Abstract
In the experimental Candida albicans intravenous challenge model, the kidney is one of the main organs involved in disease. In progressive infection, fungal burdens are found to increase over time, with rapid increases occurring from 24 h postinfection. Renal transcriptional responses were analyzed at this time in the kidneys of mice infected by either a virulent or an attenuated C. albicans strain, allowing comparison of host responses in progressive and nonprogressive infection. The results of this study demonstrate that both infections share a common transcriptional response, consisting of functions associated with the acute-phase reaction. In addition, challenge with the virulent strain led to a massively increased expression of cytokine genes, other innate response genes and genes suggestive of initiation of the adaptive immune response. This immune response to C. albicans infection, which occurs only in progressive infection, may contribute to development of sepsis and, ultimately, host death.
Keywords: Candida albicans, experimental model, systemic infection, kidney, transcript profiling, innate immune response
Introduction
Systemic fungal infections remain a significant cause of morbidity and mortality, with Candida albicans being the most common causative agent (Odds et al., 2007; Bougnoux et al., 2008; Caggiano et al., 2008; Playford et al., 2008;).
Intravenous C. albicans challenge in mice is a well-characterized model of severe clinical disseminated infection (MacCallum & Odds, 2005; Spellberg et al., 2005;), used to investigate fungal virulence factors (Navarro-Garcia et al., 2001; Brand et al., 2004;) and host immune responses (Ashman, 2008; Romani, 2008; MacCallum et al., 2009;). In this model, within minutes of entering the bloodstream, C. albicans is detectable in all major organs (MacCallum & Odds, 2005), with the kidneys and brain being the main target organs (Louria et al., 1963; MacCallum & Odds, 2005;), reflecting the situation in the human host (Parker et al., 1976; Odds, 1988;). In infection initiated by the virulent strain SC5314, renal fungal burdens increase from the time of infection, with rapid increases occurring from 24 h postinfection. Although the kidneys are the most heavily colonized organs, and some loss of renal function has been shown (Leunk & Moon, 1979; Spellberg et al., 2005;), renal failure is not the main cause of death. Host deterioration, and eventually death, is due to a progressive sepsis, reflecting the clinical situation (Spellberg et al., 2005).
Because of similarities to severe human disseminated infection, the mouse intravenous challenge model is ideal for studying the transcriptional response of the kidney to infection by C. albicans. DNA microarrays have been used previously to investigate the transcriptional responses of host cells (Prigneau et al., 2003; Ishibashi et al., 2004; Mullick et al., 2004a; Barker et al., 2005, 2008; Kim et al., 2005b; Fradin et al., 2006; Muller et al., 2007) and fungal cells (Fradin et al., 2003, 2005; Lorenz et al., 2004; Sandovsky-Losica et al., 2006; Sohn et al., 2006; Fernandez-Arenas et al., 2007; Thewes et al., 2007; Zakikhany et al., 2007; Walker et al., 2009) during their interaction. Research has focused mainly on examining the responses of single host cell types, such as macrophages (Prigneau et al., 2003; Lorenz et al., 2004; Barker et al., 2005; Kim et al., 2005b;), neutrophils (Fradin et al., 2005, 2006) and endothelial cells (Muller et al., 2007; Barker et al., 2008;). The aim of this study was to investigate the transcriptional responses of the whole kidney to infection by C. albicans, and to examine how these responses may relate to disease development, including progression of sepsis.
Materials and methods
Candida albicans strains and growth conditions
Candida albicans strains NGY152 (wild-type CAI4+CIp10, Brand et al., 2004) and NGY355 (pmr1Δ null mutant, Bates et al., 2005) were maintained routinely on Sabouraud agar at 4 °C, and stored long term in glycerol stocks at −80 °C. For the mouse intravenous challenge, inocula were prepared as described previously (Bates et al., 2005).
Experimental infection model
Female BALB/c mice (Harlan, UK) (average weight 20 g) were infected intravenously with an inoculum of 4.6 × 104 CFU g−1 body weight. This inoculum level leads to a survival time of approximately 5 days for mice infected with the virulent strain NGY152 (MacCallum & Odds, 2005), but all animals infected with the attenuated strain NGY355 survived to the end of the 28-day experiment (Bates et al., 2005). Groups of mice (N=3) were infected with C. albicans NGY152 (virulent), NGY355 (attenuated) or saline (controls). Actual inocula levels were determined by viable counts.
Mice were humanely terminated at 24 h. Kidneys were dissected and processed to provide portions for RNA extraction (flash-frozen in liquid nitrogen), histological analysis (formalin-fixed) and burden determination (homogenized in saline). Other organs (lung, liver, spleen and brain) were sampled and processed for organ burden determination as described previously (MacCallum & Odds, 2005). All work involving experimental animals was performed under UK Home Office licenses and regulations.
RNA extraction
Frozen kidney pieces (approximately 50 mg) were ground to a fine powder in a Mikro-dismembrator U homogenizer (Sartorius Ltd, UK). The powder was resuspended in 1 mL TRIzol (Invitrogen, UK) and RNA extracted as per the manufacturer's instructions. RNA samples were DNase treated and purified using RNeasy mini cleanup columns (Qiagen, UK), the concentration was determined by spectrophotometry (Nanodrop ND-1000; Labtech International, UK) and the quality of the RNA was analyzed by a Bioanalyzer (Agilent Technologies, UK).
Microarrays
For microarray analysis, pools of RNA for each condition were produced. Briefly, equal quantities (8 μg) of the three independent samples were mixed and made up to 24 μL. Each pool was then divided into three aliquots. For each treatment, two aliquots were used in the production of biotin-labeled cRNA. Standard Affymetrix protocols and kits (one-cycle cDNA synthesis, first strand synthesis, second strand synthesis, double-stranded DNA clean-up, GeneChip IVT synthesis and clean-up of biotin-labeled cRNA) were used to produce the labeled cRNA samples (http://www.affymetrix.com). From each cRNA sample, 20 μg was fragmented (1 × fragmentation buffer; 94 °C for 35 min), and 15 μg of this was used to produce a hybridization cocktail. Control noneukaryotic biotinylated and fragmented cRNAs (bioB, C and D from Escherichia coli and cre from bacteriophage P1) were added to the hybridization cocktail.
Cocktails for all six samples were hybridized to Affymetrix mouse genome 430A 2.0 arrays according to the manufacturer's instructions in an Affymetrix GeneChip hybridization oven 640. Arrays were washed in an Affymetrix GeneChip fluidics station and scanned with an Affymetrix GeneChip scanner 3000.
Microarray gene expression data analysis
For each Affymetrix array, the CEL file was loaded into Genespring (Agilent Technologies). Data were normalized on a Per Chip basis (normalized to the 50th percentile) and on a Per Gene basis (normalized to the mean). Finally, data for Candida-infected kidneys were normalized on a Per Gene basis to control (saline-infected) samples.
Normalized data were analyzed by significance analysis of microarrays (http://www-stat.stanford.edu) to produce lists of genes with statistically significantly different expression levels, based on a false discovery rate value of 0. Gene lists were further trimmed to include only genes whose expression level differed at least 1.5-fold compared with uninfected controls. Gene trees were constructed in Genespring, with commonly regulated genes grouped together. Regulated genes were further divided into three large clusters with the three-cluster K-means function of Genespring.
Microarray data files [raw (CEL) and normalized files] were deposited in ArrayExpress (http://www.ebi.ac.uk/microarray-as/aer), accession number: E-MEXP-1458.
cDNA synthesis and quantitative reverse transcriptase (QRT)-PCR
qRT-PCR with the Universal Probe Library system (Roche, UK) was used to verify differences in gene expression observed in microarray analysis.
Superscript II (Invitrogen) was used to synthesize cDNAs from 3 μg of each of the three independent RNA samples extracted from infected or control kidneys as per the manufacturer's instructions.
The oligonucleotide primers (Invitrogen) and Universal probes (Roche) used for qRT-PCR are listed in Table 1. PCR reactions (20 μL) contained 3 μL cDNA, 0.2 μL Universal probe, 0.2 μM forward primer, 0.2 μM reverse primer, 1 × probes Mastermix (Roche) and 1 × murine GAPDH endogenous control (PE Applied Biosystems, UK). Reactions were run on a LightCycler 480 (Roche); initial activation was carried out at 95 °C for 5 min, 50 amplification cycles of 95 °C for 10 s and 60 °C for 30 s and then a final cooling step at 40 °C for 10 s. For each gene, the copy number was determined from a standard curve and normalized to the GAPDH control. qRT-PCR reactions were carried out in triplicate for each of the three independent cDNA samples from each treatment group. Expression levels were compared using nonparametric statistical tests (Kruskal–Wallis and Mann–Whitney U-tests) due to unequal variances for the different groups.
Table 1.
qRT-PCR oligonucleotide primers and Universal probes
| Primer name | Sequence (5′–3′) | Universal probe |
|---|---|---|
| Tlr2F | TGCCCAGATGGCTAGTGG | 51 |
| Tlr2R | CAGAAACTATGATTGCGGACAC | |
| Tlr4F | GGACTCTGATCATGGCACTG | 51 |
| Tlr4R | CTGATCCATGCATTGGTAGGT | |
| Tlr13F | CTATGTGCTAGGAGCTTCTGAGAG | 6 |
| Tlr13R | TTCATCCTTCAAGCATCAGTGTA | |
| Il6F | GCTACCAAACTGGATATAATCAGGA | 6 |
| Il6R | CCAGGTAGCTATGGTACTCCAGAA | |
| Myd88F | TGACTTCCAGACCAAGTTTGC | 80 |
| Myd88R | GAATCAGTCGCTTCTGTTGGA | |
| Il1bF | TGTAATGAAAGACGGCACACC | 78 |
| Il1bR | TCTTCTTTGGGTATTGCTTGG | |
| Clec4nF | CAGTGAAGGGACTATGGTGTCA | 78 |
| Clec4nR | GCTCCAGAAGTTCTCCTTGGT | |
| Cxcl12F | TCCTCTTGCTGTCCAGCTCT | 81 |
| Cxcl12R | CAGGCTGACTGGTTTACCG |
Bioinformatics: biological function and pathway analysis
Biological functions and pathways associated with the gene lists obtained from expression analysis were identified by means of Ingenuity Pathway Analysis (Ingenuity Systems; http://www.ingenuity.com) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov) (Dennis et al., 2003) bioinformatic resources. Both resources determine association by P-values, found by comparing genes on a list with the total number of genes for a pathway, and use knowledge databases that are continually updated from a number of sources.
Histology
Sections (5 μm) were cut from paraffin wax blocks of formalin-fixed kidney portions. Sections were stained by periodic acid-Schiff to show the fungal cells clearly and poststained with hematoxylin to illustrate kidney morphology and immune infiltrates.
Results
Reduced virulence of strain NGY355 is evident at 24 h postinfection
In the murine model of systemic candidiasis, C. albicans strain NGY152 is virulent, attaining high renal fungal burdens and rapidly causing fatal infection (Brand et al., 2004; MacCallum & Odds, 2005;). The second C. albicans strain, NGY355 (pmr1Δ), a mutant that lacks N- and O-linked mannan side chains in the cell wall, has been shown to be severely attenuated in virulence, both in terms of mouse survival times and of fungal organ burdens (Bates et al., 2005; MacCallum et al., 2009;).
Mice infected with either NGY152 (virulent) or NGY355 (attenuated) were sampled 24 h postinfection, when a rapid increase in kidney fungal burden is known to occur in kidneys infected by the virulent strain (MacCallum & Odds, 2005). In contrast, little change in kidney fungal burden was found at this time for the attenuated (pmr1Δ) strain (MacCallum et al., 2009).
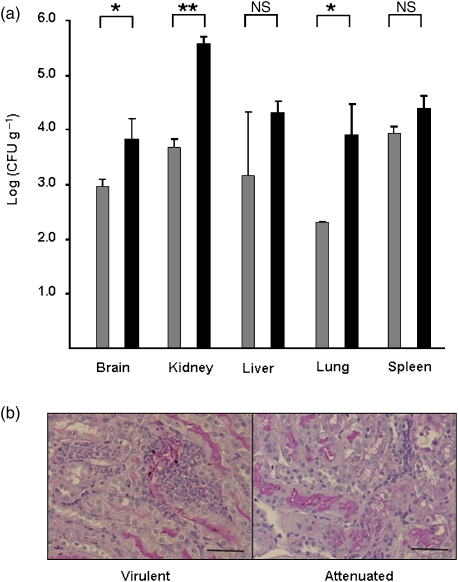
In this study, virulence differences between the two strains were evident at this early time, in terms of significantly higher burdens in the kidney, brain and lung (Fig. 1a) in mice infected with the virulent strain, with the greatest difference (approximately 100-fold) observed for the kidney.
Fig. 1.
Disease pathology at 24 h postinfection is more severe in virulent strain infected mice. Groups of mice (N=3) were infected with either Candida albicans NGY152 (virulent), C. albicans NGY355 (attenuated) or saline as control, and sampled at 24 h postinfection. Disease progression was measured in terms of organ burdens and histopathology. (a) Higher fungal burdens were measured in the brain, lung and kidneys in mice infected with the virulent strain. Results represent the mean ± SD. Attenuated infection is represented by gray bars, virulent infection by black bars. Means were compared by Students t-test: *P<0.05, **P<0.01 and NS, significant. (b) Renal histopathology. Sections (5 μm) were stained with periodic acid-Schiff stain (fungal cells stained pink) and poststained with hematoxylin (shows tissue structure and stain immune cells blue/purple). Fungal cells (indicated by arrowheads) are obvious only in the kidneys of mice infected by the virulent strain. Scale bar represents 100 μm.
The kidneys of virulent strain-infected mice contained filamentous fungal cells associated with leukocyte infiltrates (Fig. 1b), but there was little evidence of similar fungal lesions or immune cell infiltration in the kidneys of mice infected with the attenuated fungal strain. Control kidneys from saline-infected mice showed no evidence of fungal cells or immune cell infiltrates (data not shown).
Renal gene expression changes associated with C. albicans infection
Because infection by the different C. albicans strains led to different renal phenotypes, i.e. immune infiltrate and lesion development, global transcript profiling of kidneys 24 h postinfection was used to reveal the local host responses underlying these different responses. Whole genome DNA microarrays were used to compare expression profiles of kidneys infected with either the virulent or the attenuated C. albicans strain relative to control, uninfected kidneys.
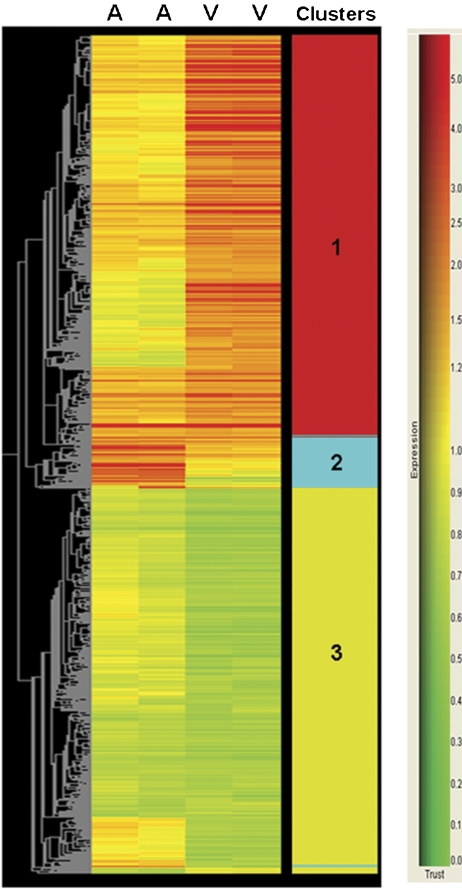
Results demonstrated that 2099 (5.4%) murine genes were upregulated in the kidneys of mice infected with NGY152 (virulent strain), compared with only 557 (1.4%) genes in the kidneys of mice infected with NGY355 (attenuated strain). The numbers of downregulated genes were similar to those upregulated, with 2069 (5.3%) for the virulent strain infection and 393 (1.0%) for the attenuated strain. There were 273 common upregulated and 286 common downregulated genes, representing the majority of those with altered expression levels in NGY355 infection. These commonly regulated genes are suggested to represent a core transcriptional response of the kidney to C. albicans infection.
All infection-regulated genes were grouped into thee large sets (Fig. 2) using clustering analysis based on K-means. Regulated genes common to both infections, and representative of the core response, were found in cluster 2, while genes more upregulated in the virulent strain infection formed cluster 1. Cluster 3 represented genes that were commonly downregulated or downregulated specifically in virulent strain infection.
Fig. 2.
Genes with altered renal expression in systemic candidiasis form three distinct clusters. Differentially regulated genes in the kidneys of infected mice were analyzed by the Gene Tree function of Genespring to group genes regulated in a similar manner. Genes are colored by their fold regulation relative to control, as shown by the key on the right. K-means clustering separated the genes into three large clusters as numbered in the clusters column. Each column contains data from single microarray hybridizations, with V representing infection by the virulent strain and A representing infection by the attenuated strain.
Biological functions and pathways associated with C. albicans infection
To identify biological functions and/or pathways associated with genes with altered expression in infected kidneys, results were analyzed by Ingenuity Pathway Analysis (http://www.ingenuity.com) and the DAVID (http://david.abcc.ncifcrf.gov) resource.
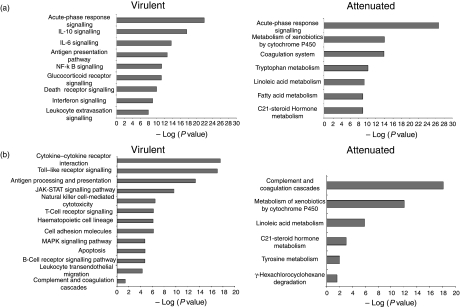
Results demonstrated that both the attenuated and the virulent strains induced changes in gene expression associated with the acute-phase response (a generalized response to disturbances in physiological homeostasis) and complement and coagulation cascades (Fig. 3a and b). These responses represent the core response of the kidney to C. albicans infection, with their expression regulated in response to infection with either strain (cluster 2, Fig. 2).
Fig. 3.
Infection by virulent or attenuated Candida albicans strains induces different responses in the kidney. Gene lists were analyzed by Ingenuity Pathway Analysis (included fold regulation) (a) or via the DAVID website (b). The most significantly regulated biological pathways or functions were determined by P-value.
In addition to the core response, infection with the attenuated strain induced regulation of genes in some metabolic pathways, whereas infection with the virulent C. albicans strain was strongly associated with regulation of genes involved in induced immune responses (Fig. 3a and b). Functions most strongly associated with virulent strain infection included cytokine–cytokine receptor interactions and a number of immune signaling pathways, such as interleukin (IL)-6, IL-10 and nuclear factor κB. In addition, gene expression changes linked to leukocyte movement were also associated with infection by the virulent strain. There was also evidence for initiation of an adaptive immune response with gene expression changes associated with antigen processing and presentation, and also signaling from both T-cell and B-cell receptors in the kidneys of mice infected with the virulent strain (Fig. 3b). Genes associated with the above functions were found in cluster 1 (Fig. 2), with little change in the expression of these same genes in kidneys infected by the attenuated strain.
Genes downregulated during infection, found in cluster 3 (Fig. 2), were associated with pyruvate metabolism, fatty acid metabolism, degradation of valine, leucine and isoleucine, as well as tryptophan metabolism. However, these associations were based on only one-fifth of the genes, with the majority of downregulated genes not yet assigned to functions or pathways.
Cytokine and pattern recognition receptor (PRR) gene expression during infection
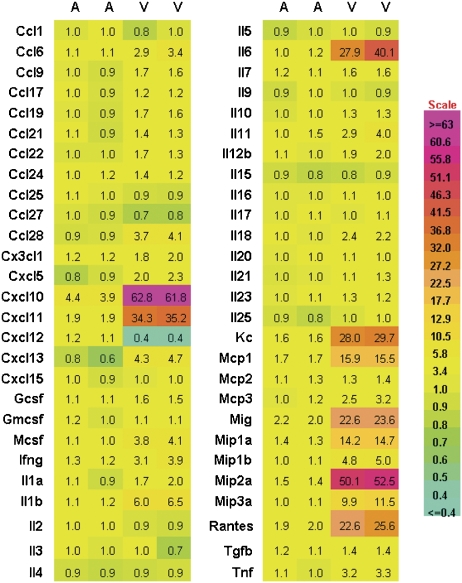
Because of the differences in renal gene expression associated with cytokine interactions and signaling in the different infections, a more detailed examination of cytokine gene expression was performed (Fig. 4). Results demonstrated a clear induction of the proinflammatory cytokine genes Il1a and b, Il6 and Tnf, which have also been linked to the acute-phase response, in the kidneys of mice infected with the virulent strain, whereas little induction of these genes was seen in kidneys infected with the attenuated strain. Among 54 cytokine genes examined, 32 were significantly altered in virulent strain infection compared with control kidneys (Fig. 4). The majority of these genes were upregulated, but a single cytokine gene, Cxcl12, was downregulated in the renal response to the virulent C. albicans strain and unchanged in attenuated strain infection.
Fig. 4.
Cytokine/chemokine gene expression in infected kidneys. For each cytokine, the gene expression levels obtained by microarray analyses are shown relative to control, where A represents infection by the attenuated strain and V represents infection by the virulent strain. Expression level (-fold) change is colored as shown on the scale.
In the kidneys of mice infected with the attenuated strain, the only cytokine/chemokine genes upregulated were Cxcl10, Cxcl11, Kc, Mcp1, Mig, Mip1a, Mip2a and Rantes. These genes, along with Il6, were also the most upregulated in virulent strain infection, with expression levels at least 10-fold higher than those of mice infected with the attenuated strain (Fig. 4).
The large differences in cytokine gene expression in response to C. albicans infection prompted an examination of the expression of PRR genes, the signaling from which influences cytokine production. PRRs known to be involved in the recognition of C. albicans include Toll-like receptors (TLRs) (Netea et al., 2006a,b; Carpenter & O'Neill, 2007;) and C-type lectins (Willment & Brown, 2008).
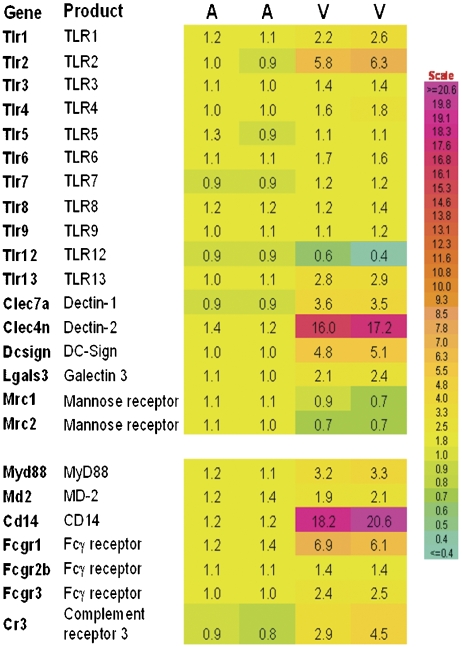
There was little change in PRR gene expression in kidneys infected with the attenuated strain (Fig. 5), but in the kidneys of mice infected with the virulent strain there was an increased expression of genes encoding TLRs (Tlr1, Tlr2, Tlr4, Tlr6 and Tlr13) and their associated adaptor molecules: Myd88, Cd14 and Md2 (Fig. 5). The greatest increase in TLR gene expression was seen for Tlr2. In addition, a mouse-specific TLR gene, Tlr12, was downregulated during virulent strain infection, but was unaltered in attenuated strain infection.
Fig. 5.
PRR gene expression in infected kidneys. The expression levels of various pattern recognition receptors and some adaptor molecules are shown, where A represents infection by the attenuated strain and V represents infection by the virulent strain. Expression levels are normalized to controls. Expression level (-fold) change is colored as shown on the scale.
Although the majority of non-TLR PRR genes examined showed increased expression in the kidneys of virulent strain-infected mice, the expression of mannose receptor genes was reduced, and the greatest increase in non-TLR PRR gene expression was seen for dectin-2 (Fig. 5). Similarly, expression of FCγ receptor genes, whose products are required for dectin-2 signaling (Sato et al., 2006), were also upregulated in response to virulent strain infection.
Confirmation of transcript profiling results by qRT-PCR
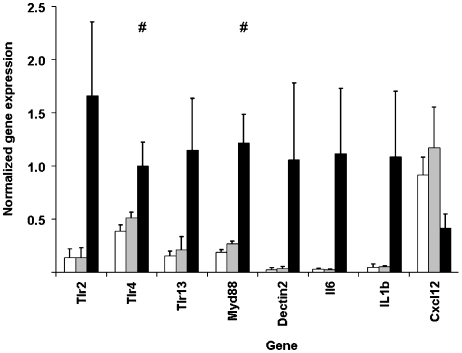
In order to confirm the gene expression differences found by microarray analysis, qRT-PCR was used to analyze expression levels of several genes of interest. Immune response-associated genes, with altered expression in virulent strain infection, were selected for further investigation: Tlr2, Tlr4, Tlr13, Myd88, Il1b, Il6, dectin-2 and Cxcl12 (Fig. 6).
Fig. 6.
qRT-PCR confirms microarray gene expression differences. qRT-PCR was used to analyze gene expression of immune-related genes in individual kidney samples. mRNA copy number for each gene was normalized to a GAPDH control. Data represent the mean of three individual samples (± SD) for each treatment group. White bars represent control kidneys; gray bars, attenuated strain-infected kidneys and black bars, virulent strain-infected kidneys. Means were compared by Kruskal–Wallis and Mann–Whitney-U-tests. For all genes, P<0.001 for control vs. virulent and attenuated vs. virulent. For control vs. attenuated, #P<0.05 for control vs. attenuated.
Results obtained by qRT-PCR for the individual kidney RNA samples showed good correlation with the transcript profiling results for RNA pools (r=0.93, data not shown), with statistically significant differences found for all genes when renal responses to the virulent strain were compared with those of mice infected with the attenuated strain or of control mice (Fig. 6). These results confirmed the changes in innate immune response-related gene expression seen by microarray analysis in kidneys in response to the virulent strain, but that only occurred to a limited extent in response to the attenuated strain.
Surprisingly, qRT-PCR also demonstrated a small, but significant, increase in Tlr4 gene expression in response to infection by the attenuated strain that was not seen by microarray analysis (Fig. 6).
Discussion
The mouse intravenous challenge model is frequently used to characterize the virulence potential of C. albicans isolates and strains and is a good model of severe human disseminated infection. From previous studies, we have shown that kidney burdens in virulent strain-infected mice continue to increase until the mice become severely ill (MacCallum & Odds, 2005). In contrast, in infection initiated by the attenuated strain, kidney burdens remain roughly constant over 28 days of infection (Bates et al., 2005; MacCallum et al., 2009;). These results demonstrate that the virulent strain initiates a progressive infection, while infection initiated by the attenuated strain is controlled. Analysis of organ burdens at 24 h postinfection demonstrated virulence differences between the two strains, even at this early time in infection. Kidney burdens were significantly higher for mice infected with the virulent strain and infiltrating immune cells were already evident in the kidneys of these mice. These results clearly indicate that early renal responses to the two C. albicans strains must differ.
In this study, the transcriptional responses of the kidney at 24 h postinfection were analyzed to determine how responses to known virulent and attenuated C. albicans strains differ. Microarray gene expression patterns were confirmed by qRT-PCR, confirming that the use of RNA pools for transcript profiling is a valid approach for this experimental infection model.
Results clearly demonstrated that the kidney responds to C. albicans infection by inducing a core response: the acute-phase response and complement and coagulation cascades. Induction of the acute-phase response was not surprising as this reaction is a response to physiological alterations and it is usually activated during inflammation. Although previously associated with the liver (Kushner, 1982), renal proximal tubular epithelial cells have also been shown to generate a strong acute-phase response (Luyckx et al., 2009), suggesting that this cell type may be involved in the core response.
While the core response appeared sufficient to prevent overgrowth of the attenuated strain within the first 24 h of infection, reducing fungal burdens to levels similar to those found at 28 days postinfection (Bates et al., 2005), it was insufficient to control the virulent strain. Deficiencies in the core response to C. albicans infection have previously been shown to lead to increased susceptibility to systemic candidiasis (Gelfand et al., 1978; Odds, 1988; Mullick et al., 2004b; Held et al., 2008;), and may allow normally attenuated strains to initiate progressive infections.
In progressive renal infection initiated by the virulent strain, the fungus proliferated in the kidneys, accompanied by a massive transcriptional change, mostly genes associated with innate and adaptive immune responses. This demonstrates a clear link between induction of host immune responses in the kidney and progression of C. albicans infection. Immune responses occurring in the kidney have previously been linked to C. albicans infection outcome (Brieland et al., 2001; Spellberg et al., 2003; Cao et al., 2005; MacCallum et al., 2009;).
The majority of cytokines and chemokines genes examined showed increased expression in response to infection by the virulent strain, their products thus leading to attraction and activation of immune cells. In contrast, the attenuated strain showed little induction of proinflammatory gene expression in the kidney, correlating with the lack of infiltrating immune cells in the kidney (Fig. 1b) and also a previous study where the attenuated strain induced much lower levels of tumor necrosis factor (TNF)-α and IL-6, due to its defect in cell wall protein mannosylation (Netea et al., 2006b). However, although the defect in TNF-α and IL-6 induction is due to the lack of mannosylation in the attenuated pmr1Δ mutant1 (Netea et al., 2004), we have recently demonstrated that attenuated clinical isolates, in general, induce lower renal levels of chemokines and proinflammatory cytokines compared with virulent isolates (MacCallum et al., 2009). This confirms that upregulation of cytokines and chemokines is a general response of kidneys exposed to virulent C. albicans strains (MacCallum et al., 2009).
Proinflammatory cytokines, for example TNF-α and interferon (IFN)-γ, are required for normal responses to C. albicans infection (Steinshamn & Waage, 1992; Kullberg et al., 1993; Louie et al., 1994; Brieland et al., 2001;). However, previous studies have demonstrated that the fatal outcome of C. albicans infection is linked to the development of T-helper type 2 (Th2) and IL-10 responses in the kidney (Brieland et al., 2001; Spellberg et al., 2003; Cao et al., 2005;). IL-10 is known to have strong inhibitory effects on defense against C. albicans (Romani et al., 1994), with IL-10 knockout mice more resistant to C. albicans infection (Vazquez-Torres et al., 1999). We found, in agreement with others (Brieland et al., 2001; Spellberg et al., 2003;), that renal expression of Il10 was induced in progressive infection. Cytokine profiles, however, showed evidence of both Th1 and Th2 responses in infected kidneys, similar to the results obtained by Bellocchio et al. (2004). Differences between this study and the study of Cao et al. (2005), who clearly found that a Th2 response in the kidney was linked to a lethal outcome, may be due to the use of different C. albicans strains, or due to different kinetics of infection progression, as mice in this study had lower survival times than those in the study of Cao et al. (2005).
The renal transcriptional response to C. albicans progressive infection also included regulation of PRR gene expression. Previously, a number of PRRs have been demonstrated to be involved in host recognition of C. albicans (Netea et al., 2006a; Willment & Brown, 2008;), particularly TLR2, TLR4 (Netea et al., 2002, 2008; Villamon et al., 2004b; Blasi et al., 2005) and dectin-1 (Brown et al., 2003; Gantner et al., 2005; Gow et al., 2007; Ferwerda et al., 2008;). In this study, the kidney cells upregulating expression of the PRRs were not identified but, in response to renal injury, TLR2 and TLR4 levels are increased on tubular epithelial cells (Wolfs et al., 2002; Kim et al., 2005a; Leemans et al., 2005; Shigeoka et al., 2007; Wu et al., 2007;). This again suggests that these cells may play a role in renal responses to C. albicans infection, especially as these cells are capable of producing numerous cytokines and chemokines (reviewed in Daha & van Kooten 2000), and have been shown to respond to other infections with induction of inflammatory cascades (Ross et al., 2006). However, the microarray results in this study also show good agreement with those found for endothelial cells (Muller et al., 2007), indicating that transcriptional changes may be due to multiple cell types in the kidney. Examination of the transcriptional responses of kidney cell subsets to C. albicans infection will be the subject of future studies.
In response to infection by the attenuated strain, there was little change in PRR gene expression in the kidneys by microarray analysis. However, qRT-PCR indicated a small significant increase in Tlr4 expression in these kidneys. This demonstrated that, as shown previously (Allanach et al., 2008), qRT-PCR is more sensitive in detection of subtle changes in gene expression and is more suitable for investigating small gene expression differences.
The PRR genes most highly upregulated in response to infection by the virulent strain were Tlr2 and a C-type lectin gene, dectin-2, which is involved in the recognition of C. albicans hyphae (McGreal et al., 2006; Sato et al., 2006;). TLR2 has also been implicated in the recognition of C. albicans hyphae (van der Graaf et al., 2005) and plays an important role in C. albicans infection (Netea et al., 2002; Blasi et al., 2005; Gil & Gozalbo, 2006; Murciano et al., 2007;). Many of the cytokine/chemokine genes involved in the response to virulent strain infection have been shown to require signaling from TLR2 in combination with dectin-1 (Brown et al., 2003; van der Graaf et al., 2005; Netea et al., 2006b; Ferwerda et al., 2008;). However, in this study, although dectin-1 was upregulated in response to infection by the virulent strain, dectin-2 was much more highly upregulated. Differences in the expression of C-type lectins between the different studies are most likely due to the host cells analyzed, with whole kidneys analyzed in this study and isolated immune cells analyzed in others.
One of the major questions arising from this study is why infection by the virulent strain in the kidney is not controlled when there is an obvious inflammatory response occurring. This may be due to induction of regulatory responses in the kidneys. Although TLR2 signaling can induce the production of proinflammatory mediators (macrophage inflammatory protein-2, KC (keratinocyte-derived cytokine), TNF-α, IL-1β and IFN-γ) (Netea et al., 2002, 2004; Bellocchio et al., 2004; Villamon et al., 2004a, b; van der Graaf et al., 2005; Gil & Gozalbo, 2006; Murciano et al., 2007; De Filippo et al., 2008), it can also induce IL-10 production, suppressing immune responses to C. albicans (Romani et al., 1994, 2004; Netea et al., 2004). TLR2 is also involved in controlling the expansion and function of regulatory T cells (Netea et al., 2004; Sutmuller et al., 2006;), with fewer regulatory T cells found in TLR2 knockout mice (Netea et al., 2004). Therefore, increased expression of Tlr2, leading to increased levels of IL-10 and increased survival of regulatory T cells, may dampen the immune response against C. albicans, even in the presence of immune infiltrates in the kidney, and allow infection to progress.
Amplification of immune responses to infection in the kidney may also occur, as the proinflammatory cytokines TNF-α and IFN-γ have been shown to induce renal expression of Tlr2 and Tlr4 (Wolfs et al., 2002). This may lead to continued production of both proinflammatory and immunosuppressive mediators in response to fungal cells. Continued inflammatory responses, where the fungal growth is not controlled, can be regarded as inappropriate, and have previously been shown to contribute to development of sepsis (reviewed in Sriskandan & Altmann, 2008), which leads to host deterioration and eventually death in the mouse model of systemic C. albicans infection (Spellberg et al., 2005).
This study has several limitations, including study of a single time point during infection and the use of single examples of virulent and attenuated C. albicans strains; however, it does serve as a basis for future expansion in this area of research, examining the renal responses temporally during infection progression, in different mouse strains (including mouse strains with defined gene knockouts) and responses in organs other than the kidney, where infection by C. albicans is known to be controlled.
Conclusion
This study has characterized the transcriptional profiles of kidneys responding to infection by virulent and attenuated C. albicans strains. A core response was identified, which consisted of the acute-phase response, including the complement and coagulation cascades. Progressive infection initiated by the virulent C. albicans strain was characterized by a strong inflammatory transcriptional response, with massive induction of cytokine and chemokine gene expression, accompanied by an influx of immune cells into the infected kidney. The induced renal immune response did not control growth of the virulent C. albicans strain in vivo, but possibly contributes to the development of sepsis, causing host death. In the future, therapies targeting the immune response to progressive infection may be effective in treating systemic C. albicans infection.
Acknowledgments
Many thanks are due to Professor Frank Odds, University of Aberdeen, for helpful discussions on this manuscript. Thanks are also due to Mrs Diane Stewart of the IMS Microarray Core Facility (IMCF), University of Aberdeen, for skilled technical assistance and to the IMS Histology and EM facility, University of Aberdeen, for preparation of the paraffin organ blocks. Bioanalyzer analyses were carried out by the Genomics section, Rowett Institute of Nutrition and Health, University of Aberdeen, UK. This study was funded by the IMCF, University of Aberdeen, and D.M.M. was funded by grant no. 080088/Z/06/Z from the Wellcome Trust.
Statement
Re-use of this article is permitted in accordance with the Terms and Conditions set out at: http://www3.interscience.wiley.com/authorresources/onlineopen.html
References
- Allanach K, Mengel M, Einecke G, Sis B, Hidalgo LG, Mueller T, Halloran PF. Comparing microarray versus RT-PCR assessment of renal allograft biopsies: similar performance despite different dynamic ranges. Am J Transplant. 2008;8:1006–1015. doi: 10.1111/j.1600-6143.2008.02199.x. [DOI] [PubMed] [Google Scholar]
- Ashman RB. Protective and pathologic immune responses against Candida albicans infection. Front Biosci. 2008;13:3334–3351. doi: 10.2741/2929. [DOI] [PubMed] [Google Scholar]
- Barker KS, Liu T, Rogers PD. Coculture of THP-1 human mononuclear cells with Candida albicans results in pronounced changes in host gene expression. J Infect Dis. 2005;192:901–912. doi: 10.1086/432487. [DOI] [PubMed] [Google Scholar]
- Barker KS, Park H, Phan QT, Xu L, Homayouni R, Rogers PD, Filler SG. Transcriptome profile of the vascular endothelial cell response to Candida albicans. J Infect Dis. 2008;198:193–202. doi: 10.1086/589516. [DOI] [PubMed] [Google Scholar]
- Bates S, MacCallum DM, Bertram G, Munro CA, Hughes HB, Buurman ET, Brown AJ, Odds FC, Gow NA. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J Biol Chem. 2005;280:23408–23415. doi: 10.1074/jbc.M502162200. [DOI] [PubMed] [Google Scholar]
- Bellocchio S, Montagnoli C, Bozza S, Gaziano R, Rossi G, Mambula SS, Vecchi A, Mantovani A, Levitz SM, Romani L. The contribution of the Toll-like/IL-1 receptor superfamily to innate and adaptive immunity to fungal pathogens in vivo. J Immunol. 2004;172:3059–3069. doi: 10.4049/jimmunol.172.5.3059. [DOI] [PubMed] [Google Scholar]
- Blasi E, Mucci A, Neglia R, et al. Biological importance of the two Toll-like receptors, TLR2 and TLR4, in macrophage response to infection with Candida albicans. FEMS Immunol Med Mic. 2005;44:69–79. doi: 10.1016/j.femsim.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Bougnoux ME, Kac G, Aegerter P, d'Enfert C, Fagon JY CandiRea Study Group. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intens Care Med. 2008;34:292–299. doi: 10.1007/s00134-007-0865-y. [DOI] [PubMed] [Google Scholar]
- Brand A, MacCallum DM, Brown AJ, Gow NA, Odds FC. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot Cell. 2004;3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieland J, Essig D, Jackson C, Frank D, Loebenberg D, Menzel F, Arnold B, DiDomenico B, Hare R. Comparison of pathogenesis and host immune responses to Candida glabrata and Candida albicans in systemically infected immunocompetent mice. Infect Immun. 2001;69:5046–5055. doi: 10.1128/IAI.69.8.5046-5055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano G, Iatta R, Laneve A, Manca F, Montagna MT. Observational study on candidaemia at a university hospital in southern Italy from 1998 to 2004. Mycoses. 2008;51:123–128. doi: 10.1111/j.1439-0507.2007.01452.x. [DOI] [PubMed] [Google Scholar]
- Cao F, Li J, Yan X, Wu Y, Zhang D. Relationship between host survival and the type of immune response in different organs during disseminated candidiasis. J Huazhong Univ Sci Technolog Med Sci. 2005;25:141–143. doi: 10.1007/BF02873560. 184. [DOI] [PubMed] [Google Scholar]
- Carpenter S, O'Neill LA. How important are Toll-like receptors for antimicrobial responses? Cell Microbiol. 2007;9:1891–1901. doi: 10.1111/j.1462-5822.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- Daha MR, van Kooten C. Is the proximal tubular cell a proinflammatory cell? Nephrol Dial Transpl. 2000;15(suppl 6):41–43. doi: 10.1093/ndt/15.suppl_6.41. [DOI] [PubMed] [Google Scholar]
- De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol. 2008;180:4308–4315. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60.01–R60.11. [PubMed] [Google Scholar]
- Fernandez-Arenas E, Cabezon V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, Gil C. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics. 2007;6:460–478. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- Ferwerda G, Meyer-Wentrup F, Kullberg BJ, Netea MG, Adema GJ. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell Microbiol. 2008;10:2058–2066. doi: 10.1111/j.1462-5822.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d'Enfert C, Hube B. Stage-specific gene expression of Candida albicans in human blood. Mol Microbiol. 2003;47:1523–1543. doi: 10.1046/j.1365-2958.2003.03396.x. [DOI] [PubMed] [Google Scholar]
- Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- Fradin C, Mavor AL, Weindl G, Schaller M, Hanke K, Kaufmann SH, Mollenkopf H, Hube B. The early transcriptional response of human granulocytes to infection with Candida albicans is not essential for killing, but reflects cellular communications. Infect Immun. 2006;75:1493–1501. doi: 10.1128/IAI.01651-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner BN, Simmons RM, Underhill DM. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 2005;24:1277–1286. doi: 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand JA, Hurley DL, Fauci AS, Frank MM. Role of complement in host defense against experimental disseminated candidiasis. J Infect Dis. 1978;138:9–16. doi: 10.1093/infdis/138.1.9. [DOI] [PubMed] [Google Scholar]
- Gil ML, Gozalbo D. TLR2, but not TLR4, triggers cytokine production by murine cells in response to Candida albicans yeasts and hyphae. Microbes Infect. 2006;8:2299–2304. doi: 10.1016/j.micinf.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Gow NA, Netea MG, Munro CA, et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J Infect Dis. 2007;196:1565–1571. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held K, Thiel S, Loos M, Petry F. Increased susceptibility of complement factor B/C2 double knockout mice and mannan-binding lectin knockout mice to systemic infection with Candida albicans. Mol Immunol. 2008;45:3934–3941. doi: 10.1016/j.molimm.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Miura NN, Adachi Y, Ogura N, Tamura H, Tanaka S, Ohno N. DNA array analysis of altered gene expression in human leukocytes stimulated with soluble and particulate forms of Candida cell wall beta-glucan. Int Immunopharmacol. 2004;4:387–401. doi: 10.1016/j.intimp.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Kim BS, Lim SW, Li C, Kim JS, Sun BK, Ahn KO, Han SW, Kim J, Yang CW. Ischemia–reperfusion injury activates innate immunity in rat kidneys. Transplantation. 2005a;79:1370–1377. doi: 10.1097/01.tp.0000158355.83327.62. [DOI] [PubMed] [Google Scholar]
- Kim HS, Choi EH, Khan J, et al. Expression of genes encoding innate host defense molecules in normal human monocytes in response to Candida albicans. Infect Immun. 2005b;73:3714–3724. doi: 10.1128/IAI.73.6.3714-3724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg BJ, van 't Wout JW, Hoogstraten C, van Furth R. Recombinant interferon-gamma enhances resistance to acute disseminated Candida albicans infection in mice. J Infect Dis. 1993;168:436–443. doi: 10.1093/infdis/168.2.436. [DOI] [PubMed] [Google Scholar]
- Kushner I. The phenomenon of the acute phase response. Ann NY Acad Sci. 1982;389:39–48. doi: 10.1111/j.1749-6632.1982.tb22124.x. [DOI] [PubMed] [Google Scholar]
- Leemans JC, Stokman G, Claessen N, Rouschop KM, Teske GJ, Kirschning CJ, Akira S, van der Poll T, Weening JJ, Florquin S. Renal-associated TLR2 mediates ischemia/reperfusion injury in the kidney. J Clin Invest. 2005;115:2894–2903. doi: 10.1172/JCI22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leunk RD, Moon RJ. Physiological and metabolic alterations accompanying systemic candidiasis in mice. Infect Immun. 1979;26:1035–1041. doi: 10.1128/iai.26.3.1035-1041.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A, Baltch AL, Smith RP, Franke MA, Ritz WJ, Singh JK, Gordon MA. Tumor necrosis factor alpha has a protective role in a murine model of systemic candidiasis. Infect Immun. 1994;62:2761–2772. doi: 10.1128/iai.62.7.2761-2772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louria DB, Brayton RG, Finkel G. Studies on the pathogenesis of experimental Candida albicans infections in mice. Sabouraudia. 1963;2:271–283. [Google Scholar]
- Luyckx VA, Cairo LV, Compston CA, Phan WL, Mueller TF. The oncostatin M pathway plays a major role in the renal acute phase response. Am J Physiol-Renal. 2009;296:F875–F883. doi: 10.1152/ajprenal.90633.2008. [DOI] [PubMed] [Google Scholar]
- MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005;48:151–161. doi: 10.1111/j.1439-0507.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- MacCallum DM, Castillo L, Brown AJP, Gow NAR, Odds FC. Early-expressed chemokines predict kidney immunopathology in experimental disseminated Candida albicans infections. PLoS One. 2009;4:e6420. doi: 10.1371/journal.pone.0006420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGreal EP, Rosas M, Brown GD, Zamze S, Wong SY, Gordon S, Martinez-Pomares L, Taylor PR. The carbohydrate-recognition domain of dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology. 2006;16:422–430. doi: 10.1093/glycob/cwj077. [DOI] [PubMed] [Google Scholar]
- Muller V, Viemann D, Schmidt M, Endres N, Ludwig S, Leverkus M, Roth J, Goebeler M. Candida albicans triggers activation of distinct signaling pathways to establish a proinflammatory gene expression program in primary human endothelial cells. J Immunol. 2007;179:8435–8445. doi: 10.4049/jimmunol.179.12.8435. [DOI] [PubMed] [Google Scholar]
- Mullick A, Elias M, Harakidas P, et al. Gene expression in HL60 granulocytoids and human polymorphonuclear leukocytes exposed to Candida albicans. Infect Immun. 2004a;72:414–429. doi: 10.1128/IAI.72.1.414-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullick A, Elias M, Picard S, et al. Dysregulated inflammatory response to Candida albicans in a C5-deficient mouse strain. Infect Immun. 2004b;72:5868–5876. doi: 10.1128/IAI.72.10.5868-5876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murciano C, Yanez A, Gil ML, Gozalbo D. Both viable and killed Candida albicans cells induce in vitro production of TNF-alpha and IFN-gamma in murine cells through a TLR2-dependent signaling. Eur Cytokine Netw. 2007;18:38–43. doi: 10.1684/ecn.2007.0085. [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F, Sanchez M, Nombela C, Pla J. Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol Rev. 2001;25:245–268. doi: 10.1111/j.1574-6976.2001.tb00577.x. [DOI] [PubMed] [Google Scholar]
- Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis. 2002;185:1483–1489. doi: 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- Netea MG, Sutmuller R, Hermann C, Van der Graaf CA, Van der Meer JW, van Krieken JH, Hartung T, Adema G, Kullberg BJ. Toll-like receptor 2 suppresses immunity against Candida albicans through induction of IL-10 and regulatory T cells. J Immunol. 2004;172:3712–3718. doi: 10.4049/jimmunol.172.6.3712. [DOI] [PubMed] [Google Scholar]
- Netea MG, Ferwerda G, van der Graaf CAA, Van der Meer JWM, Kullberg BJ. Recognition of fungal pathogens by toll-like receptors. Curr Pharm Design. 2006a;12:4195–4201. doi: 10.2174/138161206778743538. [DOI] [PubMed] [Google Scholar]
- Netea MG, Gow NA, Munro CA, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006b;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, van de Veerdonk F, Verschueren I, van der Meer JW, Kullberg BJ. Role of TLR1 and TLR6 in the host defense against disseminated candidiasis. FEMS Immunol Med Mic. 2008;52:118–123. doi: 10.1111/j.1574-695X.2007.00353.x. [DOI] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. London: Bailliere Tindall; 1988. [Google Scholar]
- Odds FC, Hanson MF, Davidson AD, Jacobsen MD, Wright P, Whyte JA, Gow NA, Jones BL. One year prospective survey of Candida bloodstream infections in Scotland. J Med Microbiol. 2007;56:1066–1075. doi: 10.1099/jmm.0.47239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JC, Jr, McCloskey JJ, Knauer KA. Pathobiologic features of human candidiasis. A common deep mycosis of the brain, heart and kidney in the altered host. Am J Clin Pathol. 1976;65:991–1000. doi: 10.1093/ajcp/65.6.991. [DOI] [PubMed] [Google Scholar]
- Playford EG, Marriott D, Nguyen Q, Chen S, Ellis D, Slavin M, Sorrell TC. Candidemia in nonneutropenic critically ill patients: risk factors for non-albicans Candida spp. Crit Care Med. 2008;36:2034–2039. doi: 10.1097/CCM.0b013e3181760f42. [DOI] [PubMed] [Google Scholar]
- Prigneau O, Porta A, Poudrier JA, Colonna-Romano S, Noel T, Maresca B. Genes involved in beta-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast. 2003;20:723–730. doi: 10.1002/yea.998. [DOI] [PubMed] [Google Scholar]
- Romani L. Cell mediated immunity to fungi: a reassessment. Med Mycol. 2008;46:515–529. doi: 10.1080/13693780801971450. [DOI] [PubMed] [Google Scholar]
- Romani L, Puccetti P, Mencacci A, Cenci E, Spaccapelo R, Tonnetti L, Grohmann U, Bistoni F. Neutralization of IL-10 up-regulates nitric oxide production and protects susceptible mice from challenge with Candida albicans. J Immunol. 1994;152:3514–3521. [PubMed] [Google Scholar]
- Romani L, Montagnoli C, Bozza S, Perruccio K, Spreca A, Allavena P, Verbeek S, Calderone RA, Bistoni F, Puccetti P. The exploitation of distinct recognition receptors in dendritic cells determines the full range of host immune relationships with Candida albicans. Int Immunol. 2004;16:149–161. doi: 10.1093/intimm/dxh012. [DOI] [PubMed] [Google Scholar]
- Ross MJ, Fan C, Ross MD, Chu TH, Shi Y, Kaufman L, Zhang W, Klotman ME, Klotman PE. HIV-1 infection initiates an inflammatory cascade in human renal tubular epithelial cells. J Acq Imm Def Syndr. 2006;42:1–11. doi: 10.1097/01.qai.0000218353.60099.4f. [DOI] [PubMed] [Google Scholar]
- Sandovsky-Losica H, Chauhan N, Calderone R, Segal E. Gene transcription studies of Candida albicans following infection of HEp2 epithelial cells. Med Mycol. 2006;44:329–334. doi: 10.1080/13693780500434701. [DOI] [PubMed] [Google Scholar]
- Sato K, Yang XL, Yudate T, Chung JS, Wu J, Luby-Phelps K, Kimberly RP, Underhill D, Cruz PD, Ariizumi K. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor {gamma} chain to induce innate immune responses. J Biol Chem. 2006;281:38854–38866. doi: 10.1074/jbc.M606542200. [DOI] [PubMed] [Google Scholar]
- Shigeoka AA, Holscher TD, King AJ, Hall FW, Kiosses WB, Tobias PS, Mackman N, McKay DB. TLR2 is constitutively expressed within the kidney and participates in ischemic renal injury through both MyD88-dependent and -independent pathways. J Immunol. 2007;178:6252–6258. doi: 10.4049/jimmunol.178.10.6252. [DOI] [PubMed] [Google Scholar]
- Sohn K, Senyurek I, Fertey J, Konigsdorfer A, Joffroy C, Hauser N, Zelt G, Brunner H, Rupp S. An in vitro assay to study the transcriptional response during adherence of Candida albicans to different human epithelia. FEMS Yeast Res. 2006;6:1085–1093. doi: 10.1111/j.1567-1364.2006.00130.x. [DOI] [PubMed] [Google Scholar]
- Spellberg B, Johnston D, Phan QT, Edwards JE, Jr, French SW, Ibrahim AS, Filler SG. Parenchymal organ, and not splenic, immunity correlates with host survival during disseminated candidiasis. Infect Immun. 2003;71:5756–5764. doi: 10.1128/IAI.71.10.5756-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B, Ibrahim AS, Edwards JE, Jr, Filler SG. Mice with disseminated candidiasis die of progressive sepsis. J Infect Dis. 2005;192:336–343. doi: 10.1086/430952. [DOI] [PubMed] [Google Scholar]
- Sriskandan S, Altmann DM. The immunology of sepsis. J Pathol. 2008;214:211–223. doi: 10.1002/path.2274. [DOI] [PubMed] [Google Scholar]
- Steinshamn S, Waage A. Tumor necrosis factor and interleukin-6 in Candida albicans infection in normal and granulocytopenic mice. Infect Immun. 1992;60:4003–4008. doi: 10.1128/iai.60.10.4003-4008.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutmuller RP, den Brok MH, Kramer M, Bennink EJ, Toonen LW, Kullberg BJ, Joosten LA, Akira S, Netea MG, Adema GJ. Toll-like receptor 2 controls expansion and function of regulatory T cells. J Clin Invest. 2006;116:485–494. doi: 10.1172/JCI25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, Hube B. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol. 2007;63:1606–1628. doi: 10.1111/j.1365-2958.2007.05614.x. [DOI] [PubMed] [Google Scholar]
- van der Graaf CA, Netea MG, Verschueren I, van der Meer JW, Kullberg BJ. Differential cytokine production and Toll-like receptor signaling pathways by Candida albicans blastoconidia and hyphae. Infect Immun. 2005;73:7458–7464. doi: 10.1128/IAI.73.11.7458-7464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Wagner RD, Warner T, Balish E. Early resistance of interleukin-10 knockout mice to acute systemic candidiasis. Infect Immun. 1999;67:670–674. doi: 10.1128/iai.67.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamon E, Gozalbo D, Roig P, O'Connor JE, Ferrandiz ML, Fradelizi D, Gil ML. Toll-like receptor 2 is dispensable for acquired host immune resistance to Candida albicans in a murine model of disseminated candidiasis. Microbes Infect. 2004a;6:542–548. doi: 10.1016/j.micinf.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Villamon E, Gozalbo D, Roig P, O'Connor JE, Fradelizi D, Gil ML. Toll-like receptor-2 is essential in murine defenses against Candida albicans infections. Microbes Infect. 2004b;6:1–7. doi: 10.1016/j.micinf.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Walker LA, MacCallum DM, Bertram G, Gow NA, Odds FC, Brown AJ. Genome-wide analysis of Candida albicans gene expression patterns during infection of the mammalian kidney. Fungal Genet Biol. 2009;46:210–219. doi: 10.1016/j.fgb.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willment JA, Brown GD. C-type lectin receptors in antifungal immunity. Trends Microbiol. 2008;16:27–32. doi: 10.1016/j.tim.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Wolfs TG, Buurman WA, van Schadewijk A, de Vries B, Daemen MA, Hiemstra PS, van 't Veer C. In vivo expression of Toll-like receptor 2 and 4 by renal epithelial cells: IFN-gamma and TNF-alpha mediated up-regulation during inflammation. J Immunol. 2002;168:1286–1293. doi: 10.4049/jimmunol.168.3.1286. [DOI] [PubMed] [Google Scholar]
- Wu H, Chen G, Wyburn KR, Yin J, Bertolino P, Eris JM, Alexander SI, Sharland AF, Chadban SJ. TLR4 activation mediates kidney ischemia/reperfusion injury. J Clin Invest. 2007;117:2847–2859. doi: 10.1172/JCI31008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]