‐Src downstream of PDGF signaling have not been tyrosine phosphorylated if c‐Src is rendered inactive.
Figure 1.
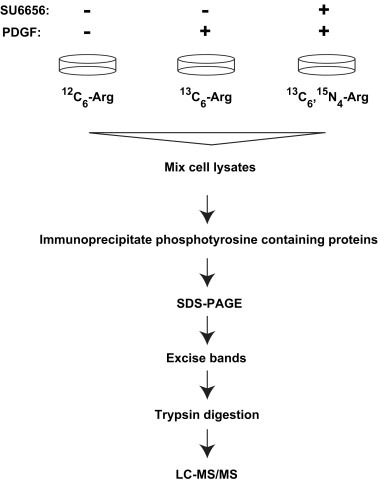
A schematic illustration of the approach to identify c‐Src kinase substrates in PDGF receptor signaling using SILAC. Mouse embryonic fibroblasts (NIH 3T3) were grown in three different stable isotope‐labeled media. One population of cells growing in 12C6‐arginine‐containing medium was left unstimulated, another population of cells growing in 13C6‐arginine‐containing medium was stimulated with 100ng/ml PDGF‐BB ligand and the third population of cells growing in 13C6–15N4‐arginine‐containing medium was pre‐treated with SU6656 for 1h and then stimulated with 100ng/ml PDGF‐BB ligand. The cells were lyzed and tyrosine‐phosphorylated proteins were enriched and analyzed as described in Materials and methods.
Figure 2.
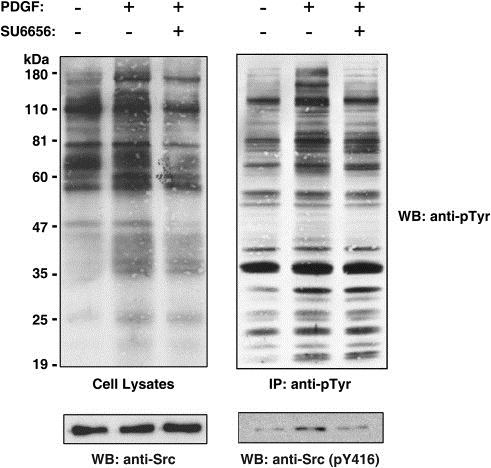
Phosphotyrosine profile of NIH/3T3 cells upon treatment with SU6656 and stimulation with PDGF‐BB. Untreated NIH/3T3 cells, PDGF treated cells and SU6656+PDGF treated NIH/3T3 cells were lysed and cell extracts were subjected to affinity purification of tyrosine‐phosphorylated proteins. Cell lysates and immunoprecipitates were then resolved by SDS‐PAGE and transferred to a nitrocellulose membrane. The membranes were probed with anti‐phosphotyrosine antibodies and re‐probed with anti‐Src or anti‐Src (pY416) antibodies as shown.
2.2. Quantitation of phosphorylation events reveals known and novel c‐Src substrates as well as PDGF signaling pathway components
Mixing of light, medium and heavy isotope‐labeled cell lysates followed by immunoaffinity purification of tyrosine‐phosphorylated proteins allowed us to systematically compare the profile of proteins from all three conditions in a single MS experiment. The ratio of the intensity of the medium isotope versus the light isotope containing peptides provided the information about enrichment of the peptide from a tyrosine‐phosphorylated protein and its degree of phosphorylation and involvement in PDGF signaling pathway. Thus, the greater the extent of phosphorylation of a protein in PDGF signaling pathway, the higher should be its abundance in anti‐phosphotyrosine antibody immunoprecipitates. On the other hand, the ratio of heavy to light provides information about the protein's phosphorylation being under direct regulation of c‐Src in the context of PDGF signaling. If the heavy/light ratio was almost as much as the medium/light ratio, we concluded that the proteins phosphorylation was not under the control of c‐Src but if the medium/light ratio was higher than the heavy/light ratio, the protein was being modified on its tyrosine residues by Src kinase in PDGF signaling. Peptides with little increase in intensity indicate that the protein was not different in abundance in the different states being compared. Such proteins were not investigated further as they are likely to be non‐specifically bound proteins that are not of interest or are basally phosphorylated in a manner that does not change with PDGF treatment.
We identified a total of 169 proteins from the anti‐phosphotyrosine immunoprecipitates. Out of these, 43 proteins were candidate c‐Src kinase substrates based on the fold‐changes observed with PDGF treatment. We also searched HPRD (Keshava Prasad et al., 2009; Peri et al., 2003) to see if the proteins identified in this experiment were already shown to be involved in PDGF receptor or c‐Src signaling. We classified these proteins into 4 categories (1, 2, 3, 4) as discussed in the following sections.
Table 1.
A list of known PDGF signaling intermediates identified in this study that are also known c‐Src kinase substrates.
| Accession # | Protein | Fold increase (medium/light±SDa) | Fold increase (heavy/light±SDa) | Reference | |
|---|---|---|---|---|---|
| 1 | NP_001020126 | PI 3 kinase, p85 alpha | 3.3±1.3 | 2.2±0.8 | (Auger et al., 1989; Carpenter et al., 1993) |
| 2 | NP_035945 | FYN binding protein | 2.9 | 1.7 | (Koga et al., 2005) |
| 3 | NP_032328 | Heat shock protein 1, beta | 2.4±0.2 | 0.9±0.1 | (Hutchison et al., 1992; Luo et al., 2008) |
| 4 | NP_067255 | Phospholipase C, gamma 1 | 2.1 | 1.8 | (Khare et al., 1997; Roche et al., 1996) |
| 5 | NP_032835 | PDGF Receptor, beta | 2±0.4 | 1.5±0.2 | (Hansen et al., 1996) |
| 6 | NP_034200 | Dok1 | 1.8±0.1 | 0.9±0.05 | (Zhao et al., 2006) |
| 7 | NP_035332 | Shp2 | 1.7 | 1.2 | (Feng et al., 1993) |
| 8 | NP_033536 | Ezrin | 1.6 | 1.2 | (Srivastava et al., 2005; Zhang and Hutchins, 1997) |
| 9 | NP_038744 | RasGAP SH3‐domain binding protein | 1.5 | 1 | (Amanchy et al., 2008; Schlesinger et al., 1999) |
| 10 | NP_001002011 | Lamin A | 1.3±0.1 | 1±0.2 | (Fields et al., 1990; Qiao et al., 2006) |
| 11 | NP_001003908 | Clathrin, heavy polypeptide | 1.3±0.07 | 1±0.1 | (Luo et al., 2008; Qiao et al., 2006) |
| 12 | NP_034963 | Moesin | 1.3±0.2 | 0.9±0.1 | (Hugo et al., 1996; Thorn et al., 1999) |
| 13 | NP_079555 | hnRNPK | 1.3 | 0.8 | (Nagano et al., 2006; Ostareck‐Lederer et al., 2002) |
| 14 | NP_032895 | Cytosolic phospholipase A2 | 1.3 | 0.8 | (Shankar et al., 2006) |
| 15 | NP_031829 | Cortactin | 1.3 | 0.6 | (Amanchy et al., 2008; Kanner et al., 1990; Wu et al., 1991) |
| 16 | NP_075373 | Vps35 | 1.5±0.1 | 1.1±0.1 | (Courtneidge, 2003; Korolchuk et al., 2007) |
SD: Standard deviation calculated from the ratio of peptides quantitated by SILAC.
Table 2.
A list of novel PDGF signaling intermediates identified in this study that are known c‐Src kinase substrates.
| Accession # | Protein | Fold increase (medium/light±SDa) | Fold increase (heavy/light±SDa) | Reference | |
|---|---|---|---|---|---|
| 1 | NP_598862 | Proteasome 26S non‐ATPase subunit 2 | 1.7±0.4 | 1.1±0.05 | (Qiao et al., 2006) |
| 2 | NP_058028 | Cytidine 5′‐triphosphate synthase | 1.4±0.1 | 1.1±0.04 | (Luo et al., 2008) |
| 3 | NP_034743 | KH‐type splicing regulatory protein | 1.3±0.07 | 1.1±0.3 | (Amanchy et al., 2008) |
| 4 | NP_476513 | FUSE binding protein 1 | 1.3 | 1.0 | (Amanchy et al., 2008) |
| 5 | NP_033529 | Valosin containing protein | 1.3±0.05 | 0.9±0.1 | (Li et al., 2008) |
| 6 | NP_742012 | Isoleucine‐tRNA synthetase | 1.3±0.1 | 0.9±0.07 | (Luo et al., 2008) |
| 7 | NP_035229 | Pyruvate kinase 3 | 1.3±0.7 | 0.8±0.3 | (Eigenbrodt et al., 1992) |
SD: Standard deviation calculated from the ratio of peptides quantitated by SILAC.
Table 3.
A list of novel potential c‐Src kinase substrates identified in this study that are known PDGF receptor signaling intermediates.
| Accession # | Protein | Fold increase (medium/light±SDa) | Fold increase (heavy/light±SDa) | Reference | |
|---|---|---|---|---|---|
| 1 | NP_032867 | PI 3 kinase, p85 beta | 2.4±1.0 | 1.8±0.6 | (Herbst et al., 1995) |
| 2 | NP_034610 | Heat shock protein 1, alpha | 1.5±0.1 | 1.0±0.04 | (Barati et al., 2006) |
| 3 | NP_032080 | Fyn | 1.5 | 1.1 | (Hansen et al., 1997) |
| 4 | NP_112442 | Heat shock protein 8 | 1.5±0.2 | 1.1±0.2 | (Cobreros et al., 2008) |
| 5 | NP_035500 | Cytoplasmic FMR1 interacting protein 1 | 1.4 | 1.2 | (Castets et al., 2005) |
SD: Standard deviation calculated from the ratio of peptides quantitated by SILAC.
Table 4.
A list of novel potential c‐Src kinase substrates identified in this study with no prior known involvement in PDGF receptor signaling pathway.
| Accession # | Protein | Fold increase (medium/light±SDa) | Fold increase (heavy/light±SDa) | |
|---|---|---|---|---|
| 1 | NP_031969 | Eps15 | 2.5±0.4 | 1.0±0.3 |
| 2 | NP_035718 | Tripartite motif protein 28 | 2.4 | 1.0 |
| 3 | NP_001028465 | UAP1 like‐1 | 2 | 1.4 |
| 4 | NP_035853 | Xanthine dehydrogenase | 1.8 | 0.9 |
| 5 | NP_035449 | Seryl‐aminoacyl‐tRNA synthetase 1 | 1.7 | 1.0 |
| 6 | NP_033924 | Calpain 2 | 1.5±0.2 | 1.0±0.08 |
| 7 | NP_919323 | Unc‐84 homolog | 1.5 | 0.8 |
| 8 | NP_034611 | Heat shock protein 9A | 1.4±0.07 | 0.9±0.3 |
| 9 | NP_149065 | Threonyl‐tRNA synthetase | 1.4 | 0.9 |
| 10 | NP_058017 | Stress‐induced phosphoprotein 1 | 1.3±0.06 | 1.0±0.01 |
| 11 | NP_001028472 | Guanine monophosphate synthetase | 1.3 | 0.9 |
| 12 | NP_031933 | eEF 2 | 1.3±0.1 | 0.9±0.2 |
| 13 | NP_031623 | Calnexin | 1.3±0.1 | 0.9±0.1 |
| 14 | NP_598798 | ATP citrate lyase | 1.2±0.05 | 0.8±0.01 |
| 15 | NP_034607 | Heat shock protein 1 (chaperonin) | 1.3±0 .05 | 0.9±0.1 |
SD: Standard deviation calculated from the ratio of peptides quantitated by SILAC.
2.2.1. Known PDGF signaling pathway intermediates which are also known c‐Src kinase substrates
We identified 16 proteins in this category (Table 1) that have been shown to be c‐Src substrates and to be involved in PDGF signaling. However, for most of these proteins, it has not been shown that their tyrosine phosphorylation is mediated by PDGF‐activated Src kinase.
PDGF receptor has been shown to be associated with an active Src kinase and is known to be phosphorylated on tyrosine 934 located within its kinase domain (Hansen et al., 1996). The regulatory subunit of PI 3 kinase has been demonstrated to be tyrosine phosphorylated in PDGF signaling (Auger et al., 1989; Soltoff et al., 1992). Its subunits, p85 and p110, have both been shown to be tyrosine phosphorylated by the Src kinase (Auger et al., 1989; Carpenter et al., 1993). Here we show that this protein can be a Src substrate in the PDGF signaling pathway (Figure 3A). As can be seen in Figure 3A and Table 1, the tyrosine phosphorylation of p85 is not reduced to basal levels upon Src kinase inhibition which implies that other kinases may also be responsible for the tyrosine phosphorylation of this kinase.
Figure 3.
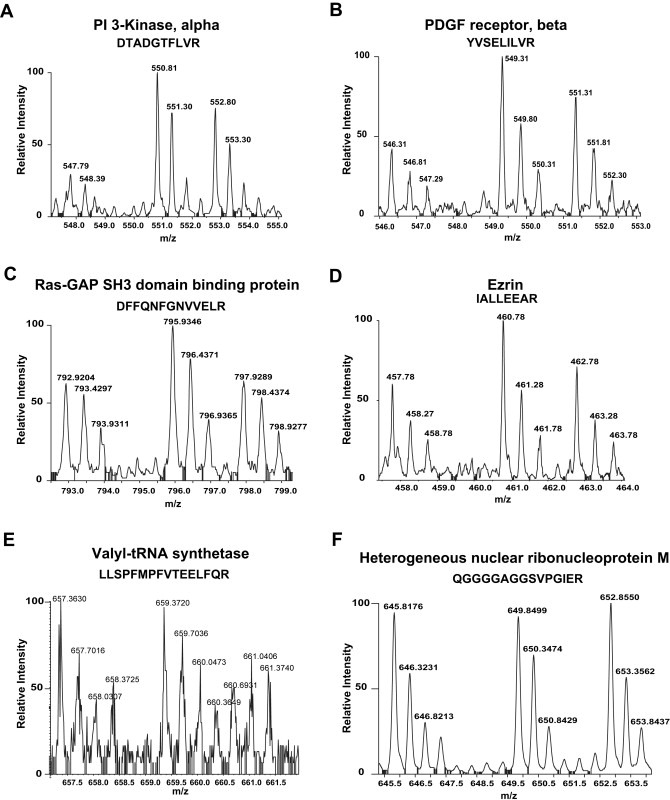
MS spectra of proteins showing different profiles identified in our SILAC screen. The 3 spectral peaks in each figure represent the mass shift of the same peptide. (A) A doubly charged peptide with the sequence DTADGTFLVR, from PI 3 kinase, alpha (1:3.3:2.2); (B) A doubly charged peptide with the sequence YVSELILVR from PDGF receptor, beta (1:2:1.5); (C) A doubly charged peptide with the sequence DFFQNFGNVVELR from Ras‐GAP SH3‐domain‐containing protein (1:1.5:1); (D) A doubly charged peptide with the sequence IALLEEAR from Ezrin (1:1.6:1.2); (E) A triply charged peptide with the sequence LLSPFMPFVTEELFQR from valyl‐tRNA synthetase (1:1.03:0.74); (F) A doubly charged peptide with the sequence from QGGGGAGGSVPGIER from heterogeneous nuclear ribonucleoprotein M (1:1.1:1.1). While some of the proteins (A–D) show involvement in PDGF signaling pathway as c‐Src substrates, some proteins do not show any involvement in PDGF signaling pathway (E) or do not show characteristic spectra as that of c‐Src substrates (F). The ratios shown in parentheses reflect ratio of abundance of peptides across three states (light:medium:heavy).
Cortactin was first identified as a tyrosine‐phosphorylated protein in v‐Src‐transformed chicken embryo fibroblasts (Kanner et al., 1990). Later several studies confirmed this as a c‐Src substrate (Amanchy et al., 2008; Courtneidge, 2003; Luo et al., 2008) along with its participation in growth factor induced signaling and cell adhesion events (Weed and Parsons, 2001). Stimulation of Swiss 3T3 cells with PDGF increased the cytosolic phospholipase A2 (cPLA2) activity (Domin and Rozengurt, 1993) and PDGF induced the expression and activity of phospholipase 2 in a time‐dependent manner in vascular smooth muscle cells (Yellaturu and Rao, 2003). Though there was no direct evidence of the role of c‐Src kinase in the PDGF‐induced phospholipase 2 activity, its activity and phosphorylation in platelets were shown to be dependent on Src kinase activity (Shankar et al., 2006). Our results integrate the evidence for the role of phospholipase 2 downstream of c‐Src in PDGF signaling.
Phospholipase C gamma (PLCγ) has been shown to interact with PDGF receptor and to be required for PDGF‐induced DNA synthesis (Roche et al., 1996). Using another Src kinase inhibitor, PP1, phosphorylation of RasGAP and docking protein 1 (Dok1) have been shown to be dependent on Src kinase activity in PDGF signaling (Shah and Vincent, 2005). SFKs have been shown to play an important role in the phosphorylation of PDGF receptor interacting proteins, including PLCγ and RasGAP, upon PDGF stimulation (DeMali and Kazlauskas, 1998). SH2 domain‐containing protein tyrosine phosphatase‐2 (Shp2) has previously been shown to bind to autophosphorylated platelet‐derived growth factor (PDGF) receptors through its SH2 domains. It is phosphorylated on tyrosine residues transiently in PDGF‐ and EGF‐stimulated cells and constitutively in v‐Src‐transformed cells (Feng et al., 1993).
FYN binding protein (Fyb), also termed adhesion and degranulation promoting adaptor protein (ADAP) and SLP‐76‐associated phosphoprotein (SLAP130), was originally identified as a PDGF receptor interacting protein (Roche et al., 1998) and later also as c‐Src binding partner in bone marrow cells (Koga et al., 2005). Fyb was phosphorylated on Y807 by c‐Src and was found to be a negative regulator of c‐Src activity and PDGF‐induced mitogenesis. Though its involvement in PDGF signaling downstream of c‐Src has been well described (Sirvent et al., 2008), here we present the evidence for its phosphorylation being Src‐dependent in PDGF signaling pathway.
Vacuolar protein sorting 35 (Vps35) was identified as a Src kinase substrate in a screen where mammalian cDNA libraries were inserted into phage vectors for protein production and plaques proteins were screened for upstream kinase (Courtneidge, 2003). Vps35 showed involvement in receptor endocytosis, and actin cytoskeletal organization but it is the first time Vps35 has been shown to be involved as a Src kinase substrate in PDGF signaling pathway.
Lamin A was found to be phosphorylated in response to PDGF (Fields et al., 1990) and it has also been identified as a tyrosine‐phosphorylated protein in NIH/3T3 cells by chemically inducing Src kinase activity (Qiao et al., 2006). Clathrin has also been shown as a c‐Src substrate in Src‐transformed (Luo et al., 2008) and Src activated fibroblasts (Qiao et al., 2006) and as a tyrosine kinase substrate in PDGF stimulated vascular smooth cells (Furge et al., 1999). Ezrin and moesin regulate cell morphology and plasma membrane dynamics by anchoring actin filaments to integral membrane proteins. Both Moesin and Ezrin/villin 2/cytovillin have previously been reported as Src substrates (Thorn et al., 1999) and to be involved in PDGF signaling (Hugo et al., 1996). We present here the evidence for these two proteins as Src substrates in the context of PDGF receptor signaling. While heterogeneous nuclear ribonucleoprotein K (hnRNPK) is a known c‐Src kinase substrate with a role in translational activation (Ostareck‐Lederer et al., 2002), reports have also shown that it has a role in the regulation of actin cytoskeleton in PDGF signaling pathway (Nagano et al., 2006). We have shown earlier that hnRNPK is tyrosine phosphorylated in cells overexpressing an active Src kinase (Amanchy et al., 2008). Here, we present evidence to support that it is a Src substrate in PDGF signaling pathway. Although the above mentioned proteins are known c‐Src substrates and also identified as components in PDGF signaling, this is the first study implicating them as phosphorylated substrates of c‐Src in PDGF signaling.
2.2.2. Known c‐Src substrates identified as novel PDGF signaling intermediates
We identified 7 known c‐Src kinase substrates in this category (Table 2), which have not been previously reported as intermediates in PDGF signaling. Proteasome 26S non‐ATPase subunit 2 has been shown to be tyrosine phosphorylated in a screen where a mutant Src kinase was chemically induced (Qiao et al., 2006). Cytidine 5′‐triphosphate (CTP) synthase has been shown to be regulated by phosphorylation on multiple sites (Huang and Graves, 2003). CTP synthase and isoleucine‐tRNA synthetase have been identified as tyrosine phosphorylated in Src‐transformed cells (Luo et al., 2008). We have shown earlier that KH‐type splicing regulatory protein and FUSE binding protein 1 are tyrosine phosphorylated upon expression of a constitutively active c‐Src (Amanchy et al., 2008). Pyruvate kinase 3 (type M2) is a known substrate of the Src tyrosine kinase, which showed a lower affinity for its substrate, phosphoenolpyruvate, after Src mediated phosphorylation (Eigenbrodt et al., 1992). We have previously identified valosin containing protein (VCP) as a tyrosine kinase substrate in pervanadate treated HeLa cells (Amanchy et al., 2005a) and VCP has also been described as a c‐Src substrate with a role in proteolytic degradation of misfolded proteins (Li et al., 2008).
2.2.3. Novel c‐Src substrates with known involvement in PDGF signaling pathway
We identified 5 known PDGF signaling intermediates as novel c‐Src kinase substrates (Table 3). Phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 2 (p85 beta) has previously been reported as a PDGF receptor interacting protein (Herbst et al., 1995; Inukai et al., 2001). Cytoplasmic FMR1 interacting protein 1 co‐localizes with actin and helps in cytoskeletal reorganization on PDGF stimulation (Castets et al., 2005). It has previously reported that heat shock protein 8 associates with PDGF and VEGF (Vascular endothelial growth factor) receptors and regulates border cell migration (Cobreros et al., 2008). It has also been shown in a proteomic analysis that a group of chaperone proteins – Heat shock proteins (Hsp) Hsp70, Hsp90 alpha, Hsp90 beta, Grp78, Grp94, and protein disulfide isomerase (PDI) – are potential Akt substrates in PDGF signaling pathway in mesangial cells (Barati et al., 2006). Fyn, a member of c‐Src kinase family, was found to be phosphorylated on tyrosine 28 located within its N‐terminal region by PDGF receptor (Hansen et al., 1997).
2.2.4. Novel Src substrates with unknown role in PDGF signaling pathway
We identified 15 proteins as novel c‐Src kinase substrates and novel components in PDGF signaling (Table 4). Epidermal growth factor receptor pathway substrate 15 (Eps15) was implicated in the endocytic pathway during receptor internalization (Torrisi et al., 1999). Tripartite motif protein 28 (Trim28), also known as transcriptional intermediary factor 1, beta (TIF1β), belongs to a family of conserved Transcriptional Intermediary Factor 1 genes (TIF1α, β, γ), of which TIF1α has been shown to be a protein kinase (Fraser et al., 1998). Recent human kinome annotation efforts led to identification of all three genes as atypical protein kinases, atypical because they do not have a typical kinase domain (Manning et al., 2002). Calpain 2 can interact with the c‐Src kinase in the formation of invadopodia and breast cancer invasion (Cortesio et al., 2008) and functions as a downstream effector in v‐Src‐transformed myoblasts (Ciuffini et al., 2008). Isopeptidase T plays a regulatory role in proteasomal and/or lysosomal degradation of growth factor receptors, although its mechanism and relation to PDGF receptor have not yet been elucidated (Kato et al., 2000). ATP citrate lyase activation in IL‐3 (Bauer et al., 2005), transforming growth factor (TGF) beta and epidermal growth factor (EGF) signaling (Reinhart and Roehrig, 1987) indicates that this protein could be regulating glucose‐dependent lipogenesis as well as growth and transformation. Recently, it has been shown that selective inhibition of ATP citrate lyase resulted in inhibition of tumor cell growth in vitro and in vivo, which suggests that it may be a novel therapeutic target in lung cancers (Migita et al., 2008). Calnexin, the endoplasmic reticulum‐specific protein, was found to be associated with SH2 containing protein tyrosine phosphatase‐2 (Shp2) and phospholipase C gamma (PLCγ) (Wang et al., 2005), which were also identified in our screen as Src substrates. UAP1 like‐1, Xanthine dehydrogenase, aminoacyl synthetases, GMP synthetase are all metabolic enzymes whose roles in cellular signaling pathways have not yet been unveiled.
2.3. Bioinformatics analysis of the identified proteins
We carried out a systematic bioinformatics analysis of all Src substrates identified in this study for their primary subcellular localization based on gene ontology terms (Ashburner et al., 2000). Two‐thirds of the identified proteins were cytosolic proteins, which are expected because c‐Src is primarily localized to the cytosol. However, we also found proteins that were localized to the plasma membrane where c‐Src is found when it gets myristylated (Garber et al., 1985). Hence the membrane proteins could bind to the myristylated membrane‐bound form of c‐Src or could associate with the membrane upon phosphorylation by c‐Src. 12% of the proteins identified were nuclear proteins which corroborates with our earlier published reports where we have identified nuclear associated proteins as c‐Src substrates (Amanchy et al., 2008) as well as by other reports that c‐Src also localizes to the nucleus (Redmond et al., 1992). We also performed a bioinformatics analysis of these proteins for their biological process. One‐thirds of the proteins identified in this screen are proteins involved in cell signaling and communication, 25% of the proteins are proteins with a protein processing and transport function and 11% of the proteins are involved in maintaining cellular organization while 23% of the proteins are metabolic enzymes and proteins involved in transcription and translation represent 8% of the identified proteins.
2.4. Experimental validation of a subset of novel proteins as Src substrates
Because antibodies against most of proteins identified in our analysis are not commercially available, we could only validate the role of a subset of the identified Src substrates in PDGF signaling by directly examining their tyrosine phosphorylation in NIH/3T3 cells upon PDGF stimulation in the presence or absence of the Src kinase inhibitor. Cortactin, SH2 containing protein tyrosine phosphatase‐2 (Shp2), Fyn binding protein (FYB) and cytosolic phospholipase 2 (cPLA2) were immunoprecipitated with specific antibodies followed by Western blotting with anti‐phosphotyrosine antibodies (Figure 4A). Trim28, Ezrin, Calpain 2 and Epidermal growth factor receptor substrate 15 (Eps15) were immunoprecipitated with anti‐phosphotyrosine antibodies followed by Western blotting with specific antibodies against these proteins (Figure 4B). The total amount of each protein in the whole cell lysates was similar across different conditions. As expected, we observed that all of these proteins were phosphorylated in NIH/3T3 cells upon addition of PDGF. Importantly, their phosphorylation was decreased in the cells pre‐treated with a c‐Src kinase inhibitor (Figure 4). All of these Western blotting results also substantiate our quantitative phosphoproteomic strategy in identification of c‐Src kinase substrates in PDGF receptor signaling.
Figure 4.
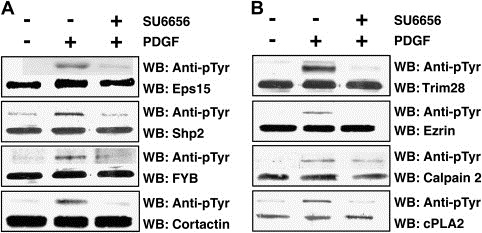
Experimental validation of a subset of novel substrates identified in this study by Western blotting. NIH/3T3 cells have been grown to 80% confluence and serum‐starved for 12h followed by either no treatment, stimulation with PDGF‐BB (100ng/mL for 5min) or PDGF stimulation after preinculation with SU6656 (2μM for 1h prior to lysis or stimulation). (A) Cell lysates were subjected to immunoprecipitation using antibodies against Eps15, Cortactin, Shp2, Fyb or cPLA2 and probed by Western blotting with anti‐phosphotyrosine antibodies. (B) Cell lysates were subjected to immunoprecipitation using anti‐phosphotyrosine antibodies and probed by Western blotting with antibodies against Trim28, Ezrin, Calpain 2, respectively. The total amount of proteins in whole cell lysates across different conditions was monitored by Western blotting with specific antibodies against these proteins.
In this study, we have successfully employed the SILAC strategy combined with chemical inhibitors and phosphoproteome enrichment to identify and catalog Src phosphoproteome – specifically in PDGF receptor signaling pathway – from NIH/3T3 cells. Though we have not focused on identification of phosphorylated peptides, future studies can be aimed at specific enrichment of phosphopeptides that are specific to Src enzymatic activity. Furthermore, a different mass spectrometry‐based technique such as multiple reaction monitoring (MRM) could be used to monitor the abundance of specific phosphorylated peptides. Further studies focusing on the biological importance of these molecules need to be done to expand our understanding of the roles of c‐Src and its novel substrates in PDGF signaling. We believe that these studies will impact future experiments to dissect cellular and cancer signaling pathways.
3. Experimental procedures
3.1. Chemicals and antibodies
Stable isotope containing amino acids, 13C6‐arginine and 13C6‐15N4‐arginine, were purchased from Cambridge Isotope Labs (Andover, MA). Anti‐Flag M2 monoclonal antibody, sodium orthovanadate and hydrogen peroxide were purchased from Sigma–Aldrich (St. Louis, MO), Complete protease inhibitor cocktail tablets were purchased from Roche (Indianapolis, IN) and 10mM (500μg/135μl) solution of SU6656 (Cat. No. 572635, Gibbstown, NJ) in DMSO was purchased from Calbiochem. Anti‐phosphotyrosine antibodies (4G10) agarose‐conjugate and streptavidin–agarose beads were purchased from Upstate Biotechnology (Lake Placid, NY). Anti‐phosphotyrosine RC20 biotin conjugate was purchased from BD transduction laboratories (Lexington, KY). Sequencing grade trypsin was purchased from Promega (Madison, WI). Rabbit polyclonal antibodies against Src (pY416) were purchased from Calbiochem (EMD biosciences, Gibbstown, NJ). Rabbit polyclonal antibodies against epidermal growth factor receptor susbtrate 15/EPS15 (sc‐534), rabbit polyclonal antibodies against SH2 domain‐containing protein tyrosine phosphatase‐2 (Shp2) (sc‐280), goat polyclonal antibodies against fyn binding protein (FYB) (sc‐7105), mouse monoclonal antibodies against Cortactin (sc‐55579), goat polyclonal antibodies against Tripartite motif protein 28 (Trim28) (sc‐19168), rabbit polyclonal antibodies against Ezrin (sc‐20773), goat polyclonal antibodies against Calpain 2 (sc‐7532), rabbit polyclonal antibodies against c‐Src (sc‐19), goat polyclonal antibodies against cytosolic phospholipase A2 (cPLA2) (sc‐14463) were obtained from Santa Cruz Biotechnology, Inc (Santa Cruz, CA).
3.2. Stable isotope labeling with amino acids in cell culture (SILAC)
NIH/3T3 cells were grown in DMEM containing either 12C6‐arginine or 13C6‐arginine or 13C6‐15N4‐arginine supplemented with 10% dialyzed FBS and antibiotics. The NIH/3T3 cells were adapted to the SILAC medium as described earlier (Amanchy et al., 2008). The cells were grown to 80% confluence in 15 of 150mm dishes per condition. Cells were then serum‐starved for 12h before treatment. The cells grown in 13C6‐arginine‐containing media were stimulated with PDGF‐BB (100ng/ml) for 5min. The cells grown in 13C6‐15N4‐arginine‐containing medium were pre‐treated with Src kinase inhibitor SU6656 (2μM in DMSO) for 1h followed by stimulation of cells with PDGF‐BB (100ng/ml in DMEM) for 5min. Cells were lysed in modified RIPA buffer (50mM Tris–HCl, pH 7.4, 150mM NaCl, 1mM EDTA, 1% Nonidet P‐40, 0.25% sodium deoxycholate, and 1mM sodium orthovanadate (tyrosine phosphatase inhibitor) in the presence of protease inhibitors) and centrifuged at 12,000×g for 20min at 4°C. Protein concentration of the lysates was determined.
3.3. Immunoaffinity purification of tyrosine‐phosphorylated proteins
Equal amount of supernatants from cell lysates were mixed (60mg of protein per condition), precleared with protein A‐agarose and incubated with anti‐phosphotyrosine antibodies (400μg of 4G10 agarose‐conjugate, 75μg of biotin‐conjugated RC20 antibody and 100μg of streptavidin–agarose beads) overnight at 4°C in the presence of protease and phosphatase inhibitor cocktail. Immunoprecipitated tyrosine‐phosphorylated proteins were washed three times with the lysis buffer and eluted three times with 100mM phenyl phosphate at 37°C. The eluted phosphoproteins were dialyzed and resolved by SDS‐PAGE. The gel was stained using a colloidal blue staining kit from Invitrogen.
3.4. LC–MS/MS and data analysis
The stained gel was excised into bands and each band was digested with trypsin as described previously (Amanchy et al., 2005b). Each fraction from the in‐gel digestion was analyzed using reversed phase liquid chromatography (RP‐HPLC) coupled to a QSTAR Pulsar mass spectrometer (Applied Biosystems). Samples were injected using an Agilent 1100 auto sampler (Agilent Technologies, Palo Alto, CA) with 8μL loop. For LC separation, we used an Eskigent nanoLC‐2D (Eskigent, Dublin, CA) to generate the gradient by mixing solvent A (0.1% formic acid) and solvent B (0.1% formic acid, 90% acetonitrile). Tryptic peptides was loaded with a flow of 5μL/min at 1% solvent B onto a trap column (75μm×3cm, C18 material 5–10μm, 120Å, YMC, Japan) followed by separation with a gradient of 10–40% of solvent B on an analytical column (75μm×10cm, C18 material 5μm, 120Å, YMC, Japan). An electrospray emitter with an 8μm tip was used for ESI (New Objective, Woburn, MA). MS spectra were acquired from m/z 350 to 1200 in a data‐dependent mode and the three most abundant ions were selected for MS/MS. An exclusion time of 45s was used. LC–MS/MS data were acquired as instrument raw (*.wiff) files using Analyst QS 1.1 (MDS Sciex, Concord, Ontario, Canada) and processed to mascot generic format (mgf) files using the mascot.dll data import filter script in Mascot Daemon (version 2.2.2) (Matrixscience, Manchester, UK). The merged data were searched using Mascot search engine (version 2.2.0) (Matrixscience, Manchester, UK) against the mouse RefSeq database (version 26). The search parameters were as follows: enzyme – trypsin; maximum allowed missed cleavages – 2; fixed modification – carbamidomethylation of cysteine residues; variable modifications – oxidation of methionine, phosphorylation of tyrosine, arginine‐13C6 labeling, and arginine‐13C6‐15N4 labeling. Mass tolerance was set to 0.3Da for precursors and 0.3Da for fragment ions. The false discovery rate (FDR) of identified peptides was calculated by the number of total peptide hits in reverse database divided by the number of total peptide hits in forward and reverse databases above the same score. Proteins with at least two reliable peptides (rank 1; unique; individual score higher than or equal to 43 (i.e. better than 1% FDR)) were considered as positively identified proteins. Relative quantitation of stable isotope‐labeled peptides was performed using MSQuant (v1.4.3a39) downloaded from http://msquant.sourceforge.net. MSQuant quantitates individual peptide ratios at different time points at chromatographic peaks and calculates protein ratios. Mascot search results in .html format were parsed with the raw data file in MSQuant. Quantitative as well as qualitative data were manually inspected using MSQuant.
Supporting information
Supplementary Table 1. A list of c‐Src kinase substrates identified by SILAC
Acknowledgements
AP is supported by grants from the National Institutes of Health (CA106424 and U54 RR020839), National Heart, Lung, and Blood Institute, National Institutes of Health, under contract number HV‐28180 and Department of Defense Era of Hope Scholar award (W81XWH‐06‐1‐0428). JZ is supported by grant from Department of Defense Breast Cancer Research Program (W81XWH‐05‐1‐0304). We thank the help of Raghunath Reddy for the bioinformatics analysis.
Appendix. Supplementary data 1.
Supplementary data associated with this article can be found, in the online version, at doi: 10.1016/j.molonc.2009.07.001.
Amanchy Ramars, Zhong Jun, Hong Rosa, Kim James H., Gucek Marjan, Cole Robert N., Molina Henrik, Pandey Akhilesh, (2009), Identification of c‐Src tyrosine kinase substrates in platelet‐derived growth factor receptor signaling, Molecular Oncology, 3, doi: 10.1016/j.molonc.2009.07.001.
References
- Amanchy, R. , Kalume, D.E. , Iwahori, A. , Zhong, J. , Pandey, A. , 2005. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC). J. Proteome Res.. 4, 1661–1671. [DOI] [PubMed] [Google Scholar]
- Amanchy, R. , Kalume, D.E. , Pandey, A. , 2005. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci. STKE. 2005, pl2 [DOI] [PubMed] [Google Scholar]
- Amanchy, R. , Zhong, J. , Molina, H. , Chaerkady, R. , Iwahori, A. , Kalume, D.E. , Gronborg, M. , Joore, J. , Cope, L. , Pandey, A. , 2008. Identification of c-Src tyrosine kinase substrates using mass spectrometry and peptide microarrays. J. Proteome Res.. 7, 3900–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C.A. , Blake, J.A. , Botstein, D. , Butler, H. , Cherry, J.M. , Davis, A.P. , Dolinski, K. , Dwight, S.S. , Eppig, J.T. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet.. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger, K.R. , Serunian, L.A. , Soltoff, S.P. , Libby, P. , Cantley, L.C. , 1989. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 57, 167–175. [DOI] [PubMed] [Google Scholar]
- Bagrodia, S. , Chackalaparampil, I. , Kmiecik, T.E. , Shalloway, D. , 1991. Altered tyrosine 527 phosphorylation and mitotic activation of p60c-src. Nature. 349, 172–175. [DOI] [PubMed] [Google Scholar]
- Bain, J. , McLauchlan, H. , Elliott, M. , Cohen, P. , 2003. The specificities of protein kinase inhibitors: an update. Biochem. J.. 371, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barati, M.T. , Rane, M.J. , Klein, J.B. , McLeish, K.R. , 2006. A proteomic screen identified stress-induced chaperone proteins as targets of Akt phosphorylation in mesangial cells. J. Proteome Res.. 5, 1636–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, D.E. , Hatzivassiliou, G. , Zhao, F. , Andreadis, C. , Thompson, C.B. , 2005. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 24, 6314–6322. [DOI] [PubMed] [Google Scholar]
- Blagoev, B. , Ong, S.E. , Kratchmarova, I. , Mann, M. , 2004. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol.. 22, 1139–1145. [DOI] [PubMed] [Google Scholar]
- Blake, R.A. , Broome, M.A. , Liu, X. , Wu, J. , Gishizky, M. , Sun, L. , Courtneidge, S.A. , 2000. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol.. 20, 9018–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon, T.J. , Eck, M.J. , 2004. Structure and regulation of Src family kinases. Oncogene. 23, 7918–7927. [DOI] [PubMed] [Google Scholar]
- Carpenter, C.L. , Auger, K.R. , Duckworth, B.C. , Hou, W.M. , Schaffhausen, B. , Cantley, L.C. , 1993. A tightly associated serine/threonine protein kinase regulates phosphoinositide 3-kinase activity. Mol. Cell. Biol.. 13, 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets, M. , Schaeffer, C. , Bechara, E. , Schenck, A. , Khandjian, E.W. , Luche, S. , Moine, H. , Rabilloud, T. , Mandel, J.L. , Bardoni, B. , 2005. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet.. 14, 835–844. [DOI] [PubMed] [Google Scholar]
- Ciuffini, L. , Castellani, L. , Salvati, E. , Galletti, S. , Falcone, G. , Alema, S. , 2008. Delineating v-Src downstream effector pathways in transformed myoblasts. Oncogene. 27, 528–539. [DOI] [PubMed] [Google Scholar]
- Cobreros, L. , Fernandez-Minan, A. , Luque, C.M. , Gonzalez-Reyes, A. , Martin-Bermudo, M.D. , 2008. A role for the chaperone Hsp70 in the regulation of border cell migration in the Drosophila ovary. Mech. Dev.. 125, 1048–1058. [DOI] [PubMed] [Google Scholar]
- Cooper, J.A. , Gould, K.L. , Cartwright, C.A. , Hunter, T. , 1986. Tyr527 is phosphorylated in pp60c-src: implications for regulation. Science. 231, 1431–1434. [DOI] [PubMed] [Google Scholar]
- Cortesio, C.L. , Chan, K.T. , Perrin, B.J. , Burton, N.O. , Zhang, S. , Zhang, Z.Y. , Huttenlocher, A. , 2008. Calpain 2 and PTP1B function in a novel pathway with Src to regulate invadopodia dynamics and breast cancer cell invasion. J. Cell Biol.. 180, 957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtneidge, S.A. , 2003. Isolation of novel Src substrates. Biochem. Soc. Trans.. 31, 25–28. [DOI] [PubMed] [Google Scholar]
- DeMali, K.A. , Kazlauskas, A. , 1998. Activation of Src family members is not required for the platelet-derived growth factor beta receptor to initiate mitogenesis. Mol. Cell. Biol.. 18, 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMali, K.A. , Godwin, S.L. , Soltoff, S.P. , Kazlauskas, A. , 1999. Multiple roles for Src in a PDGF-stimulated cell. Exp. Cell Res.. 253, 271–279. [DOI] [PubMed] [Google Scholar]
- Dobreva, I. , Fielding, A. , Foster, L.J. , Dedhar, S. , 2008. Mapping the integrin-linked kinase interactome using SILAC. J. Proteome Res.. 7, 1740–1749. [DOI] [PubMed] [Google Scholar]
- Domin, J. , Rozengurt, E. , 1993. Platelet-derived growth factor stimulates a biphasic mobilization of arachidonic acid in Swiss 3T3 cells. The role of phospholipase A2. J. Biol. Chem.. 268, 8927–8934. [PubMed] [Google Scholar]
- Eigenbrodt, E. , Reinacher, M. , Scheefers-Borchel, U. , Scheefers, H. , Friis, R. , 1992. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit. Rev. Oncog.. 3, 91–115. [PubMed] [Google Scholar]
- Feng, G.S. , Hui, C.C. , Pawson, T. , 1993. SH2-containing phosphotyrosine phosphatase as a target of protein–tyrosine kinases. Science. 259, 1607–1611. [DOI] [PubMed] [Google Scholar]
- Fields, A.P. , Tyler, G. , Kraft, A.S. , May, W.S. , 1990. Role of nuclear protein kinase C in the mitogenic response to platelet-derived growth factor. J. Cell Sci.. 96, (1) 107–114. [DOI] [PubMed] [Google Scholar]
- Fraser, R.A. , Heard, D.J. , Adam, S. , Lavigne, A.C. , Le Douarin, B. , Tora, L. , Losson, R. , Rochette-Egly, C. , Chambon, P. , 1998. The putative cofactor TIF1alpha is a protein kinase that is hyperphosphorylated upon interaction with liganded nuclear receptors. J. Biol. Chem.. 273, 16199–16204. [DOI] [PubMed] [Google Scholar]
- Furge, L.L. , Chen, K. , Cohen, S. , 1999. Annexin VII and annexin XI are tyrosine phosphorylated in peroxovanadate-treated dogs and in platelet-derived growth factor-treated rat vascular smooth muscle cells. J. Biol. Chem.. 274, 33504–33509. [DOI] [PubMed] [Google Scholar]
- Garber, E.A. , Cross, F.R. , Hanafusa, H. , 1985. Processing of p60v-src to its myristylated membrane-bound form. Mol. Cell. Biol.. 5, 2781–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, K.L. , Hunter, T. , 1988. Platelet-derived growth factor induces multisite phosphorylation of pp60c-src and increases its protein–tyrosine kinase activity. Mol. Cell. Biol.. 8, 3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi, I. , Yamaguchi, N. , Suda, J. , Iwama, A. , Hirao, A. , Hashiyama, M. , Aizawa, S. , Suda, T. , 1996. Analysis of CSK homologous kinase (CHK/HYL) in hematopoiesis by utilizing gene knockout mice. Biochem. Biophys. Res. Commun.. 224, 172–179. [DOI] [PubMed] [Google Scholar]
- Hansen, K. , Johnell, M. , Siegbahn, A. , Rorsman, C. , Engstrom, U. , Wernstedt, C. , Heldin, C.H. , Ronnstrand, L. , 1996. Mutation of a Src phosphorylation site in the PDGF beta-receptor leads to increased PDGF-stimulated chemotaxis but decreased mitogenesis. EMBO J.. 15, 5299–5313. [PMC free article] [PubMed] [Google Scholar]
- Hansen, K. , Alonso, G. , Courtneidge, S.A. , Ronnstrand, L. , Heldin, C.H. , 1997. PDGF-induced phosphorylation of Tyr28 in the N-terminus of Fyn affects Fyn activation. Biochem. Biophys. Res. Commun.. 241, 355–362. [DOI] [PubMed] [Google Scholar]
- Herbst, R. , Shearman, M.S. , Jallal, B. , Schlessinger, J. , Ullrich, A. , 1995. Formation of signal transfer complexes between stem cell and platelet-derived growth factor receptors and SH2 domain proteins in vitro. Biochemistry. 34, 5971–5979. [DOI] [PubMed] [Google Scholar]
- Huang, M. , Graves, L.M. , 2003. De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell. Mol. Life Sci.. 60, 321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo, C. , Hugo, C. , Pichler, R. , Gordon, K. , Schmidt, R. , Amieva, M. , Couser, W.G. , Furthmayr, H. , Johnson, R.J. , 1996. The cytoskeletal linking proteins, moesin and radixin, are upregulated by platelet-derived growth factor, but not basic fibroblast growth factor in experimental mesangial proliferative glomerulonephritis. J. Clin. Invest.. 97, 2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, K.A. , Brott, B.K. , De Leon, J.H. , Perdew, G.H. , Jove, R. , Pratt, W.B. , 1992. Reconstitution of the multiprotein complex of pp60src, hsp90, and p50 in a cell-free system. J. Biol. Chem.. 267, 2902–2908. [PubMed] [Google Scholar]
- Inukai, K. , Funaki, M. , Anai, M. , Ogihara, T. , Katagiri, H. , Fukushima, Y. , Sakoda, H. , Onishi, Y. , Ono, H. , Fujishiro, M. , 2001. Five isoforms of the phosphatidylinositol 3-kinase regulatory subunit exhibit different associations with receptor tyrosine kinases and their tyrosine phosphorylations. FEBS Lett.. 490, 32–38. [DOI] [PubMed] [Google Scholar]
- Kanner, S.B. , Reynolds, A.B. , Vines, R.R. , Parsons, J.T. , 1990. Monoclonal antibodies to individual tyrosine-phosphorylated protein substrates of oncogene-encoded tyrosine kinases. Proc. Natl. Acad. Sci. USA. 87, 3328–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , Miyazawa, K. , Kitamura, N. , 2000. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem.. 275, 37481–37487. [DOI] [PubMed] [Google Scholar]
- Keshava Prasad, T.S. , Goel, R. , Kandasamy, K. , Keerthikumar, S. , Kumar, S. , Mathivanan, S. , Telikicherla, D. , Raju, R. , Shafreen, B. , Venugopal, A. , 2009. Human protein reference database – 2009 update. Nucleic Acids Res.. 37, D767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare, S. , Bolt, M.J. , Wali, R.K. , Skarosi, S.F. , Roy, H.K. , Niedziela, S. , Scaglione-Sewell, B. , Aquino, B. , Abraham, C. , Sitrin, M.D. , 1997. 1,25 Dihydroxyvitamin D3 stimulates phospholipase C-gamma in rat colonocytes: role of c-Src in PLC-gamma activation. J. Clin. Invest.. 99, 1831–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiecik, T.E. , Johnson, P.J. , Shalloway, D. , 1988. Regulation by the autophosphorylation site in overexpressed pp60c-src. Mol. Cell. Biol.. 8, 4541–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, S. , Yogo, K. , Yoshikawa, K. , Samori, H. , Goto, M. , Uchida, T. , Ishida, N. , Takeya, T. , 2005. Physical and functional association of c-Src and adhesion and degranulation promoting adaptor protein (ADAP) in osteoclastogenesis in vitro. J. Biol. Chem.. 280, 31564–31571. [DOI] [PubMed] [Google Scholar]
- Korolchuk, V.I. , Schutz, M.M. , Gomez-Llorente, C. , Rocha, J. , Lansu, N.R. , Collins, S.M. , Wairkar, Y.P. , Robinson, I.M. , O'Kane, C.J. , 2007. Drosophila Vps35 function is necessary for normal endocytic trafficking and actin cytoskeleton organisation. J. Cell Sci.. 120, 4367–4376. [DOI] [PubMed] [Google Scholar]
- Kratchmarova, I. , Blagoev, B. , Haack-Sorensen, M. , Kassem, M. , Mann, M. , 2005. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 308, 1472–1477. [DOI] [PubMed] [Google Scholar]
- Kruger, M. , Kratchmarova, I. , Blagoev, B. , Tseng, Y.H. , Kahn, C.R. , Mann, M. , 2008. Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl. Acad. Sci. USA. 105, 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Zhao, G. , Schindelin, H. , Lennarz, W.J. , 2008. Tyrosine phosphorylation of ATPase p97 regulates its activity during ERAD. Biochem. Biophys. Res. Commun.. 375, 247–251. [DOI] [PubMed] [Google Scholar]
- Luo, W. , Slebos, R.J. , Hill, S. , Li, M. , Brabek, J. , Amanchy, R. , Chaerkady, R. , Pandey, A. , Ham, A.J. , Hanks, S.K. , 2008. Global impact of oncogenic Src on a phosphotyrosine proteome. J. Proteome Res.. 7, 3447–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, G. , Whyte, D.B. , Martinez, R. , Hunter, T. , Sudarsanam, S. , 2002. The protein kinase complement of the human genome. Science. 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- Migita, T. , Narita, T. , Nomura, K. , Miyagi, E. , Inazuka, F. , Matsuura, M. , Ushijima, M. , Mashima, T. , Seimiya, H. , Satoh, Y. , 2008. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res.. 68, 8547–8554. [DOI] [PubMed] [Google Scholar]
- Nagano, K. , Bornhauser, B.C. , Warnasuriya, G. , Entwistle, A. , Cramer, R. , Lindholm, D. , Naaby-Hansen, S. , 2006. PDGF regulates the actin cytoskeleton through hnRNP-K-mediated activation of the ubiquitin E3-ligase MIR. EMBO J.. 25, 1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, M. , Nada, S. , Yamanashi, Y. , Yamamoto, T. , Nakagawa, H. , 1991. CSK: a protein–tyrosine kinase involved in regulation of src family kinases. J. Biol. Chem.. 266, 24249–24252. [PubMed] [Google Scholar]
- Ostareck-Lederer, A. , Ostareck, D.H. , Cans, C. , Neubauer, G. , Bomsztyk, K. , Superti-Furga, G. , Hentze, M.W. , 2002. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol.. 22, 4535–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri, S. , Navarro, J.D. , Amanchy, R. , Kristiansen, T.Z. , Jonnalagadda, C.K. , Surendranath, V. , Niranjan, V. , Muthusamy, B. , Gandhi, T.K. , Gronborg, M. , 2003. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res.. 13, 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y. , Molina, H. , Pandey, A. , Zhang, J. , Cole, P.A. , 2006. Chemical rescue of a mutant enzyme in living cells. Science. 311, 1293–1297. [DOI] [PubMed] [Google Scholar]
- Ralston, R. , Bishop, J.M. , 1985. The product of the protooncogene c-src is modified during the cellular response to platelet-derived growth factor. Proc. Natl. Acad. Sci. USA. 82, 7845–7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond, T. , Brott, B.K. , Jove, R. , Welsh, M.J. , 1992. Localization of the viral and cellular Src kinases to perinuclear vesicles in fibroblasts. Cell Growth Differ.. 3, 567–576. [PubMed] [Google Scholar]
- Reinhart, G. , Roehrig, K. , 1987. Effect of transforming growth factor beta (TGF-beta) on ATP citrate lyase in isolated hepatocytes. Mol. Cell. Biochem.. 77, 121–125. [DOI] [PubMed] [Google Scholar]
- Roche, S. , McGlade, J. , Jones, M. , Gish, G.D. , Pawson, T. , Courtneidge, S.A. , 1996. Requirement of phospholipase C gamma, the tyrosine phosphatase Syp and the adaptor proteins Shc and Nck for PDGF-induced DNA synthesis: evidence for the existence of Ras-dependent and Ras-independent pathways. EMBO J.. 15, 4940–4948. [PMC free article] [PubMed] [Google Scholar]
- Roche, S. , Alonso, G. , Kazlauskas, A. , Dixit, V.M. , Courtneidge, S.A. , Pandey, A. , 1998. Src-like adaptor protein (Slap) is a negative regulator of mitogenesis. Curr. Biol.. 8, 975–978. [DOI] [PubMed] [Google Scholar]
- Roskoski, R. , 2005. Src kinase regulation by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun.. 331, 1–14. [DOI] [PubMed] [Google Scholar]
- Schlesinger, T.K. , Demali, K.A. , Johnson, G.L. , Kazlauskas, A. , 1999. Platelet-derived growth factor-dependent association of the GTPase-activating protein of Ras and Src. Biochem J. 344, (2) 519–526. [PMC free article] [PubMed] [Google Scholar]
- Shah, K. , Vincent, F. , 2005. Divergent roles of c-Src in controlling platelet-derived growth factor-dependent signaling in fibroblasts. Mol. Biol. Cell.. 16, 5418–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar, H. , Kahner, B.N. , Prabhakar, J. , Lakhani, P. , Kim, S. , Kunapuli, S.P. , 2006. G-protein-gated inwardly rectifying potassium channels regulate ADP-induced cPLA2 activity in platelets through Src family kinases. Blood. 108, 3027–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy, S. , Chackalaparampil, I. , Bagrodia, S. , Lin, P.H. , Shalloway, D. , 1992. Role of p34cdc2-mediated phosphorylations in two-step activation of pp60c-src during mitosis. Proc. Natl. Acad. Sci. USA. 89, 7237–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirvent, A. , Leroy, C. , Boureux, A. , Simon, V. , Roche, S. , 2008. The Src-like adaptor protein regulates PDGF-induced actin dorsal ruffles in a c-Cbl-dependent manner. Oncogene. 27, 3494–3500. [DOI] [PubMed] [Google Scholar]
- Soltoff, S.P. , Rabin, S.L. , Cantley, L.C. , Kaplan, D.R. , 1992. Nerve growth factor promotes the activation of phosphatidylinositol 3-kinase and its association with the trk tyrosine kinase. J. Biol. Chem.. 267, 17472–17477. [PubMed] [Google Scholar]
- Srivastava, J. , Elliott, B.E. , Louvard, D. , Arpin, M. , 2005. Src-dependent ezrin phosphorylation in adhesion-mediated signaling. Mol. Biol. Cell. 16, 1481–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, M.P. , Rush, J. , Macneill, J. , Ren, J.M. , Sprott, K. , Nardone, J. , Yang, V. , Beausoleil, S.A. , Gygi, S.P. , Livingstone, M. , 2007. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. USA. 104, 19855–19860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.M. , Brugge, J.S. , 1997. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol.. 13, 513–609. [DOI] [PubMed] [Google Scholar]
- Thorn, J.M. , Armstrong, N.A. , Cantrell, L.A. , Kay, B.K. , 1999. Identification and characterisation of Xenopus moesin, a Src substrate in Xenopus laevis oocytes. Zygote. 7, 113–122. [DOI] [PubMed] [Google Scholar]
- Torrisi, M.R. , Lotti, L.V. , Belleudi, F. , Gradini, R. , Salcini, A.E. , Confalonieri, S. , Pelicci, P.G. , Di Fiore, P.P. , 1999. Eps15 is recruited to the plasma membrane upon epidermal growth factor receptor activation and localizes to components of the endocytic pathway during receptor internalization. Mol. Biol. Cell.. 10, 417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy, L. , Boulon, S. , Lam, Y.W. , Urcia, R. , Boisvert, F.M. , Vandermoere, F. , Morrice, N.A. , Swift, S. , Rothbauer, U. , Leonhardt, H. , Lamond, A. , 2008. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol.. 183, 223–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veracini, L. , Franco, M. , Boureux, A. , Simon, V. , Roche, S. , Benistant, C. , 2006. Two distinct pools of Src family tyrosine kinases regulate PDGF-induced DNA synthesis and actin dorsal ruffles. J. Cell Sci.. 119, 2921–2934. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Huang, L. , 2008. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell. Proteomics. 7, 46–57. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Downey, G.P. , Herrera-Abreu, M.T. , Kapus, A. , McCulloch, C.A. , 2005. SHP-2 modulates interleukin-1-induced Ca2+ flux and ERK activation via phosphorylation of phospholipase Cgamma1. J. Biol. Chem.. 280, 8397–8406. [DOI] [PubMed] [Google Scholar]
- Weed, S.A. , Parsons, J.T. , 2001. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 20, 6418–6434. [DOI] [PubMed] [Google Scholar]
- Wu, H. , Reynolds, A.B. , Kanner, S.B. , Vines, R.R. , Parsons, J.T. , 1991. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol. Cell. Biol.. 11, 5113–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaturu, C.R. , Rao, G.N. , 2003. Cytosolic phospholipase A2 is an effector of Jak/STAT signaling and is involved in platelet-derived growth factor BB-induced growth in vascular smooth muscle cells. J. Biol. Chem.. 278, 9986–9992. [DOI] [PubMed] [Google Scholar]
- Zhang, F.X. , Hutchins, J.B. , 1997. Protein phosphorylation in response to PDGF stimulation in cultured neurons and astrocytes. Brain Res. Dev. Brain Res.. 99, 216–225. [DOI] [PubMed] [Google Scholar]
- Zhao, M. , Janas, J.A. , Niki, M. , Pandolfi, P.P. , Van Aelst, L. , 2006. Dok-1 independently attenuates Ras/mitogen-activated protein kinase and Src/c-myc pathways to inhibit platelet-derived growth factor-induced mitogenesis. Mol. Cell. Biol.. 26, 2479–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. A list of c‐Src kinase substrates identified by SILAC


