Abstract
Human microRNAs (miRNAs) are potent regulators of gene expression and thus involved in a broad range of biological processes. The objective of this study was to update the properties of human miRNAs and to search for SNPs and CNVs with potential effects on them. Based on the latest miRBase 13.0 database, we identified 380 (53.9%) precursor miRNAs (pre-miRNAs) embedded in gene loci that are enriched in biological processes such as “Neuronal activities”, “Cell Cycle” and “Protein phosphorylation” (Bonferroni p < 0.05). Gene lengths of the pre-miRNA host genes are significantly larger than other genes in the genome (p < 2.2E-16). Using data mining public resources, we performed a genome-scale search for the regulatory polymorphisms in the loci of pre-miRNAs and their related genes. Altogether, we found 187 SNPs in the pre-miRNAs, 497 consensus SNPs in the seed-matching untranslated regions of target genes, 385 CNVs harboring pre-miRNA precursors and 9 CNVs covering important miRNA processing genes. We also noticed that minimum free energy changed by pre-miRNA-residing SNPs could be ranked by the order from low to high as the SNPs in the loop domain, the SNPs in the adjacent stem and basal stem domains, and the SNPs in mature miRNA and its complementary sequence domains (p = 0.0065). With a full list of miRNA-related polymorphisms, this study will facilitate future association studies between the genetic polymorphisms in miRNA targets or pre-miRNAs and the disease susceptibility or therapeutic outcome.
Keywords: microRNA, miRNA, gene, pathway, gene ontology, SNP, CNV
Introduction
As a new class of abundantly distributed small non-coding RNA molecules, miRNAs are initially transcribed as primary miRNA (pri-miRNAs) that are further processed through stem-loop pre-miRNAs into a single-stranded mature form.1 Generally, miRNAs partially complement the 3′-untranslated regions (UTR) of target mRNAs, and subsequently invoke a series of posttranscriptional silencing events on the target genes. These include intervention of translational initiation and elongation, induction of deadenylation and interruption of mRNA and protein synthesis.2,3 The core of miRNA’s pairing sequence is termed “seed”, which is usually conserved across multiple species.4 Since this seed sequence is on average 8 nucleotides in length and critical for the miRNA-target binding, the miRNAs are estimated to have anywhere from several to thousands of targets.4 Therefore, mature miRNAs as potent regulators of gene expression have been implicated in various biological development processes and disease progression.5,6
SNPs within the sequences of human miRNAs and their targets have been shown to have impact on various phenotypes including blood pressure,7 drug resistance,8 outcomes of therapeutic intervention,9 abnormal psychiatry disorder,10 the development of gastric mucosal atrophy,11 the risk of diseases that consist of asthma,12 diabetes,13 Parkinson,14 cancers of colorectal,15 breast,16 bladder,17 papillary thyroid,18,19 lung20,21 and esophageal.22 Moreover, miRNAs and their target genes may be located at the genomic regions of high instability, a feature often observed in cancer and other genetic diseases. To note, specific deletions of key enzymes such as Dicer1 (dicer 1, ribonuclease type III) may cause global impairment of miRNA processing leading to severe abnormality.23
As shown in Table 1, several researchers have uncovered links between genomic variations and miRNAs.9,15,16,18–22,24–36 However, a systematic update of the genetic factors that influence miRNA activities according to the most recent miRBase is not yet in place. Taking advantage of the latest miRNA databases and bioinformatics tools, we analyzed human miRNA for their biological properties and performed a genome-wide scan for functional genetic variations that may potentially affect human miRNA processing and targeting.
Table 1.
The studies on genetic variants related to miRNAs in humans
| Discoveries of genetic variants related to miRNAs | Reference |
|---|---|
| 10 SNPs in miRNA precursors | 24 |
| 339 SNPs in conserved seed-matching regions of target gene | 25 |
| 483 SNPs in conserved seed-matching regions of target gene | 26 |
| 1 SNP in the seed-matching region of target gene | 27 |
| A database with 22758 SNPs in the miRNA target sites or with potential ability to creat novel miRNA target sites | 28 |
| 12 SNPs in miRNA precursors | 29 |
| 1 SNP in the seed-matching region of target gene | 30 |
| About 400 SNPs in the miRNA target sites; 250 SNPs with the potential ability to creat novel miRNA target sites | 31 |
| 265 SNPs in the miRNA target sites | 32 |
| 79 SNPs in the seed-matching regions of target genes; 7 SNPs in miRNA precursors; 1 SNP in the mature miRNA | 33 |
| 57 SNPs in the seed-matching regions of target genes | 15 |
| 1 SNP in the seed-matching region of target gene | 8 |
| A joint database with SNPs in the miRNAs and their targets | 35 |
| One SNP (rs11614913) in hsa-mir-196a2 | 9, 20 |
| One functional SNP in a miRNA target site. | 16 |
| 41 genetic variations in 26 microRNA-related genes. | 22 |
| One SNP in a let-7 microRNA complementary site. | 21 |
| 23 SNPs in 11 genes in the miRNA biogenesis pathway, 7 SNPs in 7 pre-miRNAs, and 10 SNPs in 8 pri-miRNAs | 74 |
| One SNP (rs2910164) in pre-miR-146a | 18, 19 |
| 27 SNPs in hairpin structures of pre-miRNAs | 36 |
| 188 SNPs in the miRNA precursors; 497 consensus SNPs in the seed-matching regions of target genes; 385 CNVs harboring miRNA precursors; 9 CNVs covering important miRNA processing genes. | Present study |
Results
Human miRNAs
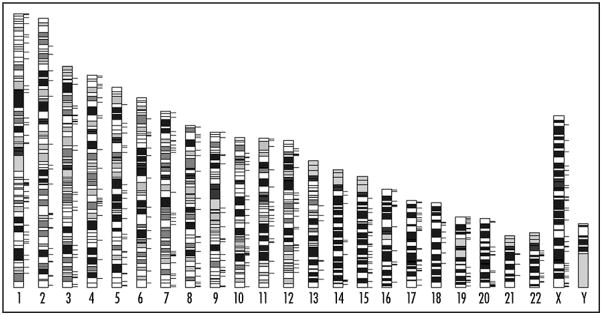
As shown in the Figure 1, there are a total of 718 loci for human pre-miRNA genes in the genome based on the miRBase 13.0. The pre-miRNAs with multiple copies include mir-1184 (3 copies), mir-1233 (2 copies), mir-1244 (4 copies), mir-1302-2 (4 copies), mir-1972 (2 copies), mir-1974 (2 copies), mir-1977 (2 copies) and mir-1978 (2 copies). These 706 pre-miRNAs can be processed into 885 mature miRNAs (including mirR*products) with lengths ranging from 17 to 27 nucleotides. Based on the common seed sequence of mature miRNA products, the pre-miRNAs may be further grouped into families. In the current released version of miRBase, there are altogether 381 miR-families (644 members) comprising of up to 42 members in humans. The largest pre-miRNA families include mir-515 (n = 42), mir-548 (n = 31), mir-154 (n = 19), mir-506 (n = 18) and let-7 (n = 12).
Figure 1.
Genomic distribution of the pre-miRNAs in humans. The ticks in the right of ideogram are the locations of pre-miRNAs. The darker bands in the ideogram are AT-rich, while the lighter bands are GC-rich.
Pre-miRNA host genes
There are 380 human pre-miRNA genes residing in the loci of 340 protein-coding-genes (PCGs) (Suppl. Table 1). The genes hosting the most pre-miRNAs include HTR2C (5 pre-miRNAs), RTL1 (5 pre-miRNAs) and LARP7 (5 pre-miRNAs). By comparing the length of gene-spanning regions between pre-miRNA host PCGs and other PCGs in humans, our results showed that the pre-miRNA-host PCGs have significant larger gene-spanning regions (Fig. 2A, Kolmogorov-Smirnov test, D = 0.38, p < 2.2E-16). In addition, the host genes were analyzed for enriched categories in the Gene Ontology (GO)37 and Kyoto Encyclopedia of Genes and Genomes (KEGG)38 pathways. As shown in Table 2, the host genes are significantly enriched in the pathways of “Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade” and “Coenzyme A biosynthesis”, the biological processes of “Neuronal activities”, “Protein phosphorylation”, “Cell cycle” and “Cell structure and motility”, and the molecular function of “Kinase” (Bonferroni corrected p < 0.05). Among these pre-miRNA host genes, only 33 genes are not annotated and these are significantly less than those found in all NCBI annotated genes (p < 0.0001, df = 1, χ2 = 67.98, OR = 0.25). In addition, the category of “unclassified” for both the pathway and GO analysis are significantly underrepresented for miRNA host genes (Bonferroni p < 0.05), implying that important functions are implicated for miRNA host genes. To note, the pre-miRNA host PCGs trend to have larger gene length than the rest PCGs in human genome, thus they are more likely (by chance) to harbor miRNAs. And this may potentially cause some biased observations for certain GO terms.
Figure 2.
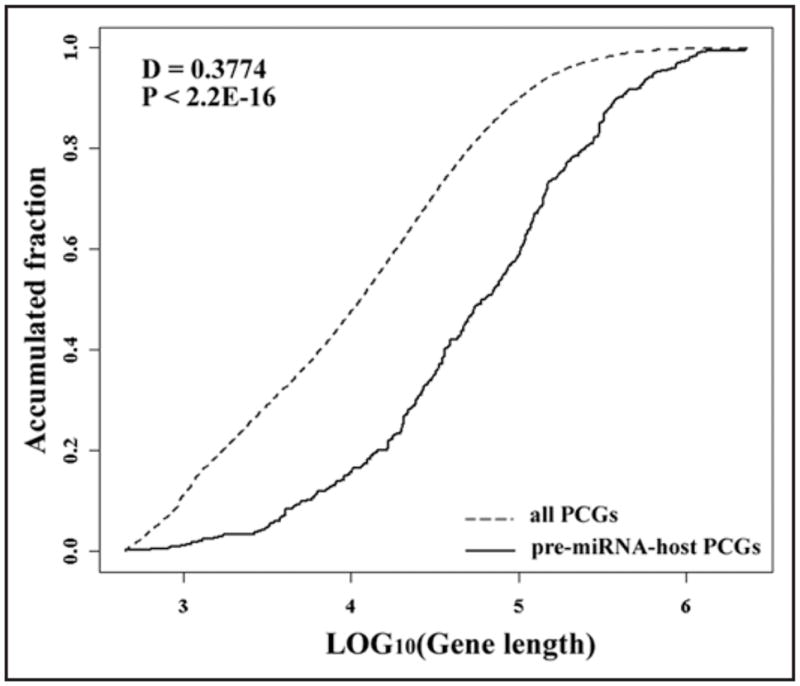
Gene length comparison between all PCGs and miRNA host PCGs.
Table 2.
Pathways and GO analysis of pre-miRNA host genes
| Categories | Total | Observed | Expected | +/− | Bonferroni P |
|---|---|---|---|---|---|
| Pathways | |||||
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 49 | 8 | 0.58 | + | 0.0000306 |
| Coenzyme A biosynthesis | 7 | 3 | 0.08 | + | 0.0149 |
| Unclassified | 22436 | 246 | 267.32 | − | 0.0418 |
| Biological Process | |||||
| Biological process unclassified | 11321 | 99 | 134.89 | − | 0.00053 |
| Neuronal activities | 569 | 17 | 6.78 | + | 0.0178 |
| Protein phosphorylation | 660 | 20 | 7.86 | + | 0.0316 |
| Cell cycle | 1009 | 24 | 12.02 | + | 0.0366 |
| Cell structure and motility | 1148 | 26 | 13.68 | + | 0.0459 |
| Molecular Function | |||||
| Molecular function unclassified | 10934 | 96 | 130.27 | − | 0.00102 |
| Kinase | 684 | 18 | 8.15 | + | 0.048 |
SNPs and CNVs in pre-miRNAs
In the present study, we performed a genome-scale search for both SNPs and CNVs that may potentially affect miRNA processing or targeting. Our results revealed that only 188 SNPs are located at 138 pre-miRNA regions (Table 3). In contrast, on average there are over 300 SNPs per 100 bps in the flanking regions of all the pre-miRNAs (Fig. 2B). This observation agrees with the previous finding based on the miRNAs in the earlier miRBase released version.31,36 Among the SNPs in the hairpins of pre-miRNAs, there are 16 SNPs in adjacent stem, 77 SNPs in the basal stem, 17 SNPs in the loop, 44 SNPs in mature miRNA, 54 SNPs in the complementary sequence of mature miRNA (Table 3). We use RNAfold web server to determine the minimum free energy (MFE) of hairpin structures for all the SNP-residing pre-miRNAs.39 The changes of minimum free energy (MFE) by the SNPs in the pre-miRNAs are also given in the Table 3. Significantly more MFE changes are caused by SNPs in mature miRNA and its complementary sequence domains than the SNPs in the adjacent stem and basal stem domains that are followed by SNPs in the loop domain (Fig. 3, Kruskal-Wallis test, χ2 = 14.25, df = 4, p = 0.0065).
Table 3.
SNPs in the hairpin structures of pre-miRNAs
| pre_name | SNP | Allele | SNP_position | Change of MFE (% of changes)a | Location | Reference mature_miR |
|---|---|---|---|---|---|---|
| mir-663b | rs62165009 | A/G | chr2: 132731049 | 2.5 (5.20%) | Adjacent_stem | miR-663b |
| mir-559 | rs58450758 | C/T | chr2: 47458370 | 1.8 (3.05%) | Adjacent_stem | miR-559 |
| mir-1324 | rs7614638 | C/T | chr3: 75762634 | 0.1 (0.31%) | Adjacent_stem | miR-1324 |
| mir-1324 | rs3008994 | C/G | chr3: 75762635 | 3 (10.17%) | Adjacent_stem | miR-1324 |
| mir-1303 | rs34889453 | -/A | chr5: 154045576 | 1.9 (4.52%) | Adjacent_stem | miR-1303 |
| mir-1303 | rs33982250 | -/A | chr5: 154045578 | 3.55 (8.80%) | Adjacent_stem | miR-1303 |
| mir-183 | rs41281222 | C/T | chr7: 129202042 | 1.8 (4.49%) | Adjacent_stem | miR-183 |
| mir-1299 | rs62555121 | A/T | chr9: 68292091 | 2.32 (7.25%) | Adjacent_stem | miR-1299 |
| mir-612 | rs12803915 | A/G | chr11: 64968555 | 0.8 (1.59%) | Adjacent_stem | miR-612 |
| mir-548l | rs11020790 | C/T | chr11: 93839358 | 0.4 (0.96%) | Adjacent_stem | miR-548l |
| mir-620 | rs5801168 | -/AT | chr12: 115070809 | 0.7 (2.50%) | Adjacent_stem | miR-620 |
| mir-300 | rs12894467 | C/T | chr14: 100577480 | 0 (0.00%) | Adjacent_stem | miR-300 |
| mir-1233 | rs347882 | C/G | chr15: 32461618 | 0.7 (1.53%) | Adjacent_stem | miR-1233 |
| mir-1233 | rs347882 | C/G | chr15: 32607839 | 0.7 (1.53%) | Adjacent_stem | miR-1233 |
| mir-1282 | rs11269 | G/T | chr15: 41873201 | 0.5 (1.27%) | Adjacent_stem | miR-1282 |
| mir-27a | rs11671784 | A/G | chr19: 13808296 | 0.8 (2.13%) | Adjacent_stem | miR-27a |
| mir-1302-2 | rs11266858 | A/G | chr1: 20239 | 0.5 (0.81%) | basal_stem | miR-1302 |
| mir-1302-2 | rs4248191 | G/T | chr1: 20274 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-1302-2 | rs422363 | C/T | chr1: 20352 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-1977 | rs9783068 | C/T | chr1: 556068 | 0.1 (0.49%) | basal_stem | miR-1977 |
| mir-1977 | rs41453547 | A/G | chr1: 556069 | 2.2 (12.02%) | basal_stem | miR-1977 |
| mir-1302-3 | rs2441622 | A/G | chr2: 114057020 | 0.1 (0.18%) | basal_stem | miR-1302 |
| mir-1302-3 | rs7589328 | C/T | chr2: 114057098 | 1.1 (1.99%) | basal_stem | miR-1302 |
| mir-1302-3 | rs6542147 | A/G | chr2: 114057133 | 0.5 (0.89%) | basal_stem | miR-1302 |
| mir-149 | rs2292832 | C/T | chr2: 241044176 | 3.9 (7.51%) | basal_stem | miR-149 |
| mir-558 | rs35999329 | -/TGTG | chr2: 32610742 | 3.8 (9.31%) | basal_stem | miR-558 |
| mir-216a | rs41291179 | A/T | chr2: 56069594 | 1.5 (3.83%) | basal_stem | miR-216a |
| mir-548i-1 | rs34864809 | -/G | chr3: 126992064 | 0 (0.00%) | basal_stem | miR-548i |
| mir-1324 | rs3008993 | A/G | chr3: 75762696 | 0 (0.00%) | basal_stem | miR-1324 |
| mir-577 | rs34115976 | C/G | chr4: 115797446 | 4.9 (11.11%) | basal_stem | miR-577 |
| mir-943 | rs3034718 | -/CT | chr4: 1957986 | 2.6 (6.09%) | basal_stem | miR-943 |
| mir-943 | rs35401110 | -/CT | chr4: 1957987 | 0.6 (1.43%) | basal_stem | miR-943 |
| mir-943 | rs1077020 | C/T | chr4: 1957991 | 0.9 (2.12%) | basal_stem | miR-943 |
| mir-1289-2 | rs35296450 | C/G | chr5: 132791194 | 1.3 (3.78%) | basal_stem | miR-1289 |
| mir-1289-2 | rs35731356 | -/G | chr5: 132791203 | 0.7 (2.07%) | basal_stem | miR-1289 |
| mir-585 | rs62376934 | A/G | chr5: 168623190 | 2.9 (5.28%) | basal_stem | miR-585 |
| mir-9-2 | rs41265488 | A/T | chr5: 87998503 | 0.7 (1.78%) | basal_stem | miR-9 |
| mir-583 | rs10697860 | -/ATAAA | chr5: 95440607 | 0 (0.00%) | basal_stem | miR-583 |
| mir-339 | rs13232101 | G/T | chr7: 1029100 | 0 (0.00%) | basal_stem | miR-339-3p |
| mir-595 | rs4909237 | C/T | chr7: 158018264 | 0.6 (1.90%) | basal_stem | miR-595 |
| mir-489 | rs35930643 | -/A | chr7: 92951259 | 3.2 (8.99%) | basal_stem | miR-489 |
| mir-1208 | rs56863230 | C/G | chr8: 129231548 | 0 (0.00%) | basal_stem | miR-1208 |
| mir-1208 | rs2648841 | G/T | chr8: 129231615 | 3.5 (15.84%) | basal_stem | miR-1208 |
| mir-486 | rs59908561 | -/G | chr8: 41637116 | 0 (0.00%) | basal_stem | miR-486-3p |
| mir-1302-2 | rs11266858 | A/G | chr9: 20154 | 0.5 (0.81%) | basal_stem | miR-1302 |
| mir-1302-2 | rs4248191 | G/T | chr9: 20189 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-1302-2 | rs422363 | C/T | chr9: 20267 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-202 | rs12355840 | C/T | chr10: 134911103 | 0.3 (0.51%) | basal_stem | miR-202 |
| mir-605 | rs2043556 | C/T | chr10: 52729412 | 2.6 (4.97%) | basal_stem | miR-605 |
| mir-1908 | rs174561 | C/T | chr11: 61339284 | 3.2 (7.08%) | basal_stem | miR-1908 |
| mir-194-2 | rs11231898 | A/G | chr11: 64415412 | 1.9 (3.85%) | basal_stem | miR-194 |
| mir-612 | rs550894 | A/C | chr11: 64968516 | 0.2 (0.39%) | basal_stem | miR-612 |
| mir-548c | rs17120527 | A/G | chr12: 63302567 | 0 (0.00%) | basal_stem | miR-548c-3p |
| mir-141 | rs34385807 | -/C | chr12: 6943605 | 4.6 (9.35%) | basal_stem | miR-141 |
| mir-617 | rs12815353 | C/G | chr12: 79750457 | 0 (0.00%) | basal_stem | miR-617 |
| mir-492 | rs2289030 | C/G | chr12: 93752417 | 1.7 (4.27%) | basal_stem | miR-492 |
| mir-622 | rs59274393 | C/T | chr13: 89681529 | N.A. | basal_stem | miR-622 |
| mir-18a | rs41275866 | C/G | chr13: 90801010 | 0.3 (1.36%) | basal_stem | miR-18a* |
| mir-329-1 | rs34557733 | -/A | chr14: 100562882 | 0 (0.00%) | basal_stem | miR-329 |
| mir-1185-2 | rs11844707 | A/G | chr14: 100580366 | 3.3 (9.37%) | basal_stem | miR-1185 |
| mir-624 | rs57264777 | A/T | chr14: 30553694 | N.A. | basal_stem | miR-624 |
| mir-1260 | rs28909969 | -/T | chr14: 76802381 | 3.1 (17.67%) | basal_stem | miR-1260 |
| mir-1302-2 | rs422363 | C/T | chr15: 100318199 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-1302-2 | rs4248191 | G/T | chr15: 100318277 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-1302-2 | rs11266858 | A/G | chr15: 100318312 | 0.5 (0.81%) | basal_stem | miR-1302 |
| mir-211 | rs34520022 | -/G | chr15: 29144537 | 0.8 (1.77%) | basal_stem | miR-211 |
| mir-147b | rs56073218 | C/G | chr15: 43512547 | 6 (21.05%) | basal_stem | miR-147b |
| mir-7-2 | rs41276930 | C/T | chr15: 86956077 | 1.7 (3.62%) | basal_stem | miR-7 |
| mir-140 | rs7205289 | A/C | chr16: 68524506 | 2.4 (4.56%) | basal_stem | miR-140-3p |
| mir-423 | rs6505162 | A/C | chr17: 25468309 | 0 (0.00%) | basal_stem | miR-423-3p |
| mir-1253 | rs7217038 | A/T | chr17: 2598127 | 0 (0.00%) | basal_stem | miR-1253 |
| mir-193a | rs60406007 | G/T | chr17: 26911146 | 4 (8.55%) | basal_stem | miR-193a-5p |
| mir-365-2 | rs35143473 | -/T | chr17: 26926632 | 1.5 (3.89%) | basal_stem | miR-365 |
| mir-187 | rs41274312 | A/G | chr18: 31738790 | 0.81 (1.72%) | basal_stem | miR-187 |
| mir-639 | rs35149836 | A/G | chr19: 14501439 | 0 (0.00%) | basal_stem | miR-639 |
| mir-1302-2 | rs11266858 | A/G | chr19: 22983 | 0.5 (0.81%) | basal_stem | miR-1302 |
| mir-1302-2 | rs4248191 | G/T | chr19: 23018 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-1302-2 | rs422363 | C/T | chr19: 23096 | 0.1 (0.16%) | basal_stem | miR-1302 |
| mir-1283-1 | rs57111412 | A/G | chr19: 58883555 | 4.8 (12.24%) | basal_stem | miR-1283 |
| mir-521-2 | rs13382089 | G/T | chr19: 58911666 | 4.3 (14.14%) | basal_stem | miR-521 |
| mir-516b-2 | rs10583889 | -/TT | chr19: 58920586 | 0.9 (2.41%) | basal_stem | miR-516b* |
| mir-518a-1 | rs61636451 | A/G | chr19: 58926153 | N.A. | basal_stem | miR-518a-3p |
| mir-516a-1 | rs2569389 | A/G | chr19: 58951814 | 4.1 (8.99%) | basal_stem | miR-516a-3p |
| mir-220b | rs1053262 | C/G | chr19: 6447045 | 0.2 (0.53%) | basal_stem | miR-220b |
| mir-663 | rs7266947 | A/C | chr20: 26136912 | 1.1 (2.19%) | basal_stem | miR-663 |
| mir-499 | rs7267163 | C/T | chr20: 33041937 | 2.2 (3.66%) | basal_stem | miR-499-3p |
| mir-646 | rs6513496 | C/T | chr20: 58316929 | 2.4 (7.02%) | basal_stem | miR-646 |
| mir-1-1 | rs6122014 | C/T | chr20: 60561960 | 0.9 (3.00%) | basal_stem | miR-1 |
| mir-941-1 | rs56202554 | C/T | chr20: 62021238 | 1.9 (3.63%) | basal_stem | miR-941 |
| mir-941-1 | rs55795631 | C/T | chr20: 62021321 | 0 (0.00%) | basal_stem | miR-941 |
| mir-941-1 | rs6089780 | A/G | chr20: 62021324 | 4.1 (8.17%) | basal_stem | miR-941 |
| mir-650 | rs5996397 | C/G | chr22: 21495340 | 0.8 (2.22%) | basal_stem | miR-650 |
| mir-548j | rs4822739 | C/G | chr22: 25281185 | 3.1 (6.13%) | basal_stem | miR-548j |
| mir-146a | rs61270459 | C/G | chr5: 159844984 | N.A. | loop | miR-146a |
| mir-1274a | rs318039 | C/T | chr5: 41511523 | 3.5 (17.95%) | loop | miR-1274a |
| mir-581 | rs788517 | A/G | chr5: 53283143 | 0 (0.00%) | loop | miR-581 |
| mir-96 | rs41274239 | A/G | chr7: 129201810 | 1 (2.91%) | loop | miR-96 |
| mir-1307 | rs7911488 | A/G | chr10: 105144079 | 0 (0.00%) | loop | miR-1307 |
| mir-2110 | rs17091403 | C/T | chr10: 115923895 | 1.5 (4.40%) | loop | miR-2110 |
| mir-1265 | rs11259096 | C/T | chr10: 14518624 | 1 (2.00%) | loop | miR-1265 |
| mir-620 | rs10549054 | -/TA | chr12: 115070798 | 0.3 (1.09%) | loop | miR-620 |
| mir-656 | rs58834075 | C/T | chr14: 100602846 | 0.6 (2.60%) | loop | miR-656 |
| mir-1233 | rs347881 | C/T | chr15: 32461601 | 1.6 (3.44%) | loop | miR-1233 |
| mir-1233 | rs347881 | C/T | chr15: 32607822 | 1.6 (3.44%) | loop | miR-1233 |
| mir-27a | rs895819 | C/T | chr19: 13808292 | 0 (0.00%) | loop | miR-27a* |
| mir-639 | rs45556632 | C/G | chr19: 14501403 | 1.7 (4.09%) | loop | miR-639 |
| mir-516b-2 | rs10670323 | -/AAAGA | chr19: 58920554 | 2.1 (5.65%) | loop | miR-516b |
| mir-516b-2 | rs33953969 | -/AAAGA | chr19: 58920555 | 2.1 (5.65%) | loop | miR-516b* |
| mir-521-1 | rs2561251 | A/G | chr19: 58943749 | N.A. | loop | miR-521 |
| mir-650 | rs11558654 | A/T | chr22: 21495309 | 0 (0.00%) | loop | miR-650 |
| mir-92b | rs12759620 | C/G | chr1: 153431668 | 3.7 (5.60%) | mature_mirR | miR-92b |
| mir-1977 | rs2854138 | A/G | chr1: 556124 | 1.8 (9.63%) | mature_mirR | miR-1977 |
| mir-34a | rs35301225 | A/C/T | chr1: 9134389 | 4.7 (10.22%) | mature_mirR | miR-34a |
| mir-1978 | rs55723650 | A/G | chr2: 149355840 | 1.8 (13.14%) | mature_mirR | miR-1978 |
| mir-1978 | rs56489998 | C/T | chr2: 149355844 | 0 (0.00%) | mature_mirR | miR-1978 |
| mir-568 | rs28632138 | G/T | chr3: 115518077 | 1.5 (4.92%) | mature_mirR | miR-568 |
| mir-1324 | rs10155043 | C/T | chr3: 75762664 | 0 (0.00%) | mature_mirR | miR-1324 |
| mir-1255b-1 | rs6841938 | A/G | chr4: 36104443 | 2.5 (17.01%) | mature_mirR | miR-1255b |
| mir-585 | rs62376935 | C/T | chr5: 168623213 | 7.1 (14.85%) | mature_mirR | miR-585 |
| mir-581 | rs1694089 | G/T | chr5: 53283151 | 0.7 (1.84%) | mature_mirR | miR-581 |
| mir-581 | rs810917 | A/G | chr5: 53283157 | 6.2 (19.07%) | mature_mirR | miR-581 |
| mir-449b | rs10061133 | A/G | chr5: 54502301 | 0 (0.00%) | mature_mirR | miR-449b |
| mir-590 | rs6971711 | C/T | chr7: 73243535 | 2.4 (8.16%) | mature_mirR | miR-590-3p |
| mir-25 | rs41274221 | C/T | chr7: 99529136 | 1.8 (5.00%) | mature_mirR | miR-25 |
| mir-1322 | rs59878596 | C/T | chr8: 10720304 | 0.1 (0.89%) | mature_mirR | miR-1322 |
| mir-596 | rs61388742 | C/T | chr8: 1752832 | 2.8 (8.31%) | mature_mirR | miR-596 |
| mir-608 | rs58078477 | C/G | chr10: 102724756 | N.A. | mature_mirR | miR-608 |
| mir-608 | rs4919510 | C/G | chr10: 102724768 | 1.4 (4.23%) | mature_mirR | miR-608 |
| mir-938 | rs12416605 | C/T | chr10: 29931266 | 1.1 (3.05%) | mature_mirR | miR-938 |
| mir-606 | rs34610391 | -/A | chr10: 76982290 | 0 (0.00%) | mature_mirR | miR-606 |
| mir-548l | rs13447640 | A/G | chr11: 93839369 | 2.5 (6.39%) | mature_mirR | miR-548l |
| mir-431 | rs12884005 | A/G | chr14: 100417161 | 0 (0.00%) | mature_mirR | miR-431* |
| mir-379 | rs61991156 | A/G | chr14: 100558167 | 0.6 (2.31%) | mature_mirR | miR-379 |
| mir-299 | rs41286566 | C/T | chr14: 100559898 | 2.9 (7.71%) | mature_mirR | miR-299-5p |
| mir-412 | rs61992671 | A/G | chr14: 100601607 | 1.6 (4.61%) | mature_mirR | miR-412 |
| mir-208b | rs2754157 | A/T | chr14: 22957059 | 1.5 (5.03%) | mature_mirR | miR-208b |
| mir-154 | rs41286570 | A/G | chr14:100595880 | 0.4 (1.1%) | mature_mirR | miR-154 |
| mir-1268 | rs28599926 | C/T | chr15: 20014635 | 0.7 (2.73%) | mature_mirR | miR-1268 |
| mir-627 | rs2620381 | A/C | chr15: 40279140 | 4.7 (9.07%) | mature_mirR | miR-627 |
| mir-1276 | rs34381260 | -/T | chr15: 84114798 | 4.1 (13.95%) | mature_mirR | miR-1276 |
| mir-940 | rs35356504 | -/C | chr16: 2261821 | 0 (0.00%) | mature_mirR | miR-940 |
| mir-662 | rs9745376 | A/G | chr16: 760250 | 0.5 (1.11%) | mature_mirR | miR-662 |
| mir-548h-3 | rs9913045 | A/G | chr17: 13387649 | 2.5 (4.85%) | mature_mirR | miR-548h |
| mir-423 | rs61093106 | A/C | chr17: 25468297 | N.A. | mature_mirR | miR-423-3p |
| mir-122 | rs41292412 | C/T | chr18: 54269338 | 2.5 (5.67%) | mature_mirR | miR-122* |
| mir-125a | rs12975333 | G/T | chr19: 56888340 | 6.1 (14.52%) | mature_mirR | miR-125a-5p |
| mir-520c | rs7255628 | C/G | chr19: 58902546 | 6.6 (14.77%) | mature_mirR | miR-520c-5p |
| mir-518e | rs34416818 | -/A | chr19: 58924924 | 4.4 (10.24%) | mature_mirR | miR-518e* |
| mir-499 | rs3746444 | A/G | chr20: 33041912 | 0.4 (0.65%) | mature_mirR | miR-499-3p |
| mir-646 | rs6513497 | G/T | chr20: 58317000 | 0.8 (2.40%) | mature_mirR | miR-646 |
| mir-124-3 | rs34059726 | G/T | chr20: 61280352 | 6.5 (19.23%) | mature_mirR | miR-124 |
| mir-941-2 | rs34604519 | C/G | chr20: 62021613 | 3.6 (6.63%) | mature_mirR | miR-941 |
| mir-941-3 | rs35544770 | A/G | chr20: 62021716 | 3.6 (6.63%) | mature_mirR | miR-941 |
| mir-941-3 | rs12625454 | C/G | chr20: 62021725 | 3.6 (6.63%) | mature_mirR | miR-941 |
| mir-1302-2 | rs11266859 | A/G | chr1: 20279 | 0.4 (0.64%) | mirR_complementary | miR-1302 |
| mir-1302-2 | rs422582 | A/C | chr1: 20287 | 6 (10.60%) | mirR_complementary | miR-1302 |
| mir-1977 | rs9701099 | C/T | chr1: 556076 | 1.1 (5.37%) | mirR_complementary | miR-1977 |
| mir-1302-3 | rs2441621 | G/T | chr2: 114057085 | 6 (10.62%) | mirR_complementary | miR-1302 |
| mir-1244 | rs1804520 | A/G | chr2: 232286288 | 1.3 (7.78%) | mirR_complementary | miR-1244 |
| mir-217 | rs41291173 | A/G | chr2: 56063644 | 0.4 (1.20%) | mirR_complementary | miR-217 |
| mir-570 | rs9860655 | C/T | chr3: 196911485 | 4.1 (10.25%) | mirR_complementary | miR-570 |
| mir-564 | rs2292181 | C/G | chr3: 44878438 | 1.9 (3.61%) | mirR_complementary | miR-564 |
| mir-1324 | rs28620398 | G/T | chr3: 75762617 | 3.1 (10.54%) | mirR_complementary | miR-1324 |
| mir-1255a | rs28664200 | C/T | chr4: 102470524 | 0.3 (0.49%) | mirR_complementary | miR-1255a |
| mir-1244 | rs1804520 | A/G | chr5: 118338200 | 1.3 (7.78%) | mirR_complementary | miR-1244 |
| mir-1294 | rs13186787 | A/G | chr5: 153706962 | 0.7 (1.01%) | mirR_complementary | miR-1294 |
| mir-146a | rs2910164 | C/G | chr5: 159844996 | 2.8 (6.95%) | mirR_complementary | miR-146a |
| mir-1229 | rs2291418 | C/T | chr5: 179157930 | 0 (0.00%) | mirR_complementary | miR-1229 |
| mir-548a-1 | rs12197631 | G/T | chr6: 18680035 | 2.51 (7.59%) | mirR_complementary | miR-548a-3p |
| mir-1206 | rs2114358 | A/G | chr8: 129090361 | 0.5 (2.65%) | mirR_complementary | miR-1206 |
| mir-939 | rs35486628 | -/G | chr8: 145590185 | 0 (0.00%) | mirR_complementary | miR-939 |
| mir-1234 | rs2291134 | C/G | chr8: 145596344 | 4.2 (8.38%) | mirR_complementary | miR-1234 |
| mir-1302-2 | rs11266859 | A/G | chr9: 20194 | 0.4 (0.64%) | mirR_complementary | miR-1302 |
| mir-1302-2 | rs422582 | A/C | chr9: 20202 | 6 (10.60%) | mirR_complementary | miR-1302 |
| mir-603 | rs11014002 | C/T | chr10: 24604659 | 1.8 (4.44%) | mirR_complementary | miR-603 |
| mir-604 | rs2368393 | A/G | chr10: 29874004 | 0.1 (0.37%) | mirR_complementary | miR-604 |
| mir-604 | rs2368392 | A/G | chr10: 29874009 | 0.6 (2.25%) | mirR_complementary | miR-604 |
| mir-607 | rs12778876 | A/T | chr10: 98578485 | 4.6 (6.79%) | mirR_complementary | miR-607 |
| mir-607 | rs12780546 | A/T | chr10: 98578486 | 4.5 (6.64%) | mirR_complementary | miR-607 |
| mir-1304 | rs2155248 | G/T | chr11: 93106514 | 6.5 (9.69%) | mirR_complementary | miR-1304 |
| mir-619 | rs34651680 | -/G | chr12: 107754861 | 0 (0.00%) | mirR_complementary | miR-619 |
| mir-620 | rs3043743 | -/T/TA | chr12: 115070818 | 0.4 (1.65%) | mirR_complementary | miR-620 |
| mir-620 | rs34380284 | -/C | chr12: 115070819 | 2.9 (11.60%) | mirR_complementary | miR-620 |
| mir-1178 | rs7311975 | C/T | chr12: 118635876 | 0 (0.00%) | mirR_complementary | miR-1178 |
| mir-1244 | rs1804520 | A/G | chr12: 12156173 | 1.3 (7.78%) | mirR_complementary | miR-1244 |
| mir-196a-2 | rs11614913 | C/T | chr12: 52671866 | 4.6 (9.87%) | mirR_complementary | miR-196a |
| mir-618 | rs2682818 | A/C | chr12: 79853667 | 3.5 (10.12%) | mirR_complementary | miR-618 |
| mir-1244 | rs1804520 | A/G | chr12: 9283394 | 1.3 (7.78%) | mirR_complementary | miR-1244 |
| mir-92a-1 | rs9589207 | A/G | chr13: 90801590 | 0 (0.00%) | mirR_complementary | miR-92a |
| mir-453 | rs56103835 | C/T | chr14: 100592309 | 0.3 (1.16%) | mirR_complementary | miR-453 |
| mir-625 | rs12894182 | A/C | chr14: 65007642 | 6.5 (9.50%) | mirR_complementary | miR-625 |
| mir-1302-2 | rs422582 | A/C | chr15: 100318264 | 6 (10.60%) | mirR_complementary | miR-1302 |
| mir-1302-2 | rs11266859 | A/G | chr15: 100318272 | 0.4 (0.64%) | mirR_complementary | miR-1302 |
| mir-631 | rs5745925 | -/CT | chr15: 73433019 | 6.5 (20.63%) | mirR_complementary | miR-631 |
| mir-1826 | rs62030476 | A/G | chr16: 33873090 | 0 (0.00%) | mirR_complementary | miR-1826 |
| mir-1972 | rs57629257 | C/T | chr16: 68621762 | 2.7 (7.62%) | mirR_complementary | miR-1972 |
| mir-633 | rs17759989 | A/G | chr17: 58375343 | 0.6 (1.80%) | mirR_complementary | miR-633 |
| mir-1181 | rs2569788 | C/G | chr19: 10375159 | 2.2 (5.66%) | mirR_complementary | miR-1181 |
| mir-1302-2 | rs11266859 | A/G | chr19: 23023 | 0.4 (0.64%) | mirR_complementary | miR-1302 |
| mir-1302-2 | rs422582 | A/C | chr19: 23031 | 6 (10.60%) | mirR_complementary | miR-1302 |
| mir-520h | rs56013413 | A/G | chr19: 58937600 | 4.1 (12.57%) | mirR_complementary | miR-520h |
| mir-663 | rs28670321 | C/T | chr20: 26136824 | 0 (0.00%) | mirR_complementary | miR-663 |
| mir-663 | rs2019798 | G/T | chr20: 26136846 | 0.5 (1.01%) | mirR_complementary | miR-663 |
| mir-645 | rs35645123 | -/A | chr20: 48635761 | 0 (0.00%) | mirR_complementary | miR-645 |
| mir-941-1 | rs7268785 | C/G | chr20: 62021250 | 3.6 (6.63%) | mirR_complementary | miR-941 |
| mir-941-1 | rs2427556 | A/G | chr20: 62021268 | 4.1 (8.17%) | mirR_complementary | miR-941 |
| mir-941-3 | rs12625445 | C/G | chr20: 62021669 | 3.6 (6.63%) | mirR_complementary | miR-941 |
| mir-548j | rs12161068 | C/T | chr22: 25281215 | 0.4 (0.75%) | mirR_complementary | miR-548j |
N.A. stands for inconsistent allele reports existed.
Figure 3.
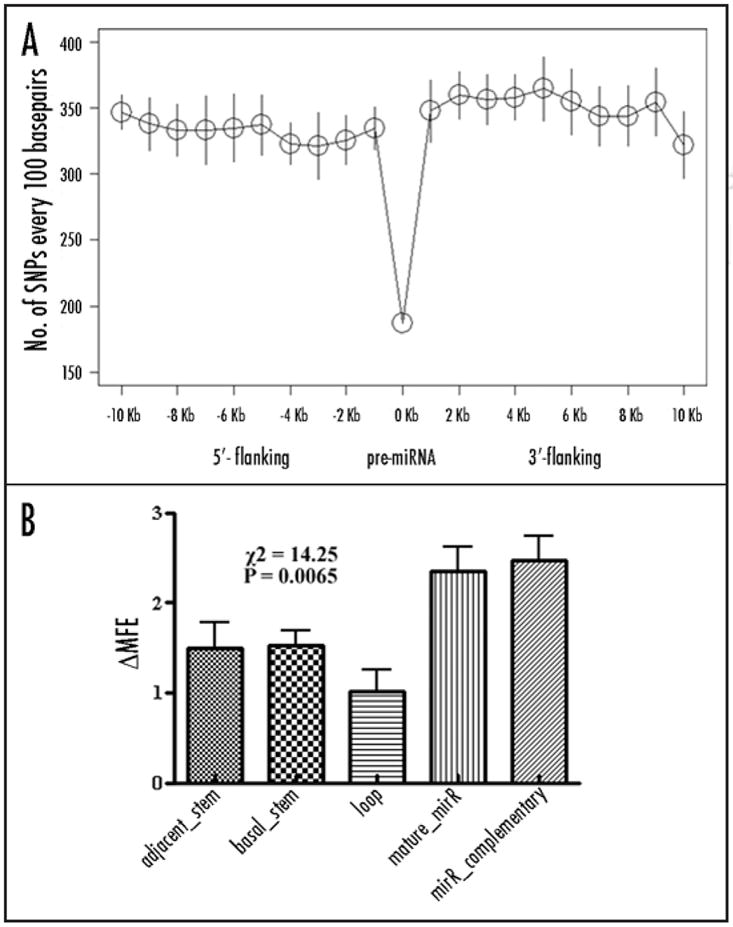
(A) SNP densities in the human pre-miRNA loci. (B) MFE changes in miRNA domains.
We also evaluated the known CNV coverage of human pre-miRNA genes using the CNVs deposited in the Database of Genomic Variants (DGV).40 We found that 193 pre-miRNAs were located in the regions covered by 385 CNV markers (Table 4 and Suppl. Table 2). No significant difference for the distribution of pre-miRNAs in PCGs (n = 109) and in the intergenic regions (n = 84, χ2 = 0.71, df = 1, p = 0.39).
Table 4.
Representative pre-miRNAs in the CNV regions
| pre-miRNA ID | pre-miRNA location (hg18) | Host gene | CNV ID | CNV position (hg18) | Observed CNVs |
|---|---|---|---|---|---|
| mir-200b | chr1:1092347-1092441(+) | - | Variation_30362 | chr1:702445-1697636 | 11 gains |
| mir-200a | chr1:1093106-1093195(+) | - | Variation_30362 | chr1:702445-1697636 | 11 gains |
| mir-429 | chr1:1094248-1094330(+) | - | Variation_30362 | chr1:702445-1697636 | 11 gains |
| mir-320b-1 | chr1:117015894-117015972(+) | - | Variation_4243 | chr1:116927828-117128034 | 11 gains |
| mir-555 | chr1:153582765-153582860(−) | ASH1L | Variation_6789 | chr1:153489907-154184585 | 39 losses |
| mir-1302-2 | chr1:20229-20366(+) | - | Variation_30360 | chr1:1794-115824 | 25 gains |
| mir-663b | chr2:132731009-132731123(−) | LOC100133239 | Variation_31014 | chr2:132726460-132763489 | 19 gains |
| mir-570 | chr3:196911452-196911548(+) | - | Variation_2491 | chr3:196584076-196965419 | 188 gains |
| mir-566 | chr3:50185763-50185856(+) | SEMA3F | Variation_4335 | chr3:50173490-50368468 | 18 losses |
| mir-1324 | chr3:75762604-75762699(+) | - | Variation_2462 | chr3:75474768-76085726 | 3 gains, 13 losses |
| mir-218-1 | chr4:20138996-20139105(+) | SLIT2 | Variation_4380 | chr4:20111993-20290054 | 9 gains, 18 losses |
| mir-95 | chr4:8057928-8058008(−) | ABLIM2 | Variation_4373 | chr4:8009599-8099960 | 10 losses |
| mir-548i-2 | chr4:9166887-9167035(−) | - | Variation_2069 | chr4:9010036-9203157 | 42 losses |
| mir-1236 | chr6:32032595-32032696(−) | RDBP | Variation_4492 | chr6:31995533-32055579 | 36 losses |
| mir-548o | chr7:101833194-101833307(−) | PRKRIP1 | Variation_4553 | chr7:101767725-102083105 | 42 losses |
| mir-1183 | chr7:21477201-21477289(+) | SP4 | Variation_4527 | chr7:21455625-21654541 | 20 gains |
| mir-939 | chr8:145590172-145590253(−) | CPSF1 | Variation_4613 | chr8:145536611-145740218 | 16 losses |
| mir-1234 | chr8:145596284-145596367(−) | CPSF1 | Variation_4613 | chr8:145536611-145740218 | 16 losses |
| mir-548i-3 | chr8:7983873-7984021(−) | - | Variation_2116 | chr8:7917018-8067760 | 25 gains, 34 losses |
| let-7a-1 | chr9:95978060-95978139(+) | - | Variation_4645 | chr9:95873863-96081830 | 18 gains |
| let-7f-1 | chr9:95978450-95978536(+) | - | Variation_4645 | chr9:95873863-96081830 | 18 gains |
| let-7d | chr9:95980937-95981023(+) | LOC158257 | Variation_4645 | chr9:95873863-96081830 | 18 gains |
| mir-202 | chr10:134911006-134911115(−) | - | Variation_2896 | chr10:134868158-135282675 | 20 gains, 1 loss |
| mir-1268 | chr15:20014593-20014644(−) | - | Variation_3070 | chr15:18403665-21241985 | 204 gains, 24 losses |
| mir-1233 | chr15:32461562-32461643(−) | GOLGA8A, GOLGA8B | Variation_7058 | chr15:29769358-32654590 | 47 gains, 48 losses |
| mir-657 | chr17:76713671-76713768(−) | AATK | Variation_5036 | chr17:76600967-76762177 | 13 losses |
| mir-338 | chr17:76714278-76714344(−) | AATK | Variation_5036 | chr17:76600967-76762177 | 13 losses |
| mir-1250 | chr17:76721591-76721703(−) | AATK | Variation_5036 | chr17:76600967-76762177 | 13 losses |
| mir-199a-1 | chr19:10789102-10789172(−) | DNM2 | Variation_5087 | chr19:10728678-10897044 | 11 losses |
| mir-1909 | chr19:1767158-1767237(−) | REXO1 | Variation_7191 | chr19:1271267-1950204 | 13 gains, 10 losses |
| mir-1270 | chr19:20371080-20371162(−) | FLJ44894 | Variation_3183 | chr19:20360525-20566187 | 35 losses |
| mir-1227 | chr19:2185061-2185148(−) | PLEKHJ1 | Variation_5068 | chr19:2138091-2323221 | 17 losses |
| mir-220c | chr19:53755341-53755423(−) | SULT2B1 | Variation_32261 | chr19:53097718-55337070 | 18 losses |
| mir-150 | chr19:54695854-54695937(−) | LOC100128528 | Variation_5111 | chr19:54643858-54765745 | 25 losses |
| mir-663 | chr20:26136822-26136914(−) | LOC284801 | Variation_31037 | chr20:26136626-26139184 | 10 gains |
| mir-124-3 | chr20:61280297-61280383(+) | - | Variation_5147 | chr20:61234049-61347722 | 70 losses |
| mir-185 | chr22:18400662-18400743(+) | C22orf25 | Variation_2261 | chr22:18259187-18435258 | 10 losses |
| mir-649 | chr22:19718465-19718561(−) | - | Variation_5170 | chr22:19664133-19854524 | 12 gains |
| mir-650 | chr22:21495270-21495365(+) | IGL@ | Variation_2268 | chr22:21394879-21570697 | 35 gains, 32 losses |
| mir-1912 | chrX:113792275-113792354(+) | HTR2C | Variation_7758 | chrX:113724807-114050880(+) | 4 gains, 9 losses |
| mir-1264 | chrX:113793386-113793454(+) | HTR2C | Variation_7758 | chrX:113724807-114050880(+) | 4 gains, 9 losses |
| mir-1298 | chrX:113855906-113856017(+) | HTR2C | Variation_7758 | chrX:113724807-114050880(+) | 4 gains, 9 losses |
| mir-1911 | chrX:113904000-113904079(+) | HTR2C | Variation_7758 | chrX:113724807-114050880(+) | 4 gains, 9 losses |
| mir-448 | chrX:113964273-113964383(+) | HTR2C | Variation_7758 | chrX:113724807-114050880(+) | 4 gains, 9 losses |
Polymorphisms with potential effects on miRNA targeting
Using the predicted targets by TargetScanS41 and PITA,42 we found 1,238 and 4,235 SNPs located in the putative seed-matching regions of targeting genes (Suppl. Tables 3 and 4). As shown in Table 5, eleven 3′-UTR SNPs may disrupt the miRNA-target regulation that has been supported by experimental evidence maintained in TarBase.43 A total of 497 overlapping SNPs are found in the same seed-matching regions of 434 target genes by both TargetScanS and PITA (Suppl. Table 5). As shown in Table 6, the 434 overlapping target genes are significantly enriched in the KEGG pathways of “Angiogensis” (Bonferroni p = 4 × 10−6) and “T cell activation” (Bonferroni p = 0.004) as well as the GO biological processes of “Developmental processes” (Bonferroni p = 3 × 10−21), “Signal transduction” (Bonferroni p = 5 × 10−12), “mRNA transcription regulation” (Bonferroni p = 2 × 10−6), “Neurogenesis” (Bonferroni p = 2 × 10−6) and “Oncogenesis” (Bonferroni p = 8 × 10−5). Focusing on both the pre-miRNA host genes and the miRNA target genes with SNPs in the consensus target sites, the pathway and GO analysis show some similar pathways and GO categories for these two lists, such as “Neuronal activities”, “Cell cyle”, “Protein phosphorylation”, and “Cell structure and motility” in the biological processes, and the “kinase” in the shared molecular function. These suggest a potential involvement of miRNAs in these biological activities.
Table 5.
SNPs in the seed-matching regions of miRNA target genes
| SNPa | miRNA | Target | Prediction method |
|---|---|---|---|
| rs56788643 | let-7g | HMGA2 | PITA |
| rs35180728 | miR-1 | ARCN1 | TargetScanS |
| rs36076633 | miR-1 | TAGLN2 | TargetScanS |
| rs8829 | miR-101 | EZH2 | PITA |
| rs3783620 | miR-126 | VCAM1 | PITA |
| rs34335657 | miR-129 | CAMTA1 | PITA |
| rs41286082 | miR-141 | KLF5 | PITA |
| rs1621 | miR-199a* | MET | PITA |
| rs34954531 | miR-30a-3p | VEZT | PITA |
| rs1051780 | miR-34 | VAMP2 | TargetScanS |
| rs17620927 | miR-373 | MKRN1 | PITA |
| rs56788643 | miR-98 | HMGA2 | PITA |
SNPs with frequency report are in bold font.
Table 6.
Pathways and GO analysis of genes with SNPs in miRNA target sites
| Categories | Total | Observed | Expected | +/− | Bonferroni P |
|---|---|---|---|---|---|
| Pathway | |||||
| Angiogenesis | 229 | 19 | 3.87 | + | 0.00000402 |
| Unclassified | 22436 | 340 | 379.36 | − | 0.0000077 |
| T cell activation | 111 | 10 | 1.88 | + | 0.00426 |
| PDGF signaling pathway | 189 | 13 | 3.2 | + | 0.00459 |
| Biological Process | |||||
| Developmental processes | 2152 | 104 | 36.39 | + | 3.47E-21 |
| Biological process unclassified | 11321 | 107 | 191.42 | − | 7.92E-16 |
| Signal transduction | 3406 | 115 | 57.59 | + | 5.33E-12 |
| Intracellular signaling cascade | 871 | 46 | 14.73 | + | 2.54E-09 |
| mRNA transcription | 1914 | 67 | 32.36 | + | 0.0000021 |
| Neurogenesis | 587 | 32 | 9.93 | + | 0.00000232 |
| mRNA transcription regulation | 1459 | 56 | 24.67 | + | 0.00000251 |
| Mesoderm development | 551 | 30 | 9.32 | + | 0.00000506 |
| Ectoderm development | 692 | 34 | 11.7 | + | 0.00000702 |
| Nucleoside, nucleotide and nucleic acid metabolism | 3343 | 93 | 56.53 | + | 0.0000265 |
| Cell proliferation and differentiation | 1028 | 40 | 17.38 | + | 0.0000398 |
| Oncogenesis | 472 | 24 | 7.98 | + | 0.0000825 |
| Protein modification | 1157 | 42 | 19.56 | + | 0.000559 |
| Neuronal activities | 569 | 25 | 9.62 | + | 0.000601 |
| Muscle contraction | 198 | 13 | 3.35 | + | 0.00137 |
| Other intracellular signaling cascade | 225 | 15 | 3.8 | + | 0.00194 |
| Cell structure and motility | 1148 | 38 | 19.41 | + | 0.00248 |
| Receptor protein tyrosine kinase signaling pathway | 211 | 14 | 3.57 | + | 0.00402 |
| Protein phosphorylation | 660 | 27 | 11.16 | + | 0.00612 |
| Cell cycle | 1009 | 32 | 17.06 | + | 0.0187 |
| Cell communication | 1213 | 38 | 20.51 | + | 0.0346 |
| Cell surface receptor mediated signal transduction | 1638 | 47 | 27.7 | + | 0.0466 |
| Molecular Function | |||||
| Molecular function unclassified | 10934 | 105 | 184.88 | − | 2.18E-14 |
| Transcription factor | 2052 | 69 | 34.7 | + | 0.00000117 |
| Nucleic acid binding | 2850 | 83 | 48.19 | + | 0.0000187 |
| Voltage-gated ion channel | 145 | 13 | 2.45 | + | 0.000276 |
| Ion channel | 357 | 18 | 6.04 | + | 0.00151 |
| Other miscellaneous function protein | 427 | 21 | 7.22 | + | 0.00293 |
| Other transcription factor | 349 | 18 | 5.9 | + | 0.00631 |
| Miscellaneous function | 866 | 28 | 14.64 | + | 0.0286 |
| Other DNA-binding protein | 331 | 16 | 5.6 | + | 0.0345 |
| Membrane traffic protein | 359 | 15 | 6.07 | + | 0.0415 |
| Kinase | 684 | 23 | 11.57 | + | 0.0485 |
Variations at important miRNA-processing genes
Given these delicate processes in miRNA biogenesis (Fig. 4), any alterations of the key proteins involved in the miRNA processing and targeting will potentially lead to a global deregulation of the miRNA-mediated posttranscriptional silencing. Altogether, we found 3,921 SNPs in these miRNA-processing genes. A total of 83 SNPs may change the coding sequence of protein products. However, there are only 35 SNPs with allele frequency reports (Suppl. Table 6). Their contributions to gene functions remain to be discovered. A further analysis between CNVs and 13 miRNA-related genes shows that there are deletion in the loci of SNIP1, RNASEN, DICER1 and DGCR8, suggesting a potentially disrupted miRNA-processing pathway in those CNV carriers (Table 7).
Figure 4.
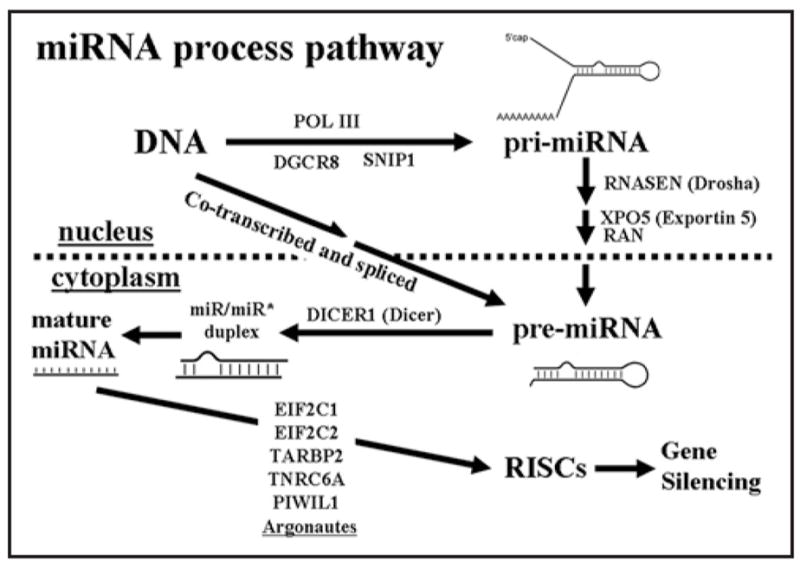
miRNA process pathway. The figure is recreated according to refs.2,3,64–69,73
Table 7.
Important miRNA-processing genes in the CNV regions
| Gene | Entrez ID | Gene location (hg18) | CNV ID | CNV position (hg18) | Observed CNVsa |
|---|---|---|---|---|---|
| SNIP1 | 79753 | chr1:37774729-37792490(−) | Variation_0006 | chr1:37714745-37826968 | 1 loss |
| RNASEN | 29102 | chr5:31436358-31568039(−) | Variation_3550 | chr5:31332034-31505885 | N.A. |
| RNASEN | 29102 | chr5:31436358-31568039(−) | Variation_47879 | chr5:31412880-31908908 | 3 losses |
| XPO5 | 57510 | chr6:43598050-43651642(−) | Variation_9530 | chr6:43583452-43604640 | 1 gain |
| PIWIL1 | 9271 | chr12:129388567-129422826(+) | Variation_3000 | chr12:128577929-129659406 | 1 gain |
| PIWIL1 | 9271 | chr12:129388567-129422826(+) | Variation_3901 | chr12:128512417-129681529 | N.A. |
| PIWIL1 | 9271 | chr12:129388567-129422826(+) | Variation_8740 | chr12:128578742-129654380 | 1 gain |
| PIWIL1 | 9271 | chr12:129388567-129422826(+) | Variation_35062 | chr12:129417091-129445428 | 1 gain |
| DICER1 | 23405 | chr14:94622319-94693512(−) | Variation_5766 | chr14:94638002-94644051 | 1 loss |
| GEMIN4 | 50628 | chr17:594411-602251(−) | Variation_3136 | chr17:595817-897708 | 2 gains |
| GEMIN4 | 50628 | chr17:594411-602251(−) | Variation_4021 | chr17:568336-1008155 | N.A. |
| GEMIN4 | 50628 | chr17:594411-602251(−) | Variation_5332 | chr17:60543-963131 | 1 gain |
| DGCR8 | 54487 | chr22:18447834-18479400(+) | Variation_4117 | chr22:18406493-18689447 | N.A. |
| DGCR8 | 54487 | chr22:18447834-18479400(+) | Variation_5168 | chr22:18267966-18449970 | 6 losses |
| DGCR8 | 54487 | chr22:18447834-18479400(+) | Variation_31071 | chr22:17399088-19383198 | 6 gains |
N.A. stands for not available.
Discussions
In the present study, we evaluated the properties of human pre-miRNAs based on the miRBase database (release 13). Our survey shows that 53.9% of pre-miRNAs are located in PCGs. We found that pre-miRNA host genes have longer spanning regions than other genes in the genome and pre-miRNA host genes are more likely to be annotated with functional descriptions as compared with other genes in the genome (p < 0.0001, OR = 4.05), although this may be biased by a trend in the published studies of pre-miRNAs. A total of 193 pre-miRNAs (27.4%) are located in regions with genome instability. Interestingly, there are 10 out of 12 let-7 family members found in CNV regions (Suppl. Table 2). Given that let-7 plays a role in tumorigenesis, this finding suggests a non-random connection between pre-miRNA, CNVs and cancer development which is in agreement with previous findings.44
PCGs may host multiple pre-miRNAs and thus have potential to network with others. For example, HTR2C, a host gene for 5 pre-miRNAs, is a G-protein coupled receptor and mediates the signaling of neuronal activities.45 RTL1, a host gene of 5 pre-miRNAs is a reverse transcriptase and aspartic protease46 that is involved in angiogenesis, apoptosis and pathway-mitogen activated protein kinase kinase/MAP kinase cascade. Harboring 5 pre-miRNAs, LARP7 is a ribonucleoprotein and plays an important role in tRNA metabolism.47
Given the extensive variation found in the human genome, miRNA-mediated functions may be affected by polymorphisms in the miRNA target loci, pre-miRNA gene loci and/or the miRNA regulating gene loci.48 The polymorphism-driven alterations of miRNA activity are observed in numerous association studies.43 Several studies have catalogued the SNPs at the human pre-miRNAs and mature miRNA binding sites based on the earlier miRBase version (Table 1). In the present study, we also evaluated the roles of genomic variants, including both SNPs and CNVs, which may influence the miRNA-associated biological functions. Using the 718 genomic coordinates of 705 human pre-miRNAs (not including mir-941-4), we found 188 SNPs in the 138 pre-miRNAs with various numbers of SNPs in different pre-miRNA domains. The SNP-residing domains with the trend of MFE changes from low to high are loop, stem (including adjacent stem and basal stem), mature miRNA and their complementary sequences (Fig. 3). SNPs in pre-miRNAs could potentially change the stem-loop structures and thus may influence the miRNA processing and maturation. SNPs in the mature miRNAs, especially in the seed regions, are likely to affect the specificity for gene silencing.
Bioinformatics databases along with experimental evidence implicate eleven SNPs in miRNA target sites (Table 5). In our analysis, we found three SNPs with allele frequencies reported in NCBI dbSNP, including rs3783620, rs1621 and rs17620927, that may affect miRNA:gene regulation recorded in TarBase v5.0.49 For example, the miR-126 was reported to repress the translational level of VCAM1 by binding to the 3′-UTR of the gene50 and this regulation might be influenced by SNP rs3783620 in the seed-matching region on the 3′-UTR of VCAM1. To note, VCAM1 is an important gene in regulating leukocyte trafficking to sites of inflammation.51 In addition, Kim et al. reported that miR-199a* mediated the downregulation of MET gene by targeting at the 3′-UTR.52 However, SNP rs1621 in the seed-matching sequence of MET could affect this activity. Another example is miR-373 and MKRN1 gene. Using a microarray assay, the expression of miR-373 was identified to be inversely associated with the expression of MKRN1.53 SNP rs17620927, located in the miR-373 target site for MKRN1 likely affects miRNA-mediated gene silencing.
It is notable that the pre-miRNAs with multiple genomic copies, including mir-1244 (4 copies) and mir-1302-2 (4 copies) reside in CNV regions. This implies that the multiple copies of these pre-miRNAs are likely to be generated by genomic instability. The copy number changes may affect both the miRNAs and their host genes. Among the pre-miRNAs and their host genes in the CNV regions, some of them are implicated with important biological functions. Among them, mir-200b was reported to mediate gene silencing of ZFHX1B, a gene that is important in the TGFbeta signaling pathway.54 As shown in the Table 4, mir-555 is found to be located in a CNV loss region and it is embedded in ASH1L, a gene that is identified as a histone methyltransferase regulating gene transcription.55 AATK, a tyrosine kinase involved in apoptosis,56 is also located in the CNV loss region and harbors three pre-miRNAs including mir-657, mir-338 and mir-1250. These structural variants of the pre-miRNAs may be implicated with important biological effects in humans. Eleven out of 95 individuals were found to carry a deletion that covers mir-199a-1 and its host PCG (DNM2). DNM2 encodes a member of GTPases and it was a candidate gene of dominant intermediate Charcot-Marie-Tooth disease57 and autosomal dominant centronuclear myopathy.58 HTR2C that harbors 5 pre-miRNAs was found to be located in a CNV with genotypes of 4 gains and 9 losses. HTR2C gene encodes a serotonin receptor that is associated with mental disorders59,60 and side effects induced by antipsychotic drugs.61,62 Since little evidence is available about functional connection between the pre-miRNAs and their host PCGs, the current study provides researchers with a comprehensive list for future analysis.
As shown in Figure 4, pre-miRNAs have been shown to be produced through either alternative splicing from their host genes or processing by Drosha (RNASEN, nuclear ribonuclease type III).2 Subsequently, these intermediate precursors are exported by Exportin-5 (XPO5) and Ran-GTP (RAN) from nucleus to the cytoplasm, where Dicer (DICER1) excises stem-loop pre-miRNAs into single-stranded mature miRNA.2,3,63 These mature miRNA will be mediated by the argonaute family proteins (EIF2C1, EIF2C2)64 and other proteins (TARBP2, TNRC6A, PIWIL1)65,66 to evoke a series of posttranscriptional silencing of target genes. Besides these well known genes, recent evidence shows that SNIP1,67 and DGCR8,68 along with Drosha are involved in the initiation of miRNA biogenesis. Gemin3 (DDX20) and Gemin4 (GEMIN4) are two other important genes for miRNA function. They can form novel complex ribonucleoproteins to perform gene silencing function with miRNA and eIF2C2, a member of the Argonaute protein family.69
In this study, we evaluated the contribution of CNVs to 13 miRNA-processing pathway genes. SNIP1 and DGCR8 are important proteins in the initiation of the pri-miRNA transcription. EIF2C2 is in the Argonaut family of genes that play an important role in gene silencing. The CNVs with genomic loss suggest that other proteins may rescue their biological functions. Interestingly, we also noticed there is a 350 bp loss in the DICER1 loci, which will cause a truncated form of DICER1. Given the severe abnormality caused by the Dicer1 gene deletion,23 we speculate that this truncated form of DICER maintains the function of DICER1. Since the CNV frequencies at most genes except DGCR8 are relatively low, more evidence is needed for an in-depth exploration.
In summary, we have performed an in-depth analysis of human miRNAs based on multiple association studies. Compared with previous studies, we focused on both SNPs and CNV information that may potentially affect miRNA processing and maturation. Since aberrant miRNA expressions have been implicated in oncogenesis and other diseases,70 the miRNA-related polymorphisms provided by us will facilitate future studies and increase our understanding of the role of miRNAs in human gene regulation.
Materials and Methods
Searching for host genes for the pre-miRNAs
The chromosomal coordinates for 705 (not including mir-941-4) pre-miRNAs and over 30,000 human genes were obtained from the miRBase version 13.0 and NCBI dbGene. We compared the genomic start and end positions of over 30,000 genes with those of 705 pre-miRNAs. A host gene of a pre-miRNA is defined when their loci overlap with each other. All of the genomic coordinates in the present study were based on hg18 (March, 2006).
Searching for SNPs in the pre-miRNAs, mature miRNAs and miRNA target sites
The genomic coordinates of mature miRNAs were inferred from the pre-miRNAs by the sequence matching. The miRNA target seed-matching regions predicted by TargetScan v4.1 and PITA were downloaded from the UCSC genome browser.71 Genomic positions of over 10 million SNPs were retrieved from NCBI dbSNP129 and then they were used to search for SNPs within the start and end positions of pre-miRNAs, mature miRNAs and miRNA target sites.
Searching for CNVs covering pre-miRNAs
The genomic coordinates of CNVs (after excluding inversions) were downloaded from Database of Genomic Variants (DGV) version 7.40 The genomic start and end positions of over 20,000 CNVs with those of 705 pre-miRNAs were compared to see whether there were overlapping regions between them.
GO and KEGG pathway analysis
PANTHER database72 was used to identify enriched functional annotation categories for pre-miRNA host genes and the genes with SNPs in the miRNA target sites. KEGG pathway and two GO terms (biological process and molecular function) were evaluated. Uncorrected p < 0.01 was considered statistically significant.
Web resources
NCBI dbSNP: http://www.ncbi.nih.gov/SNP/
Database of Genomic Variants: http://projects.tcag.ca/variation/
UCSC genome browser: http://genome.ucsc.edu/
miRBase: http://microrna.sanger.ac.uk/sequences/
TarBase: http://diana.cslab.ece.ntua.gr/tarbase/
TargetScanS miRNA target database: http://www.targetscan.org/vert_42/
PITA miRNA target database: http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html
RNAfold web server: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi
Supplementary Material
Acknowledgments
This Pharmacogenetics of Anticancer Agents Research (PAAR) Group (http://pharmacogenetics.org/) study was supported by NIH/NIGMS grant UO1GM61393 and data deposits are supported by UO1GM61374 (http://pharmgkb.org/). This study was also supported by P50 CA125183 University of Chicago Breast Cancer SPORE grant.
Footnotes
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–21. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Croce CM, Calin GA. miRNAs, cancer and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Bicker S, Schratt G. microRNAs: Tiny Regulators of Synapse Function in Development and Disease. J Cell Mol Med. 2008;12:1466–76. doi: 10.1111/j.1582-4934.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–13. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA. 2007;104:13513–8. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatry. 2009;14:381–9. doi: 10.1038/mp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, et al. A polymorphism of microRNA 27a genome region is associated with the development of gastric mucosal atrophy in Japanese male subjects. Dig Dis Sci. 2007;52:1691–7. doi: 10.1007/s10620-006-9648-5. [DOI] [PubMed] [Google Scholar]
- 12.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–34. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv K, Guo Y, Zhang Y, Wang K, Jia Y, Sun S. Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/deletion polymorphism with type 2 diabetes. Biochem Biophys Res Commun. 2008;374:101–5. doi: 10.1016/j.bbrc.2008.06.102. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–9. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 16.Brendle A, Lei H, Brandt A, Johansson R, Enquist K, Henriksson R, et al. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29:1394–9. doi: 10.1093/carcin/bgn126. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–7. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 18.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA. 2009;106:1502–5. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, et al. A Functional Genetic Variant in microRNA-196a2 Is Associated with Increased Susceptibility of Lung Cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–7. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 21.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–40. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila Pa) 2008;1:460–9. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 24.Iwai N, Naraba H. Polymorphisms in human pre-miRNAs. Biochem Biophys Res Commun. 2005;331:1439–44. doi: 10.1016/j.bbrc.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 25.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–6. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 26.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 27.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 28.Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, et al. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:51–4. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–31. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 30.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, et al. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates micror-na-155 binding. J Biol Chem. 2007;282:24262–9. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–41. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landi D, Gemignani F, Barale R, Landi S. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol. 2008;27:35–43. doi: 10.1089/dna.2007.0650. [DOI] [PubMed] [Google Scholar]
- 34.Mishra PJ, Mishra PJ, Banerjee D, Bertino JR. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853–8. doi: 10.4161/cc.7.7.5666. [DOI] [PubMed] [Google Scholar]
- 35.Reumers J, Conde L, Medina I, Maurer-Stroh S, Van Durme J, Dopazo J, et al. Joint annotation of coding and non-coding single nucleotide polymorphisms and mutations in the SNPeffect and PupaSuite databases. Nucleic Acids Res. 2008;36:825–9. doi: 10.1093/nar/gkm979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quach H, Barreiro LB, Laval G, Zidane N, Patin E, Kidd KK, et al. Signatures of purifying and local positive selection in human miRNAs. Am J Hum Genet. 2009;84:316–27. doi: 10.1016/j.ajhg.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–48. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 41.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 43.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn809. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Nat Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drago A, Serretti A. Focus on HTR2C: A possible suggestion for genetic studies of complex disorders. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30864. In press. [DOI] [PubMed] [Google Scholar]
- 46.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nature Genet. 2003;34:261–2. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 47.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29:588–99. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borel C, Antonarakis SE. Functional genetic variation of human miRNAs and phenotypic consequences. Mamm Genome. 2008;19:503–9. doi: 10.1007/s00335-008-9137-6. [DOI] [PubMed] [Google Scholar]
- 49.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–7. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Nat Acad Sci USA. 2008;105:1516–21. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mebius RE. Organogenesis of lymphoid tissues. Nat Rev Immunol. 2003;3:292–303. doi: 10.1038/nri1054. [DOI] [PubMed] [Google Scholar]
- 52.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–66. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 53.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 54.Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–8. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27:8466–79. doi: 10.1128/MCB.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaozza E, Baker SJ, Vora RK, Reddy EP. AATYK: a novel tyrosine kinase induced during growth arrest and apoptosis of myeloid cells. Oncogene. 1997;15:3127–35. doi: 10.1038/sj.onc.1201575. [DOI] [PubMed] [Google Scholar]
- 57.Zuchner S, Noureddine M, Kennerson M, Verhoeven K, Claeys K, De Jonghe P, et al. Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nature Genet. 2005;37:289–94. doi: 10.1038/ng1514. [DOI] [PubMed] [Google Scholar]
- 58.Bitoun M, Maugenre S, Jeannet PY, Lacene E, Ferrer X, Laforet P, et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nature Genet. 2005;37:1207–9. doi: 10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- 59.Tecott LH, Sun LM, Akana SF, Strack AM, Lowenstein DH, Dallman MF, et al. Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature. 1995;374:542–6. doi: 10.1038/374542a0. [DOI] [PubMed] [Google Scholar]
- 60.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–56. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 61.Ji SP, Zhang Y, Van Cleemput J, Jiang W, Liao M, Li L, et al. Disruption of PTEN coupling with 5-HT2C receptors suppresses behavioral responses induced by drugs of abuse. Nat Med. 2006;12:324–9. doi: 10.1038/nm1349. [DOI] [PubMed] [Google Scholar]
- 62.Reynolds GP, Zhang ZJ, Zhang XB. Association of antipsychotic drug-induced weight gain with a 5-HT2C receptor gene polymorphism. Lancet. 2002;359:2086–7. doi: 10.1016/S0140-6736(02)08913-4. [DOI] [PubMed] [Google Scholar]
- 63.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 64.Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–92. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 65.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–23. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 66.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Nat Acad Sci USA. 2006;103:13415–20. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Nat Acad Sci USA. 2008;105:10073–8. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 69.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:284–8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–85. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–62. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.