INTRODUCTION
The evolution of our understanding of protein folding can be credited in part to the advancement of investigative techniques that probe protein dynamics across many orders of magnitude of time. Information from folding studies on long time scales (milliseconds to seconds) has identified processes leading to the aggregation of misfolded proteins implicated in disorders such as Parkinson’s disease and Creutzfeldt-Jakob disease. At the early end of the time window accessible to kinetic experiments, events on the nanosecond and microsecond time scales have provided insights into fundamental elements of protein folding previously buried in the “burst” phase of classical and stopped-flow mixing methods. Increased access to kinetic information has been made possible not only by the increased capacity of experimenters to trigger protein folding over fast time scales, but also by the coupling of folding trigger methods to optical probes, such as infrared, circular dichroism, resonance Raman, and fluorescence spectroscopies, that are each sensitive to particular aspects of biomolecular structure. By itself, each probe can provide valuable information about protein folding dynamics. In combination, however, the different optical techniques can provide a clearer and more structurally detailed description of the changes that occur during the process of folding or unfolding.
Such structural dynamics are significant for understanding not only the folding/unfolding mechanisms of proteins, but also their function in physiologically relevant biochemical processes. The high time resolution afforded by optical spectroscopies has generally been an important complement to the atomic-level detail revealed by structural methods such as x-ray crystallography, extended X-ray absorption fine structure, and nuclear magnetic and electron spin resonance spectroscopies in understanding protein dynamical processes. This complementary relationship is now growing closer with the increasingly structure-specific information available from time-resolved optical methods and recent improvements in the time-resolution of traditional structural methods [1, 2]. These exciting advances will together provide unprecedented details about the mechanisms of both folding and functional reactions in biochemical systems.
Optical spectroscopy encompasses a variety of techniques that measure the absorption, emission, or scattering of light by biomolecules. After briefly mentioning several more familiar time-resolved techniques (fluorescence, infrared, and Raman spectroscopy) that have been instrumental in advancing our understanding of protein folding, we will focus here on nanosecond time-resolved polarization methods, specifically circular dichroism (CD), magnetic circular dichroism (MCD) and optical rotatory dispersion (ORD) techniques, and their application to the study of protein folding in ferrous cytochrome c.
NANOSECOND OPTICAL SPECTROSCOPY
Time-resolved absorption spectroscopy is a relatively straightforward optical method for monitoring the overall kinetic behavior of protein folding mechanisms. However, for the assignment of specific protein conformations to intermediate species, more structure-sensitive probes are necessary. Resonance Raman spectroscopy, for example, can offer information about the ligation state of heme proteins. Structural details about the protein backbone and the amino acid side chains can be obtained from the protein vibrations upon coupling UV-excitation with the resonance Raman technique. In addition, specific chromophores can be probed selectively with the choice of different excitation wavelengths. Overall, this method imparts a structure-specificity that is similar to infrared measurements, but without the complications of water absorbance. The complementary technique of infrared spectroscopy focuses closely on the C=O and N-H stretching vibrations of the amide bands [3]. Vibrational absorptions arise from transitions that are relatively localized on a molecule. For complex molecules, there is the danger of oversimplification in the assignment of a large number of potentially overlapping IR absorptions to the stretch or bend of a specific bond. However, for proteins, the regularity of specific structural motifs (α-helix, β-sheet) can simplify the IR spectrum and offer a highly sensitive probe of local molecular structures that are IR active. In fluorescence studies the use of either intrinsic tryptophan residues or extrinsic fluorophores has been used to follow tertiary structural dynamics [4]. This method is particularly sensitive to polypeptide collapse, as is small angle X-ray spectroscopy, which has been used as a probe of the global dimensions of a macromolecule [5].
Circular dichroism (CD) in the far-UV spectral region is probably the most widely recognized application of polarized absorption spectroscopy to the study of biomolecules [6]. The relatively intense far-UV CD associated with the n → π* transitions of the polypeptide backbone reflects elements of secondary structure such as α helices and β sheets. The variety of chromophores found in biomolecules means that CD bands of interest may also be present in other regions of the optical spectrum. CD bands in the near-UV region reflect asymmetries in the environments of the aromatic amino acid residues phenylalanine, tyrosine, and tryptophan imposed by protein tertiary structure. The asymmetric protein environment may also induce a CD in the absorption bands of otherwise achiral prosthetic group chromophores, such as heme. Thus, CD data measured over a broad spectral range (far-UV to visible regions) will often give a wide range of global information about the structure of a biomolecule.
Since it was initially coupled with stopped-flow and flash photolysis methods in 1974, the time resolution of optical CD measurements has improved from milliseconds to picosecond [7–13]. The evolution of nanosecond CD (TRCD) ellipsometry from single-wavelength to multi-wavelength measurements and the extension of these measurements into the far-UV spectral region and to time-resolved magnetic circular dichroism (TRMCD) have been particularly useful for the study of protein folding. The subsequent development of nanosecond time-resolved optical rotatory dispersion (ORD) and magnetic ORD (MORD) methods provided a further signal-to-noise advantage that permits the measurement of kinetic data on the fastest folding time scales, with the trade-off that contributions from the different absorption bands of a given protein are more overlapped spectrally than in TRCD and TRMCD data.
Nanosecond Circular Dichroism Spectroscopy
Ellipsometric methods are the most sensitive way to detect CD and thus provide a natural starting point for developing high time-resolution instrumentation. However, the traditional null-detection method in which a sample is placed between crossed linear polarizers is typically compromised by interference from the circular birefringence (CB = 2·ORD) properties of the sample, as well as the presence of any linear dichroism or linear birefringence in the sample or any other optical elements (e.g., windows, lenses) between the crossed polarizers. Note that such artifacts are not typically a problem for conventional CD instruments, in which the measuring beam is modulated between left and right circular polarizations (LCP and RCP). Measuring the typical CD signal magnitude of one part in 104 directly from an LCP – RCP absorption difference measurement requires a high signal-to-noise ratio, which is accomplished by using a photoelastic modulator (PEM) and phase-locked detection. However, the 1–100 kHz resonant frequencies of PEMs limit the time-resolution of conventional CD instruments to milliseconds.
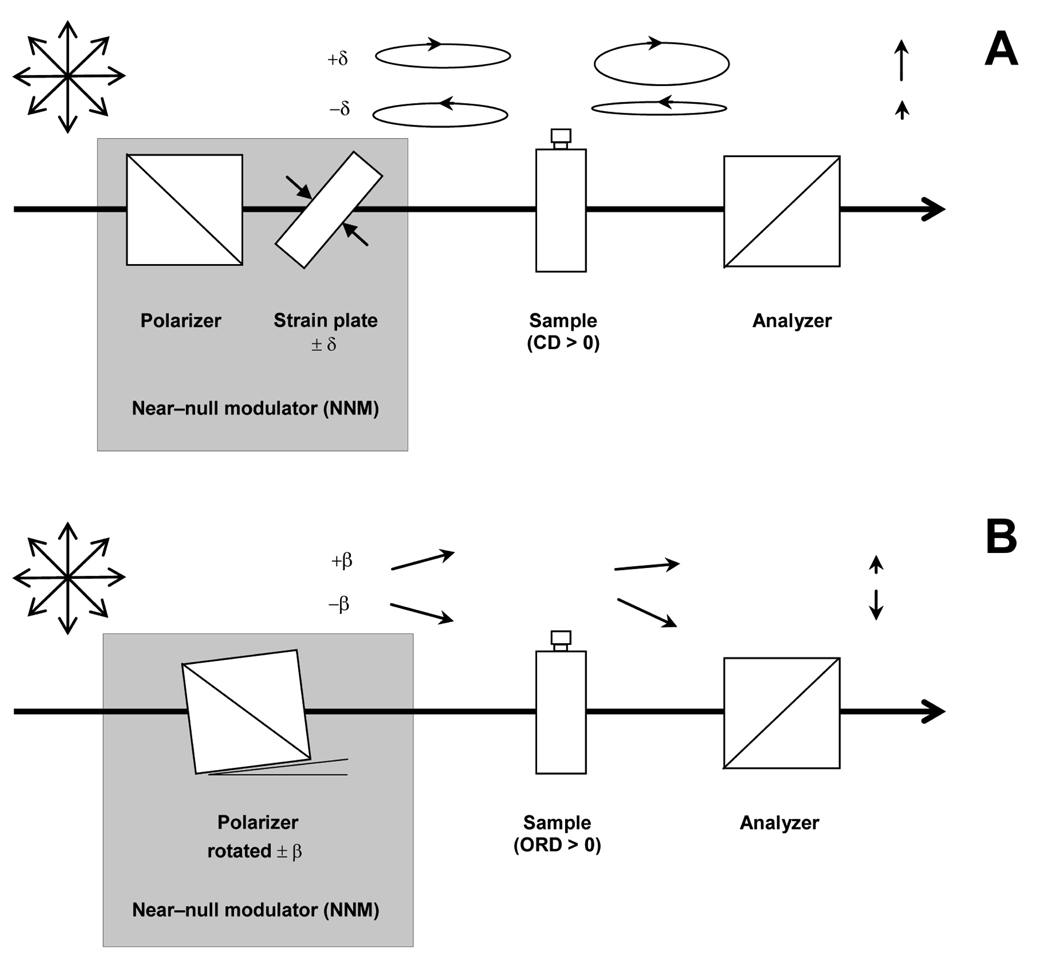
Nanosecond time resolution is achieved in CD measurements by using a modification of the early ellipsometric method (Figure 1a). In this quasi-null approach, changes in the beam polarization caused by the CD of the sample are compared with a small reference ellipticity. This reference is implemented by mechanically straining a fused silica plate to produce a slight linear birefringence of known phase retardance, δ. This retardance converts the linear polarization of the incident light to highly eccentric left or right elliptical polarization (LEP, REP), depending on the orientation of the fast retardance axis of the plate with respect to the beam polarization. Because elliptically polarized light comprises left and right circularly polarized components of different amplitudes, the differential absorption of these components by a circularly dichroic sample will result in changes in the net ellipticity of the beam. In particular, a positive CD in the sample causes an increase or decrease in the eccentricity of an LEP or REP light beam, respectively. These changes are monitored with an analyzing polarizer, oriented with its polarization axis perpendicular to the first polarizer, that passes the minor-axis linear component of the LEP and REP light to a detector. Normalizing the difference of the LEP and REP minor-axis detected intensities to their sum gives an ellipsometric signal, S = (IR − IL) / (IR + IL), that is proportional to CD/δ. Note that the CB accompanying the CD of the sample will rotate the axes of the ellipse in the same direction for both LEP and REP, and thus the effect of CB will cancel to first order from the ellipsometric signal S. Thus, the differential enhancing or disenhancing effect of a sample’s CD on reference LEP and REP ellipticities induced in the probe beam by a strain plate permits CD measurements with high sensitivity without the interference from the intrinsic CB of the sample that limited traditional ellipsometric measurements.
Figure 1.
A Near-null modulator (NNM) is used for both the time-resolved CD (A) and ORD (B) systems. For CD the NNM comprises a polarizer and strain plate (fused silica plate), whereas the NNM in the ORD apparatus is a polarizer mounted on a rotation stage. The CD of the sample adds to or subtracts from the reference ellipticity that is produced by the strain plate and the ORD of the sample adds to or subtracts from the reference rotation that is produced by the polarizer.
TRCD measurements can be used to probe changes in protein secondary structure, the relative orientations of dipole-dipole coupled chromophores, or chromophore-protein interactions, to name some examples. Since the first measurements of far-UV TRCD data in 1993 [13], the method has been applied to the study of a variety of biochemical systems such as the phytochrome photoreceptor [14] and myoglobin [15]. The first application in far-UV TRCD studies of protein folding was a 1998 investigation of rapid helix formation in the cytochrome c-CO photolysis system [16] developed previously by Roder and coworkers [17]. The use of multi-channel detection in these experiments was advantageous in that it was possible to obtain the spectral signature of the secondary structure changes that occurred during the biochemical reaction or during protein folding. However, the time resolution of these experiments was limited to about 100 ns. The time resolution of the TRCD measurements was improved (∼2 ns) when an upconverted argon-pumped titanium sapphire laser was coupled to the TRCD apparatus [18]. Using the generated second, third, and fourth harmonics of the fundamental output, which was tunable from 780 to 910 nm, it is possible to measure single-wavelength TRCD signals over the spectral range of 205–910 nm.
Magnetic Circular Dichroism Spectroscopy
Whereas natural TRCD measurements are limited to optically active chromophores, the introduction of a magnetic field to the TRCD apparatus can induce an additional CD signal that is independent of natural chirality. Magnetic circular dichroism (MCD) may be considered a generalization of the Zeeman effect, wherein application of a magnetic field leads to the splitting of degenerate energy levels. This splitting gives rise to a CD signal because the electric dipole transitions involving Zeeman levels are circularly polarized. Generally, MCD is sensitive to structural features within a biomolecule to the extent that they perturb the electronic states of a chromophore that is MCD active, i.e., contains orbital or spin degeneracies or near degeneracies. For example, in heme proteins such structural features include the axial ligation, spin, and oxidation states of the heme iron atom, all of which can strongly affect the MCD of the heme spectral bands. Since the first nanosecond time-resolved MCD (TRMCD) measurements were reported for the lowest excited triplet state of zinc tetraphenylporphine [19], the TRMCD method has been applied largely to the study of photodissociation intermediates of heme protein-ligand complexes (e.g., cytochrome aa3, cytochrome ba3, cytochrome c3, myoglobin and hemoglobin) and, more recently, to the protein folding problem in cytochrome c [20, 21]. In heme proteins, the MCD spectra in the Soret and visible regions are particularly sensitive to the oxidation and ligation state of the heme iron. These spectral markers have been useful in protein folding studies, wherein nonnative intermediate species have been identified during the protein folding process.
Optical Rotatory Dispersion Spectroscopy
Nanosecond time scale optical rotatory dispersion (ORD) changes were first measured in a 1995 study of hemoglobin [22] by adapting a quasi-null polarimetric method introduced previously by Keston and Lospalluto for high-sensitivity static ORD measurements [23]. In principle, time-resolved ORD (TRORD), Figure 1b, and TRCD report identical information because ORD signals and CD spectra are Kramers-Kronig transform mates [24]. In practice, CD signals are localized to absorption bands and thus generally easier to interpret in applications such as the analysis of protein secondary structure. In contrast, ORD signals can be measured outside of absorption bands, giving the ORD method a signal-to-noise advantage in light-limited kinetic measurements. TRORD and time-resolved magnetic ORD (TRMORD) studies have been performed on protein folding in cytochrome c and the kinetics of ligand rebinding in myoglobin and hemoglobin.
PROTEIN FOLDING TRIGGERS
Other significant advances in protein folding/unfolding studies have addressed the challenge of rapidly triggering folding/unfolding reactions. Because biomolecules are dynamic structures, time resolution is of particular importance to mechanistic studies. Nanosecond time-resolved detection methods would be of limited use in protein folding studies without a means to trigger protein folding/unfolding reactions on an equally fast or faster time scale. There are currently two general classes of rapid protein folding initiation methods: those that trigger folding in proteins/peptides with endogenous or exogenous photoactive groups and those that trigger folding in proteins/peptides without a photosensitive chromophore.
Laser-Induced Temperature-Jump
To study a broader range of proteins, more general triggers falling in the latter class have been developed. A laser-induced temperature-jump (T-jump) is probably the most widely used method to rapidly shift the equilibrium between folded and unfolded states of proteins. Early T-jump methods used a capacitive electrical discharge across the sample cell, from either a conventional or a coaxial cable capacitor, to achieve T-jumps within a microsecond or 50 ns, respectively ([25] and references within). A different approach to the T-jump method used the absorption of high-powered laser pulses by a dye [26] or by water itself [27] to produce a rapid temperature increase. However, the use of a dye can potentially interfere with the folding/unfolding/biochemical reaction of interest, as can the high salt concentrations required for capacitive discharge methods. Consequently, the most common way to generate a T-jump takes advantage of the absorption of laser light from 1.4 µm to 2 µm by an aqueous sample, whereby a 10–30K T-jump can be induced within the ∼10 ns pulse width of a laser. The T-jump initiation method has been coupled with time-resolved absorption, fluorescence, resonance Raman, infrared and, more recently, ORD detection methods [28].
Rapid Mixing
In 1985, Regenfuss et al. introduced a microflow stopped-flow apparatus that took advantage of highly turbulent flow conditions to completely mix two solutions within tens of microseconds [29]. This very general fast-triggering method improves upon the millisecond mixing dead time typical of stopped-flow experiments. More recent attempts to improve the instrumental dead time have reported achieving mixing times that ranged from ∼100 ns [30, 31] to 45 µs [32]. In the latter study, Shastry et al. addressed many technical issues, such as stability, geometry, and artifacts, that surround use of the jet mixing device. In 2004, a microfluidic mixer achieved a mixing time of < 10 µs [33] by using an optimization of a turbulence-free mixing method described by Knight et al. [34] wherein four channels, etched to a depth of 10 µm, taper in width and intersect at the center of a mixing chip. The side flow (buffer) hydrodynamically focuses the inlet flow (denatured protein) into a stream as narrow as 50 nm. Because of the small length scales, the buffer molecules rapidly diffuse across the inlet stream and fast mixing is achieved. This rapid mixing system has been applied to fast protein folding measurements using Förster resonance energy-transfer (FRET) with acyl-CoA binding protein [33] and using UV fluorescence spectroscopy with cytochrome c, apomyoglobin, and lysozyme [35].
Photoexcitation Events
Laser-induced T-jump or rapid denaturant dilution can be used to trigger folding/unfolding in either iron oxidation state of cyt c, although in practice these methods have been applied mainly to the ferric state. The coupling of heme-residue binding with the folding stability of cyt c offers the further possibility of using photodissociation or photo-initiated electron transfer (ET) reactions to trigger folding in the ferrous form of the unfolded protein. Building on flash photolysis techniques introduced in the 1940s [36], folding reactions have been initiated by the photodissociation of an exogenous ligand bound to ferrous heme or by the photo-initiated reduction of oxcyt c. In 1993, Jones et al. presented a CO ligand photolysis method that prepared an unfolded pentacoordinated-heme form of redcyt c under folding conditions within the time resolution of their nanosecond absorption apparatus [17]. This initiation method relies on the difference in the folding free energies of CO-bound and CO-unbound redcyt c. For example, in the presence of 4.6-M GuHCl denaturant (which destabilizes the protein enough to permit heme-CO binding), redcyt c is largely folded in the absence of CO, whereas CO-bound redcyt c is largely unfolded [16, 17]. Similarly, folding free energy differences also exist between the oxidized and reduced forms of cyt c [37, 38]. Thus, at GuHCl concentrations between 2 and 4 M, oxcyt c is largely unfolded and redcyt c is largely folded. Therefore, either photoreduction (e.g., by photoexcitation of an exogenous ET donor such as NADH) or photodissociation can be used with the appropriate amount of denaturant to produce an immediate photoproduct that is redcyt c in a (metastable) unfolded state under folding conditions. However, any folding pathways initiated by the photolysis trigger will be interrupted by the diffusive recombination reaction of the ligand, which limits the duration of the monitoring time window to hundreds of microseconds at 1 atm CO. An advantage of using photoreduction triggers is that a longer window of time that includes the appearance of the natively folded state (after tens to hundreds of milliseconds) can be conveniently observed with the proper choice of photoreductant.
HALLMARKS OF EARLY EVENTS IN REDCYT C FOLDING
In a collaboration with Tony Fink in 1998, we coupled the photolysis trigger system of Jones et al. [17] with the ns time-resolved circular dichroism detection method in order to follow very early changes in the secondary structure of cyt c [16]. This work became the seed for several subsequent papers that further probed these early events, in the process finding kinetic evidence that both connected them to specific structural features of the protein and pointed to their broader significance in the context of the energy landscape model of protein folding [20, 21, 39–42]. In particular, these studies found the first kinetic evidence pointing to slow conformational diffusion as the source of kinetically isolated folding subpopulations, i.e., multiple folding pathways, in a folding-competent protein sequence. The remainder of this chapter will focus on these results and further directions that redcyt c folding studies employing nanosecond TRCD, TRORD, TRMCD, and TRMORD polarization spectroscopies have taken in the last ten years.
In the CO photolysis studies mentioned above, the unfolded five-coordinate heme protein (His18 remains bound to the heme iron) that is the immediate product of photodissociation goes on to reform a six-coordinate heme species by binding the native Met80 moiety (and perhaps nonnative Met65 to some extent as well) with a very rapid 2-µs time constant. Then, in an apparent retrogression on the way to the natively folded state, an intermediate is formed with a 50-µs time constant in which the sixth axial position is occupied predominantly by a nonnative histidine (His33 or 26) ligand. The eventual displacement of the nonnative His ligands and formation of the native folded structure were expected to occur on time scales much greater than 1 ms. However, in this photolysis-triggered folding system, as in other heme protein ligand photolysis experiments, the rebinding of exogenous ligand is expected to proceed on a tens of microseconds to milliseconds time scale. In this case, CO rebinding effectively ends the protein’s conformational evolution before reaching the native state and returns it to its unfolded starting point. Indeed, CO dilution studies indicated that the slowest process observable in this system (observed time constants of 200 and 900 µs at 1 atm CO) was bimolecular CO rebinding [16].
Although kinetic competition from CO rebinding and nonnative heme-residue ligation effectively prevented the production of natively folded protein in this system, intriguingly, the far-UV TRCD data nevertheless showed the formation of ∼10% native-like secondary structure within 2 µs of CO photodissociation [16]. (Note that the detection of this small amplitude process is a challenge in ns secondary and tertiary structure-sensitive measurements, requiring a higher signal to noise ratio than was available, for instance, in the Trp fluorescence results reported previously for this system [43].) The observation of this very fast folding intermediate raised questions such as: What is its relation to the “burst phase” intermediate observed in stopped-flow CD studies? Is it a productive intermediate on the way to native state folding? How native-like or molten globule-like is its tertiary structure? What can it tell us about the overall mechanism of folding in cyt c? While at least partial answers to some of these questions are discussed below, perhaps the most fundamental question touched upon by this observation was that of the conformational dynamics of the unfolded chains.
To appreciate the importance of the unfolded dynamics, recall its connection to a form of heterogeneity in folding kinetics that is a distinguishing characteristic of the “new view” energy landscape funnel model. In this view, the top of the funnel represents the unfolded protein conformations, which travel down the length of the funnel in a free-energy biased form of configurational diffusion to achieve the lower conformational entropy of the native state. The extent of equilibration of the unfolded conformers at the periphery of the funnel, a consequence of the conformational diffusion time around the funnel, governs the type of kinetic regime under which the protein folding reaction takes place. If the peripheral diffusion time is slower than (free energy-biased diffusional) downhill folding, then the different unfolded conformations can fold along kinetically isolated pathways and exhibit heterogeneous kinetics. Conversely, if interconversion between the unfolded conformers is much faster than the formation of the native state, then such configurational equilibrium leads to homogeneous kinetics that often follow the classical view involving passage over a single transition state. The kinetic heterogeneity that arises from slow equilibration between unfolded polypeptide chains is most likely to affect the earliest events and intermediates in protein folding, even if its influence may not necessarily persist long enough to affect the late-time folding processes producing the native state.
Just how long the unfolded chains require to equilibrate has been an open question. However, the microsecond time scale of the fast helix formation process suggested that it might well overlap with the unfolded equilibration time and thus lie in the energy landscape regime just described. Indeed, the earliest evidence in this regard for the cyt c CO photolysis system was interpreted with an analysis that implied an equilibration time of at least 40 µs [44], in which case a great deal of conformation-based kinetic heterogeneity could be frozen in during fast helix formation. However, the early TROD and near- and far-UV TRCD data were ambiguous in this regard in that they could be interpreted with either heterogeneous or homogeneous kinetic models [16].
A later TRMCD study gave the first qualitative indication of kinetic heterogeneity stemming from slow equilibration between unfolded conformational ensembles in this system [20]. That TRMCD analysis considered two limiting-case scenarios for heme-residue binding after CO photolysis. In the first, the frozen-conformation scenario, the five-coordinate photoproduct species was divided into two non-communicating unfolded protein subensembles, one with a propensity to bind the Met ligand (presumably corresponding to protein backbone conformations placing Met80 or Met65 near the distal heme site for facile binding) and the other to bind the His ligand. In the second, equilibrated-conformation scenario, these subensembles were considered to rapidly interconvert. The data supported the first scenario in that the resulting fit to the MCD spectrum of the bis-His heme-ligated transient species was better than in the modeling analysis that did not account for conformational diffusion of early folding kinetic heterogeneity. This provided kinetic evidence for multiple pathways in redcyt c folding in that the Met and His binding reactions appeared to proceed separately to at least some extent in kinetically isolated conformational ensembles, implying limited conformational diffusion on the time scale of the fast helix folding process identified in the far-UV TRCD data.
Further evidence of a similar kinetic heterogeneity in the fast helix formation process followed from redcyt c folding studies in which the far-UV TRCD/TRORD probe was coupled with the electron transfer trigger method [39–41]. Redcyt c folding was probed in various concentrations of GuHCl (2.7, 3, 3.3, and 4 M), with the results identifying two folding phases: a very fast (hundreds of nanoseconds to tens of microseconds) and a slower (> 1 ms) phase. The fast phase, similar in time scale and amplitude to the fast folding observed in the CO photolysis system, represents a time-resolved view of the “burst” phase of conventional stopped-flow CD studies. Taking advantage of the long time window provided by the photoreduction system, we were also able to observe the slow phase in which redcyt c folds to its native structure. A comparison of the time constants for these folding phases obtained in different denaturant concentrations is shown in Table 1. For the slow phase, increasing the denaturant concentration increased the folding time constant. This observation was consistent with the usual expectation that folding slows in higher denaturant concentrations because of a linear dependence of activation free energy changes on changes in the free energy driving folding [38].
Table 1.
Observed Lifetimes for Redcyt c Folding
| [GdnHCl], M | τ1 (µs) | τ2 (ms) |
|---|---|---|
| Horse heart | ||
| 2.7 | 12 ± 2 | 100 ± 10 |
| 3 | 5 ± 3 | 120 ± 20 |
| 3.3 | 2.2 ± 0.5 | 160 ± 10 |
| 4 | ≤ 0.4 | 380 ± 30 |
| Tuna | ||
| 3.3 | 40 | 185 |
| 4 | 28 | 430 |
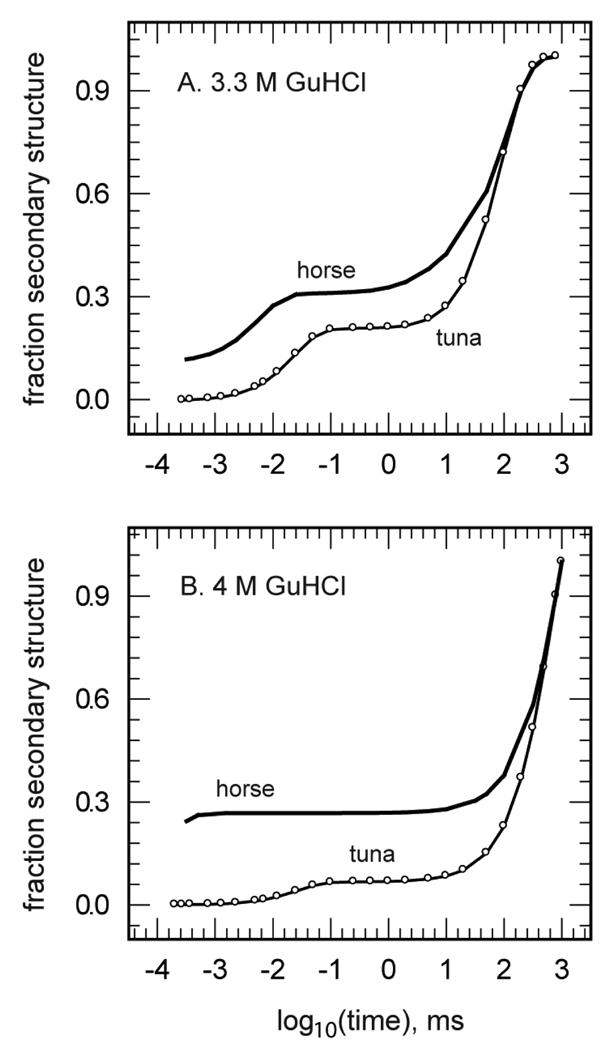
The dynamics of the fast phase of folding, however, exhibited unusual kinetics in that the time constant became much faster with increasing concentrations of GuHCl. An example of this trend in the secondary structure dynamics is shown in Figure 2, which shows representative data taken at 3.3 and 4.0 M GuHCl. The observed variation in folding kinetics with denaturant concentration was monotonic and ranged from the formation of no apparent secondary structure until after 5 µs in 2.7 M GuHCl to the formation of about 20% secondary structure within several hundred nanoseconds in 4.0 M GuHCl. Even more surprisingly, the latter time was actually much shorter than the average time constant for photoreduction in this system, ∼5 µs. In other words, this reduction-triggered folding process proceeded faster than the bulk of the reduction reaction used to trigger folding. A resolution of this apparent paradox was found in the heterogeneous natures of both the reductant and the protein. Among the mixture of reductants produced by NADH photolysis were very reactive solvated electrons and among the unfolded chains were a subset of conformations that were inferred to react quickly with this reductant while remaining kinetically isolated from the bulk of chain conformations by slow conformational diffusion. Combined with the TRMCD results from the redcyt c-CO photolysis study, the far-UV TRCD and TRORD data suggested the presence of conformational heterogeneity in the initial unfolded protein ensembles, an approximately 20% subpopulation that was configurationally inclined to fast folding. This result also further supported the interpretation that the very fast helix process observed in both the CO photolysis and photoreduction experiments formed near-native amounts of secondary structure in a small subset of protein conformers, rather than a small amount of secondary structure throughout the bulk of the protein sample.
Figure 2.
The TRORD kinetic results of horse and tuna redcyt c folding in 3.3 M (A) and 4 M (B) GuHCl. These traces show the dramatic differences in the kinetic traces between horse and tuna redcyt c, despite the 80% sequence homology between the two proteins. The differences in the kinetic data are suggested to be due to the substitution of the His ligand at position 33 in horse redcyt c with a Trp moiety.
We next looked for structural correlates of the fast-folding kinetic phase by examining species and mutational variants of cyt c in photoreduction-triggered folding studies. Specifically, we looked at variants lacking one or both of the His33 and His26 nonnative ligands identified by other workers as dominant in coordinating the ferric heme in the horse heart oxcyt c starting material used in the earlier TRCD studies [45]. The question to be addressed was would the earliest folding events in redcyt c be affected by changes in the partial constraint on the backbone conformations of the unfolded chains presented by this His ligation? This question was first addressed in far-UV TRORD studies of tuna heart redcyt c [39]. Although the sequence homology (80%) and structural similarity of the horse and tuna proteins are high, a marked difference was observed between their fast-folding processes, as gauged by TRORD, that was presumably due to the replacement of His33 by tryptophan (which does not bind to heme iron) in the tuna sequence. Figure 2a and 2b show the differences in early-phase secondary structure formation kinetics for the two redcyt c proteins in 3.3 and 4 M GuHCl, respectively. Secondary structure formation was markedly slower and weaker in amplitude in tuna redcyt c at each denaturant concentration, with essentially no formation occurring before 5 µs. These data suggested that His33 is somehow important for rapid formation of secondary structure in the fast folding subpopulation, although they did not address whether His18-Fe-His33 coordination actually facilitates fast folding or whether it slows folding to a lesser degree than does His18-Fe-His26 coordination. They also indicated that even if His33 heme ligation is a necessary condition for ultrafast folding in a small subset of the unfolded chains, it is not necessarily sufficient because the majority of the unfolded polypeptide chains of the initial oxcyt c sample had this heme ligation. (Note that these results were also significant in ruling out the heterogeneity of His misligation in horse cyt c, about 80% His33 and 20% His26 in the ferric form [45], as a possible trivial explanation for the kinetic heterogeneity of its unfolded chains, i.e., the ∼20% subset that rapidly folded to helix. That explanation would have predicted a marked increase in the amplitude of microsecond and submicrosecond folding in tuna vs horse, contrary to observation.)
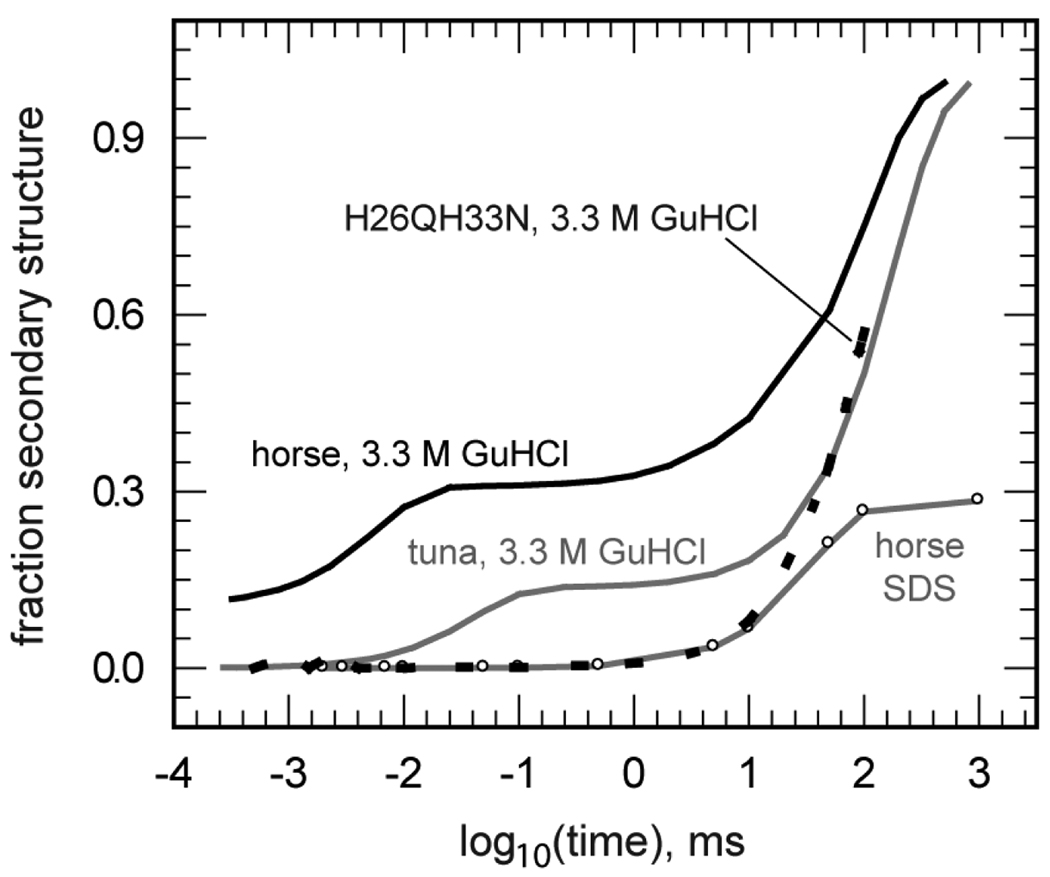
The roles of His33 and His26 were further explored with far-UV TRORD measurements of redcyt c folding in a horse heart double mutant wherein both histidine residues were replaced with residues of low heme affinity (H26QH33N) [42]. The results of denaturant titrations indicated that double mutation significantly reduced the folding stability of redcyt c H26QH33N relative to the WT protein, but not the ferric form. This left a relatively narrow GuHCl concentration range centered at 3.3 M within which photoreduction-triggered folding studies could be conducted in the mutant. The heme ligation state of the unfolded mutant was probed with MCD measurements in the Soret and visible regions, as the removal of both histidines making up the dominant ligation of unfolded WT horse cyt c left several other candidates, including Met80/65, Lys72/73/79, and the non-acetylated N-terminal group, as potential ligands at the sixth axial heme site. The MCD studies confirmed the presence of methionine ligation in the folded state of the ferrous double mutant and suggested that the axial ligand in the unfolded ferric form was a lysine residue. Altogether, these results implied that the 3.3-M GuHCl H26QH33N oxcyt c photoreduction experiment passed through a His18-Fe(II)-Lys ligated instantaneous photoproduct that required displacement of the lysine residue by Met80 to produce the natively folded ferrous protein. Focusing in the submillisecond time regime (the fast folding phase), the far-UV TRORD studies observed a kinetic trace for the electron-transfer triggered folding of the double mutant that was markedly different from that measured for the WT horse heart protein (Figure 3). The fast phase of folding observed in the WT kinetics (τ ∼ 5 µs) was absent for H26QH33N, leaving only the slow phase observed after 1 ms. Thus, these results indicated that the His18-Fe-His33 heme coordination facilitates folding in an absolute sense and that it does so more than does His18-Fe-His26 heme ligation.
Figure 3.
Comparison of kinetic traces obtained from far-UV TRORD experiments on horse and tuna redcyt c folding. The data were obtained for horse (black line) and tuna (grey line) WT redcyt c in 3.3 M GuHCl, horse H26QH33N redcyt c in 3.3 M GuHCl (dashed black line), and horse WT redcyt c in SDS (grey line, open circles). The kinetic traces present an average of the multi-wavelength TRORD data measured between 228 and 236 nm.
The combined results of the far-UV TRORD studies on fast folding of redcyt c focused attention on the structural basis for the kinetic effects of His33 and His26 heme axial ligation in the unfolded state. The dramatic differences in the kinetic traces for fast folding in the tuna and horse WT redcyt c proteins suggested that heme-His26 ligation was unable to provide the structural requirements for very fast folding provided by His33 ligation, whatever those might be. His26 bridges two Ω-loops (20s and 40s) via two hydrogen bonds to Pro44 and Asn31, forming a triad H-bond network. According to modeling studies of WT horse cyt c and the H26Q single mutant (46) and NMR studies of the WT horse and tuna cyt c [47], that triad H-bond network is maintained despite glutamine substitution of His26 and GuHCl concentrations of 7 M. His33 is associated with a β-turn (residues 21–24) and the carboxy-terminal residue of the protein backbone, which are linked to the histidine by van der Waals interactions. Thus, the rigidity of the His26 residue induced by hydrogen bonding may inhibit its ability to form a productive heme-His26 loop to compensate for the absence of His33. In contrast, His33 may be better able to facilitate folding because it is ‘anchored’ more weakly by van der Waals interactions.
Although the greater flexibility of the His33 residue appears to enhance formation of heme-His33 coordination, the sequence positions of His33 and His26 relative to the heme group may also play a role. Based on the amino acid composition of the loop formed between the heme group and His33 versus His26, there is a greater likelihood of loop formation by His33 because of the presence of one more glycine and a proline in the His18-His33 loop [48–51]. According to stopped-flow absorption studies of the rates of formation and of breakage of loops, there is a general correlation between increasing rates of formation and decreasing loop sizes [52]. Extrapolation of the stopped-flow data to a loop size of 9 (the smallest loop size studied was 10) gives a rate of formation that is greater for the heme-His26 than the heme-His18 loop. Although it might be expected that the rate of loop breakage is an important factor that determines whether an initial loop will persist and proceeds towards productive folding, Kurchan et al. do not observe a regular trend for loop sizes less than 37. In this light, it appears that for the WT tuna heart TRORD data the small loop size formed between the heme and His26 may be only one explanation for why it might have a smaller driving force for helix formation during this phase. Another factor that has been proposed to contribute to the lower affinity of His26 for the ferric heme group is the presence of surrounding Lys residues (Lys25 and Lys27) that lead to charge repulsion with the iron [49].
Conformational Diffusion
In tentatively assigning the kinetic isolation of the fast folding subensemble from the bulk of the unfolded protein to slow conformational diffusion, an alternative explanation for the low amplitude of the fast process was first discounted. The possible existence of a strong (unfolding) back reaction was deemed unlikely because the amplitude of the fast phase did not change as expected with denaturant concentration. That is, increasing the denaturant concentration would be expected to further decrease an (unfavorable) equilibrium between unfolded and partly unfolded states, contrary to observation. The most likely explanation for the kinetically isolated fast folding process was slow conformational diffusion of the unfolded polypeptide chains. With this parameter, which has been suggested to occur with a time constant on the microsecond time scale for ∼100-residue protein chains [53], it is possible to explain the observations of heterogeneous and homogeneous folding kinetics. That is, experimental work probing folding with millisecond time resolution would encounter fast equilibration of the unfolded chains such that the overall picture of folding could be described with classical, homogeneous kinetics. In contrast, if the time resolution of the experiment is faster than the time constant of conformational diffusion, then it is possible to probe the earliest windows of folding, where heterogeneity is greatest, and to detect heterogeneous folding processes.
A quantitative measurement of the conformational diffusion time in unfolded cytochrome c was approached using the CO photolysis trigger to initiate folding of the horse and tuna WT proteins and several histidine mutants (H33N, H26Q, H33NH26Q) of the horse heart protein [21]. After photolysis of the CO ligand, the His and Met residue axial binding reactions were probed with Soret region MCD spectroscopy. The conformational diffusion parameter was approximated as a first-order time constant (τd) for the exchange between the conformational subpopulations that offer the His26, His33, or Met residues for binding at the heme axial site. The TRMCD data obtained for all of the proteins were fit simultaneously to a kinetic model for heme-residue binding that included the effect of this diffusional parameter, a procedure that yielded a τd value of 3 ± 2 µs. This value was considered to represent an aggregation of intrachain diffusion processes (such as the intrachain diffusion of Met80, Met65, His33, and His26 to and from the heme group) that are intrinsic to the transitions between conformational subensembles. This value may be compared with intrachain diffusion time constants reported for several other protein and polypeptide systems: a 15-residue loop in unfolded Ru(NH3)53+-modified Zn-substituted cyt c (τ = 250 ns) [54], the residue 6–85 fragment of λ-repressor protein (τ = 2 µs) [55], and non-folding Xaa-Ser repeat polypeptides [53]. Although differences in factors such as denaturing solvent conditions, the presence of the covalently attached heme prosthetic group, non-folding versus folding-competent peptide sequences, and temperature contribute to uncertainty in making these comparisons, a rough extrapolation to a 62-residue loop, the size of the Met80-His18 segment contained in the loop formed by Met80-heme binding, shows broadly similar values. That is, the different measurements of conformational diffusion times appear to converge near 1 µs for loop sizes of 60–100 residues. The time constant for exchange between conformational subensembles observed for unfolded cyt c (3 ± 2 µs) lay at the fast edge of the broad range inferred from our previous TRMCD study [20] and was considerably faster than the previous estimate of ∼40 µs implied by a value assigned by previous workers to the unimolecular rate constant for Met binding [17]. This divergence between present and previous estimates for cyt c, in which the present value appears more consistent with the results from other protein systems, was attributed at least in part to the tighter constraint on possible outcomes of the kinetic modeling of the Met and His binding rates that was afforded by the TRMCD data for the histidine variants.
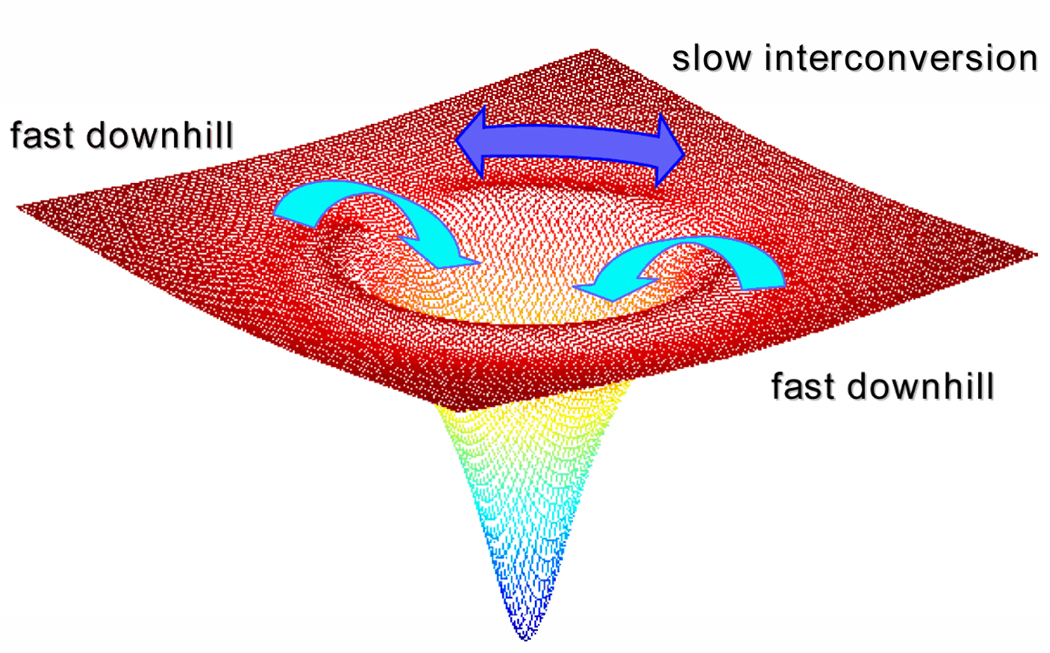
The convergence of the TRMCD results for cyt c with those from other systems also provided additional support to the suggestion by Gruebele and coworkers that the time scale of conformational diffusion in the unfolded chains represents a more general kinetic limit for classical folding [55]. Folding on slower time scales would imply that the unfolded state(s) and the transition state are in conformational equilibrium, as required by classical transition state theory (TST), whereas faster folding would be better described by the downhill folding scenario of the energy landscape model (Figure 4). Furthermore, the 3-µs interconversion time constant reported by Abel et al. [21] was slow enough to support the previous assignment of the kinetic isolation of the very fast-folding conformational ensemble in the horse heart protein to slow conformational diffusion.
Figure 4.
Conformational diffusion is a determining factor of whether protein folding exhibits heterogeneous versus homogeneous kinetics. Slow conformational interconversion around the top of the folding funnel leads to biased diffusion of unfolded protein subpopulations down kinetically isolated pathways towards the native folded state. If diffusion between conformational subpopulations is faster than folding to the native state, then folding follows homogeneous kinetics, or kinetics that appear to follow the classical transition state theory view.
Nonnative tertiary structural states
One question prompted by these natural and magnetic TRCD/TRORD studies is what is the biological significance of nonnative tertiary structural states in proteins, e.g., the state with His18-Fe-His33 heme ligation observed in cyt c under denaturing conditions? Besides their significance in folding studies discussed further below, many studies have looked at the physiological occurrence and roles of structurally disordered states. Some studies have tailored the experimental solution conditions in order to mimic the environment of the mitochondrial membrane using, for example, a solution of 30% acetonitrile/70% water having a dielectric constant (ε = 67) similar to that of the mitochondrial membrane (lipid vesicle ε = 30–60) [56]. Redcyt c exhibited a substantial decrease in the solvent accessible surface area and a significant conformational difference in His33 (in contrast to His26) in this medium relative to an aqueous medium. The conformational rearrangement of His33 in the membrane-like medium may have reflected the protein’s response to a membrane environment. For its role in cell respiration, the conformational adjustments in cyt c may help to preserve the specificity of the electron-transfer pathways and, in apoptosis, changes in secondary structure are likely to assist cyt c in crossing the mitochondrial membrane into the cytosol [57–59].
Conformational rearrangements away from the native protein structure are not uncommon in the cellular environment. Membrane translocation, as well as other protein functions such as ligand binding or signal transduction, relies on the flexibility of the protein structure [60–66]. Traditionally, because of the importance of the structure-function relationship, proteins have largely been discussed in terms of their native geometry. However, it has become clear that the unfolded and partially unfolded structures have critical functional roles. For example, to traverse the channels that run along the axes of the proteasome in the process of protein degradation by ATP-dependent proteases, the protein needs to be in the unfolded state [64]. Signal transduction involves chromophore and domain-specific polypeptide backbone conformational changes of the sensory and signaling proteins before they can interact with a partner protein [65]. And many proteins with little secondary structure only compact to the folded state after interaction with partner proteins or after binding to a substrate [66].
An important partly unfolded structure is the molten globule (MG), with compact native-like secondary structure and fluctuating tertiary structure [67–71]. In addition to transmembrane translocation, the MG species has been associated with the chaperone machinery and in human disease [72–82]. It is becoming clear that the unfolded (U), the native (N), and the MG and other partly unfolded states can each be important to functional success on the cellular level, as well as to understanding folding itself.
Folding of horse heart redcyt c MG
The important roles that the partially unfolded states of proteins appear to play in cellular processes make it even more critical to understand the dynamics of the interconversion between these states and the more rigid native conformations. This was one motivation leading us to use far-UV TRORD spectroscopy to study the kinetics of folding from the molten globule state of redcyt c. Another motivation was to investigate the still controversial mechanistic connection between this particular folding process, which proceeds under milder, more physiological conditions, and the U → N folding process, which proceeded under the harsher GuHCl denaturant conditions used in our earlier photoreduction studies. In particular, would the results of far-UV TRORD studies on redcyt c MG folding support the hypothesis proposed previously that redcyt c folds via a MG intermediate? As mentioned above, the fast phase (< 1 ms) of redcyt c folding was assigned to formation of an MG species and the slow phase (> 1 ms) was correlated with formation of N from MG [42]. The MG far-UV TRORD study thus addressed the question: if the initial species was a molten globule, would its folding dynamics (MG → N) be comparable to the slow phase that was observed previously for U → MG → N folding [83]?
An equilibrium model for the MG intermediate species was formed for these experiments by the addition of sodium dodecyl sulfate (SDS) to a solution of oxcyt c. Below the critical micelle concentration (CMC, 2.2 mM in 45 mM NaP [84]), interaction with SDS monomers led to the partial unfolding of oxcyt c, whereas binding of SDS micelles above the CMC facilitated the recovery of that secondary structure [84–87]. The addition of 0.5 mM SDS decreased the secondary structure content of oxcyt c by 30%, as assessed by far-UV CD. In contrast, redcyt c began unfolding only at SDS concentrations above 0.7 mM. At an intermediate SDS concentration, 0.65 mM, far- and near-UV CD and Soret and visible band MCD spectral analyses confirmed the presence of a MG state in the oxidized species. The near-UV CD spectra indicated that the native tertiary contacts were compromised and the MCD spectra indicated a perturbation of the heme ligation, giving largely His18-Fe-His coordination (85%) with a minor His18-Fe-Met component. Thus, the equilibrium data indicated that photoreduction of oxcyt c in 0.65 mM SDS could be expected to produce a prompt redcyt c photoproduct with the MG structure of the initial oxidized species, which would then fold to the native state. Indeed, the far-UV TRORD data showed a single, slow folding phase (τ ≈ 50 ms), with no evidence of the fast (< 1 ms) phase that was observed in the GuHCl photoreduction studies (Figure 3). Using TRORD data from a second SDS concentration, 0.5 mM, the time constant for folding was extrapolated to ∼1 ms in zero denaturant. A corresponding extrapolation of the GuHCl data gave a time constant of ∼ 5.5 ms in zero denaturant for the slow phase (MG → N) of the overall U → MG → N folding reaction. Remarkably, the ratio of these observed time constants was consistent with the hypothesis that they arose from the same underlying MG → N rate constant when one accounts for the 20–30% fractional content of MG intermediate that forms in the fast phase (U → MG) of U → MG → N folding in GuHCl. These results thus supported the hypothesis that the folding intermediate observed in the photoreduction studies of cyt c fully unfolded in GuHCl is indeed a productive MG intermediate.
CONCLUSIONS
A combination of time-resolved natural and magnetic CD/ORD studies of the early events in Fe(II) cytochrome c folding has identified a very fast (∼1 µs) helix formation process in this system that has been difficult to detect by other methods. These studies have further established that this process proceeds in what may be called the energy-landscape time regime. This is a time regime that precedes the full equilibration of unfolded chain conformers characterizing the classical pathway time regime in which folding kinetics may be described simply by a pre-exponential factor and activation free energy for passage over a transition state. These studies found qualitative and quantitative evidence that this folding process can proceed faster than conformational diffusion among the unfolded states of the polypeptide backbone (τd ≈ 3 µs). This result thus places this very fast folding process in a non-classical time regime in which folding may be better described by biased diffusion over a free energy landscape than by activated passage between the broad minima of transition state theory. This result also helps to resolve concerns that the folding kinetics of cytochrome c are somehow anomalous by showing that its unfolded chain dynamics are in fact reasonably consistent with those of other polypeptides of similar size [88]. More generally, this result bolsters the notion that an expression for the TST prefactor of protein folding, a fundamental parameter that remains an open question after decades of kinetic studies, may be derived from τd.
The mechanism of folding in cyt c remains controversial, particularly with regard to the existence and nature of possible pathways involving kinetic intermediates. Most fundamentally, our measurement of the unfolded conformational diffusion time tells us that we can now be more confident that such classical pathway thinking is indeed appropriate for events happening at times t >> τd. This certainly includes the milliseconds to seconds time scale over which the slow kinetic phase leading to the native fold proceeds.
What, then, does the very fast helix folding phase identified in our ns studies, strongly overlapped as it is with the non-classical regime t ≤ τd, tell us about possible classical folding pathways for this system? First, the observation of bona fide secondary structure formation on nanosecond to microsecond time scales in these constant denaturing solvent experiments undercuts the view advanced by some researchers that previous observations of cyt c kinetic intermediates in rapid denaturant-dilution experiments were artifacts of the solvent jump method, e.g., a simple polymer collapse unconnected to the secondary or tertiary structural transitions specific to polypeptides. Second, having strengthened the case for an observable kinetic intermediate in cyt c folding, what can we say about its nature? Molten globule states have long been considered promising candidates for kinetic folding intermediates in proteins, but recent fluorescence energy transfer measurements of structure in equilibrium molten globules have been interpreted as discounting this possibility in cyt c [89]. However, the noncanonical denaturant dependence of the fast folding phase and the correspondence between the slow folding rate in U → I → N and the folding rate from an SDS-prepared MG form discussed above together tend to suggest that I is a productive folding intermediate with molten globule character that is an obligatory step on the pathway to N. While the equilibrium molten globule forms of cyt c available to researchers may not be perfect models for the kinetic intermediate, the results reviewed here suggest that some type of collapsed state with near-native secondary structure and nonnative tertiary structure remains an important and productive candidate for future study.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant EB02056.
References
- 1.Srajer V, Royer WE., Jr Time-resolved x-ray crystallography of heme proteins. Methods Enzymology. 2008;437:379–395. doi: 10.1016/S0076-6879(07)37019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu K, Vaughn JL, Sienkiewicz A, Scholes CP, Fetrow JS. Kinetics and motional dynamics of spin-labeled yeast iso-1-cytochrome c: 1. Stopped-flow electron paramagnetic resonance as a probe for protein folding/unfolding of the C-terminal helix spin-labeled at cysteine 102. Biochemistry. 1997;36:2884–2897. doi: 10.1021/bi962155i. [DOI] [PubMed] [Google Scholar]
- 3.Callender RH, Dyer RB, Gilmanshin R, Woodruff WH. Fast events in protein folding: the time evolution of primary processes. Annu Rev Phys Chem. 1998;49:173–202. doi: 10.1146/annurev.physchem.49.1.173. [DOI] [PubMed] [Google Scholar]
- 4.Millar DP. Time-resolved fluorescence spectroscopy. Curr Opin Struct Biol. 1996;6:637–642. doi: 10.1016/s0959-440x(96)80030-3. [DOI] [PubMed] [Google Scholar]
- 5.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annual Review of Biophysics and Biomolecular Structure. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 6.Berova N, Nakanishi K, Woody R. Circular dichroism: Principles and applications. 2nd ed. New York: Wiley-VCH; 2000. [Google Scholar]
- 7.Lewis JW, Goldbeck RA, Kliger DS, Xie XL, Dunn RC, Simon JD. Time-Resolved Circular Dichroism Spectroscopy-Experiment, Theory, and Applications to Biological-Systems. Journal of Physical Chemistry. 1992;96:5243–5254. [Google Scholar]
- 8.Anson M, Bayley PM. Measurement of Circular Dichroism at Millisecond Time Resolution-Stopped-Flow Circular Dichroism System. Journal of Physics E-Scientific Instruments. 1974;7:481–486. [Google Scholar]
- 9.Ferrone FA, Hopfield JJ, Schnatterly SE. Measurement of Transient Circular Dichroism-New Kinetic Technique. Review of Scientific Instruments. 1974;45:1392–1396. [Google Scholar]
- 10.Anson M, Martin SR, Bayley PM. Transient CD Measurements at Submillisecond Time Resolution Application to Studies of Temperature-Jump Relaxation of Equilibria of Chiral Biomolecules. Review of Scientific Instruments. 1977;48:953–962. [Google Scholar]
- 11.Lewis JW, Tilton RF, Einterz CM, Milder SJ, Kuntz ID, Kliger DS. New technique for measuring circular dichroism changes on a nanosecond time scale. Application to (carbonmonoxy)myoglobin and (carbonmonoxy)hemoglobin. J. Phys. Chem. 1985;89:289–294. [Google Scholar]
- 12.Xie XL, Simon JD. Picosecond Time-Resolved Circular-Dichroism Spectroscopy-Experimental Details and Applications. Review of Scientific Instruments. 1989;60:2614–2627. [Google Scholar]
- 13.Zhang CF, Lewis JW, Cerpa R, Kuntz ID, Kliger DS. Nanosecond Circular Dichroism Spectral Measurements-Extension to the Far-Ultraviolet Region. Journal of Physical Chemistry. 1993;97:5499–5505. [Google Scholar]
- 14.Chen EF, Parker W, Lewis JW, Song PS, Kliger DS. Time-Resolved UV Circular Dichroism of Phytochrome A: Folding of the N-Terminal Region. Journal of the American Chemical Society. 1993;115:9854–9855. [Google Scholar]
- 15.Chen EF, Kliger DS. Time-resolved near UV circular dichroism and absorption studies of carbonmonoxymyoglobin photolysis intermediates. Inorganica Chimica Acta. 1996;242:149–158. [Google Scholar]
- 16.Chen E, Wood MJ, Fink AL, Kliger DS. Time-resolved circular dichroism studies of protein folding intermediates of cytochrome c. Biochemistry. 1998;37:5589–5598. doi: 10.1021/bi972369f. [DOI] [PubMed] [Google Scholar]
- 17.Jones CM, Henry ER, Hu Y, Chan CK, Luck SD, Bhuyan A, Roder H, Hofrichter J, Eaton WA. Fast events in protein folding initiated by nanosecond laser photolysis. Proc Natl Acad Sci U S A. 1993;90:11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen YX, Chen EF, Lewis JW, Kliger DS. Nanosecond time-resolved circular dichroism measurements using an upconverted Ti:sapphire laser. Review of Scientific Instruments. 1996;67:3010–3016. [Google Scholar]
- 19.Goldbeck RA, Dawes TD, Milder SJ, Lewis JW, Kliger DS. Measurement of Magnetic Circular Dichroism (MCD) on a Nanosecond Timescale. Chemical Physics Letters. 1989;156:545–549. [Google Scholar]
- 20.Goldbeck RA, Thomas YG, Chen E, Esquerra RM, Kliger DS. Multiple pathways on a protein-folding energy landscape: kinetic evidence. Proc Natl Acad Sci U S A. 1999;96:2782–2787. doi: 10.1073/pnas.96.6.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abel CJ, Goldbeck RA, Latypov RF, Roder H, Kliger DS. Conformational equilibration time of unfolded protein chains and the folding speed limit. Biochemistry. 2007;46:4090–4099. doi: 10.1021/bi0622930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro DB, Goldbeck RA, Che D, Esquerra RM, Paquette SJ, Kliger DS. Nanosecond optical rotatory dispersion spectroscopy: application to photolyzed hemoglobin-CO kinetics. Biophys J. 1995;68:326–334. doi: 10.1016/S0006-3495(95)80191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keston A, Lospalluto J. Simple ultrasensitive spectropolarimeters. Fed. Proc. 1953;12:229. [Google Scholar]
- 24.Moscowitz A. Theoretical Aspects of Optical Activity .1. Small Molecules. Advances in Chemical Physics. 1962;4:67–112. [Google Scholar]
- 25.Hoffman GW. Nanosecond Temperature-Jump Apparatus. Review of Scientific Instruments. 1971;42:1643–1647. [Google Scholar]
- 26.Phillips CM, Mizutani Y, Hochstrasser RM. Ultrafast thermally induced unfolding of RNase A. Proc Natl Acad Sci U S A. 1995;92:7292–7296. doi: 10.1073/pnas.92.16.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner DH, Sutin N, Beitz JV, Flynn GW. Laser Raman Temperature-Jump Study of Kinetics of Triiodide Equilibrium-Relaxation Times in 10−8-10−7 Second Range. Journal of the American Chemical Society. 1972;94 1554-&. [Google Scholar]
- 28.Chen EF, Wen YX, Lewis JW, Goldbeck RA, Kliger DS, Strauss CEM. Nanosecond laser temperature-jump optical rotatory dispersion: Application to early events in protein folding/unfolding. Review of Scientific Instruments. 2005;76 083120–083127. [Google Scholar]
- 29.Regenfuss P, Clegg RM, Fulwyler MJ, Barrantes FJ, Jovin TM. Mixing Liquids in Microseconds. Review of Scientific Instruments. 1985;56:283–290. [Google Scholar]
- 30.Chan CK, Hu Y, Takahashi S, Rousseau DL, Eaton WA, Hofrichter J. Submillisecond protein folding kinetics studied by ultrarapid mixing. Proc Natl Acad Sci U S A. 1997;94:1779–1784. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi S, Yeh SR, Das TK, Chan CK, Gottfried DS, Rousseau DL. Folding of cytochrome c initiated by submillisecond mixing. Nat Struct Biol. 1997;4:44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 32.Shastry MC, Luck SD, Roder H. A continuous-flow capillary mixing method to monitor reactions on the microsecond time scale. Biophys J. 1998;74:2714–2721. doi: 10.1016/S0006-3495(98)77977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hertzog DE, Michalet X, Jager M, Kong X, Santiago JG, Weiss S, Bakajin O. Femtomole mixer for microsecond kinetic studies of protein folding. Anal Chem. 2004;76:7169–7178. doi: 10.1021/ac048661s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knight JB, Vishwanath A, Brody JP, Austin RH. Hydrodynamic focusing on a silicon chip: Mixing nanoliters in microseconds. Physical Review Letters. 1998;80:3863–3866. [Google Scholar]
- 35.Lapidus LJ, Yao S, McGarrity KS, Hertzog DE, Tubman E, Bakajin O. Protein hydrophobic collapse and early folding steps observed in a microfluidic mixer. Biophys J. 2007;93:218–224. doi: 10.1529/biophysj.106.103077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter G. Flash Photolysis and Spectroscopy a New Method for the Study of Free Radical Reactions. Proceedings of the Royal Society of London Series A-Mathematical and Physical Sciences. 1950;200:284–300. [Google Scholar]
- 37.Pascher T, Chesick JP, Winkler JR, Gray HB. Protein folding triggered by electron transfer. Science. 1996;271:1558–1560. doi: 10.1126/science.271.5255.1558. [DOI] [PubMed] [Google Scholar]
- 38.Mines GA, Pascher T, Lee SC, Winkler JR, Gray HB. Cytochrome c folding triggered by electron transfer. Chem Biol. 1996;3:491–497. doi: 10.1016/s1074-5521(96)90097-6. [DOI] [PubMed] [Google Scholar]
- 39.Chen E, Goldbeck RA, Kliger DS. The earliest events in protein folding: A structural requirement for ultrafast folding in cytochrome c. J Am Chem Soc. 2004;126:11175–11181. doi: 10.1021/ja0400077. [DOI] [PubMed] [Google Scholar]
- 40.Chen EF, Wittung-Stafshede P, Kliger DS. Far-UV time-resolved circular dichroism detection of electron-transfer-triggered cytochrome c folding. Journal of the American Chemical Society. 1999;121:3811–3817. [Google Scholar]
- 41.Chen EF, Goldbeck RA, Kliger DS. Earliest events in protein folding: Submicrosecond secondary structure formation in reduced cytochrome c. Journal of Physical Chemistry A. 2003;107:8149–8155. [Google Scholar]
- 42.Chen E, Abel CJ, Goldbeck RA, Kliger DS. Non-native heme-histidine ligation promotes microsecond time scale secondary structure formation in reduced horse heart cytochrome c. Biochemistry. 2007;46:12463–12472. doi: 10.1021/bi7007273. [DOI] [PubMed] [Google Scholar]
- 43.Chan CK, Hofrichter J, Eaton WA. Optical triggers of protein folding. Science. 1996;274:628–629. doi: 10.1126/science.274.5287.628. [DOI] [PubMed] [Google Scholar]
- 44.Hagen SJ, Hofrichter J, Szabo A, Eaton WA. Diffusion-limited contact formation in unfolded cytochrome c: Estimating the maximum rate of protein folding. Proc Natl Acad Sci U S A. 1996;93:11615–11617. doi: 10.1073/pnas.93.21.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colon W, Wakem LP, Sherman F, Roder H. Identification of the predominant non-native histidine ligand in unfolded cytochrome c. Biochemistry. 1997;36:12535–12541. doi: 10.1021/bi971697c. [DOI] [PubMed] [Google Scholar]
- 46.Taler G, Navon G, Becker OM. The interaction of borate ions with cytochrome c surface sites: A molecular dynamics study. Biophysical Journal. 1998;75:2461–2468. doi: 10.1016/S0006-3495(98)77690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto Y. A 1H NMR study of structurally relevant inter-segmental hydrogen bond in cytochrome c. Biochim Biophys Acta. 1997;1343:193–202. doi: 10.1016/s0167-4838(97)00109-x. [DOI] [PubMed] [Google Scholar]
- 48.Hammack BN, Smith CR, Bowler BE. Denatured state thermodynamics: residual structure, chain stiffness and scaling factors. J Mol Biol. 2001;311:1091–1104. doi: 10.1006/jmbi.2001.4909. [DOI] [PubMed] [Google Scholar]
- 49.Godbole S, Hammack B, Bowler BE. Measuring denatured state energetics: deviations from random coil behavior and implications for the folding of iso-1-cytochrome c. J Mol Biol. 2000;296:217–228. doi: 10.1006/jmbi.1999.3454. [DOI] [PubMed] [Google Scholar]
- 50.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 51.Pierce MM, Nall BT. Coupled kinetic traps in cytochrome c folding: His-heme misligation and proline isomerization. J Mol Biol. 2000;298:955–969. doi: 10.1006/jmbi.2000.3700. [DOI] [PubMed] [Google Scholar]
- 52.Kurchan E, Roder H, Bowler BE. Kinetics of loop formation and breakage in the denatured state of iso-1-cytochrome c. J Mol Biol. 2005;353:730–743. doi: 10.1016/j.jmb.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 53.Krieger F, Fierz B, Bieri O, Drewello M, Kiefhaber T. Dynamics of unfolded polypeptide chains as model for the earliest steps in protein folding. J Mol Biol. 2003;332:265–274. doi: 10.1016/s0022-2836(03)00892-1. [DOI] [PubMed] [Google Scholar]
- 54.Chang IJ, Lee JC, Winkler JR, Gray HB. The protein-folding speed limit: intrachain diffusion times set by electron-transfer rates in denatured Ru(NH3)5(His-33)-Zn-cytochrome c. Proc Natl Acad Sci U S A. 2003;100:3838–3840. doi: 10.1073/pnas.0637283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 56.Sivakolundu SG, Mabrouk PA. Structure-function relationship of reduced cytochrome c probed by complete solution structure determination in 30% acetonitrile/water solution. Journal of Biological Inorganic Chemistry. 2003;8:527–539. doi: 10.1007/s00775-002-0437-0. [DOI] [PubMed] [Google Scholar]
- 57.Keilin D. The history of cell respiration and cytochrome. Cambridge U.P: Cambridge; 1966. [Google Scholar]
- 58.Pettigrew GW, Moore GR. Cytochromes c : Biological aspects. Berlin, New York: Springer-Verlag; 1987. [Google Scholar]
- 59.Jiang XJ, Wang XD. Cytochrome c-mediated apoptosis. Annual Review of Biochemistry. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 60.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. Journal of Molecular Biology. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 61.Ptitsyn OB. Molten globule and protein folding. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 62.Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins-Structure Function and Genetics. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 63.Tompa P. Intrinsically unstructured proteins. Trends in Biochemical Sciences. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 64.Prakash S, Matouschek A. Protein unfolding in the cell. Trends Biochem Sci. 2004;29:593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 65.Cusanovich MA, Meyer TE. Photoactive yellow protein: a prototypic PAS domain sensory protein and development of a common signaling mechanism. Biochemistry. 2003;42:4759–4770. doi: 10.1021/bi020690e. [DOI] [PubMed] [Google Scholar]
- 66.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 67.Ptitsyn OB. How the Molten Globule Became. Trends in Biochemical Sciences. 1995;20:376–379. doi: 10.1016/s0968-0004(00)89081-7. [DOI] [PubMed] [Google Scholar]
- 68.Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov S, Ptitsyn OB. α-Lactalbumin: compact state with fluctuating tertiary structure? FEBS Lett. 1981;136:311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- 69.Ohgushi M, Wada A. Molten globule state - A compact form of globular proteins with mobile side chains. FEBS Letters. 1983;164:21–24. doi: 10.1016/0014-5793(83)80010-6. [DOI] [PubMed] [Google Scholar]
- 70.Dolgikh DA, Abaturov LV, Bolotina IA, Brazhnikov EV, Bychkova VE, Gilmanshin RI, Lebedev YO, Semisotnov GV, Tiktopulo EI, Ptitsyn OB. Compact state of a protein molecule with pronounced small-scale mobility-Bovine α-Lactalbumin. European Biophysics Journal with Biophysics Letters. 1985;13:109–121. doi: 10.1007/BF00256531. [DOI] [PubMed] [Google Scholar]
- 71.Fink AL. Molten globule. Encycl. Life Sci. 2001:1–6. [Google Scholar]
- 72.Martin J, Langer T, Boteva R, Schramel A, Horwich AL, Hartl FU. Chaperonin-mediated protein folding at the surface of groEL through a 'molten globule'-like intermediate. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 73.Hartl FU, Hlodan R, Langer T. Molecular chaperones in protein-folding-the art of avoiding sticky situations. Trends in Biochemical Sciences. 1994;19:20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 74.Lu H, Golovanov AP, Alcock F, Grossmann JG, Allen S, Lian LY, Tokatlidis K. The structural basis of the TIM10 chaperone assembly. Journal of Biological Chemistry. 2004;279:18959–18966. doi: 10.1074/jbc.M313046200. [DOI] [PubMed] [Google Scholar]
- 75.Rajaraman K, Raman B, Ramakrishna T, Rao CM. The chaperone-like α-crystallin forms a complex only with the aggregation-prone molten globule state of α-lactalbumin. Biochemical and Biophysical Research Communications. 1998;249:917–921. doi: 10.1006/bbrc.1998.9242. [DOI] [PubMed] [Google Scholar]
- 76.Horwich A. Protein aggregation in disease: a role for folding intermediates forming specific multimeric interactions. Journal of Clinical Investigation. 2002;110:1221–1232. doi: 10.1172/JCI16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 78.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 79.Uversky VN, Fink AL. Protein misfolding, aggregation and conformational diseases: Part A, protein aggregation and conformational diseases. New York, N.Y: Springer Science+Business Media; 2006. [Google Scholar]
- 80.Uversky VN, Fink AL. Protein misfolding, aggregation, and conformational diseases. New York: Springer; 2007. [Google Scholar]
- 81.Almstedt K, Lundqvist M, Carlsson J, Karlsson M, Persson B, Jonsson BH, Carlsson U, Hammarstrom P. Unfolding a folding disease: Folding, misfolding and aggregation of the marble brain syndrome-associated mutant H107Y of human carbonic anhydrase II. Journal of Molecular Biology. 2004;342:619–633. doi: 10.1016/j.jmb.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 82.Lindgren M, Sorgjerd K, Hammarstrom P. Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophysical Journal. 2005;88:4200–4212. doi: 10.1529/biophysj.104.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen E, Van Vranken V, Kliger DS. The folding kinetics of the SDS-induced molten globule form of reduced cytochrome c. Biochemistry. 2008;47:5450–5459. doi: 10.1021/bi702452u. [DOI] [PubMed] [Google Scholar]
- 84.Takeda K, Takahashi K, Batra PP. Kinetic aspects of the interaction of horse heart cytochrome c with sodium sodecyl sulfate. Archives of Biochemistry and Biophysics. 1985;236:411–417. doi: 10.1016/0003-9861(85)90641-1. [DOI] [PubMed] [Google Scholar]
- 85.Das TK, Mazumdar S, Mitra S. Characterization of a partially unfolded structure of cytochrome c induced by sodium dodecyl sulphate and the kinetics of its refolding. European Journal of Biochemistry. 1998;254:662–670. doi: 10.1046/j.1432-1327.1998.2540662.x. [DOI] [PubMed] [Google Scholar]
- 86.Hiramatsu K, Yang JT. Cooperative binding of hexadecyltrimethylammonium chloride and sodium dodecyl sulfate to cytochrome c and the resultant change in protein conformation. Biochimica Et Biophysica Acta. 1983;743:106–114. doi: 10.1016/0167-4838(83)90423-5. [DOI] [PubMed] [Google Scholar]
- 87.Oellerich S, Wackerbarth H, Hildebrandt P. Conformational equilibria and dynamics of cytochrome c induced by binding of sodium dodecyl sulfate monomers and micelles. European Biophysics Journal with Biophysics Letters. 2003;32:599–613. doi: 10.1007/s00249-003-0306-y. [DOI] [PubMed] [Google Scholar]
- 88.Winkler JR. Cytochrome c folding dynamics. Current Opinion in Chemical Biology. 2004;8:169–174. doi: 10.1016/j.cbpa.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 89.Pletneva EV, Gray HB, Winkler JR. Nature of the cytochrome c molten globule. Journal of the American Chemical Society. 2005;127:15370–15371. doi: 10.1021/ja0555318. [DOI] [PubMed] [Google Scholar]