Abstract
Proper, graded communication between different cell types is essential for normal development and function. In the nervous system, heart, and for some cancer cells, part of this communication requires signaling by soluble and membrane-bound factors produced by the NRG1 gene. We have previously shown that glial-derived neurotrophic factors activate a rapid, localized release of soluble neuregulin from neuronal axons that can, in turn promote proper axoglial development (Esper, R. M., and Loeb, J. A. (2004) J. Neurosci. 24, 6218–6227). Here we elucidate the mechanism of this localized, regulated release by implicating the delta isoform of protein kinase C (PKC). Blocking the PKC delta isoform with either rottlerin, a selective antagonist, or small interference RNA blocks the regulated release of neuregulin from both transfected cells and primary neuronal cultures. PKC activation also leads to the rapid phosphorylation of the pro-NRG1 cytoplasmic tail on serine residues adjacent to the membrane-spanning segment, that, when mutated markedly reduce the rate of NRG1 activity release. These findings implicate this specific PKC isoform as an important factor for the cleavage and neurotrophin-regulated release of soluble NRG1 forms that have important effects in nervous system development and disease.
The neuregulins (NRGs)2 are a family of growth and differentiation factors with a broad range of functions during development and in the adult. NRGs are necessary for glial and cardiac development and participate in a wide range of biologic processes ranging from proper formation of peripheral nerves and the neuromuscular junction to tumor growth (2–9). The NRGs have also been implicated as both potential mediators and therapeutic targets for a number of human diseases including cancer, schizophrenia, and multiple sclerosis (10–12). NRGs function as mediators of cell-to-cell communication through a multitude of alternatively spliced isoforms arising from at least four distinct genes that bind to and activate members of the epidermal growth factor receptor family HER-2/3/4 (ErbB-2/3/4) (13–19).
Although all known isoforms of the NRG1 gene have an epidermal growth factor-like domain sufficient to bind to and activate its receptors (20), products of this gene are divided into three classes based on structurally and functionally different N-terminal regions (21) The type I and II forms have a unique N-terminal, heparin-binding Ig-like domain (22–26). This Ig-like domain potentiates the biological activities of soluble NRG1 forms and leads to their highly selective tissue distributions through its affinity for specific cell-surface heparan sulfates (12, 20, 27, 28). These forms are first expressed as transmembrane precursors (pro-NRG1) that undergo proteolytic cleavage to release their soluble ectodomains. The type III NRG1 forms, on the other hand, are not typically released from cells, because their N-terminal domain consists of a cysteine-rich domain that can serve as a membrane tether making this form ideal for juxtacrine signaling. This form has been strongly implicated to be important peripheral nerve myelination (29–31).
While many of the biological functions of type I/II NRG1 forms are less clear, their ability to be released from axons in the peripheral and central nervous systems in a regulated manner provides the potential for long range cell-cell communication not possible from membrane-bound forms. Studies examining the regulation of type I NRG1 release from neuronal axons have implicated protein kinase C (PKC) as a mediator of NRG1 release from pro-NRG1 in transfected cell lines (32). Subsequent studies in intact neurons found that PKC activation was sufficient to release NRG1 from sensory and motor neuron axons and that NRG1 could also be released by Schwann cell-derived neurotrophic factors, such as BDNF and GDNF (1). Recently, the β-secretase protease BACE1 has been suggested to cleave these NRG1 forms so that when it is knocked out in mice, deficits similar to those seen in NRG1 knockouts are seen (33, 34). These findings suggest that reciprocal communication between NRG1s and neurotrophins could be an important mechanisms for local axoglial communication that is critical for normal peripheral nerve development. Consistently, PKC has been implicated as a key mediator for the electrically mediated release of NRG1 from cultured cerebellar granule cells and pontine nucleus neurons (35).
The PKC family consists of 10 serine/threonine kinases isoforms (α, βI, βII, γ, δ, ϵ, ζ, θ, λ, and η) each with a unique cellular distribution, target specificity, mechanism of activation, and function (36). One of these functions promotes the cleavage and release of soluble signaling proteins that are initially synthesized as membrane-spanning precursors. In addition to NRG1, other proteins released upon PKC activation include epidermal growth factor, transforming growth factor-α, amyloid precursor protein, l-selectin, and interleukins (1, 37–43). We hypothesize that neurotrophic factors induce the cleavage and release of NRG1 from pro-NRG1 through PKC activation. This hypothesis seems reasonable, because neurotrophin binding to the Trk family of neurotrophin receptor tyrosine kinases, but not the low affinity neurotrophin receptor p75 (44), activates phospholipase Cγ-mediated conversion of membrane-bound phosphatidylinositol bisphosphate to inositol triphosphate and diacylglycerol, which in turn, can activate PKC (45–48). Although this can be achieved using phorbol 12-myristate 13-acetate (PMA), a diacylglycerol analog sufficient to activate most PKC isozymes (48), the exact PKC isoform and mechanism by which this occurs is not known. Here, we demonstrate NRG1 is released from cells through direct activation of the PKCδ isoform using siRNA and PKC isoform-specific inhibitors in transfected Chinese hamster ovary (CHO) cells, PC12, and primary neuronal cultures. We further demonstrate that PKC activation induces rapid phosphorylation of the cytoplasmic tail of pro-NRG1 on specific serine residues that are required for efficient NRG1 activity release. These findings provide mechanistic insights into how highly localized, reciprocal signaling occurs along neuronal axons, which has important implications for normal development and disease.
EXPERIMENTAL PROCEDURES
Neuron Cultures, Stable NRG1-expressing Cell Lines, and Transient Transfection
Sensory neurons were cultured from E12 chicken dorsal root ganglia as previously described in serum-free media (L15, penicillin/streptomycin, 0.6% glucose, 2 mm glutamine, 0.06% NaHCO3) supplemented with N2, B27, and 10 ng/ml NGF (Invitrogen) (1, 49). The cultures were treated with 1.75 mm Ara-C (Sigma) on alternate days (24 h on, 24 h off, 24 h on, then 24 h off) to remove contaminating fibroblasts and glia. PC12TrkB cells were a generous gift from Dr. Moses Chao. This rat pheochromocytoma cell line expresses both the NGF receptor (TrkA) and the BDNF receptor (TrkB), but no detectable endogenous neuregulin, and can be induced to differentiate into neuron-like cells with neurotrophic factor stimulation.
Stably-transfected CHO cell lines were prepared using the Flp-In system CHOFRT cell line from Invitrogen (Carlsbad, CA). The gene encoding chicken pro-NRG1 was amplified by PCR from pCDNAI/λ12.7 to introduce flanking restriction sites and cloned into the multiple cloning site of the pCDNA5/FRT vector (Invitrogen). The new pro-NRG1 vector was then co-transfected with the Flip Recombinase vector (pOG44) into the CHOFRT cells to create stable pro-NRG1-expressing fibroblast cell lines, which were selected with 400 μg/ml hygromycin, according to the manufacturer's instructions. The pro-NRG1 vectors were transiently transfected into PC12TrkB cells using Lipofectamine/PLUS (Invitrogen).
Measurement of NRG1 Activity Release from Cultured Neurons
NRG1 activity released from cells was measured with a sensitive and highly specific biological assay as previously described (1). In brief, L6 myoblasts were plated into 48-well plates (Corning Costar), incubated at 37 °C, and allowed to fuse and differentiate for 7–8 days. The L6 culture media was removed and replaced with the test sample, and the cells were incubated for 45 min at 37 °C, after which the cells were placed on ice and lysed. Following immunoprecipitation with erbB2 and erbB3 antibodies (Neomarkers), proteins were resolved on a 6% reducing acrylamide gel and transferred to a polyvinylidene difluoride membrane. Western blots were performed using an antibody against phosphotyrosine (pY, 4G10) as described previously (28) and then stripped and re-probed with a mixture of the erbB2 and erbB3 antibodies to measure the total amount of erbB protein present in each lysate. Specificity has been shown by blocking this activity with NRG antibodies, heparin, and a soluble NRG1 antagonist. Band intensity was quantified on a flatbed scanner with a transparency adapter using Metamorph image analysis software as previously described (28). The calculated ratio of tyrosine-phosphorylated erbB protein to total erbB protein provides a quantitative linear measurement of NRG1 concentration.
Neurotrophic Factors, Antibodies, Antagonists, and PKC Inhibitors
Recombinant NGF was purchased from Invitrogen, and recombinant human BDNF was a gift from Regeneron. Recombinant NRGβ1-(1–246) was from Amgen. Antibodies specific for pro-NRG1 (sc348), PKCδ (sc937), and erbB (sc284 and sc285) were from Santa Cruz Biotechnology (Santa Cruz, CA), and the β-actin antibody (AC-15) was from Sigma. A second anti-NRG1 antibody that recognizes the truncated cytoplasmic tail of the c isoform, 1310, was provided by Amgen (27). Additional erbB antibodies were from Neomarkers (Fremont, CA). For Western blots, each antibody was used at 1:5000 dilution, and the pro-NRG1 antibody was used at 1:100 for immunoprecipitation. Rottlerin and PMA were from Sigma. Go6976 and GF109203X (bisindolylmaleimide I) were from Calbiochem. Multiple PKCδ siRNAs and non-silencing control siRNA were from Qiagen (Valencia, CA) and designed by Internet-based software provided by Qiagen.
To knockdown the delta (δ) isoform of PKC, 2.5 μg of siRNA was transfected into CHO or PC12 cell lines, or primary chicken dorsal root ganglia sensory neurons (in 12-well plates at 4.0 × 105 cells/well) with 5 μl of Lipofectamine2000 (Invitrogen). The cells were incubated for 4 days, which was required for complete protein knockdown as assessed by Western blot of cell lysates.
[32P]Phosphate Labeling of Pro-NRG1 and TLC
[32P]orthophosphate (H3PO4) was purchased from MP Biomedicals (formerly ICN, Irvine, CA), and used for in vitro phosphoprotein labeling as described (50). Briefly, CHO fibroblasts expressing pro-NRG1 were plated in 12-well plates (Costar) at a density of 1.25 × 105 cells/dish and allowed to grow overnight. The next day, the media was changed, and the cells were washed three times in 1 ml of phosphate-free DMEM, and incubated in phosphate-free DMEM for 1 h at 37 °C. The cells were washed twice in phosphate-free DMEM, and incubated with 250 μCi/ml [32P]orthophosphate in 500 μl of phosphate-free DMEM for 2 h at 37 °C. Some cells were treated with 12.5 μm rottlerin for 15 min prior to PMA treatment. PMA (10 nm) was added to the cells for variable periods of time before placing the cells on ice. The radioactive media was removed and discarded, and the cells were washed in 1 ml of ice-cold DMEM, lysed in 1 ml of ice-cold radioimmune precipitation assay buffer (1% Nonidet P-40, 0.5% sodium deoxycholic acid, 0.1% SDS, 0.15 m NaCl, 0.05 m Tris, 0.05 m EDTA, 5 μg/ml leupeptin, 1 μg/ml pepstatin, and 10 mm sodium o-vanadate), and immunoprecipitated overnight with the pro-NRG1 antibody (sc348) at 1:100 dilution with Protein-A-Sepharose beads (Sigma). The immunoprecipitated proteins were resolved by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane, which was then exposed to Kodak XAR-5 film at −80 °C with an enhancer screen. Parallel wild-type and pro-NRG1-expressing CHO cell cultures were lysed in 2× dithiothreitol sample buffer and run on the same gel without immunoprecipitation to serve as controls. A Western blot was performed by probing the membrane with the pro-NRG1 antibody (sc348) to confirm the identity of the 32P-incorporated protein bands. This antibody is directed against the C-terminal region of pro-NRG1 and detects both pro-NRG1 and cleaved tail fragments.
TLC for phosphoamino acid analysis was performed essentially as described (50). Specifically, protein bands corresponding to the pro-NRG1 precursor and the proteolytic tail fragments were excised from the membrane and hydrolyzed with 6 n HCl at 110 °C for 1 h, and concentrated in a SpeedVac to near dryness. Samples were reconstituted to 10 μl in pH 1.9 TLC buffer (25:87:897 formic acid, glacial acetic acid, water) and separated on 100-μm cellulose TLC plates (Fisher, Pittsburgh, PA) with non-radioactive phosphoamino acid standards (Sigma) in PAA Buffer (75:50:15:60 n-butanol, pyridine, glacial acetic acid, water), followed by amine staining with ninhydrin (Fisher). TLC plates were then exposed to Kodak XAR-5 film at −80 °C with enhancer screens.
Site-directed Mutagenesis and Assay of Pro-NRG Cleavage
Specific serine residues of the pro-NRGβ1c cytoplasmic tail were mutated to alanine using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA), per the manufacturer's instructions. PCR primers for the specific regions to be mutated were designed using the Stratagene Codon Replacement strategy. After PCR amplification of the mutated pro-NRG DNA sequences, they were inserted into the pCDNA5/FRT vector (Invitrogen) and sequenced for verification. The new pro-NRG1 vectors were then co-transfected with the Flip Recombinase vector (pOG44) into the CHOFRT cells to create stable pro-NRG1-expressing fibroblast cell lines, which were selected with 400 μg/ml hygromycin, according to the manufacturer's instructions. Additional site-specific mutant pro-NRGβ1c sequences in the pCDNA/FRT vector were purchased from Genscript (Piscataway, NJ) and co-transfected with the Flip Recombinase vector to create stable CHOFRT cell lines.
The pro-NRG1-expressing CHO cells were plated into 12-well plates and treated with 10 nm PMA for increasing amounts of time. The cells were placed on ice and lysed. Cell lysates were separated on a 10% polyacrylamide gel. Western blot of the resulting membranes was done with the 1310 antibody, specific for the pro-NRG cytoplasmic tail.
RESULTS
PKC Activation with PMA Promotes Rapid Pro-NRG1 Cleavage and Release of NRG1 Activity
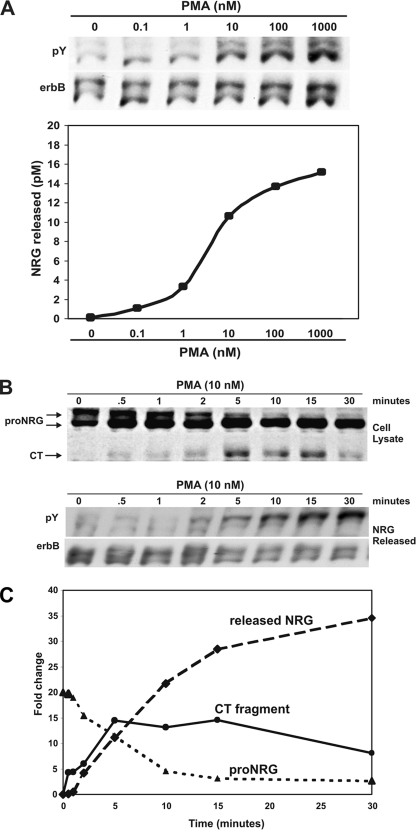
Activation of PKC with the phorbol ester, PMA, causes the release of NRG1 activity from neurons and transfected cells (1, 32). To determine the optimal concentration of PMA to promote NRG1 activity release from the Type I (β1a) chicken pro-NRG1 precursor, stably transfected CHO cells treated with increasing concentrations of PMA, and released NRG1 activity was measured with a sensitive and specific biological assay measuring phosphorylation of erbB receptors in cultured myotubes (Fig. 1A). This quantitative bioassay was used instead of Western blotting due to the absence of sufficiently sensitive and specific antibodies for soluble NRG1 (1). Increasing amounts of NRG1 activity were released from these cells in response to increasing concentrations of PMA, with a maximal level achieved at 1 μm. 10 nm PMA was determined to be approximately half-maximal to promote NRG1 activity release from these cells.
FIGURE 1.
PKC activation promotes pro-NRG1 cleavage and release of NRG1 activity. A, CHO fibroblasts stably transfected with pro-NRGβ1a were treated with increasing concentrations of PMA for 10 min, and NRG1 activity released was approximated by erbB phosphorylation in L6 myotubes. 10 nm PMA stimulated approximately half-maximal amounts of NRG1activity release. B, stably transfected CHO cells expressing pro-NRGβ1a were treated with 10 nm PMA for increasing periods of time. Total cell lysates were collected, and pro-NRG1 and cytoplasmic tail fragments (CT) were detected by Western blot analysis (sc348). CT fragments were detected within 30 s of PKC activation. On Western blots, pro-NRG1 appeared as two distinct bands, due to variable N-linked glycosylation of the ectodomain. NRG1 activity release was measured by erbB phosphorylation. C, quantitation of the relative amounts of each protein band in the NRG1 Western blot indicated that the CT fragments appeared within 30 s of PKC activation, rose sharply, and reached a plateau (solid line, circles). Within 2 min of PKC activation, there was a dramatic (∼75%) reduction in the amount of the upper pro-NRG1 band (dotted line, triangles) as the amount of released NRG1 activity increased in parallel (dashed line, diamonds). The entire process appears to be completed by 15–20 min after PMA addition.
The kinetics of PKC-mediated NRG1 activity release was determined using 10 nm PMA (Fig. 1, B and C). The amount of pro-NRG1 and its cleavage products were determined by Western blot of total cell lysates using an antibody against the cytoplasmic tail of pro-NRG1. pro-NRG1 appears as two distinct bands on Western blots due to variable N-linked glycosylation of the extracellular domain (51). Within 30 s of PKC activation, pro-NRG1 cytoplasmic tail fragments (CT) begin to appear and reach a maximum concentration by 5 min after PKC activation before decreasing to an approximately half-maximal steady level (Fig. 1C). This observation correlated with the disappearance of mature (upper/higher glycosylated) pro-NRG1 from the cells and the appearance of soluble NRG1 activity in the culture media, as detected by erbB receptor phosphorylation in L6 myotubes. Thus, PKC activation was sufficient to promote the cleavage of pro-NRG1 and subsequent release of soluble NRG1 activity in a process that is rapid and saturable.
PKCδ Is Necessary for Pro-NRG1 Cleavage and Release of Soluble NRG1 Activity from CHO Fibroblasts
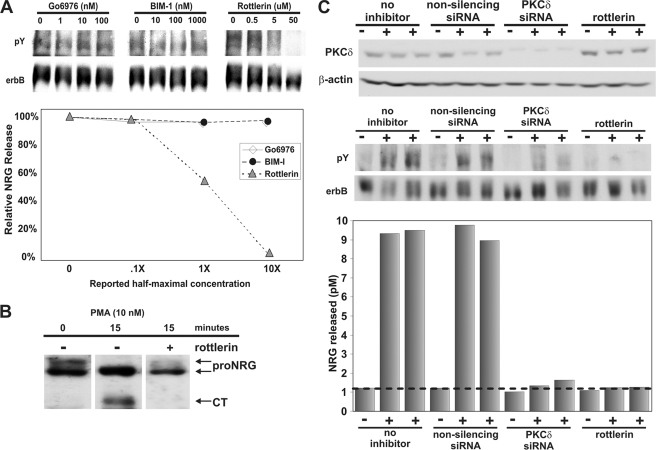
To determine the specific isoform of PKC responsible for pro-NRG1 cleavage and release, we tested a series of isoform-specific PKC antagonists, because PMA activates all isoforms of PKC. GF109203X (bisindolylmaleimide I) is a broad spectrum antagonist with strongest activity against the α, βI, βII, γ, and ϵ, but not δ forms, whereas Go6976 is more specific against the α, βI, and βII forms (52). Rottlerin is a selective antagonist for the δ isoform (53–55). In dose-response studies with each of these PKC inhibitors only rottlerin blocked PMA-stimulated NRG1 activity release from transfected fibroblasts, demonstrating a potentially specific role for PKCδ in the release of NRG1 (Fig. 2A). When CHO cells expressing pro-NRG1 were pre-treated with the PKCδ antagonist, rottlerin, stimulation with PMA failed to promote pro-NRG1 cleavage as shown by Western blot in Fig. 2B. Although there are loading differences, this clearly shows a significantly reduced ratio of the cleaved cytoplasmic tail, which we have repeated in three other experiments (not shown). Consistently, the more heavily glycosylated upper pro-NRG1 band is only minimally reduced with the antagonist (ratio of 0.5 to 0.4, with the antagonist). This strongly suggests that the δ isoform of PKC works in part through promoting the pro-NRG1 cleavage event, which is a prerequisite for the release of soluble NRG1. As a complementary approach to using pharmacological inhibitors, we asked whether three distinct siRNAs against PKCδ would have a similar effect on pro-NRG1 cleavage and release. Each of these siRNAs that was first shown to effectively reduce the levels of PKCδ by Western blotting markedly reduced NRG1 activity release from transfected CHO cells following PKC activation (data shown only for one of these siRNAs) (Fig. 2C). Thus, using a combination of pharmacologic and genetic approaches, we demonstrate that the δ isoform of PKC is necessary for the release of NRG1 activity by PMA.
FIGURE 2.
PKCδ is necessary for NRG1 activity release from CHO cells. A, CHO cells transfected with pro-NRGβ1a were treated with increasing concentrations of various PKC inhibitors before treatment with 10 nm PMA. X indicates the half-maximal inhibitory concentration reported in the literature for each inhibitor. After 10 min of PMA stimulation, NRG1 activity release was measured by erbB phosphorylation in L6 myotubes and quantified in the graph below the gel. Only the PKCδ antagonist, rottlerin, was able to inhibit NRG1 activity release. B, following 10 nm PMA for 15 min with or without pre-treatment with 12.5 μm rottlerin, a Western blot of the cell lysate (sc348) revealed that rottlerin completely blocked the cleavage of pro-NRG1 and accumulation of the cytoplasmic tail fragment following PKC activation. C, CHO-pro-NRGβ1a cells were treated with either non-silencing control siRNA, siRNA against PKCδ, or 12.5 μm rottlerin before treatment with 10 nm PMA. siRNA knockdown was determined by collecting total cell lysate and performing a Western blot against PKCδ. The membrane was reprobed for β-actin to determine equal protein loading. NRG1 activity release from these cells was measured by erbB phosphorylation in L6 myotubes and quantified in the graph below. PKCδ siRNA and rottlerin both inhibited PMA-stimulated NRG1 activity release in this and replicate experiments.
PKCδ Is Necessary for NRG1 Activity Release from Neurons and PC12 Cells in Response to Neurotrophins
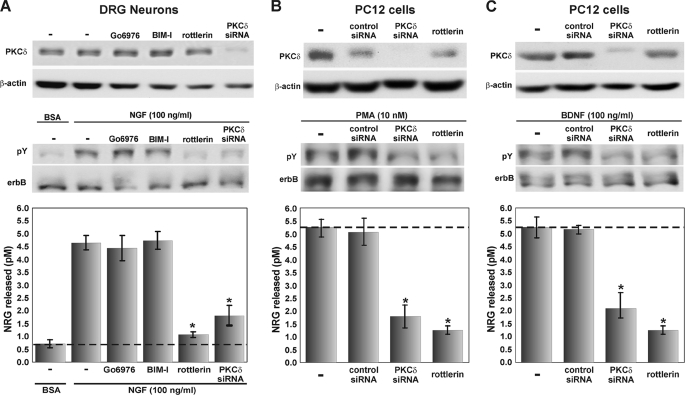
Because neurotrophins promote NRG1 activity release from axons as well as activate PKC through Trk receptors (44, 56, 57), we asked whether PKCδ is required for NRG1 activity release after NGF stimulation of primary dorsal root ganglia sensory neurons (Fig. 3A). Using the same PKCδ siRNAs and pharmacologic inhibitors, we found that only inhibition of the δ isoform of PKC effectively reduced the amount of soluble NRG1 activity released after PKC activation with NGF. Similarly, PKCδ activity is necessary for the release of NRG1 from PC12TrkB cells that express both pro-NRG1 and the BDNF receptor, trkB, either after stimulation with PMA (Fig. 3B) or BDNF (Fig. 3C), as documented by a significant reduction of NRG1 activity release after inhibition of PKCδ with either siRNA or rottlerin, but not with inhibition of other isoforms of PKC (data not shown), nor by the neurotrophic factors by themselves (1). Taken together, these data strongly suggest that neurotrophins promote the rapid release of NRG1 from neurons through a mechanism that requires the δ isoform of PKC.
FIGURE 3.
The δ isoform of PKC is necessary for the neurotrophin-mediated release of NRG1 from neurons. A, primary cultures of chick dorsal root ganglia sensory neurons were treated with 100 ng/ml NGF in the absence or presence of PKCδ inhibition with either siRNA knockdown (for 4 days) or rottlerin. PKCδ inhibition significantly reduced the neurotrophin-stimulated release of endogenous NRG1 (error bars represent ±S.E. from triplicates; * = p < 0.01 Student's t test). B and C, PC12TrkB cells expressing pro-NRGβ1a were transfected with siRNA against PKCδ and incubated for 4 days for complete knockdown. Parallel non-transfected cells were treated with 12.5 μm rottlerin for 30 min, and the cells were exposed to either 10 nm PMA (B) or 100 ng/ml BDNF (C). Culture media was assayed for release of soluble NRG1, and Western blots of CHO cell lysate showed specific knockdown of PKCδ. Inhibition of PKCδ with siRNA or rottlerin significantly reduced NRG1 activity release from cells after PKC activation with PMA or stimulation with BDNF (error bars represent ±S.E. from triplicates; * = p < 0.01 Student's t test comparing of these individually with the NGF alone control).
Specific Serine Residues on the Cytoplasmic Tail of Pro-NRG1 Are Phosphorylated and Required for Efficient NRG1 Activity Release
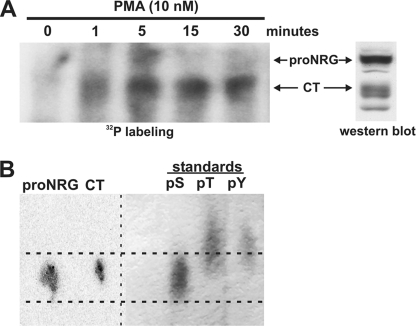
The exact mechanism of how PKC activation leads to rapid cleavage and release of NRG1 from pro-NRG1 is not known. One possibility is that PKC activates this process by phosphorylating pro-NRG1 on its cytoplasmic tail. To determine if this is correct, we treated CHO cells expressing pro-NRG1 with 10 nm PMA in the presence of [32P]ATP. Parallel non-radioactive CHO cell cultures served as controls. Non-transfected CHO cell lysates were separated on the same gel to identify the radiolabeled bands. 32P incorporation into the cytoplasmic tail of pro-NRG1 was detected within 1 min and remained for at least 30 min after PKC activation (Fig. 4A). The kinetics of the labeling reaction showed maximal incorporation of 32P into both the intact pro-NRG1 precursor and the cleaved cytoplasmic tail at 5 min but then complete disappearance of the intact pro-NRG1 by 15 min. These kinetics findings suggest PKC-induced phosphorylation of the cytoplasmic tail results in a rapid cleavage event that is complete within minutes after phosphorylation.
FIGURE 4.
PKC activation induces phosphorylation of serine residues on the pro-NRG1 cytoplasmic tail. A, CHO-pro-NRGβ1a cells were grown with [32P]orthophosphate prior to treatment with 10 nm PMA for increasing periods of time. Pro-NRG1 and cleaved cytoplasmic tail fragments (CT) were immunoprecipitated from cell lysates with an antibody directed against a portion of the pro-NRG1 cytoplasmic tail (sc348). The lysates were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane that was exposed to film. The membrane was probed with sc348 to identify the 32P-incorporated proteins (far right). After 1 min of PKC activation, there was dramatic incorporation of 32P into the cytoplasmic tail (CT) of pro-NRG1, suggesting rapid pro-NRG1 cleavage following phosphorylation. B, 32P-incorporated pro-NRG1 and the cleaved cytoplasmic tail from the 5-min time point were excised from the membrane and hydrolyzed in 6 n HCl and separated on a 100-μm TLC plate; cold phosphoamino acid standards were run in parallel, and the plate was exposed to film. Phosphorylated serine was the predominant phosphoamino acid detected in both pro-NRG1 and the cleaved cytoplasmic tail.
To determine the specific amino acids phosphorylated, protein bands corresponding to pro-NRG1 and the cleaved tail fragments (CT) from the 5-min time point were subjected to phosphoamino acid TLC. Non-radioactive phosphoserine, phosphothreonine, and phosphotyrosine were used as running standards. The 32P signal from the hydrolyzed pro-NRG1 and cytoplasmic tail bands aligned only with the phosphoserine standard, suggesting that PKC activation specifically led to phosphorylation on serine residues on the cytoplasmic tail of pro-NRG1 (Fig. 4B).
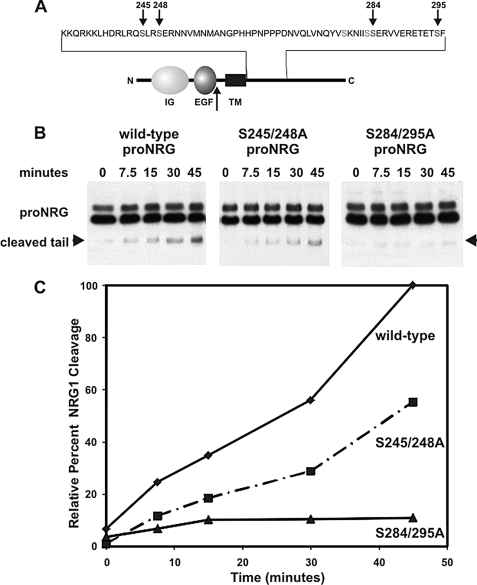
To determine the contributions of specific serine residues on the cytoplasmic tail for NRG1 cleavage, the pro-NRGβ1c form was examined with specific serine residues changed to alanines for PMA-induced pro-NRG1 cleavage. This is a different splice form than that used for previous experiments. The reason is that this particular pro-NRG1 splice form has the shortest cytoplasmic tail that still undergoes regulated cleavage (β1c isoform) (58). Two constructs shown in Fig. 5A were prepared by site-directed mutagenesis converting pairs of serine residues to alanines to compare the relative importance of proximal and more distal regions of the cytoplasmic tail. These particular pairs of serines were chosen from 25 possible serine residues based on known consensus sequences for PKCδ phosphorylation (59, 60). Each construct was stably transfected into CHO cells and compared with the wild-type construct. The cells were treated with 10 nm PMA for increasing periods of time, and the amount of pro-NRG1 cleavage, as evidenced by the appearance of the C-terminal cytoplasmic fragment, was assessed by Western blotting. It is worth noting that, even without any mutations, this alternatively spliced form with a shorter cytoplasmic tail is cleaved at a slower rate than the full-length cytoplasmic domain form shown in Fig. 1. Although the cleaved cytoplasmic domain was readily produced by PMA in the wild-type construct, mutant forms missing serines 284 and 295 had substantially less PKC-mediated cleavage of pro-NRGβ1c. Interestingly, independent loss of serines 245 and 249 that are closer to the membrane-spanning domain had a more modest reduction in cleavage rate of pro-NRG1. Taken together, these results suggest that, although a number of cytoplasmic serine residues near the membrane-spanning segment of pro-NRG1 can contribute to the regulated release of NRG1, the more distally located serine residues at 284 and 295 appear to be more important.
FIGURE 5.
Serine residues 284 and 295 of the pro-NRG1 cytoplasmic tail are necessary for PKC-mediated cleavage. A, site-directed mutagenesis was used to determine the requirement for pairs of specific serine residues on the pro-NRGβ1c cytoplasmic tail. CHO cells expressing wild-type or mutant forms of pro-NRGβ1c with Ser-Ala amino acid substitutions at either proximal (S245A/S249A) or more distal sites (S284A/S295A) that correspond to consensus sequences were treated with 10 nm PMA for increasing periods of time. The arrow indicates the site of pro-NRG1 cleavage. B, cell lysates were separated on SDS-PAGE, and Western blot with the 1310 antibody identified bands corresponding to pro-NRG1 and the cleaved cytoplasmic tail fragment. Mutation of serines 284 and 295 to alanines significantly blocked PKC-mediated cleavage of pro-NRG1 more so than mutations closer to the membrane-spanning region at serines 245 and 249. This experiment was quantified in C and expressed relative to 100% of the signal seen with the wild-type construct at 45 min and repeated once showing the same results.
DISCUSSION
The complexity of the nervous system comes from a dynamic system of communication between billions of cells that are arranged in a functionally appropriate way. One of the great mysteries is how this communication occurs at specific subcellular regions, such as at synapses or at specific segments of axons or dendrites. Here, we have tied closer together two important intercellular signaling systems, the neurotrophins and the neuregulins, by elucidating how soluble forms of NRG1 can be released at specific subcellular locations through PKCδ-activated neurotrophin signaling as summarized in the model in Fig. 6. This mechanism is independent of transcription and therefore does not require nuclear communication, which would necessitate a mechanism that distinguishes the activation site from an enormous number of possible dendritic and axonal regions. PKCδ signaling could serve this function well as it becomes adherent to local membranes after activation and thereby can keep signaling at these local regions where NRG1 activity release is needed (46, 48, 61, 62). Once activated, PKCδ directly or indirectly induces the rapid phosphorylation of the cytoplasmic domain of pro-NRG1 that appears to result in the rapid proteolytic cleavage and release of the soluble ectodomain. Finally, once released at these local sites, soluble forms of NRG1 remain restricted in their localization through specific heparan sulfate proteoglycans where they can exert important effects (27, 51).
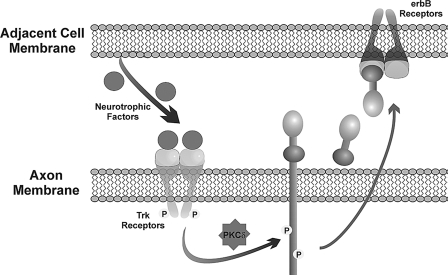
FIGURE 6.
A model for bidirectional axoglial signaling involving NRG1, neurotrophic factors, and PKC. In this model, cells that surround neuronal axons, such as glial or muscle cells, secrete specific neurotrophic factors that activate the Trk receptors on the axon. This results in the activation of PKCδ that rapidly induces the phosphorylation and cleavage of pro-NRG1 providing a reciprocal signal back to the neurotrophin-producing cell. Once released, NRG1 binds to and activates erbB receptors on the adjacent cell resulting in important, localized communication.
Previous work from our laboratory has demonstrated that Schwann cell-derived neurotrophic factors, especially BDNF and GDNF, promote the rapid release of NRG1 from axons through a bidirectional signaling system at the axoglial interface (1), and that muscle-derived neurotrophic factors may play a similar role at the neuromuscular junctions (63–66). Here we have extended this work to implicate PKCδ as a primary mediator of how neurotrophic factors can produce these highly localized signaling events. We postulate that PKCδ activation leads to the phosphorylation of specific serine residues on the pro-NRG1 cytoplasmic tail, which rapidly promotes its cleavage and subsequent release of soluble NRG1 from the axon at sites near where it is activated. Once released, NRG1 can then bind and activate its receptors on the adjacent cell, which, in the case of developing Schwann cells, is critical for their survival and differentiation (67). PKC activation has been shown to be essential for the regulated release of many other transmembrane signaling proteins that release their active ectodomains raising the possibility that similar, localized signaling mechanism could result in their processing and release as well (42, 68–70). Consistently, the Alzheimer precursor protein has been shown to be phosphorylated in response to PKC activation (71, 72).
Previous studies have demonstrated that the CT of pro-NRG1 is critical for the efficient processing and release of the soluble NRG1 ectodomain (73). A minimum of 13 amino acids of the cytoplasmic tail are needed for NRG1 activity release, and deletion studies have shown that two critical regions of the pro-NRG1 cytoplasmic tail are needed for the efficient cleavage and release of NRG1 (58). Within these regions are 25 conserved serine residues, several of which match known phosphorylation consensus sequences (59, 60). Our findings here show that the loss of some of these serine residues by site-directed mutagenesis results in significant reductions in PKC-induced NRG1 activity release suggesting that their phosphorylation is the trigger that promotes pro-NRG1 cleavage and release. Of these 25 possible phosphorylation sites, either Ser-284 and/or Ser-295 appear to be critical. Mechanistically, phosphorylation could induce a conformational change in the protein that exposes the extracellular, cleavage region to a protease or produces vesicular movements that bring pro-NRG1 in contact with the protease. Other regions of the cytoplasmic tail may also be important as we generally see reduced efficiency in cleavage with the shorter forms (compare Figs. 1 and 5). In either case, the time between phosphorylation and cleavage and release is very rapid with little intracellular accumulation of phosphorylated pro-NRG1 or the cleaved ectodomain (32), but a large pool of phosphorylated cleaved cytoplasmic domain.
Previous pulse-chase labeling studies suggest that pro-NRG1 is first expressed on the cell surface after which it is rapidly processed and released or degraded, depending on the types of extracellular signals it receives, and this process requires vesicular acidification (32). Our results here also show that the slower migrating, more heavily glycosylated pro-NRG1 is more efficiently cleaved than the faster migrating band. Our earlier study also demonstrated that this slower migrating band is selectively expressed on the cell surface, suggesting that the glycosylation state may determine which forms of pro-NRG1 are cleaved. Thus, the slight delay observed between phosphorylation and cleavage and release could be due to vesicular transport of more highly glycosylated phosphorylated forms of pro-NRG1 from the cell surface to an intracellular compartment where the cleaving protease resides. Finally, as discussed above, further complexity in the regulation of ectodomain cleavage and release can come from alternatively spliced forms of pro-NRG1 that have cytoplasmic tails of three different lengths.
The δ isoform of PKC appears well suited for this role. Using a green fluorescent protein-tagged PKCδ molecule, it has been demonstrated that PMA causes PKCδ activation and translocation from the cytoplasm to the cell membrane (62). Furthermore, PKCδ has been shown to be involved in neurite extension in cultured PC12 cells (74), and NGF signaling stimulates the PKCδ pathway in several neuronal cell systems (75). The identity and location of the protease that cleaves pro-NRG1 is currently unknown, although one particular protease called BACE1 has recently been implicated, there is conflicting data as to whether this is the exclusive enzyme that cleaves pro-NRG1 in vivo (33, 34). Although mice deficient in BACE1 have a similar demyelinating phenotype as NRG1 knockouts, a drug that specifically blocks BACE1 leading to reduced Alzheimer precursor protein processing has no effect on pro-NRG1 cleavage (76). Consistently, other proteases have been suggested to play a role in the regulated cleavage of pro-NRG1, including TACE, ADAMs, and Meltrin-b (37, 38, 40, 69, 77–81).
Although many studies have demonstrated the ability of PKC activation to stimulate regulated ectodomain release of transmembrane precursors, for NRG1 we have identified neurotrophins as biologically relevant extracellular signals that can induce NRG1 activity release through PKCδ signaling. The neurotrophin family of soluble signaling factors has many broad roles in development and disease throughout the nervous system (82–84). Binding of neurotrophins to their trk receptors results in the activation a plethora of signaling pathways, including PKC, that lead to diverse biological effects ranging from survival to differentiation, raising the possibility that some of the functions attributed to neurotrophins may in fact be indirectly due to NRG1 activity release. Because NRG1 signaling has been implicated to have important roles in normal development and various human diseases, including multiple sclerosis, schizophrenia, Parkinsons disease, and cancer (85–88), the results of this study may be of value in understanding the mechanisms and possible treatment of these and other disorders.
Acknowledgment
PC12TrkB cells were a generous gift from Dr. Moses Chao.
This work was supported by National Multiple Sclerosis Society Grants RG-3410-A-2 and RG-3410B3 (to J. A. L.).
- NRG
- neuregulin
- pro-NRG1
- transmembrane precursor of NRG1
- PKC
- protein kinase C
- NGF
- nerve growth factor
- BDNF
- brain-derived neurotrophic factor
- GDNF
- glial cell-derived neurotrophic factor
- BACE1
- β-site Alzheimer precursor protein-cleaving enzyme 1
- PMA
- phorbol 12-myristate 13-acetate
- CHO
- Chinese hamster ovary
- CT
- cytoplasmic tail
- siRNA
- small interference RNA
- DMEM
- Dulbecco's modified Eagle's medium.
REFERENCES
- 1.Esper R. M., Loeb J. A. (2004) J. Neurosci. 24, 6218–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adlkofer K., Lai C. (2000) Glia 29, 104–111 [DOI] [PubMed] [Google Scholar]
- 3.Atlas E., Cardillo M., Mehmi I., Zahedkargaran H., Tang C., Lupu R. (2003) Mol. Cancer Res. 1, 165–175 [PubMed] [Google Scholar]
- 4.Calaora V., Rogister B., Bismuth K., Murray K., Brandt H., Leprince P., Marchionni M., Dubois-Dalcq M. (2001) J. Neurosci. 21, 4740–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong Z., Brennan A., Liu N., Yarden Y., Lefkowitz G., Mirsky R., Jessen K. R. (1995) Neuron 15, 585–596 [DOI] [PubMed] [Google Scholar]
- 6.Hippenmeyer S., Shneider N. A., Birchmeier C., Burden S. J., Jessell T. M., Arber S. (2002) Neuron 36, 1035–1049 [DOI] [PubMed] [Google Scholar]
- 7.Sandrock A. W., Jr., Dryer S. E., Rosen K. M., Gozani S. N., Kramer R., Theill L. E., Fischbach G. D. (1997) Science 276, 599–603 [DOI] [PubMed] [Google Scholar]
- 8.Meyer D., Birchmeier C. (1995) Nature 378, 386–390 [DOI] [PubMed] [Google Scholar]
- 9.Negro A., Brar B. K., Lee K. F. (2004) Recent Prog. Horm. Res. 59, 1–12 [DOI] [PubMed] [Google Scholar]
- 10.Loeb J. A. (2007) Neurology 68, S38–S42; discussion S43–S54 [DOI] [PubMed] [Google Scholar]
- 11.Esper R. M., Pankonin M. S., Loeb J. A. (2006) Brain Res. Rev. 51, 161–175 [DOI] [PubMed] [Google Scholar]
- 12.Pankonin M. S., Sohi J., Kamholz J., Loeb J. A. (2009) Brain Res. 1258, 1–11 [DOI] [PubMed] [Google Scholar]
- 13.Carraway K. L., 3rd, Weber J. L., Unger M. J., Ledesma J., Yu N., Gassmann M., Lai C. (1997) Nature 387, 512–516 [DOI] [PubMed] [Google Scholar]
- 14.Chang H., Riese D. J., 2nd, Gilbert W., Stern D. F., McMahan U. J. (1997) Nature 387, 509–512 [DOI] [PubMed] [Google Scholar]
- 15.Fischbach G. D., Rosen K. M. (1997) Annu. Rev. Neurosci. 20, 429–458 [DOI] [PubMed] [Google Scholar]
- 16.Harari D., Tzahar E., Romano J., Shelly M., Pierce J. H., Andrews G. C., Yarden Y. (1999) Oncogene 18, 2681–2689 [DOI] [PubMed] [Google Scholar]
- 17.Ishiguro H., Higashiyama S., Yamada K., Ichino N., Taniguchi N., Nagatsu T. (1998) Nihon Shinkei Seishin Yakurigaku Zasshi 18, 137–142 [PubMed] [Google Scholar]
- 18.Zhang D., Sliwkowski M. X., Mark M., Frantz G., Akita R., Sun Y., Hillan K., Crowley C., Brush J., Godowski P. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9562–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falls D. L. (2003) Exp. Cell Res. 284, 14–30 [DOI] [PubMed] [Google Scholar]
- 20.Loeb J. A., Fischbach G. D. (1995) J. Cell Biol. 130, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinthorsdottir V., Stefansson H., Ghosh S., Birgisdottir B., Bjornsdottir S., Fasquel A. C., Olafsson O., Stefansson K., Gulcher J. R. (2004) Gene 342, 97–105 [DOI] [PubMed] [Google Scholar]
- 22.Holmes W. E., Sliwkowski M. X., Akita R. W., Henzel W. J., Lee J., Park J. W., Yansura D., Abadi N., Raab H., Lewis G. D., et al. (1992) Science 256, 1205–1210 [DOI] [PubMed] [Google Scholar]
- 23.Wen D., Peles E., Cupples R., Suggs S. V., Bacus S. S., Luo Y., Trail G., Hu S., Silbiger S. M., Levy R. B., et al. (1992) Cell 69, 559–572 [DOI] [PubMed] [Google Scholar]
- 24.Corfas G., Falls D. L., Fischbach G. D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1624–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falls D. L., Rosen K. M., Corfas G., Lane W. S., Fischbach G. D. (1993) Cell 72, 801–815 [DOI] [PubMed] [Google Scholar]
- 26.Wen D., Suggs S. V., Karunagaran D., Liu N., Cupples R. L., Luo Y., Janssen A. M., Ben-Baruch N., Trollinger D. B., Jacobsen V. L., et al. (1994) Mol. Cell Biol. 14, 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeb J. A., Khurana T. S., Robbins J. T., Yee A. G., Fischbach G. D. (1999) Development 126, 781–791 [DOI] [PubMed] [Google Scholar]
- 28.Li Q., Loeb J. A. (2001) J. Biol. Chem. 276, 38068–38075 [DOI] [PubMed] [Google Scholar]
- 29.Michailov G. V., Sereda M. W., Brinkmann B. G., Fischer T. M., Haug B., Birchmeier C., Role L., Lai C., Schwab M. H., Nave K. A. (2004) Science 304, 700–703 [DOI] [PubMed] [Google Scholar]
- 30.Taveggia C., Zanazzi G., Petrylak A., Yano H., Rosenbluth J., Einheber S., Xu X., Esper R. M., Loeb J. A., Shrager P., Chao M. V., Falls D. L., Role L., Salzer J. L. (2005) Neuron 47, 681–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinkmann B. G., Agarwal A., Sereda M. W., Garratt A. N., Muller T., Wende H., Stassart R. M., Nawaz S., Humml C., Velanac V., Radyushkin K., Goebbels S., Fischer T. M., Franklin R. J., Lai C., Ehrenreich H., Birchmeier C., Schwab M. H., Nave K. A. (2008) Neuron 59, 581–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loeb J. A., Susanto E. T., Fischbach G. D. (1998) Mol. Cell Neurosci. 11, 77–91 [DOI] [PubMed] [Google Scholar]
- 33.Hu X., Hicks C. W., He W., Wong P., Macklin W. B., Trapp B. D., Yan R. (2006) Nat. Neurosci. 9, 1520–1525 [DOI] [PubMed] [Google Scholar]
- 34.Willem M., Garratt A. N., Novak B., Citron M., Kaufmann S., Rittger A., DeStrooper B., Saftig P., Birchmeier C., Haass C. (2006) Science 314, 664–666 [DOI] [PubMed] [Google Scholar]
- 35.Ozaki M., Itoh K., Miyakawa Y., Kishida H., Hashikawa T. (2004) J. Neurochem. 91, 176–188 [DOI] [PubMed] [Google Scholar]
- 36.Goekjian P. G., Jirousek M. R. (1999) Curr. Med. Chem. 6, 877–903 [PubMed] [Google Scholar]
- 37.Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K. A., Gerhart M., Davis R., Fitzner J. N., Johnson R. S., Paxton R. J., March C. J., Cerretti D. P. (1997) Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 38.Le Gall S. M., Auger R., Dreux C., Mauduit P. (2003) J. Biol. Chem. 278, 45255–45268 [DOI] [PubMed] [Google Scholar]
- 39.Kamal A., Stokin G. B., Yang Z., Xia C. H., Goldstein L. S. (2000) Neuron 28, 449–459 [DOI] [PubMed] [Google Scholar]
- 40.Hinkle C. L., Mohan M. J., Lin P., Yeung N., Rasmussen F., Milla M. E., Moss M. L. (2003) Biochemistry 42, 2127–2136 [DOI] [PubMed] [Google Scholar]
- 41.Lanni C., Mazzucchelli M., Porrello E., Govoni S., Racchi M. (2004) Eur. J. Biochem. 271, 3068–3075 [DOI] [PubMed] [Google Scholar]
- 42.Thabard W., Collette M., Bataille R., Amiot M. (2001) Biochem. J. 358, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arribas J., Massague J. (1995) J. Cell Biol. 128, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plo I., Bono F., Bezombes C., Alam A., Bruno A., Laurent G. (2004) J. Neurosci. Res. 77, 465–474 [DOI] [PubMed] [Google Scholar]
- 45.Patapoutian A., Reichardt L. F. (2001) Curr. Opin. Neurobiol. 11, 272–280 [DOI] [PubMed] [Google Scholar]
- 46.Kikkawa U., Matsuzaki H., Yamamoto T. (2002) J. Biochem. 132, 831–839 [DOI] [PubMed] [Google Scholar]
- 47.Huang E. J., Reichardt L. F. (2003) Annu. Rev. Biochem. 72, 609–642 [DOI] [PubMed] [Google Scholar]
- 48.Shirai Y., Saito N. (2002) J. Biochem. 132, 663–668 [DOI] [PubMed] [Google Scholar]
- 49.Banker G. G. K. (1998) Culturing Nerve Cells, 2nd Ed., MIT Press, Cambridge [Google Scholar]
- 50.Boyle W. J., van der Geer P., Hunter T. (1991) Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 51.Pankonin M. S., Gallagher J. T., Loeb J. A. (2005) J. Biol. Chem. 280, 383–388 [DOI] [PubMed] [Google Scholar]
- 52.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gschwendt M., Muller H. J., Kielbassa K., Zang R., Kittstein W., Rincke G., Marks F. (1994) Biochem. Biophys. Res. Commun. 199, 93–98 [DOI] [PubMed] [Google Scholar]
- 54.Jayasuriya H., McChesney J. D., Swanson S. M., Pezzuto J. M. (1989) J. Nat. Prod. 52, 325–331 [DOI] [PubMed] [Google Scholar]
- 55.Susarla B. T., Robinson M. B. (2003) J. Neurochem. 86, 635–645 [DOI] [PubMed] [Google Scholar]
- 56.Arevalo J. C., Wu S. H. (2006) Cell Mol. Life Sci. 63, 1523–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zirrgiebel U., Ohga Y., Carter B., Berninger B., Inagaki N., Thoenen H., Lindholm D. (1995) J. Neurochem. 65, 2241–2250 [DOI] [PubMed] [Google Scholar]
- 58.Liu X., Hwang H., Cao L., Wen D., Liu N., Graham R. M., Zhou M. (1998) J. Biol. Chem. 273, 34335–34340 [DOI] [PubMed] [Google Scholar]
- 59.Aris J. P., Basta P. V., Holmes W. D., Ballas L. M., Moomaw C., Rankl N. B., Blobel G., Loomis C. R., Burns D. J. (1993) Biochim. Biophys. Acta 1174, 171–181 [DOI] [PubMed] [Google Scholar]
- 60.Li W., Yu J. C., Shin D. Y., Pierce J. H. (1995) J. Biol. Chem. 270, 8311–8318 [DOI] [PubMed] [Google Scholar]
- 61.O'Driscoll K. R., Teng K. K., Fabbro D., Greene L. A., Weinstein I. B. (1995) Mol. Biol. Cell 6, 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q. J., Bhattacharyya D., Garfield S., Nacro K., Marquez V. E., Blumberg P. M. (1999) J. Biol. Chem. 274, 37233–37239 [DOI] [PubMed] [Google Scholar]
- 63.Zhan W. Z., Mantilla C. B., Sieck G. C. (2003) Sheng Li Xue Bao 55, 617–624 [PubMed] [Google Scholar]
- 64.Loeb J. A., Fischbach G. D. (1997) J. Neurosci. 17, 1416–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loeb J. A. (2003) J. Neurocytology, (in press) [DOI] [PubMed] [Google Scholar]
- 66.Loeb J. A., Hmadcha A., Fischbach G. D., Land S. J., Zakarian V. L. (2002) J. Neurosci. 22, 2206–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winseck A. K., Caldero J., Ciutat D., Prevette D., Scott S. A., Wang G., Esquerda J. E., Oppenheim R. W. (2002) J. Neurosci. 22, 4509–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edwards D. R., Handsley M. M., Pennington C. J. (2008) Mol. Aspects Med. 29, 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diaz-Rodriguez E., Esparis-Ogando A., Montero J. C., Yuste L., Pandiella A. (2000) Biochem. J. 346, 359–367 [PMC free article] [PubMed] [Google Scholar]
- 70.Hahn D., Pischitzis A., Roesmann S., Hansen M. K., Leuenberger B., Luginbuehl U., Sterchi E. E. (2003) J. Biol. Chem. 278, 42829–42839 [DOI] [PubMed] [Google Scholar]
- 71.Hung A. Y., Selkoe D. J. (1994) EMBO J. 13, 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Isohara T., Horiuchi A., Watanabe T., Ando K., Czernik A. J., Uno I., Greengard P., Nairn A. C., Suzuki T. (1999) Biochem. Biophys. Res. Commun. 258, 300–305 [DOI] [PubMed] [Google Scholar]
- 73.Liu X., Hwang H., Cao L., Buckland M., Cunningham A., Chen J., Chien K. R., Graham R. M., Zhou M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13024–13029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wooten M. W., Seibenhener M. L., Heikkila J. E., Mischak H. (1998) Cell Signal 10, 265–276 [DOI] [PubMed] [Google Scholar]
- 75.Corbit K. C., Foster D. A., Rosner M. R. (1999) Mol. Cell Biol. 19, 4209–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sankaranarayanan S., Price E. A., Wu G., Crouthamel M. C., Shi X. P., Tugusheva K., Tyler K. X., Kahana J., Ellis J., Jin L., Steele T., Stachel S., Coburn C., Simon A. J. (2008) J. Pharmacol. Exp. Ther. 324, 957–969 [DOI] [PubMed] [Google Scholar]
- 77.Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., Black R. A. (1998) Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 78.Montero J. C., Yuste L., Diaz-Rodriguez E., Esparis-Ogando A., Pandiella A. (2000) Mol. Cell Neurosci. 16, 631–648 [DOI] [PubMed] [Google Scholar]
- 79.Gschwind A., Hart S., Fischer O. M., Ullrich A. (2003) EMBO J. 22, 2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shirakabe K., Wakatsuki S., Kurisaki T., Fujisawa-Sehara A. (2001) J. Biol. Chem. 276, 9352–9358 [DOI] [PubMed] [Google Scholar]
- 81.Han B., Fischbach G. D. (1999) J. Biol. Chem. 274, 26407–26415 [DOI] [PubMed] [Google Scholar]
- 82.Farinas I. (1999) Microsc. Res. Tech. 45, 233–242 [DOI] [PubMed] [Google Scholar]
- 83.Ernfors P. (2001) Cell Mol. Life Sci. 58, 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang E. J., Reichardt L. F. (2001) Annu. Rev. Neurosci. 24, 677–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gilmour L. M., Macleod K. G., McCaig A., Sewell J. M., Gullick W. J., Smyth J. F., Langdon S. P. (2002) Clin. Cancer Res. 8, 3933–3942 [PubMed] [Google Scholar]
- 86.Cannella B., Hoban C. J., Gao Y. L., Garcia-Arenas R., Lawson D., Marchionni M., Gwynne D., Raine C. S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10100–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Franklin R. J. (2002) Nat. Rev. Neurosci. 3, 705–714 [DOI] [PubMed] [Google Scholar]
- 88.Stefansson H., Sigurdsson E., Steinthorsdottir V., Bjornsdottir S., Sigmundsson T., Ghosh S., Brynjolfsson J., Gunnarsdottir S., Ivarsson O., Chou T. T., Hjaltason O., Birgisdottir B., Jonsson H., Gudnadottir V. G., Gudmundsdottir E., Bjornsson A., Ingvarsson B., Ingason A., Sigfusson S., Hardardottir H., Harvey R. P., Lai D., Zhou M., Brunner D., Mutel V., Gonzalo A., Lemke G., Sainz J., Johannesson G., Andresson T., Gudbjartsson D., Manolescu A., Frigge M. L., Gurney M. E., Kong A., Gulcher J. R., Petursson H., Stefansson K. (2002) Am. J. Hum. Genet. 71, 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]