Abstract
Mu opioid receptors (MOP) are transducers of the pharmacological effects of many opioid drugs, including analgesia and tolerance/dependence. Previously, we observed increased MOP signaling during postnatal development that was not associated with increased MOP or G protein expression. A yeast two-hybrid screen of a human brain cDNA library using the MOP C-terminus as bait identified RanBPM as a potential MOP-interacting protein. RanBPM has been recognized as a multi-functional scaffold protein that interacts with a variety of signaling receptors/proteins. Co-immunoprecipitation studies in HEK293 cells indicated that RanBPM constitutively associates with MOP. Functionally, RanBPM had no effect on MOP-mediated inhibition of adenylyl cyclase, yet reduced agonist-induced endocytosis of MOP. Mechanistically, RanBPM interfered with βarrestin2-GFP translocation stimulated by MOP but not α1B-adrenergic receptor activation, indicating selectivity of action. Our findings suggest that RanBPM is a novel MOP-interacting protein that negatively regulates receptor internalization without altering MOP signaling through adenylyl cyclase.
Keywords: Mu opioid receptor, RanBPM, internalization, scaffold protein, Beta-arrestin, yeast two-hybrid screen
Introduction
Mu opioid receptors (MOP) mediate the actions of most clinically important opioid drugs. Agonist-occupied MOP promote GTP binding to the Gi/o family of G proteins leading to numerous physiological responses including analgesia, decreased GI activity, respiratory depression, and euphoria [15]. MOP signaling is terminated when receptors are uncoupled from G proteins by phosphorylation and recruitment of βarrestins (βarrs) [1] and their associated endocytotic machinery [13]. Interestingly, peptide and alkaloid agonists, such as DAMGO and etorphine, rapidly induce receptor phosphorylation and endocytosis, whereas morphine, an alkaloid partial agonist, does not [12;25]. Considerable evidence indicates altered regulation of MOP internalization contributes to opioid tolerance and dependence [32].
Accessory/scaffolding proteins are important modulators of MOP agonist signaling, either by inhibiting MOP-stimulated G protein activation [7], altering agonist-induced internalization of MOP [22], or by association with signal transduction machinery and additional accessory proteins [9]. Previously our laboratory observed dramatic increases in the efficiency of G protein coupling to MOP in the rat brain during postnatal development [26]. These changes could not be explained by changes in either receptor expression or agonist binding characteristics. To identify proteins that might differentially modulate MOP signal transduction during development, we screened an adult human brain cDNA library by yeast two-hybrid methodology using the MOP C-terminus as bait. We identified the scaffold protein RanBP9/RanBPM (hereafter RanBPM) as a MOP-interacting protein.
RanBPM is a 90-kDa protein enriched in brain that is primarily cytoplasmic or membrane-bound [6;18;20]. Several lines of evidence suggest that RanBPM may serve a scaffolding role to modulate cell signaling [17]. RanBPM binds to and modulates the activity of a diverse group of proteins, including the LFA-1 integrin receptor [6], the cyclin-dependent kinase CDK11p46 [16], the receptor tyrosine kinases p75NTR [2] and MET [28], and other signaling modulators such as the de-ubiquitinating enzyme USP11 [10]. In this study we show that RanBPM endogenously associates with MOP and that over-expression of RanBPM in HEK293 cells effectively blocks agonist-induced endocytosis of MOP without altering inhibition of adenylyl cyclase. Our results suggest that RanBPM interacts with and modulates the activity of the MOP.
Materials and Methods
Yeast Two-hybrid Studies
The BD Matchmaker Two-Hybrid System 3 (BD Biosciences Clontech, Palo Alto, CA) was used according to the manufacturer’s protocol. cDNA encoding Asn332-Pro399 of the MOP C-terminal tail subcloned into the pGBKT7-GAL4 DNA-binding domain vector was used to screen a pre-transformed human brain cDNA library (Clontech) constructed in the pACT2-GAL4 activation domain vector. DNA from positive clones was sequenced, tested for autonomous growth, and positive interactions were confirmed using vectors encoding for the GAL4 binding domain and the GAL4 activation domain/RanBPM fusion protein as well as the GAL4 binding domain/MOP C-terminus fusion protein with the GAL4 activation domain.
Cell Culture and Transient Transfection
HEK293 cells were maintained in high glucose DMEM (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum and penicillin/streptomycin. Transfections used calcium phosphate precipitation method. FLAG-tagged GPCR cDNA constructs were gifts from: Wolfgang Sadee (Ohio State University) – MOP, Kenneth Minneman (Emory University) – α1BAR. Human RanBPM cDNA was a gift from Takeharu Nishimoto (Kyushu University, Japan). βarr2-GFP cDNA was a gift from Jeffrey Benovic (Thomas Jefferson University). HEK293 cells stably expressing FLAG-MOP were a gift from Wolfgang Sadee (Ohio State University).
Immunoprecipitation and Immunoblot Analysis
Confluent cells were lysed in 4°C solubilization buffer (1% NP-40, 150 mM NaCl, 1 mM EGTA, pH 7.4 containing Mammalian Protease/Phosphatase Inhibitor Cocktails I & II [Sigma-Aldrich, St. Louis, MO]) and supernatants were collected by centrifugation (30 min; 10,000xg). A total of 750 μg of protein was pre-cleared and incubated with anti-FLAG or anti-RanBPM antibodies followed by incubation with Protein G-Sepharose (Sigma-Aldrich), all at 4°C. Sepharose beads were pelleted, washed with solubilization buffer, and re-suspended in 2X SDS-sample buffer (10 mM Tris, 15 mM SDS, 20 mM DTT, 20% glycerol, 0.02% bromphenol blue, pH 6.8). Proteins were resolved by SDS-PAGE, transferred to Immobilon PVDF membranes (Millipore, Bedford, MA), and immunoblotted with anti-FLAG M2 (Sigma-Aldrich) or anti-RanBPM antibodies [6].
Radioligand Binding Assays
MOP and MOP/RanBPM HEK293 cell membranes were prepared as described previously [4]. MOP in cell membranes were assessed by [3H]naloxone (59.5 Ci/mmol; PerkinElmer, Boston, MA) binding (1 hr, RT). Homologous competition studies used 1 nM [3H]naloxone + unlabeled naloxone (0.01 nM – 200 nM), with 10 μM naltrexone to determine non-specific binding. Kd and Bmax values were calculated using Prism (GraphPad Inc, San Diego, CA) [5].
Adenylyl Cyclase Assay
Confluent cells were labeled with 2 μ Ci [3H]adenine (24.4 Ci/mmol; Perkin Elmer) per 35-mm dish, stimulated with agonist for 20 min, and adenylyl cyclase activity was measured as previously described [14]. Data are expressed as percent conversion of [3H]ATP to [3H]cAMP.
Confocal Microscopy
Cells were grown on glass coverslips to 60–75% confluency, stimulated with agonist for 30 min and fixed in 3.7% paraformaldehyde with 0.5% Triton-X100 for 10 min. Adherent cells were incubated with anti-FLAG M2-Cy3 conjugate (Sigma-Aldrich) followed by washing with PBS. Cells were dried briefly and mounted onto glass slides using Vectashield (Vector Labs, Burlingame, CA). FLAG-tagged MOP were visualized by immunofluorescence using a Zeiss LSM410 confocal laser scanning microscope.
Sucrose Density Gradient Assay
Cells were stimulated with and without 10 μM DAMGO for 30 min at 37°C to induce receptor internalization. Sub-cellular fractionation was performed as described previously [30]. MOP in each fraction were quantified by [3H]naloxone binding as described above.
βarr2-GFP Translocation Assay
Cells transfected with βarr2-GFP and FLAG-MOP, FLAG-α1B ARs, and/or RanBPM were grown for 24 hrs, harvested, and plated onto glass coverslips. Cells were stimulated with and without agonist (10 μM DAMGO for MOP; 1 μM phenylephrine for α1BAR) for 5 min at 37 °C 24 hrs after plating, fixed in 3.7% paraformaldehyde, washed with PBS, dried and mounted onto glass slides using Vectashield. βarr-GFP was visualized by confocal microscopy.
Results
Characterization of RanBPM binding to MOP
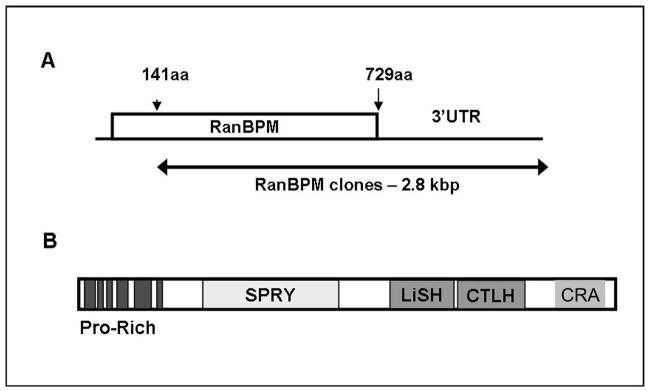
The yeast two-hybrid system was used to identify proteins that interact with MOP. In screening over 2.2 × 106 cDNAs, two positive clones were found to contain 2.8-kbp fragments identical to a contiguous segment encoding for RanBPM (GenBank Accession no. NM_005493), including 1.8 kbp within the open reading frame (Asn141-His729) and 1.0 kbp of 3′ untranslated region (Fig. 1A). Domain architecture analysis by SMART (Simple Modular Architecture Research Tool) [23] showed that RanBPM contains a SPRY domain (SP1a/Ryanodine receptor) that is implicated in protein-protein interactions; a LiSH/CTLH motif (Lissencephaly type-1-like homology/C-terminal to LisH) that mediates association with microtubular structures; and a proline-rich N-terminus that contains multiple putative SH3 domain-binding sites (Fig. 1B).
Figure 1.
RanBPM lacking its N-terminal region interacts with the C-terminal tail of MOP. (A) Interactions between RanBPM and the MOP C-terminus were detected using yeast two-hybrid methodology. Two cDNA clones contained 1.0 kbp of 3′ untranslated region (UTR) and lacked approximately 420 bp of the 5′ region immediately following the start codon that contains a high GC content (~81%) and encodes a proline-rich segment. (B) The relative location the putative proline-rich SH3-binding motifs, the SP1a/Ryanodine receptor domain (SPRY), Lissencephaly type-1-like homology/C-terminal to LisH (LiSH/CTLH), and the CT11-RanBPM (CRA) motifs are shown.
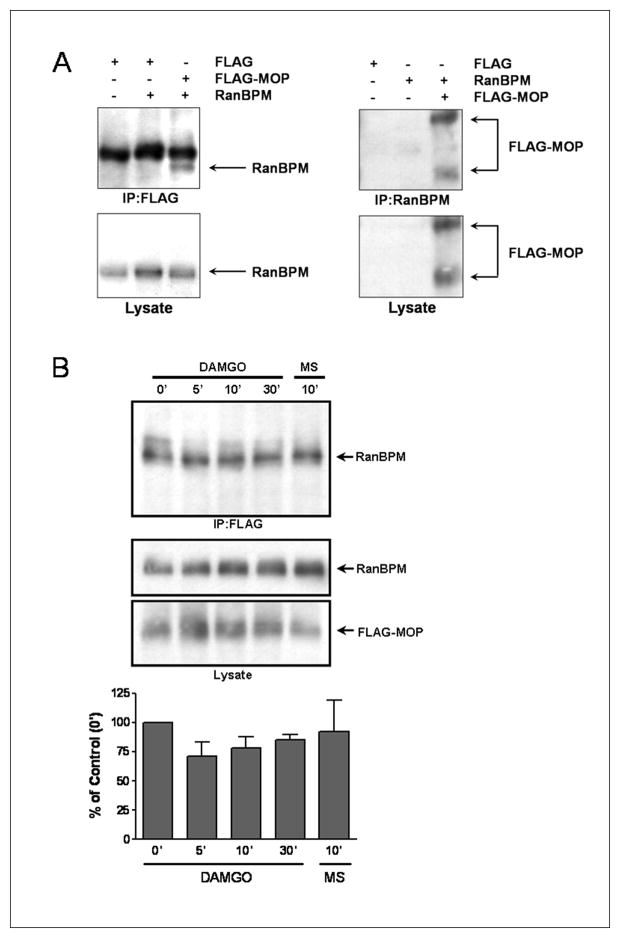
To determine whether full-length MOP interacts with the full-length RanBPM in intact cells, we used co-immunoprecipitation studies with transiently transfected HEK293 cells. RanBPM was detected when MOP were immunoprecipitated from the lysates of HEK293 cells co-transfected with FLAG-MOP and RanBPM but not from cells transfected with the FLAG peptide alone or with FLAG peptide+RanBPM (Fig. 2A; left panel). Likewise, FLAG-MOP co-immunoprecipitated with RanBPM, but only when cells were co-transfected with both proteins (Fig. 2A; right panel), indicating that the full-length MOP interacts with the full-length RanBPM in mammalian cells. Densitometric analysis showed that the relative amount of RanBPM co-immunoprecipitated with FLAG-MOP did not change following treatment with DAMGO or morphine (Fig. 2B). These data suggest that RanBPM constitutively binds MOP in intact mammalian cells independent of receptor activation, even by compounds that promote differing conformations of the MOP C-terminus, such as morphine and DAMGO [12;31].
Figure 2.
Full-length RanBPM interacts with MOP in mammalian cells. HEK293 cells were transiently transfected with pGADT7-RanBPM and/or pcDNA3.1-FLAG MOP or with pcDNA3.1-FLAG as control. Immunoblots were probed with anti-FLAG or anti-RanBPM antibodies. (A) RanBPM co-immunoprecipitated with FLAG-MOP only in cells co-expressing RanBPM and FLAG-MOP (lanes 1–3). FLAG-MOP co-immunoprecipitated with RanBPM only in cells co-expressing both proteins (lanes 4–6). (B) Cells were treated with 10 μM DAMGO or morphine (MS) or were untreated (0′ time point). Lysates were immunoprecipitated using anti-FLAG antibodies (upper panel) or immunoblotted directly using anti-RanBPM or anti-FLAG antibodies (middle and lower panels, respectively). . Densitometric analyses are relative optical density of RanBPM immunoreactivity in immunoprecipitates, expressed as a percentage of RanBPM immunoreactivity observed in untreated cells and are the means ± S.E.M. from three experiments. No significant difference (p > 0.05) was observed between agonist treatment and control as determined by ANOVA.
RanBPM has no effect on MOP agonist affinity and inhibition of cAMP accumulation by MOP
MOP signal transduction was characterized in HEK293 cells stably expressing FLAG-MOP ± RanBPM. RanBPM over-expression had no effect on binding affinity of the MOP antagonist [3H]naloxone or on MOP levels in either cell line, as indicated by comparable Ki and Bmax values (Table 1). Similarly, RanBPM over-expression had no effect on the EC50 of DAMGO–mediated inhibition of forskolin-stimulated [3H]cAMP accumulation (Table 1). These data suggest MOP expression/ligand binding properties are unaffected by RanBPM over-expression and that RanBPM has no functional consequence on MOP signaling at the level of adenylyl cyclase.
Table 1.
Receptor density, ligand binding, and inhibition of forskolin-stimulated cAMP accumulation by MOP are unaltered by RanBPM.
| Stable Cell Lines | Kd (nM) | Bmax (fmol/mg) | DAMGO EC50 (μM) | Maximal Inhibition of cAMP Accumulation (%) |
|---|---|---|---|---|
| MOP | 2.6 ± 0.8 | 54 ± 5.4 | 3.4 ± 1.7 | 78 ± 7.8 |
| MOP+RanBPM | 2.8 ± 0.7 | 49 ± 6.2 | 2.1 ± 0.8 | 79 ± 4.3 |
The binding characteristics (Kd and Bmax) for [3H]naloxone were determined on membranes from HEK293 cells stably transfected with FLAG-MOP ± RanBPM and analyzed by computerized curve fitting (see Materials and Methods). DAMGO-mediated inhibition of cAMP accumulation was assessed as described in the same cells by [3H]adenine labeling and determining percent conversion of [3H]ATP to [3H]cAMP in the presence of 10 μM forskolin and 200 μM IBMX. Data shown are mean ± S.E.M. of three independent experiments each performed in duplicate.
RanBPM inhibits agonist-induced internalization of MOP
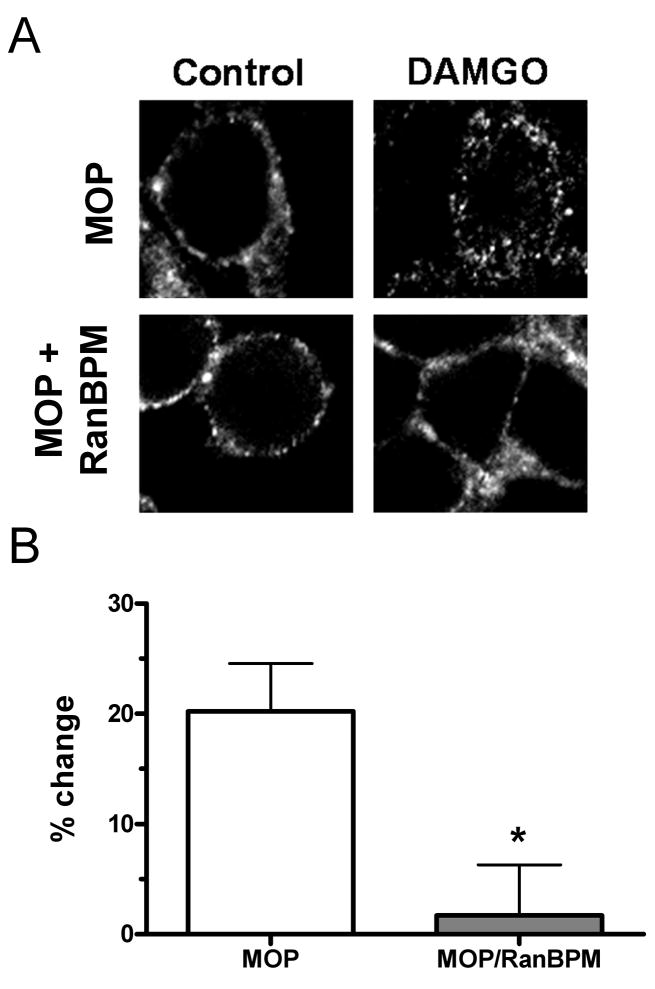
MOP trafficking was evaluated using immunofluorescence and sucrose density gradients to assess subcellular distribution. Following DAMGO stimulation, FLAG-MOP (Cy3-labeled) fluorescence was detected as punctate labeling in the intracellular space (Fig. 3A), characteristic of receptor internalization [25]. In contrast, DAMGO treatment did not lead to a redistribution of fluorescence to the intracellular space in MOP/RanBPM cells. Sucrose density gradient experiments used [3H]naloxone binding to assess the distribution of MOP in fractions associated with the plasma membrane (heavy peak) or with intracellular vesicles (light peak). Treatment with DAMGO resulted in a shift of receptors from the heavy peak fraction to the light vesicle fraction in MOP cells, (20 ± 4 %) but not in MOP/RanBPM cells (1.7 ± 4 %) (Fig 3B). These data provide evidence that RanBPM over-expression inhibits the agonist-induced internalization of MOP by blocking MOP redistribution from the plasma membrane to intracellular vesicles.
Figure 3.
RanBPM alters the agonist-induced subcellular redistribution of MOP. (A) HEK293 cells stably expressing FLAG-MOP ± RanBPM (MOP and MOP/RanBPM) were treated with vehicle (left panels) or 10 μM DAMGO (right panels) for 30 min. FLAG-MOP was visualized with Cy3-conjugated anti-FLAG antibody followed by confocal microscopy. (B) DAMGO-induced internalization of FLAG-MOP was quantified by sucrose density gradient centrifugation assays. See Methods for details. Data are expressed as percent change of receptors in the plasma membrane fraction that shifted to the light vesicle fraction from the untreated heavy fraction following DAMGO treatment. Data are mean ± S.E.M. from at least three experiments. Statistical significance was determined using unpaired Students t-test (*p < 0.05 compared to control).
MOP-mediated βarr2-GFP translocation is blocked by RanBPM
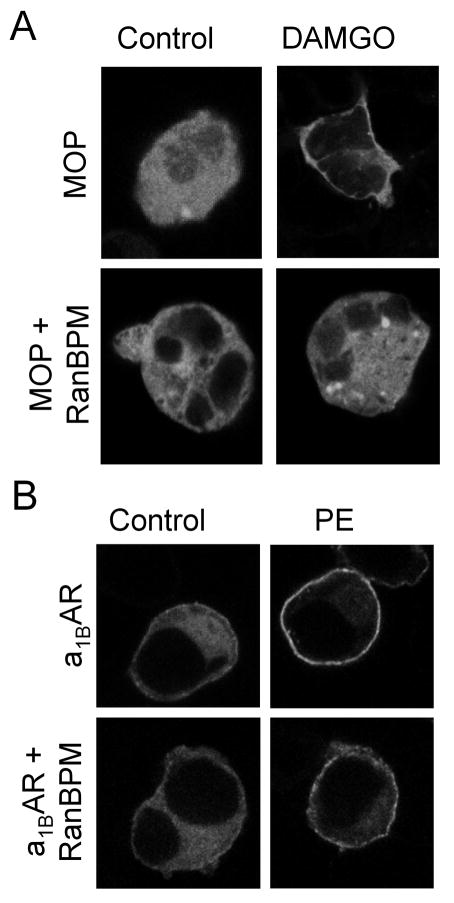
MOP internalization involves βarr adapter proteins [32]. βarr exhibits a cytosolic distribution in the basal state and is translocated to the plasma membrane following agonist-induced phosphorylation of the C-terminal tail of many GPCRs. Therefore, βarr2-GFP was co-expressed with MOP ± RanBPM in HEK293 cells. As expected, untreated cells expressing only MOP exhibited a diffuse cellular fluorescence, indicating a cytosolic distribution of βarr2-GFP (Fig. 4A). Treatment with DAMGO led to a rapid translocation of βarr2-GFP to the plasma membrane, as indicated by ring-like fluorescence associated with the plasma membrane. Untreated cells co-expressing MOP and RanBPM exhibited a cytosolic distribution of βarr2-GFP similar to those expressing only MOP. However, in MOP/RanBPM cells agonist treatment failed to induce translocation of βarr2-GFP. In contrast, RanBPM had no effect on α1BAR -stimulated βarr2-GFP translocation (Fig. 4B), indicating RanBPM selectively alters agonist-induced internalization of MOP.
Figure 4.
MOP-stimulated translocation of βarr2-GFP is selectively inhibited by RanBPM. (A) Agonist-mediated recruitment of βarr2-GFP was visualized by confocal microscopy of HEK293 cells transiently transfected with βarr2-GFP and FLAG-MOP without (MOP) and with RanBPM (MOP/RanBPM). Cells were treated with either 10 μM DAMGO or vehicle for 10 min. (B) Agonist-mediated recruitment of βarr2-GFP was visualized by confocal microscopy of HEK293 cells transiently transfected with βarr2-GFP and FLAG-α1BAR without (α1BAR) and with RanBPM (α1BAR/RanBPM). Cells were treated with either 1 μM phenylephrine (PE) or vehicle for 10 min. Agonist stimulation resulted in the redistribution of βarr2-GFP from the cytosol to the plasma membrane in both MOP and α1BAR cells (panels A & B). No translocation of βarr2-GFP was observed following MOP-stimulation in cells over-expressing RanBPM (panel A, DAMGO), whereas α1BAR-mediated βarr2-GFP translocation was not altered by RanBPM over-expression (panel B, PE).
Discussion
Our data indicate that RanBPM associates with and modulates the function of MOP in mammalian cells. We provide the first evidence that RanBPM alters agonist-induced internalization of MOP but not MOP-mediated inhibition of AC. Due to its diverse domain architecture, RanBPM may provide an important link between MOP signal transduction and regulatory proteins that influence opioid signaling.
The proline-rich N-terminus of RanBPM contains six discrete poly-proline sequences that provide low-affinity, transient docking sites for protein interaction domains, including the SH3 domain present in many signal transduction/cytoskeletal proteins [11]. Several proteins thought to mediate MOP stimulation of ERK/MAPK contain SH3 domains, including the Src, Grb2, and the PI3 kinase regulatory subunit. Of these, Src and Grb2 are predicted to bind with high affinity to multiple sequences within the proline-rich N-terminus of RanBPM (iSPOT; [3]). Moreover, endogenous RanBPM co-precipitates with the adapter protein Sos, a constitutive binding partner of Grb2, in a manner that requires the RanBPM N-terminus [28;29]. These observations, together with evidence suggesting RanBPM interacts with several receptor tyrosine kinases, point to a potential direct link between MAPK regulation, receptor tyrosine kinases, and the mu opioid receptor [27].
DAMGO and morphine differentially regulate MOP internalization, possibly by inducing different conformations of the C-terminal tail, one of which favors rapid phosphorylation/βarr recruitment (DAMGO-induced) and one which does not (morphine-induced) [31]. Receptor activation did not appreciably change the extent of RanBPM binding to MOP; however, high levels of RanBPM altered DAMGO-stimulated trafficking of MOP to resemble those reported for morphine-activated MOP, namely limited βarr2 recruitment [32] and decreased receptor internalization [12;25]. Thus, RanBPM may play a role in determining specificity of MOP agonist effects. Future studies will determine whether RanBPM alters adaptive changes associated with chronic opioid exposure such as MOP activation of ERK and other components of the MAPK pathway. Furthermore, because RanBPM interacts with MOP constitutively, the relative expression levels of RanBPM in specific neuroanatomical areas within the CNS may be important in determining the extent of its regulation of MOP in vivo.
In this study we provide the first evidence that RanBPM, a putative scaffolding protein, alters agonist-induced internalization of MOP, but not MOP-mediated inhibition of adenylyl cyclase. Recent work supports the functional interaction of RanBPM with GPCR. RanBPM was shown to bind to multiple metabotropic glutamate receptors, including mGlu2 and mGlu8 [24], which have been implicated in modulating morphine and related opioid drug effects [8;19;21] . Detailed analysis of the sites of interaction between RanBPM will lead to new tools for further investigation of the functional consequences of RanBPM interactions with MOP and other GPCR. Their interactions could provide novel targets for therapeutic interventions to selectively modulate GPCR-mediated processes.
Acknowledgments
This research was supported by the National Institutes of Health DA016346 (LCM), the Bower, Bennet, & Bennet Endowed Chair Research Award (JNT), the American Foundation for Pharmaceutical Education New Investigator Program (JNT), and the University of Nebraska Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arden JR, Segredo V, Wang Z, Lameh J, Sadee W. Phosphorylation and agonist-specific intracellular trafficking of an epitope-tagged mu-opioid receptor expressed in HEK 293 cells. J Neurochem. 1995;65:1636–1645. doi: 10.1046/j.1471-4159.1995.65041636.x. [DOI] [PubMed] [Google Scholar]
- 2.Bai D, Chen H, Huang BR. RanBPM is a novel binding protein for p75NTR. Biochem Biophys Res Commun. 2003;309:552–557. doi: 10.1016/j.bbrc.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 3.Brannetti B, Zanzoni A, Montecchi-Palazzi L, Cesareni G, Helmer-Citterich M. iSPOT: a web tool for the analysis and recognition of protein domain specificity. Comparative and Functional Genomics. 2001;2:314–318. doi: 10.1002/cfg.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bylund DB, Deupree JD, Toews ML. Radioligand-binding methods for membrane preparations and intact cells. Methods Mol Biol. 2004;259:1–28. doi: 10.1385/1-59259-754-8:001. [DOI] [PubMed] [Google Scholar]
- 5.Coulter CL, Happe HK, Murrin LC. Dopamine transporter development in postnatal rat striatum: an autoradiographic study with [3H]WIN 35,428. Brain Res Dev Brain Res. 1997;104:55–62. doi: 10.1016/s0165-3806(97)00135-1. [DOI] [PubMed] [Google Scholar]
- 6.Denti S, Sirri A, Cheli A, Rogge L, Innamorati G, Putignano S, Fabbri M, Pardi R, Bianchi E. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem. 2004;279:13027–13034. doi: 10.1074/jbc.M313515200. [DOI] [PubMed] [Google Scholar]
- 7.Feng GJ, Kellett E, Scorer CA, Wilde J, White JH, Milligan G. Selective interactions between helix VIII of the human -opioid receptors and the C terminus of periplakin disrupt G protein activation. J Biol Chem. 2003;278:33400–33407. doi: 10.1074/jbc.M305866200. [DOI] [PubMed] [Google Scholar]
- 8.Fischer BD, Miller LL, Henry FE, Picker MJ, Dykstra LA. Increased efficacy of micro-opioid agonist-induced antinociception by metabotropic glutamate receptor antagonists in C57BL/6 mice: comparison with (−)-6-phosphonomethyl-deca-hydroisoquinoline-3-carboxylic acid ( LY235959) Psychopharmacology (Berl) 2008;198:271–278. doi: 10.1007/s00213-008-1130-y. [DOI] [PubMed] [Google Scholar]
- 9.Georgoussi Z, Leontiadis L, Mazarakou G, Merkouris M, Hyde K, Hamm H. Selective interactions between G protein subunits and RGS4 with the C-terminal domains of the mu- and delta-opioid receptors regulate opioid receptor signaling. Cell Signal. 2006;18:771–782. doi: 10.1016/j.cellsig.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Ideguchi H, Ueda A, Tanaka M, Yang J, Tsuji T, Ohno S, Hagiwara E, Aoki A, Ishigatsubo Y. Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. Biochem J. 2002;367:87–95. doi: 10.1042/BJ20011851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 12.Keith DE, Murray SR, Zaki PA, Chu PC, Lissin DV, Kang L, Evans CJ, von Zastrow M. Morphine activates opioid receptors without causing their rapid internalization. J Biol Chem. 1996;271:19021–19024. doi: 10.1074/jbc.271.32.19021. [DOI] [PubMed] [Google Scholar]
- 13.Kovoor A, Nappey V, Kieffer BL, Chavkin C. Mu and delta opioid receptors are differentially desensitized by the coexpression of beta-adrenergic receptor kinase 2 and beta-arrestin 2 in xenopus oocytes. J Biol Chem. 1997;272:27605–27611. doi: 10.1074/jbc.272.44.27605. [DOI] [PubMed] [Google Scholar]
- 14.Kreps DM, Whittle SM, Hoffman JM, Toews ML. Lysophosphatidic acid mimics serum-induced sensitization of cyclic AMP accumulation. FASEB J. 1993;7:1376–1380. doi: 10.1096/fasebj.7.14.8224610. [DOI] [PubMed] [Google Scholar]
- 15.Law PY, Wong YH, Loh HH. Molecular mechanisms and regulation of opioid receptor signaling. Annu Rev Pharmacol Toxicol. 2000;40:389–430. doi: 10.1146/annurev.pharmtox.40.1.389. [DOI] [PubMed] [Google Scholar]
- 16.Mikolajczyk M, Shi J, Vaillancourt RR, Sachs NA, Nelson M. The cyclin-dependent kinase 11(p46) isoform interacts with RanBPM. Biochem Biophys Res Commun. 2003;310:14–18. doi: 10.1016/j.bbrc.2003.08.116. [DOI] [PubMed] [Google Scholar]
- 17.Murrin LC, Talbot JN. RanBPM, a scaffolding protein in the immune and nervous systems. Journal of Neuroimmune Pharmacology. 2007;2:290–295. doi: 10.1007/s11481-007-9079-x. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura M, Masuda H, Horii J, Kuma K, Yokoyama N, Ohba T, Nishitani H, Miyata T, Tanaka M, Nishimoto T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J Cell Biol. 1998;143:1041–1052. doi: 10.1083/jcb.143.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen DA, Ji F, Yuferov V, Ho A, Chen A, Levran O, Ott J, Kreek MJ. Genotype patterns that contribute to increased risk for or protection from developing heroin addiction. Mol Psychiatry. 2008;13:417–428. doi: 10.1038/sj.mp.4002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishitani H, Hirose E, Uchimura Y, Nakamura M, Umeda M, Nishii K, Mori N, Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa M, Miyakawa T, Nakamura K, Kitano J, Furushima K, Kiyonari H, Nakayama R, Nakao K, Moriyoshi K, Nakanishi S. Altered sensitivities to morphine and cocaine in scaffold protein tamalin knockout mice. Proc Natl Acad Sci U S A. 2007;104:14789–14794. doi: 10.1073/pnas.0706945104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onoprishvili I, Andria ML, Kramer HK, Ancevska-Taneva N, Hiller JM, Simon EJ. Interaction between the opioid receptor and filamin A is involved in receptor regulation and trafficking. Mol Pharmacol. 2003;64:1092–1100. doi: 10.1124/mol.64.5.1092. [DOI] [PubMed] [Google Scholar]
- 23.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seebahn A, Rose M, Enz R. RanBPM is expressed in synaptic layers of the mammalian retina and binds to metabotropic glutamate receptors. FEBS Lett. 2008;582:2453–2457. doi: 10.1016/j.febslet.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Sternini C, Spann M, Anton B, Keith DE, Jr, Bunnett NW, von Zastrow M, Evans C, Brecha NC. Agonist-selective endocytosis of mu opioid receptor by neurons in vivo. Proc Natl Acad Sci U S A. 1996;93:9241–9246. doi: 10.1073/pnas.93.17.9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talbot JN, Happe HK, Murrin LC. Mu opioid receptor coupling to Gi/o proteins increases during postnatal development in rat brain. J Pharmacol Exp Ther. 2005;314:596–602. doi: 10.1124/jpet.104.082156. [DOI] [PubMed] [Google Scholar]
- 27.von Zastrow M, Svingos A, Haberstock-Debic H, Evans C. Regulated endocytosis of opioid receptors: cellular mechanisms and proposed roles in physiological adaptation to opiate drugs. Curr Opin Neurobiol. 2003;13:348–353. doi: 10.1016/s0959-4388(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Li Z, Messing EM, Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J Biol Chem. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Li Z, Schoen SR, Messing EM, Wu G. A novel MET-interacting protein shares high sequence similarity with RanBPM, but fails to stimulate MET-induced Ras/Erk signaling. Biochem Biophys Res Commun. 2004;313:320–326. doi: 10.1016/j.bbrc.2003.11.124. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Wang L, Zheng J, Anderson JL, Toews ML. Identification of distinct carboxyl-terminal domains mediating internalization and down-regulation of the hamster alpha(1B)- adrenergic receptor. Mol Pharmacol. 2000;57:687–694. doi: 10.1124/mol.57.4.687. [DOI] [PubMed] [Google Scholar]
- 31.Whistler JL, Chuang HH, Chu P, Jan LY, von Zastrow M. Functional dissociation of mu opioid receptor signaling and endocytosis: implications for the biology of opiate tolerance and addiction. Neuron. 1999;23:737–746. doi: 10.1016/s0896-6273(01)80032-5. [DOI] [PubMed] [Google Scholar]
- 32.Whistler JL, von Zastrow M. Morphine-activated opioid receptors elude desensitization by beta-arrestin. Proc Natl Acad Sci U S A. 1998;95:9914–9919. doi: 10.1073/pnas.95.17.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]