Abstract
Tyrosyl-DNA phosphodiesterase 1 (Tdp1) is an enzyme that catalyzes the hydrolysis of 3'-phosphotyrosyl bonds. Such linkages form in vivo when topoisomerase I (Top1) processes DNA. For this reason, Tdp1 has been implicated in the repair of irreversible Top1-DNA covalent complexes. Tdp1 inhibitors have been regarded as potential therapeutics in combination with Top1 inhibitors, such as the camptothecin derivatives, topotecan and irinotecan, which are used to treat human cancers. Using a novel high-throughput screening assay, we have identified the C21-substituted progesterone derivative, NSC 88915 (1), as a potential Tdp1 inhibitor. Secondary screening and cross-reactivity studies with related DNA processing enzymes confirmed that compound 1 possesses specific Tdp1 inhibitory activity. Deconstruction of compound 1 into discrete functional groups reveals that both components are required for inhibition of Tdp1 activity. Moreover, the synthesis of analogues of compound 1 has provided insight into the structural requirements for the inhibition of Tdp1. Surface plasmon resonance shows that compound 1 binds to Tdp1, whereas an inactive analogue fails to interact with the enzyme. Based on molecular docking and mechanistic studies, we propose that these compounds are competitive inhibitors, which mimics the oligonucleotide-peptide Tdp1 substrate. These steroid derivatives represent a novel chemotype and provide a new scaffold for developing small molecule inhibitors of Tdp1.
Keywords: Topoisomerase I, Tyrosyl DNA Phosphodiesterase, steroid, inhibitor
Introduction
Topoisomerase I (Top1) inhibitors represent an important class of chemotherapeutic drugs with distinct mechanisms of damaging DNA (for review, see 1). Among these are the camptothecin (CPT) derivatives, topotecan and irinotecan, which are used clinically in cancer chemotherapy. Currently, newer CPT derivatives, such as gimatecan (Novartis®),2 S39625 (Servier®),3 and diflomotecan4 are in clinical development. In addition, several non-CPT Top1 inhibitors including the indenoisoquinolines, the phenanthrolines, and the indolocarbazoles, have been identified and are currently undergoing clinical development (for reviews, see 5–7).
Mechanistically, Top1 inhibitors selectively bind to the Top1-DNA interface and damage DNA by trapping the cleavage complex between the Top1 catalytic tyrosine and the 3'-end of the broken DNA.8, 9 Likewise, Top1 cleavage complexes have also been shown to accumulate at preexisting DNA lesions (for reviews, see 10–12), such as strand breaks, abasic sites, base mismatches, and specific oxidized or modified bases. Top1-DNA cleavage complexes caused by DNA lesions have the propensity to self-sufficiently yield abortive Top1 cleavage complexes, whereas the reversible drug-stabilized Top1 cleavage complexes require conversion to Top1-linked DNA strand breaks by collision of DNA and RNA polymerases during replication and transcription, respectively (for reviews, see 1, 10). Consequently, these irreversible Top1-DNA lesions confer a unique barrier for the DNA repair machinery since the DNA strand break is encumbered with a 3'-protein adduct.
Tyrosyl-DNA phosphodiesterase (Tdp1) has been associated with the repair of Top1 cleavage complexes by virtue of its ability to hydrolyze the phosphodiester linkage between a tyrosine residue and a DNA 3'-phosphate.13, 14 Besides the Top1-derived phosphotyrosyl bond, Tdp1 has been shown to hydrolyze other covalently linked 3'-blocking lesions, although less efficiently than 3′-phosphotyrosyl ends.15 For example, Tdp1 has been shown to cleave 3'-terminal phosphoglycolate diester linkages, which are commonly generated by oxidative DNA damage.16 Interestingly, cells harboring the disease-associated Tdp1 SCAN1 (spinocerebellar ataxia with axonal neuropathy-1) mutation are hypersensitive to both CPT and oxidative stress (i.e. H2O2 and ionizing radiation).17–20 Cell extracts from SCAN1 cells have also been shown to be deficient in processing 3'-phosphoglycolates.21, 22 Moreover, CPT-treated skin fibroblasts from SCAN1 patients have been shown to accumulate Tdp1-DNA intermediates wherein the mutant form of Tdp1 (H493R) becomes covalently linked to DNA, which provides in vivo evidence for the involvement of Tdp1 in the removal of drug-induced Top1-DNA cleavage complexes.23
In addition to studies performed with the physiologically relevant SCAN1 Tdp1 mutant, the recent generation of Tdp1 knockout mice further establishes the function of Tdp1 in the repair of Top1-DNA cleavage complexes and oxidative DNA damage. Specifically, primary neural cells from Tdp1−/− mice have been shown to accrue more total DNA strand breaks than wild-type cells after treatment with CPT, H2O2, and ionizing radiation.24 Both Tdp1−/− mice and cells derived from Tdp1−/− mice are hypersensitive to the Top1 inhibitors.23, 24 Taken together, these studies demonstrate that a single defect in Tdp1 activity is sufficient for Top1 inhibitor hypersensitivity.
In corroboration, two independent studies have shown that overexpression of wild-type Tdp1 in human cells protects against CPT-induced cell death,25, 26 whereas the catalytically inactive Tdp1 mutant does not.25 A recent study has also observed an increase in expression and activity of Tdp1 in greater than 50% of the non-small celpl lung cancer tissue samples analyzed compared to non-neoplastic tissues.27 Thus, the presence and activity of Tdp1 is consistent with a role for the enzyme in protecting cells against the cytotoxic effects of Top1 inhibitors. It is therefore logical to develop inhibitors of Tdp1 to counteract the inherited resistance to Top1 inhibitors caused by the Tdp1-mediated repair of Top1-DNA lesions. Tdp1 inhibitors may possibly augment current radiotherapy as well.
At present, only a small number of Tdp1 inhibitors have been characterized. Although unattractive as pharmacological inhibitors of Tdp1, both vanadate and tungstate, which inhibit Tdp1 at millimolar concentrations, have been useful in co-crystallization studies of Tdp1.28, 29 The aminoglycoside antibiotic, neomycin B, has also examined as a potential Tdp1 inhibitor based on its ability to target members of the phospholipase D superfamily.30 In addition, recent high-throughput screening efforts have identified furamidine31 as well as several phosphotyrosine mimetics as Tdp1 inhibitors.32
In this report, we characterize a new chemotype of fully synthetic small molecule inhibitors of Tdp1 that were originally identified in a high-throughput screen.31, 33 We demonstrate that the lead compound (1, see Figure 1A) blocks the formation of the initial Tdp1-DNA covalent intermediate through the use of the SCAN1 Tdp1 mutant enzyme. In addition, molecular docking of the inhibitor into the active site of Tdp1 suggests that it competes for binding by mimicking the Tdp1 substrate.
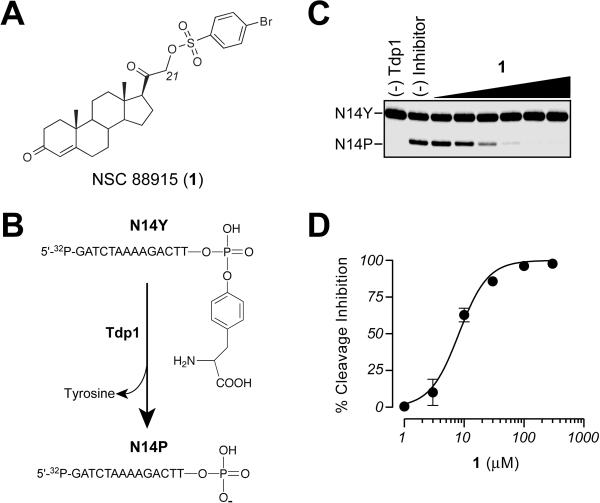
Figure 1.
Tdp1 inhibition by 1. A) Chemical structure of 1. B) Schematic representation of the Tdp1 gel-based biochemical assay. Tdp1 hydrolyzes the 3′-phosphotyrosine bond and converts N14Y to an oligonucleotide containing a 3′phosphate (N14P). C) Representative gel demonstrating dose-dependent inhibition of Tdp1 by 1. D) Graphical representation of the percent inhibition of Tdp1 by 1. Each point represents the mean ± SEM for three independent experiments.
Experimental Section
Chemistry
General procedures and techniques were identical with those previously reported.34 1H NMR spectra were recorded in CDCl3 or DMSO d6 at 500 MHz with Me4Si as an internal standard using a Varian Inova 500 MHz spectrometer. High-resolution mass spectra (HRMS) were determined on a Bruker 12Tesla APEX-Qe FTICR-MS by positive ion ESI mode by Ms. Susan A Hatcher, Facility Director, College of Sciences Major Instrumentation Cluster, Old Dominion University, Norfolk, VA. Deoxycorticosterone and compound 7 were obtained from Steraloids, Inc, Newport, RI, USA. All other reagents were purchased from Sigma-Aldrich. Compounds were stored in an atmosphere of argon and in the cold (−20 or −80 °C).
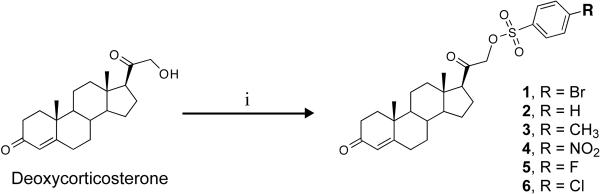
General method for the preparation of deoxycorticosterone-21-sulfonates (1–6).35, 36
Compounds 1–6 were synthesized from commercially available deoxycorticosterone using a simple general procedure as outlined in Scheme 1. To a 0.1M solution of the deoxycorticosterone in anhydrous tetrahydrofuran at 0 °C, was added 1.5 equivalents of triethylamine and 1.5 equivalents of 4-substituted benzenesulfonyl chloride was added over 10 minutes. The reaction mixture (solution) was allowed to stand at 4–8 °C for 24 hours. The cold solution was evaporated to dryness and to this was added water containing sodium bicarbonate and extracted with ethyl acetate (50 × 2 ml). The extract was washed with brine and dried over sodium sulfate. The solvent was evaporated and crude product was purified using flash column chromatograpy [FCC, pet. Ether/EtOAc, (7:3, v/v)] to obtain the various sulfonates in 60 – 70% yields.
Scheme 1.
Synthesis of deoxycorticosterone benzene sulfonates (1–6).a
aReagents and conditions: (i) THF/Et3N, ArSO2Cl, 0 °C, then 4–8 °C, 24 hr
4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesufonate) (1)
Mp: 147–149 °C (lit. 148–150 °C). IR (CHCl3): 1731, 1661, 1576, 1376, 1188, 609 cm−1. 1H NMR (CDCl3) δ 0.65 (s, 3H, 19-CH3), 1.18 (s,3H,18-CH3), 2.59 (m, 2H, 2-CH2), 4.56 (m, 2H, 21-CH2), 5.73 (s, 1H, C4-H), 7.71 (d, 2H, J = 8.5 Hz, 3'5'Ar-Hs), 7.81 ( d, 2H, J = 8.6 Hz, 2'6'Ar-Hs). HRMS calcd 571.1124 (C27H33BrO5SNa+), found 571.1115.
4-Pregnen-21-ol-3,20-dione-21-(4-benzenesufonate) (2)
Mp: 166–168 °C. IR (CHCl3): 1730, 1660, 1579, 1380, 1177 cm−1. 1H NMR (CDCl3) δ 0.64 (s, 3H, 18 -CH3), 1.17 (s,3H,19-CH3), 2.63 (m, 2H, 2-CH2), 4.58 (m, 2H, 21-CH2), 5.73 (s, 1H, C4-H), 7.59 (m, 2H, 3'5'Ar-Hs), 7.69 (s, 1H, 4'Ar-H), 7.95 ( d, 2H, J = 7.0 Hz, 2'6'Ar-Hs). HRMS calcd 493.2019 (C27H34O5SNa+), found 493.2005.
4-Pregnen-21-ol-3,20-dione-21-(4-methyl-benzenesufonate) (3)
Mp: 160–161 °C, IR (CHCl3): 1731, 1660, 1570, 1384, 1191 cm−1. 1H NMR (CDCl3) δ 0.64 (s, 3H, 18 -CH3), 1.17 (s,3H,19-CH3),2.45 (s, 3H,4'-CH3), 2.65(m, 2H, 2-CH2), 4.58 (m, 2H, 21-CH2), 5.73 (s, 1H, C4-H), 7.37 (d, 2H, J = 8.5 Hz, 3'5'Ar-Hs), 7.83 (d, 2H, J = 8.0 Hz, 2'6'Ar-Hs). HRMS calcd 507.2175 (C28H36O5SNa+), found 507.2168.
4-Pregnen-21-ol-3,20-dione-21-(4-nitro-benzenesufonate) (4)
Mp: 137–138 °C, IR (CHCl3): 1733, 1657, 1569, 1373, 1197 cm−1. 1H NMR (CDCl3) δ 0.66 (s, 3H, 18 -CH3), 1.18 (s,3H,19-CH3), 2.56 (m, 2H, 2-CH2), 4.78 (m, 2H, 21-CH2), 5.73 (s, 1H, C4-H), 8.17 (d, 2H, J = 7.5 Hz, 3'5'Ar-Hs), 8.42 (d, 2H, J = 8.0 Hz, 2'6'Ar-Hs), HRMS calcd 538.1869(C27H33NO7SNa+), found 538.1857.
4-Pregnen-21-ol-3,20-dione-21-(4-flouro-benzenesufonate) (5)
Mp: 156–157 °C. IR (CHCl3): 1729, 1665, 1566, 1379, 1177, 1011 cm−1. 1H NMR (CDCl3) δ 0.65 (s, 3H, 18 -CH3), 1.18 (s,3H,19-CH3), 2.60 (m, 2H, 2-CH2), 4.66 (m, 2H, 21-CH2), 5.73 (s, 1H, C4-H), 7.23 (d, 2H, J = 8.5 Hz, 3'5'Ar-Hs), 8.00 (d, 2H, J = 7.5 Hz, 2'6'Ar-Hs). HRMS calcd 511.1924 (C27H33FO5SNa+), found 511.1912.
4-Pregnen-21-ol-3,20-dione-21-(4-chloro-benzenesufonate) (6)
Mp: 134–135 °C. IR (CHCl3): 1730, 1663, 1577, 1372, 1178, 679 cm−1. 1H NMR (CDCl3) δ 0.65 (s, 3H, 18 -CH3), 1.18 (s,3H,19-CH3), 2.59 (m, 2H, 2-CH2), 4.66 (m, 2H, 21-CH2), 5.73 (s, 1H, C4-H), 7.55 (d, 2H, J = 8.5 Hz, 3'5'Ar-Hs), 7.90 (d, 2H, J = 8.5 Hz, 2'6'Ar-Hs). HRMS calcd 499.2818 (C31H40O4Na+), found 499.2822.
Oligonucleotides
The N14Y oligonucleotide (5′-GATCTAAAAGACTTY-3′), which contains a 3′-phosphotyrosine (Y), was synthesized by Midland Certified Reagents Company (Midland, TX). All of the other DNA oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
End-labeling and preparation of DNA substrates
Oligonucleotides were 5'-end labeled using T4 polynucleotide kinase and [γ-32P] ATP. Unincorporated radioactive nucleotides were removed using a mini Quick Spin Oligo column (Roche Diagnostics, Indianapolis, IN, USA) after inactivation of the kinase by heating for 5 min at 95° C. For construction of double-stranded DNA substrates, labeled single-stranded oligonucleotides were annealed with a complementary strand by heating for 5 min at 95 °C and slowly cooling to 25° C.
Expression and purification of Tdp1
Wild-type and mutant (H493R) human Tdp1 enzymes were expressed in E. coli BL21 (DE3) cells and purified as described previously.31
Tdp1 gel-based assay
One nanomolar of 5′-32P-labeled DNA substrate was incubated with 0.1 nM recombinant Tdp1 in the absence or presence of inhibitor for 20 min at 25° C in a reaction buffer containing 50 mM Tris-HCl (pH 7.5), 80 mM KCl, 2 mM EDTA, and 40 μg/ml bovine serum albumin (BSA). Reactions were terminated by the addition of two volumes of gel loading buffer (96% (v/v) formamide, 10 mM EDTA, 1% (w/v) xylene cyanol and 1% (w/v) bromphenol blue). The samples were subsequently heated to 95° C for 5 min and subjected to 18% sequencing gel electrophoresis. A concentration of 100 nM was used when employing the SCAN1 mutant H493R. In addition, H493R reactions were divided in half. One-half of the reaction was run on a sequencing gel, while the other half was analyzed by 4–20% SDS-PAGE electrophoresis. Imaging and quantification were preformed using the Typhoon 8600 and ImageQuant software (Molecular Dynamics), respectively.
Counter-screen assays
a) APE1 assay. Twenty nanomolar of 5'-32P labeled 21-mer double-stranded DNA containing a single AP site was incubated with 0.1 units of APE1 (New England Biolabs, Ipswich, MA) in NEBuffer 4 [20 mM Tris-acetate (pH 7.9), 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9] for 30 min at 25° C in a total volume of 10 μl. Reactions were terminated and run analogous to the Tdp1 gel-based assay described above. Cleavage at the AP site by APE1 was detected by the appearance of a predominant second band of higher mobility on the gel. b) Top1 relaxation assay. Recombinant Top1 was incubated with 0.25 μg of supercoiled ΦX174 DNA for 30 min at 37° C in a reaction buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 15 μg/ml BSA. Reactions were terminated by the addition of 0.5% SDS and 0.5 mg/ml proteinase K (final concentrations), followed by incubation for 30 min at 50° C. Next, loading dye was added and the samples were subjected to 1% agarose gel electrophoresis. After electrophoresis, the gels were stained with 1X buffer solution containing 10 μg/ml of ethidium bromide and the DNA was visualized by transillumination with UV light.
Surface plasmon resonance
Direct binding experiments were performed on a Biacore 2000 instrument (GE, Piscatawy, NJ). Recombinant Tdp1 was coupled to the carboxylmethylated dextran surface of a CM5 chip (GE, Piscatawy, NJ) using standard amine coupling chemistry. Briefly, the surface was activated with 0.1 M N-hydroxysuccinimide and 0.4 M N-ethyl-N'-(3dimethylaminopropyl)carbodiimide at a flow rate of 20 μl/min. Tdp1 was diluted in 10 mM sodium acetate (pH 4.5) and injected until a density of 1000 RU was attached. Activated amine groups were quenched with an injection of 1 M ethanolamine (pH 8.0). A reference surface was prepared in the same manner excluding Tdp1. Inhibitors were diluted in running buffer [10 mM sodium phosphate (pH 7.5), 150 mM NaCl, and 1% DMSO (v/v)] and injected at 20 μl/min at 25° C. A solution of 50 mM NaOH was used to regenerate the surface after each injection. Each cycle of inhibitor injection was followed by a running buffer cycle for referencing purposes. A DMSO calibration curve was included to correct for refractive index mismatches between the running buffer and inhibitor dilution series.
Molecular Docking
The X-ray crystal structure of human Tdp1 in complex with vanadate, DNA, and a human Top1-derived peptide [PDB accession code 1NOP 29] was obtained from the Protein Data Bank.37 Before the docking procedure, the X-ray structure was manipulated with the program Maestro (Schrödinger, Inc., Portland, OR). The water molecules and the co-crystallized DNA-peptide substrate were deleted from the structure. Hydrogen atoms were assigned, active site residues (K265 and K495) were neutralized, and a series of minimizations were performed using the OPLS2005 force field. The lowest energy conformation of each inhibitor was located by using the Monte Carlo stochastic dynamics (MCMM) conformational search routine in Macromodel (Version 9.6). In each case, all rotatable bonds were selected, 50,000 conformations were generated and unique conformations within 2 kcal/mol of the lowest energy conformation were retained. The initial geometry of the structures was optimized using the OPLS 2005 force field performing 1000 steps of conjugate minimization. Docking was performed using the program Glide (version 5.0, Schrödinger Inc., New York, NY) with Extra Precision mode running on a Silicon Graphics workstation. The inhibitors were docked into a 16 Å cavity, which was defined around the Tdp1 active site.
Free energy of binding calculation
Theoretical free energy of binding (FEB) was calculated utilizing the eMBrAcE procedure developed by Schrödinger Inc. as part of the MacroModel package.38 For each inhibitor, the enzyme-inhibitor complex (Ecomplex), the free enzyme (Eenzyme), and the free inhibitor (Einhibitor) were all subjected to energy minimization using the OPLS 2001 force field with a dielectric constant of 1.0. A conjugate gradient minimization protocol with default values was used in all minimization. eMBrAcE minimization calculations were performed using an interaction mode. The energy difference is then calculated using the equation: ΔE = Ecomplex − Eenzyme − Einhibitor
Results
Identification and confirmation of 4-Pregnen-21-ol-3,20-dione-21-(4-bromobenzenesufonate) (NSC 88915) as a Tdp1 inhibitor
We recently implemented a high-throughput electrochemiluminescence assay to identify small molecule inhibitors of Tdp1.31, 33 In screening the 1981 compounds from the `diversity set' of the NCI-Developmental Therapeutics Program, we identified a C21-substituted progesterone derivative, 1 (NSC 88915) (Figure 1A), which exhibited inhibitory activity against Tdp1 at low micromolar concentrations.31 The inhibitory activity was confirmed using a gel-based assay, which utilizes a 5'-[32P]-labeled single-stranded oligonucleotide comprising a 3'-phosphotyrosine (N14Y). Recombinant Tdp1 hydrolyzes the phosphodiester linkage between the tyrosine and the 3'-end of this DNA substrate, resulting in the generation of an oligonucleotide with a free 3'-phosphate end (N14P) (Figures 1B and C).13 Through the analysis of concentration versus percentage inhibition curves, we estimated the IC50 value for 1 as 7.7 ± 0.8 μM. (Figure 1D).
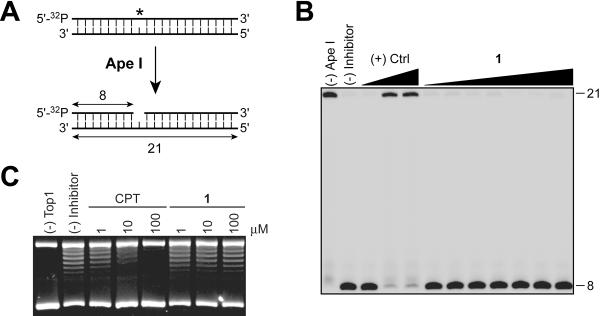
To evaluate the selectivity of 1 for Tdp1, we counter-screened 1 against two different enzymes that interact and process DNA. No evidence for inhibition of the DNA repair enzyme, apurinic/apyrimidinic endonuclease (APE1), or human Top1 was achieved in the presence of 1 (up to 100 μM, Figure 2). These data suggest that the inhibitory action of 1 on Tdp1 is independent of DNA interactions.
Figure 2.
Counter screening of 1 against apurinic/apyrimidinic endonuclease enzyme (APE1) and topoisomerase I (Top1). A) Schematic representation of the APE1 gel-based biochemical assay. APE1 hydrolyzes the DNA phosphodiester backbone 5′ of the abasic site (*) generating an 8-mer oligonucleotide. B) Representative gel showing that 1 is inactive against APE1. C) Representative gel showing that 1 is inactive in a Top1 DNA relaxation assay.
Importance of both the steroid and the phenyl sulfonyl ester moieties
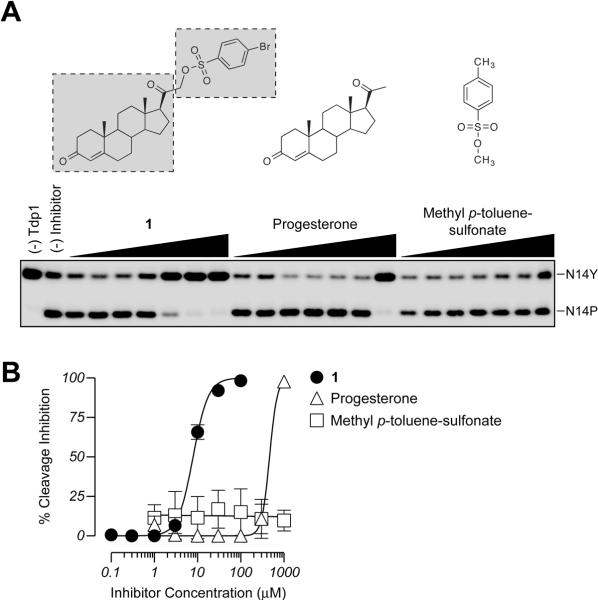
To establish the relative contribution of specific substructures toward the overall Tdp1 inhibitory activity, we deconstructed 1 into two available fragments. Progesterone and methyl p-toluene sulfonate were used to mimic the steroid and phenyl sulfonyl ester moieties, respectively (see structures in Figure 3A). While progesterone was weakly active, methyl p-toluene sulfonate remained inactive even at 1 mM concentration, failing to give an IC50 value (Figures 3A and B). Testosterone was also assayed, yet showed similar activity to that of progesterone (data not shown). Thus, both the combination of the steroid and phenyl sulfonyl ester fragments is essential for the activity of 1 against Tdp1.
Figure 3.
Both the steroid and phenyl sulfonyl ester moieties of 1 are required for Tdp1 inhibition. A) Representative gel demonstrating dose-dependent inhibition of Tdp1 by 1 and its two parts: progesterone and methyl p-toluene sulfonate (chemical structures shown above gel). B) Graphical representation of the percent inhibition of Tdp1 by 1, progesterone, and methyl p-toluene sulfonate. Each point represents the mean ± SEM for three independent experiments.
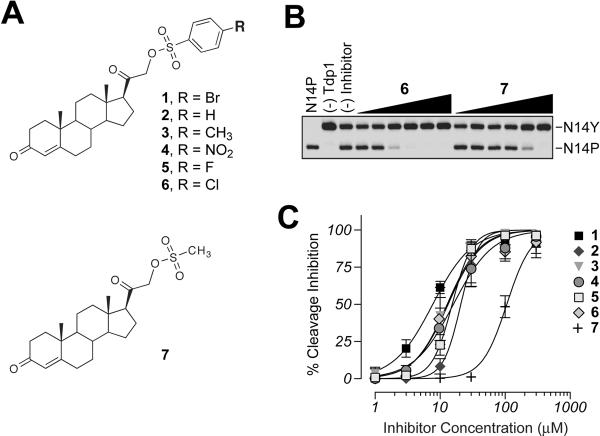
The importance of the phenyl sulfonyl ester moiety became more evident following structure activity relationship studies. Initial replacement of the para-bromide in 1 by various substituents (R = H, CH3, NO2, F, and Cl, see 2′6 in Figure 4A) resulted in minimal changes in the Tdp1 inhibition (< 3-fold change in IC50 values, Figures 4B and C). More interestingly, the mesylate analogue (7) (Figure 4A) was minimally active (Figures 4B and C). These results indicate that Tdp1 inhibition is dependent on the presence of the phenyl ring attached via sulfonyl ester bond at the C21-position of these steroid derivatives.
Figure 4.
Structure-activity relationship of analogues of 1. A) Chemical structures of the analogues tested. B) Representative gel demonstrating dose-dependent inhibition of Tdp1 by 6 and 7. C) Graphical representation of the percent inhibition of Tdp1 by 1–7. Each point represents the mean ± SEM for three independent experiments.
Inhibition of Tdp1 processing of both single- and double-stranded DNA substrates
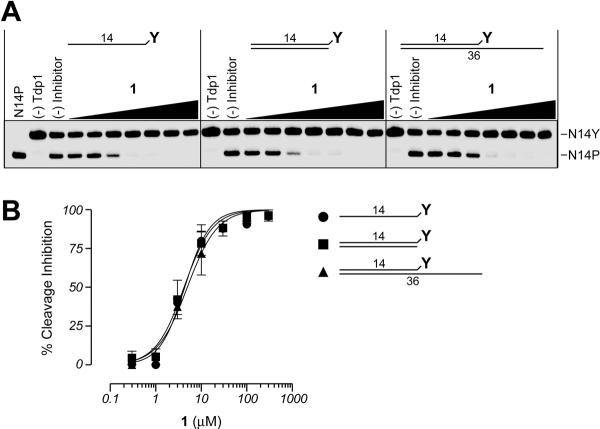
As reported previously, Tdp1 can utilize both single- and double-stranded DNA substrates 39, 40. Thus, in addition to the single-stranded N14Y substrate (Figures 1 and 3), we assayed the activity of 1 using double-stranded DNA substrates bearing a blunt or 22-base recessed 3'-phosphotyrosine terminus (Figure 5A). As demonstrated in Figures 5A and B, Tdp1 inhibition was similar with both the single- and double-stranded DNA substrates. These findings provide additional evidence that the steroid derivatives do not exert their Tdp1 inhibitory effects through binding or intercalating with the DNA substrate.
Figure 5.
The processing of both single-stranded and duplex substrates by Tdp1 is inhibited by 1. A) Representative gels showing the inhibition of Tdp1 activity by 1 using single-stranded, blunt end duplex, or 5'-overhang duplex substrates. B) Graphical representation of the percent inhibition of Tdp1 by 1 using the 3 different DNA substrates shown in A. Each point represents the mean ± SEM for three independent experiments.
Direct biophysical analysis of inhibitor binding to recombinant Tdp1 using surface plasmon resonance
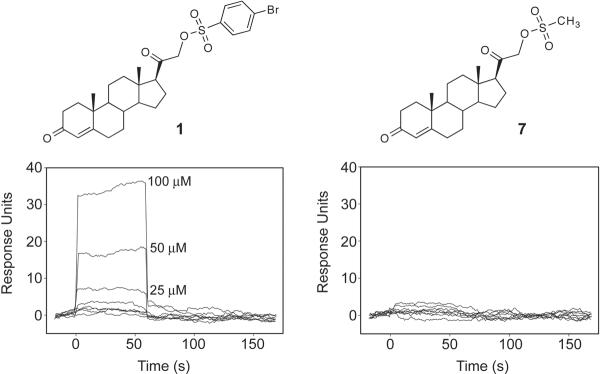
To better understand the Tdp1-inhibitor interactions and measure the affinity of 1 to Tdp1, we used surface plasmon resonance (SPR). Tdp1 was amine-coupled to a sensor chip, and the drug binding was determined in a Biacore 2000. In Figure 6 (left), the increase in resonance units from the initial baseline represents the binding of 1 to the surface-bound Tdp1. The plateau corresponds to the steady-state phase of the Tdp1-inhibitor interaction, while the decrease in response units represents the dissociation or reversibility of the interaction. No interaction with Tdp1 was detected for the weakly active inhibitor 7 (Figure 6, right). These experiments demonstrate direct binding of 1 to Tdp1.
Figure 6.
Differential binding of 1 and 7 to Tdp1 using surface plasmon resonance (SPR). SPR sensorgrams showing drug interactions with Tdp1. Ten successive 2-fold dilutions from 100 μM were tested for each inhibitor.
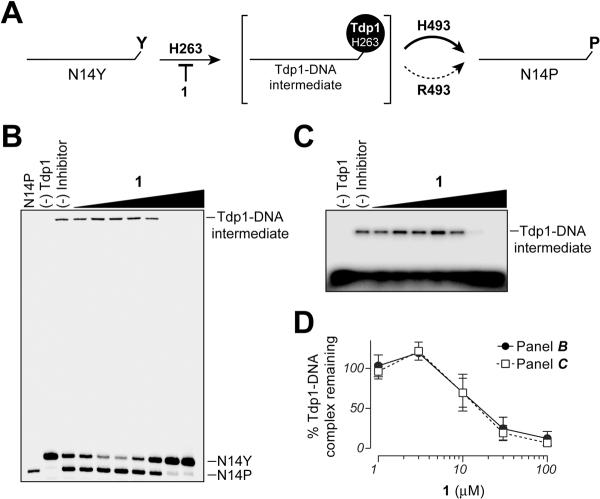
Mechanistic insights of Tdp1 inhibition by the steroid derivatives using the SCAN1 mutant
In human Tdp1 cleavage reactions two active site histidines (H263 and H493) perform two successive nucleophilic attacks to hydrolyze the phosphotyrosyl Top1-DNA bond. The first step involves an enzyme-substrate intermediate between H263 and the 3'-DNA end.19, 28, 41 The second histidine, H493 is critical for hydrolysis of this Tdp1-DNA covalent intermediate (Figure 7A). The homozygous H493R mutation associated with the recessive neurodegenerative disease SCAN1 has been shown to preferentially disrupt the second step of the Tdp1 reaction [i.e. the hydrolysis of the phosphohistidine (H263) intermediate] resulting in an accumulation of the Tdp1-DNA covalent intermediate (Figure 7A).19, 23, 42 We took advantage of the catalytic properties of the Tdp1 H493R mutant to elucidate the mode of inhibition by 1. In the absence of the Tdp1 inhibitor, the H493R mutant accumulates Tdp1-DNA complexes, as demonstrated by the appearance of DNA-labeled material in the wells of DNA sequencing gels (Figure 7B) and as a DNA-labeled slowly migrating band in protein SDS-PAGE gels (Figure 7C).32 In the presence of 1, a concentration-dependent decrease of the Tdp1-DNA complexes was observed both in DNA sequencing and protein SDS-PAGE gels (Figure 7D). Inhibition in the formation of Tdp1-DNA covalent intermediate indicates that 1 interferes with the binding of the DNA substrate to the Tdp1 active site (Figure 7A).
Figure 7.
Use of the SCAN1 Tdp1 mutant (H493R) to assess the ability of 1 to inhibit the binding of Tdp1 to DNA. A) Schematic representation of covalent intermediate accumulation of the H493R SCAN1 mutant. B and C) Representative denaturing DNA-PAGE gel and protein-SDS-PAGE gel, respectively. Both show inhibition of Tdp1-DNA complex formation by 1. D) Graphical representation of percent Tdp1 binding to DNA as shown in (B) and (C). Each point and bar represents the mean ± SEM for three independent experiments.
Molecular docking of steroid derivatives into the active site of Tdp1 and the correlation of binding free energies with inhibitory activities
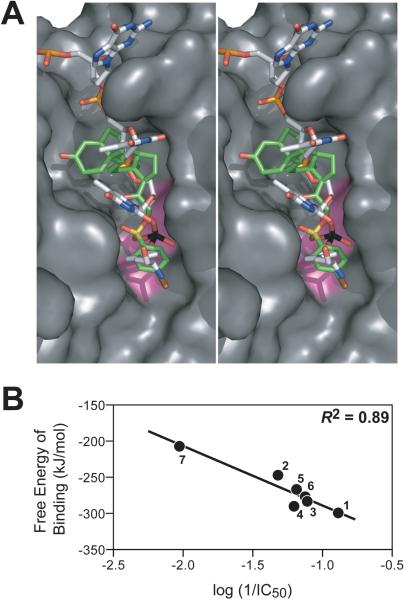
To investigate the binding mode of the steroid derivatives to Tdp1 at the molecular level, we performed docking analysis using the high-resolution structure of Tdp1, co-crystallized with a peptide-vanadate-DNA substrate mimic (PDB accession code 1NOP).29 After constructing a molecular model from 1NOP (see Materials and Methods), the eight steroid derivatives were docked into the substrate binding pocket of Tdp1. Figure 8A shows a stereo view representation of 1 and the tyrosyl-vanadate-DNA substrate mimic overlapped within the active site of Tdp1. Based on the model, the inhibitor appears to mimic the substrate/Tdp1 interaction. The vanadate participating in the 3'-phosphotyrosyl linkage of the Tdp1 substrate is mimicked by the sulfonyl ester moiety of 1, while the steroid and phenyl bromide moieties of the inhibitor are substituted for the DNA and tyrosyl portions of the Tdp1 substrate, respectively. In addition, Figure 8B provides evidence for a linear relationship between the calculated free energy of binding (docking) and the IC50 values for 1–7 (correlation coefficient R2 of 0.89).
Figure 8.
Molecular docking of inhibitors into the active site of Tdp1. A) Overlay of Tdp1 substrate and the best potential docking pose of 1 within the active site of Tdp1. The surface of the protein is shown in grey with the active site residues (H263, K265, H493, and K495) highlighted in pink; the Tdp1 substrate is colored by atom type: carbon in white, nitrogen in blue, oxygen in red, phosphorous in orange, and vanadate in black; and the inhibitor is colored by atom type: carbon in green, oxygen in red, sulfur in yellow, and bromine in brown. B) Linear correlation between the experimental activities (log 1/IC50) attained from the Tdp1 gel-based assay and the calculated free energy of binding from docking for 1–7 (see Figure 4).
Discussion
Cells have developed multiple pathways to repair damaged DNA. Dysregulation of these pathways is a primary cause of genomic instability and carcinogenesis; yet, when functioning properly, their action can reduce the effectiveness of conventional DNA-damage based anticancer therapies. For example, increased DNA repair has been shown to be a clinically important mechanism of resistance to platinum-based therapies (for review, see 43). Accordingly, the pharmacological inhibition of specific DNA repair proteins has emerged as a promising strategy to both combat the resistance and potentiate the cytotoxicity of current anticancer treatments. DNA repair inhibitors also have the potential to be used as monotherapies given that most cancer cells are deficient in one or more of the DNA damage response pathways, making them hyperdependent upon the remaining functional pathways (for reviews, see 44, 33, 45). Thus, inactivation of these compensatory repair pathways may lead to an increase of endogenous tumor-specific DNA lesions and selective killing of cancer cells, while normal cells would remain resistant owing to the redundancy in their DNA repair systems. The most noteworthy examples of this concept are the PARP inhibitors, which have shown significant therapeutic potential in BRCA1 and BRCA2-deficient tumors46, 47 and in combination with gemcitabine 48 and Top1 inhbititors.49
Cellular sensitivity or resistance to Top1 inhibitors is likely determined by the loss or increased activity of specific DNA repair pathways, respectively.50 Thus, a viable therapeutic approach to enhance the pharmacological action of Top1 inhibitors is inactivation of the pathways involved in the repair of drug-induced Top1-mediated DNA damage. Top1-DNA covalent complexes or the presumed cytotoxic lesions generated by the Top1 inhibitors are the primary substrates for Tdp1.14, 15 Tdp1 has also been shown to process 3'-phosphoglycolates, although less efficiently than 3'-phosphotyrosyl moieties.16 Thus, Tdp1 inhibitors are envisioned to enhance the antipoliferative effects of Top1 inhibitors as well as ionizing radiation. The observation that Tdp1 knockout mice are hypersensitive to camptothecins and bleomycin provides proof of principle for such strategies.23, 24 In addition, the viability and mild phenotype of Tdp1 knockout mice is indicative of a lack of severe side effects when applying Tdp1 inhibitors.23, 24 However, an alternative checkpoint-mediated nucleolytic pathway involving at least three endonuclease complexes [yMus81/yMms4 (hMus81/hEme1); yMre11/yRad50/yXrs2 (hMre11/hRad50/hNbs1); yRad1/yRad10 (hXPF/hERCC1)] has also been demonstrated through genetic analysis in yeast cells.40, 51–54 Although redundancy exists in the repair of Top1-DNA lesions, Tdp1 inhibitors have the potential to provide an increased selectivity and a broader therapeutic window for Top1 inhibitors given that defects in one or more DNA damage repair pathways are a common characteristic of cancer cells, as previously mentioned. Thus, in an effort to extend this new paradigm of targeting DNA repair for anticancer therapy, we have initiated a search for selective inhibitors of Tdp1.31, 32
To date, only a limited number of Tdp1 inhibitor chemotypes have been reported.30–33 Here, we describe the identification, synthesis, and in vitro biochemical evaluation of a new chemical family of steroidal Tdp1 inhibitors, which were identified in a high-throughput screen.31 The initial positive hit, 1, inhibits Tdp1 activity at low micromolar concentrations (Figure 1). We also demonstrate the selectivity of 1 for Tdp1 by counterscreening against other DNA processing enzymes (Figure 2). Initially, we explored the structure-activity relationship of 1 by asking whether parsing the compound into two fragments, which were related components of the larger molecule (i.e. progesterone and methyl p-toluene sulfonate, see Figure 3), maintained Tdp1 inhibitory activity. As shown in Figure 3, we found that the composite structure was much more active than the isolated fragments. Another key structural modification we examined was the incorporation of functional groups at the para-position of the phenyl ring. Our results demonstrate that a wide variety of substituents were tolerated at this position without seriously reducing the activity against Tdp1, whereas complete elimination of the para-substituent resulted in a moderate decrease in activity (~3-fold) (Figure 4). More interestingly, we found a significant reduction in the activity of the mesylate analogue (7), which suggests that the phenyl ring portion is important to the activity of this series of compounds. Overall, the para-bromide compound (1) or the initial hit from the screen was determined to be most active.
Cellular studies have been performed with compound 1 in combination with CPT in HCT116 cells. Compound 1 exhibited limited activity by itself (up to 25 μM) and only an additive effect was observed with camptothecin under these conditions (data not shown). In addition, we did not observe an enhancement in the amount of CPT-induced Top1-DNA cleavage complexes by 1 using the immunocomplex of enzyme (ICE) bioassay (data not shown). It is likely that limited cytotoxicity and lack of detectable Tdp1 inhibition by 1 in a cellular environment may be due to limited drug uptake and possibly “off target” effects. Thus, compound 1 represents a lead molecule for development of next generation agents against Tdp1 in vivo.
There are several possible sites at which a Tdp1 inhibitor could exert its inhibitory action including: (1) the DNA substrate; (2) the Tdp1 active site; (3) the interface of the Tdp1-DNA intermediate. The comparable IC50 values of 1 to inhibit Tdp1 activity toward both single- and double-stranded substrates (Figure 5), strongly suggest that the inhibitor does not target the DNA. In contrast, we provide evidence for direct binding of 1 to recombinant Tdp1 (Figure 6), yet the SPR results do not exclude the possibility of interfactial inhibition. Thus, in an effort to gain a more detailed understanding of the mechanistic basis for inhibition of Tdp1 by 1, we utilized the SCAN1 mutant. If 1 stabilizes the Tdp1-DNA intermediate, we would have detected an increase in the slower migrating bands in Figures 7B and C. However, we observed a reduction in the formation of the Tdp1-DNA complex, which is indicative of competition between the inhibitor and the substrate for the Tdp1 active site.
The active site topology of Tdp1 has been revealed by co-crystal structures of Tdp1 bound to the transition state mimic vanadate.28, 29 This structure allowed us to investigate the binding modes of the discovered inhibitors. Docking of 1 into the active site of Tdp1 results in an orientation similar to that of the vanadate-peptide-DNA substrate mimic (Figure 8). The primary interaction of 1 with the Tdp1 is through the sulfonyl ester moiety of the inhibitor, which mimics the vanadate in the crystal structure (see Figure 8, shown in yellow and black, respectively). In addition, the overlay of the phenyl ring of the inhibitor and the tyrosine residue of the Tdp1 substrate further validates its importance for Tdp1 inhibition. In contrast, the steroid portion of the inhibitor seems unorganized in the DNA binding pocket of Tdp1, which may be proposed as a reliable location for modifications in next generation inhibitors. Lastly, the binding free energies of steroidal analogues to Tdp1 were found to have a good correlation with the experimental inhibitory activities (Figure 8B). These results provide insight into the structural requirements for the activity of this class of inhibitors and will be useful in designing new steroidal derivatives as Tdp1 inhibitors. In addition, a well-defined pharmacophore has been identified on the basis of this series of inhibitors presented here, as well as other positive hits from the high-throughput screen, for the identification of new chemotypes that target Tdp1.
Supplementary Material
Acknowledgements
The wild-type Tdp1 and SCAN1 mutant Tdp1 constructs were generous gifts from Drs. J.J. Champoux (University of Washington) and Dr. H. Nash (Laboratory of Molecular Biology, National Institute of Mental Health, NIH), respectively. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations
- Tdp1
tyrosyl-DNA phosphodiesterase
- Top1
topoisomerase I
- CPT
camptothecin
- SCAN1
spinocerebellar ataxia with axonal neuropathy-1
- IC50
half maximal (50%) inhibitory concentration
Footnotes
Supporting Information Available. HPLC chromatograms and high-resolution mass spectral data of compounds 1–6. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 2.Pratesi G, Beretta GL, Zunino F. Gimatecan, a novel camptothecin with a promising preclinical profile. Anticancer Drugs. 2004;15:545–552. doi: 10.1097/01.cad.0000131687.08175.14. [DOI] [PubMed] [Google Scholar]
- 3.Takagi K, Dexheimer TS, Redon C, Sordet O, Agama K, Lavielle G, Pierre A, Bates SE, Pommier Y. Novel E-ring camptothecin keto analogues ( S38809 and S39625) are stable, potent, and selective topoisomerase I inhibitors without being substrates of drug efflux transporters. Mol Cancer Ther. 2007;6:3229–3238. doi: 10.1158/1535-7163.MCT-07-0441. [DOI] [PubMed] [Google Scholar]
- 4.Kroep JR, Gelderblom H. Diflomotecan, a promising homocamptothecin for cancer therapy. Expert Opin Investig Drugs. 2009;18:69–75. doi: 10.1517/13543780802571674. [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y. DNA Topoisomerase I Inhibitors: Chemistry, Biology, and Interfacial Inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pommier Y, Cushman M. The indenoisoquinoline noncamtothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther. 2009;8:1008–1014. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teicher BA. Next generation topoisomerase I inhibitors: Rationale and biomarker strategies. Biochem Pharmacol. 2008;75:1262–1271. doi: 10.1016/j.bcp.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 8.Marchand C, Antony S, Kohn KW, Cushman M, Ioanoviciu A, Staker BL, Burgin AB, Stewart L, Pommier Y. A novel norindenoisoquinoline structure reveals a common interfacial inhibitor paradigm for ternary trapping of the topoisomerase I-DNA covalent complex. Mol Cancer Ther. 2006;5:287–295. doi: 10.1158/1535-7163.MCT-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pommier Y, Cherfils J. Interfacial protein inhibition: a nature's paradigm for drug discovery. Trends Pharmacol Sci. 2005;28:136–145. doi: 10.1016/j.tips.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, Agama K, Nitiss JL, Redon C. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pommier Y, Redon C, Rao A, Seiler JA, Sordet O, Takemura H, Antony S, Meng L-H, Liao ZY, Kohlhagen G, Zhang H, Kohn KW. Repair of and Checkpoint Response to Topoisomerase I-Mediated DNA Damage. Mutat Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Pourquier P, Pommier Y. Topoisomerase I-mediated DNA damage. Adv Cancer Res. 2001;80:189–216. doi: 10.1016/s0065-230x(01)80016-6. [DOI] [PubMed] [Google Scholar]
- 13.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 14.Yang SW, Burgin AB, Jr., Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci U S A. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J Biol Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3' overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem. 2002;277:27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 17.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 18.El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst) 2007;6:1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst) 2006;5:1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins AJ, Subler MA, Akopiants K, Wiley JL, Taylor SM, Rice AC, Windle JJ, Valerie K, Povirk LF. In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation. DNA Repair (Amst) 2009;8:654–663. doi: 10.1016/j.dnarep.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3'-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–297. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano R, Interthal H, Huang C, Nakamura T, Deguchi K, Choi K, Bhattacharjee MB, Arimura K, Umehara F, Izumo S, Northrop JL, Salih MA, Inoue K, Armstrong DL, Champoux JJ, Takashima H, Boerkoel CF. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007;26:4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, McKinnon PJ. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barthelmes HU, Habermeyer M, Christensen MO, Mielke C, Interthal H, Pouliot JJ, Boege F, Marko D. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J Biol Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 26.Nivens MC, Felder T, Galloway AH, Pena MM, Pouliot JJ, Spencer HT. Engineered resistance to camptothecin and antifolates by retroviral coexpression of tyrosyl DNA phosphodiesterase-I and thymidylate synthase. Cancer Chemother Pharmacol. 2004;53:107–115. doi: 10.1007/s00280-003-0717-6. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Zhou S, Begum S, Sidransky D, Westra WH, Brock M, Califano JA. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer. Lung Cancer. 2007;55:303–311. doi: 10.1016/j.lungcan.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies DR, Interthal H, Champoux JJ, Hol WG. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J Mol Biol. 2002;324:917–932. doi: 10.1016/s0022-2836(02)01154-3. [DOI] [PubMed] [Google Scholar]
- 29.Davies DR, Interthal H, Champoux JJ, Hol WG. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide. Chem Biol. 2003;10:139–147. doi: 10.1016/s1074-5521(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 30.Liao Z, Thibaut L, Jobson A, Pommier Y. Inhibition of human tyrosyl-DNA phosphodiesterase by aminoglycoside antibiotics and ribosome inhibitors. Mol Pharmacol. 2006;70:366–372. doi: 10.1124/mol.105.021865. [DOI] [PubMed] [Google Scholar]
- 31.Antony S, Marchand C, Stephen AG, Thibaut L, Agama KK, Fisher RJ, Pommier Y. Novel high-throughput electrochemiluminescent assay for identification of human tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1. Nucleic Acids Res. 2007;35:4474–4484. doi: 10.1093/nar/gkm463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchand C, Lea WA, Jadhav A, Dexheimer TS, Austin CP, Inglese J, Pommier Y, Simeonov A. Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay. Mol Cancer Ther. 2009;8:240–248. doi: 10.1158/1535-7163.MCT-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dexheimer TS, Antony S, Marchand C, Pommier Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med Chem. 2008;8:381–389. doi: 10.2174/187152008784220357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handratta VD, Vasaitis TS, Njar VC, Gediya LK, Kataria R, Chopra P, Newman D, Jr., Farquhar R, Guo Z, Qiu Y, Brodie AM. Novel C-17-heteroaryl steroidal CYP17 inhibitors/antiandrogens: synthesis, in vitro biological activity, pharmacokinetics, and antitumor activity in the LAPC4 human prostate cancer xenograft model. J Med Chem. 2005;48:2972–2984. doi: 10.1021/jm040202w. [DOI] [PubMed] [Google Scholar]
- 35.Carroll FI, Philip A, Ferguson AM, Wall MF. Steroid derivatives of purine-6-(1H)-thione (1a–c) J Heterocycl Chem. 1968;5:805–811. [Google Scholar]
- 36.Simons SS, Pons M, Johnson DF. α-Keto mesylate: A reactive, thiol-specific functional group. J Org Chem. 1980;45:3084–3088. [Google Scholar]
- 37.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohamadi N, Richards GJ, Guida WC, Liskamp R, Lipton M, Caulfield C, Chang G, Hendrickson T, Still WC. Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem. 1990;11:440–467. [Google Scholar]
- 39.Raymond AC, Staker BL, Burgin AB., Jr. Substrate specificity of tyrosyl-DNA phosphodiesterase I (Tdp1) J Biol Chem. 2005;280:22029–22035. doi: 10.1074/jbc.M502148200. [DOI] [PubMed] [Google Scholar]
- 40.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 41.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci U S A. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 43.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 44.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 46.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 47.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 48.Jacob DA, Bahra M, Langrehr JM, Boas-Knoop S, Stefaniak R, Davis J, Schumacher G, Lippert S, Neumann UP. Combination therapy of poly (ADP-ribose) polymerase inhibitor 3-aminobenzamide and gemcitabine shows strong antitumor activity in pancreatic cancer cells. J Gastroenterol Hepatol. 2007;22:738–748. doi: 10.1111/j.1440-1746.2006.04496.x. [DOI] [PubMed] [Google Scholar]
- 49.Smith LM, Willmore E, Austin CA, Curtin NJ. The novel poly(ADP-Ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks. Clin Cancer Res. 2005;11:8449–8457. doi: 10.1158/1078-0432.CCR-05-1224. [DOI] [PubMed] [Google Scholar]
- 50.Pommier Y, Barcelo J, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao Z, Seiler J, Zhang H, Marchand C, Agama K, Redon C. Repair of topoisomerase I-mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vance JR, Wilson TE. Repair of DNA strand breaks by the overlapping functions of lesion-specific and non-lesion-specific DNA 3' phosphatases. Mol Cell Biol. 2001;21:7191–7198. doi: 10.1128/MCB.21.21.7191-7198.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci U S A. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Pouliot JJ, Nash HA. The role of TDP1 from budding yeast in the repair of DNA damage. DNA Repair (Amst) 2004;3:593–601. doi: 10.1016/j.dnarep.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Deng C, Brown JA, You D, Brown JM. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.