Abstract
Identification of thyroid hormone receptor (TR) co-regulators has enhanced our understanding of thyroid hormone (TH) action. However, it is likely that many other co-regulators remained unidentified, and unbiased methods are required to discover these proteins. We have previously demonstrated that the yeast Saccharomyces cerevisiae is an excellent system in which to study TR action, and that defined TR signaling complexes in a eukaryotic background devoid of complicating influences of mammalian cell co-regulators can be constructed and analyzed for endogenous yeast genes, many of which are conserved in mammals. Here, a modified synthetic genetic array analysis was performed by crossing a yeast strain that expressed TRβ1 and the co-activator GRIP1/SRC2 with 384 yeast strains bearing deletions of known genes. Eight genes essential for TH action were isolated, of which 4 are conserved in mammals. Examination of one, the yeast CCR4 and its human homolog CCR4/NOT6 (hCCR4), confirmed that (i) transfected CCR4 potentiates a TH response in cultured cells more efficiently than established TR co-activators and (ii) knockdown of CCR4 expression strongly inhibited a TH response (>80%). TH treatment promoted rapid and sustained hCCR4 recruitment to the TH-responsive deiodinase 1 promoter and TR co-localizes with hCCR4 in the nucleus and interacts with hCCR4 in 2-hybrid and pull-down assays. These findings indicate that a modified yeast synthetic genetic array strategy is a feasible method for unbiased identification of conserved genes essential for TR and other nuclear receptor hormone functions in mammals.
Keywords: nuclear receptor co-activators, conserved nuclear receptor signaling genes
Thyroid hormone (TH) receptors (TRs) are members of the nuclear receptor (NR) family (1, 2). TRs bind TH response elements (TREs) located at variable distances from the transcription initiation site of genes with roles in growth, development, and metabolism. Unliganded TRs are transcriptionally active, either stimulating or inhibiting transcription. Hormone binding (mostly triiodothyronine) generally reverses these effects; active repression by unliganded TRs becomes transcriptional enhancement and vice versa. These changes in transcriptional response are a consequence of hormone-dependent alterations in the conformation of the receptor C-terminal ligand binding domain, which result in release of cognate co-regulators and sequential recruitment of other co-regulator complexes (3).
TR co-activators have been identified on the basis of their direct interactions with TRs and other hormone-dependent NRs (4, 5). Among the best known are the steroid receptor co-activators SRC1, SRC2 (TIF2/GRIP1), and SRC3 (pCIP/RAC3/AIB1/ACTR), which are implicated in chromatin modification. SRC-1 and SRC-3 possess intrinsic histone acetyl-transferase activity and all SRCs bind auxiliary factors with histone acetyl-transferase activity, including p300, CREB-binding protein (CBP), and p300/CBP-associated factor P/CAF. Another cofactor, the 220-kDa TR associated protein (TRAP220/DRIP205), is part of the mediator/SMCC complex, which contacts the basal transcription machinery (6). Other co-activators with potential roles in NR signaling include components of the CCR-NOT complexes (7). The well characterized CCR-NOT subunit is called carbon catabolite repressor 4 (CCR4) in yeast and NOT6 in higher eukaryotes. CCR4/NOT6 (hCCR4) resembles the ExoIII family of nuclease/phosphatases (8) and is implicated in mRNA deadenylation in the cytoplasm (9). Although identification of TR interacting cofactors has provided great insights into the mechanism of transcriptional activation, questions about the identity, function, and interactions of many TR co-activators remain unanswered. Potential roles of cofactors with alternate contact modes, or cofactors that lie completely downstream of direct TR contacts, are less clear. New unbiased techniques are needed to systematically identify and categorize key TR co-activators.
We previously proposed that the yeast Saccharomyces cerevisiae is an excellent system in which to study NR signaling (10). Yeast lack conventional NRs and SRCs, but contain homologs of key mammalian enzymes required for chromatin modification and general transcription factors. Thus, it is possible to reconstruct defined NR signaling complexes in the absence of confounding effects of other NR cofactors present in mammalian cells. We reconstituted TH-dependent gene activation at TRE-driven β-gal reporters in yeast that express TR, with and without the heterodimer partner retinoid X receptor (RXR) and GRIP1/SRC2 (11–14), and used similar approaches to investigate TR interactions with adenovirus E1A and truncated versions of the co-repressor N-CoR, co-activators that bind TRs in the typical co-repressor mode (15, 16). Our previous studies showed that SWI/SNF, GCN5, and ADA2 (SAGA) multi-component complexes are also required for activity of thyroid hormone action (13, 16). This represents proof of concept that yeast genetic analysis can reveal cofactor requirements for TR signaling and, given strong homologies between yeast and mammalian genes, suggests that similar approaches could be used in a more comprehensive manner to identify factors needed for TR action that are also important in mammalian cells.
In the current report, we used a synthetic genetic analysis (SGA) to test requirements for yeast genes in TR signaling. Standard SGA relies on the fact that >80% of the 6,200 predicted yeast genes are non-essential and that strains bearing deletions of these genes are viable (17). A modified version of SGA was devised in which a yeast strain that contains a TRE-driven β-gal reporter and expresses TRβ1 and GRIP1/SRC2 is crossed with strains that bear deletions of 384 different genes implicated in transcription or chromatin remodeling. We find that 8 of these genes are needed for optimal TR activity and present evidence that one, hCCR4, is also essential for TR signaling in cultured HeLa cells. These observations indicate that the proposed modified yeast SGA strategy is a feasible method for the systematic identification of mammalian genes essential for NR function.
Results
Yeast SGA Screen.
To test the possibility that SGA could reveal novel factors that are required for TR activity, we obtained sample plates with yeast strains bearing individual deletions of 384 genes. The genes included are listed in Table S1; the panel corresponds to a portion of the complete ordered array of yeast mutant strains (17). It does not include genes that we have previously shown to be needed for TR signaling in yeast—ADA2, GCN5 and swi/snf 2, 3, and 6—but represents a reasonable fraction of known yeast genes and contains multiple genes with potential roles in transcription and chromatin remodeling.
The screen was performed in accordance with the previously described SGA procedures, which were modified as outlined in Fig. 1A and described in Materials and Methods. A typical plate screen is illustrated in Fig. 1B. One hundred twenty-five clones were selected on the basis of suspected reductions in blue coloration. Each was grown in liquid culture for direct β-gal assays, and 31 clones with >60% loss of function in the test SGA strain were selected for further study. We also verified requirements for these genes by purchasing commercially available yeast deletion mutants from the American Type Culture Collection (ATCC), which were prepared in a slightly different genetic background. Eight of the 31 selected clones continued to exhibit >60% loss of function by a β-gal assay.
Fig. 1.
SGA screening reveals yeast genes essential for hTRβ1 action. (A) Strategy: MATα:Y5563 yeast strain co-transformants that expressed hTRβ1 and GRIP1 and contained a TRE-F2 × 1 β-gal reporter were crossed with an ordered array of MATa gene deletion mutants. To select for growth of mutant meiotic progeny expressing TR, GRIP1, and TRE-F2 × 1 β-gal reporter, the MATa meiotic progeny was transferred to medium that contains kanamycin and lacks leucine. (B) Yeast X-gal agarose overlay of SGA deletion mutants: the cultures from A were spotted an agarose plate (6% dimethylformamide, 0.1% SDS, 0.1 mg/mL X-Gal, 0.5% agarose, 10−7 M Triac or nil) grown overnight at 30 °C. A blue color indicated that the deleted gene was not required for a TH-dependent activation of the TRE- reporter, whereas white indicated a failure of gene activation.
Of the 8 identified genes (Table S2), 4 have known human homologs. These include CCR4/NOT6, the major component of the CCR4/NOT complex as previously described (7, 9). In addition, TR also exhibited strong requirements for SAC3 (18), a nuclear pore-associated protein; SHP1 (19), a protein involved in ubiquitin dependent protein-degradation; and the SWI4 subunit of the swi/snf chromatin remodeling complex (20). Four genes that were needed for TR action were specific to yeast without a known human homolog: YBR175W, YJL175W, SRB2, and LGE1. For further details on their known functional properties, see Table S2.
Validation of yCCR4 as a TR Regulator.
To validate our modified SGA method, we focused on one putative essential yeast TR interacting protein with a known human homolog, namely CCR4 (9, 21). We compared TR activity at the TRE driven reporter in the WT 4741 strain versus the ΔyCCR4 deletion mutant generated in the SGA screen in the absence and presence of GRIP1/SRC2 (Fig. 2A). In accordance with previous results, hTRβ1 exhibited moderate activity at the F2-driven β-gal reporter with L-triiodothyroacetic acid (Triac), and this was potentiated ≈5-fold by GRIP1/SRC2. TR activity was severely diminished (>90%) in the ΔyCCR4 mutant strain in the absence and presence of GRIP1/SRC2. Exogenous yCCR4 (expressed as a LexA fusion) did not potentate TR activity in WT yeast but did rescue the signaling defect in the CCR4 deletion mutant strain (Fig. 2B). Thus, TR action shows a strong requirement for yCCR4, and this is not absolutely dependent on exogenous GRIP1/SRC2.
Fig. 2.
Validation of yCCR4 as a gene needed for TR action in yeast. (A) Comparison of hTRβ1 activation in WT and CCR4 deletion strains. Results of β-gal assays performed with commercial (ATCC) WT yeast strain YBR175W BY4741 (4741-WT) and CCR4 deletion mutant yeast strain YAL021C BY4741 (4741-ΔyCCR4). This strain was transformed with a single copy of the chicken lysozyme (F2) TRE, hTRβ1, GRIP1, or empty vector. Cultures were treated with vehicle (empty bars) or 10−7 M Triac (solid bars). β-Gal activities were expressed as Miller units per milligram protein. Data shown were pooled from 3 independent experiments and calculated as mean ± SE. β-Gal assays were performed as described as outlined in Materials and Methods. (B) CCR4 replacement restores TH-dependent gene activation in the CCR4 deletion strain. Experimental procedures and β-gal assays were performed as described in A except that the LexA plasmid was substituted for GRIP1/SRC2 with the same selection marker restrictions.
Other NRs also exhibited reduced activity in the ΔyCCR4 deletion strain (Fig. S1 A–D). CCR4 was required for all 4 NRs tested [TR, ER androgen receptor (AR), retinoic acid receptor (RAR), and RXR]. However, the magnitude of this effect was smaller for ERs ≈60%), RXRs (≈60–70%), and ARs (≈50%) than for TRs (>90%). We conclude that yCCR4 is required for optimal activity of exogenous NRs in yeast, but that this factor plays an especially prominent role in TR action.
hCCR4-NOT6 Is Essential for hTRβ1 Function in Human Cells.
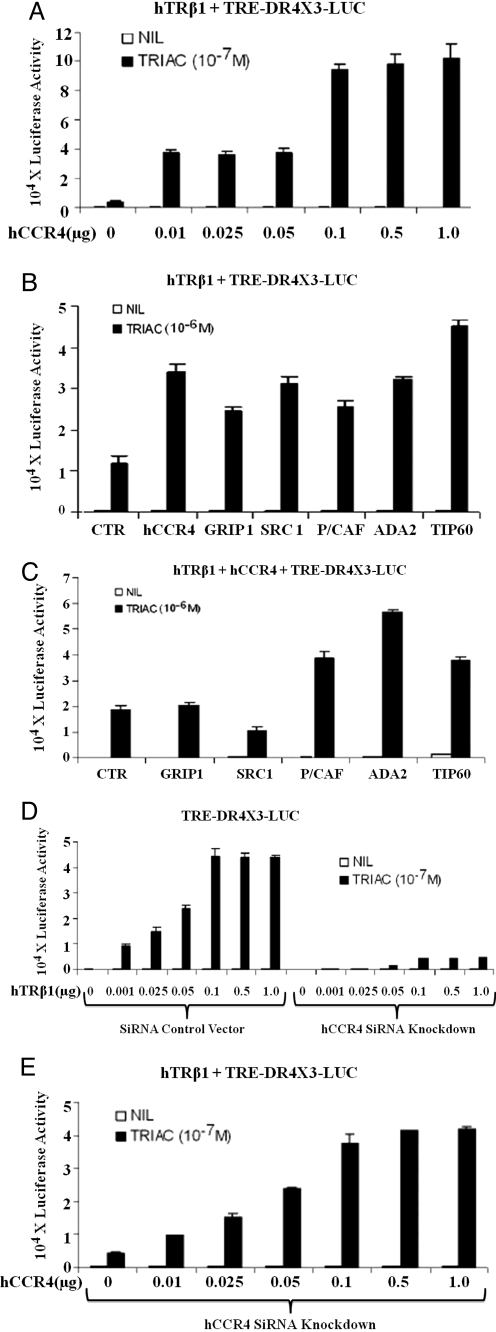
To determine whether the human homolog of yCCR4 (hCCR4/NOT6) is required for TR signaling in mammalian cells, we determined effects of hCCR4 transfection of TRβ1 activity in HeLa cells. Fig. 3A shows that co-expression of hTRβ1 with increasing concentrations of hCCR4 potentiated TH response at a standard TRE-driven reporter (DR-4 × 3-LUC). This effect was larger than effects obtained with GRIP1/SRC2 and comparable to that obtained with several other TR co-activators (SRC1, P/CAF, ADA2, TIP60; Fig. 3B). Interestingly, hCCR4 did not synergize with SRC1 or GRIP1/SRC2 in TR co-activation; GRIP1/SRC2 failed to potentiate TR activity in the presence of exogenous hCCR4 and SRC1 weakly suppressed TR activity in these conditions (Fig. 3C). By contrast, P/CAF, ADA2, and TIP60 continued to enhance TR activity in the presence of exogenous hCCR4.
Fig. 3.
hCCR4 is needed for hTRβ1 action in mammalian cells. (A) hCCR4 enhances hTRβ1 TH-dependent gene activation. Co-transfection of increasing DNA concentrations of hCCR4 with 0.1 μg of hTRβ1 progressively enhances TH-dependent hTRβ1 reporter gene activation. (B) hCCR4 compared with other co-activators: co-transfection of TRE-DR4 × 3-LUC reporter with a comparable quantity of hCCR4 or CTR (control vector), GRIP1, SRC1, P/CAF, ADA2, and TIP60. Experiments and luciferase assays were performed as described in A. (C) Potency of co-activators on TH-dependent reporters with co-transfected hCCR4 as in B. Note that reporter activation by P/CAF, ADA2, and TIP60, but not GRIP1 or SRC1, was further enhanced by hCCR4. (D) Detection of impaired co-activator function in hCCR4 siRNA knockdown. Illustrated are the results comparing the luciferase reporter gene responses for WT HeLa cells (Left) compared with siRNA hCCR4-knockdown HeLa cells (Right) using standard techniques (see Materials and Methods). (E) Restoration of hCCR4 co-activator function by a WT replacement. hCCR4 siRNA knockdown HeLa cells were co-transfected with hTRβ1, reporter TRE-DR4 × 3-LUC, and increasing doses of hCCR4. Methods are otherwise similar to D.
We next examined effects of hCCR4 knockdown on TR action. We verified that parental HeLa cells express hCCR4 transcripts by Northern blot and RT-PCR (Fig. S2 A and B). We created a retrovirus that expresses an siRNA specific for hCCR4 and obtained stable HeLa cell lines that express the hCCR4 siRNA or contain a control retrovirus vector. The absence of hCCR4 expression in the hCCR4 siRNA expressing cell line was confirmed by RT-PCR (Fig. S2 C and D). Transfection analysis revealed that TRβ1 action was severely curtailed in this background but not in the control cell line (Fig. 3D). Moreover, this deficit was rescued by exogenous hCCR4; a transfected hCCR4 expression vector enhanced response ≈8-fold (Fig. 3E) and restored TR signaling to levels comparable to those with TR and transfected hCCR4 in control HeLa cells. Co-expression of transfected TR and hCCR4 did not affect expression levels of the other protein (Fig. S2E). Thus, TR activity shows a very strong requirement for hCCR4. Similar observations were obtained in HeLa cells for ER and AR, although the requirement for hCCR4 appeared strongest with TRs (Fig. S2 F–I).
TR Interacts with hCCR4 in Vivo and in Vitro.
To determine whether TRβ recruits hCCR4 to a target promoter in cultured cells, we performed ChIP analysis on extracts from a HeLa cell derivative that stably expresses hTRβ1 fused to a tandem affinity purification (tap) tag (Fig. 4).
Fig. 4.
Enhanced binding of hCCR4 to the DIO1 gene promoter. Shown are the results of ChIP assays for HeLa cells transfected with hCCR4 and treated with vehicle (lane 1) or Triac (10−7 M) for different times: 0 min (lane 1), 5 min (lane 2), 10 min (lane 3), 60 min (lane 4), 120 min (lane 5), 360 min (lane 6), and overnight (lane 7). Following formaldehyde cross-linking, lysates were immunoprecipitated with antibodies against hTRβ1 or pre-immune serum as indicated. Primers specific for the DIO1 promoter and the coding region of GAPDH were used to amplify the DNA associated with hTRβ1 and hCCR4 in vivo.
As expected, ChIP analysis confirmed that hTRβ1 is recruited to the TRE in the human deiodinase 1 (DIO1) promoter in the absence of ligand, and also revealed that TRβ interactions were modestly elevated at several times after TH treatment (Fig. 4). TH treatment also promoted very rapid recruitment of hCCR4 to the target DIO1 promoter; hCCR4 was detected at the DIO1 TRE within 5 min of TH treatment (Fig. 4). hCCR4 was maintained for several hours after TH induction, unlike SRCs and TRAP220, which tend to cycle on and off TR-regulated promoters. Thus, we conclude that hCCR4 is rapidly and stably recruited to a TRβ1 target promoter after TH treatment.
TR/hCCR4 interactions were also observed in immunofluorescence co-localization experiments. Studies performed in HeLa cells in the absence of co-expressed hCCR4 and hTRβ1 revealed weak endogenous signal for hCCR4 in the cytoplasm and nucleus and no signal for TRβ1 in the absence or presence of TH (Fig. S2J). Compartmental localization of hCCR4 was unchanged in the presence or absence of TH. By contrast, strong signals were detected in the presence of co-expressed hCCR4 and hTRβ1 (Fig. 5). Detection of hCCR4 in the nuclear and cytoplasmic compartments is consistent with roles in transcriptional response and cytoplasmic mRNA modification (9, 21). This suggests that TRs interact with hCCR4 in living cells and that TH alters the distribution of TR/hCCR4 complexes between nucleus and cytoplasm.
Fig. 5.
hCCR4 co-localizes with hTRβ1. Co-transfection of hCCR4 and hTRβ1 in HeLa cells whereas only control empty vectors were transfected as control (Fig. S2J). The cells were treated with vehicle (−TH; Upper) or 10−6 M Triac (+TH; Lower). The green and red colors show localization of CCR4 and hTRβ1, respectively. The merged image shows co-localization of CCR4 and hTRβ1. Blue and white arrows indicate the cytoplasm and nucleus, respectively.
Additionally, the binding mode by which TR interacts directly with hCCR4 was also studied by using mammalian 2-hybrid (Fig. S2K) and GST pull-down studies (Fig. S2L). These studies demonstrated that TR binds directly to hCCR4 through its hormone binding domain, and these interactions are enhanced by TH. Moreover, further mutational studies have shown that TH-dependent reduction in binding for the hCCR4200–558 mutant is likely localized in part to its N-terminal 100–200 aa sequences—a leucine-rich repeat domain (Fig. S2L).
Discussion
In the current report, we have confirmed, by using a modified yeast SGA analysis method, that one of the 4 genes with a known human homolog, i.e., hCCR4, is an important TR co-activator in mammalian cells. Transfected hCCR4 in HeLa cells potentiates TR action with a magnitude equal to or better than in previous studies with defined TR cofactors such as SRC1 and SRC2/GRIP1 (22). The overall magnitude of deleting hCCR4 is similar to that obtained in mouse embryo fibroblasts with a targeted deletion of the TRAP220 gene (23), suggesting that hCCR4 plays a role in TH action that is as important as other established TR co-regulators. CHIP analysis reveals rapid recruitment of hCCR4 to the promoter of an endogenous TR target gene (DIO1) in response to TH. As reported here, the fact that the major site of TR/CCR4 co-localization changes from the cytoplasm to the nucleus upon TH treatment leads us to suspect a role for hCCR4 in trafficking of TRs between different cellular compartments. All of these findings suggest that hCCR4 plays a very important role in TR signaling that is distinct from other known TR co-activators.
Our analysis also suggests that hCCR4 plays a major role not only in TR action but also in other hormone-dependent NRs; we observed requirements for yCCR4 in ER, AR, RXR, and RAR activity in reconstituted transcription activation assays in yeast cells (Fig. S1), irrespective of the presence or absence of GRIP1/SRC2, and in HeLa cells (Fig. S2 F–I). Subsequent to our earlier report (24) on the important role of hCCR4 as a NR co-activator, other workers reached a similar conclusion on its effects on TRα and other NRs by amplifying the activity of NR ligand binding domain in the absence of direct contacts (25). Effects of hCCR4 on TRα in this study (25) did not appear as robust (≈3-fold for TRα compared with as high as 30-fold for TRβ1 in our work), and siRNA knockdown had an even more modest effect on NR action (i.e., 50% reduction vs. >10-fold for TRβ1 in our experiments). Moreover, we demonstrated direct TR binding to hCCR4 in three ways: GST pull-downs, immunofluorescent nuclear co-localization experiments in HeLa cells, and mammalian 2-hybrid assays. Association of hCCR4 with TR in vivo was further suggested by ChIP assays. The reasons for differences between the previously published results (25) and our studies are not clear, but could be related to the fact that the other group (25) used a shorter hCCR4 cDNA than the full-length cDNA used in our experiments. Nevertheless, their study (25) supports our observations that hCCR4 is an important TR co-activator and a general NR co-regulator.
In conclusion, it is encouraging that the yeast SGA approach described herein has identified at least one TR co-factor that is important for TR activity in cultured cells and appears to display unique activities and binding modes. Studies using similar methodology to that described in the current report for CCR4 are in progress to validate the other genes with human homologs identified in the SGA analysis. Thus, of 384 yeast genes analyzed, eight (2%) were of major importance (i.e., >60% loss of function; Table S2) for TR action. Our results therefore validate the feasibility of applying an SGA strategy for systematic identification of conserved yeast genes with human homologs essential for TR and other hormone-dependent NRs.
Materials and Methods
Modified Yeast SGAs.
The SGA array was a gift from C. Boone (Toronto, ON, Canada). The yeast strain MATa:Y5563 (MATa, lys1D his3D1 leu2D0 ura3D0 met15D0 LYS2+, can1D::MFA1pr-HIS3) was used as a mating strain. Using the available selection markers of LEU2 (hTRβ1), NATR (GRIP1), and URA3 (TRE-F2x1 β-gal reporter), YEp56-hTRβ1 (LEU2), pRS426N-GRIP1 (NATR; nourseothricin; clonNAT; Werner BioAgents), and the pC2-TRE-F2 × 1-β-gal reporter (URA3) were transformed into the MATa:Y5563 yeast strain, and these strains were crossed with a MATa strain carrying gene deletions linked to the kanamycin resistance marker (17). The MATa meiotic progeny were transferred to medium that contains kanamycin and lacks leucine to select for growth of double-mutant meiotic progeny expressing hTRβ1, GRIP1, and the TRE-F2x1 β-gal reporter in a background containing the gene deletion of choice. Mutations that affect hTRβ1/TH-dependent β-gal activity were detected by a standard yeast X-gal agarose overlay assay with or without TH (Fig. 1A).
Yeast clones that had reduced blue color were retested by selecting them from a replicate plate and growing them in culture for standard liquid β-gal assay. For clones with an impaired TH-dependent response, commercial (i.e., ATCC) WT yeast strain (4741-WT) were compared with deletion mutants in the 4741 background. All experiments had suitable controls for determining specific transcriptional effects for hTRβ1 and GRIP1/SRC2 as previously described (11, 26).
Yeast Reporter Assays.
Yeast transformants were grown in minimal medium, YEp56-hTRβ1, pRS423-GRIP1/SRC2, and the β-gal reporter plasmids TRE-F2x1 and β-gal assays were performed as described previously (13). LexA-yCCR4 (HIS3) replacement was constructed as a LexA-CCR4 fusion plasmid in which full-length yeast CCR4 (residues 1–837) have been fused in-frame with LexA at the EcoRI-BamHI polylinker site of pLexA1–87, a 2-μm-based plasmid described previously (15, 16).
Expression, Interaction, and Silencing Vectors.
The hTRβ1 was cloned into BamH1/EcoR1 sites of pcDNA3.1 HisC, pHIS6-CBP-SBP, pGEX 2TK, and pM vectors. hCCR4 (CCR4/NOT6, AB033020) was cloned using appropriate primers from MegaMan Transcriptome Library (Stratagene) and cloned into BamHI site of pcDNA 3.1 HisC and pVP16 vectors. All plasmids were analyzed by restriction digestion and sequenced to confirm the inserted cDNAs. The hCCR4/NOT6 cDNA sequence was analyzed using siRNA converter program (Ambion), and siRNA sequences between 47% and 50% GC content were chosen for optimal knockdown effects. Two sets of oligonucleotides with flanking 5′ApaI and 3′EcoR1 were synthesized, annealed, and ligated into pMSCvneo-SIL vector with unique ApaI and EcoR1 sites.
Transfections and Reporter and Mammalian 2-Hybrid Assays.
HeLa cells were transferred to 6-well plates and grown to 70% confluence, and cells were transfected with DR4-LUC, hTRβ1, and/or vector alone with other factors or carrier DNA and pCMV-β-Gal (up to 0.3 μg of total plasmid DNA) in MEM supplemented with 10% charcoal-treated serum. Twelve hours later, cells were washed with PBS solution and supplemented with hormone or vehicle in hormone-free MEM-charcoal treated FBS medium. Cells were harvested after overnight incubation and whole cell extracts were prepared by 3 cycles of freezing and thawing in cell extraction buffer containing complete protease inhibitor mixture (Roche). β-Gal activities were determined in 10-μL aliquots to normalize the transfection efficiency. Samples containing 10 U of β-gal activity were used for determining luciferase activity using a commercial system (Promega). Two-hybrid protein-protein interaction assays were also performed in HeLa, with a reporter plasmid (pGL4.14-G5-Luc) containing 5 GAL4 binding elements up-stream of the thymidine kinase promoter region fused to the luciferase reporter gene. Total light emission during the initial 20 s of the reaction was measured with a luminometer (Lumat LB 9501; Berthold).
hCCR4 siRNA Knockdown.
Purified plasmids were used for transfection of PT67 cells (Becton-Dickinson) for viral packaging and to select transfectants with G418 resistance. Six colonies were chosen for developing the viral supernatants. Identical experiments were performed using vector without the insertion of duplex oligonucleotides as control. The viral supernatants were used for the infection of HeLa cells and selected using G418. Transformants were tested for absence of the selected gene by RT-PCR with appropriate primers, by Northern blot and transcription activation function of hTRβ1. Replacement of the hCCR4 siRNA knockdown in HeLa cells was performed by transient co-transfection of pcDNA3.1-HisC-CCR4 expression vector. Appropriate loss of transcriptional CCR4 expression in the siRNA knockdown was monitored by comparison to WT cell lines and cells transformed with control virus supernatants.
CHIP Assay.
HeLa cells were engineered to stably express TAP-tagged hTRb1. pTAP-TRβ1 plasmid was used to transfect exponentially growing HeLa cells by calcium phosphate precipitation. Transfectants were selected using G418 and colonies were collected after several weeks. hTRβ was verified by dot blot and immunoprecipitation using rabbit polyclonal antibodies raised against GST-hTRβ1 and peroxidase coupled goat anti-rabbit IgGs. One HeLa cell clone expressing high levels of TAP-hTRβ1 was selected for ChIP. The hTRβ1 expressing HeLa cells were transfected with an hCCR4 expression vector. WT HeLa cells were used as a negative control. These cells were treated with vehicle or 10−7 M Triac for 5, 10, 60, 120, and 360 min, and overnight, and the chromatin was cross-linked using formaldehyde for 10 min at room temperature. ChIP assays were performed using a ChIP-IT Express kit from Active Motif. One hundred fifty milligrams of total protein determined by Bio-Rad reagent and 2 mL of polyclonal antibodies were raised against GST-hTRb1. DNA from both IP input and IP bound fractions were amplified by PCR using pairs of DIO1 primers and the coding region of the GAPDH gene as internal control. The input DIO1 PCR product was measured in an aliquot of the cross-linked chromatin without IP using 0.5% of material used for hTRβ1 ChIP or 1% for CCR4 ChIP. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining. ChIP was repeated at least 3 times using 3 different chromatin preparations with identical results. The primer sequences for the human iodothyronine DIO1 promoter were identical to a previous report (27). The human GAPDH gene primers were used as controls.
Immunofluorescence and Microscopy.
HeLa cells were seeded on glass coverslips and grown for 24 h and maintained in 5% CO2 at 37°C to 70% confluence. They were transfected with hCCR4 and/or hTRβ1 or carrier DNA (up to 0.3 μg of total plasmid DNA) in MEM supplemented with 10% charcoal-treated serum. Twelve hours later, cells were washed with PBS solution and supplemented with hormone or vehicle in hormone-free MEM-charcoal treated FBS medium for 24 h. For immunofluorescence staining, cells were fixed for 20 min at room temperature in 4% paraformaldehyde in PBS solution, permeabilized in methanol for 2 min, and washed 4 times with PBS solution and probed with primary and secondary antibodies. Primary rabbit polyclonal anti-hCCR4 (1:10; produced by our laboratory) and mouse monoclonal anti-hTRβ1 (1:100; Santa Cruz Biotechnology) antibodies were used. Secondary antibodies were FITC-labeled anti-rabbit (1:00) and TRITC-labeled anti-mouse (1:100) antibodies (Santa Cruz Biotechnology). Imaging was performed using a Leica 2.00 build 0871 confocal laser system.
Pull-Down Assays.
GST-hTRβ1, GST-hTRβ1 DNA binding domain, or GST-hTRβ1-hormone binding domain was expressed in Escherichia coli BL21 cells and prepared by standard methods. Full-length or deletional mutants of [35S]methionine-labeled factors were synthesized by in vitro transcription and translation (TNT; Promega). Equivalent amounts of GST or GST-fusion protein were used for in vitro binding assays as previously described. hCCR4, hCCR4100–558, or hCCR4200–558 was cloned into pcDNA1 AR for transcription and translation to produce 35S-Met-labeled hCCR4s.
Supplementary Material
Acknowledgments.
We acknowledge the excellent technical assistance of Mr. Y.-F. Yang and Ms. R. Wang, and the collaborative assistance and advice for SGA experiments of Drs. C. Boone and N. Krogan of the Banting and Best Department of Medical Research, University of Toronto. This work was supported in part by a Diana Meltzer Abramsky Research Fellowship from the Thyroid Foundation of Canada (to X.M.); Canadian Institutes of Health Operational Grant MOP-49448 (to P.G.W.); the Mount Sinai Hospital Foundation of Toronto and Department of Medicine Research Funds; the Julius Kuhl and Temmy Latner/Dynacare Family Foundations; the Joseph and Mildred Sonshine Family Centre for Head and Neck Diseases (to P.G.W.); and National Institutes of Health Grants GM 78078 and DK51281 (to C.L.D and J.D.B.).
Footnotes
Presented in part at the 77th Annual Meeting of the American Thyroid Association, Phoenix, October 11–15, 2006.
Conflict of interest statement: J.D.B. is a consultant to and shareholder of Karo Bio AB, which has commercial interests in thyroid hormone mimetics.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910134106/DCSupplemental.
References
- 1.Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Lazar MA. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–466. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]
- 3.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 4.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 6.Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- 7.Morel AP, et al. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J Cell Sci. 2003;116:2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- 8.Dlakic M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 9.Tucker M, et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 10.Meng X, et al. Corepressor/coactivator paradox: potential constitutive coactivation by corepressor splice variants. Nucl Recept Signal. 2006;4:e022. doi: 10.1621/nrs.04022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walfish PG, et al. Yeast hormone response element assays detect and characterize GRIP1 coactivator-dependent activation of transcription by thyroid and retinoid nuclear receptors. Proc Natl Acad Sci USA. 1997;94:3697–3702. doi: 10.1073/pnas.94.8.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butt TR, Walfish PG. Human nuclear receptor heterodimers: opportunities for detecting targets of transcriptional regulation using yeast. Gene Expr. 1996;5:255–268. [PMC free article] [PubMed] [Google Scholar]
- 13.Anafi M, et al. GCN5 and ADA adaptor proteins regulate triiodothyronine /GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol Endocrinol. 2000;14:718–732. doi: 10.1210/mend.14.5.0457. [DOI] [PubMed] [Google Scholar]
- 14.Velasco LF, et al. Thyroid hormone response element organization dictates the composition of active receptor. J Biol Chem. 2007;282:12458–12466. doi: 10.1074/jbc.M610700200. [DOI] [PubMed] [Google Scholar]
- 15.Meng X, et al. E1A and a nuclear receptor corepressor splice variant (N-CoRI) are thyroid hormone receptor coactivators that bind in the corepressor mode. Proc Natl Acad Sci USA. 2005;102:6267–6272. doi: 10.1073/pnas.0501491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng X, et al. Cellular context of coregulator and adaptor proteins regulates human adenovirus 5 early region 1A-dependent gene activation by the thyroid hormone receptor. Mol Endocrinol. 2003;17:1095–1105. doi: 10.1210/me.2002-0294. [DOI] [PubMed] [Google Scholar]
- 17.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 18.Jones AL, et al. SAC3 may link nuclear protein export to cell cycle progression. Proc Natl Acad Sci USA. 2000;97:3224–3229. doi: 10.1073/pnas.050432997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuberth C, Richly H, Rumpf S, Buchberger A. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 2004;5:818–824. doi: 10.1038/sj.embor.7400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan AH, Neely KE, Vignali M, Reese JC, Workman JL. Promoter targeting of chromatin-modifying complexes. Front Biosci. 2001;6:D1054–D1064. doi: 10.2741/hassan. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng W, et al. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 23.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 24.Meng X, et al. Characterization of CCR4-NOT6 coactivator action on the human thyroid receptor. Thyroid. 2007;17(suppl 1):128. [Google Scholar]
- 25.Garapaty S, Mahajan MA, Samuels HH. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting Factor NIF-1. J Biol Chem. 2008;283:6806–6816. doi: 10.1074/jbc.M706986200. [DOI] [PubMed] [Google Scholar]
- 26.Shah V, et al. Complex actions of thyroid hormone receptor antagonist NH-3 on gene promoters in different cell lines. Mol Cell Endocrinol. 2008;296:69–77. doi: 10.1016/j.mce.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters. in vivo. Proc Natl Acad Sci USA. 2002;99:7934–7939. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.