Abstract
We present a new evidence taxonomy that, when combined with a set of inclusion criteria, enable drug experts to specify what their confidence in a drug mechanism assertion would be if it were supported by a specific set of evidence. We discuss our experience applying the taxonomy to representing drug-mechanism evidence for 16 active pharmaceutical ingredients including six members of the HMG-CoA-reductase inhibitor family (statins). All evidence was collected and entered into the Drug Interaction Knowledge Base (DIKB); a system that can provide customized views of a body of drug-mechanism knowledge to users who do not agree about the inferential value of particular evidence types. We provide specific examples of how the DIKB’s evidence model can flag when a particular use of evidence should be re-evaluated because its related conjectures are no longer valid. We also present the algorithm that the DIKB uses to identify patterns of evidence support that are indicative of fallacious reasoning by the evidence-base curators.
Keywords: knowledge representation, drug safety, drug-drug interactions, evidence taxonomy, evidentialism
1 Introduction
A 2006 report from the US Institute of Medicine estimates that over 1.5 million preventable adverse drug events (ADEs) occur each year in America [1]. Preventable ADEs include situations where a patient is harmed because a clinician fails to avoid, or properly manage, an interacting drug combination. Multiple studies indicate that these drug-drug interactions DDIs are a significant source of preventable ADEs [2,3].
Factors contributing to the occurrence of preventable DDIs include a lack of knowledge of the patient’s concurrent medications and inaccurate or inadequate knowledge of drug interactions by health care providers [4,5]. Information technology, especially electronic prescribing systems with clinical decision support features, can help address each of these factors to varying degrees and there is currently a great deal of interest from both government and private organizations in expanding the use of information technology during medication prescribing and dispensing [1,6]. Unfortunately, studies have found the DDI components of a wide variety of clinical decision-support tools to be sub-optimal in both the accuracy of their predictions and the timeliness of their knowledge [7–9].
What all of the systems in these studies have in common is that they rely upon some representation of drug knowledge to infer DDIs; what we refer to in this paper as a “drug-interaction knowledge base” (KB). Currently, a handful of large drug information databases are used as drug-interaction KBs in a large range of drug interaction alerting products and electronic prescribing tools [6]. The basic service most drug-interaction KBs provide is to catalog drug pairs found to interact in clinical trials or reported as such in clinician-submitted case reports. One major limitation of this approach is that it constrains drug-interaction KBs, and the tools that utilize them, to covering only interacting drug pairs that KB maintainers find in the literature and think important to include. Clinicians often must infer the potential risk of an adverse event between medication combinations that have not been studied together in a clinical trial [5]. Systems that only catalog DDI studies involving drug pairs can offer little or no support in these situations.
Some contemporary drug interaction KBs supplement their DDI knowledge by generalizing interactions involving some drug to all other drugs within its therapeutic class [10]. While clinically relevant class-based interactions exist (for example, the SSRIs and NSAIDs), this approach has been criticized for leading to some DDI predictions that are either false or are likely to have little clinical relevance [11,12]. The main reason class-based prediction can lead to false alerts is because drugs within a therapeutic class do not necessarily share the same pharmacokinetic properties. False predictions can have a negative effect on electronic prescribing systems by triggering false or irrelevant DDI alerts that can markedly impede the work-flow of care providers [13]. A high rate of irrelevant alerting is a potential barrier to widespread adoption of Computerized Physician Order Entry systems with clinical decision support [14] and stands as a major obstacle to improving patient safety.
Part of pre-clinical drug development is the use of mechanism-based DDI prediction to predict interactions between a new drug candidate and drugs currently on the market [15]. The same knowledge that is useful for predicting DDIs in the premarket setting can help clinicians in the post-market setting assess the possibility of a DDI occurring between two drugs that have never been studied together in clinical trials [11]. However, little research has been done on how to best represent and synthesize drug-mechanism knowledge to support clinical decision making. Our research attempts to fill this knowledge gap by focusing on how to best utilize drug-mechanism knowledge to help drug-interaction KBs expand their coverage beyond what has been tested in clinical trials while avoiding prediction errors that occur when individual drug differences are not recognized.
A pilot experiment that we conducted helped identify three major challenges to representing drug-mechanism knowledge [16]. First, there is often considerable uncertainty behind claims about a drug’s properties and this uncertainty affects the confidence that someone knowledgeable about drugs places on mechanism-based DDI predictions. Another challenge is that mechanism knowledge is sometimes missing; a fact that can make it diffcult to assess the validity of some claims about a drug’s mechanisms. Finally, mechanism knowledge is dynamic and any repository for drug-mechanism knowledge is faced with the non-trivial task of staying up to date with science’s rapid advances.
We have previously reported on the design and implementation of a novel knowledge-representation approach that we hypothesized could overcome these challenges [17]. The approach was implemented in a new system called the Drug Interaction Knowledge Base (DIKB); a system that enables knowledge-base curators to link each assertion about a drug property to both supporting and refuting evidence. DIKB maintainers place evidence for, or against, each assertion about a drug’s mechanistic properties in an evidence-base that is kept current through an editorial board approach. Maintainers attach to each evidence item entered into the evidence base a label describing its source and study type. Users of the system can define specific belief criteria for each assertion in the evidence-base using combinations of these evidence-type labels. The system has a separate knowledge-base that contains only those assertions in the evidence-base that meet belief criteria. The DIKB’s reasoning system uses assertions in this knowledge-base and so only makes DDI predictions using those facts considered current by the system’s maintainers and believable by users.
An intriguing feature of this evidential approach to knowledge representation is that the system can provide customized views of a comprehensive body of drug-mechanism knowledge to expert users who have differing opinions about what combination of evidence justifies belief in a biomedical assertion. Another intriguing feature is that researchers can test the empirical prediction accuracy of a rule-based theory using many sub-sets of a given body of evidence. The results of such tests can suggest which combination of evidence enables the theory to make the most optimal set of predictions in terms of accuracy and coverage of a validation set. We have found this feature useful for integrating basic science and clinical research for the purpose of predicting DDIs and discuss it in more detail in Part II of this two-paper series.
A key component of the DIKB is a new evidence taxonomy that, when combined with a set of inclusion criteria, enables drug experts to specify what their confidence in a drug mechanism assertion would be if it were supported by a specific set of evidence. The primary focus of this paper is the design and application of the new evidence taxonomy. The next section summarizes the requirements that the new taxonomy was designed to meet and contrasts them with other biomedical evidence taxonomies. The section after presents the current version of the taxonomy and details on its implementation. Then follows a discussion of our experience applying the taxonomy to representing drug-mechanism evidence for 16 active pharmaceutical ingredients including six members of the HMG-CoA-reductase inhibitor family (statins). This discussion includes mention of extensions to our previously reported work on the DIKB’s evidence model [17] including a new algorithm that the DIKB uses to identify patterns of evidence support that are indicative of fallacious reasoning by the evidence-base curators.
2 Considerations for an evidence taxonomy oriented toward confidence assignment
The DIKB’s method for modeling and computing with evidence depends on an evidence taxonomy oriented toward confidence assignment. The evidence taxonomy must have suffi-cient coverage of all the kinds of evidence that might be relevant including various kinds of experiments, clinical trials, observation-based reports, and statements in product labeling or other resources. Another important requirement for the taxonomy is that users must be able to assess their confidence in each type either by itself or in combination with other types. Only a handful of biomedical informatics systems exist that attempt to label or categorize evidence; these include the PharmGKB’s categories of pharmacogenetics evidence [18], Medical Subject Headings’ Publication Types [19], Gene Ontology’s evidence codes [20], and Pathway Tools’ evidence ontology [21]. The next few sections summarize these biomedical evidence taxonomies and contrast them with the DIKB’s requirements.
2.1 PharmGKB’s “Categories of pharmacogenetics evidence”
The PharmGKB is a Web-based knowledge repository for pharmacogenetics and pharma-cogenomics research. Scientists upload into the system data supporting phenotype relationships among drugs, diseases, and genes. All data in the PharmGKB is tagged with labels from one or more of five non-hierarchical categories called categories of pharmacogenetics evidence [18]. The categories of pharmacogenetics evidence are different from the DIKB’s evidence types because the latter represent specific sources of scientific inference such as experiments and clinical trials while the former are designed to differentiate the various kinds pharmacogenetic gene-drug findings by the specific phenotypes they cover (e.g. clinical, pharmacokinetic, pharmacodynamic, genetic, etc). In other words, the categories are oriented toward data integration rather than confidence assignment. The designers of the PharmGKB used this approach because they hypothesized that it would be capable of coalescing the results of a range of methods and study types within the field of pharmacogenetics into a single data repository that would be useful to all researchers in the field [22].
2.2 Medical Subject Headings “Publication Types”
One of the most used biomedical evidence taxonomies is the publication-type taxonomy that is a component of the Medical Subject Headings (MeSH) controlled vocabulary [23]. The MeSH controlled vocabulary is a set of over 20,000 terms used to index a very broad spectrum of medical literature for the National Library of Medicine’s PubMed database (formerly MEDLINE). Each article in PubMed is manually indexed with several MeSH terms and additional descriptors including the article’s publication type. The MeSH publication type taxonomy is designed to provide a general classification for the very wide range of articles indexed in PubMed. Hence, the taxonomy is very broad but relatively shallow. For example, publication types in the 2008 MeSH taxonomy [19] include types as varied as Controlled Clinical Trial and Sermons but only one type, In Vitro, that represents all kinds of in vitro studies including those using non-human tissue.
In knowledge representation terms, the coverage by MeSH publication types of the evidence types relevant for validating drug-mechanism knowledge is too coarse-grained. This is because the design of some in vitro experiments makes them better suited for supporting some drug-mechanism assertions more than others. For example, a recent FDA guidance to industry on drug interaction studies distinguishes three different in vitro experimental methods for identifying which, if any, specific Cytochrome P-450 enzymes metabolize a drug [15]. The three experiment types are different from the in vitro experiment type that the FDA suggests is appropriate for identifying if a drug inhibits a drug metabolizing enzyme. The next two sections will discuss two systems whose coverage of in vitro evidence is less coarse than MeSH publication types – the Gene Ontology evidence codes [20] and the Pathway Tools’ evidence ontology [21].
2.3 Gene Ontology “Evidence Codes” and the need for inclusion criteria
The Gene Ontology (GO) is a system of three separate ontologies defining relationships between biological objects in micro- and cellular biology [24]. GO is a consortium-based effort that has gained wide acceptance in the bioinformatics community because it supports consistent descriptions of the cellular location of a gene product, the biological process it participates in, and its molecular function. Authors of GO annotations are expected to specify an evidence code that indicates how a particular annotation is supported. GO evidence codes [20] are labels representing the kinds of support that a biologist might use to annotate the molecular function, cellular component, or biological process (s)he is assigning to a gene or gene product. GO has over a dozen evidence codes including codes that indicate that a biological inference is supported by experimental evidence, computational analysis, traceable and non-traceable author statements, or the curators’ judgement based on other GO annotations.
In the DIKB, the user’s confidence in an assertion rests on some arrangement of one or more evidence types. This means that the user must trust the validity of each instance of evidence that the system uses to meet the belief criteria without necessarily reviewing the evidence for his or herself. In contrast with these requirements, the authors of the GO evidence codes are very clear that the codes cannot be used as a measure of the validity of a GO annotation:
Evidence codes are not statements of the quality of the annotation. Within each evidence code classification, some methods produce annotations of higher confidence or greater specificity than other methods, in addition the way in which a technique has been applied or interpreted in a paper will also affect the quality of the resulting annotation [20].
This quote from GO evidence code documentation mentions two possible characteristics of GO evidence codes that preclude them from serving as a measure of the justification for biological annotations. First, GO evidence codes seem to represent evidence types that vary in terms of their appropriateness for justifying hypotheses. Like MeSH publication types, GO evidence codes are too coarse-grained for use as a tool for confidence assignment. Second, GO evidence codes do not address the fact that there are many possible problems with studies, experiments, author statements, and other types of evidence that can effect their validity. In other words, even if GO evidence codes were granular enough for decision support, the user would have to assess the quality of each evidence item directly or else place faith in the annotator’s judgment.
According to the The Agency for Healthcare Research and Quality, there are three components of a study that contribute or detract from its quality - its design, how it is conducted, and how its results are analyzed ([25], p.1). While it is possible to create meta-data labels that accurately reflect a study’s design, it is intractable to abstract the full range of issues that affect a study’s conduct and analysis. Our approach to ensuring user confidence in abstract evidence types is to develop and consistently apply inclusion criteria for each type of evidence in the DIKB. Inclusion criteria help ensure that all evidence within a collection meet some minimum standard in terms of quality. They are complimentary to evidence type definitions which should represent evidence classes that are fairly homogeneous in terms of their appropriateness for justifying hypotheses. The criteria are designed to help answer the kinds of methodology questions that expert users have when told that an evidence item is of a certain type.
2.4 The Pathway Tools’ “Evidence Ontology” and confirmation bias
One other currently used biomedical evidence taxonomy is found in the Pathway Tools system of pathway/genome databases (PGDBs) [21]. The Pathway Tools evidence ontology is both a computable evidence taxonomy and a set of data-structures designed so that PGDB maintainers can attach 1) the types of evidence that support an assertion in the PGDB, 2) the source of each evidence item, and 3) a numerical representation of the degree of confidence a scientist has in an assertion. The taxonomy component of the evidence ontology shares several of the types defined in GO evidence codes (Section 2.3) but adds a number of subtypes that define more specific kinds of experiments and assays than GO. The data-structure component of the “evidence ontology” enables PGDB maintainers to record the source of an evidence item, the accuracy of a given method for predicting specific hypotheses, and the scientist’s confidence in a PGDB assertion given the full complement of evidence supporting an assertion.
PGDB users are presented with a visual summary of the kinds of evidence support for a given assertion in the form of icons representing top-level evidence-types from the Pathway Tools’ evidence taxonomy (e.g. “computational” or “experimental”). Users can click on the icons to view more detailed information of the specific evidence items represented by the top-level icons including the sources of each item and its specific evidence type. This approach enables Pathway Tools to provide an overview of the kinds of evidence support for an assertion so that users might make their own judgements on the amount of confidence they should have in a PGDB assertion.
While Pathway Tools’ evidence types serve a similar function as DIKB evidence types by helping users assess their confidence in knowledge-base assertions, PGDB maintainers use them to represent only supporting evidence. We hypothesize that this approach could contribute to a form of bias called confirmation bias that can undermine attempts by users of a knowledge-base to assess the validity of its assertions.
Griffin in his review of research in the domain probability judgement calibration [26] lists several robust findings from a considerable body of research exploring biases people have when estimating the likelihood of uncertain hypotheses. Among them is the finding that people tend to exhibit various forms of over-confidence when estimating the probability that some hypothesis is true. One possible explanation for this tendency is that over-confidence is a result of confirmation bias – “…people tend to search for evidence that supports their chosen hypothesis” [26]. Under this model, confidence estimations should be more accurate when people consider situations where their hypotheses might not be true. Griffin reports that the results of some research studies are consistent with this model but that confirmation bias does not seem to be the sole cause of over-confidence during probability judgement.
We applied these results to the DIKB by requiring that maintainers seek both supporting and refuting evidence for each drug-mechanism assertion. The intent of this arrangement is to help maintainers avoid any tendency to collect evidence that only supports knowledge-base assertions and to help expert users create unbiased criteria for judging their confidence in the system’s assertions.
2.5 Curator inferences and default assumptions
In both GO evidence codes and the Pathways Tools’ evidence ontology there is an evidence type called Inferred by Curator that curators use for knowledge they infer from other assertions or annotations in the respective systems [20,21]. Inferred by Curator is not really an evidence type; rather it is a label indicating that a particular assertion exists within a knowledge-base as a result of judgement of some curator. An evidence code of this type will not work in the DIKB because its users must map their confidence in the system’s assertions to combinations of evidence codes. A user viewing an assertion tagged with Inferred by Curator might apply the level of trust that they have for the knowledge source based on previous experiences. Unfortunately, whatever judgement the user makes will be more about the knowledge-curation system rather than the specific scientific proposition in question. Alternatively, the expert might attempt to explicitly trace the curators’ judgement so as to decide for themselves if the inference was reasonable. This process might be straightforward or confusing depending on the complexity of the logic used by the curator when making the inference in question.
In constructing the DIKB we have also found situations where it was desirable to assert some knowledge element based on our knowledge of other assertions in the system. As a trivial example, when evidence in the DIKB supports the assertion that some enzyme, E, is responsible for 50% or more of some drug or drug metabolite’s total clearance from the body, then the system should also contain an assertion that more than 50% of a drug’s clearance is by metabolism. A more complex example can be found in the rules that the DIKB uses to infer a drug or drug metabolite’s metabolic clearance pathway (Appendix E, supplementary material). In such cases, new rules are added to the DIKB so that it will automatically add the needed assertions to its knowledge-base. The system’s links each automatically-inferred assertion to the assertions and rules from which it was inferred. A programmer can write code that leverages the data structures used to create the DIKB’s evidence-base to generate a report showing the logic and evidence support for any automatically inferred assertions.1
The advantage of the DIKB’s approach becomes apparent when one considers that the construction and maintenance of a large knowledge-base is a collaborative effort. GO and the PGDBs in the Pathway Tools system require curation by many domain experts and we think it reasonable to expect that, in spite of the best of intentions, curators will sometimes make mistakes or not be entirely consistent in how they enter knowledge or assign evidence. Furthermore, as a knowledge-based system grows it becomes less tractable for curators to know all of the inferences supported directly by other knowledge in the system. In contrast, once a rule is added to the DIKB that makes an assertion based on other assertions present in the system, it will always be applied consistently and across all possible instances where it is applicable.
It turns out that there are other occasions where an evidence type like Inferred by Curator might seem applicable within the DIKB. The system’s curators sometimes face situations where they are justified in entering an assertion without linking it to evidence. Such an event can occur when the curator is unable to find evidence for an assertion or when (s)he decides that an assertion does not need to be justified by evidence. In both cases the DIKB curator can decide to enter it as a default assumption. A default assumptions is a special kind of assertion considered justified by default but retractable either manually by curators or automatically by the system as it proceeds with inference.
2.6 Summary of Considerations
In summary, none of the evidence taxonomies that we reviewed have sufficient coverage of all the kinds of evidence that might be relevant for representing drug mechanism knowledge. Also, none are designed so that users can assess their confidence in each type either by itself or in combination with other types. The next section discusses the new evidence taxonomy that meets these requirements.
3 The DIKB Evidence Taxonomy and Inclusion Criteria
The current DIKB evidence taxonomy (shown in Table 3) contains 36 evidence types arranged under seven groupings representing evidence from retrospective studies, clinical trials, metabolic inhibition identification, metabolic catalysis identification, statements, reviews, and observational reports.
Table 3.
The version of the DIKB’s evidence taxonomy used in the reported study
|
We developed the taxonomy iteratively by collecting evidence for the drugs and drug metabo-lites shown in Figure 1, identifying the attributes of each evidence item, and deciding on evidence-type definitions. We were able to incorporate some definitions from WordNet [27], MeSH [23], and NCI Thesaurus [28] but the majority of the taxonomy consists of new definitions. The structure of the taxonomy and granularity of its definitions is similar to the Pathway Tools’ evidence ontology [21] though the only definitions that the two resources share are for traceable and non-traceable author statements. Also, we deliberately excluded the “Inferred by Curator” evidence type present in the Pathway Tools’ evidence ontology [21] and Gene Ontology’s evidence codes [20] for the reasons discussed in Section 2.5.
Fig. 1.
The 16 drugs and 19 drug metabolites chosen for DIKB experiments
We implemented the taxonomy in the OWL-DL language [29]; a description logic that provides a formal semantics for representing taxonomic relationships in a manner that can be automatically checked to ensure consistent classification. We used the Prot’eg’e ontology editor2 to create the taxonomy and the RACER inference engine [30] to test it for consistent type definitions. The evidence taxonomy is integrated into the structured vocabulary used by DIKB and is available on the Web [31].
We designed the set of seven inclusion criteria shown in Appendix B (supplementary material) to compliment a sub-set of evidence type definitions from the DIKB’s evidence taxonomy. Like the evidence taxonomy, we developed the inclusion criteria iteratively during the early stages of collecting evidence for the drugs and drug metabolites shown in Figure 1. This meant that changes to inclusion criteria would sometimes require that evidence previously thought acceptable be discarded. The criteria became stable after making progress collecting evidence on several drugs. In their current form, the seven criteria define the minimum quality standards for 21 evidence types in the taxonomy.
There were a total of 12 evidence types for which we did not define inclusion criteria. Seven of these are general evidence types: Statement, Non-traceable Statement, An observation-based report, An observation-based ADE report, A clinical trial, A DDI clinical trial, and A retrospective study. We used more specific evidence types within the taxonomic sub-hierarchies that these five types resided in and so defined inclusion criteria accordingly.
The other five evidence types with no inclusion criteria represent classes of evidence that we decided not to include for this study. We excluded the two types of author statements in the taxonomy (A traceable author statement and A traceable drug-label statement) because our evidence collection policy requires that curators retrieve and evaluate the evidence source that an author’s statement refers to rather than rely strictly on the author’s interpretation of that evidence source. We excluded the type A retrospective population pharmacokinetic study because we thought evidence of this class would be difficult to acquire and interpret. We also neglected to define inclusion criteria for the type A retrospective DDI study because we did not come across evidence of this type while defining inclusion criteria. Finally, the evidence collection process that we describe in Section 4.1 did not include public adverse-event reporting databases so we did not define inclusion criteria for the type An observation-based adverse-drug event report in a public reporting database.
4 Using the DIKB’s evidence taxonomy to represent a body of drug-mechanism evidence
We applied the novel evidence taxonomy to the task of representing drug-mechanism evidence for six members of a family of drugs called HMG-CoA reductase inhibitors (statins) and ten drugs with which they are sometimes co-prescribed. Members of the statin drug family are very commonly used to help treat dyslipidemia. While statins have a relatively wide therapeutic range, patients taking a drug from this class are at a higher risk for a muscle disorder called myopathy if they take another drug that reduces the statin’s clearance [32]. The sixteen drugs we chose are all currently sold on the US market, popularly prescribed by physicians, and have been the subject of numerous in vivo and in vitro pharmacokinetic studies. Many of them are known to be cleared, at least partly, by drug metabolizing enzymes that are susceptible to inhibition.
DDIs that occur by metabolic inhibition can affect the concentration of active or toxic drug metabolites in clinically relevant ways. For example, both lovastatin and simvastatin are administered in lactone forms that have little or no HMG-CoA reductase inhibition activity but that are readily converted by the body to pharmacodynamically active metabolites [33,34]. Clinical trial data indicates that metabolism by CYP3A4 is a clinically relevant clearance pathway for these metabolites [35,36]. Similarly, in vitro evidence indicates that CYP3A4 is the primary catalyst for the conversion of the HMG-CoA reductase inhibitor atorvastatin into its two active metabolites [37]. For this reason, we also collected and entered drug-mechanism evidence for 19 active metabolites of the drugs we had chosen. Figure 1 lists the 16 drugs and 19 drug metabolites we chose to represent in the DIKB.
4.1 The Evidence Collection Process
The quality and coverage of the DIKB’s drug-mechanism knowledge depends a great deal on the process used to collect and maintain evidence. We attempted to apply a process geared toward building a coherent body of knowledge that has minimal bias and is up-to-date. One informaticist (RB) and two drug-experts (CC and JH) formed an evidence board that was responsible for collecting and entering all evidence into the DIKB.3 The evidence-collection process was iterative for the first few months while evidence types and inclusion criteria were being developed. The board would choose a particular drug to model then collect a set of journal articles, drug labels, and authoritative statements that seemed relevant to each of the various drug-mechanism assertions in the DIKB’s rule-based theory of drug-drug interactions. The evidence board would then meet together and discuss each evidence item and the issues that affected its use in the DIKB. By the time all members of the evidence board committed to using the evidence types (Table 3) and inclusion criteria (Appendix B, supplementary material) the following evidence collection process had become routine:
The evidence board chose a particular drug to model.
The informaticist then received from each drug expert references to specific evidence sources that they thought would support or rebut one or more drug-mechanism assertions.
The informaticist did his own search of the literature that included seeking information from primary research articles in PubMed, statements in drug product labeling or FDA guidances, and various drug information references including Goodman & Gilman’s [38]. One of the drug experts was affliated with the proprietary University of Washington Metabolism and Transport Drug Interaction Database4 and performed searches of that resource then forwarded the results to the informaticist.
The informaticist would then summarize all evidence items from each source, classify their evidence types, and check if they met inclusion criteria. The evidence board would then meet and decide as a group whether each evidence item should enter the DIKB’s evidence-base or be rejected as support or rebuttal for a specific assertion.
The informaticist would enter accepted evidence items into the DIKB using the DIKB’s Web interface. He also entered rejected evidence items into a simple database used by the DIKB during evidence validation tests.
The DIKB performed several validation tests on each new evidence entry before it was stored in the system’s evidence-base. These included checking if an evidence entry was redundant or had previously been rejected by DIKB curators as support or rebuttal for the assertion it was being linked to. The system also checked if a new evidence item would create an evidence pattern that was indicative of circular reasoning by evidence-base curators. This last test was possible because we made sure to explicitly represent any conjectures behind a specific application of evidence. The next section describes the motivation for representing conjectures and the novel algorithm used to identify circular reasoning.
4.2 Conjectures and knowledge-base maintenance
Interpreting the results of a scientific investigation as support for a particular assertion can sometimes require making conjectures that scientific advance might later prove to be invalid. If such conjectures are later shown to be false, it is important to re-consider how much support the scientific investigation lends to any assertion it was once thought to support. One unique feature of the DIKB is that it can represent the conjectures behind a specific application of evidence. These representations are called evidence-use assumptions and they facilitate keeping knowledge in the system both current and consistent.
4.2.1 Evidence-use assumptions help keep knowledge current
To illustrate how evidence-use assumptions help keep knowledge current suppose that a pharmacokinetic clinical trial involving healthy patients finds a significant increase in the systemic concentration of drug-A in the presence of drug-B. If the study meets inclusion criteria, and it is thought that that drug-B is a selective inhibitor of the ENZ enzyme in humans, then an evidence-base curator might apply this evidence as support for the assertion (drug-A substrate-of ENZ). This particular application of the hypothetical study would depend on the conjecture that drug-B is an in vivo selective inhibitor of the ENZ. Otherwise, alternative explanations for the observed increase in the systemic concentration of drug-A remain quite feasible. In this situation it will be important to reconsider this use of evidence if future work reveals that drug-B increases patient exposure to drug-A by some other mechanism than reducing ENZ’s catalytic function such as modulation of the function of an alternate drug-metabolizing enzyme or an efflux transport protein.
Unlike systems that just cite evidence, the DIKB’s formal model of evidence enables it to flag when a conjecture has become invalid and alert knowledge-base curators to the need to reassess their original interpretation of what assertions a piece of evidence supports. Currently, DIKB curators make an evidence-use assumption known to the DIKB by first identifying the label of an assertion in the knowledge-base that represents the evidence-use assumption. They then add the label to a list of assumptions that is contained in the data structure used to represent the specific evidence item that they are viewing.
In our experience, evidence-use assumptions are an attribute of a particular type of evidence. For example, pharmacokinetic DDI studies, like the one mentioned in the previous hypothetical example, often depend on the assumption that the precipitant has no measurable effect on any other clearance route of the object drug. This is an evidence-use assumption that applies to all pharmacokinetic drug-drug interaction studies using selective inhibitors. Based on this observation, we have attempted to define evidence-use assumptions for each new evidence type that is added to the DIKB’s evidence taxonomy. These assumptions are written as general statements that apply to one or more evidence types and are added to inclusion criteria documentation so that curators will know what specific assumption(s) to declare when adding an item of evidence to the system. After curators have approved an evidence item, they identify assertions within the DIKB that match each specific evidence-use assumption. In many cases, a suitable assertion will not be present in the DIKB. If so, curators must add the new assertion to the DIKB then link it as an evidence-use assumption for the evidence item.
4.2.2 Evidence-use assumptions help keep knowledge consistent
Evidence-use assumptions can also help identify a pattern, called a circular line of evidence support, that is indicative of fallacious reasoning by evidence-base curators. A hypothetical example should help clarify the kind of situation we are describing and its implications.
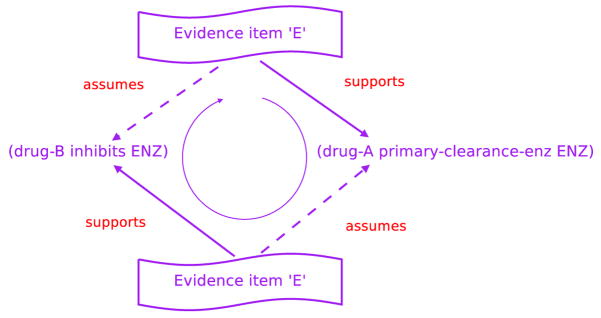
Assume some evidence item, E, exists in the evidence-base as support for the assertion (drug-B inhibits ENZ) and that (drug-A primary-clearance-enzyme ENZ) is an evidence-use assumption for this application of E. In addition, assume that E also acts as support for (drug-A primary-clearance-enzyme ENZ) and that this other use of E depends on the validity of the assertion (drug-B inhibits ENZ). If there is no evidence against either assertion and E meets both assertions’ supporting belief criteria, then the system will consider both assertions to be valid.
Figure 2 makes apparent the dilemma – the conjecture, (drug-A primary-clearance--enzyme ENZ), is necessary for evidence item E to act as support for the assertion (drug-B inhibits ENZ) but is being justified by the same evidence item, E, that assumes the same proposition E is supposed to justify. Intriguingly, the same unsound reasoning would be present even if evidence item E is being used to refute the assertion (drug-B inhibits ENZ). Neither kind of circular reasoning should be allowed in the DIKB’s evidence-base.
Fig. 2.
A circular line of evidence support that indicates circular reasoning within the evidence-base
We have designed and implemented the following algorithm in the DIKB for detecting when an new evidence item would cause a circular line of evidence support:
Let E be an evidence item that is being considered as evidence for or against some assertion, A. Assume that the use of E as evidence for or against A is contingent on the validity of one or more other assertions in the set AL = as1, as2, …, asn. The set of assertions in AL are the evidence-use assumptions for E. If E is currently being used as evidence for or against some assertion, asi, in AL and the use of E to support or refute asi depends on assuming A, then the use of E to support or refute A would create a circular line of evidence support.
The DIKB will not allow a curator to enter an evidence item that passes this test into itsevidence base.
Circular reasoning might be present in the evidence-base anytime an evidence-use assumption is supported by the same evidence item that the assumption is linked to. We can create an algorithm to identify when this form of circular evidence support is present in the knowledge-base by simplifying the previous algorithm.
Let E be an evidence item and let the set AL = as1, as2, …, asn be the set of evidence-use assumptions for E. If E is currently being used as evidence for or against some assertion, asi, in AL, then circular reasoning might be present in the evidence-base.
The DIKB does not currently implement this algorithm in its validation tests but will in future versions.
5 Results
Work on the evidence-base stopped in January 2008. In its present state it consists of 257 evidence items from 102 unique sources applied as evidence for or against 207 drug-mechanism assertions.
5.1 The Classification of Evidence within the Evidence-base
The evidence board used only one-third of the 36 types in the evidence taxonomy to classify all the 257 evidence items. Some evidence types were not used because of specific evidence collection policies while other types were not used because no acceptable evidence in their class could be found. For example, even though the evidence board collected numerous case reports describing adverse drug events in patients taking two or more of the drugs in our study, none of the five observation-based evidence types were entered into the system. This was because none of the reports that were found measured the systemic concentrations of the purported object drug in a way that satisfied the inclusion criteria for supporting or refuting an assertion about a drug’s metabolic properties.
The 12 evidence types that were used to classify evidence items are shown in Table 1 along with the number of supporting or refuting evidence items each type was assigned to. It is clear from Table 1 that some evidence types are present in the evidence-base much more often than other types even though the experiments they represent have relatively similar purposes. For example, the evidence-base has almost eight-fold more evidence items of the type A CYP450, human microsome, metabolic enzyme inhibition experiment then the type A CYP450, recombinant, metabolic enzyme inhibition experiment even though the purpose of both kinds of experiments is to test a drug or drug metabolite’s ability to inhibit some enzyme in vitro. Similarly, the system has three-fold more items of the type A CYP450, recombinant, drug metabolism identification experiment with possibly NO probe enzyme inhibitor(s) than the type A CYP450, human microsome, drug metabolism identification experiment using chemical inhibitors even though both experiments attempt to identify the CYP450 enzymes capable of metabolizing a drug or drug metabolite in vitro.
Table 1.
The evidence board used only one-third of the 36 types in the evidence taxonomy to classify all the 257 non-redundant evidence items. The 12 evidence types are shown in this table along with the number of supporting or refuting evidence items each type was assigned to. Indented evidence types are sub-types of the type in the previous row
| Clinical trial types | ||
|---|---|---|
| Evidence type | Evidence for | Evidence against |
| A pharmacokinetic clinical trial | 31 | 0 |
| A genotyped pharmacokinetic clinical trial | 5 | 1 |
| A randomized DDI clinical trial | 49 | 11 |
| A non-randomized DDI clinical trial | 8 | 0 |
| A parallel groups DDI clinical trial | 4 | 0 |
|
| ||
| Total | 97 | 12 |
| in vitro experiment types | ||
|
| ||
| Evidence type | Evidence for | Evidence against |
|
| ||
| A CYP450, recombinant, metabolic enzyme inhibition experiment | 2 | 0 |
| A CYP450, human microsome, metabolic enzyme inhibition experiment | 13 | 2 |
| A drug metabolism identification experiment | 4 | 0 |
| A CYP450, recombinant, drug metabolism identification experiment with possibly NO probe enzyme inhibitor(s) | 31 | 6 |
| A CYP450, human microsome, drug metabolism identification experiment using chemical inhibitors | 8 | 4 |
|
| ||
| Total | 58 | 12 |
| Non-traceable statement types | ||
|
| ||
| Evidence type | Evidence for | Evidence against |
|
| ||
| A non-traceable, but possibly authoritative, statement | 22 | 0 |
| A non-traceable drug-label statement | 52 | 4 |
|
| ||
| Total | 74 | 4 |
Generally-defined evidence types were often used when an evidence item did not fit one of the more specific evidence-types within a particular sub-hierarchy. Eleven of the twelve types shown in Table 1 are sub-types of some other, more general, evidence types within the greater evidence taxonomy. One exception was the most general in vitro evidence type A drug metabolism identification experiment that is assigned four times in the current DIKB evidence-base. All four uses of the evidence type were to classify metabolite identification experiments that could not be classified using the more specific types within the hierarchy.
5.2 Observed biases
One can calculate from Table 1 that evidence types assignments in the current DIKB slightly favor clinical trial types (42%) over in vitro studies (27%) and non-traceable statements in drug labeling and FDA guidance documents (30%). The distribution of evidence types among individual assertion types is much more diverse than that of the evidence-base as a whole (Tables 3 and 4 in supplementary material). For example, all 15 evidence items linked to inhibition-constant assertions are from in vitro evidence types while no in vitro evidence is currently linked to a maximum-concentration assertion. Likewise, two-thirds of the evidence items linked to maximum-concentration assertions are instances of clinical trial types while the one-third are instances of non-traceable statement types. Approximately the opposite distribution of evidence types is present in items linked to bioavailability assertions (38% clinical trial types and 62% non-traceable statements).
While the evidence board attempted, where appropriate, to collect both supporting and refuting evidence for each assertion, the current evidence-base is strongly biased toward supporting evidence. Eighty-two percent of the 102 evidence sources provide evidence items that are used strictly as support for one or more assertions. In comparison, only 3% of sources provide strictly refuting evidence items and only 15% of sources provide both supporting and refuting evidence items. Of the 257 non-redundant evidence items, 229 (89%) support, and 28 (11%) refute, some drug mechanism assertion. In terms of the 20 assertions types that the DIKB currently represents, only four (20%) have any assertions with refuting evidence; substrate-of, inhibits, increases-auc, and primary-metabolic-enzyme.
5.3 The use of default assumptions
The evidence-board labeled approximately one-fifth (39) of the assertions in the DIKB default assumptions. Nearly half (17) of the default assumptions were entered because of a DIKB policy that treated certain information in FDA guidances as completely authoritative.5 The 17 assertions are linked to evidence items that refer to the FDA guidance that prompted the evidence-board’s decision to make them default assumptions.
Another 17 assertions are labeled default assumptions but have no evidence items linked to them at all. Five of these were entered by the evidence board because of actions specified in the inclusion criteria for pharmacokinetic DDI studies (defined in Appendix B of supplementary material). The remaining 12 were entered without evidence based on the knowledge of one or more members of the evidence-board. These were entered as default assumptions out of convenience with the intent that a DIKB curator would seek evidence for and against the assertions at a later time.
5.4 The application of evidence-use assumptions
Nearly one-quarter (23%) of the evidence items in the current evidence-base have at least one evidence-use assumption. Table 2 provides a sample of five of these evidence items. Fifty-three evidence items are linked to one evidence-use assumption and five evidence items are linked to two bringing the total number of evidence-use assumptions in the current DIKB to 63. Only twenty-three (11%) of the 207 assertions in the DIKB comprise all 63 evidence-use assumptions. The number of times the evidence board used any specific assertion as an evidence-use assumption ranged from once to nine times (mean: 2.7, median: 1.0).
Table 2.
A sample of five of the 58 evidence items in the DIKB’s evidence-base that were entered with evidence-use assumptions
| Source | Assertion evidence is supporting | Evidence type | Evidence-use assumption(s) |
|---|---|---|---|
| [45] | diltiazem inhibits CYP3A4 | A randomized DDI clinical trial | triazolam’s primary-total-clearance enzyme is CYP3A4 |
| [36] | simvastatin is a substrate-of CYP3A4 | A randomized DDI clinical trial | itraconazole is a selective inhibitor of CYP3A4 in vivo |
| [46] | alprazolam is a substrate-of CYP3A5 | A genotyped pharmacokinetic clinical trial | CYP3A5 has multiple drug- metabolizing phenotypes |
| [47] | clarithromycin is a substrate-of CYP3A4 | A CYP450, human mi- crosome, drug metabolism identification experiment using chemical inhibitors | ketoconazole is a selective inhibitor of CYP3A4 in vitro |
| [35] | lovastatin’s primary-total-clearance enzyme is CYP3A4 | A randomized DDI clinical trial |
|
6 Discussion and Conclusion
We successfully used the DIKB’s new evidence taxonomy to integrate drug mechanism evidence from a variety of sources including in vitro experiments, clinical trials, and statements from drug product labels. The evidence taxonomy and related inclusion criteria were instrumental to ensuring that the evidence entered into the DIKB was of high quality. All 257 evidence items in the DIKB are labeled by their type from the novel evidence taxonomy and meet the inclusion criteria for their assigned type. The taxonomy was also used extensively in a set of tests used to ensure that the current evidence-base has no redundant entries, rejected evidence items, or applications of evidence that were the result of circular reasoning by the co-investigators.
6.1 Limitations of the current evidence-base
The DIKB is designed so that expert users can define belief criteria using abstract evidence types. Incorrect classification of an evidence item’s type could cause the system ability to falsely appear as if it has satisfied the user’s belief criteria. One limitation of the current evidence-base is that we did not independently evaluate how accurately and consistently the evidence-board classified evidence. The evidence board employed some internal consistency checks such as reviewing each evidence item multiple times before it was entered into the DIKB and using double-entry methods to track an evidence item’s progress through the evidence collection process. However, it would be desirable to acquire independent verification that the evidence-board’s classifications were accurate and consistent across all entries.
While the evidence board attempted to collect both supporting and refuting evidence for each assertion, the current evidence-base is strongly biased toward supporting evidence (see Section 5.2). It is possible that this bias in the DIKB’s evidence-base is a reflection of a more general bias in the scientific literature towards publishing studies that confirm hypotheses. Our methods are not capable of answering this question definitively because we do not claim to have collected an exhaustive set of evidence within any of our evidence classifications. It is unclear at this time if this observed bias will hinder the system’s ability to help users overcome any tendency toward confirmation bias (see Section 2.4).
Another limitation of the current evidence-base is that the evidence-board did not search for evidence in the FDA Summary Basis for Approval for each drug or the EMBASE6, Web of Science®, Cochrane Library7, CINHAL8 publication database. It is possible that these resources might have contained important evidence that is now missing in the DIKB. Future work on the evidence-base should include comprehensive searches of these sources as well as other possible sources such as The Medical Letter9.
6.2 Future Work on the Taxonomy
While the taxonomy provides coverage of a broad range of possible evidence types relevant for support drug mechanism assertions, it is likely that many other evidence types are yet to be defined and included. Most evidence items in the DIKB (98%) are classified using relatively specific types within the taxonomy. However, as Section 5.1 notes, there is a need for additional types to more specifically classify metabolite identification experiments. Future work will address this need and the need for a detailed evaluation of the taxonomy to test its coverage and determine if there is good agreement in the evidence classifications made by drug experts using the taxonomy. Future work should also test our hypotheses that 1) expert users should be able to assess their confidence in the system’s assertions relatively quickly once they are familiar with evidence type definitions and their associated inclusion criteria and 2) that this process should involve less effort and be more consistent than requiring the expert to review the original sources for each evidence item.
6.3 Future Work to Support Evidence Collection
Some assertions in the evidence-base have numerous pieces of evidence to support them of many different types. For example, as of this writing, the assertion (itraconazole inhibits CYP3A4) can be supported by at least three randomized clinical trials [39–41], drug product labeling [42], and an FDA guidance [15]. An interesting question in this case is – when should one stop collecting evidence for an assertion?
DIKB curators are charged with collecting a minimally-biased body of relevant evidence from which customized views of drug-mechanism knowledge can be created. Since the belief criteria of different expert users will not necessarily be known in advance, curators must attempt to collect all available items of each evidence type that is relevant for supporting or refuting each assertion. The evidence collection approach that we used (Section 4.1) went a long way toward achieving this ideal. However, time constraints and an over-abundance of evidence for some assertions, meant that we did not collect all relevant evidence items of each evidence type. Achieving the ideal will certainly require the use of advanced informatics tools to ease the curators task. Research in machine learning and artificial intelligence provides several examples of machine classifiers that accurately identify relevant articles from indexed research abstracts [43] and automatically extract biomedical relationships [44]. We think that human curators should always make the final decision as to how to apply a given item of evidence but automated tools have the potential to greatly ease their task.
6.4 Conclusion
We have presented a novel drug-mechanism evidence taxonomy that, when combined with a set of inclusion criteria, enables drug experts to specify what their confidence in a drug mechanism assertion would be if it were supported by a specific set of evidence. While it is likely that many other evidence types are yet to be defined and included in the taxonomy, the current version was instrumental to ensuring that the 257 drug-mechanism evidence items entered into the DIKB’s current evidence-base were of high quality.
We have also highlighted features of the evidential knowledge-representation approach implemented in the DIKB that should be useful for representing knowledge in other biomedical domains where knowledge is dynamic, sometimes missing, and often uncertain. Rather than provide expert users with a static view of knowledge within a specific domain, the evidential approach enables them to construct customized views of a comprehensive body of knowledge based on their own, subjective, interpretation of evidence. An even more powerful feature of an evidential system is that it can iterate through a large number of possible evidence-type combinations to determine which combination of evidence enables a model or theory to make the most optimal set of predictions in terms of accuracy and coverage of a validation set. Part II of this series provides a complete description an experiment we conducted to explore this if the method could be used to make accurate and clinically relevant DDI predictions.
Supplementary Material
Acknowledgments
This project was funded in part by a National Library of Medicine Biomedical and Health Informatics Training Program grant (T15 LM07442) and an award from the Elmer M. Plein Endowment Research Fund (University of Washington School of Pharmacy). The DIKB ontology and evidence taxonomy were developed using the Prot’eg’e resource, which is supported by grant LM007885 from the United States National Library of Medicine. The author expresses sincere appreciation to Drs. Tom Hazlet and John Gennari for their comments during the course of this project.
Footnotes
More specifically, the DIKB uses declarative rules and Truth Maintenance System (TMS) jus-tifications to automatically add the needed assertions to knowledge-base [17]. The system’s TMS links each automatically-inferred assertion to the assertions and rules from which it was inferred.
The professional role of each co-investigator during the evidence collection process is mentioned throughout this section to convey to the reader the interdisciplinary approach used to construct the evidence base.
Specifically, DIKB curators assumed the validity of drugs or chemicals listed as selective inhibitors or probe substrates of certain drug-metabolizing enzymes in an FDA guidance to industry on drug-interaction studies [15].
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Committee on Identifying and Preventing Medication Errors, Preventing medication errors, Tech. rep., Institute of Medicine, 0309102685 (2006).
- 2.Gurwitz J, Field T, Judge J, Rochon P, Harrold L, Cadoret C, Lee M, White K, LaPrino J, Erramuspe-Mainard J, DeFlorio M, Gavendo L, Auger J, Bates D. The incidence of adverse drug events in two large academic long-term facilities. Am J Med. 2005;118:251–258. doi: 10.1016/j.amjmed.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Juurlink DN, Mamdani M, Kopp A, Laupacis A, Redelmeier DA. Drug-drug interactions among elderly patients hospitalized for drug toxicity. JAMA. 2003;289(13):1652–1658. doi: 10.1001/jama.289.13.1652. [DOI] [PubMed] [Google Scholar]
- 4.Chen YF, Avery A, Neil K, Johnson C, Dewey M, Stockly I. Incidence and possible causes of prescribing potential hazardous/contraindicated drug combinations in general practice. Drug Safety. 2005;28:67–80. doi: 10.2165/00002018-200528010-00005. [DOI] [PubMed] [Google Scholar]
- 5.Preskorn SH. How drug-drug interactions can impact managed care. The American Journal of Managed Care. 2004;10(6 Suppl):S186–S198. [PubMed] [Google Scholar]
- 6.Miller R, Gardner R, Johnson K, Hripscak G. Clinical decision support and electronic prescribing systems: a time for responsible thought and action. JAMIA. 2005;12(4):365–76. doi: 10.1197/jamia.M1830. pubMed ID:15905481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazlet T, Lee TA, Hansten P, Horn JR. Performance of community pharmacy drug interaction software. J Am Pharm Assoc. 2001;41(2):200–204. doi: 10.1016/s1086-5802(16)31230-x. pubMed ID:11297332. [DOI] [PubMed] [Google Scholar]
- 8.Min F, Smyth B, Berry N, Lee H, Knollmann B. American Society for Clinical Pharmacology and Therapeutics. Vol. 75. 2004. Critical evaluation of hand-held electronic prescribing guides for physicians. [Google Scholar]
- 9.Smith WD, Hatton RC, Fann AL, Baz MA, Kaplan B. Evaluation of drug interaction software to identify alerts for transplant medications. Ann Pharmacother. 2005;39(1):45–50. doi: 10.1345/aph.1E331. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh TC, Kuperman G, Jaggi T, Hojnowski-Diaz P, Fiskio J, Williams D, Bates D, Gandhi T. Characteristics and consequences of drug allergy alert overrides in a computerized physician order entry system. JAMIA. 2004;11(6):482–91. doi: 10.1197/jamia.M1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansten P. Drug interaction management. Pharmacy World and Science. 2003;25(3):94–97. doi: 10.1023/a:1024077018902. [DOI] [PubMed] [Google Scholar]
- 12.Bergk V, Haefeli W, Gasse C, Brenner H, Martin-Facklam M. Information deficits in the summary of product characteristics preclude an optimal management of drug interactions: a comparison with evidence from the literature. Eur J Clin Pharmacol. 2005;61(5–6):327–35. doi: 10.1007/s00228-005-0943-4. [DOI] [PubMed] [Google Scholar]
- 13.van der Sijs H, Aarts J, Vulto A, Berg M. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc. 2006;13(2):138–147. doi: 10.1197/jamia.M1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reichley R, Seaton T, Resetar E, Micek S, Scott K, Fraser V, Dunagan C, Bailey T. Implementing a commercial rule base as a medication order safety net. JAMIA. 2005;12(4):383–389. doi: 10.1197/jamia.M1783. pubMed ID:15802481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Internal Authorship. [Last accessed 09/25/2006 (September 2006)];FDA guideline: Drug interaction studies – study design, data analysis, and implications for dosing and labeling. doi: 10.1038/sj.clpt.6100054. Internet, http://www.fda.gov/Cber/gdlns/interactstud.htm. [DOI] [PubMed]
- 16.Boyce R, Collins C, Horn J, Kalet I. Qualitative pharmacokinetic modeling of drugs; Proceedings of the AMIA; 2005. pp. 71–75. [PMC free article] [PubMed] [Google Scholar]
- 17.Boyce R, Collins C, Horn J, Kalet I. Modeling Drug Mechanism Knowledge Using Evidence and Truth Maintenance. IEEE Transactions on Information Technology in Biomedicine. 2007;11(4):386–397. doi: 10.1109/titb.2007.890842. [DOI] [PubMed] [Google Scholar]
- 18.Rubin DL, Carrillo M, Woon M, Conroy J, Klein TE, Altman RB. A resource to acquire and summarize pharmacogenetics knowledge in the literature. Medinfo. 2004;11(Pt 2):793–797. [PubMed] [Google Scholar]
- 19.Internal Authorship. [Last accessed 05/14/2008 (2008)];Medical Subject Headings Publication Types. Internet, http://www.nlm.nih.gov/mesh/pubtypes2008.html.
- 20.Gene Ontology Consortium. [Last accessed 05/12/2008 (2008)];The GO Evidence Code Guide. Internet, http://www.geneontology.org/GO.evidence.shtml.
- 21.Karp P, Paley S, Krieger C, Zhang P. An evidence ontology for use in pathway/genome databases. Pacific Symposium on Biocomputing. 2004:190–201. doi: 10.1142/9789812704856_0019. [DOI] [PubMed] [Google Scholar]
- 22.Altman RB, Flockhart DA, Sherry ST, Oliver DE, Rubin DL, Klein TE. Indexing pharmacogenetic knowledge on the World Wide Web. Pharmacogenetics. 2003;13(1):3–5. doi: 10.1097/00008571-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Coletti MH, Bleich HL. Medical Subject Headings Used to Search the Biomedical Literature. J Am Med Inform Assoc. 2001;8(4):317–323. doi: 10.1136/jamia.2001.0080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Research. 34 doi: 10.1093/nar/gkj021. database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West S, King V, Carey T, Lohr K, McKoy N, Sutton S, Lux L. Tech. Rep. 02-E016. Agency for Healthcare Research and Quality; 2002. Systems to rate the strength of scientific evidence. [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin D, Brenner L. The Blackwell Handbook of Judgement and Decision Making, Blackwell. Ch. Perspectives On Probability Judgment 2004 [Google Scholar]
- 27.Miller GA. WordNet: a lexical database for English. Commun ACM. 1995;38(11):39–41. [Google Scholar]
- 28.de Coronado S, Haber MW, Sioutos N, Tuttle MS, Wright LW. NCI Thesaurus: using science-based terminology to integrate cancer research results. Medinfo. 2004;11(Pt 1):33–37. [PubMed] [Google Scholar]
- 29.World Wide Web Consortium. [Last accessed 02/14/2007];Web Ontology Languave (OWL) 2007 Internet, http://www.wff. org/2004/OWL/
- 30.Haarslev V, M’oller R. Racer: A core inference engine for the semantic web. 2003:27–36. [Google Scholar]
- 31.Boyce R, Collins C, Gennari J, Horn J, Kalet I. [Last accessed 09/12/2008];DIKB Ontology. 2007 Internet, http://www.pitt.edu/~rdb20/data/DIKB_evidence_ontology_v1.0.owl.
- 32.Herman RJ. Drug interactions and the statins. CMAJ. 1999;161(10):1281–1286. [PMC free article] [PubMed] [Google Scholar]
- 33.Merck, zocor (simvastatin) tablet, film coated, FDA-approved drug product labeling, last accessed on DailyMed 04/19/2008 (07 2007).
- 34.Merck, mevacor (lovastatin) tablet, FDA-approved drug product labeling, last accessed on DailyMed 04/19/2008 (01 2008).
- 35.Neuvonen PJ, Jalava KM. Itraconazole drastically increases plasma concentrations of lovastatin and lovastatin acid. Clin Pharmacol Ther. 1996;60(1):54–61. doi: 10.1016/S0009-9236(96)90167-8. [DOI] [PubMed] [Google Scholar]
- 36.Neuvonen PJ, Kantola T, Kivisto KT. Simvastatin but not pravastatin is very susceptible to interaction with the CYP3A4 inhibitor itraconazole. Clin Pharmacol Ther. 1998;63(3):332–341. doi: 10.1016/S0009-9236(98)90165-5. [DOI] [PubMed] [Google Scholar]
- 37.Jacobsen W, Kuhn B, Soldner A, Kirchner G, Sewing KF, Kollman PA, Benet LZ, Christians U. Lactonization is the critical first step in the disposition of the 3-hydroxy-3-methylglutaryl-coa reductase inhibitor atorvastatin. Drug Metab Dispos. 2000;28(11):1369–1378. [PubMed] [Google Scholar]
- 38.Brunton L. Ch. Design and Optimization of Dosage Regimens: Pharmacokinetic Data. McGraw-Hill; New York: 2006. Goodman & Gilman’s the Pharmacological Basis of Therapeutics. [Google Scholar]
- 39.Olkkola KT, Backman JT, Neuvonen PJ. Midazolam should be avoided in patients receiving the systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;55(5):481–485. doi: 10.1038/clpt.1994.60. [DOI] [PubMed] [Google Scholar]
- 40.Varhe A, Olkkola KT, Neuvonen PJ. Oral triazolam is potentially hazardous to patients receiving systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;56(6 Pt 1):601–607. doi: 10.1038/clpt.1994.184. [DOI] [PubMed] [Google Scholar]
- 41.Yasui N, Kondo T, Otani K, Furukori H, Kaneko S, Ohkubo T, Nagasaki T, Sugawara K. Effect of itraconazole on the single oral dose pharmacokinetics and pharmacodynamics of alprazolam. Psychopharmacology (Berl) 1998;139(3):269–273. doi: 10.1007/s002130050715. [DOI] [PubMed] [Google Scholar]
- 42.Janssen, sporanox (itraconazole) capsule, FDA-approved drug product labeling, last accessed on DailyMed 05/16/2008 (01 2008).
- 43.Rubin DL, Thorn CF, Klein TE, Altman RB. A statistical approach to scanning the biomedical literature for pharmacogenetics knowledge. J Am Med Inform Assoc. 2005;12(2):121–129. doi: 10.1197/jamia.M1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pustejovsky J, Castano J, Zhang J, Kotecki M, Cochran B. Robust relational parsing over biomedical literature: extracting inhibit relations; Pacific Symposium on Biocomputing; 2002. pp. 362–373. [DOI] [PubMed] [Google Scholar]
- 45.Kosuge K, Nishimoto M, Kimura M, Umemura K, Nakashima M, Ohashi K. Enhanced effect of triazolam with diltiazem. Br J Clin Pharmacol. 1997;43(4):367–372. doi: 10.1046/j.1365-2125.1997.00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park JY, Kim KA, Park PW, Lee OJ, Kang DK, Shon JH, Liu KH, Shin JG. Effect of CYP3A5*3 genotype on the pharmacokinetics and pharmacodynamics of alprazolam in healthy subjects. Clin Pharmacol Ther. 2006;79(6):590–599. doi: 10.1016/j.clpt.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Rodrigues AD, Roberts EM, Mulford DJ, Yao Y, Ouellet D. Oxidative metabolism of clarithromycin in the presence of human liver microsomes. Major role for the cytochrome P4503A (CYP3A) subfamily. Drug Metab Dispos. 1997;25(5):623–630. [PubMed] [Google Scholar]
- 48.Internal Authorship, FDA guidance for industry - population pharmacokinetics, Tech. rep., Federal Drug Administration (1999).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.