Abstract
Mechanisms contributing to disease-associated trinucleotide repeat instability are poorly understood. DNA ligation is an essential step common to replication and repair, both potential sources of repeat instability. Using derivatives of DNA ligase I (hLigI)-deficient human cells (46BR.1G1), we assessed the effect of hLigI activity, overexpression, and its interaction with proliferating cell nuclear antigen (PCNA) upon the ability to replicate and repair trinucleotide repeats. Compared with LigI+/+, replication progression through repeats was poor, and repair tracts were broadened beyond the slipped-repeat for all mutant extracts. Increased repeat instability was linked only to hLigI overexpression and expression of a mutant hLigI incapable of interacting with PCNA. The endogenous mutant version of hLigI with reduced ligation activity did not alter instability. We distinguished the DNA processes through which hLigI contributes to trinucleotide instability. The highest levels of repeat instability were observed under the hLigI overexpression and were linked to reduced slipped-DNAs repair efficiencies. Therefore, the replication-mediated instability can partly be attributed to errors during replication but also to the poor repair of slipped-DNAs formed during this process. However, repair efficiencies were unaffected by expression of a PCNA interaction mutant of hLigI, limiting this instability to the replication process. The addition of purified proteins suggests that disruption of LigI and PCNA interactions influences trinucleotide repeat instability. The variable levels of age- and tissue-specific trinucleotide repeat instability observed in myotonic dystrophy patients and transgenic mice may be influenced by varying steady state levels of DNA ligase I in these tissues and during different developmental windows.
More than 40 hereditary diseases are caused by gene-specific repeat instability (1). Changes at trinucleotide repeats (TNRs)3 constitute the largest component of this group, causing at least 15 different human diseases, including myotonic dystrophy (DM1), Huntington disease, and fragile X syndrome (FRAXA). Repeat changes in humans are expansion-biased and occur both in parent-to-offspring transmissions and in somatic tissues. The formation of unusual DNA structures during DNA replication and/or aberrant repair of these intermediates has been postulated as the likely source for the development of repeat tract changes (1–3), although the exact molecular mechanisms are unclear. Various proteins have been identified as players in the mutagenic process of TNR instability, including FEN1 (4–6), OGG1 (7), and some mismatch repair factors, such as MSH2, MSH3, and PMS2 (8–13). All processes suggested to be involved in repeat instability require a nick located within or proximal to the repeat tract, which ultimately must be ligated. Importantly, many proposed mechanisms of repeat instability involve slippage at the nick (1, 2, 7).
Ligation is an essential step in DNA replication, repair, and recombination (14, 15). Human DNA ligase I (hLigI) is considered the main replicative ligase and plays an important role in the joining of Okazaki fragments during lagging strand synthesis (16–18). hLigI is also involved in repair processes including base excision repair (18–23), nucleotide excision repair (24, 25), and possibly in mismatch repair (26). In both replication and repair, hLigI modulates DNA polymerase activity (21, 27, 28). Thus, the recruitment of hLigI to specific replication and repair processes plays an important role in DNA metabolism and might accommodate particular requirements to DNA stability.
In a yeast model for CTG/CAG repeat instability that is prone to contractions, disruption of the hLigI homologue (cdc9 gene) further increased this effect (5, 29–31). Although these yeast studies did not reveal whether the effect of LigI was via DNA replication or repair, it highlighted a potentially active role of the enzyme in TNR instability. Some of these studies suggested that a proper LigI-PCNA interaction is required.
LigI activity is strongly linked to PCNA. The interaction between both factors is essential for the recruitment of hLigI to replication foci and sites of DNA damage (22). In addition, this interaction indirectly up- and down-regulates DNA synthesis by polymerases δ and ϵ, respectively (21, 27), and is crucial for coordinating molecular events during Okazaki fragment processing and long patch base excision repair (18).
Mutations in the human LIGI gene have been described in a patient with symptoms similar to Bloom syndrome, including growth delay, immune deficiency, and hypersensitivity to sunlight (32). The mutant allele expressed in SV40-immortalized fibroblasts established from this patient (46BR.1G1) encodes a version of hLigI (hemizygous or homozygous for the Arg-771 to Trp) which maintains only 3–5% of ligase activity compared with the non-mutant hLigI (16). The hLigI-deficient 46BR.1G1 cells are hypomutable by DNA damage (33) but are hypersensitive to killing by DNA alkylating agents (34–39). In addition, these cells exhibit abnormal DNA processing mechanisms, such as replication fork errors, slowed Okazaki fragment joining, and reduced double strand breaks repair (17, 18, 40–42). In this study we have used derivatives of the deficient 46BR.1G1 cells expressing wild type and mutant versions of hLigI to gain insight into the role of this factor in regulating stability of TNRs.
EXPERIMENTAL PROCEDURES
Cell Culture, Extract Preparation, and DNA Extraction
Three different 46BR.1G1 derivative cell lines (46BRLigI) were used in our in vitro assays, created from stable transfected pRC/RSV plasmids (Invitrogen) into the original patient cell line (18); they are (i) 46BRLigIm/m carrying an empty vector, (ii) 46BRLigIm/m,wt expressing a wild type hLigI cDNA mutant, and (iii) 46BRLigIm/m,wt-PCNA expressing a hLigI cDNA mutant in PCNA binding. 46BRLigIm/m;wt and 46BRLigIm/m;wt-PCNA are complemented hLigI cell lines but not truly corrected because the endogenous hLigI mutation is still present in all derivative cell lines. Additional attempts to produce more isogenic 46BR variants have not succeeded. Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 300 μg/ml Geneticin as described (18). Functional protein extracts were prepared as described (43). DNA extractions from 46BRLigI cell lines were performed by proteinase K lysis and phenol/chloroform/ethanol procedure from 2–4 million cells. As a LigI+/+ control, the proficient HeLa S3 human cell line was used (43, 44).
Arg-771 to Trp Mutation and cDNA hLigI Sequencing
Sequencing the endogenous Arg-771 to Trp mutation in the 46BRLigI cells was performed using a pair of primers specific for the intronic portions of the LIGI gene (Gene ID 3978; forward in intron 23 and reverse in intron 24) to amplify only genomic hLigI DNA and to avoid amplification from hLigI-transfected cDNAs. Sequencing from a non-46BR.1G1 derivative DNA was used as a control. To sequence hLigI cDNA from transfected cell lines, we designed four pairs of primers over the exonic hLigI gene sequence. Expected PCR fragments from cDNA were discriminated from genomic DNA products. Our internal PCR control was no amplification from the empty hLigI cDNA-transfected cell line (46BRLigIm/m). Primer sequences and amplification conditions are described in the supplemental “Experimental Procedures.”
Protein Level and Phosphorylation Status of hLigI
Protein analysis of LigI+/+ and 46BRLigI functional extracts was performed by Western blot. Protein levels were compared by running on 7.5 or 12% acrylamide SDS-PAGE gels based on protein molecular weight, transferred onto a nitrocellulose filter, and probed depending on the blot performed against LigI (5H5, MBL; 1:5,000), actin (Ab-5, BD Biosciences; 1:5,000), PCNA (PC-10, Santa Cruz; 1:40,000), FEN-1 (B-4, Santa Cruz; 1:500), MSH2 (Ab-2, Calbiochem; 1:1,000), and MSH6 (clone 44, BD Biosciences; 1:500). Equal protein loading was performed in each blot. Phosphorylation status of hLigI was assessed by prior dephosphorylation by alkaline phosphatase (New England Biolabs) treatment of the functional extracts (4 h at 37 °C). Samples before and after the alkaline phosphatase treatment were then compared by running on 7.5% acrylamide SDS-PAGE gel and probed with anti-ligase I antibody (5H5, MBL; 1:5,000). To ensure reproducibility, each Western blot analysis was performed three times using independent extract preparations.
Replication Templates, in Vitro Replication, and Replication Efficiencies
The non-repeat template (pKN16), the (CTG)79· (CAG)79 templates (pDM79EF and pDM79HF), and the in vitro replication reactions were performed as described (44). In brief, DNA templates (150 ng) were replicated in reactions containing final concentration of dATP, dGTP, dTTP, and dCTP (100 μm each), GTP, UTP, and CTP (200 μm each), ATP (4 mm), creatine phosphate (40 mm) (Roche Applied Science); creatine kinase, (100 mg/ml; Roche Applied Science); l mg of SV40 T-antigen (Chimerx); 150 μg of protein extract. For direct analysis of the replication products, 0.033 μm = 0.099 Ci of [α-32P]dCTP (3000 Ci/mmol, PerkinElmer Life Sciences) was included in each 50-μl reaction. Reactions were incubated for 4 h at 37 °C and terminated with 50 μl of stop solution (2 mg/ml proteinase K, 2% SDS, and 50 mm EDTA, pH 8.0) with a further incubation of 1 h at 37 °C. Carrier tRNA (1.5 mg) was added, and protein was extracted twice with phenol/chloroform. Replication products were ethanol-precipitated, resuspended in water, and divided in two aliquots. Equal quantities of reaction products were digested with BamHI only or with BamHI+DpnI to discriminate between total and completely replicated material, respectively. Finally, digestion products were resolved by electrophoresis on a 15-cm 1% agarose gels. Gels were run for 16 h at 4 V/cm in Tris-buffered EDTA buffer, dried and exposed to Kodak film. Replication amounts were determined by autoradiography and quantified using ImageQuant GE Healthcare Version 1.2 software. Replication efficiency results were based on the average of three independent experiments.
Mutation Analysis (STRIP Assay)
Mutation analysis was performed using the stability of trinucleotide repeats by individual product analysis (STRIP assay) as described in detail (44). Replication reaction mix was the same as used to measure replication efficiency but without including radioactive nucleotides. Replication products were digested with DpnI to eliminate un-replicated parental and partially replicated material. DpnI-digested material was transformed into Escherichia coli DH5α cells, and individual bacterial colonies picked, each representing an individual product of completed primate replication, were cultured for less than 6 h. Repeat length analysis was performed on high resolution 4% polyacrylamide gels. Stable/unstable molecule frequencies were based on two independent experiments for each template-extract pair, each analyzed separately (there were no significant differences between these) and then pooled (a total of 100–200 colonies were scored) to facilitate presentation. Instability differences were quantified by Fisher's exact test (p < 0.05).
Slipped-DNA Substrates and Repair Reactions
A set of previously characterized human DM1 mutagenic intermediates containing pure (CTG)x·(CAG)y repeats where x = 30 or 50 and y = 50 or 30 have been used to perform slipped-strand repair reactions (50 μl) as described (43). Briefly, 5–10 ng of each slipped substrate was incubated in 30 mm Hepes-KOH, pH 7.8, 100 μg/ml creatine kinase, 40 mm creatine phosphate, 4 mm ATP, 200 μm CTP, GTP, and UTP, 33 mm each cold dNTP, and 150 μg of extract at 37 °C for 30 min. Extracts, reaction termination, and protein extraction were performed as described in the replication assay. Finally, after releasing the repeat-containing fragment (EcoRI/HindIII digestion) and resolving on native 4% acrylamide gels, products were electrotransferred to membrane and hybridized with a radiolabeled EcoRI/HindIII fragment-containing 17 repeats. Repair efficiencies were determined on a molar level by Southern blot probing of reaction products that had been generated in the presence of nonradioactive dNTPs and measured as the proportion of radio intensity of the repeat-containing product (ImageQuant GE Healthcare Version 1.2) relative to all repeat-containing fragments. All starting unprocessed substrates were assessed simultaneously. Repair levels for each substrate were based on the average of three independent experiments. Repair tract sizes were assessed by mapping radio-incorporation levels in slipped-strand reaction products derived with radioactive dNTPs and a final EcoRI-DdeI digestion (43). To determine the radiographic intensity of EcoRI-DdeI fragments, specific incorporation is reported as relative amounts of incorporation per base pair normalized to the juxtaposed 540-bp fragment (ImageQuant GE Healthcare 1.2).
Protein Purification
Human DNA ligase I and PCNA proteins were prepared and purified as per published protocols (45, 46).
RESULTS
Different hLigI Protein Backgrounds in 46BRLigI Extracts
Toward assessing a potential role of hLigI in the processing of CTG/CAG repeats, we first characterized the three derivatives of hLigI-deficient 46BR.1G1 cells (18). DNA sequence, protein levels, and post-translation modifications were analyzed.
Sequence polymorphisms of the hLigI gene, even synonymous variations, could affect ligation activity, varying the transcription, transcript stability, or translation of hLigI and, in turn, influence its DNA replication or repair capacities (47). Sequencing the endogenous LIGI gene from genomic DNA confirmed that all 46BR.1G1 variant cells contained the described homozygous/hemizygous 46BR mutation (Arg-771 to Trp; CGG to TGG) (16) (Fig. 1). However, we found that hLigI cDNAs used to complement the original low activity 46BR.1G1 cell line had differences among them, depending on the transfection source. Sequencing the integrated hLigI cDNA sequences in the stably transfected 46BRLigIm/m;wt and 46BRLigIm/m;wt-PCNA cell lines revealed two (640 and 802 amino acids) and one base pair change (802 amino acids), respectively, relative to LIGI gene sequence (GeneID 3978) (Fig. 1). Although the 802 variant (rs20581) has previously been identified (snp500cancer.nci.nih.gov) and may predispose to lung cancer (47), both DNA changes did not alter the hLigI amino acid sequence. Finally, we also confirmed the presence of the DNA sequence changes that replace adjacent phenylalanine residues with alanine residues within the conserved PCNA interaction motif encoded by the mutant hLigI cDNA from 46BRLigIm/m;wt-PCNA cell line (Fig. 1) (18).
FIGURE 1.
hLigI mutations and polymorphisms between different 46BR.1G1 derivatives. Upper, human hLigI protein schematic outlining various functional motifs including the PCNA-interacting protein box (PIP box) that mediates the interaction with PCNA. Ser-51, -66, -76, and -91 are the amino acid positions phosphorylated during the cell cycle (50–53). Lower, sequencing results. Broken arrow, intronic primers (46BR-F and 46BR-R) were used to check the Arg-771 to Trp 46BR mutation at genomic hLigI sequence (16). Solid arrows, a set of four pairs of exonic primers were used to check possible polymorphisms in cloned wild type hLigI cDNAs used to stably supplement 46BR.1G1 cells (18). The 8–9-amino acid mutation in the 46BRLigIm/m;wt-PCNA cell line that abolish binding to PCNA was also confirmed (18).
Because it has been suggested that the steady state levels of hLigI protein may affect replication or repair pathways (40) and hLigI is frequently overexpressed in human malignant processes (48), we also examined its levels in the three 46BRLigI cell lines (18). Interestingly, different levels of hLigI were observed (Fig. 2A). The amount of hLigI in the mutant lines 46BRLigIm/m and 46BRLigIm/m;wt-PCNA was similar but lower than that present in LigI+/+ cells, which contains only wild type hLigI. The 46BRLigIm/m;wt cells contained considerably higher levels of hLigI protein than either of the other cell lines; specifically it harbored 8- and 14-fold more hLigI protein than its precursor 46BRLigIm/m or its sister line 46BRLigIm/m;wt-PCNA and 3-fold more than the LigI+/+ cells. The variability in hLigI protein expression between the hLigI-transfected cell lines is likely because of variable gene copy number and/or transcriptional activity arising from different hLigI-vector integration sites (18, 49). The altered protein expression appeared to be limited to hLigI, as the levels of other proteins, including PCNA, FEN-1, MSH2, or MSH6, were identical between the extracts (Fig. 2A). Thus, in addition to being a 46BR.1G1 line variant expressing a fully functional wild type hLigI, the 46BRLigIm/m;wt extract contained considerable increased amounts of hLigI protein (overexpression feature). The hLigI protein levels were comparable in the 46BRLigIm/m and 46BRLigIm/m;wt-PCNA lines, both containing the defective hLigI, with the latter also containing a functional hLigI that is incapable of interacting with PCNA.
FIGURE 2.
Western analysis of functional cell extracts. A, hLigI, MSH2, MSH6, FEN-1, PCNA, and actin proteins levels are shown. B, phosphorylation status of hLigI of the same extracts shown in A with and without calf intestinal alkaline phosphatase (CIP) treatment. Protein amount was equally loaded in all cases. Blots are representative examples of three independent analyses.
The levels of protein phosphorylation were also assessed, as this hLigI post-translation modification has been linked to variable levels of replication and repair activity (50–53). It is unknown if the mutant 46BR.1G1 protein displays an altered ability to be phosphorylated, an effect that may contribute to its altered ligation activity. The levels of hLigI phosphorylation did not vary between any of the 46BR.1G1 derivatives and were identical to those in the LigI+/+ cells (Fig. 2B). Thus, the ability of hLigI to be phosphorylated is unaltered by coding mutations that reduce ligation activity. Additionally, this phosphorylation is not dependent upon an interaction with PCNA (51). Together, our 46BRLigI cell line characterization results established that in addition to the previously genetically defined variables, including the severely reduced ligation activity (46BRLigIm/m) and the wild type hLigI, which is incapable of interacting with PCNA (46BRLigIm/m;wt-PCNA) (18), the overexpressed hLigI levels of the complemented 46BRLigIm/m;wt line must also be considered as an experimental variable.
Replication Efficiency Is Reduced on CTG/CAG Templates with a Defective hLigI
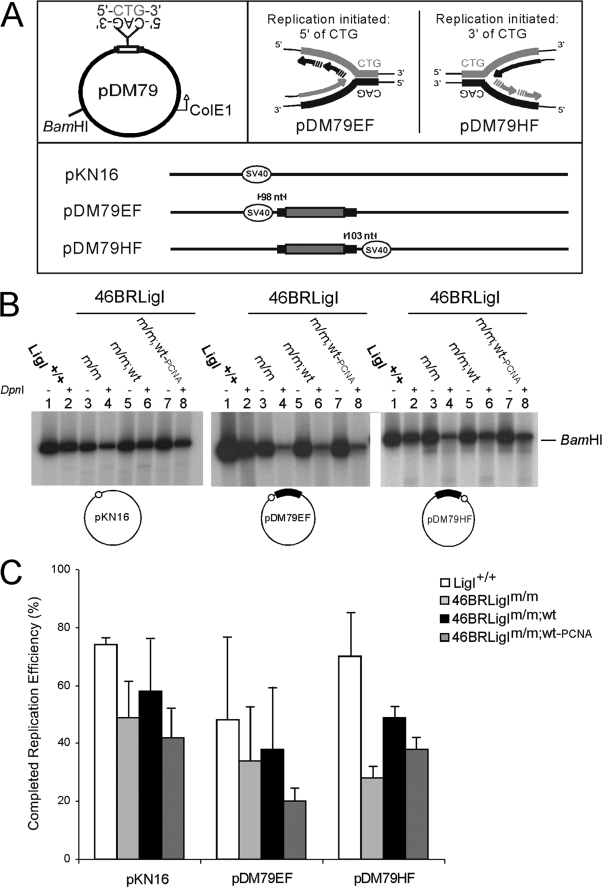
Based upon the critical role of hLigI in DNA metabolism and taking advantage of the functional extracts prepared from the three distinct hLigI backgrounds present in the 46BRLigI cell lines, we first assessed their efficiencies to replicate through repeat DNAs. We combined the SV40 in vitro DNA replication system (see “Experimental Procedures”) with plasmid templates that either contained (CTG)79·(CAG)79 repeats (pDM79EF and pDM79HF) or a template without TNRs (pKN16) (Fig. 3A). Replication efficiencies were quantitated as the proportion of completely replicated (DpnI-resistant) material relative to the total amount of incorporation (before DpnI digestion). To facilitate visualization, equal amounts of replication reactions were linearized with BamHI only or with BamHI + DpnI and electrophoretically resolved on agarose gels. A representative gel is shown in Fig. 3B, and the quantification of replication efficiencies is plotted in Fig. 3C.
FIGURE 3.
Replication efficiencies under different hLigI backgrounds. A, schematic diagram of replication templates (44). pDM79 circular plasmids contain the DM1 (CTG)79·(CAG)79 repeats in the stable orientation relative to the unidirectional bacterial origin of replication (ColE1). The bold lanes indicate flanking non-repetitive sequences from the human DM1 locus. Location of SV40-ori determines the replication direction and which strand will serve as the leading or lagging strand template. The direction of replication fork progression for pDM79E initiates 3′ of the CAG and pDM79H initiates 5′ of the CAG tract. The pKN16 template contains the SV40-ori but no the repeat tract. B, total (lanes 1, 3, 5, and 7) and complete replication (lanes 2, 4, 6, and 8) templates under different hLigI conditions. Templates were resolved in a 1% agarose gels after digestion with BamHI or both BamHI and DpnI, and radio-incorporation was detected by autoradiography. C, complete replication efficiencies are presented as the ratio of completely replicated (BamHI and DpnI) material as a percentage of the total incorporation (BamHI only) for each template and extract. Efficiencies of multiple independent experiments were quantified using ImageQuant software.
Replication efficiency producing a fully replicated product (DpnI-resistant material) was clearly affected by the alteration of the normal hLigI status in all templates, either containing or not-containing CTG/CAG repeats (Fig. 3B, even lanes). Thus, extracts from all 46BR.1G1 derivatives exhibited reduced replication compared with LigI+/+ extract (Fig. 3C). Efficiencies from the altered hLigI extracts were also sensitive to the DNA template, as the presence of the repeat tract led to lower replication levels (Fig. 3C). In general, a greater reduction of replication efficiency by all ligase I variant extracts compared with the Lig+/+ extract was observed for the template using CAG as the lagging strand template (Fig. 3C). Together the results showing the presence of a slower replication process indicate that the ability to yield completely replicated material was influenced mainly by changes in hLigI. In general, the low hLigI activity extract (46BRLigIm/m) and the extract containing the PCNA interaction-defective version of hLigI (46BRLigIm/m;wt-PCNA) had the lowest replication efficiencies. The extract containing high levels of wild type hLigI (46BRLigIm/m;wt) approached the replication efficiency of the HeLa (LigI+/+).
Increased Instability after Replication with 46BRLigIm/m;wt and 46BRLigIm/m;wt-PCNA Extracts
The observed DNA replication disruption under different hLigI conditions may be linked to varying (CTG)x·(CAG)x instability levels. Replication-mediated repeat instability was assessed using an established assay (44) and the same SV40 in vitro system and templates with 79 CTG/CAG repeats used to assess replication efficiencies (STRIP assay). This assay determines the repeat instability incurred in fully replicated material (even lanes in Fig. 3B, see “Experimental Procedures” for significance analysis).
The instability observed after replication using HeLa (LigI+/+) extracts is sensitive to replication direction, with both orientations yielding instability (Fig. 4). The template using CTG as the lagging-strand template yielded more expansions relative to the length heterogeneity in the starting parental templates, whereas the template using CAG as the lagging-strand template yielded more contractions. These results confirm what we previously published with LigI+/+ (HeLa) extracts (44). We next proceeded to address the effect of ligase in this instability after 100–200 replicated molecules counted for each template/extract pair.
FIGURE 4.
Repeat template stability levels mediated by replication under different hLigI conditions. Schematic diagram showing molecule frequencies with repeat traces that are less than 79, 79, more than 79 after STRIP assay using the two 79-repeat templates (pDM79EF and pDM79HF). Parental frequencies (bottom bar in each panel) represent the starting repeat tract length distribution in the replication templates assessed after transformation in the DH5α bacteria. LigI+/+ (HeLa) extract showed the contraction (pDM79EF) and expansion (pDM79HF) biases previously published with these templates (44). Significant statistical differences (*) compared with LigI+/+ frequencies were obtained in the 46BRLigIm/m;wt and 46BRLigIm/m;wt-PCNA extracts (Fisher test, p < 0.05) after 100–200 colonies scored in total for each template-extract pair experiment.
For templates with 79 repeats, mutation frequencies did not show differences between a full activity hLigI extract (LigI+/+) and the reduced ligase activity extract (46BRLigIm/m) (p = 0.21 and p = 0.87 in pDM79EF and pDM79HF, respectively). These results indicate that low ligation activity does not affect CTG/CAG instability regardless of replication direction (Fig. 4). Moreover, they separate alterations in the efficiency to yield fully replicated molecules from the levels of repeat instability they will incur.
In contrast to the absence of altered instability by the reduced ligase extract, replication mediated by the ligase overexpression or by the PCNA-interacting mutant presented significantly increased repeat instability mutation profiles compared with LigI+/+ (46BRLigIm/m;wt, p = 1.16 × 10−5 and p = 0.0012, in pDM79EF and pDM79HF, respectively; 46BRLigIm/m;wt-PCNA, p = 0.0055 and p = 0.0045, in pDM79EF and pDM79HF, respectively) (Fig. 4). Increased levels of expansions and contractions were observed for both these extracts regardless of replication direction. Mutation profiles were also significantly different in three of four cases compared with 46BRLigIm/m results (46BRLigIm/m;wt, p = 0.0086 and p = 0.016, respectively; 46BRLigIm/m;wt-PCNA, p = 0.32 and p = 0.016, respectively) with a strong degree of confidence, as the three cell lines were derived from the same 46BR.1G1 source (18). Quantification showed drops from 8 to 19% in the number of 46BRLigIm/m;wt and 46BRLigIm/m;wt-PCNA stable molecules, yielding significant gains in contracted (less than 79 repeats) and expanded (greater than 79 repeats) molecules (Fig. 4). Changes in the repeat tract lengths were mostly limited to short repeat gains or losses (supplemental Fig. 1). The repeat stability modulation by hLigI seems to be limited to long repeat-disease tract lengths, as analysis with short repeat templates containing only 17 CTG/CAG repeats (non-disease length) did not show altered instability in any case (data not shown).
Together, these results suggest that increased replication-mediated instability was linked specifically to hLigI overexpression and expression of a PCNA-interaction mutant of hLigI but not by reduced ligase activity. These effects were observed as increased expansions and contractions in fully replicated molecules for both replication directions without an obvious relation to replication efficiency.
Repair Synthesis Tract Size Is Affected by LigI
We cannot exclude a possible contribution of DNA repair to the perturbed instabilities obtained after replication, as repair can occur simultaneously in the in vitro replication reactions. For instance, the processing of slipped-DNAs formed during replication through repeat sequences may cause mutations. Such repair processes may be perturbed by altered ligase deficiencies, particularly at excision and closing of excision tracts, as shown previously (17, 18, 40–42).
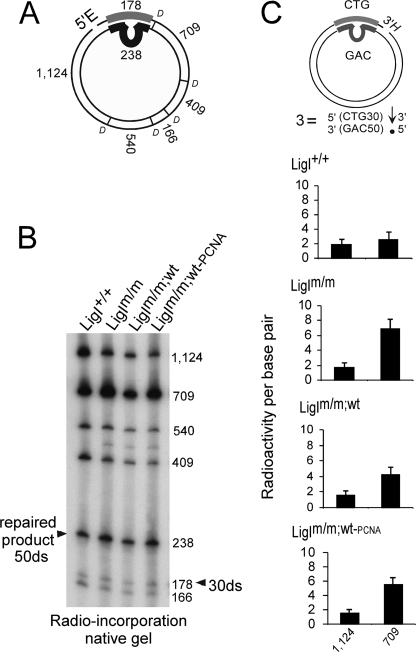
Toward addressing an effect of ligase upon slipped-DNA repair, we first assessed if the repair excision and synthesis process was affected by the different hLigI environments on slipped-DNAs. We mapped repair incorporation on eight different slipped-CTG/CAG repeat substrates, which lack the SV40-ori and, thus, cannot replicate (supplemental Fig. 2). These substrates have been structurally characterized and modeled as intermediates of contraction and expansion events formed during DNA metabolism (54). Expansion substrates contained nicks in the same strand as the slip-out, whereas contraction substrates contained nicks in the opposite strand (supplemental Fig. 2). All three altered hLigI extracts spread the radio-incorporation out to considerable lengths into regions beyond the slip-out to be repaired, extending from the nick away from the slip-out, in most substrates (Fig. 5, supplemental Fig. 3). In some cases the amount of repair incorporation into the downstream flank was even greater than that occurring in the repeat tract (data not shown), indicating that the excision gap (and hence synthesis tract) was considerably larger into the flank than required to repair the slip-out. This repair-gap widening was consistently observed for altered LigI extracts but never when repair was mediated by the LigI+/+ extract (Fig. 5, supplemental Fig. 3). Thus, similar to the perturbed replication efficiencies detected above, we also observed that DNA repair excision tract size was altered by all the different 46BRLigI extracts. Results reproducibility was achieved by three independent experiments in some chosen substrates (Fig. 5, supplemental Fig. 3). Anomalously long repair patch sizes have previously been observed in other studies for 46BR.1G1 but not with added functional hLigI (17, 55). Such altered repair tract sizes for slipped-DNAs may reflect aberrant repair that may lead to instability.
FIGURE 5.
Repair patch sizes detected under different hLigI backgrounds. A, EcoRI-DdeI (E and D) map of slipped-DNA substrates, with fragment sizes (nucleotides) indicated. B, reaction products of substrate 3 produced by the different extracts were digested with EcoRI-DdeI and resolved on native acrylamide, with each fragment indicated. C, assessment of the relative incorporation/bp into the fragments upstream (1124 bp) and downstream (709 bp) of the repeat tract, normalized to the 540-bp fragment (opposite the repeat) by densitometric analysis. Three independent experiments were performed for this substrate. The same analysis for all eight slipped-DNA substrates is shown in supplemental Fig. 3.
Role of Repair in Repeat Stability in the 46BR Extracts
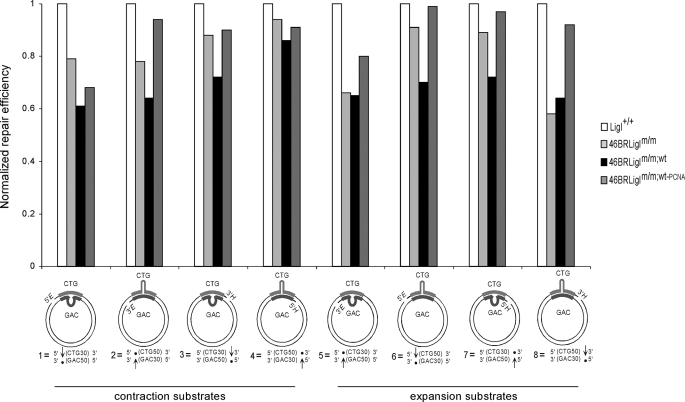
The length mutational changes in (CTG)79·(CAG)79 templates arising by in vitro replication can occur through errors of repair. We were able to distinguish repair-mediated instability from replication-mediated events by assessing repair levels from the same set of eight slipped-CTG/CAG repeat substrates used to assess repair tract lengths (supplemental Fig. 2). Repair efficiencies using the same extract preparations and reaction conditions were quantified on a molar level by identifying repaired products by Southern blotting. The various slipped-DNA substrates showed the same relative repair behavior as previously reported (43), with CAG slip-outs repaired better than CTG slip-outs and with 5′-nicked substrates repaired better than 3′-nicked substrates (supplemental Fig. 4). Three independent experiments for each substrate/extract pair assured the reproducibility of the results (supplemental Fig. 4). Assessment of the effect of distinct hLigI variations in repair levels was facilitated by comparing results normalized against LigI+/+ levels (Fig. 6). Interestingly, both extracts that initially had shown increased repeat instability after replication (46BRLigIm/m;wt and 46BRLigIm/m;wt-PCNA) now expressed different slipped-DNA repair efficiencies. The overexpressed extract (46BRLigIm/m;wt) was the worst at repairing most substrates (7 of 8 cases), with a mean of a 30% reduction compared with LigI+/+ levels (Fig. 6). Conversely, the inability of hLigI to interact with PCNA (46BRLigIm/m;wt-PCNA extract) showed similar repair levels compared with LigI+/+ (Fig. 6). The generally poor repair by the overexpressing 46BRLigIm/m;wt extract suggests that many slipped-DNAs escaping repair and that DNA repair may also be involved in the increased number of length changes observed in replication assays. The absence of an effect of the PCNA binding mutant on repair efficiency indicates that the increased repeat instability in the replication assay by these extracts is limited to replication fork events.
FIGURE 6.
Normalized slipped-DNA repair levels under different hLigI backgrounds. Repair levels are reported relative to the repair efficiency mediated by the LigI+/+ (HeLa) extract for each template.
LigI and PCNA Protein Levels Change the Adjustment of CTG/CAG Template Processing
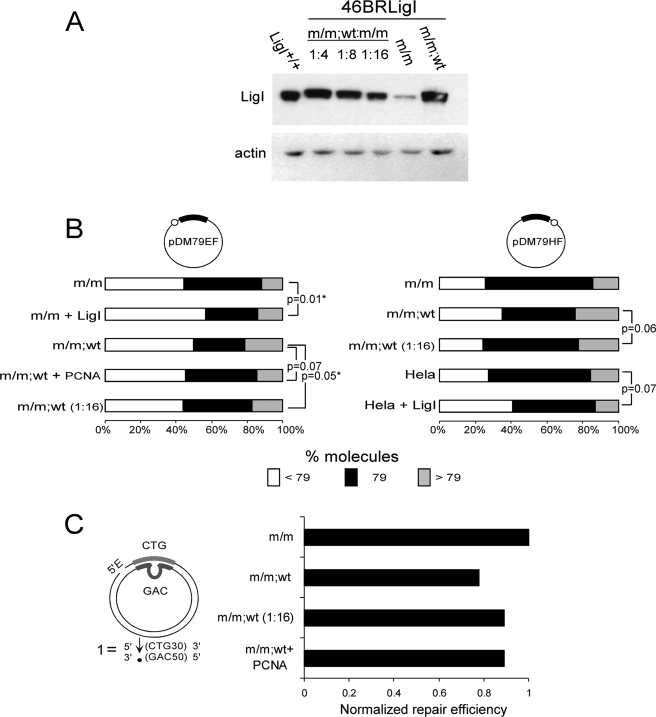
The above results indicate that the increased steady state level of wild type hLigI present in the 46BRLigIm/m;wt cell line can contribute to altered repeat instability by both replication- and repair-dependent processes. However, the altered levels of instability between the overexpressing line 46BRLigIm/m;wt and the wild type (LigI+/+) HeLa line may be because of other genetic or regulatory differences between the cells. To directly address these possibilities, we took two approaches, which were diluting the overexpression feature and mimicking it through the addition of purified hLigI or PCNA. First, we modulated the level of hLigI in the 46BRLigIm/m,wt context by diluting this extract with 46BRLigIm/m extract until hLigI protein levels were comparable with the LigI+/+ extract (Fig. 7A). In this manner we eliminated the anomalously high levels of hLigI while maintaining both the level and genetic isoforms of other proteins known to be involved in repeat instability (MSH2, MSH6, FEN1). This “hLigI-diluted” extract was used to measure replication-mediated instability and repair efficiency. The results showed levels of CTG/CAG stability and repair similar to the 46BRLigIm/m extract (p = 0.73 and p = 0.27; pDM79EF and pDM79HF, respectively), closer to the patterns yielded by the LigI+/+ extract and distinct from the initial overexpressed 46BRLigIm/m;wt levels (p = 0.05 and p = 0.06; pDM79EF and pDM79HF, respectively) (Fig. 7B). Likewise, after dilution, repair efficiency increased close to levels observed in 46BRLigIm/m extract (Fig. 7C). The hLigI-diluted extracts yielded levels of replication and repair that only approached the levels attained by the 46BRLigIm/m extract even though they contained levels of functional wild type hLigI similar to those in the HeLa cells. This incomplete recovery may allude to other effects (see “Discussion”).
FIGURE 7.
Repeat length distribution and repair levels after changing LigI or PCNA amounts into the in vitro reactions. A, Western blot confirms dilution of hLigI levels in 46BRLigIm/m;wt diluted with 46BRLigIm/m extract at a ratio of 1:16. This dilution was used for in vitro replication and repair reactions. B, repeat tract length distributions after replication (*, statistical significance, p < 0.05, Fisher's exact test) after 100–200 colonies scored in total for each experiment. C, repair levels after 46BRLigIm/m;wt dilution or added hPCNA protein. Repair results were normalized against the 46BRLigIm/m levels.
We followed an alternative approach to eliminate possible cell-type differences between the hLig overexpressing 46BRLigIm/m;wt and control LigI+/+ HeLa extracts; the addition of high levels of purified hLigI protein to either HeLa or 46BRLigIm/m extracts would be expected to have parallel effects upon replication- or repair-mediated CTG/CAG instability as the overexpressing 46BRLigIm/m;wt extracts. We added titrated amounts of purified hLigI (data not shown) to HeLa and 46BRLigIm/m extracts to resemble the hLigI overexpression feature observed in the 46BRLigIm/m;wt extract and used these for replication. Supplementing these extracts with high levels (0.9 μg) of wild type hLigI increased replication-mediated instability (p = 0.07 and p = 0.01, LigI+/+ and 46BRLigIm/m, respectively) to levels similar to the overexpressing 46BRLigIm/m;wt extract (Fig. 7B). Thus, the overexpression of hLigI protein does in fact appear to be responsible for the altered instability. This “add back protein” approach allowed us to draw similar conclusions in different cell types (HeLa and 46BR.1G1), suggesting it as a general phenomenon affecting CTG/CAG repeat stability independently of the cell source used. Finally, the use of a specific in vitro inhibitor of hLigI activity (56) able to further reduce hLigI activity in 46BRLigI backgrounds (supplemental Fig. 5A) did not cause significant changes in replication-mediated instability nor in repair efficiency levels (supplemental Fig. 5, B and C), consistent with the absence of a link between ligation activity and instability, as mentioned above. Together these results support the notion that perturbation of instability by hLigI overexpression may not be because of altered ligase activity but may be the result of altered protein dynamics due to saturating protein interactions by the overexpressed hLigI (possibly with PCNA).
To directly address a protein-protein dynamic effect for PCNA, whose free levels might be depleted by saturating interactions with the overexpressed ligase, we added high levels (1.8 μg) of purified hPCNA to the extract with this feature and assessed its capacity for instability. Instability was reduced by supplementing the hLigI-overexpressed (46BRLigIm/m;wt)-mediated replication with hPCNA (Fig. 7B). The effect approached a level of significance (p = 0.07). Additional PCNA availability in the reaction, restoring the LigI-PCNA balance, appeared to recover repeat- instability to levels of the non-overexpressing state. Similarly, increased repair efficiencies were observed when hPCNA was added to the hLigI-overexpressed reactions (Fig. 7C).
Overall, the modification of hLigI and available hPCNA protein levels in functional protein extracts demonstrated the importance of LigI, linked to its proper interaction and/or balance with PCNA, in repeat instability modulation. The importance of this is supported by either hLigI overexpression (46BRLigIm/m;wt) or loss of LigI-PCNA interaction (46BRLigIm/m;wt-PCNA).
DISCUSSION
hLigI is not only the major ligase active at DNA replication forks but also participates in various forms of DNA repair. The involvement of DNA nicks in replication, repair, and recombination and the potential for repeat strand slippage at nicks present this step as one through which DNA mutations at repeat sequences may occur. Cell lines harboring distinct hLigI backgrounds (18, 32) lead to anomalous DNA replication and repair processes. In agreement with published studies, DNA replication efficiency was dependent upon having enough ligation activity and the ability of hLigI to interact with PCNA (16–18, 33, 57–59). Expression of wild type hLigI mostly corrected the replication defect of hLigI-deficient 46BRLigIm/m extract (17, 18, 40). These results are also consistent with PCNA coordinating the action of hLigI during Okazaki fragment processing and ligation and the essential role of PCNA in efficient fork progression (17, 40). At the same time, replication was slightly hindered by the presence of CTG/CAG repeats in the template, an effect further exacerbated by hLigI-altered backgrounds. Reduced replication across repeats may reflect frequent replication pauses and increased duration of mutant hLigI upon the DNA due to the presence of highly stable secondary structures formed by CTG/CAG repeats (60–62). Likewise, hLigI altered backgrounds resulted in larger, polar repair tracts that, unlike those generated by HeLa (LigI+/+), extended beyond the slip-out lesion. Similar increases in repair tracts resulting from hLigI deficiency have been described for base excision repair, nucleotide excision repair, and double-strand-break repair (17, 21, 55) and in yeast extracts deficient for the cdc9 gene (20). The widened and polar repair tracts are likely the result of strand displacement DNA synthesis under hLigI-deficient backgrounds (17). hLigI inhibits the strand displacement activity of DNA polymerase δ in long-patch base excision repair through an interaction with PCNA (21, 27). Similarly, polymerase δ strand displacement can be modulated in Okazaki fragment processing by PCNA, FEN-1, and RPA (64). Coordination of the ligation step with DNA repair excision and synthesis and replication fork progression clearly involve proper ligation activity and PCNA binding. A contribution of the mutant 46BR hLigI to the altered metabolism cannot be discarded, as this mutation was present in all 46BRLigI cell lines. This hypothesis is supported by the incompletely recovered replication-mediated instabilities by the hLigI-diluted extracts, which only approached the levels attained by the 46BRLigIm/m extract. It is also supported by the disconnect between the aberrant tract sizes and the altered repair efficiencies by all 46BR.1G1 cell variants. This might suggest the possibility that the endogenous mutant hLigI (Arg-771 to Trp mutation) (32) may have a dominant negative effect on these processes (53).
The contribution of hLigI to genetic instability may not be linked to its DNA sealing activity but, instead, to its involvement in dynamic protein-protein interactions. We did not observe any significant change in CTG/CAG repeat instability because of reduced hLigI activity even after further reduction of this activity by drug inhibition. In accordance with our results, high levels of DNA instability have not been linked to deficiencies in ligase I activity (patient cells, transgenic mice, or yeast) (16–18, 30–41, 57–59). Only limited chromosomal genetic instability was observed in the 46BR mouse model, characterized by unusual epithelial tumors rarely seen in mice (59). In contrast, CTG/CAG instability was markedly increased in a replication direction-independent manner by extracts containing either high levels of wild type hLigI or a mutant version that is unable to bind PCNA.
On several points our results agree with previous yeast studies (30, 31) and extend them by revealing specific contributions of replication- or post-replication repair to repeat instability; (i) mutant low activity ligase did not affect instability, (ii) overexpression of wt-LigI increased instability, and (iii) a dependence upon PCNA interaction was observed in both studies. Although the yeast study observed a replication direction dependence on these effects, we observed effects for both directions. These differences may be because of the different types or levels of instability occurring in our primate system compared with the yeast model used. Although we did not test an effect of overexpression of a PCNA-interacting mutant ligase, as done in the yeast studies, we were able to distinguish the processes through which the PCNA-interacting mutant and the overexpressed functional ligase contribute to instability; the PCNA-interacting mutant contributes primarily through replication, whereas ligase overexpression was through repair and possibly replication. Although the yeast study did not distinguish these processes, the effects they observed may be similar to those we observe. Replication-induced repeat instability through loss of PCNA binding may be the result of a failure of hLigI to localize properly at replication foci (18, 46, 65–67) and/or modulate the appropriate DNA polymerases (21, 27).
hLigI and PCNA form a functional complex (68, 69) critical for efficient ligation of DNA (70) as well as other DNA processes (18, 22, 41, 71). The coordination between both factors is extremely complex, as secondary LigI-PCNA interactions points outside the consensus sequence (PCNA interaction protein box (PIP box)) (46, 67) have been described (69, 70, 72, 73). In addition, changes in DNA structure might be crucial in switching among the protein factors bound to PCNA (74). Many variables affecting the dynamics of the switching process between PCNA factors remain unknown at the molecular level. Specifically, the binding of PCNA with hLigI is exclusive with its interaction with either FEN-1 (31, 46, 69, 72, 75) or DNA polymerase δ (76, 77), both of which have been implicated in CTG/CAG instability (5, 6, 31, 78, 79). Polymerase δ is involved in replication, synthesizing Okazaki fragments (81), and in various forms of DNA repair including damaged and unpaired DNAs (82–84). Inhibition of polymerases with aphidicolin treatment in DM1 patient cells with large expansions causes enhanced CTG expansions (85). On the other hand, overexpression of hLigI either by internal steady high expression of wild type hLigI or simulating the overexpression feature by the addition of purified hLigI to in vitro reactions increased repeat instability frequencies. This anomalous hLigI condition involved both replication and post-replication repair processes, displaying a more severe impact in repeat size changes (higher instability frequencies) than observed by a hLigI mutation that prevents PCNA binding. Recovered stability and repair levels after adding hPCNA in reactions with 46BRLigIm/m;wt extract suggested that hLigI overexpression disturbs processes coordinated by PCNA by limiting the availability of this factor for necessary protein-protein interactions with other partners such as FEN-1 and polymerase δ. At the same time, overexpression might also disturb the interaction of other important hLigI partners (23, 86–88). To provide further support for this hypothesis, we were able to change both CTG/CAG-length distributions and repair levels, similar to observed when hLigI binding to PCNA is intact (LigI+/+ and 46BRLigIm/m), by reducing the relative internal level of hLigI in the overexpressed extract.
Several other studies support our imbalance proposal. (i) In yeast, abolishing the interaction between LigI and PCNA increased TNR instability levels (30). Likewise, overexpression of yeast LigI also increased repeat instability. This increase was dependent upon the presence of a functional PCNA interaction motif but not linked to ligation activity (31). (ii) An excess of hLigI, either full-length or its amino-terminal region (required for PCNA interaction) but not the catalytic domain, inhibited in vitro SV40 DNA replication by HeLa cell extracts (40). This disruption by the non-catalytic domain of hLigI was interpreted to be because of an imbalanced availability of hLigI-interacting partners, like PCNA. This highlights the problems that might arise in conditions of hLigI overexpression. (iii) Deficiencies of PCNA or genes encoding enzymes that interact directly with this factor in coordinated processes, such as FEN-1 or DNA polymerases, yield similar unstable repeat phenotypes (4, 5, 29, 89–92). The similar effects upon repeat instability suggest that these factors work together to maintain or mutate repeat lengths. (iv) The process of V(D)J recombination is critical to immunoglobin diversity and, like TNR instability, is a tissue- and site-specific genetic alteration. Cellular disruptions in the expression levels of various proteins including down- and up-regulation of hLigI and polymerase δ coincided with advanced steps of V(D)J recombination in pre-B lymphocytes (93). Interestingly, in a cell-free V(D)J recombination system, the addition of hLigI, but not other ligases, enhanced mutagenesis by increasing V(D)J coding junction diversity by affecting the levels of insertions and deletions (94). Both the coincident protein expression changes and the sensitivity of the genetic outcome of the V(D)J system reflect upon the results we observed for hLigI overexpression upon TNR instability. Whether similar alterations in protein expression and activity coincide with repeat expansions is unknown.
In conclusion, our results show that repeat instability is not linked to perturbed DNA metabolic processes widely detected under different hLigI alterations. However, they indicated that maintaining CTG/CAG repeat stability is linked specifically to the functional coordination existing between LigI and PCNA factors, as shown in the model proposed (Fig. 8). Our data indicate that hLigI protein levels may be important for maintaining CTG/CAG stability. Other findings suggest that the amounts of hLigI protein can increase under distinct cellular situations, as differentiating processes (63), human tumors (48), or UV irradiation (63, 80). Considering the LigI concentration sensitivity in CTG/CAG instability with the high levels of tissue-specific CTG/CAG instability in TNR patients and mouse (Ref. 1 and references therein), we hypothesize that variations in hLigI levels among different tissues or cell types as well as at distinct developmental windows could be linked to the high degree of tissue-specific somatic repeat instability observed in some of the TNR diseases. Mice show strong tissue-specific variations in LigI protein levels (data not shown). Curiously, the tissue in which increased genetic instability was reported (spleen) in the 46BR mouse model (59) was one that normally shows high LigI levels (data not shown). It would be of interest to know if tissue-specific hLigI protein levels correlate with tissue-specific and development-specific levels of CTG/CAG instability.
FIGURE 8.
Model proposed. A, during replication, the coordination between LigI, PCNA, and other factors (i.e. DNA polymerase (red circle)/FEN-1 (blue circle)) is not affected by normal levels of wt-LigI or low activity LigI (HeLa or 46BRLigIm/m (yellow circle)). However, the inability to bind with PCNA (46BRLigIm/m;wt-PCNA (yellow square)) in a ligation replication step may affect LigI-PCNA coordination, as the presence of LigI would be demanding by the process of Okazaki fragments to be sealed. Similarly, high levels of LigI (46BRLigIm/m;wt) may hinder the coordination with other factors. B, repair coordination is not affected by the binding between LigI and PCNA (46BRLigIm/m;wt-PCNA) either by a low demanding nick ligation step (covered by the LigIm/m presence) or by alternative repair pathways involving additional LigI partners (i.e. 9-1-1 sliding clamp), not affected by the specific LigI-PCNA mutation. On the contrary, LigI overexpression may affect PCNA-other partner repair-coordination by sequestration, but at the same time may also disturb the alternative repair pathways where LigI has a role. Pink background, altered repeat DNA processing steps. Blue and red lines, repeat tracts.
Supplementary Material
Acknowledgments
We thank Kerrie Nichol, Rachel Lau, and Gagan Panigrahi for technical assistance and Alessandra Montecucco for scientific discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM57479 (to A. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- TNR
- trinucleotide repeat
- PCNA
- proliferating cell nuclear antigen
- hPCNA
- human PCNA
- hLigI
- human DNA ligase I
- STRIP assay
- stability of trinucleotide repeats by individual product analysis
- DMEM
- Dulbecco's modified Eagle's medium
- wt
- wild type.
REFERENCES
- 1.Pearson C. E., Nichol Edamura K., Cleary J. D. (2005) Nat. Rev. Genet. 6, 729–742 [DOI] [PubMed] [Google Scholar]
- 2.Mirkin S. M. (2005) Nat. Struct. Mol. Biol. 12, 635–637 [DOI] [PubMed] [Google Scholar]
- 3.Mirkin S. M. (2006) Curr. Opin. Struct. Biol. 16, 351–358 [DOI] [PubMed] [Google Scholar]
- 4.Freudenreich C. H., Kantrow S. M., Zakian V. A. (1998) Science 279, 853–856 [DOI] [PubMed] [Google Scholar]
- 5.Schweitzer J. K., Livingston D. M. (1998) Hum. Mol. Genet. 7, 69–74 [DOI] [PubMed] [Google Scholar]
- 6.Yang J., Freudenreich C. H. (2007) Gene 393, 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovtun I. V., Liu Y., Bjoras M., Klungland A., Wilson S. H., McMurray C. T. (2007) Nature 447, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manley K., Shirley T. L., Flaherty L., Messer A. (1999) Nat. Genet. 23, 471–473 [DOI] [PubMed] [Google Scholar]
- 9.van den Broek W. J., Nelen M. R., Wansink D. G., Coerwinkel M. M, te Riele H., Groenen P. J., Wieringa B. (2002) Hum. Mol. Genet. 11, 191–198 [DOI] [PubMed] [Google Scholar]
- 10.Savouret C., Brisson E., Essers J., Kanaar R., Pastink A., te Riele H., Junien C., Gourdon G. (2003) EMBO J. 22, 2264–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savouret C., Garcia-Cordier C., Megret J., te Riele H., Junien C., Gourdon G. (2004) Mol. Cell. Biol. 24, 629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler V. C., Lebel L. A., Vrbanac V., Teed A., te Riele H., MacDonald M. E. (2003) Hum. Mol. Genet. 12, 273–281 [DOI] [PubMed] [Google Scholar]
- 13.Gomes-Pereira M., Fortune M. T., Ingram L., McAbney J. P., Monckton D. G. (2004) Hum. Mol. Genet. 13, 1815–1825 [DOI] [PubMed] [Google Scholar]
- 14.Timson D. J., Singleton M. R., Wigley D. B. (2000) Mutat. Res. 460, 301–318 [DOI] [PubMed] [Google Scholar]
- 15.Martin I. V., MacNeill S. A. (2002) Genome Biology 3, 3005.1–3005.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes D. E., Tomkinson A. E., Lehmann A. R., Webster A. D., Lindahl T. (1992) Cell 69, 495–503 [DOI] [PubMed] [Google Scholar]
- 17.Prigent C., Satoh M. S., Daly G., Barnes D. E., Lindahl T. (1994) Mol. Cell. Biol. 14, 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin D. S., McKenna A. E., Motycka T. A., Matsumoto Y., Tomkinson A. E. (2000) Curr. Biol. 10, 919–922 [DOI] [PubMed] [Google Scholar]
- 19.Prasad R., Singhal R. K., Srivastava D. K., Molina J. T., Tomkinson A. E., Wilson S. H. (1996) J. Biol. Chem. 271, 16000–16007 [DOI] [PubMed] [Google Scholar]
- 20.Wu X., Braithwaite E., Wang Z. (1999) Biochemistry 38, 2628–2635 [DOI] [PubMed] [Google Scholar]
- 21.Pascucci B., Stucki M., Jónsson Z. O., Dogliotti E., Hübscher U. (1999) J. Biol. Chem. 274, 33696–33702 [DOI] [PubMed] [Google Scholar]
- 22.Mortusewicz O., Rothbauer U., Cardoso M. C., Leonhardt H. (2006) Nucleic Acids Res. 34, 3523–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balakrishnan L., Brandt P. D., Lindsey-Boltz L. A., Sancar A., Bambara R. A. (2009) J. Biol. Chem. 284, 15158–15172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araújo S. J., Tirode F., Coin F., Pospiech H., Syväoja J. E., Stucki M., Hübscher U., Egly J. M., Wood R. D. (2000) Genes Dev. 14, 349–359 [PMC free article] [PubMed] [Google Scholar]
- 25.Mocquet V., Lainé J. P., Riedl T., Yajin Z., Lee M. Y., Egly J. M. (2008) EMBO J. 27, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., Li G. M. (2005) Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 27.Mossi R., Ferrari E., Hübscher U. (1998) J. Biol. Chem. 273, 14322–14330 [DOI] [PubMed] [Google Scholar]
- 28.Bhagwat A. S., Sanderson R. J., Lindahl T. (1999) Nucleic Acids Res. 27, 4028–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ireland M. J., Reinke S. S., Livingston D. M. (2000) Genetics 155, 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Refsland E. W., Livingston D. M. (2005) Genetics 171, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian J., Vijayakumar S., Tomkinson A. E., Arnheim N. (2005) Genetics 171, 427–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webster A. D., Barnes D. E., Arlett C. F., Lehmann A. R., Lindahl T. (1992) Lancet 339, 1508–1509 [DOI] [PubMed] [Google Scholar]
- 33.Henderson L. M., Arlett C. F., Harcourt S. A., Lehmann A. R., Broughton B. C. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 2044–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Squires S., Johnson R. T. (1983) Carcinogenesis 4, 565–572 [DOI] [PubMed] [Google Scholar]
- 35.Teo I. A., Arlett C. F., Harcourt S. A., Priestley A., Broughton B. C. (1983) Mutat. Res. 107, 371–386 [DOI] [PubMed] [Google Scholar]
- 36.Teo I. A., Broughton B. C., Day R. S., James M. R., Karran P., Mayne L. V., Lehmann A. R. (1983) Carcinogenesis 4, 559–564 [DOI] [PubMed] [Google Scholar]
- 37.Poirier V., James M. R., Arlett C. F., Lehmann A. R. (1985) Carcinogenesis 6, 837–841 [DOI] [PubMed] [Google Scholar]
- 38.Lehmann A. R., Willis A. E., Broughton B. C., James M. R., Steingrimsdottir H., Harcourt S. A., Arlett C. F., Lindahl T. (1988) Cancer Res. 48, 6343–6347 [PubMed] [Google Scholar]
- 39.Lönn U., Lönn S., Nylen U., Winblad G. (1989) Carcinogenesis 10, 981–985 [DOI] [PubMed] [Google Scholar]
- 40.Mackenney V. J., Barnes D. E., Lindahl T. (1997) J. Biol. Chem. 272, 11550–11556 [DOI] [PubMed] [Google Scholar]
- 41.Goetz J. D., Motycka T. A., Han M., Jasin M., Tomkinson A. E. (2005) DNA Repair 4, 649–654 [DOI] [PubMed] [Google Scholar]
- 42.Soza S., Leva V., Vago R., Ferrari G., Mazzini G., Biamonti G., Montecucco A. (2009) Mol. Cell. Biol. 29, 2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panigrahi G. B., Lau R., Montgomery S. E., Leonard M. R., Pearson C. E. (2005) Nat. Struct. Mol. Biol. 12, 654–662 [DOI] [PubMed] [Google Scholar]
- 44.Panigrahi G. B., Cleary J. D., Pearson C. E. (2002) J. Biol. Chem. 277, 13926–13934 [DOI] [PubMed] [Google Scholar]
- 45.Chen X., Pascal J., Vijayakumar S., Wilson G. M., Ellenberger T., Tomkinson A. E. (2006) Methods Enzymol. 409, 39–52 [DOI] [PubMed] [Google Scholar]
- 46.Levin D. S., Bai W., Yao N., O'Donnell M., Tomkinson A. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12863–12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee Y. C., Morgenstern H., Greenland S., Tashkin D. P., Papp J., Sinsheimer J., Cao W., Hashibe M., You N. C., Mao J. T., Cozen W., Mack T. M., Zhang Z. F. (2008) Int. J. Cancer 122, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun D., Urrabaz R., Nguyen M., Marty J., Stringer S., Cruz E., Medina-Gundrum L., Weitman S. (2001) Clin. Cancer Res. 7, 4143–4148 [PubMed] [Google Scholar]
- 49.Somia N. V., Jessop J. K., Melton D. W. (1993) Mutat. Res. 294, 51–58 [DOI] [PubMed] [Google Scholar]
- 50.Rossi R., Villa A., Negri C., Scovassi I., Ciarrocchi G., Biamonti G., Montecucco A. (1999) EMBO J. 18, 5745–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrari G., Rossi R., Arosio D., Vindigni A., Biamonti G., Montecucco A. (2003) J. Biol. Chem. 278, 37761–37767 [DOI] [PubMed] [Google Scholar]
- 52.Vitolo B., Lidonnici M. R., Montecucco C., Montecucco A. (2005) Eur. J. Histochem. 49, 349–354 [DOI] [PubMed] [Google Scholar]
- 53.Vijayakumar S., Dziegielewska B., Levin D. S., Song W., Yin J., Yang A., Matsumoto Y., Bermudez V. P., Hurwitz J., Tomkinson A. E. (2009) Mol. Cell. Biol. 29, 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson C. E., Tam M., Wang Y. H., Montgomery S. E., Dar A. C., Cleary J. D., Nichol K. (2002) Nucleic Acids Res. 30, 4534–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vispé S., Satoh M. S. (2000) J. Biol. Chem. 275, 27386–27392 [DOI] [PubMed] [Google Scholar]
- 56.Yang S. W., Huang P., Plunkett W., Becker F. F., Chan J. Y. (1992) J. Biol. Chem. 267, 2345–2349 [PubMed] [Google Scholar]
- 57.Bentley D., Selfridge J., Millar J. K., Samuel K., Hole N., Ansell J. D., Melton D. W. (1996) Nat. Genet. 13, 489–491 [DOI] [PubMed] [Google Scholar]
- 58.Bentley D. J., Harrison C., Ketchen A. M., Redhead N. J., Samuel K., Waterfall M., Ansell J. D., Melton D. W. (2002) J. Cell Sci. 115, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 59.Harrison C., Ketchen A. M., Redhead N. J., O'Sullivan M. J., Melton D. W. (2002) Cancer Res. 62, 4065–4074 [PubMed] [Google Scholar]
- 60.Veeraraghavan J., Rossi M. L., Bambara R. A. (2003) J. Biol. Chem. 278, 42854–42866 [DOI] [PubMed] [Google Scholar]
- 61.Kang S., Ohshima K., Shimizu M., Amirhaeri S., Wells R. D. (1995) J. Biol. Chem. 270, 27014–27021 [DOI] [PubMed] [Google Scholar]
- 62.Ohshima K., Wells R. D. (1997) J. Biol. Chem. 272, 16798–16806 [DOI] [PubMed] [Google Scholar]
- 63.Montecucco A., Biamonti G., Savini E., Focher F., Spadari S., Ciarrocchi G. (1992) Nucleic Acids Res. 20, 6209–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maga G., Villani G., Tillement V., Stucki M., Locatelli G. A., Frouin I., Spadari S., Hübscher U. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14298–14303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levin D. S., Vijayakumar S., Liu X., Bermudez V. P., Hurwitz J., Tomkinson A. E. (2004) J. Biol. Chem. 279, 55196–55201 [DOI] [PubMed] [Google Scholar]
- 66.Jónsson Z. O., Hindges R., Hübscher U. (1998) EMBO J. 17, 2412–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montecucco A., Rossi R., Levin D. S., Gary R., Park M. S., Motycka T. A., Ciarrocchi G., Villa A., Biamonti G., Tomkinson A. E. (1998) EMBO J. 17, 3786–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tom S., Henricksen L. A., Park M. S., Bambara R. A. (2001) J. Biol. Chem. 276, 24817–24825 [DOI] [PubMed] [Google Scholar]
- 69.Vijayakumar S., Chapados B. R., Schmidt K. H., Kolodner R. D., Tainer J. A., Tomkinson A. E. (2007) Nucleic Acids Res. 35, 1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pascal J. M., Tsodikov O. V., Hura G. L., Song W., Cotner E. A., Classen S., Tomkinson A. E., Tainer J. A., Ellenberger T. (2006) Mol. Cell 24, 279–291 [DOI] [PubMed] [Google Scholar]
- 71.Sporbert A., Domaing P., Leonhardt H., Cardoso M. C. (2005) Nucleic Acids Res. 33, 3521–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chapados B. R., Hosfield D. J., Han S., Qiu J., Yelent B., Shen B., Tainer J. A. (2004) Cell 116, 39–50 [DOI] [PubMed] [Google Scholar]
- 73.Mayanagi K., Kiyonari S., Saito M., Shirai T., Ishino Y., Morikawa K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4647–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanov I., Chapados B. R., McCammon J. A., Tainer J. A. (2006) Nucleic Acids Res. 34, 6023–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T. (2005) EMBO J. 24, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garg P., Burgers P. M. (2005) Crit. Rev. Biochem. Mol. Biol 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 77.Riva F., Savio M., Cazzalini O., Stivala L. A., Scovassi I. A., Cox L. S., Ducommun B., Prosperi E. (2004) Exp. Cell Res. 293, 357–367 [DOI] [PubMed] [Google Scholar]
- 78.Freudenreich C. H., Stavenhagen J. B., Zakian V. A. (1997) Mol. Cell. Biol. 17, 2090–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Callahan J. L., Andrews K. J., Zakian V. A., Freudenreich C. H. (2003) Mol. Cell. Biol. 23, 7849–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nocentini S., Mezzina M. (1981) Chromosome Damage and Repair ( Seeberg E., Kleppe K. eds) pp. 329–333, Plenum Press, New York [Google Scholar]
- 81.Nick McElhinny S. A., Gordenin D. A., Stith C. M., Burgers P. M., Kunkel T. A. (2008) Mol. Cell 25, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Longley M. J., Pierce A. J., Modrich P. (1997) J. Biol. Chem. 272, 10917–10921 [DOI] [PubMed] [Google Scholar]
- 83.Burgers P. M., Gerik K. J. (1998) J. Biol. Chem. 273, 19756–19762 [DOI] [PubMed] [Google Scholar]
- 84.Corrette-Bennett S. E., Borgeson C., Sommer D., Burgers P. M., Lahue R. S. (2004) Nucleic Acids Res. 32, 6268–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Z., Lau R., Marcadier J. L., Chitayat D., Pearson C. E. (2003) Am. J. Hum. Genet. 73, 1092–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smirnova E., Toueille M., Markkanen E., Hübscher U. (2005) Biochem. J. 389, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang W., Lindsey-Boltz L. A., Sancar A., Bambara R. A. (2006) J. Biol. Chem. 281, 20865–20872 [DOI] [PubMed] [Google Scholar]
- 88.Song W., Levin D. S., Varkey J., Post S., Bermudez V. P., Hurwitz J., Tomkinson A. E. (2007) J. Biol. Chem. 282, 22721–22730 [DOI] [PubMed] [Google Scholar]
- 89.Kokoska R. J., Stefanovic L., Buermeyer A. B., Liskay R. M., Petes T. D. (1999) Genetics 151, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schweitzer J. K., Livingston D. M. (1999) Genetics 152, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baida A., López A., Marcos R., Velázquez A. (2003) DNA Repair 2, 827–833 [DOI] [PubMed] [Google Scholar]
- 92.López A., Xamena N., Marcos R., Velázquez A. (2005) Mutat. Res. 570, 253–265 [DOI] [PubMed] [Google Scholar]
- 93.Jessberger R., Schär P., Robins P., Ferrari E., Riwar B., Hübscher U. (1997) Nucleic Acids Res. 25, 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramsden D. A., Paull T. T., Gellert M. (1997) Nature 388, 488–491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.