Abstract
During development, Met signaling regulates a range of cellular processes including growth, differentiation, survival and migration. The Met gene encodes a tyrosine kinase receptor, which is activated by Hgf (hepatocyte growth factor) ligand. Altered regulation of human MET expression has been implicated in autism. In mouse, Met signaling has been shown to regulate cerebellum development. Since abnormalities in cerebellar structure have been reported in some autistic patients, we have used the zebrafish to address the role of Met signaling during cerebellar development and thus further our understanding of the molecular basis of autism. We find that zebrafish met is expressed in the cerebellar primordium, later localizing to the ventricular zone (VZ), with the hgf1 and hgf2 ligand genes expressed in surrounding tissues. Morpholino knockdown of either Met or its Hgf ligands leads to a significant reduction in the size of the cerebellum, primarily as a consequence of reduced proliferation. Met signaling knockdown disrupts specification of VZ-derived cell types, and also reduces granule cell numbers, due to an early effect on cerebellar proliferation and/or as an indirect consequence of loss of signals from VZ-derived cells later in development. These patterning defects preclude analysis of cerebellar neuronal migration, but we have found that Met signaling is necessary for migration of hindbrain facial motor neurons. In summary, we have described roles for Met signaling in coordinating growth and cell type specification within the developing cerebellum, and in migration of hindbrain neurons. These functions may underlie the correlation between altered MET regulation and Autism Spectrum Disorders.
Keywords: autism, Met, Hgf, zebrafish, hindbrain, cerebellum, facial motor neuron, migration, morphogenesis, proliferation
INTRODUCTION
Autism and autism spectrum disorders (ASDs) affect increasing numbers of children yet the developmental basis of these disorders remains unclear. Genetic linkage analysis has identified vulnerability loci on several different chromosomes (Yang and Gill, 2007). One commonly reproduced ASD linkage region spans chromosome 7q21-q36, which includes the MET locus (Gupta and State, 2007). A recent paper (Campbell et al., 2006) has reported that a specific variant of the MET receptor gene is shared by a significant number of individuals with ASDs. The variant allele, a G to C substitution 20 base-pairs upstream of the MET transcriptional start site, has a highly significant P-value (p=0.000005) in a large sample of 743 families. Campbell et al. (2006) showed that the variant allele is further enriched in “multiplex” families with more than one autistic child. In vitro reporter experiments showed that the variant regulatory elements drive only 50% of normal transcription levels in mouse neural cells. Further, postmortem studies of autistic patients revealed a significant reduction of MET protein levels within the cerebral cortex (Campbell et al., 2007), suggesting that disruption of MET signaling contributes to ASD. The levels of the MET ligand HGF are altered in plasma and cerebrospinal fluid of autistic patients, providing additional support for a specific dysregulation of the MET signaling pathway (Sugihara et al., 2007; Vargas et al., 2005). In addition to changes in the forebrain, alterations in hindbrain structures have also been associated with autism; specifically, imaging and postmortem studies have revealed changes in cerebellar structure, and disrupted cerebellar gene expression has also been associated with ASD (Bauman, 1996; Fatemi et al., 2002a; Fatemi et al., 2008; Ritvo et al., 1986). Interestingly, in vitro studies have shown that the Met ligand Hgf protects cultured rat cerebellar neurons from cell death (Zhang et al., 2000), while in vivo analysis of a mouse conditional Met hypomorph has revealed a role in postnatal cerebellar development (Ieraci et al., 2002). Here we have used the zebrafish model to investigate how Met signaling functions during embryonic development of the cerebellum.
The cerebellum develops from the dorsal aspect of rhombomere (r)1 of the hindbrain, through a series of inductive and patterning interactions that appear to be conserved between mammals and zebrafish (Chizhikov et al., 2006; Foucher et al., 2006; McFarland et al., 2008; Millen and Gleeson, 2008; Sillitoe and Joyner, 2007; Wurst and Bally-Cuif, 2001). In mammals the cerebellum forms a laminated structure composed of an outer molecular layer of interneurons and glia, an adjacent monolayer of Purkinje cells, a large internal granular layer, and a core containing the deep cerebellar nuclei (Goldowitz and Hamre, 1998). The basic structure and the cell types of the cerebellum are largely conserved from mammals to zebrafish, although there is evidence that deep cerebellar nuclei cells are functionally replaced by eurydendroid cells in teleosts, including the zebrafish (Bae et al., 2009; McFarland et al., 2008; Castro et al., 2006; Ito 2006; Ikenaga et al., 2006; Ikenaga et al., 2005; Ito 2002). In both mammals and zebrafish the cerebellar cell types initially arise in multiple temporal waves from two distinct germinative neuroepithelia: the ventricular zone (VZ) and the upper rhombic lip (URL). In mouse, the VZ cells express the transcription factor Ptf1a and line the dorsal aspect of the r1 4th ventricle (Hoshino et al., 2005); they give rise to GABAergic cells, including deep cerebellar nuclei cells, Purkinje cells, interneurons and glia (Sillitoe and Joyner, 2007). Postmitotic Purkinje cells migrate along VZ radial glial fibers to settle within the central monolayer of the cerebellum (Wang and Zoghbi, 2001). In zebrafish, VZ progenitor cells also appear to express ptf1a (Bae et al., 2009; Volkmann et al., 2008) and, although there is no direct evidence, Purkinje cell precursors likely also originate from the VZ (Bae et al., 2009; Mione et al., 2008).
The URL cells express Math1 in mouse, and lie in r1 at the interface between the dorsal VZ and the roof plate (Wingate, 2001). The mouse URL gives rise to various non-cerebellar neurons as well as cerebellar glutamatergic deep cerebellar nuclei cells, unipolar brush cells, and the granule cell progenitors (Englund et al., 2006; Fink et al., 2006; Machold and Fishell, 2005; Wang et al., 2005). Granule cell progenitors migrate tangentially over the surface of the cerebellar primordium to form the mitotically active external granule layer (Wingate and Hatten, 1999), they then exit the cell cycle and migrate radially inward to form the internal granule layer. This proliferation and inward migration of granule cells is in part regulated by signals from Purkinje neurons (Sillitoe and Joyner, 2007). The zebrafish URL is spatially patterned in a similar fashion to mammals (Sgaier et al., 2007; Sgaier et al., 2005), expresses the Math1 ortholog atoh1a (Koster and Fraser, 2001), and produces similar cell types to mammalian URL (Koster and Fraser, 2006; Rieger et al., 2008; Volkmann et al., 2008). Recently, using a stable zebrafish transgenic line that expresses gata1-GFP in cerebellar granule cell precursors, Volkmann et al. (2008) demonstrated that granule cell precursors begin migrating from the URL at 48 hours post fertilization (hpf) and continue to migrate over several days. In summary, the development of the zebrafish cerebellum shares many features with that of mammals, and zebrafish therefore provides a viable model to investigate the molecular basis of cerebellar development.
In this study we have focused on the role of the Met/HGF signaling pathway in zebrafish cerebellar development to begin to uncover the developmental basis of autism. Hgf (Hepatocyte growth factor) is a pleiotropic growth factor that signals through its membrane bound tyrosine kinase receptor Met (proto-oncogene associated with cellular metastatic cancer; (Bottaro et al., 1991; Rubin et al., 1993)). Analyses of mouse Met and Hgf mutants and in vitro studies, have identified multiple roles for Met signaling during neural development (Birchmeier et al., 2003). For example, Hgf can act as a chemoattractant, a survival factor, or as a mitogen, for discrete neuronal subtypes (Cacci et al., 2003; Ebens et al., 1996; Garzotto et al., 2008; Giacobini et al., 2007; Ieraci et al., 2002; Maina et al., 1998; Maina et al., 1997; Maina and Klein, 1999; Powell et al., 2001). Limited functional analysis of zebrafish Met has revealed additional roles in motorneuron neurotransmitter selection (Tallafuss and Eisen, 2008), in muscle cell progenitor migration, in neuromast deposition (Haines et al., 2004), and in control of liver size (Latimer and Jessen, 2008).
Here we show that zebrafish met is an early marker of the cerebellar primordium, and is later expressed specifically in the VZ, while its hgf1 and hgf2 ligand genes are expressed ventral to the primordium. Using morpholino knockdown we show that Met signaling is required for normal specification of VZ progenitor cells. Disruption of Met signaling also causes defective granule cell generation, possibly as an indirect consequence of VZ defects. Importantly, we demonstrate that proliferation within the developing cerebellum is significantly reduced in Met and Hgf morphants, which in turn may influence the generation of various cerebellar cell types. As alterations in neuronal migration have been implicated in autism, and Met has been shown to regulate various cell migratory pathways, we investigated the role of Met in migration of a well-known hindbrain neuronal population. We find that met is expressed specifically in facial branchiomotor neurons (FMNs) and is required for their normal tangential migration within the hindbrain. In summary, we show that Met signaling is critical for cerebellar morphogenesis, including normal growth and cell type specification, and plays an important role in hindbrain cell migration. These findings validate further consideration of zebrafish as a model for studying human neurodevelopmental disorders.
MATERIALS and METHODS
Zebrafish embryo maintenance
Wild-type fish strain *AB, HuC-GFP transgenic line (Park et al., 2000), Ptf1a-GFP line (Godinho et al., 2005), and the Islet1-GFP transgenic line (Higashijima et al., 2000) were used in this study. Embryos were raised at 28.5°C and stage-matched based on morphological criteria (Kimmel et al., 1995). To inhibit pigmentation embryos were transferred to a solution of embryo medium containing 0.02 mM phenlythiourea (PTU, Sigma) between 18 and 24hpf.
Hgf2 ligand cloning
A novel zebrafish hgf2 was identified by analysis of genomic sequence of Danio rerio obtained from the Ensembl database: http://www.ensembl.org/Danio_rerio (Ensembl Gene ID: ENSDARG00000063316 found on chromosome 18). A 2003-bp PCR fragment was amplified from 48hpf embryos and cloned into pGEM-T (Invitrogen, San Diego): hgf2-forward: GACATGCTCTGCAAGACTACCA; hgf2-reverse: GTAGAAGGCCACGTTCACAAA.
Antisense morpholino oligonucleotide microinjections
Embryos were microinjected with 1.5-3nl of antisense morpholino oligonucleotides (MO: GeneTools, OR) as described (Nasevicius and Ekker, 2000). MOs targeted to the met untranslated region (Met-UTR), and to the met translation start site (Met-ATG), were as described (Haines et al., 2004). We also designed a MO to block met mRNA splicing at position exon8 - intron8 (Met-E8I8: 5’-GGATTTGTAAACATGCAAACCTCTG-3’), leading to a truncated Met protein. We used MOs targeted to the hgf1 translation start site (Hgf1-ATG: 5’-CATCCACATAATGTCCGATCTCATG-3’) and to block hgf1 mRNA splicing at position exon6 - intron6 (Hgf1-E6I6: 5’-AGATGAGGTGATGTCTTACCATCTG-3’), leading to a truncated Hgf1 protein. We also used MO to block hgf2 mRNA splicing at position exon4 -intron4 (Hgf2-E4I4: 5’-CTGGTAAACAATAAACTCACCCTCC-3’), leading to a truncated Hgf2 protein.
Supplemental Table S1 summarizes the concentration of each MO used (individual and together) to elicit a dose response that leads to facial motor neuron (FMN) migration phenotypes. This initial analysis was used to establish appropriate injection parameters for further phenotypic analysis (see Supplemental Table S2). For the migration phenotypic analysis we used the concentration of each individual Met, Hgf1 and Hgf2 MO that gave the highest percentage of embryos with FMN migration defects. For the cerebellum phenotypic analysis, Met-UTR and Met-E8I8 MO were injected together at concentration at which each MO alone had no effect (2.5mg/ml each; referred to throughout this study as Met MO-injected embryos or Met morphants). In addition, for the cerebellum study, individual Hgf1 or Hgf2 MO injections did not cause a phenotype, while co-injection of Hgf1-E6I6 (5mg/ml) and Hgf2-E4I4 (5mg/ml) MO resulted in cerebellar phenotypes similar to those obtained with the Met MO (referred to throughout as Hgf1+Hgf2 MO, double Hgf MO, or HGF morphants). At these concentrations, a range of cerebellar phenotypic consequences were observed, and we have indicated the number of embryos showing a severe phenotype where appropriate in the Results sections. The most severe phenotypic defects may have resulted in a slight delay in development, although we attempted to stage match control and morphant embryos based on morphological criteria.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed as described (Prince et al., 1998). Whole mount two-color fluorescent in situ hybridization used FITC- and cy3-plus tyramide reagents (PerkinElmer) following incubation with anti-Dig-HRP (Roche) or anti-Fluorescein-HRP (PerkinElmer) antibodies. The met plasmid (a gift from Dr. Peter Currie) contains a partial met gene. The hgf1 plasmid (a gift from Dr. Dae-Gwon Ahn) contains the full-length hgf1 gene. hgf2 plasmid was digested with SacII and antisense riboprobe synthesized using Sp6 RNA polymerase (Promega). Other riboprobes were used as described: krox20 (Oxtoby and Jowett, 1993), ptf1a (Volkmann et al., 2008), rora2 (Katsuyama et al., 2007), coe2 (Bally-Cuif et al., 1998), fgf8 (Foucher et al., 2006), phox2a (Guo et al., 1999), olig2 (McFarland et al., 2008), shh (Krauss et al., 1993), atoh1a and atoh1b (Adolf et al., 2004), reelin (Costagli et al., 2002), pax6a (Volkmann et al., 2008). Embryos were mounted in glycerol and photographed using a Zeiss Axioskop microscope and Nikon D1 camera.
Acridine Orange staining and Immunohistochemistry
Acridine Orange (Sigma) staining for cell death was performed as described (Elsen et al., 2008). Whole-mount immunohistochemistry was performed as described (McClintock et al., 2001). Primary antibodies used were: anti-GFP (monoclonal, 1:500, Molecular Probes), anti-Zn5 (monoclonal, 1:10, Molecular Probes, Eugene), anti-phospho Histone3 (pH3, polyclonal, 1:500, Upstate Biotechnology), anti-EphA4 (polyclonal, 1:1000, kind gift from Dr. David Wilkinson). Secondary antibodies were conjugated to Alexa Fluor-488, -546 (Molecular Probes). Embryos were mounted in glycerol and confocal images acquired using a Zeiss LSM-510 confocal microscope, and further image processing done with ImageJ. Transverse sections were obtained by embedding stained embryos in Durcupan/Araldite and sectioning at 5μm intervals. In addition, sections of about 50μm were cut manually with micro-scissors or vibratome sectioned (embedded in 3% agarose).
Imaging, Mitotic Index and Cell Counting
Live HuC-GFP fish were mounted in 0.2% agarose in dorsal view, and horizontal Z-sections were obtained using a Zeiss LSM510 confocal microscope. Whole-mount embryos labeled with anti-pH3 antibody to detect proliferating cells were counterstained with the nuclear marker Sytox-green (50μM, Molecular Probes). Embryos were mounted in dorsal view and z-sections of the dorsal hindbrains collected at 3μm intervals. Total number of mitotic cells was calculated through the 60μm depth covering the cerebellum on both sides of the cerebellum (dorsal r1). Mitotic index (proliferating cells/total number of cells) was then calculated for each embryo. Each individual count of proliferating and total cells was made from three consecutive 3μm sections using ImageJ software as previously described (Elsen et al., 2008), from half of the cerebellum (r1). Between 22 and 32 specimens were assayed for each experimental condition. ANOVA tests for comparison of multiple groups were performed using GraphPad Prism 4.03 software; P<0.05 was considered significant.
RESULTS
Met receptor and Hgf ligands are expressed in the cerebellar primordia
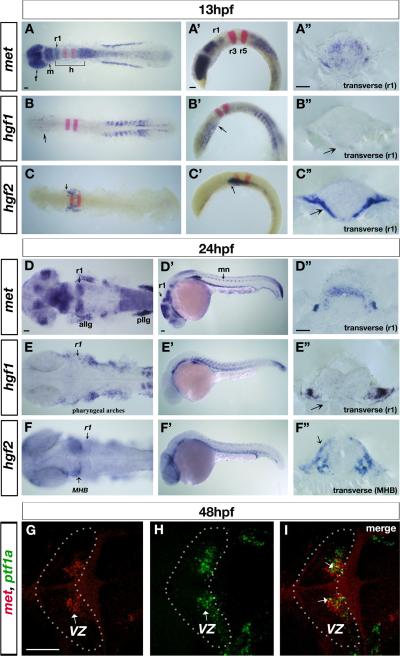
To investigate potential roles of Met signaling in the nervous system, we analyzed expression of met and its ligands, hgf1 and hgf2, from gastrulation through neurulation. We found that met is expressed during gastrulation in the anterior neuroectoderm (data not shown), while hgf1 and hgf2 expression is excluded from the neuroectoderm at these early stages (data not shown). By early neurulation, at 13hpf, met is expressed at high levels in the forebrain, dorsal midbrain, and in a stripe in medial r1 of the hindbrain (the dorsal part of which will became the future cerebellum), as well as at low levels throughout the rest of the hindbrain, and in a gradient from r7 decreasing along the spinal cord (Fig. 1A-A”). It is also expressed at high levels in anterior lateral line ganglia and posterior lateral line ganglia as previously described (Haines et al., 2004). By late neurulation, at 24hpf (Fig. 1D-D” and Suppl. Fig.1A,A’,A”,B), met expression becomes more restricted to specific areas of the forebrain, dorsal midbrain, medial part of r1, and spinal cord motor neurons.
Figure 1. Zebrafish met receptor and hgf1 and hgf2 ligand expression patterns in the nervous system.
(A-F) in situ hybridization revealing expression of met receptor (A-A”; D-D”), hgf1 ligand (B-B”; E-E”) and hgf2 ligand (C-C”; F-F”) at 13hpf (A-C”) and 24hpf (D-F”) in dorsal (A-F), lateral (A’-F’), and transverse views (A”-F”). krox-20 expression (red in A-C’) marks rhombomere (r) 3 and r5. At 13hpf, met (A-A”) is expressed at high levels in the forebrain, dorsal midbrain, a stripe in r1, and in a gradient starting in r7 that tapers off towards the posterior part of the spinal cord. met is also present at low levels throughout the hindbrain. At 24hpf, met (D-D”) continues to be expressed in the forebrain regions, eyes, dorsal midbrain, dorsal r1, and in a gradient from r7 towards the posterior, as well as in the anterior and posterior lateral line ganglia, and motor neurons of the spinal cord. At 13hpf, hgf1 (B-B”) is expressed in the ventral regions of the forebrain (arrow in B), and at low levels throughout the hindbrain, in ventral regions surrounding r1 (arrow in B’ and B”), while hgf2 (C-C”) is expressed in low levels in the hindbrain and in ventral regions surrounding the hindbrain (arrow in C, C’ and C”), including r1. At 24hpf, hgf1 (E-E”) is expressed at low levels throughout the hindbrain, in ventral tissues surrounding r1, and in pharyngeal arches, while hgf2 (F-F”) is expressed in the dorsal eye, ventral tissues surrounding the hindbrain, and in tissues surrounding the MHB (arrow in F,F”). (G-I) Confocal images of double fluorescent in situ hybridization with met (G) and ptf1a (H) in dorsal view, anterior to the left, at 48hpf, showing colocalization of the two transcripts in a small number of cells (arrows in I, merged) in the VZ (arrows in G and H) of the cerebellum (outlined in G-I). r5, rhombomere5; r1, rhombomere1; r7, rhombomere7; allg, anterior lateral line ganglia; pllg, posterior lateral line ganglia; mn, motor neurons; MHB, midbrain-hindbrain boundary; VZ, ventricular zone. Scale bar, 50μm.
At 13hpf, hgf1 ligand (Fig. 1B-B”) is expressed in ventral regions of the forebrain (arrow in Fig. 1B), at very low levels throughout the hindbrain, and ventral regions underneath the r1 neuroepithelium (arrows in Fig. 1B’ and B”). Similarly, hgf2 ligand (Fig. 1C-C”) expression is present at very low levels in the hindbrain and at high levels in ventral regions surrounding the hindbrain (arrow in Fig. 1C-C”). At 24hpf, hgf1 (Fig. 1E-E”) continues to be expressed at low levels throughout the hindbrain and in ventral tissues surrounding r1 (arrow in Fig. 1E”), and at high levels in pharyngeal arches, while hgf2 (Fig. 1F-F”) is expressed in the dorsal edge of the eye, at low levels in ventral tissues surrounding the hindbrain, in high levels in pharyngeal arches, and additionally in tissues surrounding the neuroepithelium at the level of the MHB (midbrain-hindbrain boundary) (arrows in Fig. 1F,F”).
At 48hpf, met expression in r1 becomes localized to the ventricular zone (VZ) of the cerebellar primordia (Fig. 1G,H,I). Double fluorescent in situ hybridization with met (Fig. 1G) and ptf1a (Fig. 1H), which we are using as a marker for GABAergic Purkinje cell progenitors, shows some cellular colocalization (arrows in Fig. 1I), suggesting that met is expressed in the VZ including within a subpopulation of ptf1a-positive progenitor cells. Double fluorescent in situ hybridization with met and atoh1a, which we are using as a marker of glutamatergic granule cell progenitors, shows no colocalization within the URL at any of the developmental stages analyzed (data not shown). in situ hybridization with atoh1a (Suppl. Fig. 1B,B’) and atoh1b (Suppl. Fig. 1C) shows dorsal expression of these genes in the URL, while met expression is excluded from this region. We were unable to detect expression of met or hgf in the cerebellar primordium after 48hpf. The early expression patterns of zebrafish met within r1 and hgf1 and hgf2 ligands within tissue surrounding r1, suggest that Met signaling may function during early cerebellar development.
Met morphants show altered cerebellar morphogenesis and reduced neuronal differentiation
A previous functional study in mouse (Ieraci et al., 2002) has shown that Met signaling is required for the proliferation of granule cell precursors, and this proliferation defect was associated with a reduced size of the cerebellum and foliation defects. We have investigated the function of Met signaling in zebrafish cerebellum development by knocking down both Met receptor and Hgf (Hgf1 and Hgf2) ligand activity independently using antisense morpholino (MO) oligonucleotides. The morphant embryos (MO-injected) were viable at 72hpf, but died shortly thereafter, probably due to disruption of other developmental processes. Using HuC-GFP transgenic embryos to mark newly differentiated neurons (Park et al., 2000), we found that Met morphants showed a reduction in the number of newly differentiated cerebellar neurons at 48hpf (17/22), both in more dorsal positions (Fig. 2E) and mid-ventral positions (Fig. 2H), in comparison to control embryos (Fig. 2D, G, n=18). Hgf double morphants showed a similar phenotype (Fig. 2F,I). Similar reductions in differentiated neurons were observed at 72hpf (data not shown). Knockdown of Hgf1 or Hgf2 independently did not affect the generation of HuC-GFP-positive neurons, suggesting a redundant role in cerebellar development. Further phenotypic analysis was therefore performed on double Hgf1+ Hgf2 knockdown specimens (termed Hgf MO or Hgf morphants hereafter).
Figure 2. Abnormal cerebellar morphology and reduction of differentiated neurons in Met signaling morphants.
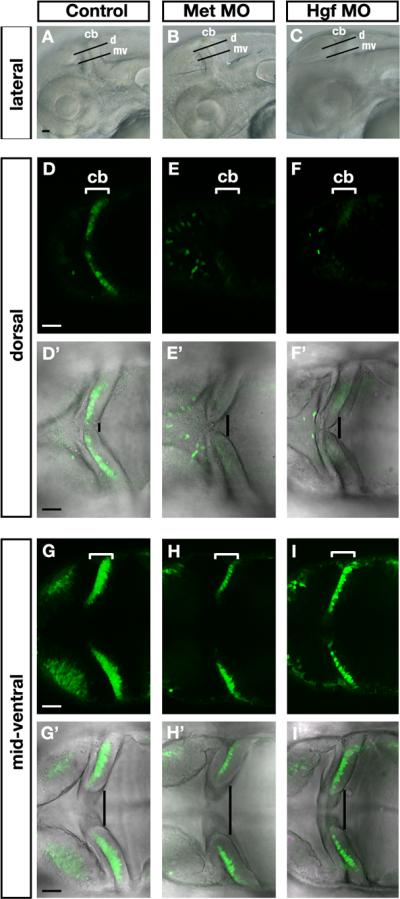
(A-C) Lateral bright-field view of 48hpf embryos illustrating the orientation of horizontal confocal sections shown in panels D-I’ (d, dorsal) and (mv, mid-ventral) in (A) control, (B) Met morphants, and (C) Hgf double morphants (Hgf1+2 MO). Anterior is to the left in all panels and dorsal up in (A-C). (D-I’) Horizontal confocal sections, dorsal view, through the midbrain and hindbrain in live 48hpf HuC-GFP-positive embryos imaged in dorsal positions (D-F’) and mid-ventral positions (G-I’) in controls (D,D’,G,G’), Met morphants (E,E’,H,H’), and Hgf1+2 double morphants (F,F’,I,I’). Bright-field and fluorescence images were merged to show the presence of Hu-positive cells away from the proliferative zone (D’,E’,F’,G’,H’,I’). The number of HuC-positive newly differentiated neurons is reduced in Met morphants (E,H) and Hgf double morphants (F,I) compared to controls (D,G). Cerebellar regions are indicated by white brackets (D-I), and dorsal cerebellar midline fusion is highlighted by a black line indicating the degree of morphological change (D’-I’) in Met morphants (E’,H’) and Hgf double morphants (F’,I’) compared to controls (D’,G’). cb, cerebellum. Scale bar, 50μm.
Morphological analysis of the cerebellar primordia in the horizontal plane at dorsal and mid-ventral positions (illustrated in Fig. 2A-C, lateral view) indicated that dorsal cerebellar folds are not completely fused in Met morphants (Fig. 2E’,H’) and to a lesser extent in Hgf morphants (Fig. 2F’,I’) at 48hpf and at 72hpf (data not shown) compared to controls (Fig. 2D’,G’). Taken together our results are consistent with the hypothesis that Hgf ligands signal through the Met receptor to control cerebellar morphogenesis and the generation of HuC-GFP-positive newly differentiated neurons derived from the URL, the VZ, or both.
Met signaling is required for specification of a subset of VZ-derived neurons
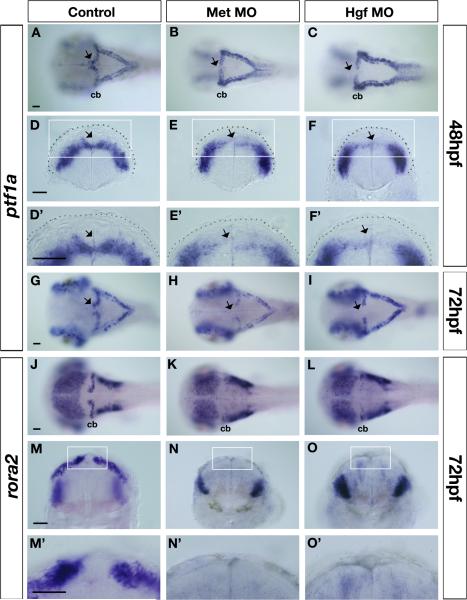
To establish the specific population(s) of cerebellar neurons affected by Met signaling disruption we used additional marker analysis. As expression of met and ptf1a show close juxtaposition and some overlap in the VZ, we investigated whether ptf1a expression was altered in Met signaling morphants. Recent studies have revealed that the mouse Ptf1a transcription factor is required for specification and survival of progenitors of all cerebellar GABAergic neurons, including Purkinje cells, in the VZ (Hoshino et al., 2005; Pascual et al., 2007). The function of zebrafish Ptf1a has not been investigated, but its expression pattern suggests a conserved role in cerebellar GABAergic neuron development. ptf1a expression in the zebrafish cerebellum starts at 24hpf in a restricted domain in the posterior lateral regions of r1, and its expression is initiated correctly in the cerebellar primordia of both Met and Hgf morphants (data not shown). By contrast, at 48hpf, when ptf1a expression extends throughout the entire VZ of r1, including the most anterior region in the dorsal midline (arrows in Fig. 3A,D), anterior ptf1a expression is specifically lost in Met morphants (23/29, arrows in Fig. 3B,E,E’) or Hgf morphants (21/28, arrows in Fig. 3C,F,F’). This loss is maintained at 72hpf, when neurons within the VZ begin to differentiate, both in Met morphants (19/21, arrow in Fig. 3H) and Hgf morphants (24/32, arrow in Fig. 3I), compared to control embryos (n=29, arrow in Fig. 3G). The expression of ptf1a within the remainder of the cerebellum at 72hpf is reduced in Met and Hgf morphants. By contrast, there is a slight increase in ptf1a expression within the remainder of the hindbrain, suggesting that Met signaling plays different roles in regulating the development of ptf1a-positive cells at different anterior-posterior levels. In summary, given the expression of met within a subpopulation of ptf1a-positive progenitor cells within the VZ, our results suggest that Met signaling is necessary for development of a subpopulation of ptf1a-positive progenitors, particularly within the medial/anterior region of the VZ.
Figure 3. Altered expression of ventricular zone derivative markers in Met signaling morphants.
Dorsal (A-C, G-L) and transverse views (D-F’,M-O’) taken at the level of the cerebellum of control (A,D,D’G,J,M,M’), Met morphants (B,E,E’H,K,N,N’), and Hgf morphants (C,F,F’,I,L,O,O’) at 48hpf (A-F’) and 72hpf (G-O’) showing expression of ventricular zone (VZ) progenitor marker ptf1a (A-I), and of Purkinje cell marker rora2 (J-O’) in the cerebellum. At 48hpf, ptf1a expression in the dorsal midline region of the cerebellum is missing in Met morphants (arrows in B,E,E’) and in Hgf morphants (arrows in C,F,F’) compared to controls (arrows in A,D,D’). Similarly, at 72hpf, ptf1a expression in the dorsal midline region of the cerebellum continues to be absent in Met morphants (arrow in H) and Hgf morphants (arrow in I) compared to controls (arrow in G). In addition, at 72hpf, expression levels of ptf1a in the rest of the cerebellum is down-regulated in Met and Hgf morphants (H,I) compared to controls (G). Neuroepithelium is outlined with a broken line in D-F’. The boxed region in D-F is shown at higher magnification in D’-F’. At 72hpf, the expression of Purkinje cell marker rora2 in the entire cerebellum, including the dorsal midline region of Met morphants (K,N,N’) and Hgf morphants (L,O,O’) is significantly reduced compared to the expression in the cerebellum of control embryos (J,M,M’). The boxed region in M-O is shown at higher magnification in M’-O’. morphants, MO (morpholino) injected embryos; cb, cerebellum. Scale bar, 50μm.
We also analyzed expression of markers of differentiated VZ-derived neurons (Katsuyama et al., 2007). We could not use the classic Purkinje cell marker ZebrinII/AldolaseC (Brochu et al., 1990; Lannoo et al., 1991a; Lannoo et al., 1991b; Meek et al., 1992) because Met signaling morphants do not survive beyond 72hpf, and the onset of expression of this marker in cerebellum is at approximately 96hpf. However, we were able to use two earlier markers of VZ progenitors, rora2 and coe2. In mouse, the RORα gene (coding for retinoic acid-related orphan nuclear receptor alpha) is expressed in postmitotic Purkinje cells. Analyses of the spontaneous mouse mutant staggerer, and of mice with a mutation of the RORα gene, show cerebellar atrophy due to death of both Purkinje and granule cells (Doulazmi et al., 2001; Dussault et al., 1998; Gold et al., 2003; Gold et al., 2007; Hamilton et al., 1996; Steinmayr et al., 1998; Vogel et al., 2000). In zebrafish, rora2 is upregulated in a stripe in the cerebellum at 72hpf, and is strongly expressed in adult Purkinje cells (Katsuyama et al., 2007), suggesting a conserved role in Purkinje cell development. Our in situ analysis of rora2 expression in Ptf1a-GFP transgenic fish confirms early expression in the VZ (Suppl. Fig. 2). We found that rora2 expression was severely reduced in the cerebellum of 72hpf Met morphants (29/37, Fig. K,N,N’) and Hgf morphants (23/33, Fig. 3L,O,O’) compared to control embryos (n=27, Fig. 3J,M,M’). Transverse sections confirm that cerebellar rora2 expression was dramatically down-regulated in Met (Fig. 3N’) and Hgf (Fig. 3O’) morphants, including in the dorsal midline region, although tissue integrity is maintained in this location. This reduction of rora2 expression reveals a loss of VZ-derived progenitor cells in embryos deficient for Met signaling.
Early B-cell factor 2 (EBF2) is one of four mammalian members of an atypical helix-loop-helix transcription factor family (COE) that is specifically expressed in the Purkinje cells. The number of Purkinje cells in Ebf2 null mice is markedly decreased, resulting in a small cerebellum (Croci et al., 2006). We find that expression of zebrafish coe2 (the zebrafish ortholog of Ebf2) in presumed Purkinje cells (Bally-Cuif et al., 1998) is also reduced in Met and Hgf morphants (Suppl. Fig. 3A-C and data not shown). By contrast, the olig2 gene marks a distinct subset of VZ neurons, thought to correspond to eurydendroid cells (Bae et al., 2009; McFarland et al., 2008), the principal efferent neurons of the teleost cerebellum. At 48hpf and 72hpf, cerebellar expression of olig2 is unaltered in Met or Hgf morphants (Suppl. Fig. 3D-F and data not shown). This finding suggests that Met signaling selectively regulates the generation of specific subtypes of VZ-derived cells and, assuming conservation of marker gene expression between Purkinje cells of mouse and zebrafish, is specifically required for Purkinje cell development.
Met signaling influences the development of cerebellar granule cells
We next wanted to determine whether Met signaling plays any role in differentiation or localization of granule cells. Cerebellar granule cell precursors arise from a restricted pool of primary progenitors in the upper rhombic lip (URL). Our expression analysis indicates that met is specifically expressed in the VZ but is excluded from the URL (Fig. 1). However, several studies in mice have shown that Purkinje cells are necessary for granule cell proliferation (Caddy and Biscoe, 1979; Herrup, 1983; Sidman et al., 1962); mouse Purkinje cells express the mitogen Shh, and genetic studies have shown that this signaling pathway is critical for granule cell proliferation and for cerebellar morphogenesis (Corrales et al., 2006; Corrales et al., 2004; Lewis et al., 2004).
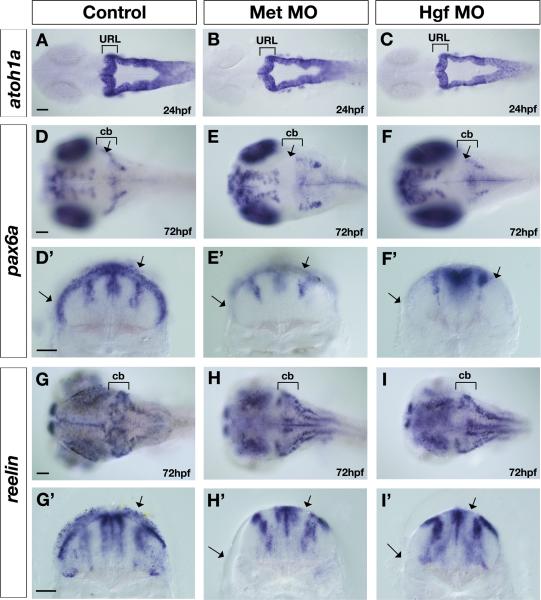
We first analyzed the expression of zebrafish atoh1a the ortholog of the mouse Math1 gene, which encodes a basic helix-loop-helix transcription factor required for specification and differentiation of granule cells (Ben-Arie et al., 1997; Machold and Fishell, 2005; Wang et al., 2005). The expression and distribution of atoh1a in the URL of Met morphants and Hgf morphants is unchanged compared to control embryos (Fig. 4A) at 24hpf (Fig. 4A-C) and 48hpf (Suppl. Fig. 4A-B). In addition, the expression and distribution of atoh1b in the URL of Met morphants is similarly unaltered compared to controls embryos at 24hpf and 48hpf (Suppl. Fig.4 C-F”). There results suggest that Met signaling is not required for the specification of URL derivatives. Further, we assayed expression of phox2a, which encodes a paired homeodomain protein required for differentiation of locus coeruleus noradrenergic neurons thought to derive from the dorsal anterior hindbrain (Guo et al., 1999; Lin et al., 2001). We found that expression of phox2a is unaffected in Met and Hgf morphants compared to controls (Suppl. Fig.5 A-F). The migration of phox2a-positive neurons from the dorsal anterior hindbrain to ventral r1 was slightly delayed at 24hpf in Met and Hgf morphants, but by 48hpf these neurons completed their migration. Thus, the URL forms properly and is able to establish URL progenitor cell types within the developing cerebellum of Met signaling deficient embryos.
Figure 4. Altered expression of upper rhombic lip derivative markers in Met signaling morphants.
Dorsal (A-F, J-L) and transverse views taken at the level of the cerebellum (D’-F’, G’-I’) of control (A,D,D’,G,G’), Met morphants (B,E,E’,H,H’), and Hgf double morphants (C,F,F’,I,I’) at 24hpf (A-C) and 72hpf (D-I’) showing expression of atoh1a (A-C), pax6a (D-F’), and reelin (G-I’) in the cerebellum (cb), marked with brackets. The expression of atoh1a in the granule cell progenitors in the URL is not affected in Met morphants (B) and Hgf morphants (C) compared to controls (A). In contrast, the expression of markers of differentiated granule cells, pax6a (D-F’) and reelin (G-I’), is severely disrupted in Met morphants (E,E’ and H,H’ respectively) and Hgf morphants (F,F’ and I,I’ respectively) compared to controls (D,D’ and G,G’ respectively). Arrows in control embryos indicate the expression in cerebellar region (D), including expression in dorsal and ventrolateral domains in transverse sections (D’,G’). In contrast, arrows in the Met morphants and Hgf morphants point to the absence of granule cell marker expression in both dorsal regions and in ventrolateral regions in dorsal (E and F respectively) and transverse sections (E’,H’ and F’,I’ respectively). The regions of the midline at the level of the 4th ventricle and the two lateral stripes of radial glia are pax6a-positive (D’-F’) and reelin-positive (G’-I’) at 72hpf and are not affected in Met signaling morphants (E’,F’,H’,I’) compared to control embryos (D’,G’). URL, upper rhombic lip; cb, cerebellum. Scale bar, 50μm.
By contrast, at 72hpf, the expression of pax6a, which encodes a homeodomain transcription factor that marks differentiated granule cells derived from the URL (Volkmann et al., 2008), is markedly reduced in both Met morphants (25/33, Fig. 4E) and Hgf morphants (17/25, Fig. 4F) compared to control embryos (n=31, Fig. 4D). Transverse sections through the cerebellum of Met morphants (Fig. 4E’) and Hgf morphants (Fig. 4F’) show the absence of both dorsal and ventral (arrows) expression of pax6a in granule cell progenitors. Similarly, the expression of the extracellular matrix molecule reelin in differentiated granule cells of the cerebellum in zebrafish (Costagli et al., 2002; Foucher et al., 2006) is reduced in Met morphants (27/35, Fig. 4H) and Hgf morphants (17/26, Fig. 4I) compared to control embryos (n= 29, Fig. 4G) at 72hpf, both in dorsal locations and in the ventral regions (arrows in transverse sections in Fig. 4G’,H’,I’). Recent time-lapse studies in zebrafish have shown that granule cell progenitors migrate tangentially out of the URL at around 48hpf towards the midbrain hindbrain boundary (MHB) (Volkmann et al., 2008). A subpopulation of these neurons, originating from a lateral location of the URL, continues to migrate along the MHB to form the ventrolateral cluster that later gives rise to eminentia granularis in the adult zebrafish (Volkmann et al., 2008). Our observation of disrupted granule marker gene expression in both dorsal and ventral regions suggests that Met signaling is necessary for the development of granule cells within all cerebellar subdivisions. The reduction in granule cell populations in Met signaling morphants may reflect an indirect role for Met signaling in proliferation, differentiation or migration of these neurons. These defects are unlikely to be a consequence of altered patterning of the cerebellum in response to MHB signals, as fgf8 expression in the MHB is unaffected in Met signaling morphants at 24hpf and 48hpf (data not shown). Rather, we suggest that, as in mouse, proper development of granule cell progenitors requires signals from VZ cells; disruption of VZ derivatives in Met signaling morphants thus leads to a disruption in granule cell development.
Cerebellar proliferation is disrupted in Met signaling morphants
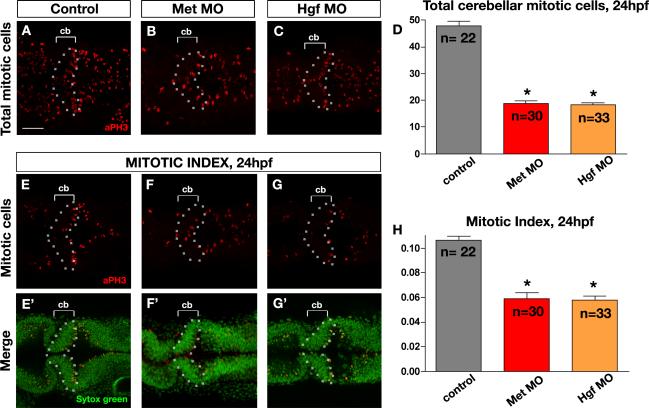
Analysis of Met mutant mice has shown that this signaling pathway regulates proliferation of granule cell progenitors in the postnatal cerebellum (Ieraci et al., 2002). Our observations of reduced numbers of HuC-positive postmitotic neurons (Fig. 2), as well as reduced numbers of VZ-derived neurons and granule cell progenitors, together with a general reduction in size of the cerebellar anlage, suggested that proliferation might similarly be deficient in Met signaling morphants. We therefore analyzed total cerebellar proliferation and rate of proliferation in the cerebellar region using pH3 (phosphohistone3) antibody, which marks cells in the M-phase of the cell cycle, and Sytox Green, which marks all nuclei. We found that at 24hpf, when cerebellar cells undergo extensive proliferation, the number of pH3-positive cells along the DV aspect of the cerebellar anlage was reduced by 60% in Met morphants (Fig. 5B,D) and by 62% in Hgf morphants (Fig. 5C,D) compared to controls (Fig. 5A,D). In addition, the rate of cerebellar proliferation (mitotic index) was reduced in Met morphants by 45% (Fig. 5F,F’,H) and in Hgf morphants by 54% (Fig. 5G,G’,H) compared to controls (Fig. 5E,E’,H). We have also noted a decrease in proliferation within the midbrain and other hindbrain regions posterior to the cerebellum, consistent with the expression of met receptor gene within these regions. We used Acridine Orange staining to assess whether cell death also contributed to the morphant phenotype, and found only a slight increase in cell death throughout the CNS of both Met and Hgf morphants (Suppl. Fig. 6A-C). We conclude that Met signaling is required for proper neural proliferation in the cerebellum, and may also play a minor general role in survival of CNS cells, as previously reported in mouse (Maina et al., 1997; Xiao et al., 2001).
Figure 5. Met signaling is required for cerebellar proliferation.
(A-C) Dorsal longitudinal confocal images (z-projections at maximum intensity) and (E-G’) single dorsal longitudinal confocal sections of posterior midbrain and anterior hindbrain of 24hpf embryos of control (A,E,E’), Met morphants (B,F,F’) and Hgf double morphants (C,G,G’) illustrating total proliferation in the cerebellum (A-C), cerebellar proliferation within a confocal section marked by anti-PH3 antibody in red (E-G), and cerebellar nuclei within a confocal section counterstained with Sytox Green (E’-G’). Compared to controls (A), Met and Hgf double morphants (B and C) showed reduced total mitotic cells in cerebellar (cb) regions marked by brackets and outlined by the broken line. The graph (D) represents the quantitative analysis of total cerebellar proliferative cells, showing a highly significant decrease in global proliferation in the morphants compared to controls. The graph (H) represents the quantitative analysis of the mitotic index (mitotic cells/total cells) in the cerebellum showing a significant decrease in the mitotic index of morphants compared to controls. Bars represent the combined results of two independent experiments. n, the number of embryos assessed per experimental condition; asterisks in panels D and H indicate significant statistical difference compared to controls. Scale bar, 50μm.
Met signaling is required for normal facial motor neuron (FMN) migration
As Met signaling in zebrafish has previously been shown to play a role in migration of both mesoderm cells and lateral line neurons (Haines et al., 2004), we wondered whether Met signaling might similarly be required for cell migration in the hindbrain. Unfortunately, the cell migratory pathways in the zebrafish cerebellum are not well characterized. Additionally, the severity of the patterning defects in the cerebellum of Met signaling morphants precludes analysis of migratory phenotypes in this region, although we cannot exclude the possibility that Met signaling may play a role in cerebellar cell migration. However, as the expression of met in the hindbrain is not limited to the cerebellum we asked whether Met signaling might play a role in migration of other migratory hindbrain cell types.
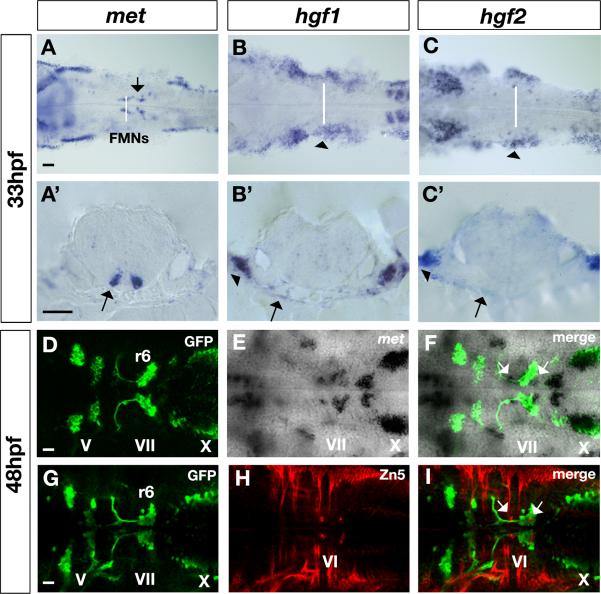
Interestingly, we found that met is expressed in the facial motor neurons (FMNs) of the VIIth nerve (arrows in Fig. 6A,A’) during stages when these neurons are migrating within the hindbrain (Chandrasekhar, 2004). In addition, hgf1 (Fig. 6B,B’) and hgf2 (Fig. 6C,C’) expression is present at low levels throughout the hindbrain, as well as in ventral tissues underlying the migrating met-expressing FMNs (arrows in Fig. 6B’ and C’, respectively). Note that hgf1 and hgf2 transcripts are also highly upregulated in the ventrolateral pharyngeal arches (arrowheads in Fig. 6B,B’ and Fig. 6C,C’, respectively). The stereotypical migration of the FMNs can be visualized using a transgenic line in which GFP is under control of islet1 regulatory elements (Islet1-GFP fish; (Higashijima et al., 2000)). FMNs are born in hindbrain rhombomere (r) 4 at around 18hpf, begin to migrate shortly thereafter, and settle in r6 by 48hpf (Fig. 6D,G). The met gene (Fig. 6E, F) is expressed at high levels within the FMNs during this migratory phase, starting at around 25hpf and continuing through 48hpf; met is also expressed in vagal motor neurons of the Xth nerve. Co-labeling with Zn5 antibody (DM-GRASP immunostaining), (Fig. 6H) revealed that met is also expressed in the stationary r5 and r6 abducens motor neurons (arrows in Fig. 6I). In summary, zebrafish met is expressed in a migratory population of cells within the hindbrain (FMNs), while hgf1 and hgf2 ligands are present in tissues surrounding these cells during their migratory phase, suggestion that their interaction regulates the migration behavior of these neurons.
Figure 6. Expression of met receptor and hgf1 and hgf2 ligand in the facial motor neurons (FMNs).
(A-C) Dorsal view and (A’-C’) transverse views at the level of r5 in the hindbrain of 33hpf embryos showing mRNA expression of met receptor (A,A’), hgf1 ligand (B,B’) and hgf2 ligand (C,C’). met receptor is expressed in the migratory FMNs (arrows in A and A’) in the ventral hindbrain, while hgf1 and hgf2 ligands are expressed in ventral tissues surrounding the hindbrain in regions where FMNs migrate towards the posterior hindbrain (arrows in B’ and C’ respectively). In addition, hgf1 and hgf2 ligands are expressed in pharyngeal arches, ventrolateral to the hindbrain (arrows in B,B’ and C,C’ respectively). White lines in A-C indicate the position at which transverse sections were obtained (A’-C’). (D-F) Dorsal confocal sections of 48hpf Islet1-GFP embryos in the hindbrain regions showing colocalization (in F) of met mRNA (E) with GFP antibody in green (D) in the VII nerve and in the X nerve. White arrows in F indicate the presence of met transcripts in other population of hindbrain cells. (G-I) Dorsal confocal sections of 48hpf Isl1-GFP embryos in the hindbrain regions double stained with GFP antibody in green (G) and Zn5 antibody, which recognizes zebrafish DM-GRASP, a cell adhesion molecule, in red (H) in the abducens motor neurons (VI nerve). Colocalization of GFP and Zn5 antibodies (in I) identifies the population of met-positive neurons as the abducens motor neurons; neurons also indicated by white arrows in F and I. FMNs, facial motor neurons; V, trigeminal motor neurons of the V nerve; VII, facial motor neurons of the VII nerve; X, vagal motor neurons of the X nerve; VI, abducens motor neurons of the VI nerve. Scale bar, 50μm.
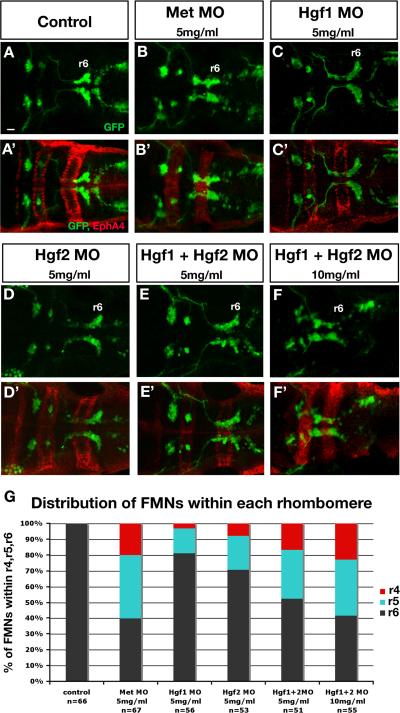
Unlike the situation in the cerebellum, where Met signaling regulates neuronal specification, disruption of Met signaling did not affect the specification of FMNs at 16hpf (data not shown) in accord with a later expression onset in these cells. Similarly, the number of neurons at 48hpf was not affected (Fig. 7). We first assayed the migration defects resulting from knockdown of Met and Hgf at various concentrations (Supp. Table 1). Next, we performed a more detailed analysis of the migration phenotypes by determining the number of FMN cell bodies present within each rhombomere (r4, r5, and r6). Supp. Table 2 and Fig. 7G summarize FMN migration phenotypes in response to Met signaling knockdown using MO at different concentrations. Fig. 7A-F’ shows double labeling of 48hpf Isl1-GFP embryos with GFP antibody to visualize the distribution of FMNs and EphA4 antibody to mark r3 and r5. In control embryos, all FMNs complete their migration to r6 (Fig. 7A,A’, Supp. Table 2, Fig. 7G). By contrast, we found that Met morphants or double Hgf morphants showed a significant block to FMN migration, with approximately 60% of neurons failing to reach r6 (Fig 7B,B’ and E-F’, Supp. Table 2, Fig. 7G). Individual knockdown of Hgf1 and Hgf2 showed a mild migration phenotype indicating a redundant function in FMN migration (Fig. 7C,C’ and D,D’, Supp. Table 2 and Fig. 7G). We conclude that similar to its roles in zebrafish lateral line and mesoderm, Met signaling is also required for proper posterior migration of hindbrain derived FMNs.
Figure 7. Met signaling is required for FMN migration.
The disposition of FMNs is visualized using 48hpf Islet1-GFP (Isl1-GFP) transgenic embryos (A-F) double labeled with GFP antibody in green (A-F) to detect branchiomotor neurons and EphA4 antibody in red (A’-F’) as a landmark of r3 and r5 to identify the position of FMNs at 48hpf. (A-F) Dorsal confocal fluorescence images of the hindbrain in dorsal view (A–F); anterior is to the left. r6 (the final destination of all FMNs at 48hpf) is labeled in all panels (A-F). In control embryos (A,A’), all FMNs have completed their migration and settled in r6. In contrast, in Met morphants (B,B’, Met MO, 5mg/ml), the majority of FMNs did not complete their migration to r6 and remain in r4 and r5. In Hgf1 morphants (C,C’, Hgf1 MO, 5mg/ml), only a few FMNs did not complete their migration, while the majority reached r6. Similarly, in Hgf2 morphants (D,D’, Hgf2 MO, 5mg/ml), some neurons completed their migration to r6, while others failed to exit r4 and r5. Simultaneous microinjection of Hgf1 and Hgf2 MO (E,E’, each at 2.5mg/ml) led to a higher percentage of FMNs that failed to complete their migration to r6 than when each Hgf MO was microinjected alone at 5mg/ml (B,B’ and C,C’). In addition, microinjection of Hgf1 and Hgf2 MO (F,F’, each at 5mg/ml) led to a more drastic migration phenotype with the majority of the FMNs failing to exit r4 and r5, a phenotype that closely resembles that observed in Met morphants (B,B’). The graph in G shows the percentage distribution of FMNs within r4, r5, and r6 for each of the conditions mentioned in A-F. Scale bar, 50μm.
DISCUSSION
Here we show that Met signaling is required for normal development of the zebrafish cerebellum. In Met signaling-deficient embryos, the size of the cerebellum is significantly reduced, reminiscent of the phenotype of Met mutant mice (Ieraci et al., 2002), and suggesting that this signaling pathway plays a role that is essentially conserved. Notably, the reduced size of the cerebellum in zebrafish embryos is associated with a reduction in the number of newly differentiated neurons and a significant decrease in early cerebellar proliferation. VZ neuronal progenitors marked by ptf1a are reduced in Met and Hgf morphants, and the majority of Purkinje cells deriving from this zone are subsequently lost. Interestingly, although atoh1a expression, which marks early URL precursors, is unaffected in the morphants, markers of URL-derived granule cell progenitors are disrupted, suggesting a secondary role of the Met signaling pathway in granule cell development. Finally, we show that Met signaling is also required for migration of FMNs from their origin in r4 to their final destination in r6.
Met signaling regulates zebrafish cerebellar morphogenesis
Previous studies in mouse and rat have shown that Met mRNA and protein are localized to the cerebellar external granule layer, which is made up of granule cell progenitors derived from the URL, while Hgf protein is present within the Purkinje cells derived from the VZ of the cerebellum (Honda et al., 1995; Ieraci et al., 2002). By contrast, in zebrafish we find that met expression initiates in presumptive forebrain during gastrulation, localizes to the cerebellar anlage during neurulation stages (11-24hpf), and becomes confined to the VZ before the onset of neuronal differentiation (48hpf). In addition, we detected expression of hgf at very low levels throughout r1 (including the cerebellum) and at higher levels in ventral regions surrounding r1. These differences in the patterns of expression of the Met receptor and Hgf ligand genes between mammals and zebrafish may reflect evolutionary differences in functions of these genes.
An obvious indication of the importance of Met signaling in zebrafish cerebellar development is the reduced size of the cerebellum in Met signaling morphants. During development of the vertebrate central nervous system, neurons are generated from progenitor cells that lie within the proliferative region of the neural tube (Lyons et al., 2003). In the cerebellum, both the VZ and the URL are sites of proliferation. We find a significant global reduction of cerebellar proliferation along the dorsoventral axis in our Met signaling morphants. However, we did not observe alterations in the numbers or distribution of atoh1a- and atoh1b-positive URL cells at 24hpf and 48hpf, suggesting that early cell fate determination of URL precursors occurs normally in Met signaling morphants, while the VZ is the primary compartment that is disrupted. Due to the lack of early molecular markers for the VZ we are unable to assess whether the VZ is the only structure in which proliferation rates are reduced. Nevertheless, consistent with disrupted VZ proliferation, we observe a reduction or loss of expression of the VZ marker ptf1a at 48hpf, especially in regions close to the midline. We suggest that the granule cell defects we find in Met signaling morphants may not result from direct disruption of URL determination, although we cannot exclude the possibility of an early cell non-autonomous effect on URL proliferation. The defects suggest a late requirement for proper signaling from VZ derivatives to granule cell progenitors.
Previous in vitro and genetic studies have shown that Met signaling is capable of protecting neurons from apoptosis (Maina et al., 1998; Maina et al., 1997; Xiao et al., 2001; Zhang et al., 2000). Consistent with these findings, we have also noted an overall increase in cell death along the length of the neural tube in Met signaling morphants, suggesting a role of Met signaling in neuronal survival. This regional nonspecific effect on neuronal survival may partially account for the reduced size of the cerebellum.
We also find in Met signaling morphants that the bilateral cerebellar structures at the dorsal midline do not fuse. We cannot determine whether these midline fusion defects persist into later stages, because Met signaling knockdown is lethal at around 72hpf. Little is known about the mechanism of midline cerebellar fusion, in either rodents or zebrafish (Broccoli et al., 1999; Foucher et al., 2006; Louvi et al., 2003), although Foucher et al. (2006) have suggested a role for Otx-expressing cells in this process in zebrafish. Our results suggest that Met signaling may also be involved in dorsal midline fusion of the cerebellar primordium.
Met signaling is required for normal VZ development and granule cell generation
At 36 and 48hpf, we find Met expression localized to the VZ, suggesting that Met signaling may play a role in the development of cells derived from this region. Limited information is currently available regarding the specific cell types derived from the zebrafish VZ. In mouse, Ptf1a is expressed in VZ progenitors and is required for the specification of all VZ GABAergic neurons, including Purkinje cells (Hoshino et al., 2005; Pascual et al., 2007). In zebrafish, ptf1a is similarly expressed in the VZ. The met gene is expressed in adjacent cells within the VZ, and a small proportion of cells co-express met and ptf1a. We found that ptf1a expression was disrupted in the absence of Met signaling, particularly in the midline. While the basis of the enhanced phenotype in the midline relative to more lateral regions is unclear, it may in part explain the lack of midline fusion discussed above. Several factors have previously been implicated in mammalian Purkinje cell migration or survival. The signaling molecule Reelin and its downstream transducer Dab1 are essential for the regulation of Purkinje cell migration (Howell et al., 1997; Sheldon et al., 1997). The atypical helix-loop-helix transcription factor Ebf2 was also found to be important for Purkinje cell migration (Croci et al., 2006). Survival of Purkinje cells is mediated by genes encoding nuclear factors Rorα (Dussault et al., 1998; Hamilton et al., 1996) and Nna1 (Fernandez-Gonzalez et al., 2002), and by neurotrophic factors (Larkfors et al., 1996; Mount et al., 1994). We find that zebrafish coe2 and rora2, homologs of the Ebf2 and Rorα markers of differentiated Purkinje neurons, are also missing or downregulated in Met signaling morphants, suggesting that Met activity is required for Purkinje cell development. We conclude that Met signaling is important for VZ progenitor cell specification and survival, despite limited expression of met transcripts within ptf1a-expressing progenitors. This may reveal the importance of signals between subtypes of VZ-derived cells during the establishment of the ptf1a-progenitor pool.
We found that Met signaling is not required for initial development of atoh1-positive cerebellar precursors in the URL at 24hpf. However, the generation of cerebellar granule cells expressing pax6a and reelin is significantly reduced in Met signaling morphants at 72hpf. In addition, we observed a decrease in global proliferation in the cerebellum, suggesting that multiple mechanisms orchestrating granule cell development may be affected in Met signaling deficient embryos. In mouse, RORα is strongly expressed in mature Purkinje cells (Hamilton et al., 1996; Ino, 2004; Nakagawa et al., 1997) and controls their expression of Sonic Hedgehog, which in turn promotes proliferation of granule cells (Dahmane and Ruiz i Altaba, 1999; Marti and Bovolenta, 2002; Wallace, 1999; Wechsler-Reya and Scott, 1999). RORα mutant mice display cerebellar atrophy, thus, RORα functions in Purkinje cells to indirectly affect the proliferation of granule cells. In zebrafish, rora2 is similarly expressed in the developing and adult cerebellum (Katsuyama et al., 2007) suggesting that it may play a similar role to its murine homolog during cerebellar development. Interestingly, Shh or Hh downstream signaling genes are not expressed during cerebellar development in zebrafish (Katsuyama et al., 2007; McFarland et al., 2008) suggesting that rora2 may regulate other unidentified factors that in turn play a role in the proliferation, migration and/or survival of granule cells. Our findings suggest that a consequence of Met signaling knockdown is a disruption in the communication between developing Purkinje cells and granule cell precursors, leading to loss or reduced number of granule cell progenitors.
In summary, our results are in accord with a dual role for Met signaling during cerebellar development. Met signaling may in parallel regulate VZ cell type specification and neural proliferation in the cerebellum. Alternatively, Met signaling may simply maintain all cerebellar cells in a proliferative state, rendering them competent to respond to unknown local factors that control cell specification. In this second scenario Met would play a permissive, rather than instructive, role in the specification of cell type. A future challenge is to identify the downstream effectors of Met signaling that regulate the various cellular morphogenesis events that shape the cerebellum.
Met signaling and neuronal migration
Previous in vivo functional studies in mice and various in vitro studies, have demonstrated a role for the Met/Hgf signaling pathway in regulating neuronal migration (Ebens et al., 1996; Garzotto et al., 2008; Giacobini et al., 2007; Powell et al., 2001; Segarra et al., 2006). These findings, taken together with previous reports of roles for zebrafish Met signaling in cell migration (Haines et al., 2004), suggest that proper migration of cerebellar precursors may similarly rely on Met function. Due to the severity of the patterning defects observed in the cerebellum of Met signaling morphants we were unable to distinguish whether migratory defects also contribute to the cerebellar phenotype. Tests of this hypothesis will be assisted by new transgenic lines marking specific cerebellar migratory neurons (Mione et al., 2008; Volkmann et al., 2008). The specific expression of the met receptor gene in a sub-population of migratory neurons within the hindbrain, however, prompted our investigation of a functional requirement for Met in neuronal migration. We have shown that Met signaling is an important component of the molecular guidance system that regulates rostrocaudal migration of FMNs in the developing hindbrain. Single knockdowns of individual hgf genes have minimal impact on FMN migration, suggesting that in this context, as in cerebellar development, the two zebrafish Hgf ligands are functionally redundant. The more drastic FMN migration defects in double Hgf morphants at higher doses phenocopy those of Met morphants, consistent with a specific Met/Hgf interaction in the migrating neurons. Further support for this idea comes from our analysis of morphant embryos in which partial knockdown of both Met receptor and Hgf ligands together (data not shown) was sufficient to induce migration defects similar to those observed in response to complete knockdown of either Met or Hgf. The variability of the phenotype observed in Met signaling morphants implies that additional signals must be involved in attracting FMNs posteriorly and/or repelling them from the anterior hindbrain in zebrafish. Several molecular pathways have been implicated in the posterior migration of r4-derived FMNs (Chandrasekhar, 2004), suggesting that Met signaling coordinates with other pathways to control FMN migratory behavior.
Met signaling and autism
Some of the proposed causes for autism relate to growth, survival and cell motility during development of several brain structures, including the cerebellum, essentially cellular events that have been shown to be mediated by Met signaling (Birchmeier et al., 2003). Postmortem studies of patients with autism have revealed cerebellar structural abnormalities that include loss of granular and Purkinje cells (Bauman and Kemper, 1985; Bauman, 1996; Fatemi et al., 2002a; Ritvo and Freeman, 1984; Whitney et al., 2008). Moreover, several studies have shown alterations in cerebella of autistic patients of key proteins related to apoptosis, BCL-2 and P53 (Araghi-Niknam and Fatemi, 2003; Fatemi and Halt, 2001; Fatemi et al., 2001); GABAergic function, GAD65, GAD67, and GABA(B) receptors (Fatemi et al., 2009; Fatemi et al., 2002b); and cell guidance, REELIN (Fatemi, 2002). Although ASD is primarily a genetic disorder involving multiple genes, insights into underlying mechanisms will require a multidisciplinary approach, including defining the molecular and cellular mechanisms by which each of these susceptibility genes mediate the development of the nervous system in model organisms (DiCicco-Bloom et al., 2006). In this study we have begun to explore how Met influences cerebellum development and have provided evidence that conserved pathways orchestrate the development of this structure in different vertebrates. Our work demonstrates that zebrafish is a viable model system for the future exploration of the underlying molecular and cellular mechanisms of various CNS disorders, including autism.
Supplementary Material
Supplemental Figure 1: Expression of the met receptor gene in medial rhombomere 1 (presumptive cerebellum)
Lateral view (A,B) and transverse sections (A’,A”,B’,C) at the level of the midbrain (A’) and cerebellum (A”,B’,C) of in situ hybridization of 24hpf embryos showing expression of the met receptor gene in purple (A,A’,A’’,B,B’), atoh1a (red in B and purple in B’) and atoh1b (purple in C). In the midbrain region, met expression is abundant in the dorsal domain (A,A’), while in r1, expression is confined to the medial domain (A,A’’) corresponding to the ventricular zone of the cerebellum. Double in situ hybridization (B) with met and atoh1a probes shows complementary expression within the cerebellar region, with met being expressed medially in the ventricular zone region and atoh1a being expressed dorsally in the upper rhombic lip region. Single in situ hybridizations with met (A”), atoh1a (B’), and atoh1b (C) also show the complementary expression domains of the met and atoh1 genes. Anterior is to the left and dorsal (transverse sections) is up in all supplemental figures. midbrain, mb; cerebellum ,cb; ventricular zone, VZ; upper rhombic lip (URL). Scale bar, 50μm.
Supplemental Figure 2: Expression of rora2 gene in the GFP-positive cells derived from the VZ of ptf1a-GFP transgenic fish
Transverse confocal sections (A) and bright field (B), and dorsal confocal sections (A’,B’) in 72hpf embryos showing GFP antibody staining of GFP-positive VZ-derived cells from ptf1a-GFP transgenic fish (A), and in situ hybridization with rora2 probe (B), and merged (C). rora2 transcript is expressed in the GFP-positive cells derived from the ptf1a-positive VZ zone cells (C,C’). Scale bar, 50μm.
Supplemental Figure 3: Met signaling regulates a subpopulation of VZ-derived cells
Dorsal view (A-F) of control embryos (A,D), Met morphants (B,E,), and Hgf morphants (C,F) at 48hpf (A-C), and 72hpf (D-F) embryos showing expression of coe2 (A-C) and olig2 (D-F) in the VZ-derived cells of the cerebellum (cb). coe2 expression is downregulated in morphants compared to controls, while olig2 expression is unchanged. Scale bar, 50μm.
Supplemental Figure 4: Expression of URL markers (atoh1a and atoh1b) is unaltered in Met signaling morphants
Dorsal view (A-F), lateral view (E’,F’), and transverse view at the level of the URL (E”,F”) in 24hpf embryos (C,D) and 48hpf embryos (A-B, E-F”) showing in situ hybridization of atoh1a (A,B) and atoh1b (C-F”) in control embryos (A,C,E,E’,E”) and Met morphants (B,D,F,F’,F”). There is no significant change in expression of atoh1a and atoh1b in Met morphants compared to control embryos. Scale bar, 50μm.
Supplemental Figure 5: Neurons derived from URL are unaltered in Met signaling morphants
Lateral view (A-F) of phox2a expression in the locus coeruleus neurons (indicated by arrows) in control (A,D), Met morphants (B,E), and Hgf morphants (C,F) of 24hpf (A-C) and 48hpf (D-F) embryos. phox2a expression in neurons derived from the URL is unchanged in morphants compared to controls at 24hpf, but there is a slight delay in the dorsal to ventral migration of these neurons. At 48hpf, these neurons have completed their migration to the ventral hindbrain region in both controls and morphants. Scale bar, 50μm.
Supplemental Figure 6: Met signaling plays a role in neuronal survival
Dorsal view (A-C) of control (A), Met morphants (B), and Hgf morphants (C) at 24hpf showing Acridine Orange staining in live embryos to detect cell death. Anterior to the left. There is an increase in cell death in both Met and Hgf morphants, not only in the cerebellar region but also throughout the rest of the hindbrain (indicated in brackets) and other CNS regions. Scale bar, 50μm.
Supplemental Table 1: Concentration dependent phenotypic responses to Met and Hgf morpholino microinjections
mg/ml: concentration of each morpholino injection and n: number of morpholino-injected Islet1-GFP transgenic embryos immunostained with GFP and EphA4 antibodies, scored at 48hpf by fluorescence microscopy according to the following criteria. “r4/r5/r6”: number of embryos with partially migrated facial motor neurons; “r6”: number of embryos whose facial motor neurons have completed their migration. Conditions that have been marked in bold were used further to assess the migration phenotype of the embryos showing the r4/r5/r6 (incomplete migration) in more detail (Fig 7G). UTR: Met morpholino blocking untranslated region; SB-E8I8: Met morpholino blocking the exon 8 - intron 8 splice donor site; SB-E6I6: Hgf1 morpholino blocking the exon 6 - intron 6 splice donor site; SB-E4I4: Hgf2 morpholino blocking the exon 4 - intron 4 splice donor site.
Supplemental Table 2: Phenotypic responses to Met and Hgf morpholino microinjections
mg/ml: concentration of each morpholino injection and n: number of morpholino-injected Islet1-GFP transgenic embryos immunostained with GFP and EphA4 antibodies, scored at 48hpf by fluorescence microscopy according to the following criteria. “r4/r5/r6”: number of embryos with partially migrated facial motor neurons; “number (%) of neurons in r4”: number of FMNs failing to migrate out of r4; ; “number (%) of neurons in r5”: number (%) of FMNs failing to migrate out of r5; “number (%) of neurons in r6”: number (%) of FMNs that have completed their migration to r6. The distribution of FMNs (% of neurons within r4,r5,r6) is illustrated in Fig. 7G.
ACKNOWLEDGMENTS
We thank Dr. Dae-Gwon Ahn for providing us with the hgf1 plasmid, Dr. Peter Currie for the met plasmid, Dr. Yu Katsuyama, for the rora2 plasmid, Dr. Marina Mione for the reelin plasmid, Dr. Steven Leach for the Ptf1a-GFP transgenic line, and Dr. David Wilkinson for the anti-EphA4 antibody. We also thank Elizabeth Sefton for excellent help with fish care. We are grateful to Drs Kathleen Millen and Elizabeth Grove for helpful discussions and critical reading of the manuscript. This work was supported by Institutional Training Grants T32GM07839 and T32HL007381, funds from the Brain Research Foundation to RKH, March of Dimes grant FY07-410 to VEP, and NRSA fellowship F31NS061425 to GEE.
REFERENCES
- Adolf B, Bellipanni G, Huber V, Bally-Cuif L. atoh1.2 and beta3.1 are two new bHLH-encoding genes expressed in selective precursor cells of the zebrafish anterior hindbrain. Gene Expr Patterns. 2004;5:35–41. doi: 10.1016/j.modgep.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Araghi-Niknam M, Fatemi SH. Levels of Bcl-2 and P53 are altered in superior frontal and cerebellar cortices of autistic subjects. Cell Mol Neurobiol. 2003;23:945–52. doi: 10.1023/B:CEMN.0000005322.27203.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae YK, Kani S, Shimizu T, Tanabe K, Nojima H, Kimura Y, Higashijima S, Hibi M. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev Biol. 2009;330:406–26. doi: 10.1016/j.ydbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Dubois L, Vincent A. Molecular cloning of Zcoe2, the zebrafish homolog of Xenopus Xcoe2 and mouse EBF-2, and its expression during primary neurogenesis. Mech Dev. 1998;77:85–90. doi: 10.1016/s0925-4773(98)00144-0. [DOI] [PubMed] [Google Scholar]
- Bauman M, Kemper TL. Histoanatomic observations of the brain in early infantile autism. Neurology. 1985;35:866–74. doi: 10.1212/wnl.35.6.866. [DOI] [PubMed] [Google Scholar]
- Bauman ML. Brief report: neuroanatomic observations of the brain in pervasive developmental disorders. J Autism Dev Disord. 1996;26:199–203. doi: 10.1007/BF02172012. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–72. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–25. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–4. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- Broccoli V, Boncinelli E, Wurst W. The caudal limit of Otx2 expression positions the isthmic organizer. Nature. 1999;401:164–8. doi: 10.1038/43670. [DOI] [PubMed] [Google Scholar]
- Brochu G, Maler L, Hawkes R. Zebrin II: a polypeptide antigen expressed selectively by Purkinje cells reveals compartments in rat and fish cerebellum. J Comp Neurol. 1990;291:538–52. doi: 10.1002/cne.902910405. [DOI] [PubMed] [Google Scholar]
- Cacci E, Salani M, Anastasi S, Perroteau I, Poiana G, Biagioni S, Augusti-Tocco G. Hepatocyte growth factor stimulates cell motility in cultures of the striatal progenitor cells ST14A. J Neurosci Res. 2003;74:760–8. doi: 10.1002/jnr.10799. [DOI] [PubMed] [Google Scholar]
- Caddy KW, Biscoe TJ. Structural and quantitative studies on the normal C3H and Lurcher mutant mouse. Philos Trans R Soc Lond B Biol Sci. 1979;287:167–201. doi: 10.1098/rstb.1979.0055. [DOI] [PubMed] [Google Scholar]
- Campbell DB, D'Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, Persico AM. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62:243–50. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, Elia M, Schneider C, Melmed R, Sacco R, Persico AM, Levitt P. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–9. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A, Becerra M, Manso MJ, Anadon R. Calretinin immunoreactivity in the brain of the zebrafish, Danio rerio: distribution and comparison with some neuropeptides and neurotransmitter-synthesizing enzymes. I. Olfactory organ and forebrain. J Comp Neurol. 2006;494:435–59. doi: 10.1002/cne.20782. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. Turning heads: development of vertebrate branchiomotor neurons. Dev Dyn. 2004;229:143–61. doi: 10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, Millen KJ. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- Cooke JE, Kemp HA, Moens CB. EphA4 is required for cell adhesion and rhombomere-boundary formation in the zebrafish. Curr Biol. 2005;15:536–42. doi: 10.1016/j.cub.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Corrales JD, Blaess S, Mahoney EM, Joyner AL. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006;133:1811–21. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 2004;131:5581–90. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- Costagli A, Kapsimali M, Wilson SW, Mione M. Conserved and divergent patterns of Reelin expression in the zebrafish central nervous system. J Comp Neurol. 2002;450:73–93. doi: 10.1002/cne.10292. [DOI] [PubMed] [Google Scholar]
- Croci L, Chung SH, Masserdotti G, Gianola S, Bizzoca A, Gennarini G, Corradi A, Rossi F, Hawkes R, Consalez GG. A key role for the HLH transcription factor EBF2COE2,O/E-3 in Purkinje neuron migration and cerebellar cortical topography. Development. 2006;133:2719–29. doi: 10.1242/dev.02437. [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Doulazmi M, Frederic F, Capone F, Becker-Andre M, Delhaye-Bouchaud N, Mariani J. A comparative study of Purkinje cells in two RORalpha gene mutant mice: staggerer and RORalpha(-/-). Brain Res Dev Brain Res. 2001;127:165–74. doi: 10.1016/s0165-3806(01)00131-6. [DOI] [PubMed] [Google Scholar]
- Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguere V. Orphan nuclear receptor ROR alpha-deficient mice display the cerebellar defects of staggerer. Mech Dev. 1998;70:147–53. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- Ebens A, Brose K, Leonardo ED, Hanson MG, Jr., Bladt F, Birchmeier C, Barres BA, Tessier-Lavigne M. Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron. 1996;17:1157–72. doi: 10.1016/s0896-6273(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Elsen GE, Choi LY, Millen KJ, Grinblat Y, Prince VE. Zic1 and Zic4 regulate zebrafish roof plate specification and hindbrain ventricle morphogenesis. Dev Biol. 2008;314:376–92. doi: 10.1016/j.ydbio.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci. 2006;26:9184–95. doi: 10.1523/JNEUROSCI.1610-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH. The role of Reelin in pathology of autism. Mol Psychiatry. 2002;7:919–20. doi: 10.1038/sj.mp.4001248. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD, Reutiman TJ, Thuras PD. Expression of GABA(B) receptors is altered in brains of subjects with autism. Cerebellum. 2009;8:64–9. doi: 10.1007/s12311-008-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR. Altered levels of Bcl2 and p53 proteins in parietal cortex reflect deranged apoptotic regulation in autism. Synapse. 2001;42:281–4. doi: 10.1002/syn.10002. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Realmuto G, Earle J, Kist DA, Thuras P, Merz A. Purkinje cell size is reduced in cerebellum of patients with autism. Cell Mol Neurobiol. 2002a;22:171–5. doi: 10.1023/a:1019861721160. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Stary JM, Kanodia R, Schulz SC, Realmuto GR. Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol Psychiatry. 2002b;52:805–10. doi: 10.1016/s0006-3223(02)01430-0. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR, Stary JM, Realmuto GM, Jalali-Mousavi M. Reduction in anti-apoptotic protein Bcl-2 in autistic cerebellum. Neuroreport. 2001;12:929–33. doi: 10.1097/00001756-200104170-00013. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD, Sidwell RW. The role of cerebellar genes in pathology of autism and schizophrenia. Cerebellum. 2008;7:279–94. doi: 10.1007/s12311-008-0017-0. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A, La Spada AR, Treadaway J, Higdon JC, Harris BS, Sidman RL, Morgan JI, Zuo J. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002;295:1904–6. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–76. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher I, Mione M, Simeone A, Acampora D, Bally-Cuif L, Houart C. Differentiation of cerebellar cell identities in absence of Fgf signalling in zebrafish Otx morphants. Development. 2006;133:1891–900. doi: 10.1242/dev.02352. [DOI] [PubMed] [Google Scholar]
- Garzotto D, Giacobini P, Crepaldi T, Fasolo A, De Marchis S. Hepatocyte growth factor regulates migration of olfactory interneuron precursors in the rostral migratory stream through Met-Grb2 coupling. J Neurosci. 2008;28:5901–9. doi: 10.1523/JNEUROSCI.1083-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacobini P, Messina A, Wray S, Giampietro C, Crepaldi T, Carmeliet P, Fasolo A. Hepatocyte growth factor acts as a motogen and guidance signal for gonadotropin hormone-releasing hormone-1 neuronal migration. J Neurosci. 2007;27:431–45. doi: 10.1523/JNEUROSCI.4979-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, Park SW, Leach SD, Wong RO. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132(22):5069–79. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Rosenfeld MG, Hamilton BA. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–31. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Gent PM, Hamilton BA. ROR alpha in genetic control of cerebellum development: 50 staggering years. Brain Res. 2007;1140:19–25. doi: 10.1016/j.brainres.2005.11.080. [DOI] [PubMed] [Google Scholar]
- Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–82. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron. 1999;24:555–66. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Gupta AR, State MW. Recent advances in the genetics of autism. Biol Psychiatry. 2007;61:429–37. doi: 10.1016/j.biopsych.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Haines L, Neyt C, Gautier P, Keenan DG, Bryson-Richardson RJ, Hollway GE, Cole NJ, Currie PD. Met and Hgf signaling controls hypaxial muscle and lateral line development in the zebrafish. Development. 2004;131:4857–69. doi: 10.1242/dev.01374. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, Kruglyak L, Lander ES. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–9. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- Herrup K. Role of staggerer gene in determining cell number in cerebellar cortex. I. Granule cell death is an indirect consequence of staggerer gene action. Brain Res. 1983;313:267–74. doi: 10.1016/0165-3806(83)90225-0. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–18. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Kagoshima M, Wanaka A, Tohyama M, Matsumoto K, Nakamura T. Localization and functional coupling of HGF and c-Met/HGF receptor in rat brain: implication as neurotrophic factor. Brain Res Mol Brain Res. 1995;32:197–210. doi: 10.1016/0169-328x(95)00075-4. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–13. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–7. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- Ieraci A, Forni PE, Ponzetto C. Viable hypomorphic signaling mutant of the Met receptor reveals a role for hepatocyte growth factor in postnatal cerebellar development. Proc Natl Acad Sci U S A. 2002;99:15200–5. doi: 10.1073/pnas.222362099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenaga T, Yoshida M, Uematsu K. Morphology and immunohistochemistry of efferent neurons of the goldfish corpus cerebelli. J Comp Neurol. 2005;487:300–11. doi: 10.1002/cne.20553. [DOI] [PubMed] [Google Scholar]
- Ikenaga T, Yoshida M, Uematsu K. Cerebellar efferent neurons in teleost fish. Cerebellum. 2006;5:268–74. doi: 10.1080/14734220600930588. [DOI] [PubMed] [Google Scholar]
- Ino H. Immunohistochemical characterization of the orphan nuclear receptor ROR alpha in the mouse nervous system. J Histochem Cytochem. 2004;52:311–23. doi: 10.1177/002215540405200302. [DOI] [PubMed] [Google Scholar]
- Ito M. Historical review of the significance of the cerebellum and the role of Purkinje cells in motor learning. Ann N Y Acad Sci. 2002;978:273–88. doi: 10.1111/j.1749-6632.2002.tb07574.x. [DOI] [PubMed] [Google Scholar]
- Ito M. Cerebellar circuitry as a neuronal machine. Prog Neurobiol. 2006;78:272–303. doi: 10.1016/j.pneurobio.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Katsuyama Y, Oomiya Y, Dekimoto H, Motooka E, Takano A, Kikkawa S, Hibi M, Terashima T. Expression of zebrafish ROR alpha gene in cerebellar-like structures. Dev Dyn. 2007;236:2694–701. doi: 10.1002/dvdy.21275. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr Biol. 2001;11:1858–63. doi: 10.1016/s0960-9822(01)00585-1. [DOI] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. FGF signaling mediates regeneration of the differentiating cerebellum through repatterning of the anterior hindbrain and reinitiation of neuronal migration. J Neurosci. 2006;26:7293–304. doi: 10.1523/JNEUROSCI.0095-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–44. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- Lannoo MJ, Brochu G, Maler L, Hawkes R. Zebrin II immunoreactivity in the rat and in the weakly electric teleost Eigenmannia (gymnotiformes) reveals three modes of Purkinje cell development. J Comp Neurol. 1991a;310:215–33. doi: 10.1002/cne.903100207. [DOI] [PubMed] [Google Scholar]
- Lannoo MJ, Ross L, Maler L, Hawkes R. Development of the cerebellum and its extracerebellar Purkinje cell projection in teleost fishes as determined by zebrin II immunocytochemistry. Prog Neurobiol. 1991b;37:329–63. doi: 10.1016/0301-0082(91)90022-s. [DOI] [PubMed] [Google Scholar]
- Larkfors L, Lindsay RM, Alderson RF. Characterization of the responses of Purkinje cells to neurotrophin treatment. J Neurochem. 1996;66:1362–73. doi: 10.1046/j.1471-4159.1996.66041362.x. [DOI] [PubMed] [Google Scholar]
- Latimer AJ, Jessen JR. Hgf/c-met expression and functional analysis during zebrafish embryogenesis. Dev Dyn. 2008;237:3904–15. doi: 10.1002/dvdy.21794. [DOI] [PubMed] [Google Scholar]
- Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol. 2004;270:393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Lin JC, Cai L, Cepko CL. The external granule layer of the developing chick cerebellum generates granule cells and cells of the isthmus and rostral hindbrain. J Neurosci. 2001;21:159–68. doi: 10.1523/JNEUROSCI.21-01-00159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Alexandre P, Metin C, Wurst W, Wassef M. The isthmic neuroepithelium is essential for cerebellar midline fusion. Development. 2003;130:5319–30. doi: 10.1242/dev.00736. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Guy AT, Clarke JD. Monitoring neural progenitor fate through multiple rounds of division in an intact vertebrate brain. Development. 2003;130:3427–36. doi: 10.1242/dev.00569. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Maina F, Hilton MC, Andres R, Wyatt S, Klein R, Davies AM. Multiple roles for hepatocyte growth factor in sympathetic neuron development. Neuron. 1998;20:835–46. doi: 10.1016/s0896-6273(00)80466-3. [DOI] [PubMed] [Google Scholar]
- Maina F, Hilton MC, Ponzetto C, Davies AM, Klein R. Met receptor signaling is required for sensory nerve development and HGF promotes axonal growth and survival of sensory neurons. Genes Dev. 1997;11:3341–50. doi: 10.1101/gad.11.24.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina F, Klein R. Hepatocyte growth factor, a versatile signal for developing neurons. Nat Neurosci. 1999;2:213–7. doi: 10.1038/6310. [DOI] [PubMed] [Google Scholar]
- Marti E, Bovolenta P. Sonic hedgehog in CNS development: one signal, multiple outputs. Trends Neurosci. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Carlson R, Mann DM, Prince VE. Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001;128:2471–84. doi: 10.1242/dev.128.13.2471. [DOI] [PubMed] [Google Scholar]
- McFarland KA, Topczewska JM, Weidinger G, Dorsky RI, Appel B. Hh and Wnt signaling regulate formation of olig2+ neurons in the zebrafish cerebellum. Dev Biol. 2008;318:162–71. doi: 10.1016/j.ydbio.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek J, Hafmans TG, Maler L, Hawkes R. Distribution of zebrin II in the gigantocerebellum of the mormyrid fish Gnathonemus petersii compared with other teleosts. J Comp Neurol. 1992;316:17–31. doi: 10.1002/cne.903160103. [DOI] [PubMed] [Google Scholar]
- Millen KJ, Gleeson JG. Cerebellar development and disease. Curr Opin Neurobiol. 2008;18:12–9. doi: 10.1016/j.conb.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mione M, Baldessari D, Deflorian G, Nappo G, Santoriello C. How neuronal migration contributes to the morphogenesis of the CNS: insights from the zebrafish. Dev Neurosci. 2008;30:65–81. doi: 10.1159/000109853. [DOI] [PubMed] [Google Scholar]
- Mount HT, Dreyfus CF, Black IB. Neurotrophin-3 selectively increases cultured Purkinje cell survival. Neuroreport. 1994;5:2497–500. doi: 10.1097/00001756-199412000-00023. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Watanabe M, Inoue Y. Prominent expression of nuclear hormone receptor ROR alpha in Purkinje cells from early development. Neurosci Res. 1997;28:177–84. doi: 10.1016/s0168-0102(97)00042-4. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–20. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–95. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, Miki N, Chitnis AB, Huh TL. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 2000;227:279–93. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci U S A. 2007;104:5193–8. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell EM, Mars WM, Levitt P. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 2001;30:79–89. doi: 10.1016/s0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Rieger S, Volkmann K, Koster RW. Polysialyltransferase expression is linked to neuronal migration in the developing and adult zebrafish. Dev Dyn. 2008;237:276–85. doi: 10.1002/dvdy.21410. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ. A medical model of autism: etiology, pathology and treatment. Pediatr Ann. 1984;13:298–305. [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, Guthrie D, Ritvo A. Lower Purkinje cell counts in the cerebella of four autistic subjects: initial findings of the UCLA-NSAC Autopsy Research Report. Am J Psychiatry. 1986;143:862–6. doi: 10.1176/ajp.143.7.862. [DOI] [PubMed] [Google Scholar]
- Rubin JS, Bottaro DP, Aaronson SA. Hepatocyte growth factor/scatter factor and its receptor, the c-met proto-oncogene product. Biochim Biophys Acta. 1993;1155:357–71. doi: 10.1016/0304-419x(93)90015-5. [DOI] [PubMed] [Google Scholar]
- Segarra J, Balenci L, Drenth T, Maina F, Lamballe F. Combined signaling through ERK, PI3K/AKT, and RAC1/p38 is required for met-triggered cortical neuron migration. J Biol Chem. 2006;281:4771–8. doi: 10.1074/jbc.M508298200. [DOI] [PubMed] [Google Scholar]
- Sgaier SK, Lao Z, Villanueva MP, Berenshteyn F, Stephen D, Turnbull RK, Joyner AL. Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development. 2007;134:2325–35. doi: 10.1242/dev.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL. Morphogenetic and cellular movements that shape the mouse cerebellum; insights from genetic fate mapping. Neuron. 2005;45:27–40. doi: 10.1016/j.neuron.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–3. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Lane PW, Dickie MM. Staggerer, a new mutation in the mouse affecting the cerebellum. Science. 1962;137:610–2. doi: 10.1126/science.137.3530.610. [DOI] [PubMed] [Google Scholar]
- Sillitoe RV, Joyner AL. Morphology, molecular codes, and circuitry produce the three-dimensional complexity of the cerebellum. Annu Rev Cell Dev Biol. 2007;23:549–77. doi: 10.1146/annurev.cellbio.23.090506.123237. [DOI] [PubMed] [Google Scholar]
- Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, Daniel H, Crepel F, Mariani J, Sotelo C, Becker-Andre M. staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc Natl Acad Sci U S A. 1998;95:3960–5. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara G, Hashimoto K, Iwata Y, Nakamura K, Tsujii M, Tsuchiya KJ, Sekine Y, Suzuki K, Suda S, Matsuzaki H, Kawai M, Minabe Y, Yagi A, Takei N, Sugiyama T, Mori N. Decreased serum levels of hepatocyte growth factor in male adults with high-functioning autism. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:412–5. doi: 10.1016/j.pnpbp.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Tallafuss A, Eisen JS. The Met receptor tyrosine kinase prevents zebrafish primary motoneurons from expressing an incorrect neurotransmitter. Neural Develop. 2008;3:18. doi: 10.1186/1749-8104-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- Vogel MW, Sinclair M, Qiu D, Fan H. Purkinje cell fate in staggerer mutants: agenesis versus cell death. J Neurobiol. 2000;42:323–37. doi: 10.1002/(sici)1097-4695(20000215)42:3<323::aid-neu4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Volkmann K, Rieger S, Babaryka A, Koster RW. The zebrafish cerebellar rhombic lip is spatially patterned in producing granule cell populations of different functional compartments. Dev Biol. 2008;313:167–80. doi: 10.1016/j.ydbio.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–8. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001;2:484–91. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–14. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Whitney ER, Kemper TL, Bauman ML, Rosene DL, Blatt GJ. Cerebellar Purkinje cells are reduced in a subpopulation of autistic brains: a stereological experiment using calbindin-D28k. Cerebellum. 2008;7:406–16. doi: 10.1007/s12311-008-0043-y. [DOI] [PubMed] [Google Scholar]
- Wingate RJ. The rhombic lip and early cerebellar development. Curr Opin Neurobiol. 2001;11:82–8. doi: 10.1016/s0959-4388(00)00177-x. [DOI] [PubMed] [Google Scholar]
- Wingate RJ, Hatten ME. The role of the rhombic lip in avian cerebellum development. Development. 1999;126:4395–404. doi: 10.1242/dev.126.20.4395. [DOI] [PubMed] [Google Scholar]
- Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- Xiao GH, Jeffers M, Bellacosa A, Mitsuuchi Y, Vande Woude GF, Testa JR. Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A. 2001;98:247–52. doi: 10.1073/pnas.011532898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MS, Gill M. A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. Int J Dev Neurosci. 2007;25:69–85. doi: 10.1016/j.ijdevneu.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Zhang L, Himi T, Morita I, Murota S. Hepatocyte growth factor protects cultured rat cerebellar granule neurons from apoptosis via the phosphatidylinositol-3 kinase/Akt pathway. J Neurosci Res. 2000;59:489–96. doi: 10.1002/(SICI)1097-4547(20000215)59:4<489::AID-JNR3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Expression of the met receptor gene in medial rhombomere 1 (presumptive cerebellum)
Lateral view (A,B) and transverse sections (A’,A”,B’,C) at the level of the midbrain (A’) and cerebellum (A”,B’,C) of in situ hybridization of 24hpf embryos showing expression of the met receptor gene in purple (A,A’,A’’,B,B’), atoh1a (red in B and purple in B’) and atoh1b (purple in C). In the midbrain region, met expression is abundant in the dorsal domain (A,A’), while in r1, expression is confined to the medial domain (A,A’’) corresponding to the ventricular zone of the cerebellum. Double in situ hybridization (B) with met and atoh1a probes shows complementary expression within the cerebellar region, with met being expressed medially in the ventricular zone region and atoh1a being expressed dorsally in the upper rhombic lip region. Single in situ hybridizations with met (A”), atoh1a (B’), and atoh1b (C) also show the complementary expression domains of the met and atoh1 genes. Anterior is to the left and dorsal (transverse sections) is up in all supplemental figures. midbrain, mb; cerebellum ,cb; ventricular zone, VZ; upper rhombic lip (URL). Scale bar, 50μm.
Supplemental Figure 2: Expression of rora2 gene in the GFP-positive cells derived from the VZ of ptf1a-GFP transgenic fish
Transverse confocal sections (A) and bright field (B), and dorsal confocal sections (A’,B’) in 72hpf embryos showing GFP antibody staining of GFP-positive VZ-derived cells from ptf1a-GFP transgenic fish (A), and in situ hybridization with rora2 probe (B), and merged (C). rora2 transcript is expressed in the GFP-positive cells derived from the ptf1a-positive VZ zone cells (C,C’). Scale bar, 50μm.
Supplemental Figure 3: Met signaling regulates a subpopulation of VZ-derived cells
Dorsal view (A-F) of control embryos (A,D), Met morphants (B,E,), and Hgf morphants (C,F) at 48hpf (A-C), and 72hpf (D-F) embryos showing expression of coe2 (A-C) and olig2 (D-F) in the VZ-derived cells of the cerebellum (cb). coe2 expression is downregulated in morphants compared to controls, while olig2 expression is unchanged. Scale bar, 50μm.
Supplemental Figure 4: Expression of URL markers (atoh1a and atoh1b) is unaltered in Met signaling morphants
Dorsal view (A-F), lateral view (E’,F’), and transverse view at the level of the URL (E”,F”) in 24hpf embryos (C,D) and 48hpf embryos (A-B, E-F”) showing in situ hybridization of atoh1a (A,B) and atoh1b (C-F”) in control embryos (A,C,E,E’,E”) and Met morphants (B,D,F,F’,F”). There is no significant change in expression of atoh1a and atoh1b in Met morphants compared to control embryos. Scale bar, 50μm.
Supplemental Figure 5: Neurons derived from URL are unaltered in Met signaling morphants
Lateral view (A-F) of phox2a expression in the locus coeruleus neurons (indicated by arrows) in control (A,D), Met morphants (B,E), and Hgf morphants (C,F) of 24hpf (A-C) and 48hpf (D-F) embryos. phox2a expression in neurons derived from the URL is unchanged in morphants compared to controls at 24hpf, but there is a slight delay in the dorsal to ventral migration of these neurons. At 48hpf, these neurons have completed their migration to the ventral hindbrain region in both controls and morphants. Scale bar, 50μm.
Supplemental Figure 6: Met signaling plays a role in neuronal survival
Dorsal view (A-C) of control (A), Met morphants (B), and Hgf morphants (C) at 24hpf showing Acridine Orange staining in live embryos to detect cell death. Anterior to the left. There is an increase in cell death in both Met and Hgf morphants, not only in the cerebellar region but also throughout the rest of the hindbrain (indicated in brackets) and other CNS regions. Scale bar, 50μm.
Supplemental Table 1: Concentration dependent phenotypic responses to Met and Hgf morpholino microinjections
mg/ml: concentration of each morpholino injection and n: number of morpholino-injected Islet1-GFP transgenic embryos immunostained with GFP and EphA4 antibodies, scored at 48hpf by fluorescence microscopy according to the following criteria. “r4/r5/r6”: number of embryos with partially migrated facial motor neurons; “r6”: number of embryos whose facial motor neurons have completed their migration. Conditions that have been marked in bold were used further to assess the migration phenotype of the embryos showing the r4/r5/r6 (incomplete migration) in more detail (Fig 7G). UTR: Met morpholino blocking untranslated region; SB-E8I8: Met morpholino blocking the exon 8 - intron 8 splice donor site; SB-E6I6: Hgf1 morpholino blocking the exon 6 - intron 6 splice donor site; SB-E4I4: Hgf2 morpholino blocking the exon 4 - intron 4 splice donor site.
Supplemental Table 2: Phenotypic responses to Met and Hgf morpholino microinjections
mg/ml: concentration of each morpholino injection and n: number of morpholino-injected Islet1-GFP transgenic embryos immunostained with GFP and EphA4 antibodies, scored at 48hpf by fluorescence microscopy according to the following criteria. “r4/r5/r6”: number of embryos with partially migrated facial motor neurons; “number (%) of neurons in r4”: number of FMNs failing to migrate out of r4; ; “number (%) of neurons in r5”: number (%) of FMNs failing to migrate out of r5; “number (%) of neurons in r6”: number (%) of FMNs that have completed their migration to r6. The distribution of FMNs (% of neurons within r4,r5,r6) is illustrated in Fig. 7G.