Abstract
Uterine leiomyosarcoma (ULMS) is a rare gynecologic malignancy with a low survival rate. Currently, there is no effective treatment for ULMS. Infrequent occurrences of human ULMS hamper the understanding of the initiation and progression of the disease, thereby limiting the ability to develop efficient therapies. In order to elucidate the roles of the p53 and BRCA1 tumor suppressor genes in gynecologic malignancies, we generated mice in which p53 and/or BRCA1 can be conditionally deleted using anti-Müllerian hormone type II receptor (Amhr2)-driven Cre recombinase. We showed that conditional deletion of p53 in mice results in the development of uterine tumors that resemble human ULMS and that concurrent deletion of p53 and BRCA1 significantly accelerates the progression of these tumors. This finding led to our hypothesis that BRCA1 may play a role in human ULMS development. Consistent with this hypothesis, we demonstrated that the BRCA1 protein is absent in 29% of human ULMS and that BRCA1 promoter methylation is the likely mechanism of BRCA1 downregulation. These data indicate that the loss of BRCA1 function may be an important step in the progression of ULMS. Our findings provide a rationale for investigating therapies that target BRCA1 deficiency in ULMS.
Keywords: BRCA1, gynecologic malignancy, mouse model, p53, uterine leiomyoma, uterine leiomyosarcoma, ULMS
Introduction
Although ULMS is a rare tumor that accounts for less than 1% of all uterine malignancies, more than 80% of patients with ULMS that has spread beyond the uterus experience tumor recurrence after initial chemotherapy (1). The etiology associated with the carcinogenesis of ULMS is largely unknown. Frequently observed mutations and overexpression of p53 in ULMS suggest that the loss of p53 function may play a critical role in the development of this cancer (2-4). Mice without a functional p53 tumor suppressor gene or with mutant p53 gain-of-function develop a spectrum of tumors. However, leiomyosarcomas that reproduce corresponding human malignancies with the same cellular origin rarely occur.
Several transgenic mouse models have been reported to give rise to leiomyosarcoma. In one mouse model, Cre-dependent activation and expression of an actin-cassette transgene encoding the T antigens of the SV40 early region resulted in the development of massive ULMS in all female mice at ~3 months of age (5). The second model was based on mouse mammary tumor virus (MMTV) promoter overexpression of Cripto-1 (CR-1). In addition to the development of mammary tumors, ULMS developed in approximately 20% of aged mice (6). Similarly, mammary tumors and ULMS arose in v-Ha-ras transgenic mice driven by the MMTV promoter (7). Disruption of Pten in the smooth muscle lineage with TagIn-Cre caused the formation of widespread smooth muscle cell hyperplasia and abdominal leiomyosarcoma but not ULMS (8).
Materials and Methods
Mouse strains
Amhr2Cre/+ mice (9) were crossed with Brca1lox/lox (10) or p53lox/lox (11) mice. Triple transgenic Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice were generated by crossing Amhr2Cre/+/Brca1lox/lox and Amhr2Cre/+/p53lox/lox mice. The resulting transgenic mice were maintained on a mixed background. All mice were genotyped by PCR using tail or ear DNA. Kaplan-Meier survival curves were drawn using GraphPad PRISM software. Mean survival time was calculated using the Log-rank test.
Confirmation of gene recombination
Genomic DNA extracted from tumors or normal tissues of the female reproductive tract was used to detect Cre-mediated recombination of the p53 and Brca1 genes. Cre-mediated deletion of p53 displayed a 612 bp PCR product amplified with primers p53-a (5′-CAC AAA AAC AGG TTA AAC CCA-3′) and p53-c (5′-GAA GAC AGA AAA GGG GAG GG-3′). PCR amplification of the recombined Brca1 gene resulted in a 621 product using the primers Brca1-d (5′-CTG GGT AGT TTG TAA GCA TCC-3′) and Brca1-g (5′-CTG CGA GCA GTC TTC AGA AAG-3′), which flanked Brca1 exon 11. The presence of wild-type Brca1 was determined by PCR using primers within exon 11 (Brca1-e: 5′-ATC AGT AGT AGA AAT CCA AGC CCA CC-3′; Brca1-f: 5′-TGC CAC TCC CAG CAT TGT TAG-3′).
Human specimens
Formalin-fixed paraffin-embedded archival human specimens were obtained from the following institutions: Massachusetts General Hospital, Boston, MA; Baylor College of Medicine, Houston, TX; Memorial Sloan-Kettering Cancer Center, New York, NY; Cedars-Sinai Medical Center, Los Angeles, CA; Olive View Medical Center, Los Angeles, CA; Inova Fairfax Hospital, Falls Church, VA; Universita Cattolica, Rome, Italy; and Istituto di Anatomia e Istologia Pathologica, Ancona, Italy.
H&E staining and immunohistochemistry
Tissues were fixed in 10% buffered formalin and embedded in paraffin. Tissue sections were deparaffinized in a graded xylene/ethanol series and used for H&E staining or immunohistochemistry with an ABC antibody staining kit (Vector Laboratories) according to the manufacturer’s instructions. After color development, the slides were counterstained with hematoxylin and mounted with mounting medium (Permount, Fisher Sciences). To determine the proliferative index of the tumors, mice were intraperitoneally injected with 100 mg/kg 5′-bromo-3′-deoxyuridine (BrdU) (Zymed Laboratories). Tissues and tumors were collected after two hours and fixed in 10% formalin overnight. Paraffin-embedded sections were deparaffinized, followed by hydrogen chloride (2N HCl) digestion, trypsinization (0.1% Trypsin), and immunohistochemistry with an ABC antibody staining kit. The following primary antibodies were used: α-smooth muscle actin (1:200 dilution, Sigma); β-catenin (H-102) (1:100 dilution, Santa Cruz); BRCA1 (Ab-1) (1:100 dilution, Calbiochem); BrdU (1:100 dilution, Vector Laboratories); p16 (M-156) (1:100 dilution, Santa Cruz); p53 (Ab-1) (1:100 dilution, Calbiochem); phospho-estrogen receptor α (Ser167) (1:100 dilution, Cell Signaling); and TROMA-1 (keratin 8) (1:25 dilution, Developmental Studies Hybridoma Bank at the University of Iowa). Hematoxylin and Eosin (H&E) and immuno-stained uterine tumor sections were reviewed by two independent observers (D.X. and E.O.).
BRCA1 promoter methylation analysis
The methylation status of ULMS specimens was determined using the EZ DNA Methylation Gold kit (Zymo Research) following manufacturer’s instructions. Ovarian cancer tissues with known BRCA1 methylation status (12) were used as a control. Bisulfite-modified DNA PCR amplification and primers have been previously described (12).
Results
Conditional deletion of p53 and Brca1 in the female mouse reproductive tract
In order to define the roles of the p53 and Brca1 tumor suppressor genes in oncogenesis of the female mouse reproductive tract, we generated mice in which p53 and Brca1 can be conditionally deleted using Cre recombinase knocked into the anti-Müllerian hormone type II receptor (Amhr2) locus (Amhr2-Cre) (Fig. S1A) (9). Three individual strains of mice, Amhr2Cre/+/p53lox/lox, Amhr2Cre/+/Brca1lox/lox, and Amhr2Cre/+/p53lox/lox/Brca1lox/lox, were generated (Figure S1B). p53 and/or Brca1 in these mice are expected to be inactivated by Cre recombinase in the Amhr2-expressing tissues, which include Müllerian duct mesenchymal cells, coelomic epithelium, and granulosa cells of the adult ovary (13). PCR of genomic DNA extracted from normal tissues of the female reproductive tract (ovary, oviduct, and uterus) was used to detect Cre-mediated recombination of the p53 (deleted exons 2-10) and Brca1 (deleted exon 11) genes. One three-month old female mouse from each genotype was selected for PCR analysis. As expected, Müllerian duct organs from Amhr2Cre/+/p53lox/lox mice harbored recombinant p53 but not recombinant Brca1, while Müllerian duct organs from Amhr2Cre/+/Brca1lox/lox mice harbored recombinant Brca1 but not recombinant p53 (Fig. S1B). Recombinant products for both Brca1 and p53 were detected in the ovaries, fallopian tubes and uteri of Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice (Fig. S1B). Primers within Brca1 exon 11 (Fig. S1A) were used to detect the presence of conditional Brca1 in various non-Amhr2-expressing cell types in the ovary, oviduct, and uterus (Fig. S1B).
Loss of p53 and Brca1 in the female mouse mesenchyme of the reproductive tract leads to the development of ULMS
Mice with deleted of p53, Brca1, or both in the Müllerian duct tissues developed normally and histopathological analyses did not reveal any specific anomalies in the Müllerian duct tissues or other organs of three month-old mice. However, uterine tumors developed in 12 of 23 (52%) of the Amhr2Cre/+/p53lox/lox female mice during the 13 month observation period. None of the 25 Amhr2Cre/+/Brca1lox/lox female mice developed uterine masses during the same time period (Fig. 1A and Table S1). However, the loss of Brca1 synergistically accelerated the formation of tumors in mice lacking p53, with 27 of 33 (82%) of the Amhr2Cre/+/p53lox/lox/Brca1lox/lox female mice developing uterine masses within 13 months (Fig. 1A and Table S1). The median time of tumor-free survival was 56 weeks for Amhr2Cre/+/p53lox/lox mice and 50 weeks for Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice. Conditional deletion of p53 and Brca1 significantly accelerated tumor development compared to inactivation of p53 alone (Log-rank Test = 13.12; p = 0.0003). At gross examination, the uterine tumors in Amhr2Cre/+/p53lox/lox and Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice looked similar although the tumors in the Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice were typically associated with more hemorrhagic necrosis (Fig. 1B). PCR was used to confirm the presence of recombinant p53 in tumors dissected from three Amhr2Cre/+/p53lox/lox mice as well as the presence of recombinant Brca1 and p53 in tumors dissected from four Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice. Representative PCR results for one tumor from each genotype are shown in Fig. 1C.
Figure 1.
Characterization of uterine tumors in mice with conditional deletion of p53 and/or Brca1 using Amhr2-driven Cre recombinase. A) Kaplan-Meier survival curves for Amhr2Cre/+/Brca1lox/lox, Amhr2Cre/+/p53lox/lox and Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice. B) Uterine tumors in Amhr2Cre/+/p53lox/lox (Tumor 1) and Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice (Tumor 2). C) Detection of Cre-mediated recombination of p53 in tumors dissected from an Amhr2Cre/+/p53lox/lox mouse (Tumor 1) and double recombination of p53 and Brca1 in a tumor dissected from an Amhr2Cre/+/p53lox/lox/Brca1lox/lox mouse (Tumor 2). Tail tissue from the Amhr2Cre/+/p53lox/lox/Brca1lox/lox mouse was used as a control.
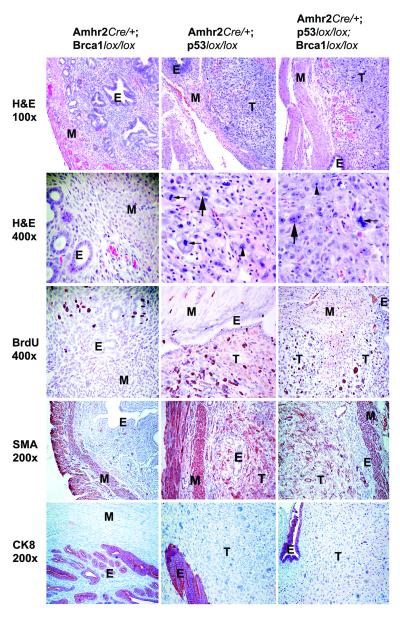
Histopathologic analysis showed that all of the tumors were ULMS, which were characterized by spindle shaped cells with hyperchromatic nuclei, prominent nucleoli, abundant mitoses and marked cytological atypia (Fig. 2). Immunohistochemical analysis (Table S2) revealed additional characteristics that are consistent with ULMS. BrdU staining indicated a high proliferation index (Fig. 2). The smooth muscle cell origin of the tumors was confirmed by the expression of smooth muscle actin and the absence of the epithelial marker Keratin 8 (Fig. 2). Several other characteristics of human ULMS, such as ERα-positivity (14), nuclear localization of β-catenin (6, 8) and overexpression of cyclin-dependent kinase inhibitor p16 (4, 15, 16), were also present in mouse ULMS (Table S2 and Fig. S2) indicating that mouse and human ULMS may arise through similar molecular pathways.
Figure 2.
Immunohistochemical analysis of normal uteri derived from Amhr2Cre/+/Brca1lox/lox mice and uterine tumors derived from Amhr2Cre/+/p53lox/lox and Amhr2Cre/+/p53lox/lox/Brca1lox/lox mice. Representative H&E staining (100x and 400x magnification). Incorporation of 5′-bromo-3′-deoxyuridine (BrdU) indicates a high proliferation index. The immunohistochemistry profile shows that the myometrium of the uterus and the uterine tumors are positive for smooth muscle actin (SMA) but negative for the epithelial marker Keratin 8 (CK8). T, tumor; E, endometrium; M, myometrium; Small arrow, abundant mitoses; Large arrow, marked cytological atypia; Arrowhead, prominent nucleoli; White asterisk, hyperchromatic nuclei.
BRCA1 expression is downregulated in human ULMS
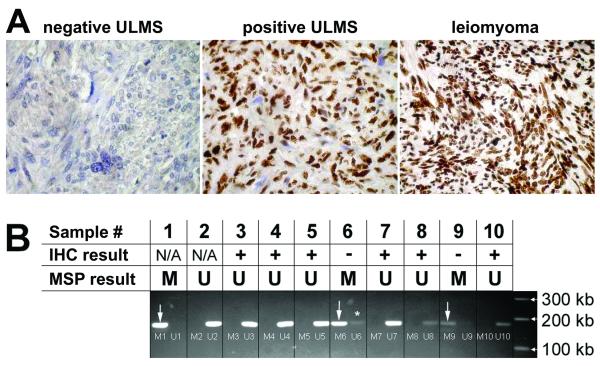
Our mouse model of ULMS indicates that BRCA1 may play a role in human ULMS carcinogenesis. This finding led to our hypothesis that BRCA1 expression may be altered in human ULMS. In order to address this question, we evaluated p53 and BRCA1 protein expression in a cohort of 85 ULMS and 76 benign uterine leiomyoma tissue specimens organized in a tissue microarray. The slides were stained with antibodies against BRCA1 and p53 using the avidin-biotin immunoperoxidase method. Nuclear positivity was scored by two independent observers and quantified as either present or absent. Results were analyzed using a two-tailed Fisher’s exact test. BRCA1 protein expression was absent in 29% (25/85) of ULMS samples and in 4% (3/76) of benign leiomyoma samples. Representative results of BRCA1 immunohistochemical detection are shown in Fig. 3A. This difference in BRCA1 protein expression between ULMS and benign leiomyoma samples was statistically significant with p<0.0001. Consistent with previous reports (17, 18), we found that p53 positivity was present in 50% (30/60) of ULMS and 0% (0/28) of benign leiomyomas (not shown). There was no significant correlation between BRCA1 and p53 staining in ULMS, suggesting that the loss of BRCA1 in ULMS may collaborate with pathways other than the p53 pathway.
Figure 3.
BRCA1 immunohistochemistry and methylation status. A) Representative immunohistochemical staining in BRCA1-negative ULMS, BRCA1-positive ULMS, and benign leiomyoma. B) Methylation-specific PCR analysis of 2 BRCA1-negative (−) and 6 BRCA1-positive (+) samples. Sample 1 = positive control for methylated BRCA1 promoter (ovary tumor in which methylation of the BRCA1 promoter has been previously demonstrated); Sample 2 = negative control for methylated BRCA1 promoter (normal male DNA); Samples 3-10 = primary human ULMS samples in which the presence (+) or absence (−) of the BRCA1 protein has been determined by immunohistochemistry (IHC). The IHC results are compared to the BRCA1 methylation status determined by Methylation-specific PCR (MSP). M = methylated product; U = unmethylated product. Both methylated and unmethylated product size = 182 bp. Asterisk indicates an unmethylated product that is probably derived from stromal cells and connective tissues within the tumor.
In order to identify a possible mechanism of BRCA1 protein downregulation in human ULMS, we selected two BRCA1-negative and six BRCA1-positive ULMS samples for which we had sufficient material to determine the BRCA1 methylation status using bisulfite-modified DNA PCR amplification (Fig. 3B). One ovarian cancer sample in which methylation of the BRCA1 promoter was previously confirmed (12) was used as a positive control, while normal male DNA was used as a negative control (Fig. 3B). BRCA1 promoter methylation was present in both samples that were BRCA1-negative as determined by immunohistochemistry and not present in the six samples that were BRCA1-positive as determined by immunohistochemistry (Fig. 3B).
Discussion
The understanding of the molecular biology of ULMS is poor due to rare occurrences of human ULMS and the lack of molecularly defined animal models. Therefore, there is a great need to generate genetically engineered mouse models that resemble the development of human ULMS. We investigated the role of p53 and Brca1 in the development and tumorigenesis of the female mouse reproductive tract based on a Cre/LoxP process in which the expression of Cre recombinase is under the control of the Amhr2 locus. Mice with p53 deletion in Amhr2-Cre expressing tissues developed ULMS, indicating that p53 may play a causative role in the formation of ULMS. In contrast, mice lacking functional Brca1 driven by Amhr2-Cre did not present any visible phenotype during the 13 month observation period. This result is consistent with the view that Brca1 plays a general role in the maintenance of genomic integrity and that a long latency is required for the activation of oncogenes and the inactivation of additional tumor suppressor genes to form Brca1-associated tumors (19, 20). Therefore, we cannot rule out the possibility that Brca1-deficient mice could develop gynecologic tumors after 13 months. Unlike human ULMS, which are highly metastatic, metastasis of mouse ULMS to other organs was not identified at the time of tumor extraction, although it is unknown whether these tumors would metastasize after 13 months.
Germline BRCA1 mutations have not been associated with a predisposition to human ULMS development, indicating that genomic alterations of BRCA1 are unlikely to play a role in the development of this disease. It is possible, however, that genetic or epigenetic somatic inactivation of BRCA1 contributes to the progression of ULMS. Our immunohistochemistry results on patient samples indicate a significant difference in BRCA1 protein expression between ULMS and benign uterine leiomyoma. Consistent with the view that BRCA1 silencing may play a role in the development or progression of ULMS, we demonstrated that the BRCA1 promoter is methylated in samples with negative BRCA1 immunohistochemical staining. Together, our findings provide a rationale for the investigation of targeted therapies that take advantage of the absence of BRCA1 expression in a subset of ULMS patients.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by grants from the National Institutes of Health (R01-CA103924), Ovarian Cancer Research Fund (OCRF), Liddy Shriver Sarcoma Initiative, LMSarcoma Direct Research Foundation, and the Sarcoma Foundation of America (SFA) to SO.
We would like to thank Robert Soslow (Memorial Sloan-Kettering Cancer Center, New York, NY); Gian Franco Zannoni (Universita Cattolica, Rome, Italy); Michele de Nictolis (Istituto di Anatomia e Istologia Pathologica, Ancona, Italy); Philip Branton (Inova Fairfax Hospital, Falls Church, VA); Christine Holschneider (Olive View Medical Center, Los Angeles, CA); and Jenny Gross (Cedars-Sinai Medical Center) for contributing human specimens; Lejla Delic (Cedars-Sinai Medical Center) for help with immunohistochemistry; Richard Behringer (University of Texas, M. D. Anderson Cancer Center) for the Amhr2-Cre mice; members of the Women’s Cancer Research Institute at Cedars-Sinai Medical Center for insightful suggestions; and Kristy J. Daniels for help in the preparation of the manuscript.
References
- 1.Dinh TA, Oliva EA, Fuller AF, Jr., Lee H, Goodman A. The treatment of uterine leiomyosarcoma. Results from a 10-year experience (1990-1999) at the Massachusetts General Hospital. Gynecol Oncol. 2004;92:648–52. doi: 10.1016/j.ygyno.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 2.de Vos S, Wilczynski SP, Fleischhacker M, Koeffler P. p53 alterations in uterine leiomyosarcomas versus leiomyomas. Gynecol Oncol. 1994;54:205–8. doi: 10.1006/gyno.1994.1194. [DOI] [PubMed] [Google Scholar]
- 3.Seki A, Kodama J, Miyagi Y, Kamimura S, Yoshinouchi M, Kudo T. Amplification of the mdm-2 gene and p53 abnormalities in uterine sarcomas. Int J Cancer. 1997;73:33–7. doi: 10.1002/(sici)1097-0215(19970926)73:1<33::aid-ijc6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill CJ, McBride HA, Connolly LE, McCluggage WG. Uterine leiomyosarcomas are characterized by high p16, p53 and MIB1 expression in comparison with usual leiomyomas, leiomyoma variants and smooth muscle tumours of uncertain malignant potential. Histopathology. 2007;50:851–8. doi: 10.1111/j.1365-2559.2007.02699.x. [DOI] [PubMed] [Google Scholar]
- 5.Politi K, Szabolcs M, Fisher P, Kljuic A, Ludwig T, Efstratiadis A. A mouse model of uterine leiomyosarcoma. Am J Pathol. 2004;164:325–36. doi: 10.1016/S0002-9440(10)63122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strizzi L, Bianco C, Hirota M, et al. Development of leiomyosarcoma of the uterus in MMTV-CR-1 transgenic mice. J Pathol. 2007;211:36–44. doi: 10.1002/path.2083. [DOI] [PubMed] [Google Scholar]
- 7.Radany EH, Hong K, Kesharvarzi S, Lander ES, Bishop JM. Mouse mammary tumor virus/v-Ha-ras transgene-induced mammary tumors exhibit strain-specific allelic loss on mouse chromosome 4. Proc Natl Acad Sci U S A. 1997;94:8664–9. doi: 10.1073/pnas.94.16.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernando E, Charytonowicz E, Dudas ME, et al. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–53. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- 9.Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat Genet. 2002;32:408–10. doi: 10.1038/ng1003. [DOI] [PubMed] [Google Scholar]
- 10.Xu X, Wagner KU, Larson D, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 11.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 12.Baldwin RL, Nemeth E, Tran H, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000;60:5329–33. [PubMed] [Google Scholar]
- 13.Teixeira J, Kehas DJ, Antun R, Donahoe PK. Transcriptional regulation of the rat Mullerian inhibiting substance type II receptor in rodent Leydig cells. Proc Natl Acad Sci U S A. 1999;96:13831–8. doi: 10.1073/pnas.96.24.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodner K, Bodner-Adler B, Kimberger O, Czerwenka K, Leodolter S, Mayerhofer K. Estrogen and progesterone receptor expression in patients with uterine leiomyosarcoma and correlation with different clinicopathological parameters. Anticancer Res. 2003;23:729–32. [PubMed] [Google Scholar]
- 15.Bodner-Adler B, Bodner K, Czerwenka K, Kimberger O, Leodolter S, Mayerhofer K. Expression of p16 protein in patients with uterine smooth muscle tumors: an immunohistochemical analysis. Gynecol Oncol. 2005;96:62–6. doi: 10.1016/j.ygyno.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Yang B. Immunohistochemical analysis of p16, p53, and Ki-67 expression in uterine smooth muscle tumors. Int J Gynecol Pathol. 2008;27:326–32. doi: 10.1097/PGP.0b013e31815ea7f5. [DOI] [PubMed] [Google Scholar]
- 17.Anderson SE, Nonaka D, Chuai S, et al. p53, epidermal growth factor, and platelet-derived growth factor in uterine leiomyosarcoma and leiomyomas. Int J Gynecol Cancer. 2006;16:849–53. doi: 10.1111/j.1525-1438.2006.00542.x. [DOI] [PubMed] [Google Scholar]
- 18.Leiser AL, Anderson SE, Nonaka D, et al. Apoptotic and cell cycle regulatory markers in uterine leiomyosarcoma. Gynecol Oncol. 2006;101:86–91. doi: 10.1016/j.ygyno.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Qiao W, Linke SP, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–71. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 20.Kim SS, Cao L, Lim SC, et al. Hyperplasia and spontaneous tumor development in the gynecologic system in mice lacking the BRCA1-Delta11 isoform. Mol Cell Biol. 2006;26:6983–92. doi: 10.1128/MCB.00796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.