Abstract
The emergence of drug-resistant human immunodeficiency virus type I (HIV-1) strains presents a challenge for the design of new drugs. Anti-HIV compounds currently in use are the subject of advanced clinical trials using either HIV-1 reverse-transcriptase, viral protease, or integrase inhibitors. Recent studies show an increase in the number of HIV-1 variants resistant to anti-retroviral agents in newly infected individuals. Targeting host cell factors involved in the regulation of HIV-1 replication might be one way to combat HIV-1 resistance to the currently available anti-viral agents. A specific inhibition of HIV-1 gene expression could be expected from the development of compounds targeting host cell factors that participate in the activation of the HIV-1 LTR promoter. Here we will discuss how targeting the host can be accomplished either by using small molecules to alter the function of the host’s proteins such as p53 or cdk9, or by utilizing new advances in siRNA therapies to knock down essential host factors such as CCR5 and CXCR4. Finally, we will discuss how the viral protein interactomes should be performed to better design therapeutics against HIV-1.
Introduction
The human immunodeficiency virus type I (HIV-1) has persisted for decades despite the use of Highly Active Anti-Retroviral Therapy (HAART) in treating infection. HAART therapy can suppress HIV-1 infections but cannot cure viral infections. In addition, long term administration of HAART therapy has highlighted just how quickly the virus can generate resistant mutants and escape the drug compounds that are currently used to target viral proteins [1–2]. The retrovirus mutates rapidly because its reverse transcriptase enzyme possesses a very low fidelity (compared to other DNA polymerases) and creates de novo mutations in approximately a third of each generation of new viruses. In fact, multiple resistant strains of HIV-1 are likely to arise within a single patient during the course of their infection. Often these resistant strains replicate better in the presence of a particular HAART compound [3]. Consequently, there is a growing effort to develop new anti-HIV-1 therapies that do not target the viral proteins, but instead target the host proteins which aid in viral replication. Currently, HAART therapies may include the drug maraviroc, which targets the host’s CCR5 surface receptor. Maraviroc forces the CCR5 receptor to misfold, which subsequently prevents gp120-mediated viral fusion. Even though resistance to maraviroc has been observed in cell culture, very few clinically isolated strains of HIV-1 have shown resistance to the drug itself. Clinical isolates instead showed a shift in tropism from R5 tropism to dual-tropism and X4 tropism [1]. This drug serves as one example of how much more difficult it is for the virus to adapt when host proteins are the target instead of viral proteins.
The purpose of this review is to discuss current and potential anti-HIV therapies that target the host instead of the virus, with the intent of making the host an unfit environment for viral replication. Targeting the host can be accomplished either by using small molecules to alter the function of the host’s proteins such as p53 or cdk9, or by utilizing new advances in siRNA therapies to knock down essential host factors such as CCR5 and CXCR4. For example, p53 and its downstream effector p21/waf1 are known to be suppressed during HIV-1 infection. Small molecules that can activate these proteins and overcome viral suppression hold promise as potent HIV-1 inhibitors [4]. Similarly, HIV-1 transcription depends on the action of cyclin-dependent kinases (cdks) such as cdk2 and cdk9. Using small molecules to block the function of these cdks can inhibit the transcription of HIV-1 [5–6]. Finally, since small molecules may eventually prove insufficient for side-stepping the problem of viral hyper-mutability, colleagues in the field have investigated the possibility of using siRNA to knock down target genes. One of the most advanced trials of anti-HIV siRNA therapy targets the CCR5 gene in an attempt to create a population of HIV-1 resistant lymphocytes [7]. Additionally, siRNA screens have been performed in search of novel essential host factors. New drugs and siRNA therapies seek to specifically target the host factors that are necessary for viral replication but not host survival [8].
2.1: p53: “The Guardian of the Genome”
The tumor suppressor protein p53 plays a central role in protecting the integrity of the genome, thus often being referred to as the “guardian of the genome” [9]. While p53 is present at low levels under unperturbed conditions, it becomes rapidly activated and stabilized upon induction by a number of stimuli, including the use of compounds that cause DNA damage[10–15]. Once it has been activated, p53 is involved in a variety of physiological events. These include inducing apoptosis by both transcription-dependent and transcription-independent mechanisms [16–21], as well as inducing cell cycle arrest at both the G1/S [22–26] and G2/M checkpoints [27–31]. Some well known transcriptional targets of p53 are p21/waf1, MDM2, 14-3-3, GADD45, p53-R2, FAS, PIG3, IGF-BP3, Killer/DR5, AIP1, which are involved in cell cycle control, modulation of DNA repair, differentiation, senescence, and control of p53 stability/activity [32–37].
Although there are numerous reports investigating p53 activation and stabilization due to cellular stress, the exact mechanism is still far from being fully understood. The classical model of p53 regulation stipulates that p53 is stabilized through phosphorylation, allowing p53 binding to its responsive promoters in a sequence specific manner, followed by the induction of target genes resulting in downstream events [38]. p53 can be phosphorylated by numerous kinases including ATM, ATR, DNA PK, and mTOR on various residues [38]. Post translational modifications of p53 are extremely important for its activation and stabilization. p53 has been shown to be phosphorylated, acetylated, methylated, ubiquitinated, sumoylated, glycosylated, ADP-ribosylated, and neddylated [38–39]. Some of these modifications are known to influence the interaction between p53 and the E3 ubiquitin ligase, MDM2, which is a major mechanism for controlling p53 through inducing proteasomal degradation. Interestingly, MDM2 is also a transcriptional target of p53, resulting in a negative feed-back loop for p53 expression [40].
Among the various post-translational modifications, phosphorylation is the most extensively characterized. There are eighteen known phosphorylation sites on p53 to date; with eleven located within the transactivation domain/Proline-rich domain, three within the DNA-binding domain, one with the tetramerization domain, and the remaining three within the C-terminal domain [39]. p53 is phosphorylated at many of these sites upon both genotoxic and non-genotoxic stresses. Of particular interest is the phosphorylation of Ser15, which is generally considered to be activated in response to different stress signals [41–45]. Ser15 phosphorylation is viewed as a priming event necessary for other post-translational modifications. This phosphorylation has been shown to stimulate the recruitment of p300, CBP, and p/CAF, which function to acetylate the C-terminus of p53, resulting in decreased ubiquitination and proteasomal degradation [46]. Other phosphorylation sites of p53 at the N-terminus such as Ser20 and Thr16 are phosphorylated subsequent to Ser15. Phosphorylation at all three of these sites induces C-terminal acetylation, which has been implicated in p53 stabilization. Stabilization of p53 is accomplished primarily via interruption of the interaction between p53 and the ubiquitin ligase MDM2 [47–51]. The sum total of these events is that p53-phospho-Ser15 is now more stable and is able to transcribe its downstream targets.
2.2: p53 and HIV-1 infection
It has been demonstrated that the p53 pathway plays an important role in HIV infection [52–53]. Wildtype p53 results in the inhibition of HIV-1 transcription from the viral Long Terminal Repeat (LTR) [54–55], which can be relieved by overexpression of Tat [55]. In contrast, mutant forms of p53 can activate LTR transcription [55–56]. p53 has not been shown to bind directly to the LTR, but rather is thought to be recruited to the promoter through its binding with TFIID [55]. It was later discovered that Tat binds directly to p53 through the p53 dimerization domain (aa 341–355) [57]. This interaction leads to two outcomes, inhibition of Tat transcription by p53 and downregulation of p53 dependent transcription by Tat [58]. Thus, it would be advantageous to keep p53 levels low in order for optimal viral transcription and replication to occur. The consequences of p53 and HIV-1 Tat binding has been thoroughly investigated by other researchers. As mentioned above, p53 acetylation influences its activation. Specifically, p300/CBP and p/CAF acetylate residues Lys373/Lys382 and Lys320 of p53 [59–61]. Tat competes with p53 binding to p300/CBP resulting in decreased p53 acetylation, p53 transcription and responsiveness to DNA damage [62]. Collectively these studies demonstrate the influence of p53 on Tat dependent transcription and also the potential implications of p53 inactivation for HIV-1 associated malignancies.
2.3: p21/waf1 and its role in HIV-1 biology
p21/waf1 is a well known transcriptional target of p53, having important roles in cell cycle checkpoints, differentiation, and cellular senescence [63–64]. Its ability to inhibit cell cycle progression is multifaceted. First, p21/waf1 is a cdk inhibitor, binding to cdk through a N-terminal domain and to its cyclin partner utilizing residues present in both its N and C-terminus [65]. p21/waf1 also associates with PCNA, blocking DNA synthesis that is required for the S phase of the cell cycle. In the cytoplasm, p21/waf1 can influence anti-apoptotic and pro-survival functions. Cytoplasmic p21/waf1 is important for the formation and stabilization of cyclin D/cdk4, 6 complexes, which are required for progression through the G1 phase of the cell cycle [66–68]. p21/waf1 inhibits Fas-mediated apoptosis through complexing with procaspase 3 and inhibiting the cleavage of procaspase 3 to its active form [69–70]. Nuclear p21/waf1 can influence transcriptional responses through acting as a transcriptional cofactor as well as regulating DNA methylation [71–72]. p21/waf1 has a relatively short half-life, which is regulated by its subcellular localization as well as its interacting partners. Interestingly, many of these partners actually share the same binding sites on p21/waf1 [65], indicating that there is a dynamic relationship between p21/waf1 protein interactions and function.
Previous research has established a model where p53 inactivation by HIV-1 Tat results in the loss of p21/waf1 induction following DNA damage, thus allowing increased cyclin E/cdk2 activity [73]. Elevated cyclin E/cdk2 activity results in loss of the G1/S checkpoint and increased virus production. In addition, pharmacological cdk inhibitors (PCIs) that mimic endogenous cdk inhibitors such as p21/waf1 inhibit HIV-1 replication [74–75]. p21/waf1 has also been investigated as a molecular barrier for HIV-1 infection of stem cells [76]. Hematopoietic stem cells were previously demonstrated to be highly resistant to HIV-1 infection [77–79]. However, when p21/waf1 was knocked down using siRNAs, the stem cells became highly susceptible to HIV-1 infection. This effect was specific as silencing of other p21/waf1 related proteins, p27 and p18 did not affect HIV-1 infection. Therefore, p21/waf1 was suggested to be a possible restriction factor, similar in function to the TRIM5 and APOBEC3G proteins [80–84]. Along these lines, high-titer infection of HIV-1 in T-cells results in a loss of p21/waf1 [73], further indicating the restrictive effect of p21/waf1 on HIV-1 infection.
2.4: p53 activating compounds
Numerous small molecules, such as leptomycin B, actinomycin D, and 9-aminoacridine (9AA) are able to efficiently reactivate p53 in some cancer cell lines [85–87]. Therefore, the restoration of p53 function may provide new ways for combating cancers and virus infection where this pathway is impaired or sequestered [4, 88–89]. There are also a number of compounds such as CP-31398, WR1065, and PRIMA-1 that have the ability to activate mutant p53, which holds promise for treatment of cancers with p53 mutations [90]. There has been an active search within the cancer community to identify small molecule compounds that would inhibit the MDM2-p53 interaction, as this would be an effective way to stabilize p53. A number of compounds have been studied including, chalcones, Chlorofusin, and the Nutlins [91]. The Nutlins are the most promising MDM2-p53 disrupting compounds as they are very selective and potent with an in vitro IC50 of 100–300 nM [92–93]. We have tested a number of p53 activating compounds including actinomycin D, Nutlins, and (9AA), but out of these tested compounds only 9AA showed a selective effect on HIV-1 infected cells. To our knowledge, 9AA is the only p53 activating compound that has been used to inhibit HIV-1 replication in infected cells [94].
9AA was originally identified as an anti-bacterial agent, but more recently has gained notice as a potential treatment for cancer, viral, and prion diseases [87, 95–96]. Unlike some of the other p53 activating compounds, such as Nutlins, the mechanism of p53 activation by 9AA is not known. It was initially thought that 9AA was toxic and induced p53 through a DNA damage response caused by DNA intercalation and possible topoisomerase II poisoning [97–99]. However, multiple studies have now demonstrated that 9AA can be utilized in a selective manner, especially for virally infected cells. In fact, no toxicity was observed in uninfected cell lines or PBMCs when up to 20 μM 9AA was utilized [100]. In addition, an independent group showed that 9AA treatment did not induce phosphorylation of histone H2A.X or activate the DNA response kinases ATM or ATR, all of which are indicators of DNA damage [87]. 9AA was not found to poison topoisomerase II as had been previously suggested [87]. Therefore, 9AA does not activate p53 through the classical DNA damage response pathway. There have been a number of studies aimed at identification of the molecular mechanism of 9AA treatment in cancer cells, such as renal carcinoma and T-cell leukemia cells. These studies have indicated that 9AA treatment results in both NF-κB inhibition and p53 activation, with NF-κB inhibition being upstream of p53 activation [87, 94, 100]. In HTLV-1 infected cells 9AA induced cell death is dependent on p53, as p53 siRNA blocks cell death [100]. Finally, a recent study from Guo et al. demonstrated alterations in the AKT/PI3K pathway upon 9AA treatment, which may contribute to p53 and NF-κB alterations [101].
Treatment with 9AA has also proven to reactivate the p53 and p21/waf1 pathway in HIV-1 infected cells [94]. This was demonstrated through increased phosphorylation of p53 on Ser15 and increased levels of p21/waf1 protein. In addition, p53 phosphorylated on Ser15 was no longer found in complex with Tat, freeing p53 from Tat inhibition. Importantly, virus replication was found to be inhibited in HIV-1 infected PBMCs by 9AA in a dose-dependent manner. Recently, we have also demonstrated that 9AA treatment inhibits Tat dependent transcription through the inhibition of cdk9 binding to the viral LTR (unpublished results). 9AA inhibition of LTR transcription was specific as other 9AA derivatives (2-aminoacridine, 4-aminoacridine, and acridine hydrochloride) did not display transcription inhibition. In addition, we observed for the first time p21/waf1 in complex with p-TEFb (cyclin T1 and cdk9) in vitro, suggesting a role of p21/waf1 in HIV-1 transcription and 9AA mediated inhibition of viral transcription (unpublished results).
While there are a number of studies focused on determining 9AA’s mechanism of action, none have identified a protein that is specifically bound by 9AA, which is critical to understanding its biological effects. One method to determine 9AA binding partners would be to couple 9AA to immobilized beads to be used as a matrix for affinity chromatography. This method has been used successfully with the cdk inhibitor, Cyc202 [102]. However, for this method to be applied, sites on 9AA that are critical for its mechanism of action must not be blocked by the attachment to the beads. We and other laboratories are currently performing studies to determine the functional groups of 9AA that are critical for its mechanism of action. Thus far, through the use of various 9AA derivatives, we determined that the amino moiety of 9AA is critical for the observed transcriptional inhibition (unpublished data). Future studies will focus on determining additional moieties that are critical for its activity and elucidating the directprotein target of 9AA.
3.1: Targeting cdk and cyclin proteins
The progression of a cell through the cell cycle is strongly regulated by the sequential activation and inhibition of a group of proteins known as cyclins and cyclin dependent kinases. When the ATP dependent serine/threonine kinase catalytic cdk subunit forms a heterodimeric complex with its regulatory cyclin subunit, the resulting complex can phosphorylate key cellular substrates. These substrates in turn regulate not only cell cycle progression, but also transcription, neuronal differentiation, and transcript splicing. Depending on the phase of the cell cycle, specific cyclins are expressed in an oscillating fashion such that the stably expressed cdks can combine with variable cyclin partners. This variable pairing of partners allows the cell to respond to molecular signals during growth and development [103]. The sequential phosphorylation events induced by cdk/cyclin complexes assist in the progression of the cell cycle through key molecular “checkpoints” found primarily at the interphase between cell cycle stages, i.e. G1/S and G2/M. In general upon entry of a cell into the cell cycle at G1, cyclin D is expressed in response to extracellular signals, and consequently complexes with cdk4. This G1 regulatory phase complex phosphorylates the Retinoblastoma protein, Rb, which acts as a tumor suppressor by binding to E2F transcription factor complexes and therefore suppressing the expression of E2F-mediated gene products [103–104]. The phosphorylation of Rb by cdk4/cyclin D inactivates Rb, therefore removing Rb from E2F dependent promoters, allowing for the binding of E2F and subsequent activation of E2F responsive gene products such cyclin A and cyclin E. The association of the newly expressed cyclin E with cdk2 results in the clearance of the G1/S cell cycle checkpoint and the subsequent passage of the cell into S phase. The expression of these cyclins and other proteins needed for DNA replication are critical for the cell’s progression into S phase [103]. G1 associated cdk/cyclin complexes also target S phase inhibitors for proteasomal degradation following ubiquitination. Additionally, multiple cellular events can trigger the arrest of the cell cycle at S phase, most of which are precipitated by DNA damage and halt cell cycle progression until the damage is repaired [103]. Progression through S phase can be controlled by cdk2/cyclin A by activating the CDC6 gene, a crucial initiator of synthesis [105]. Following successful replication, the G2/M checkpoint is regulated by the phosphorylation of the APC complex by the cdk1/cyclin B complex, which acts to break down the nuclear envelope in preparation for the mitotic prophase.
In addition to regulation of the cell cycle, cdk/cyclin complexes also play a key role in the regulation of transcription, primarily by phosphorylating RNA Polymerase II (RNA Pol II) along its C-terminal domain (CTD). Transcriptional initiation and elongation require that the CTD becomes hyperphosphorylated by various complexes that contain cdks. For example, the initiation factor TFIIH/CAK is composed of cdk7, cyclin H, and MAT1 while the elongation factor p-TEFb is composed of cdk9 and cyclin T1 [6, 106].
The highly diverse and interactive network of cdks and their associated cyclins are regulated by a large number of mechanisms, including endogenously expressed cdk inhibitors. Regulating these kinases is important in maintaining in both cell cycle progression and gene transcriptional events. To this effect, it is not surprising that a large amount of cell cycle dysregulation is seen in disease states such as cancer and viral infections, where the cell is driven towards unnatural proliferation or senescence. These types of disease states can be acquired either by the activation or inhibition of cdk/cyclin complexes which control cell cycle progression or the expression of certain genes [25, 103]. Consequently, the development of PCIs is an attractive means for controlling the specific cdks which aid either tumorigenesis or viral infection.
Endogenous cellular cdk inhibitors act as negative regulators of the cell cycle through the inhibition of the cdk/cyclin kinase activity. These cdk inhibitors can be classified into two families; the INK family which inhibits cdk4 and cdk6, and the CIP/KIP family which inhibits all cdks that associate with cyclin A, E, D, and L [107]. The INK family is composed of p16INK4a, p15INK4c, and p19INK4d and acts to specifically inhibit the association of cyclin D with either cdk4 or cd6 [108–109]. The representative member of the INK4 family is p16INK4a which has most recently has been shown to share the same transcript as p19INK4d. The CIP/KIP family, which is composed of p21Cip1/waf1, p27Kip1, and p57Kip2, acts to negatively regulate the cell cycle progression through the G1 phase [109].
3.2: Natural cdk inhibitors and pharmacological cdk inhibitors
The members of the CIP/KIP family of cdk inhibitors have been linked to cell cycle regulation and dysregulation in a variety of cancerous and infectious phenotypes. p27 is overexpressed during quiescent stages of the cell cycle and is downregulated upon entry into the cell cycle. Meanwhile p57, which is regulated by Notch/Hes1, MyoD, and p73, appears to be the only cdk inhibitor that is required for embryonic development. In contrast the protein p21/waf1 has been well characterized as a cofactor in the DNA damage pathway. Stable p53 induces the expression of p21/waf1 which then leads to cell cycle arrest. p21/waf1 is also involved in transcriptional regulation in that it can inhibit the activity of E2F1, c-Myc, and STAT3 as well as activate the p300/CREB-binding protein (CBP) complex to induce transcriptional activation of certain genes. In particular, p21/Waf1 interacts with the cdk2/cyclin E complex at the G1/S checkpoint and inhibits further progression into the S phase [73]. As a result of its functionality, p21/waf1 selectively inhibits most cdk/cyclin complexes observed during G1/S, including cdk2, 3, 4 and 6. However, p21/waf1 does not associate with cdk7/cyclin H and only weakly inhibits cdk1/cyclin B [73]. The loss of the p21/waf1 cell cycle regulator has been associated with manipulation of p53 by Tat, followed by failure to halt at the G1/S checkpoint, increased entry into S phase, and eventual apoptosis [73]. This checkpoint loss also results in increased Rb phosphorylation and increased activity of cdk2/cyclin E, which promotes unregulated viral transcription and viral progeny formation [73]. Again, all of these naturally occurring cdk inhibitors are often dysregulated by both cancer and viral infections. The study of these endogenous cdk inhibitors should allow for the development of chemicals and synthetic compounds that mimic the inhibition seen in vivo.
The frequent dysregulation of cdks and cdk inhibitors in disease states has highlighted their importance in tumorigenesis and viral infections. The development of pharmacological cdk inhibitors as therapeutic agents holds obvious potential for treating diseases. PCIs can be designed as non-specific or specific inhibitors of particular subsets of cdks. Although all PCIs are flat, heterocyclic, low molecular weight compounds, they can be classified according to their structure (purine or non-purine) as well as by their level of selectivity for the cdk of interest. These compounds are designed to structurally interfere with and competitively inhibit the catalytic-ATP binding sites of the cdk proteins. Most of the PCIs to date have been designed around the scaffold of the structure of cdk2, although the structure of cdk9 complexed with the PCI Flavopiridol was recently resolved [110]. The two most studied PCIs are Roscovitine and Flavopiridol, which inhibit cdk1, 2, 5, 7, 9 and cdk1, 2, 4, and 9 respectively [111–114]. The use of these compounds as therapeutic agents is appealing due to their low IC50, the amount of the compound needed to inhibit a target by 50%. Roscovitine is an effective inhibitor of all of its target cdks at an average IC50 of less than 1 μM. On the other hand, Flavopiridol inhibits cdk9 at an IC50 of 3 nM, a concentration approximately 10 times lower than the IC50 for the other cdks inhibited by this PCI [113–115]. Due to the success of Roscovitine and Flavopiridol, second generation PCIs with a higher potency and specificity have been developed based on these two aforementioned drugs. Although PCIs have been important in the development of cancer therapeutics, no clinical trials have been initiated to assess the effect of PCI’s on the course of HIV-1 infections.
3.3: cdks in HIV-1 infection
As described previously, Tat interacts with an RNA hairpin-like loop structure at the 5′-LTR of the viral transcript, known as the Transactivating Regulatory Element (TAR). Once bound to TAR, Tat carries out its role as the viral transactivator primarily through its interaction with cellular kinases (Tat-associated kinases, TAKs), which includes multiple cdk/cyclin complexes. cdk2/cyclin E is critical for the Tat-mediated activation of HIV-1 transcription. Specifically, the phosphorylation of cdk7 by cdk2/cyclin E in the early elongation complex is important for the cdk7-containing complex, T-cell-derived kinase (TTK). Tat recruits cdk2/cyclin E to the promoter to assist in the phosphorylation of the RNA Pol II CTD, and it has been shown that the loss of cdk2/cyclin E blocks Tat-dependent viral transcription [5, 116]. Additionally, cdk2 has also been shown to directly phosphorylate Tat both in vitro and in vivo on residues Ser6 and Ser46, both of which are critical for Tat’s transactivation activity [5, 116]. However, cdk2 is not essential for normal host cell proliferation, therefore making the inhibition of cdk2/cyclin E in HIV-1 infected cells an attractive therapeutic target.
In addition to cdk2, both cdk7 and cdk9 are known to important during HIV-1 transcription. cdk9 can phosphorylate the CTD at Ser2, whereas cdk7 can phosphorylate the CTD at Ser5. However in the presence of Tat, cdk9 can preferentially phosphorylate both residues, suggesting that cdk7 may not be necessary at all times for Tat-dependent transactivation [117]. p-TEFb is present in the cell as both a small and a large complex. The large complex contains a 7SK small nuclear RNA (snRNA) and the Hexamethylene bisacetamide-induced protein 1 (HEXIM1), both of which associate with variable cellular proteins [118]. In uninfected cells, the small p-TEFb complex associates with Brd4, which is necessary to activate transcription. However, in HIV-1 infected cells Tat can substitute for Brd4 to assist in the recruitment of p-TEFb to the viral LTR. An additional aspect of the binding of the Tat-p-TEFb complex to TAR is the autophosphorylation of cdk9 on its C-terminus, thereafter aiding in the nuclear localization of cdk9. Recent studies have also suggested that cdk9 is acetylated by the acetyltransferases GCN5 and p/CAF, resulting in a decrease in both kinase activity and transcriptional activity within HIV-1 infected cells [119].
3.4: Natural and pharmaceutical cdk inhibitors in HIV-1 infection
The first generation PCI Flavopiridol has been shown to be an effective inhibitor of cdk9 and the p-TEFb complex, especially in the context of HIV-1 infected cells [115]. As the interaction of HIV-1 Tat with the cdk9/cyclin T1 complex has been extensively characterized as critical for viral transactivation, the specific inhibition of this protein-protein interaction presents as an attractive drug target. This PCI binds to the catalytic ATP binding site on cdk9, forming a noncompetitive interaction with the enzyme and promotes a conformational change that prevents the binding of ATP by cdk9 [110, 120]. Flavopiridol inhibits HIV-1 transcription in infected cells at an IC50 of 1–10 nM and is required at a much higher concentration to inhibit normal cellular transcription, indicating specificity for viral transcription [120]. The inhibition of cdk9 in virally infected cell lines by either Flavopiridol or siRNA against both cdk9 and cyclin T1 has resulted in decreased viral replication and Tat transactivation without affecting normal phosphorylation of RNA Pol II, cellular transcription, or cellular viability [121–122]. The effectiveness of Flavopiridol was also determined in primary Peripheral Blood Lymphocytes (PBLs) infected with HIV-1, where viral replication was inhibited with minimal effects on T-cell activation and DNA synthesis [123]. The development of second generation cdk9 inhibitors based on the structure and efficiency of Flavopiridol, can effectively become potent anti-HIV-1 compounds and potential therapeutics.
Another inhibitor that binds to the ATP pocket of cdks is CYC202(R-roscovitine), a PCI derived from Roscovitine. We previously showed that CYC202 effectively inhibited wildtype and resistant HIV-1 mutants in T-cells, monocytes, and PBMCs at a low IC50. Interestingly, the cdk2/cyclin E and cdk9/cyclin T1 complexes were observed to load onto the HIV-1 genome in vivo and treatment with CYC202 was able to inhibit the uploading of these cdk/cyclin complexes onto HIV-1 DNA [124].
Finally, there is another class of cdk/cyclin inhibitors that may prove to be significant in the future; Tat peptide derivatives that bind to cdks which normally load onto HIV-1 promoter. We have previously shown that a Tat peptide (41/44) from the HIV-1 core domain can inhibit HIV-1 gene expression and act as a replication inhibitor [125]. A shorter version of this peptide was also capable of inhibiting the kinase activity of cdk/cyclin in vitro, as well as inhibiting the virus replication in vivo in a new humanized stem cell animal model. The mechanism of inhibition was attributed to dissociation of the cdk/cyclin complex needed for the transcription of HIV-1 transcripts [126].
4.1: siRNA: mechanism and therapeutic applications
Some of the most promising new experimental treatments in development are based upon the discovery of RNA-mediated interference (RNAi). In just the past few decades this discovery has blossomed into an indispensible technology for researchers and is rapidly progressing as a means of treating diseases in clinical trials [127]. This technology uses small duplexes of RNA (called siRNAs) to silence the expression of specific genes in a sequence specific manner. The mechanism behind effective RNAi therapy rests on manipulating the microRNA pathway in mammalian cells. In mammalian cells, microRNAs are small ~21bp single-stranded RNA molecules that hybridize to mRNA transcripts and direct the cell to silence genes either at the translational or transcriptional level [128]. microRNAs originate from stem-loop structures in endogenous transcripts which are processed in the nucleus by the Drosha ribonuclease. On the other hand, siRNAs are defined as originating from double-stranded RNA molecules found in the cytoplasm. Both microRNAs and siRNAs can exist as short hairpin RNA molecules prior to processing by the Dicer ribonuclease. Once Dicer processes them into small single-stranded effector molecules, they are transferred to one of the Argonaute proteins, which then use the small RNA molecule as a guide to target and suppress transcripts with complimentary sequences. However, exogenous mature siRNAs duplexes can be delivered into a cell and directly associate with Argonaute proteins without the need for processing by either Drosha or Dicer [128–130]. Presently, anti-HIV-1 therapies based on RNAi are experimental, as researchers are still investigating how best to deliver siRNAs to the cell as well as which targets are the most crucial for HIV-1 replication. The following discussions will relate the current state of RNAi based therapies in general and their possible application towards endowing patients with resistance to HIV-1 infection.
4.2: siRNA treatments for viral infections
The first topic of any experimental therapy must be its feasibility. While RNAi is an incredibly powerful tool and possesses the potential to combat congenital diseases and viral infections, there remain questions about how to safely and effectively deliver siRNA molecules to a target cell. The earliest tests of feasibility for siRNA-based therapies were carried out successfully in mouse models. Specifically, liposomes carrying siRNA duplexes were delivered via tail vein injection into mice in danger of acute liver failure [131–132]. Mice suffered from acute liver failure due to either viral infection or autoimmunity, both of which induced hepatocyte apoptosis via Fas ligand signaling. The injection of siRNAs alleviated the symptoms of liver failure and fibrosis by administering siRNA directed against Fas [132] or its downstream effector, caspase-8 [131]. Efficient knockdown of both Fas and caspase-8 prevented apoptosis of hepatocytes, thereby preventing necrosis and the subsequent invasion of fibroblasts into the lesion. A similar feasibility trial used siRNA to target the Hepatitis B virus (HBV) in acutely infected mice [133]. Coinjection of the viral plasmid alongside anti-HBV siRNAs resulted in a significant drop in HBV antigens and viral DNA found in the mouse liver and sera. Following the success of RNAi therapy in mouse models, RNAi-based therapies are currently being tested in non-human primate models and, in one case, clinical trials in humans [127]. There have already been promising results from two different trials of RNAi-based therapies in primate models [134–135]. In two independent studies, siRNA was delivered to primates via intravenous delivery of RNA-liposome complexes. In one investigation, Zimmermann et al. used siRNAs delivered by intravenous injection to knock down the endogenous ApoB protein in cynomolgus monkey liver cells [134]. Independently, Yokota et al. demonstrated that siRNAs encapsulated within cationic liposome complexes can suppress or halt the replication of GBV-B, a primate hepatitis virus found in marmosets [135]. The investigators showed that GBV-B replication fell in response to increasing doses of siRNA treatments. At the highest tested dose of injected siRNAs (5mg/kg), the investigators observed a complete absence of viral DNA in the monkeys’ sera.
Despite these early successes with RNAi-based therapies, there are still a few complications that arise when attempting to deliver double-stranded RNA molecules in vivo. The delivery of siRNAs or siRNA precursor hairpins (shRNA) can have several adverse side effects such as the induction of the type-I interferon (IFN) response [136–137], saturation of the microRNA enzymes [138], and off-target effects [139–140]. Double-stranded RNA can be detected by TLR8, TLR7, TLR3, all of which respond by initiating the cell’s innate IFN response and halting protein translation [141]. Indeed, an IFN response was observed by Yokota et al., but scrambled siRNA controls inducted greater IFN responses without suppressing viral replication [135]. The conclusion drawn was that, while the IFN response may aid in fighting infection, only targeted siRNA treatments specifically inhibited GBV-B. However, it has been demonstrated that a chemical modification (nucleoside 2′-O-methylation) to the 5′ ends of siRNAs can avoid the induction of the IFN response [137]. Once present inside the target cells, siRNAs can potentially saturate the endogenous microRNA-related proteins such as Dicer, exportin-5, and Ago2. Saturation and competition with endogenous microRNAs was suggested as a cause of mortality in a mouse model where the long term expression of shRNAs in some mice led to liver injury and death [138]. Lastly, siRNAs are known to occasionally possess off-target effects, resulting in the silencing of the target gene as well as other, unintended genes. These off-target effects are often caused by the fact that microRNAs and siRNAs do not require perfect hybridization to the target mRNA in order to functionally inhibit translation. Whereas perfect hybridization between a microRNA and its target leads to Ago2-mediated cleavage of the mRNA, an imperfect hybridization leads to silencing by inducing the sequestration of the mRNA within a P-body [142].
Another potential use for siRNA-based therapies is in the prevention of new HIV-1 infections. Palliser et al. reported that siRNA-liposome complexes can be administered intravaginally in mice in order to provide protection from Herpes simplex virus (HSV-2) infection [143]. The administration of the siRNA-liposome complex in transgenic mice showed efficient knockdown of GFP without causing inflammation or inducing an IFN response. Co-administration of a lethal dose of HSV-2 virus and seven unique anti-HSV-2 siRNAs resulted in a significant long term survival rate (75% survival in treated mice versus 25% survival in control mice) after a single acute infection. Similar survival rates were observed even when siRNA-liposome complexes were administered 6 hours after vaginal infection with HSV-2. Although DNA viruses like HSV-2 are less likely to develop resistance to siRNAs than retroviruses like HIV-1, these findings suggest that siRNAs directed against viral surface receptors (i.e. CCR5 and CXCR4) could provide protection against HIV-1 at the site of infection [143].
4.3: Current and future siRNA therapies against HIV-1
HIV-1 is known to rapidly develop resistance to anti-viral compounds [1], and its high rate of mutation also allows the virus to quickly escape siRNAs that directly target viral transcripts [144–147]. One way to reduce the chance of viral escape is to use multiple anti-HIV-1 siRNAs [148], much like a cocktail of retroviral inhibitors. Interestingly, it has been shown that the sequence of the HIV-1 TAR region remained highly conserved even after extensive use of anti-HIV-1 siRNAs. The resistant strains that did arise possessed mutations in the viral promoter that increased transcription in order to compensate for siRNA inhibition. The results indicated that the TAR nucleotide sequence itself was important for viral replication and could not be altered [149]. This observation could be explained by discovery that the TAR region is processed into two viral microRNAs (hiv-mir-TAR-5p and hiv-mir-TAR-3p) by the host Dicer enzyme [150–151]. Short TAR hairpins can be found at high levels during every stage of HIV-1 infection, and these hairpins can be directly processed by Dicer. Klase et al. demonstrated that processing of the TAR hairpin appears to influence chromatin remodeling and anti-apoptotic responses in latently infected host cells [150, 152]. Additionally, the production of TAR microRNAs resulted in transcriptional repression at the viral LTR, dependent on the presence of Dicer. Consequently, the overproduction of TAR-derived microRNA was implicated as a mechanism for inducing HIV-1 latency [150]. Two other viral microRNAs have been reported to arise from viral transcripts. One viral microRNA, hiv-mir-H1, is located in the viral LTR and has been reported to knock down the host’s AATF gene, thereby inducing apoptosis in infected PBMCs [153]. Another viral microRNA, hiv-mir-N367, originating from the Nef ORF has been implicated in knocking down the expression of Nef, thereby attenuating HIV-1 in patients who are long term non-progressors [154]. Since the TAR element is well conserved, essential to transactivation, and creates viral microRNAs, the TAR region may be the only region within HIV-1 that might be amenable to long term targeting in RNAi-based therapies.
One potential complicating factor for RNAi-based therapies of HIV-1 is the fact that the virus has the ability to suppress the RNAi pathway in infected cells [155–156]. The suppression of RNAi is carried out by the Tat protein via binding to Dicer and inhibiting its ribonuclease activity [156–157]. However, Tat-mediated suppression is less potent than other viral suppressors of RNAi such as the influenza NS1 protein [158] and may not completely inhibit RNAi in infected cells. It has been suggested that Tat suppresses Dicer activity in order to prevent the expression of certain host factors or endogenous microRNAs that would normally restrict HIV-1 replication [156, 159]. Since HIV-1 can rapidly escape siRNAs targeted at the virus (either by mutation or Tat-mediated suppression of RNAi), several laboratories have investigated the alternate possibility of identifying and targeting host factors that are essential to viral replication [8, 160–161]. This alternative approach seeks to create HIV-1 resistant lymphocytes by knocking down the host factors that are dispensable for host survival but essential for viral replication. Unlike the viral genome, the host genes will not rapidly mutate and should be well conserved, thereby making essential host factors more attractive targets for RNAi therapies against chronic HIV-1 infections.
As previously mentioned, one clinical trial is underway to test the feasibility of targeting both viral and host genes in order to halt the progression of AIDS. This trial is a joint undertaking between City of Hope National Medical Center in Durante, California and Benitec of Melbourne, Australia [127]. In this phase I trial, CD34+ hematopoietic stem cells are isolated from patients and transformed with a lentiviral vector carrying multiple HIV-1 resistance factors. The vector carries siRNAs against Tat and Rev, as well as a ribozyme designed to knock down the host’s CCR5 receptor [162]. Patients then receive autologous transplants of the transformed stem cells, which later develop into a regenerating population of HIV-1 resistant T-cells and macrophages. This therapy was shown to be effective in humanized SCID mice by Li et al. [7] and was later moved into phase I clinical trials which are still in progress [127]. While only the CCR5 surface receptor was targeted for knock down in this trial, Anderson et al. demonstrated that shRNA constructs which knock down both CCR5 and CXCR4 can confer HIV-1 resistance upon transformed PBMCs [161, 163]. Therefore, targeting both CCR5 and CXCR4 in future RNAi-based therapies would hinder the replication of both R5 and X4 tropic viruses.
While CCR5 and CXCR4 have long been known for their role in HIV-1 infection, a trio of recently published bioinformatic screens have weighed in on the question of which host factors are essential to viral replication [164–166]. These bioinformatic screens each identified two to three hundred unique host factors required during the HIV-1 life cycle, but there was little overlap between their results. The lack of overlapping hits can likely be explained by the different methods used in each screen. Zhou et al. used HeLa P4-R5 cells infected with the HXB2 strain to look at viral replication after 96 hours [164]. Brass et al. used TZM-bl HeLa cells infected with the HIV-IIIb strain to look at viral replication after 72 hours [165]. And lastly, Koenig et al. used HEK-293T cells to examine expression of a reporter gene from the HIV-1 promoter after 24 hours [166]. These three studies utilized RNAi screening libraries to identify host factors critical to HIV-1 replication. These three genomic screens were later combined and subjected to an in-depth statistical analysis in order to identify the host factors most likely to be indispensible for viral replication [8]. Only three targets (MED6, MED7, and relA) were found in all three screens, while a mere thirty-four unique targets were identified by at least two of the three screens. Fifteen of those host factors were found to be essential for a complete round of viral replication, as reported by both Brass et al [165] and Zhou et al [164]. Those genes were CD4, CXCR4, Cav-2, cyclin T1, DDX3X, AKT1, JAK1, WNK1, MED28, MED4, TCEB3, Rab26, RNF26, RGPD8, and ANKRD30A. These essential host factors fall into several function groupings such as surface receptors (CD4 and CXCR4), signaling kinases (AKT1, JAK1, WNK1), transcription components (relA, RNF26, MED4, MED6, MED7, MED28, cyclin-T1, TCEB3), nuclear pore related proteins (DDX3X and RGPD8), and vesicle related proteins (Cav-2 and Rab26).
While each of these identified host factors may be required for HIV-1 replication, some of them may be poor candidates for future RNAi-based therapies. For example, the kinases AKT1 [167], JAK1 [168], and WNK1 [169] may be poor targets for RNAi therapies since transgenic knockouts of these genes are often lethal in mice. Similarly, the loss of relA results in an embryonic lethal phenotype in mice [170]. However, while these host factors may not be suitable targets for siRNA therapies, they may still be considered as valid targets when designing drugs to inhibit HIV-1 replication. The remaining identified host factors, however, may be suitable for RNAi-based therapies. The main HIV-1 surface receptors CD4, CXCR4, and CCR5 have been tested as possible targets for siRNA-based therapies against HIV-1 infection by Anderson et al. [171] but siRNAs against CD4 were not further pursued in subsequent trials [161]. Many of the listed host factors (i.e. CD4, CXCR4, JAK1, AKT1, relA, cyclin-T1, DDX3X, TCEB3) are known to associate with HIV-1 proteins, while the remaining host factors (i.e. Cav-2, WNK1, MED28, MED4, Rab26, RNF26, RGPD8, and ANKRD30A) are novel host cell factors for HIV-1 replication [8]. Like the case of cdk9 and Flavopiridol, one or more of these novel host factors may prove to be dispensable for the host cell, but required for viral replication. Finally, these novel factors may be especially important to future therapies. While knocking down the HIV-1 surface receptors only prevents new infections, knocking down one or more of these novel host factors may confer HIV-1 resistance upon both infected and uninfected cells.
5.1: Expert Opinion
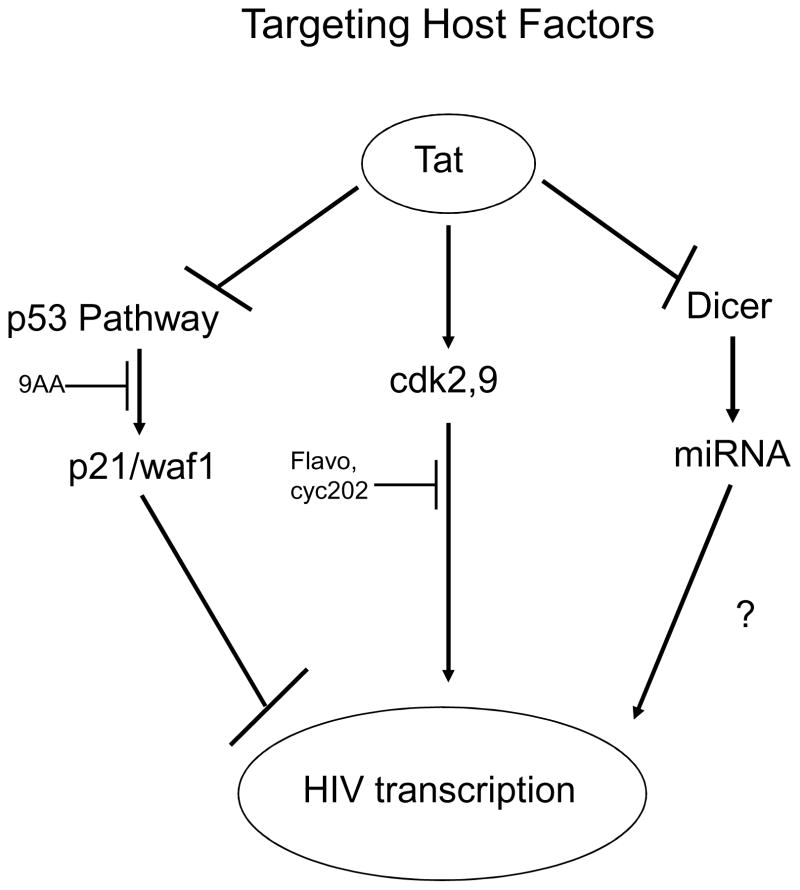
Evaluations of patients undergoing HAART treatments have revealed that the virus is able to rapidly mutate and develop resistance to drugs that directly target viral proteins [1–2]. However, HIV-1 requires many host proteins in order to carry out its replication cycle. These host proteins can be targeted in order to block HIV-1 replication. Compounds such as 9AA can activate the p53 pathway in order to overcome Tat mediated suppression and ultimately suppress viral transcription [94]. Likewise, cdk inhibitors such as Flavopiridol and Roscovitine can halt viral transcription without killing the host cells [115]. Finally, siRNAs might be directed against host proteins such as CCR5 and CXCR4 in order to rob HIV-1 of its essential host factors [163]. Figure 1 is an overview of some of the possible targets discussed in this review. By targeting these host factors, either by blocking their function or knocking them down, new therapies can be developed that are not as susceptible to viral mutation. In essence, new therapies against HIV-1 can focus on altering the host cell in order to make an environment that cannot support HIV-1 replication.
Figure 1.
HIV-1 transcription can be blocked by targeting host factors. The p53 pathway is known to be suppressed by HIV-1 and its reactivation can inhibit the replication of HIV-1. This is evident by upregulation of p21/waf1 in 9AA treated cells. Similarly, viral transcription requires the host’s cdk2 and cdk9 enzymes. Inhibiting these cdks using ATP analogs can halt viral transcription without killing the host cell. Additionally, HIV-1 possesses the ability to suppress RNAi in host cells. Although the overall downstream consequences are still being investigated, siRNAs can be directed against host factors that are essential to viral replication.
Finally, understanding what each of the viral proteins is binding to (the proteome) in specific target cells may prove to be critical for designing better inhibitors. As compared to other disease states such as diabetes or cancer, viral infections can induce two different areas of proteomic alterations, those that are related to the host and those that are related to the virus itself. In the context of the host, the proteome changes need to be implicitly focused based on cell type, T cell, monocyte/macrophage, glial cell lines, or primary cells. Due to the fundamental differences in these cell types, the virus will interact with and manipulate the expression and interaction of cellular proteins in dramatically different ways. This could be a potentially important venue for instance when studying pediatric AIDS vs. adult AIDS, where virus replication is much more pronounced and more difficult to control in the former.
Also, studies of viral protein interactomes should be performed with endogenously expressed, but not overexpressed viral proteins. Proteomic outcomes will vary significantly if a protein is overexpressed or if recombinant tagged viral proteins are used for in vitro binding assays. The overexpression of viral proteins may lead to the identification of protein interactions that are only present in the excess of viral protein and therefore not biologically significant. In addition, recombinant proteins that are expressed in a bacterial system do not exhibit the same post translational modifications and possibly do not have the correct secondary or tertiary structure, both of which can dramatically affect the detected protein-protein interactions. Therefore, the study of endogenously tagged viral proteins will provide a biological relevant model to understand protein-protein interactions.
Future studies should also investigate viral clade specific proteomes, which would investigate the protein-protein interaction differences between different HIV-1 clade isolates. As an example, it has already been shown that amino acid sequence variation between the Tat proteins of different clades can lead to the presence of more lysine residues. Acetylation of these additional residues could potentially increase the activation of the viral LTR. The importance of post translational modifications to the transcriptional state of a virally infected cell can no longer be ignored. The direct addition of an acetyl, methyl, or phospho-moiety onto Tat intensely influences the state of the viral promoter and the half-life of the protein of interest. It is therefore critical to characterize the viral proteomes in the context of the state of modification.
References
- 1.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008;10(2):67–84. [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R, Quinones-Mateu ME, Mansky LM. Drug resistance, virus fitness and HIV-1 mutagenesis. Curr Pharm Des. 2004;10(32):4065–70. doi: 10.2174/1381612043382404. [DOI] [PubMed] [Google Scholar]
- 3.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69(8):5087–94. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Q. Restoring p53-mediated apoptosis in cancer cells: new opportunities for cancer therapy. Drug Resist Updat. 2006;9(1–2):19–25. doi: 10.1016/j.drup.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Ammosova T, et al. Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology. 2006;3:78. doi: 10.1186/1742-4690-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20(8):2629–34. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li MJ, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12(5):900–9. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 8.Bushman FD, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5(5):e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–6. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 10.Janus F, et al. The dual role model for p53 in maintaining genomic integrity. Cell Mol Life Sci. 1999;55(1):12–27. doi: 10.1007/s000180050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albrechtsen N, et al. Maintenance of genomic integrity by p53: complementary roles for activated and non-activated p53. Oncogene. 1999;18(53):7706–17. doi: 10.1038/sj.onc.1202952. [DOI] [PubMed] [Google Scholar]
- 12.Somasundaram K. Tumor suppressor p53: regulation and function. Front Biosci. 2000;5:D424–37. doi: 10.2741/somasund. [DOI] [PubMed] [Google Scholar]
- 13.Striteska D. [The tumor supressor gene p53] Acta Medica (Hradec Kralove) Suppl. 2005;48(1):21–5. [PubMed] [Google Scholar]
- 14.Rezacova M, Vavrova J, Cerman J. [A cell and genotoxic stress: a reaction to double strand breaks of DNA] Cas Lek Cesk. 2005;144(Suppl 3):13–7. [PubMed] [Google Scholar]
- 15.Attardi LD. The role of p53-mediated apoptosis as a crucial anti-tumor response to genomic instability: lessons from mouse models. Mutat Res. 2005;569(1–2):145–57. doi: 10.1016/j.mrfmmm.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Yu J, Zhang L. The transcriptional targets of p53 in apoptosis control. Biochem Biophys Res Commun. 2005;331(3):851–8. doi: 10.1016/j.bbrc.2005.03.189. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Kulesz-Martin M. P53 regulation and function in normal cells and tumors. Medicina (B Aires) 2000;60(Suppl 2):9–11. [PubMed] [Google Scholar]
- 18.Erster S, et al. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24(15):6728–41. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima K, et al. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood. 2006;108(3):993–1000. doi: 10.1182/blood-2005-12-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley ST, et al. Tumor response to neoadjuvant chemoradiation therapy for rectal adenocarcinoma is mediated by p53-dependent and caspase 8-dependent apoptotic pathways. Clin Colorectal Cancer. 2005;5(2):114–8. doi: 10.3816/ccc.2005.n.023. [DOI] [PubMed] [Google Scholar]
- 21.Strachan GD, et al. E2F1 induces cell death, calpain activation, and MDMX degradation in a transcription independent manner implicating a novel role for E2F1 in neuronal loss in SIV encephalitis. J Cell Biochem. 2005;96(4):728–40. doi: 10.1002/jcb.20574. [DOI] [PubMed] [Google Scholar]
- 22.Meek DW. The role of p53 in the response to mitotic spindle damage. Pathol Biol (Paris) 2000;48(3):246–54. [PubMed] [Google Scholar]
- 23.Bartek J, Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490(3):117–22. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- 24.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13(6):738–47. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 25.Bartkova J, et al. Deregulation of the G1/S-phase control in human testicular germ cell tumours. Apmis. 2003;111(1):252–65. doi: 10.1034/j.1600-0463.2003.1110129.x. discussion 265–6. [DOI] [PubMed] [Google Scholar]
- 26.Honma M. Generation of loss of heterozygosity and its dependency on p53 status in human lymphoblastoid cells. Environ Mol Mutagen. 2005;45(2–3):162–76. doi: 10.1002/em.20113. [DOI] [PubMed] [Google Scholar]
- 27.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20(15):1803–15. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 28.Stark GR, Taylor WR. Analyzing the G2/M checkpoint. Methods Mol Biol. 2004;280:51–82. doi: 10.1385/1-59259-788-2:051. [DOI] [PubMed] [Google Scholar]
- 29.Stark GR, Taylor WR. Control of the G2/M transition. Mol Biotechnol. 2006;32(3):227–48. doi: 10.1385/MB:32:3:227. [DOI] [PubMed] [Google Scholar]
- 30.Houtgraaf JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc Revasc Med. 2006;7(3):165–72. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Sancar A, et al. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 32.Ashcroft M, Vousden KH. Regulation of p53 stability. Oncogene. 1999;18(53):7637–43. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 33.Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19(3):1751–8. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8(5):345–57. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 35.Oda Y, et al. Molecular abnormalities of p53, MDM2, and H-ras in synovial sarcoma. Mod Pathol. 2000;13(9):994–1004. doi: 10.1038/modpathol.3880180. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404(6773):42–9. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 37.Alarcon-Vargas D, Ronai Z. p53-Mdm2--the affair that never ends. Carcinogenesis. 2002;23(4):541–7. doi: 10.1093/carcin/23.4.541. [DOI] [PubMed] [Google Scholar]
- 38.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsson A, et al. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14(9):1561–75. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 40.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24(17):2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 41.Khanna KK, et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet. 1998;20(4):398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 42.Banin S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–7. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 43.Canman CE, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281(5383):1677–9. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 44.Saito S, et al. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J Biol Chem. 2003;278(39):37536–44. doi: 10.1074/jbc.M305135200. [DOI] [PubMed] [Google Scholar]
- 45.Lee JH, et al. Stabilization and activation of p53 induced by Cdk5 contributes to neuronal cell death. J Cell Sci. 2007;120(Pt 13):2259–71. doi: 10.1242/jcs.03468. [DOI] [PubMed] [Google Scholar]
- 46.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13(6):941–50. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 47.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13(1):49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Li CC, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23(11):2096–106. doi: 10.1038/sj.onc.1207411. [DOI] [PubMed] [Google Scholar]
- 49.Brooks CL, Gu W. Dynamics in the p53-Mdm2 ubiquitination pathway. Cell Cycle. 2004;3(7):895–9. [PubMed] [Google Scholar]
- 50.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21(3):307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6(14):1718–23. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- 52.Garden GA, Morrison RS. The multiple roles of p53 in the pathogenesis of HIV associated dementia. Biochem Biophys Res Commun. 2005;331(3):799–809. doi: 10.1016/j.bbrc.2005.03.185. [DOI] [PubMed] [Google Scholar]
- 53.Castedo M, et al. p53-A pro-apoptotic signal transducer involved in AIDS. Biochem Biophys Res Commun. 2005;331(3):701–6. doi: 10.1016/j.bbrc.2005.03.188. [DOI] [PubMed] [Google Scholar]
- 54.Li CY, Suardet L, Little JB. Potential role of WAF1/Cip1/p21 as a mediator of TGF-beta cytoinhibitory effect. J Biol Chem. 1995;270(10):4971–4. doi: 10.1074/jbc.270.10.4971. [DOI] [PubMed] [Google Scholar]
- 55.Duan L, et al. The tumor suppressor protein p53 strongly alters human immunodeficiency virus type 1 replication. J Virol. 1994;68(7):4302–13. doi: 10.1128/jvi.68.7.4302-4313.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subler MA, Martin DW, Deb S. Activation of the human immunodeficiency virus type 1 long terminal repeat by transforming mutants of human p53. J Virol. 1994;68(1):103–10. doi: 10.1128/jvi.68.1.103-110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Longo F, et al. A novel approach to protein-protein interaction: complex formation between the p53 tumor suppressor and the HIV Tat proteins. Biochem Biophys Res Commun. 1995;206(1):326–34. doi: 10.1006/bbrc.1995.1045. [DOI] [PubMed] [Google Scholar]
- 58.Li CJ, et al. Reciprocal modulations between p53 and Tat of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1995;92(12):5461–4. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 60.Liu L, et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19(2):1202–9. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakaguchi K, et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12(18):2831–41. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrod R, et al. Human immunodeficiency virus type-1 Tat/co-activator acetyltransferase interactions inhibit p53Lys-320 acetylation and p53-responsive transcription. J Biol Chem. 2003;278(14):12310–8. doi: 10.1074/jbc.M211167200. [DOI] [PubMed] [Google Scholar]
- 63.Ohata M, et al. Prognostic implications of p21 (Waf1/Cip1) immunolocalization in multiple myeloma. Biomed Res. 2005;26(3):91–8. doi: 10.2220/biomedres.26.91. [DOI] [PubMed] [Google Scholar]
- 64.de Oliveira RM. Klotho RNAi induces premature senescence of human cells via a p53/p21 dependent pathway. FEBS Lett. 2006;580(24):5753–8. doi: 10.1016/j.febslet.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 65.Child ES, Mann DJ. The intricacies of p21 phosphorylation: protein/protein interactions, subcellular localization and stability. Cell Cycle. 2006;5(12):1313–9. doi: 10.4161/cc.5.12.2863. [DOI] [PubMed] [Google Scholar]
- 66.LaBaer J, et al. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11(7):847–62. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 67.Cheng M, et al. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. Embo J. 1999;18(6):1571–83. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alt JR, Gladden AB, Diehl JA. p21(Cip1) Promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J Biol Chem. 2002;277(10):8517–23. doi: 10.1074/jbc.M108867200. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki A, et al. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene. 1998;17(8):931–9. doi: 10.1038/sj.onc.1202021. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki A, et al. Mitochondrial regulation of cell death: mitochondria are essential for procaspase 3-p21 complex formation to resist Fas-mediated cell death. Mol Cell Biol. 1999;19(5):3842–7. doi: 10.1128/mcb.19.5.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chuang LS, et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277(5334):1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 72.Perkins ND. Not just a CDK inhibitor: regulation of transcription by p21(WAF1/CIP1/SDI1) Cell Cycle. 2002;1(1):39–41. [PubMed] [Google Scholar]
- 73.Clark E, et al. Loss of G(1)/S checkpoint in human immunodeficiency virus type 1-infected cells is associated with a lack of cyclin-dependent kinase inhibitor p21/Waf1. J Virol. 2000;74(11):5040–52. doi: 10.1128/jvi.74.11.5040-5052.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X, Hung NJ, Costa RH. Earlier expression of the transcription factor HFH-11B diminishes induction of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001;33(6):1404–14. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- 75.de la Fuente C, et al. Pharmacological cyclin-dependent kinase inhibitors as HIV-1 antiviral therapeutics. Curr HIV Res. 2003;1(2):131–52. doi: 10.2174/1570162033485339. [DOI] [PubMed] [Google Scholar]
- 76.Zhang J, Scadden DT, Crumpacker CS. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest. 2007;117(2):473–81. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen H, et al. Intrinsic human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J Virol. 1999;73(1):728–37. doi: 10.1128/jvi.73.1.728-737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weichold FF, et al. HIV-1 protease inhibitor ritonavir modulates susceptibility to apoptosis of uninfected T cells. J Hum Virol. 1999;2(5):261–9. [PubMed] [Google Scholar]
- 79.Lee B, et al. Coreceptor/chemokine receptor expression on human hematopoietic cells: biological implications for human immunodeficiency virus-type 1 infection. Blood. 1999;93(4):1145–56. [PubMed] [Google Scholar]
- 80.Perron MJ, et al. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci U S A. 2004;101(32):11827–32. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A. 2004;101(29):10780–5. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Owens CM, et al. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J Virol. 2004;78(10):5423–37. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–53. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 84.Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 85.Hietanen S, et al. Activation of p53 in cervical carcinoma cells by small molecules. Proc Natl Acad Sci U S A. 2000;97(15):8501–6. doi: 10.1073/pnas.97.15.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shiraishi T, Nielsen PE. Down-regulation of MDM2 and activation of p53 in human cancer cells by antisense 9-aminoacridine-PNA (peptide nucleic acid) conjugates. Nucleic Acids Res. 2004;32(16):4893–902. doi: 10.1093/nar/gkh820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gurova KV, et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc Natl Acad Sci U S A. 2005;102(48):17448–53. doi: 10.1073/pnas.0508888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beraza N, Trautwein C. Restoration of p53 function: a new therapeutic strategy to induce tumor regression? Hepatology. 2007;45(6):1578–9. doi: 10.1002/hep.21789. [DOI] [PubMed] [Google Scholar]
- 89.Kastan MB, Berkovich E. p53: a two-faced cancer gene. Nat Cell Biol. 2007;9(5):489–91. doi: 10.1038/ncb0507-489. [DOI] [PubMed] [Google Scholar]
- 90.Bykov VJ, Selivanova G, Wiman KG. Small molecules that reactivate mutant p53. Eur J Cancer. 2003;39(13):1828–34. doi: 10.1016/s0959-8049(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 91.Klein C, Vassilev LT. Targeting the p53-MDM2 interaction to treat cancer. Br J Cancer. 2004;91(8):1415–9. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vassilev LT. Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3(4):419–21. [PubMed] [Google Scholar]
- 93.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 94.Wu W, et al. Drug 9AA reactivates p21/Waf1 and Inhibits HIV-1 progeny formation. Virol J. 2008;5:41. doi: 10.1186/1743-422X-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sebestik J, Hlavacek J, Stibor I. A role of the 9-aminoacridines and their conjugates in a life science. Curr Protein Pept Sci. 2007;8(5):471–83. doi: 10.2174/138920307782411400. [DOI] [PubMed] [Google Scholar]
- 96.Phuan PW, et al. Discriminating between cellular and misfolded prion protein by using affinity to 9-aminoacridine compounds. J Gen Virol. 2007;88(Pt 4):1392–401. doi: 10.1099/vir.0.82601-0. [DOI] [PubMed] [Google Scholar]
- 97.Zwelling LA. Topoisomerase II as a target of antileukemia drugs: a review of controversial areas. Hematol Pathol. 1989;3(3):101–12. [PubMed] [Google Scholar]
- 98.Zwelling LA, et al. Characterization of an amsacrine-resistant line of human leukemia cells. Evidence for a drug-resistant form of topoisomerase II. J Biol Chem. 1989;264(28):16411–20. [PubMed] [Google Scholar]
- 99.Sohn TA, et al. High-throughput measurement of the Tp53 response to anticancer drugs and random compounds using a stably integrated Tp53-responsive luciferase reporter. Carcinogenesis. 2002;23(6):949–57. doi: 10.1093/carcin/23.6.949. [DOI] [PubMed] [Google Scholar]
- 100.Jung KJ, et al. Small-molecule inhibitor which reactivates p53 in human T-cell leukemia virus type 1-transformed cells. J Virol. 2008;82(17):8537–47. doi: 10.1128/JVI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo C, et al. 9-Aminoacridine-based anticancer drugs target the PI3K/AKT/mTOR, NF-kappaB and p53 pathways. Oncogene. 2009;28(8):1151–61. doi: 10.1038/onc.2008.460. [DOI] [PubMed] [Google Scholar]
- 102.Guiffant D, et al. Identification of intracellular targets of small molecular weight chemical compounds using affinity chromatography. Biotechnol J. 2007;2(1):68–75. doi: 10.1002/biot.200600223. [DOI] [PubMed] [Google Scholar]
- 103.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 104.Ewen ME. Where the cell cycle and histones meet. Genes Dev. 2000;14(18):2265–70. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- 105.Petersen BO, et al. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. Embo J. 1999;18(2):396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fisher RP, Morgan DO. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78(4):713–24. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 107.Malumbres M. Revisiting the “Cdk-centric” view of the mammalian cell cycle. Cell Cycle. 2005;4(2):206–10. doi: 10.4161/cc.4.2.1406. [DOI] [PubMed] [Google Scholar]
- 108.Canepa ET, et al. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life. 2007;59(7):419–26. doi: 10.1080/15216540701488358. [DOI] [PubMed] [Google Scholar]
- 109.Denicourt C, Dowdy SF. Cip/Kip proteins: more than just CDKs inhibitors. Genes Dev. 2004;18(8):851–5. doi: 10.1101/gad.1205304. [DOI] [PubMed] [Google Scholar]
- 110.Baumli S, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. Embo J. 2008;27(13):1907–18. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Demidenko ZN, Blagosklonny MV. Flavopiridol induces p53 via initial inhibition of Mdm2 and p21 and, independently of p53, sensitizes apoptosis-reluctant cells to tumor necrosis factor. Cancer Res. 2004;64(10):3653–60. doi: 10.1158/0008-5472.CAN-04-0204. [DOI] [PubMed] [Google Scholar]
- 112.Schang LM, et al. Pharmacological cyclin-dependent kinase inhibitors inhibit replication of wild-type and drug-resistant strains of herpes simplex virus and human immunodeficiency virus type 1 by targeting cellular, not viral, proteins. J Virol. 2002;76(15):7874–82. doi: 10.1128/JVI.76.15.7874-7882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Knockaert M, et al. Intracellular targets of cyclin-dependent kinase inhibitors: identification by affinity chromatography using immobilised inhibitors. Chem Biol. 2000;7(6):411–22. doi: 10.1016/s1074-5521(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 114.Knockaert M, Greengard P, Meijer L. Pharmacological inhibitors of cyclin-dependent kinases. Trends Pharmacol Sci. 2002;23(9):417–25. doi: 10.1016/s0165-6147(02)02071-0. [DOI] [PubMed] [Google Scholar]
- 115.Chao SH, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275(37):28345–8. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 116.Nekhai S, et al. HIV-1 Tat-associated RNA polymerase C-terminal domain kinase, CDK2, phosphorylates CDK7 and stimulates Tat-mediated transcription. Biochem J. 2002;364(Pt 3):649–57. doi: 10.1042/BJ20011191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou M, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20(14):5077–86. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yik JH, et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12(4):971–82. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 119.Sabo A, et al. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol Cell Biol. 2008;28(7):2201–12. doi: 10.1128/MCB.01557-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou M, et al. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol. 2004;78(24):13522–33. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chiu YL, et al. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1) J Virol. 2004;78(5):2517–29. doi: 10.1128/JVI.78.5.2517-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Flores O, et al. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc Natl Acad Sci U S A. 1999;96(13):7208–13. doi: 10.1073/pnas.96.13.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Salerno D, et al. Direct inhibition of CDK9 blocks HIV-1 replication without preventing T-cell activation in primary human peripheral blood lymphocytes. Gene. 2007;405(1–2):65–78. doi: 10.1016/j.gene.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Agbottah E, et al. Antiviral activity of CYC202 in HIV-1-infected cells. J Biol Chem. 2005;280(4):3029–42. doi: 10.1074/jbc.M406435200. [DOI] [PubMed] [Google Scholar]
- 125.Agbottah E, et al. Inhibition of HIV-1 virus replication using small soluble Tat peptides. Virology. 2006;345(2):373–89. doi: 10.1016/j.virol.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 126.Van Duyne R, et al. Effect of transcription peptide inhibitors on HIV-1 replication. Virology. 2008;376(2):308–22. doi: 10.1016/j.virol.2008.02.036. [DOI] [PubMed] [Google Scholar]
- 127.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–33. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 129.Ouellet DL, et al. MicroRNAs in Gene Regulation: When the Smallest Governs It All. J Biomed Biotechnol. 2006;2006(4):69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Perron MP, Provost P. Protein components of the microRNA pathway and human diseases. Methods Mol Biol. 2009;487:369–85. doi: 10.1007/978-1-60327-547-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zender L, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci U S A. 2003;100(13):7797–802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Song E, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9(3):347–51. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 133.Giladi H, et al. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8(5):769–76. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 134.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–4. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 135.Yokota T, et al. Efficient regulation of viral replication by siRNA in a non-human primate surrogate model for hepatitis C. Biochem Biophys Res Commun. 2007;361(2):294–300. doi: 10.1016/j.bbrc.2007.06.182. [DOI] [PubMed] [Google Scholar]
- 136.Agrawal S, Kandimalla ER. Role of Toll-like receptors in antisense and siRNA [corrected] Nat Biotechnol. 2004;22(12):1533–7. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]
- 137.Gorina R, et al. Astrocytes are very sensitive to develop innate immune responses to lipid-carried short interfering RNA. Glia. 2009;57(1):93–107. doi: 10.1002/glia.20738. [DOI] [PubMed] [Google Scholar]
- 138.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 139.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Scacheri PC, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(7):1892–7. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther. 2006;14(4):463–70. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 142.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25(46):6163–9. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 143.Palliser D, et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439(7072):89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 144.Sabariegos R, et al. Sequence homology required by human immunodeficiency virus type 1 to escape from short interfering RNAs. J Virol. 2006;80(2):571–7. doi: 10.1128/JVI.80.2.571-577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Das AT, et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78(5):2601–5. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Westerhout EM, et al. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33(2):796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Boden D, Pusch O, Ramratnam B. Overcoming HIV-1 resistance to RNA interference. Front Biosci. 2007;12:3104–16. doi: 10.2741/2298. [DOI] [PubMed] [Google Scholar]
- 148.Aagaard LA, et al. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15(23):1536–49. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leonard JN, et al. HIV evades RNA interference directed at TAR by an indirect compensatory mechanism. Cell Host Microbe. 2008;4(5):484–94. doi: 10.1016/j.chom.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Klase Z, et al. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ouellet DL, et al. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 2008;36(7):2353–65. doi: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Klase Z, et al. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology. 2009;6:18. doi: 10.1186/1742-4690-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kaul D, Ahlawat A, Gupta SD. HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses to infection. Mol Cell Biochem. 2009;323(1–2):143–8. doi: 10.1007/s11010-008-9973-4. [DOI] [PubMed] [Google Scholar]
- 154.Omoto S, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bennasser Y, et al. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22(5):607–19. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 156.Triboulet R, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315(5818):1579–82. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 157.Bennasser Y, Jeang KT. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.de Vries W, Berkhout B. RNAi suppressors encoded by pathogenic human viruses. Int J Biochem Cell Biol. 2008;40(10):2007–12. doi: 10.1016/j.biocel.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 159.Liu YP, et al. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36(9):2811–24. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dolan MJ, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8(12):1324–36. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 161.Anderson J, Akkina R. HIV-1 resistance conferred by siRNA cosuppression of CXCR4 and CCR5 coreceptors by a bispecific lentiviral vector. AIDS Res Ther. 2005;2(1):1. doi: 10.1186/1742-6405-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li MJ, et al. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol Ther. 2003;8(2):196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- 163.Anderson J, et al. Safety and efficacy of a lentiviral vector containing three anti-HIV genes--CCR5 ribozyme, tat-rev siRNA, and TAR decoy--in SCID-hu mouse-derived T cells. Mol Ther. 2007;15(6):1182–8. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- 164.Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4(5):495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 165.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 166.Konig R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135(1):49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen WS, et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15(17):2203–8. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rodig SJ, et al. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93(3):373–83. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 169.Zambrowicz BP, et al. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A. 2003;100(24):14109–14. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Beg AA, et al. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376(6536):167–70. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 171.Anderson J, Banerjea A, Akkina R. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides. 2003;13(5):303–12. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]