Abstract
Diphtheria toxin T domain helps translocate the A chain of the toxin across membranes. To gain insight into translocation, the membrane topography of key residues in T domain attached to the A chain (AT protein) was compared to that in isolated T domain using fluorescence techniques. This study demonstrated that residues in T domain hydrophobic helices (TH 5-9) tended to be less exposed to aqueous solution in AT protein than in isolated T domain. Under conditions in which the loop connecting TH5 to TH6/7 locates stably on the cis (insertion) side of the membrane in isolated T domain, it moves between the cis and trans sides of the membrane in AT protein. This is indicative of the formation of a dynamic, transient transmembrane hairpin topography by TH 5-7 in the AT protein. Since TH 8-9 also forms a transmembrane hairpin, this means that TH 5-9 may form a cluster of transmembrane helices. These helices have with a non-polar surface likely to face the lipid bilayer in a helix cluster and an surface rich in uncharged hydrophilic residues which in a helix cluster would likely be inward facing (and perhaps pore-lining). This uncharged hydrophilic surface could play a crucial role in translocation, interacting transiently with the translocating A chain. A similar motif can be found in, and may be important for, other protein translocation systems.
Keywords: streptavidin-biotin, fluorescence, protein translocation, model membranes, membrane protein insertion, bacterial toxins
Diphtheria toxin, one of the best known and well-studied A-B bacterial toxins, is secreted as a proenzyme composed of a single polypeptide chain having a molecular weight of 58 kDa (1, 2). The toxin undergoes a two step maturation process. In the first step it is proteolytically nicked into two polypeptide chains: the A chain (21 kDa), which forms the N-terminal part of the toxin, and the B chain (37 kDa), which forms the C-terminal part. The two chains are joined by a single disulfide bond. The B chain is made up of the receptor-binding (R) domain and the highly helical “transmembrane” (T) domain (3-5). The R domain, which forms the C-terminal part of the B chain, contains a site which binds on the cell surface to the heparin-binding epidermal growth factor-like protein, aided by CD9 (6-8). The T domain, which forms the N-terminal half of the B chain, contains hydrophobic sequences that insert into membranes and aid the translocation of the A chain.
In the cell entry process, the receptor-bound toxin undergoes endocytosis. The T domain then undergoes low pH-triggered insertion into endosomal membranes, and at some point a second maturation step occurs, in which there is reduction of A-B disulfide bond. This releases the A chain [also called the catalytic (C) domain] into the cytoplasm, where it catalyzes ADP-ribosylation of the diphthamide residue of elongation factor 2. This shuts down protein synthesis and leads to cell death (9, 10). The last stages of translocation, including the reduction step, may be aided by cytosolic proteins (11, 12).
The detailed mechanism by which diphtheria toxin A chain is translocated across the lipid bilayer of cell membranes remains elusive. The T domain clearly has a key role in promoting translocation. At low pH the T domain forms a pore through which the A chain might pass, and it may also may function as a chaperone, interacting with the partly unfolded translocating A chain so as to prevent A chain refolding during translocation (9, 13-16).
Defining the structure of the membrane-inserted T domain is likely to provide additional insights into the mechanism of translocation. Some aspects of the conformation of membrane-inserted T domain at low pH have been characterized. It can exist both in shallowly-inserted (P state) and deeply-inserted (TM state) conformations (17-22). Formation of the TM state is promoted by a high concentration of T domain in the membrane, a thin bilayer width, and interactions with molten globule conformation proteins (13, 17-20). Both the P and TM conformations are pre-translocation states, with the A chain and N-terminal segments of T domain located on the cis side of the bilayer. The T domain can also exist in a post-translocation conformation in which the A chain and N-terminal segments of the T domain have moved to the trans side of the bilayer (23).
In the P state all T domain helices (TH1-TH9) lie near the membrane surface. In contrast, in the TM state T domain hydrophobic helices (TH5-TH9) insert deeply. The TH8-TH9 region inserts in the form of a transmembrane hairpin (18, 23-25), connected by a short loop (TL5) containing acidic residues whose protonation at low pH has been proposed to aid membrane insertion (18, 24, 26). TH8 and TH9 are critical for, and at least under some conditions sufficient for, pore formation (25, 27-29). The structure and functional role of hydrophobic helices TH 5-7 is less clear. They form two hydrophobic segments, TH5 and TH 6/7. Like TH8-9, TH 5-7 insert shallowly in the P state and deeply in the TM state (30). However, although it was speculated that they might form a transmembrane hairpin in membranes (24), they do not form a transmembrane hairpin in the TM state [the name “TM state” as used here refers to the conformation in which TH 8-9 form a transmembrane structure], and the loop connecting TH5 to TH6/7 remains exposed on the cis side of the bilayer upon insertion (27, 30). Interestingly, disruption of deep insertion of TH 8-9 by mutagenesis also disrupts deep insertion of TH 5-7(31), and disruption of insertion of TH5 affects insertion of TH 8-9 (32), indicating that TH 8-9 and TH 5-7 interact with each other in the TM state. Disruption of TH5 insertion also reduces pore formation (32).
The remaining T domain helices include amphiphilic helix TH1 and hydrophilic helices TH 2-4. Protonation of His in these segments appears to play an important role in triggering low pH induced changes in T domain structure (33). TH 1-4 shallowly insert in both the P and TM states (34). They may form a flexible tether to the A chain, although TH1 may play a more direct role in one or more stages of the translocation process (35).
The behavior of the A chain is less well understood. Isolated A chain undergoes a reversible conformational change low pH, undergoing partial unfolding to form a molten-globule like state, and gaining the ability to associate with model membranes (15, 36-39). The A chain shallowly inserts into the membrane all along its sequence (15, 39), although one study has reported possible deeply inserted segments (38).
Our group has found that at low pH the addition of proteins in a partly unfolded, molten globule-like conformation, including the A chain, converts the T domain from the P to TM state, suggesting A-T interactions are an important aspect of diphtheria toxin membrane insertion (13). In order to investigate how the interaction between the A chain and T domain alters their behavior and leads to translocation in this report we compare the topography of the isolated T domain to that when it is covalently associated as in the intact toxin (AT protein). The results show that the covalently attached A chain alters T domain topography, with TH 5 and TH 6/7 apparently forming a transient transmembrane hairpin that may play an important role in translocation. The results identify a possible translocation-aiding motif that may be conserved in other systems that translocate unfolded proteins across membranes.
EXPERIMENTAL PROCEDURES
Materials
1,2-Dioleoyl-sn-glycero-3-phospholcholine (DOPC), 1,2-dimyristoleoyl-sn-glycero-3- phosphocholine (DMoPC), and 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG) were purchased from Avanti Polar Lipids (Alabaster, AL). Lipid concentrations were determined by dry weight. N-[(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indecene-3-yl)methyl]iodoacetamide (BODIPY-FL C1-IA, BODIPY-IA), monochlorobimane, Lissamine™ rhodamine B-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine triethylammonium salt (Rho-DHPE), and rabbit anti-BODIPY-FL IgG were purchased from Invitrogen (Molecular Probes) (Eugene, OR). Pfx™ polymerase, the restriction enzymes EcoR I and Nde I, synthetic oligonucleotides, alkaline phosphatase and T4 ligase were purchased from Invitrogen (Rockville, MD). Human serum albumin (HSA) was obtained from Worthington Biochemical (Lakewood, NJ). D-biotin was purchased from Sigma-Aldrich. BODIPY-Streptavidin conjugate (BOD-SA) (discontinued except as a custom labeling product), Streptavidin (SA), N-(biotinoyl)-N’-(iodoacetyl)-ethylenediamine (biotin-IA), were purchased from Molecular Probes (Eugene, OR). Endoproteinase Arg C was from Roche Diagnostics Corporation (Indianapolis, IN). All other chemicals were reagent grade.
Site-Directed Mutagenesis
The DNA coding for AT protein contained diphtheria toxin residues 1-382 cloned into the pET-28a plasmid and containing the E148S substitution, which abolishes toxicity (40), and a N-terminal His tag used for protein purification (34). Next, the native Cys at residue 186 and 201 were replaced with Ser by mutagenesis to prepare a Cys-less template, and the nicking site (Arg190ValArgArg193) in the loop between A and T domain was mutated to GlyGlyGlyGly to abolish the sensitivity of the AT protein to degradation by proteases during isolation and storage using the procedures described previously (32). Two-step (asymmetric) PCR was used for making single cysteine mutants in AT protein as previously described (30). The mutant complementary primers, which contained the desired single amino acid mutation, were 30-33 base pairs in length. PCR was carried out on an Eppendorf Mastercycler Personal PCR System using Pfx™ polymerase. The mutations were confirmed by automated DNA sequencing (Genewiz, South Plainfield, NJ). DNA sequencing also confirmed that no undesired mutations were introduced.
Expression and Purification of AT Mutants
AT proteins were over-expressed and isolated from E. coli as described previously for the isolated T domain (20, 30), except that because pET-28a was used, kanamycin (50 μg/ml) was used to select plasmid-containing bacteria (31, 36). The mutant proteins were purified essentially as described previously (20, 27, 30). The first step involved affinity chromatography using a Talon metal affinity resin (CLONTECH, Palo Alto, CA). Generally, the E. coli extract (50 ml) was loaded onto a column containing 0.6 ml of Talon metal affinity resin followed by washing with binding buffer (5 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 8.0). Most of the AT protein eluted in three 1.0 ml aliquots of elution buffer (200mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 8.0). In the second step, proteins were treated with dithiothreitol (100mM) and then subjected to FPLC with a 3 ml Source Q anion exchange column (Amersham Bioscience, Piscataway, NJ) (27, 30, 36). AT protein was eluted using a 0-500 mM NaCl linear gradient containing 20 mM Tris-HCl, pH 8.0, with total volume of elution buffer being 10ml. Elution was carried out at a rate of 0.5 ml/min, and twenty 0.5 ml fractions were collected. AT protein generally eluted between 300 and 400 mM NaCl. The collected fractions were run on SDS-PAGE (gradient 8-25% gels) using a Phastsystem (Amersham Bioscience, Piscataway, NJ) and Coomassie Blue staining to identify the fractions containing protein and purity of the protein. Final purity appeared to be above 95% in all cases. Protein concentrations were determined by the Bio-Rad protein assay (Bradford method) (41). The concentration of the isolated AT was generally in the range 2-3 mg/ml. Protein was generally labeled right after purification. In some cases, protein was stored at 4°C, but for no more than 2 weeks. Longer storage resulted in degradation of the protein.
Fluorescence Labeling of Diphtheria Toxin Mutant Proteins
Monochlorobimane and BODIPY-IA were used to label single Cys mutants for single Cys containing AT mutant proteins similarly to as described previously (17, 20, 30). Briefly, monochlorobimane dissolved in ethanol (20:1 mol ratio probe: AT protein) or BODIPY-IA dissolved in dimethyl sulfoxide (8:1 mol ratio probe: protein to AT protein) were used. All samples (1ml containing 200-500μg/ml protein) were labeled at room temperature for 1 h followed by overnight dialysis in 5L dialysis buffer. The dialysis buffer for bimane-labeled samples was 100mM NaCl and 20mM Tris-HCl buffer pH 8.0 while for BODIPY-labeled samples the dialysis buffer was, 100mM NaCl and 20mM Tris-HCl buffer pH 8.0 containing 0.75% (v/v) dimethyl sulfoxide (DMSO). Nearly full protein recovery was obtained (17). In parallel to the Cys-containing samples, samples of Cys-less AT protein were labeled using an identical protocol, to assay non-specific labeling. Fluorescence measurements were used to compare relative labeling efficiency of mutant and Cys-less protein. To avoid interference from non-specific labeling of residues other than Cys, only preparations in which labeling efficiency was at least 15-20 times greater than that of the Cys-less protein were used.
Preparation of Small Unilamellar Vesicles (SUV)
Sonicated SUV, composed of DOPC/DOPG (7:3 in mol ratio) or DMoPC/DOPG (7:3 in mol ratio) were prepared similarly to as described previously (20, 30). Briefly, lipid mixtures dissolved chloroform were dried under nitrogen for 15 min and then under high vacuum for 1h. The dried lipids were re-suspended with 0.5-1 ml in 167 mM acetate, 6.7mM Tris-HCl, 150mM NaCl buffer pH 4.3 (low pH buffer) such that total lipid concentration was 10mM, and then the samples were sonicated to near clarity using a bath sonicator (W-220F cell disruptor, Heat System, Ultrasonics, Inc., Plainview, NY). The SUV preparations were stored at 4 °C for no more than 3d before use.
Fluorescence Measurements
Fluorescence was measured on a Spex Tau-2 Fluorolog spectrofluorimeter operating in steady state ratio mode using a semi-micro quartz cuvette (excitation path length 10 mm, emission path length 4 mm). The excitation and emission slits were set to 4.0 mm (7.2 nm bandwidth) and 5.5 mm (9.9 nm bandwidth), respectively. Tryptophan was excited at 280nm, and emission measured from 300nm to 400nm at rate of 1nm/s. Intensities at 335nm are reported. Bimane was excited at 375 nm, and emission spectra were measured from 420nm to 520nm at rate of 1 nm/s. BODIPY fluorescence was measured for 10 s with an excitation wavelength of 485 nm and emission wavelength of 515 nm. Rhodamine fluorescence was measured for 3 s with an excitation wavelength of 565nm and emission wavelength of 585nm. In all cases, background intensities from samples lacking protein were subtracted from the intensities measured in protein-containing samples. All measurements were made at room temperature unless otherwise noted. Samples were prepared in low pH buffer or 20mM Tris-Cl, 150 mM NaCl, pH 8, unless otherwise noted.
Bimane Fluorescence of Vesicle-Incorporated AT Protein
Bimane-labeled AT protein was added into SUV containing 200 μM lipid dispersed in 800 μl low pH buffer. Typically, final concentration of bimane-labeled AT protein was 4 μg/ml. In some samples additional unlabeled Cysless AT protein or human serum albumin (HSA) were then added. To do this either an aliquot (usually 4-10 μl) of purified Cysless AT protein (dissolved in elution buffer from FPLC) was added to obtain a final unlabeled AT concentration of approximately 16μg/ml (in 800μl sample), or 4μl of 1mg/ml HSA dissolved in water was added to give a final HSA concentration of 5μg/ml. After the last protein was added, the samples were incubated 30 min at room temperature and then bimane fluorescence was measured.
Quenching of BODIPY Fluorescence of Vesicle-Incorporated AT Protein by Anti-BODIPY Antibody
Anti-BODIPY antibody binding experiments were performed similarly to as described previously (20, 30). BODIPY-labeled AT proteins were incubated with SUV containing 200 μM lipid in 800 μl low pH buffer for 30 min. Typically, final concentration of BODIPY-labeled AT protein was 2μg/ml. BODIPY fluorescence was measured and then a 20 μl aliquot of anti-BODIPY antibodies (from a 3 mg/ml stock solution dissolved in phosphate buffered saline pH 7.2, 5mM azide) was added and mixed. Fluorescence intensity was remeasured after a 30 min incubation at room temperature. Samples in which additional unlabeled AT protein (Cysless AT protein, 16μg/ml) or HSA (5μg/ml) were added were prepared as described above. For each sample four fluorescence measurements were made both before and after antibody addition, mixing samples between measurements. The reported values are the average.
Preparation of Large Unilamellar Vesicles (LUV) With or Without Trapped Streptavidin
Vesicles containing trapped BODIPY-streptavidin (BOD-SA), trapped unlabeled streptavidin (SA) or no trapped protein were prepared similarly to as described previously (27, 34). A mixture containing 10mM lipid composed of 70mol% DOPC, 30mol% DOPG, 0.002mol% Rho-DHPE (a fluorescent lipid marker), 100μg/ml BOD-SA or SA, and 20mg/ml n-octyl-β-glucoside; was dissolved in 150 mM acetate and 150 mM NaCl pH 4.5 (acetate buffer) (27, 34). Sample volume of 0.5 ml. After removing n-octyl-β-glucoside by dialysis at 4 °C overnight, the samples were applied to a Sepharose CL-4B column (1cm in diameter, 50 cm in length) equilibrated in acetate buffer, to separate free BOD-SA or SA from large unilamellar vesicle (LUV)-entrapped BOD-SA or SA. After elution with acetate buffer (~1 ml per fraction), fractions containing LUV (fractions 8 and 9) were collected. Fractions containing free BOD-SA (fraction 20 to 24) were also collected for later use for external-addition to vesicles without trapped BOD-SA. The final concentration of BOD-SA, SA and/or lipid were determined by measurement of the BODIPY, tryptophan and Rho-DHPE fluorescence, respectively (27, 34), using a known dilution of stock solution as a standard. Typically, the final concentrations in the vesicle-containing fractions were 2.5-5mM lipid and 5.6-8.0μg/ml entrapped BOD-SA, in samples containing BOD-SA; 2.3-5.7mM lipid and 3.8-6.5μg/ml entrapped SA, in samples containing unlabeled SA; or 2.3-5.7 mM lipid in samples without trapped protein.
Preparation of Biotinylated Diphtheria Toxin Mutant Proteins
AT protein mutants were labeled with the cysteine-specific biotin-IA dissolved in dimethyl sulfoxide similarly to as described previously (27, 34). Generally, 2.5-3.0 mg AT protein dissolved in 0.5 ml of elution buffer was incubated with 45-55 μl of 8 mM biotin-IA for 1h at room temperature. To remove the excess unbound probe the sample was placed in dialysis tubing with a molecular weight cutoff of 8000 (Spectra/Por, address), and then dialyzed overnight (one buffer change around 6 h) against 5 L of 10 mM Tris-HCl, 150 mM NaCl, pH 8.2 at 4 °C. The biotinylated protein was separated from unbiotinylated protein using a monomeric avidin column (Pierce Biotechnology, address) similar to as described previously (27). Samples were loaded onto a 2.5ml column (1cm in diameter, 1.3 cm in length) and eluted with six separate 0.6 ml aliquots of 0.1 M phosphate, 0.15 M NaCl, 2 mM biotin pH 7.0 (biotin elution buffer). The fractions were subjected to SDS-PAGE (gradient 8-25) using the Phastsystem. The two or three fractions with the highest concentration of the biotinylated protein were combined and dialysed against 5 L of 10 mM Tris-HCl, 150 mM NaCl, pH 8.2 at 4 °C with three buffer changes to remove the free biotin. The final protein concentration (0.1 to 0.5 mg/ml) was determined using the Bradford colorimetric assay (Bio-Rad, Hercules, CA)(41).
Interaction of Biotinylated Diphtheria Toxin Mutants with Externally Added and Vesicle-Trapped BODIPY-Streptavidin
The reactivity of membrane-inserted AT protein and T domain with BOD-SA was then measured similarly to as previously (27, 34). Reactivity was measured under three different conditions: 1. with externally added BOD-SA (BOD-SAex samples), 2. with externally added BOD-SA and trapped unlabeled SA (BOD-SAex/SAtr samples) and 3. with trapped BOD-SA (BOD-SAtr samples).
For BOD-SAtr samples, LUVs containing trapped BOD-SA were diluted to just under 700 μl with low pH buffer so as to give a concentration of 0.2μg/ml entrapped BOD-SA. After dilution, the lipid concentration, which varied because trapping efficiency was variable, was in the range 100-200 μM. The initial BODIPY fluorescence was measured, and then a small aliquot (1-5 μl) of concentrated AT protein was added to give a final concentration of 0.4μg/ml biotinylated AT protein plus 3.6μg/ml unbiotinylated Cysless AT protein in a total volume of 700 μl. After incubation for 30 min. BODIPY fluorescence was then remeasured.
For the BOD-SAex and BOD-SAex/SAtr samples BODIPY fluorescence was first measured in a 630 μl sample containing BOD-SA diluted to 0.22μg/ml with low pH buffer. Then 70 μl of a concentrated AT protein-LUV mixture was added. This mixture contained 40 μg/ml AT protein (4μg/ml biotinylated plus 36μg/ml unbiotinylated Cysless AT protein) pre-incubated for 15 or 50 min at room temperature with LUVs [with or without trapped unlabeled streptavidin]. After addition to the BOD-SA containing solution, these samples had the same final AT protein and lipid concentrations as that in samples with trapped BOD-SA. Fluorescence was remeasured after a final incubation of 30 min. For T domain, the same protocol was used (with 10% of the protein being biotinylated) except that the final concentration of T domain was 2μg/ml.
For experiments in which the samples with trapped unlabeled SA were pre-incubated with AT protein or T domain for 15 min each experiment involved duplicate samples and was repeated at least n=3-14 separate times (AT residue 1, n=5; residue 163, n=4; residue 186, n=5; residue 203, n=8; residues 211, n=3; residue 293, n=14; residues 324, n=10 and residue 378, n=8, and for residue 293 in isolated T domain, n=4).
For experiments in which the samples with trapped unlabeled SA were pre-incubated with AT protein for 50 min, each experiment involved duplicate samples and was repeated at least n=2-7 times (AT residue 203, n=2; residue 288, n=7; residue 293, n=3; residue 297, n=3; residues 324, n=2)
To evaluate the % exposure of the biotinylated residues on the outside surface of the vesicles, the % external reactivity was calculated. For experiments in which the reactivity of AT protein bound to empty vesicles with externally added BOD-SA (BOD-SAex) was compared to the reactivity of vesicle-bound AT protein with BOD-SA trapped within the vesicles (BOD-SAtr) the % external reactivity was calculated from the equation: % external activity (without trapped unlabeled SA) = {(F-Fo)ex / [(F-Fo)ex + (F-Fo)tr]}×100%. Where ex refers to samples with external BOD-SA and tr refers to samples with trapped BOD-SA, Fo is BOD-SA fluorescence intensity prior to the addition of AT protein (corrected for dilution when necessary, see below), and F is BOD-SA fluorescence intensity 30 min after incubation of AT protein with the BOD-SA containing sample.
For experiments in which the reactivity of AT protein bound to vesicles containing trapped unlabeled SA with externally added BOD-SA (BOD-SAex/SAtr) was compared to the reactivity of vesicle-bound AT protein with BOD-SA trapped within the vesicles (BOD-SAtr) the % external reactivity was calculated from the equation: % external activity (with trapped unlabeled SA) = {(F-Fo)ex - SA / [(F-Fo)ex + (F-Fo)tr]}×100% where ex - SA refers to samples containing trapped unlabeled SA and externally added BOD-SA. For samples with externally added BOD-SA initial BODIPY fluorescence values were corrected to what they would be after dilution by the vesicle-containing aliquot in the absence of reaction with biotinylated AT protein. Notice that the demoninator in the case of samples with trapped unlabeled SA is the same as for the samples without trapped unlabeled SA, i.e., the denominator always contains the value for the sample with external BOD-SA lacking trapped unlabeled SA.
RESULTS
Residues and Methods Used to Probe AT Protein Topography
We studied AT protein, which contains the covalently linked A chain and T domain portions of diphtheria toxin (i.e. equivalent to diphtheria toxin lacking the receptor-binding domain, which can be deleted or replaced with retention of activity and membrane translocation (42-44)). As has been observed in both isolated A chain, isolated T domain and intact toxin (15, 36, 45, 46), AT protein undergoes a low pH induced conformational transition near pH 5 in which buried Trp become exposed, and in which it becomes hydrophobic and spontaneously inserts into model membrane vesicles (Supplemental Figures 1-3). Also like intact toxin and isolated T domain (47, 48), membrane-inserted AT protein forms pores at low pH (Supplemental Figure 4).
To determine how the covalent attachment of the A chain and T domain affects their membrane topography, labeling of single Cys residues was employed. Residues previously labeled in isolated T domain were chosen for labeling in AT protein, specifically studying residues within and around T domain hydrophobic helices TH 5-9 (276, 288, 293, 297, 311, 324, 356 and 378). Previous studies with isolated T domain have shown that when these residues are mutated to Cys and labeled, T domain maintains its native conformation at neutral pH, and undergoes the same low pH induced conformational changes and lipid binding seen in wild type, unlabeled protein (20, 30, 34, 36).
Two assays previously used to define the topography of isolated T domain were used to investigate AT protein topography (17, 20, 30, 34, 36). First, fluorescence emission λmax of bimane-labeled residues was used to evaluate the location of labeled Cys residues. We have previously shown that bimane fluorescence blue shifts upon insertion into the lipid bilayer, and that the extent of the shift is closely correlated with the depth of insertion. Second, accessibility of BODIPY-labeled residues to anti-BODIPY antibody was measured. This is sensitive to the burial of labeled residues within a membrane, but does measure exactly the same parameter as that measured by bimane because buried residues that are transiently exposed to aqueous solution (i.e. are not stably buried) can react with antibody.
These methods were used to explore the conformation and topography of the AT protein in solution at neutral and low pH, and when AT protein was membrane-inserted. The experiments with membrane-inserted protein were carried out using SUV under conditions previously shown (13, 17) to promote formation of the surface (P state) T domain conformation (i.e. in DOPC/DOPG vesicles), and under three different conditions that promote formation of the transmembrane (TM state) T domain conformation (i.e. 1. in thinner DMoPC [=di C14:1 PC]/DOPG vesicles, which form thinner bilayers than vesicles with DOPC [=di C18:1 PC], or 2. in DOPC/DOPG vesicles with a high concentration of AT protein, or 3. in DOPC/DOPG vesicles in the presence of human serum albumin a molten globule state protein). Although we mainly show data here only for the first set of conditions promoting formation of TM insertion, similar behavior was observed under all the different conditions that promote formation of the TM state (data not shown).
Topography of Hydrophobic Helices TH5-9 in Membrane-Inserted AT Protein
The membrane topography of residues within T domain hydrophobic segments (TH 5-9) of AT protein was compared to that previously observed in isolated T domain (20, 27, 30). Residues 276 and 288 (in TH5), 311 (in the TH 6/7 segment) and 356 (in the middle of TH9) were chosen for study because they within hydrophobic helices and their exposure to solution decreases when the T domain switches from the shallowly inserted P state to deeply inserted states (17, 20, 30). The behavior of residue 293 (in the hydrophilic loop between TH5 and TH6) was also studied. In isolated T domain residue 293 shows unusual behavior in which it becomes more exposed to aqueous solution when the hydrophobic segments insert more deeply.
Table 1 and Table 2 compare the behavior of these TH5-9 residues in membrane-inserted AT protein to that previously defined for membrane-inserted isolated T domain both under conditions that favor the formation of the P state and conditions that favor the formation of the TM state. A significant difference between the AT protein and isolated T domain was observed. When inserted into DOPC/DOPG vesicles, in which isolated T domain forms the P state, bimane-labeled residues within the hydrophobic helices of AT protein exhibited fluorescence that was much more blue-shifted than that of isolated T domain in DOPC/DOPG vesicles (Table 1). Instead the λmax values were similar to those observed in isolated T domain in DMoPC/DOPG vesicles, in which the isolated T domain forms the TM state.
Table 1. Comparison of bimane emission maximum (λmax) of bimane-labeled mutants in hydrophobic TH5-9 region of membrane-inserted T domain or AT protein at pH 4.3 and room temperature.
| Location | Residue Number |
Bimane emission maximum (λmax) in nm a | |||||
|---|---|---|---|---|---|---|---|
| Isolated T domainb | AT protein | T domain from Arg C digested AT proteinc |
|||||
| DOPC/ DOPG |
DMoPC/ DOPG |
DOPC/ DOPG |
DMoPC/ DOPG |
DOPC/ DOPG |
DMoPC/ DOPG |
||
| TH 5 | 276 | 468 | 461 | 463 | 461 | nd | nd |
| 288 | 472 | 468 | 465 | 464 | 470 | 467 | |
| 5-6 Loop | 293 | 459 | 470 | 462 | 466 | 459 | 472 |
| TH 6/7 | 311 | 467 | 462 | 460 | 460 | 467 | 463 |
| TH 9 | 356 | 469 | 461 | 462 | 458 | 468 | 460 |
| Average w/o 293 | 469 | 463 | 463 | 461 | — | — | |
Table 2. Comparison of the quenching percentage of BODIPY fluorescence in the TH5-9 hydrophobic region of model membrane-inserted T domain or AT protein by anti-BODIPY antibody at pH 4.3 and room temperature.
| Location | Residue Number |
% Quenching of BODIPY fluorescence a | |||
|---|---|---|---|---|---|
| Isolated T domain b | AT protein | ||||
| DOPC/ DOPG |
DMoPC/ DOPG |
DOPC/ DOPG |
DMoPC/ DOPG |
||
| TH 5 | 276 | 62.0 | 40.0 | 48.0±1.9 | 39.0±7.7 |
| 288 | 25.5 | 33.5 | 37.6±0.9 | 28.4±5.0 | |
| 5-6 Loop | 293 | 30.0 | 76.0 | 36.6±4.6 | 32.6±6.4 |
| TH 6/7 | 311 | 49.0 | 25.0 | 38.7±1.1 | 12.9±5.7 |
| TH 9 | 356 | 45.0 | 25.0 | 44.4±4.7 | 26.5±0.8 |
| Average w/o 293 | 45 | 31 | 42 | 27 | |
However, as judged by quenching, the antibody accessibility of TH 5-9 BODIPY-labeled residues in AT protein inserted into DOPC/DOPG vesicles exhibited a level of reactivity between that of the T domain in the P state and in the TM state (Table 2). This suggests that, although TH 5-9 residues in AT protein in DOPC/DOPG vesicles form a conformation similar to that of isolated T domain the TM state (as judged by bimane fluorescence), this deep insertion is not as stable as in the TM state formed by isolated T domain (as reported by anti-BODIPY quenching). Alternate possibilities are that the BODIPY-labeled residues do not insert quite as deeply as bimane-labeled ones or that there is steric blockage of labeled T domain residues from aqueous solution due to the A chain.
Bimane fluorescence λmax values for TH 5-9 residues in AT protein inserted into DMoPC/DOPG vesicles was very blue shifted, indicating that the TM state had formed, and the values were similar (but not identical) to that for isolated T domain in DMoPC/DOPG vesicles, which also forms the TM state under these conditions (Table 1). Furthermore, the antibody accessibility was low both for AT protein and isolated T domain when inserted into DMoPC/DOPG bilayers, indicative of stable deep insertion in both of these cases (Table 2).
Intriguingly, in AT protein, residue 293 in the hydrophilic loop connecting TH5 to TH6, behaved in an unexpected fashion in DMoPC/DOPG vesicles. In isolated T domain in the TM state, bimane-labeled residue 293 is very red shifted and when BODIPY-labeled is very antibody reactive ((30) and Tables 1 and 2). In contrast, in the AT protein, labeled residue 293 showed somewhat more blue-shifted bimane fluorescence (Table 1) and relatively low reactivity with anti-BODIPY antibody (Table 2). This suggests there is an important difference between the topography of this region of the protein in AT protein relative to that in isolated T domain (see below).
To confirm that the difference between the topography of isolated T domain topography and that of the AT protein were due to A-T interactions, rather than due to some undefined experimental difference between the present study and previously ones, AT proteins bimane-labeled at residues 288, 293, 311 or 356 were subjected to Arg C digestion. Arg C is an Arg-specific protease, and the T domain only has Arg residues at positions 210 and 377, near its N and C-termini, respectively. Therefore, a limiting Arg C digestion of AT protein results in the formation of a nearly intact T domain, while the A chain, which has several Arg, is digested into smaller fragments (Supplemental Figure 5). Unlike what was observed before Arg C digestion, bimane λmax values for Arg C-digested labeled AT protein were almost identical to those of the isolated T domain (Table 1). This was true both for AT protein inserted into DOPC/DOPG vesicles, and AT protein inserted into DMoPC/DOPG vesicles. This shows that the A-T interactions abolished by Arg C digestion altered the behavior of TH 5-9.
Distinguishing Exposure of AT Residues on the Cis and Trans Sides of the Bilayer Using Biotinylated Residues and BODIPY-Streptavidin
The above-noted difference between residue 293 behavior in AT protein and isolated T domain suggested that there might be an important difference between their topography in this region of the protein. To see if this difference involved whether residues were exposed on the cis side (the side from which insertion occurs) or trans side of the bilayer (the side to which translocation moves residues) the accessibility of biotinylated residues of membrane-inserted AT protein to BODIPY-labeled strepatavidin (BOD-SA) was studied. These experiments were carried out with AT protein incorporated into large unilamellar vesicles composed of DOPC/DOPG (27). In these experiments the reactivity of biotinylated residues with BOD-SA externally added to vesicles (i.e. on the cis side of the membrane) is compared to reactivity with BOD-SA that is vesicle-entrapped (i.e. on the trans side) (see schematic figure 3 in reference (27)). It should be noted that biotinylation of diphtheria toxin residues does not interfere with their translocation across a bilayer (23, 27), and that in these LUV, isolated T domain forms the TM state (27).
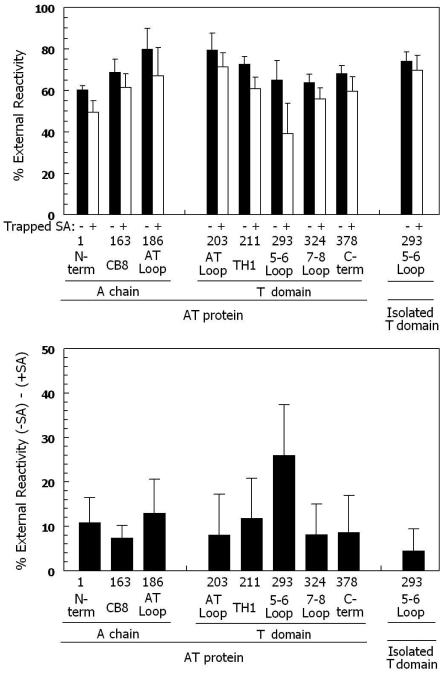
Figure 1 summarizes the reactivity of a series of biotinylated residues with BOD-SA. Most residues were more reactive with externally added BOD-SA than vesicle-entrapped BOD-SA (filled bars), such that 60-70% of each residue was exposed on the cis surface. This is in agreement with our previous studies showing that both whole toxin (49) and isolated T domain (27) insert into model membranes in a mixture of orientations, and that the orientation in which most of the protein is on the cis side of the membrane predominates. The fact that residues on both the N- and C-termini of the hydrophobic TH 6/7 sequence (i.e. residues 293 and 324) are predominantly exposed on the cis surface on the bilayer indicates that TH 5-7 does not form a stable transmembrane hairpin in AT protein, and instead forms a structure in which the 5-6 loop (connecting TH5 to TH 6/7) is exposed on the cis side of the membrane. This is in agreement with previous results on isolated T domain (27).
Figure 1.
Cis/trans surface localization of biotinylated AT protein residues in membrane-inserted AT proteins by determination of percent external reactivity. (Top) The % external reactivity (reaction with external BOD-SA/total reactivity with trapped and external BOD-SA × 100%) is shown for a series of biotinylated Cys mutants. Samples containing 4μg/ml total AT protein (0.4 μg/ml biotinylated AT protein, 3.6 μg/ml unlabeled Cys-less AT protein), 0.2 μg/ml BOD-SA, and 0.10-0.21 mM lipid (70% mol DOPC/30% mol DOPG LUV) in low pH buffer, pH 4.3. Samples in which reactivity with external BOD-SA was measured were preincubated for 15 minutes with vesicles without (filled bars) or with (open bars) 0.2 μg/ml entrapped unlabeled SA. Experiments were carried out at room temperature. In most cases, the average values from 4 or 5 experiments and standard deviations are shown. (Bottom) The difference between % external reactivity for experiments without and with trapped unlabeled SA. The value of the difference with and without trapped unlabeled SA was calculated first, and then the average and standard deviation of the differences was calculated. See Experimental Procedures for details. X-axis shows residue number and the segment of the protein in which it is located. The behavior of biotinylated residue 293 in isolated T domain is also shown as a control.
However, this does not rule out the possibility that TH 5-7 form a transient transmembrane hairpin in which the 5-6 loop is transiently located on the trans side of the membrane. To test for this, the reactivity of AT residues with externally added BOD-SA was measured using samples containing vesicle-trapped unlabeled SA (see schematic figure 3 in reference (27)). If residues are moving back-and-forth across the bilayer, as occurs when a transient transmembrane structure forms, then membrane insertion of AT protein into vesicles containing trapped unlabeled SA should allow the biotinylated group to react with the trapped unlabeled SA, and thus prevent reaction with externally added BOD-SA.
Figure 1A (open bars) shows that for almost all residues tested, including residues in the A chain and N-terminal region of the T domain, the presence of trapped unlabeled SA only slightly reduced (~10%, see Figure 1B) reaction of AT protein with externally added BOD-SA. This slight reduction might be due to some small amount of trapped SA that leaked out of the vesicles. In contrast, for residue 293 the presence of trapped unlabeled SA resulted in a 2.5 fold greater reduction of reaction with externally added BOD-SA (Figure 1B). This suggests that in a significant fraction of AT proteins, residue 293 is moving back-and-forth across the membrane.
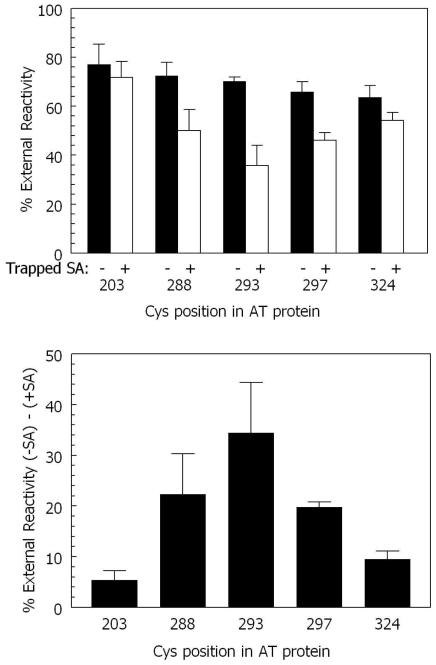
To further confirm that the 5-6 loop (which contains residue 293) was moving back-and-forth across the bilayer, this experiment was repeated using a longer preincubation time to amplify reaction with trapped unlabeled SA, and repeated with additional residues in and near the 5-6 loop (Figure 2). Increasing the preincubation time significantly increased the inhibition of reactivity by trapped unlabeled SA for residue 293 (to ~35%). Under these conditions a relatively high degree of inhibition of reactivity by trapped unlabeled SA could also be observed for residue 288 and residue 297. However, the inhibition was less than observed with residue 293, suggesting that these residues, which are at the ends of hydrophobic helices TH5 and TH6, respectively, may not be as exposed to trapped SA as residue 293 when they face the trans side of the bilayer. It should be noted that pore formation by AT protein cannot explain the inhibition of residue 293 reactivity in the presence of trapped unlabeled SA. The pores formed by diphtheria toxin are too small under our experimental conditions to allow molecules as large as SA through a lipid bilayer (48).
Figure 2.
Cis/trans surface localization of biotinylated residues in and around the 5-6 loop in membrane-inserted AT protein at pH 4.3 and room temperature. (Top) The % external reactivity for residues in and around the AT loop measured under the same conditions as in Figure 1 except that samples in which reactivity with external BOD-SA was measured were preincubated for 50 minutes with vesicles without (filled bars) or with (open bars) 0.2 μg/ml entrapped unlabeled SA. Average values and standard deviations are shown. (Bottom) The difference between % external reactivity for experiments without and with trapped unlabeled SA. The value of the difference with and without trapped unlabeled SA was calculated first, and then the average and standard deviation of the differences was calculated. X-axis shows residue number. See Experimental Procedures for details.
As a control to confirm that the difference between residue 293 behavior in AT protein and isolated T domain was reproducible, we repeated our previously published experiments using isolated T domain (27). In agreement with our previous study, the reactivity of residue 293 in isolated T domain does not show the reactivity pattern (inhibition of biotinylated AT association with BOD-SA by trapped SA) characteristic of transient translocation (Figure 1).
DISCUSSION
Comparison of TH5-9 Topology in AT Protein with that in Isolated T Domain in Model Membrane
T domain hydrophobic helices (TH 5-9) shows important changes in behavior when the A chain is present. As noted above, in membrane-inserted isolated T domain TH5-9 exist in two distinct conformations, a shallowly inserted one (which forms in DOPC/DOPG SUV), and a deeply inserted one in (which forms in DMoPC/DOPG SUV or at high T domain concentrations). As judged by bimane fluorescence and BODIPY quenching, the insertion of residues within TH 5-9 in AT protein in DOPC/DOPG vesicles was less exposed to solution than in isolated T domain. This is consistent with our previous observation that non-covalent binding of isolated A chain to isolated T domain converts the T domain from a state in which TH 5-9 residues are exposed to solution to one in which they are not (13). However, a 5-fold excess of isolated A chain was needed to induce complete T domain conversion to the less solution exposed form (13). Thus, the single covalently linked A chain in AT protein is more effective than isolated A chain in interacting with the T domain.
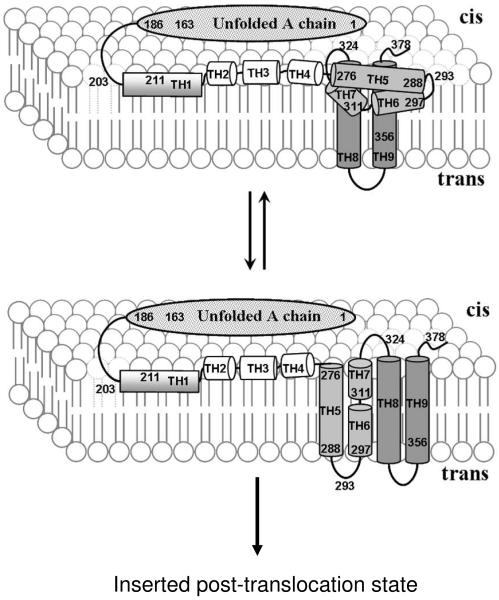
The behavior of the loop connecting TH5 to TH6 was also different in AT protein and isolated T domain. In isolated T domain, this residue 293, which is within this loop, has the unusual property of inserting shallowly when TH5-9 helix residues insert deeply, and deeply when TH5-9 helix residues insert shallowly. However, when TH 5-9 helix residues inserted deeply in AT protein, residue 293 appeared to insert more deeply than in isolated T domain. This suggests a significant difference between the insertion of the TH 5-7 helix cluster in AT protein and isolated T domain. This difference was confirmed by the BOD-SA assay, which showed that residue 293, which remains on the cis side of the membrane in isolated T domain (27), dynamically interconverts between the cis and the trans sides of membrane in a population of AT protein. It should be noted that additional studies of Cys labeled residues in the A chain in AT protein, and TH1 of the T domain in AT protein, show relatively shallow insertion, as in isolated A chain and T domain (data not shown). Combined, the experimental data suggests the model for AT protein topographies shown in Figure 3. If correct, this model would support the original suggestion (24) that TH 5-7 would form a transmembrane hairpin (with TH 5 forming one transmembrane segment and TH 6/7 forming the other), except that the transmembrane insertion of TH 5-7 would be dynamic, rather than stable. This dynamic instability is not wholly surprising because the TH 5-7 sequence has borderline hydrophobicity, being less hydrophobic than most transmembrane segments of ordinary membrane proteins and even the other hydrophobic diphtheria toxin helices, TH 8-9 (50).
Figure 3.
Schematic diagram showing pre-translocation states for AT protein in model membranes at low pH. Top: pre-translocation TM state in which TH8-9 form a transmembrane hairpin while semi-hydrophobic segments TH5-7 deeply insert without forming a transmembrane structure; Bottom: transient pre-translocation state in which both TH 8-9 and TH5-7 form a transmembrane hairpin. This conformation might then proceed to form the previously identified post-translocation inserted state (23). The A chain and hydrophilic TH1-4 region of T domain lie on or near the cis side surface of bilayer in both states. The details of how A chain interacts with T domain and the lipid bilayer are not yet defined.
Insights Into the Mechanism of Translocation
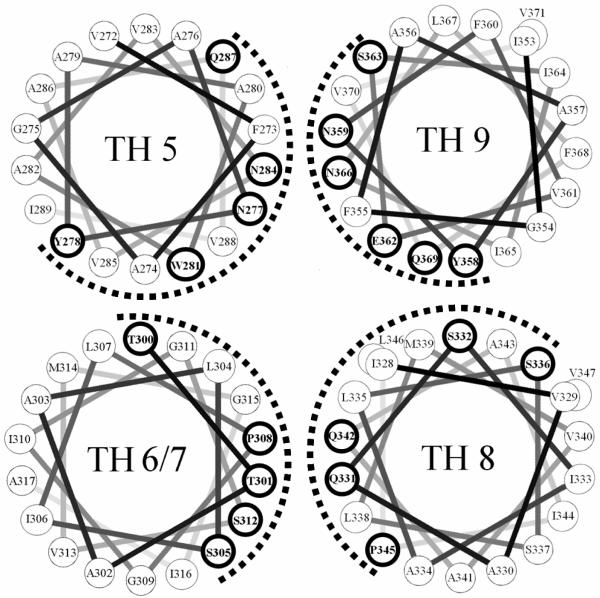
What insights do the topographical properties of TH 5-9 provide into the mechanism of translocation? One possibility is that in a fully transmembrane state TH 5-9 forms the wall of the pathway (“pore”) through which the A chain moves during translocation. In this regard, it is noteworthy that these segments have considerable amphiphilic character, and would be expected to arrange with their more polar face forming the pore/translocation pathway, as shown in Figure 4. The hydrophilic face formed by this cluster is rich in uncharged polar residues (Gln, Asn, Ser, Thr), and has no charged residues (with the possible exception of Glu 362, which might be protonated and thus uncharged at low pH). This may be significant because inspection of crystal structures shows that this type of hydrophilic/uncharged surface is seen in the pore facing surface of other protein translocators, specifically the Sec Y translocon (51) and Tol C (52), and may represent a common translocator wall motif. We speculate that the lack of charged amino acids prevents overly strong interactions between the T domain and translocating A chain. Transient interactions between the unfolded A chain and the hydrophilic surface of TH 5-9 would be consistent with the chaperone model we proposed previously for A chain-T domain interactions (13, 14).
Figure 4.
Helical wheel representation of TH 5-9. The dashed line indicates the more polar face of the hydrophobic segments. Residues with hydrogen bonding abilities are shown by bold circles. Notice that Trp and Tyr are located near the center of hydrophobic segments, which is unusual in transmembrane helices.
The dynamic equilibration of TH 5-7 might also play a role similar to that recently proposed for the Sec A protein, in which a hairpin segment of Sec A binds to the translocation substrate, and then carries it across the membrane (53, 54). If the 5-6 loop binds to the A chain, as it moves back and forth across the bilayer it could shuttle the A chain across the bilayer in a series of steps, probably aided by cytosolic proteins (11, 12). The binding and release of the A chain might be controlled by the pH gradient across the endosomal membrane, if binding to the A chain occurs at the low pH on the cis side of the membrane while dissociation of the A chain occurs at the neutral pH which is found on the trans side of the membrane.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM 31986
REFERENCES
- 1.Drazin R, Kandel J, Collier RJ. Structure and activity of diphtheria toxin. II. Attack by trypsin at a specific site within the intact toxin molecule. J Biol Chem. 1971;246:1504–1510. [PubMed] [Google Scholar]
- 2.Gill DM, Pappenheimer AM., Jr. Structure-activity relationships in diphtheria toxin. J Biol Chem. 1971;246:1492–1495. [PubMed] [Google Scholar]
- 3.Ittelson TR, Gill DM. Diphtheria toxin: specific competition for cell receptors. Nature. 1973;242:330–332. doi: 10.1038/242330b0. [DOI] [PubMed] [Google Scholar]
- 4.Uchida T, Pappenheimer AM, Jr., Harper AA. Reconstitution of diphtheria toxin from two nontoxic cross-reacting mutant proteins. Science. 1972;175:901–903. doi: 10.1126/science.175.4024.901. [DOI] [PubMed] [Google Scholar]
- 5.Uchida T, Pappenheimer AM, Jr., Harper AA. Diphtheria toxin and related proteins. 3. Reconstitution of hybrid “diphtheria toxin” from nontoxic mutant proteins. J Biol Chem. 1973;248:3851–3854. [PubMed] [Google Scholar]
- 6.Mitamura T, Iwamoto R, Umata T, Yomo T, Urabe I, Tsuneoka M, Mekada E. The 27-kD diphtheria toxin receptor-associated protein (DRAP27) from vero cells is the monkey homologue of human CD9 antigen: expression of DRAP27 elevates the number of diphtheria toxin receptors on toxin-sensitive cells. J Cell Biol. 1992;118:1389–1399. doi: 10.1083/jcb.118.6.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naglich JG, Metherall JE, Russell DW, Eidels L. Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- 8.Cha JH, Brooke JS, Ivey KN, Eidels L. Cell surface monkey CD9 antigen is a coreceptor that increases diphtheria toxin sensitivity and diphtheria toxin receptor affinity. J Biol Chem. 2000;275:6901–6907. doi: 10.1074/jbc.275.10.6901. [DOI] [PubMed] [Google Scholar]
- 9.London E. Diphtheria toxin: membrane interaction and membrane translocation. Biochimica et biophysica acta. 1992;1113:25–51. doi: 10.1016/0304-4157(92)90033-7. [DOI] [PubMed] [Google Scholar]
- 10.Pappenheimer AM., Jr. Diphtheria toxin. Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 11.Ratts R, Trujillo C, Bharti A, vanderSpek J, Harrison R, Murphy JR. A conserved motif in transmembrane helix 1 of diphtheria toxin mediates catalytic domain delivery to the cytosol. Proc Natl Acad Sci U S A. 2005;102:15635–15640. doi: 10.1073/pnas.0504937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratts R, Zeng H, Berg EA, Blue C, McComb ME, Costello CE, vanderSpek JC, Murphy JR. The cytosolic entry of diphtheria toxin catalytic domain requires a host cell cytosolic translocation factor complex. J Cell Biol. 2003;160:1139–1150. doi: 10.1083/jcb.200210028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren J, Kachel K, Kim H, Malenbaum SE, Collier RJ, London E. Interaction of diphtheria toxin T domain with molten globule-like proteins and its implications for translocation. Science. 1999;284:955–957. doi: 10.1126/science.284.5416.955. [DOI] [PubMed] [Google Scholar]
- 14.Hammond K, Caputo GA, London E. Interaction of the membrane-inserted diphtheria toxin T domain with peptides and its possible implications for chaperone-like T domain behavior. Biochemistry. 2002;41:3243–3253. doi: 10.1021/bi011163i. [DOI] [PubMed] [Google Scholar]
- 15.Zhao JM, London E. Conformation and model membrane interactions of diphtheria toxin fragment A. J Biol Chem. 1988;263:15369–15377. [PubMed] [Google Scholar]
- 16.Papini E, Schiavo G, Tomasi M, Colombatti M, Rappuoli R, Montecucco C. Lipid interaction of diphtheria toxin and mutants with altered fragment B. 2. Hydrophobic photolabelling and cell intoxication. European journal of biochemistry / FEBS. 1987;169:637–644. doi: 10.1111/j.1432-1033.1987.tb13655.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Malenbaum SE, Kachel K, Zhan H, Collier RJ, London E. Identification of shallow and deep membrane-penetrating forms of diphtheria toxin T domain that are regulated by protein concentration and bilayer width. J Biol Chem. 1997;272:25091–25098. doi: 10.1074/jbc.272.40.25091. [DOI] [PubMed] [Google Scholar]
- 18.Ren J, Sharpe JC, Collier RJ, London E. Membrane translocation of charged residues at the tips of hydrophobic helices in the T domain of diphtheria toxin. Biochemistry. 1999;38:976–984. doi: 10.1021/bi981576s. [DOI] [PubMed] [Google Scholar]
- 19.Malenbaum SE, Collier RJ, London E. Membrane topography of the T domain of diphtheria toxin probed with single tryptophan mutants. Biochemistry. 1998;37:17915–17922. doi: 10.1021/bi981230h. [DOI] [PubMed] [Google Scholar]
- 20.Kachel K, Ren J, Collier RJ, London E. Identifying transmembrane states and defining the membrane insertion boundaries of hydrophobic helices in membrane-inserted diphtheria toxin T domain. J Biol Chem. 1998;273:22950–22956. doi: 10.1074/jbc.273.36.22950. [DOI] [PubMed] [Google Scholar]
- 21.Kyrychenko A, Posokhov YO, Rodnin MV, Ladokhin AS. Kinetic intermediate reveals staggered pH-dependent transitions along the membrane insertion pathway of the diphtheria toxin T-domain. Biochemistry. 2009;48:7584–7594. doi: 10.1021/bi9009264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chenal A, Prongidi-Fix L, Perier A, Aisenbrey C, Vernier G, Lambotte S, Fragneto G, Bechinger B, Gillet D, Forge V, Ferrand M. Deciphering membrane insertion of the diphtheria toxin T domain by specular neutron reflectometry and solid-state NMR spectroscopy. J Mol Biol. 2009;391:872–883. doi: 10.1016/j.jmb.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 23.Senzel L, Gordon M, Blaustein RO, Oh KJ, Collier RJ, Finkelstein A. Topography of diphtheria Toxin’s T domain in the open channel state. J Gen Physiol. 2000;115:421–434. doi: 10.1085/jgp.115.4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choe S, Bennett MJ, Fujii G, Curmi PM, Kantardjieff KA, Collier RJ, Eisenberg D. The crystal structure of diphtheria toxin. Nature. 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 25.Huynh PD, Cui C, Zhan H, Oh KJ, Collier RJ, Finkelstein A. Probing the structure of the diphtheria toxin channel. Reactivity in planar lipid bilayer membranes of cysteine-substituted mutant channels with methanethiosulfonate derivatives. J Gen Physiol. 1997;110:229–242. doi: 10.1085/jgp.110.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaul P, Silverman J, Shen WH, Blanke SR, Huynh PD, Finkelstein A, Collier RJ. Roles of Glu 349 and Asp 352 in membrane insertion and translocation by diphtheria toxin. Protein Sci. 1996;5:687–692. doi: 10.1002/pro.5560050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosconi MP, Zhao G, London E. Analyzing topography of membrane-inserted diphtheria toxin T domain using BODIPY-streptavidin: at low pH, helices 8 and 9 form a transmembrane hairpin but helices 5-7 form stable nonclassical inserted segments on the cis side of the bilayer. Biochemistry. 2004;43:9127–9139. doi: 10.1021/bi049354j. [DOI] [PubMed] [Google Scholar]
- 28.Mindell JA, Silverman JA, Collier RJ, Finkelstein A. Structure-function relationships in diphtheria toxin channels: III. Residues which affect the cis pH dependence of channel conductance. J Membr Biol. 1994;137:45–57. doi: 10.1007/BF00234997. [DOI] [PubMed] [Google Scholar]
- 29.Silverman JA, Mindell JA, Zhan H, Finkelstein A, Collier RJ. Structure-function relationships in diphtheria toxin channels: I. Determining a minimal channel-forming domain. J Membr Biol. 1994;137:17–28. doi: 10.1007/BF00234995. [DOI] [PubMed] [Google Scholar]
- 30.Rosconi MP, London E. Topography of helices 5-7 in membrane-inserted diphtheria toxin T domain: identification and insertion boundaries of two hydrophobic sequences that do not form a stable transmembrane hairpin. J Biol Chem. 2002;277:16517–16527. doi: 10.1074/jbc.M200442200. [DOI] [PubMed] [Google Scholar]
- 31.Zhao G, London E. Behavior of diphtheria toxin T domain containing substitutions that block normal membrane insertion at Pro345 and Leu307: control of deep membrane insertion and coupling between deep insertion of hydrophobic subdomains. Biochemistry. 2005;44:4488–4498. doi: 10.1021/bi047705o. [DOI] [PubMed] [Google Scholar]
- 32.Lai B, Zhao G, London E. Behavior of the deeply inserted helices in diphtheria toxin T domain: helices 5, 8, and 9 interact strongly and promote pore formation, while helices 6/7 limit pore formation. Biochemistry. 2008;47:4565–4574. doi: 10.1021/bi7025134. [DOI] [PubMed] [Google Scholar]
- 33.Perier A, Chassaing A, Raffestin S, Pichard S, Masella M, Menez A, Forge V, Chenal A, Gillet D. Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. J Biol Chem. 2007;282:24239–24245. doi: 10.1074/jbc.M703392200. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Rosconi MP, London E. Topography of the hydrophilic helices of membrane-inserted diphtheria toxin T domain: TH1-TH3 as a hydrophilic tether. Biochemistry. 2006;45:8124–8134. doi: 10.1021/bi060587f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madshus IH. The N-terminal alpha-helix of fragment B of diphtheria toxin promotes translocation of fragment A into the cytoplasm of eukaryotic cells. J Biol Chem. 1994;269:17723–17729. [PubMed] [Google Scholar]
- 36.Hayashibara M, London E. Topography of diphtheria toxin A chain inserted into lipid vesicles. Biochemistry. 2005;44:2183–2196. doi: 10.1021/bi0482093. [DOI] [PubMed] [Google Scholar]
- 37.Hu VW, Holmes RK. Evidence for direct insertion of fragments A and B of diphtheria toxin into model membranes. J Biol Chem. 1984;259:12226–12233. [PubMed] [Google Scholar]
- 38.Wolff C, Wattiez R, Ruysschaert JM, Cabiaux V. Characterization of diphtheria toxin’s catalytic domain interaction with lipid membranes. Biochim Biophys Acta. 2004;1661:166–177. doi: 10.1016/j.bbamem.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Montecucco C, Schiavo G, Tomasi M. pH-dependence of the phospholipid interaction of diphtheria-toxin fragments. Biochem J. 1985;231:123–128. doi: 10.1042/bj2310123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbieri JT, Collier RJ. Expression of a mutant, full-length form of diphtheria toxin in Escherichia coli. Infect Immun. 1987;55:1647–1651. doi: 10.1128/iai.55.7.1647-1651.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Finkelstein A, Oh KJ, Senzel L, Gordon M, Blaustein RO, Collier RJ. The diphtheria toxin channel-forming T-domain translocates its own NH2-terminal region and the catalytic domain across planar phospholipid bilayers. Int J Med Microbiol. 2000;290:435–440. doi: 10.1016/S1438-4221(00)80059-4. [DOI] [PubMed] [Google Scholar]
- 43.Lakkis F, Steele A, Pacheco-Silva A, Rubin-Kelley V, Strom TB, Murphy JR. Interleukin 4 receptor targeted cytotoxicity: genetic construction and in vivo immunosuppressive activity of a diphtheria toxin-related murine interleukin 4 fusion protein. Eur J Immunol. 1991;21:2253–2258. doi: 10.1002/eji.1830210937. [DOI] [PubMed] [Google Scholar]
- 44.Oh KJ, Senzel L, Collier RJ, Finkelstein A. Translocation of the catalytic domain of diphtheria toxin across planar phospholipid bilayers by its own T domain. Proc Natl Acad Sci U S A. 1999;96:8467–8470. doi: 10.1073/pnas.96.15.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blewitt MG, Chung LA, London E. Effect of pH on the conformation of diphtheria toxin and its implications for membrane penetration. Biochemistry. 1985;24:5458–5464. doi: 10.1021/bi00341a027. [DOI] [PubMed] [Google Scholar]
- 46.Zhan H, Choe S, Huynh PD, Finkelstein A, Eisenberg D, Collier RJ. Dynamic transitions of the transmembrane domain of diphtheria toxin: disulfide trapping and fluorescence proximity studies. Biochemistry. 1994;33:11254–11263. doi: 10.1021/bi00203a022. [DOI] [PubMed] [Google Scholar]
- 47.Sharpe JC, Kachel K, London E. The effects of inhibitors upon pore formation by diphtheria toxin and diphtheria toxin T domain. J Membr Biol. 1999;171:223–233. doi: 10.1007/s002329900573. [DOI] [PubMed] [Google Scholar]
- 48.Sharpe JC, London E. Diphtheria toxin forms pores of different sizes depending on its concentration in membranes: probable relationship to oligomerization. J Membr Biol. 1999;171:209–221. doi: 10.1007/s002329900572. [DOI] [PubMed] [Google Scholar]
- 49.Tortorella D, Sesardic D, Dawes CS, London E. Immunochemical analysis shows all three domains of diphtheria toxin penetrate across model membranes. J Biol Chem. 1995;270:27446–27452. doi: 10.1074/jbc.270.46.27446. [DOI] [PubMed] [Google Scholar]
- 50.Zhao G, London E. An amino acid “transmembrane tendency” scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: relationship to biological hydrophobicity. Protein Sci. 2006;15:1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 52.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 53.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.