Abstract
Salutaridine reductase (SalR, EC 1.1.1.248) catalyzes the stereospecific reduction of salutaridine to 7(S)-salutaridinol in the biosynthesis of morphine. It belongs to a new, plant-specific class of short-chain dehydrogenases, which are characterized by their monomeric nature and increased length compared with related enzymes. Homology modeling and substrate docking suggested that additional amino acids form a novel α-helical element, which is involved in substrate binding. Site-directed mutagenesis and subsequent studies on enzyme kinetics revealed the importance of three residues in this element for substrate binding. Further replacement of eight additional residues led to the characterization of the entire substrate binding pocket. In addition, a specific role in salutaridine binding by either hydrogen bond formation or hydrophobic interactions was assigned to each amino acid. Substrate docking also revealed an alternative mode for salutaridine binding, which could explain the strong substrate inhibition of SalR. An alternate arrangement of salutaridine in the enzyme was corroborated by the effect of various amino acid substitutions on substrate inhibition. In most cases, the complete removal of substrate inhibition was accompanied by a substantial loss in enzyme activity. However, some mutations greatly reduced substrate inhibition while maintaining or even increasing the maximal velocity. Based on these results, a double mutant of SalR was created that exhibited the complete absence of substrate inhibition and higher activity compared with wild-type SalR.
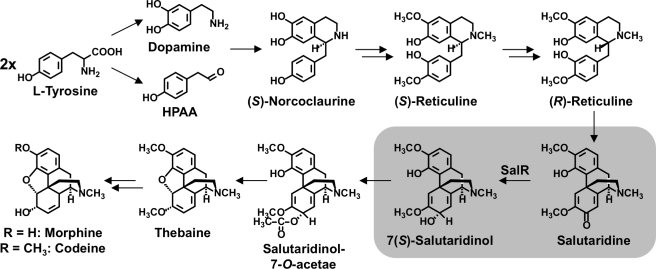
The benzylisoquinoline alkaloids (BIAs)3 comprise a large and diverse group of nitrogen-containing secondary metabolites with about 2500 compounds identified in plants (1). Among them are several important pharmaceuticals, such as the antimicrobials berberine and sanguinarine, and the vasodilator papaverine. The most prominent compounds of this class are the antitussive codeine, the analgesic morphine, and their biosynthetic precursor thebaine. The latter is used as the starting molecule for the production of a variety of semi-synthetic analgesics including oxycontin and buprenorphine. Pentacyclic morphinan alkaloids possess several chiral centers, which preclude chemical synthesis as an option for the efficient production of these widely used pharmaceuticals. Therefore, the worldwide supply of these narcotic compounds is still achieved by their isolation mainly from the opium poppy, Papaver somniferum L. With the availability of an increasing number of isolated genes encoding several pathway enzymes, recent interest has focused on the qualitative and quantitative modulation of the alkaloid profile in transgenic opium poppy plants (2–6), the production of BIAs in microbes (7, 8), and de novo synthesis by a combination of chemical and biochemical conversions. BIA biosynthesis begins with the condensation of the tyrosine-derived precursors dopamine and p-hydroxyphenylacetaldehyde to (S)-norcoclaurine (see Fig. 1) (1). Subsequent regiospecific O- and N-methylations and aromatic ring hydroxylation lead to (S)-reticuline, which is the central intermediate for almost all BIAs. For morphinan alkaloid biosynthesis, (S)-reticuline undergoes an inversion of stereochemistry to (R)-reticuline, followed by C-C phenol coupling catalyzed by a unique cytochrome P450-dependent monooxygenase to yield salutaridine. Subsequent stereospecific reduction to 7(S)-salutaridinol is required for the attachment of an acetyl moiety to produce salutaridinol-7-O-acetate, which spontaneously rearranges to thebaine (9). The O-demethylation of thebaine and the reduction of codeinone to codeine represent the penultimate steps in morphine biosynthesis. Cognate cDNAs have been isolated for all of the enzymes leading to (S)-reticuline, as well as those involved in the conversion of (R)-reticuline to salutaridine-7-O-acetate (1). Salutaridine reductase (SalR, EC 1.1.1.248) catalyzes the stereospecific, NADPH-dependent reduction of salutaridine to 7(S)-salutaridinol and is a member of the classical subgroup of the short chain dehydrogenase/reductase (SDR) protein family (10, 11). The main characteristics of this category of SDRs are the largely conserved TGXXXGhG motif for cofactor binding and the YXXXK motif, which together with an upstream Ser residue represent the catalytic center (12). In this catalytic triad, Lys forms hydrogen bonds with the ribose moiety of the cofactor, which itself is hydrogen bonded to Tyr. This hydrogen bond network is presumed to lower the pKa of the Tyr hydroxyl group, which functions as the catalytic base. Ser has been suggested to either stabilize the substrate (13, 14) or to interact with Tyr (15). Additionally, an Asn residue has been proposed to stabilize the position of the Lys residue, thereby forming a proton relay system involving water (16). Most other members of the SDR protein family are categorized into three additional subgroups (i.e. divergent, intermediate, or complex) exhibiting different overall sizes and slight amino acid sequence variations in conserved regions (17). Non-classical SDRs predominantly consist of isomerases (EC 5.-.-.-), such as galactose epimerase, and lyases (EC 4.-.-.-), such as glucose dehydratase, whereas classical SDRs encompass oxidoreductases (EC 1.-.-.-), such as SalR. Although classical SDRs are typically multimeric, SalR is a monomer because of an additional stretch of 40 amino acids preceding the YXXXK catalytic motif. In porcine testicular carbonyl reductase, these amino acids form a helix blocking the dimer interface (18) and homology modeling revealed a similar feature in SalR (19). In contrast with porcine testicular carbonyl reductase, SalR exhibits an additional stretch of 40 amino acids that have only been detected in some SDRs from plants (11, 20–22). Although attempts to obtain a crystal structure for SalR have so far been unsuccessful, homology modeling using porcine testicular carbonyl reductase as a template produced a tertiary structure in which the additional amino acids form an additional helix (19). The subsequent docking of salutaridine into the active site of this model suggested the involvement of this structural element in substrate binding, which was supported by preliminary site-directed mutagenesis.
FIGURE 1.
Selected steps in morphine biosynthesis. Double arrows indicate the involvement of more than one enzyme. The SalR reaction is highlighted. HPAA, p-hydroxyphenylacetaldehyde; SalR, salutaridine reductase.
SalR from the Persian poppy Papaver bracteatum L. shows strong substrate inhibition with a Ki around 150 μm (19). Substrate inhibition can substantially and negatively impact chemical engineering strategies by limiting the quantity of substrate that can be fed into a system, which reduces overall efficiency. The strong substrate inhibition exhibited by SalR could limit its biotechnological application in plant, microbial, or enzyme-based systems. To investigate the structural basis of substrate inhibition, we substituted all amino acids putatively involved in salutaridine binding and analyzed various kinetic parameters. Over the course of these experiments, substrate docking required modification because some mutations had unexpected consequences that did not fully agree with the original docking. The discrepancy was mainly due to the side chain arrangements of amino acids residing in the new helix. Precise prediction of this domain is difficult because of the lack of an equivalent crystal structure. In this report, we present a revised substrate docking for salutaridine into SalR, supported by comprehensive site-directed mutagenesis, which facilitated the creation of an enzyme variant devoid of substrate inhibition.
EXPERIMENTAL PROCEDURES
Chemicals
(−)-Menthone and tropinone were obtained from Fluka, cyclohexanone, 2-cyclohexen-1-one, (R)-(−)-carvone, (S)-(+)-carvone, berberine, sanguinarine, NADP+, and NADPH were obtained from Sigma, 2H-1,4-benzoxazine-3(H)-one, 2H-1,4-benzothiazine-(4H)-one, and plumbagin were obtained from Indofine (Hillsborough, NJ). Salutaridine was synthesized by bisulfite conversion of thebaine, followed by methylation with diazomethane (23, 24).
HPLC Analysis
HPLC analyses were performed on a Beckman System Gold HPLC system equipped with the 126 solvent module and the 168 diode array detector. Samples were separated on a Lichrospher RP-Select B column (5 μm particle size, 150 × 4.6 mm, Merck KGaA) at a flow rate of 1 ml/min with: solvent A, 2% (v/v) acetonitrile, 98% (v/v) water, 0.01% (v/v) phosphoric acid; and solvent B, 98% (v/v) acetonitrile, 2% (v/v) water, 0.01% (v/v) phosphoric acid. For the standard SalR enzyme assay using salutaridine as substrate, elution conditions consisted of a gradient from 2% B to 35% B over 13 min, a hold for 2 min at 35% B, and re-equilibration for 6 min at 2% B. Assays containing berberine, sanguinarine, benzoxazine, benzothiazine, and plumbagin were separated using a gradient from 2% B to 65% B over 25 min, followed by an increase in B to 100% in 1 min, subsequently holding at 100% B for 7 min, and re-equilibrating for 5 min at 2% B. The detection wavelength was set to 210 nm.
GC Analysis
Samples from menthone, cyclohexanone, 2-cyclohexen-1-one, (R)-(−)-carvone, (S)-(+)-carvone, and tropinone containing enzyme assays were analyzed on an Agilent Technologies 6890N Network GC System. Separation was performed on a HP-5 column (J & W Scientific) of 30 m × 0.32-mm inner diameter, coated with 5% phenyl methyl siloxane (0.25-μm film thickness). Helium was the carrier flow gas (flow rate of 1 ml/min), and splitless injection (injection volume of 5 μl, injection temperature 250 °C) was used. Temperature gradients were 80 °C (1 min hold), 5 °C/min to 130 °C followed by 100 °C/min to 150 °C.
Site-directed Mutagenesis and Purification of Recombinant Proteins
Primer pairs used to introduce point mutations into the SalR open reading frame are listed in supplemental Table S1. These primers were used together with the pQE31-SalR plasmid and the proofreading Pfu-DNA polymerase (Fermentas, Burlington, ON, Canada) for PCR (94 °C for 30s, followed by 12 cycles of 94 °C for 30 s, 55 °C for 1 min, 68 °C for 5 min). Subsequently, the PCR was digested with DpnI (Fermentas) for 2 h at 37 °C, and 1 μl was used to transform Escherichia coli strain XL1BlueMRF′. Plasmids were purified, sequenced, and introduced for protein overexpression into SG13009 (Qiagen, Hilden, Germany) strains. Overexpression and extraction of the recombinant proteins were performed as described (11). For protein characterization, the fraction eluting between 10 and 60 mm imidazole from the cobalt affinity column (Talon, Clontech, Mountain View, CA) was used after buffer exchange to 10 mm HEPES, pH 7.5. Purity of the enzymes was checked by SDS-PAGE (12% (w/v) polyacrylamide) according to Laemmli (25). Because the protein was purified to homogeneity according to SDS-PAGE (supplemental Fig. S1), the concentration was determined at 280 nm using the molar absorption coefficient determined on the basis of the amino acid sequence.
Enzyme Assays and Enzyme Characterizations
The reaction mixture to assay the reduction of substrates consisted of 150 mm potassium phosphate, pH 6.0, and different concentrations of NADPH, salutaridine, and protein in a total volume of 200 μl. Reactions were terminated by addition of 800 μl of methanol, centrifuged at 12,000 × g for 5 min, and 100 μl of the supernatant was subjected to HPLC analysis. For analysis by GC, reactions were extracted with 200 μl of chloroform and 5 μl of the organic phase were injected. Assays for the determination of substrate specificity contained benzoxazine, benzothiazole, plumbagine, or berberine at a concentration of 500 μm, or 250 μm for sanguinarine. All other substrates were tested at a concentration of 1000 μm (see supplemental Fig. S2 for structures). The cofactor NADPH was included at a concentration of 250 μm, and the assays were incubated for 30 min at room temperature.
Data for the calculation of the kinetic parameters were collected during the initial linear phase of the enzyme reaction showing less than 10% substrate conversion, which was either achieved by adjusting the incubation times or the amount of enzyme. Typically, the enzyme assays contained 0.5–3.9 μg of purified protein and were incubated at 30 °C between 30 s and 10 min (the incubation times and protein amounts for each mutant are shown in supplemental Table S2). Salutaridine concentrations varied between 2 and 1000 μm in the presence of 250 μm NADPH, whereas NADPH concentrations varied between 2.5 and 250 μm at constant salutaridine concentrations as indicated in Table 1. NADP+ inhibition studies were performed at a concentration range from 2.5 to 250 μm. Each measurement was conducted at least three times. The apparent kinetic data for the cofactor and the mutants exhibiting no substrate inhibition at a concentration up to 1 mm salutaridine were calculated using the Michaelis-Menten equation: v = Vmax × S/(S + Km), or when substrate inhibition was observed: v = Vmax/(1 + Km/S + S/Ki), where S denotes the substrate or cofactor concentration, respectively. Optimal velocity was determined by Vopt = Vmax/(1 + 2(Km/Ki)1/2). All calculations were performed using the FIG.P software, version 2.98 (Biosoft, Cambridge, United Kingdom).
TABLE 1.
Apparent kinetic parameters for SalR and its mutated versions
| Version | Km | Kim | Vmax/opt | kcat | kcat/Km | Km NADPH |
|---|---|---|---|---|---|---|
| μm | μm | nkat mg−1 | s−1 | s−1mm−1 | μm | |
| SalR-WT | 2.1 ± 0.6 | 184 ± 47 | 35.61 ± 2.82a | 1.306 ± 0.104 | 661.7 | 3.5 ± 0.8b |
| F104A | 31.8 ± 3.6 | 466 ± 64 | 74.38 ± 4.91a | 2.727 ± 0.180 | 86.2 | 9.7 ± 2.5c |
| V106A | 8.9 ± 2.9 | 283 ± 90 | 67.13 ± 9.03a | 2.458 ± 0.331 | 295.4 | 7.0 ± 2.3c |
| D107A | 14.8 ± 2.4 | 436 ± 78 | 16.61 ± 1.11a | 0.610 ± 0.040 | 41.7 | 30.5 ± 6.0c |
| S181A | 5.0 ± 1.9 | 199 ± 60 | 52.75 ± 6.62a | 1.933 ± 0.243 | 430.3 | 4.5 ± 1.7d |
| T182A | 4.1 ± 1.2 | 58 ± 15 | 3.43 ± 0.39a | 0.126 ± 0.014 | 32.4 | 21.3 ± 5.1d |
| L185A | 13.2 ± 1.7 | NDe | 0.44 ± 0.01f | 0.016 ± 0.001 | 1.2 | 29.7 ± 6.7g |
| Ser | 1.1 ± 0.2 | 377 ± 76 | 4.03 ± 0.15a | 0.148 ± 0.006 | 137.9 | 12.9 ± 4.1d |
| Val | 2.1 ± 0.6 | 135 ± 30 | 18.69 ± 1.34a | 0.685 ± 0.049 | 347.6 | 14.2 ± 3.8d |
| K186V | 7.8 ± 2.1 | 18 ± 8 | 27.23 ± 8.05a | 0.998 ± 0.295 | 149.3 | 3.4 ± 1.1d |
| L266A | 49.4 ± 2.2 | ND | 13.68 ± 0.17f | 0.501 ± 0.006 | 10.2 | 15.5 ± 1.3g |
| M271A | 22.5 ± 2.8 | ND | 0.16 ± 0.01f | 0.006 ± 0.001 | 0.3 | 19.8 ± 5.4g |
| N272A | 90.1 ± 5.0 | ND | 0.57 ± 0.01f | 0.021 ± 0.001 | 0.2 | 15.6 ± 3.4g |
| I275A | 33.1 ± 5.9 | 1249 ± 330 | 87.03 ± 7.58a | 3.188 ± 0.278 | 98.0 | 21.1 ± 1.9c |
| Val | 2.1 ± 0.9 | 122 ± 41 | 27.89 ± 3.01a | 1.022 ± 0.110 | 568.5 | 4.7 ± 0.7c |
| F104A/I275A | 408.6 ± 6.6 | ND | 66.85 ± 0.49f | 2.450 ± 0.017 | 5.9 | 43.0 ± 12.5h |
a Vopt.
b All kinetic parameters for salutaridine were recorded at 250 μm NADPH; salutaridine concentrations for the determination of NADPH affinity were 10 μm.
c All kinetic parameters for salutaridine were recorded at 250 μm NADPH; salutaridine concentrations for the determination of NADPH affinity were 75 μm.
d All kinetic parameters for salutaridine were recorded at 250 μm NADPH; salutaridine concentrations for the determination of NADPH affinity were 15 μm.
e ND, not detectable up to a salutaridine concentration of 1000 μm.
f Vmax.
g All kinetic parameters for salutaridine were recorded at 250 μm NADPH; salutaridine concentrations for the determination of NADPH affinity were 500 μm.
h All kinetic parameters for salutaridine were recorded at 250 μm NADPH; salutaridine concentrations for the determination of NADPH affinity were 1000 μm.
Homology Modeling and Substrate Docking
Homology modeling of SalR was based on the x-ray structure of human CBR1 (PDB code 1wma) (26) and has been described in detail (19). Because new experiments were not completely consistent with the model and a coil structure from Asn98 to Phe111 was previously uncertain, 100-ps molecular dynamic simulations at 300 K were performed for the NADPH-protein complex using the molecular modeling program MOE (Molecular Operating Environment, Chem. Comp. Group Inc., Montreal, Canada). The remaining part of the protein structure was fixed during the simulation. Subsequently, the structure was energy minimized using Charmm22 (27) and Born-Solvation (28). The resulting, slightly modified structure was used for the docking of salutaridine using PLANTS (29). For all subsequent docking arrangements the complex consisting of the ligand, NADPH, and the protein was energy optimized. The stereochemical quality of the model was checked using Procheck (30). In the Ramachandran plot, 90.4% of backbone dihedral angles were located in the most favored area, and 8.6% were found in additionally allowable regions. The only outlier occurred in a loop. PROSA II was used to analyze the native fold of the model (31). The energy plot showed that almost all residues were in the negative energy range with a combined energy z-score of −10.0, which is close to the average value of −10.9 for a protein with 320 amino acids. Altogether these analyses are consistent with a reasonable protein model.
CD Spectroscopy
CD spectra were recorded at room temperature on a Jasco J-715 spectropolarimeter in a cylindrical quartz cuvette with a path length of 0.1 cm and a volume of 400 μl consisting of 10 μm protein in 10 mm HEPES, pH 7.5. Within 185 to 260 nm the parameters were: step resolution 0.2 nm, speed 100 nm/min, response time 2.0 s, bandwidth 1.0 nm, and sensitivity 20 mdeg. All spectra represent the average of 10 scans with the background subtracted. The data were smoothed and converted to molar ellipticity using the Jasco software.
RESULTS
Modeling and Substrate Docking
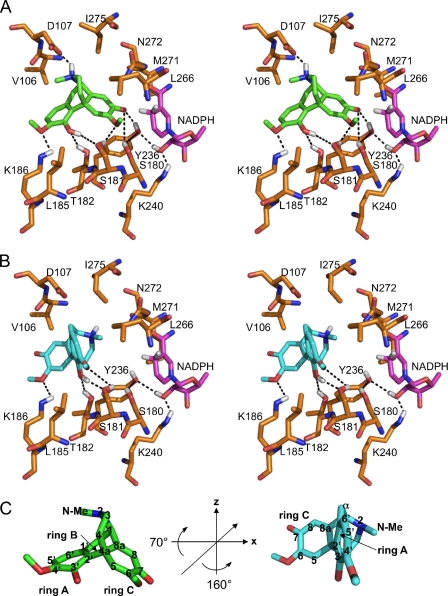
The exchange of residues, especially Asp107 and Ile275, resulted in effects on the kinetic parameters of SalR that could not be adequately explained by the original model and substrate docking. Therefore, molecular dynamic simulations were performed for the coil region between amino acids Asn98 and Phe111, which resulted in minor modifications in several side chain arrangements. Subsequently, salutaridine was docked into the active site. The major changes in the orientation of amino acid side chains occurred on a stretch of residues between β-sheet D and the new α-helix E′. The side chain of Asp107 was turned toward the substrate binding pocket, which caused a repositioning of the Val106 side chain more perpendicular to the substrate binding pocket as a means of avoiding spatial interference with Asp107 (Fig. 2A). Additionally, the side chain of Ile275 was rotated around the Cα-Cβ bond so that Cδ1 points more toward the substrate binding pocket. These new amino acid arrangements led to a change in the conformation of the substrate in the active site yielding a more realistic docking arrangement than reported previously. The revised arrangement is established by the distances between: 1) the Oδ2 of Asp107 and the isoquinoline nitrogen; 2) the hydroxyl group of Ser181 and the oxygen of the 6-methoxy group; 3) the hydroxyl group of Thr182 and the 3′-hydroxyl; and 4) the Nϵ of Lys186 and the 3′-hydroxyl as well as the oxygen of the 4′-methoxy group, all of which are within hydrogen bonding range. The catalytic residues (Ser180, Tyr236, Lys240), the nicotinamide of the cofactor NADPH, and the keto group of the substrate are all in an arrangement consistent with catalysis. Comparable with the previous docking (19), there are strong π-π interactions and shape-dependent van der Waals interactions, respectively. However, in the new docking, these interactions take place between ring A and Phe104, and between the ring C-piperidine ring system and Leu266. Further hydrophobic interactions occur between Cγ2 of Val106 and both the N-methyl and Cα, between Cδ1 of Leu185 and the 6-methoxy, and between Cγ1 and Cδ1 of Ile275 and the heterocyclic ring of the isoquinoline moiety (Fig. 2A).
FIGURE 2.
Substrate binding pocket of SalR with docked ligand. A, stereo view of salutaridine binding in the catalytic mode. B, stereo view of salutaridine binding in the non-productive mode. C, rotation of the substrate to adopt the non-productive orientation. In C, the 3-fold symmetry axis is indicated by a line in the substrate on the left, and the rotation angles are provided in the center. The carbon numbering of salutaridine is indicated when the respective atom is visible. For A–C, the colors for the carbon skeleton of each structure are gold for amino acids, magenta for NADPH, green for salutaridine binding in the catalytic mode, or turquoise for salutaridine binding in the non-productive mode. Hydrogen, nitrogen, oxygen, and sulfur atoms are shown in white, blue, red, and yellow, respectively. Only hydrogen atoms participating in hydrogen bonding are shown. Phe104 was omitted for clarity of the stereo view, and is located behind the substrate. Putative hydrogen bonds are indicated by dashed lines. Ser180, Tyr236, and Lys240 constitute the catalytic triad. Because it has been reported that the catalytic Ser might interact with the substrate or with Tyr, both scenarios are indicated in A.
Steady State Kinetic Analysis of SalR Variants
Amino acids putatively involved in substrate binding were substituted (mostly for Ala) by site-directed mutagenesis. Generally, all mutations resulted in enzymes exhibiting lower catalytic efficiencies (Table 1). Replacement of Phe104 and Leu266 with Ala had been shown previously to decrease the affinity by a factor of more than 15. This was accompanied by an increase in kcat for F104A and a decrease in kcat for L266A (19). Substantial increases in Km values were also observed for D107A, L185A, M271A, N272A, and I275A. For L185A, M271A, and N272A, the turnover number was reduced to values below 2% of the wild-type level. D107A only exhibited a 2-fold decrease in kcat, whereas I275A showed an increase of velocity by a factor of 2.3. The effect on steady-state kinetics of the latter two mutations prompted us to re-evaluate the model and the substrate binding because these mutations should not have caused the observed effects according to the previous docking. To evaluate whether Cγ1 and/or Cδ1 of Ile275 are necessary for interaction with the substrate, we created an I275V version of the enzyme. The kinetic parameters of the enzyme were almost indistinguishable from the wild-type enzyme when Ile275 was replaced by Val.
The binding pocket is relatively open toward the site where Leu185 is predicted to reside. The close proximity of this residue to the 3′-hydroxyl and the 6-methoxy groups might prevent the substrate from sliding away from the catalytic Tyr236 (Fig. 2A). Replacement of Leu185 with the smaller amino acid Ala could account for the strongly decreased maximal velocity of L185A. We tried to alleviate the effects of L185A by introducing a Ser or a Val residue at this position with the assumption that Ser might stabilize the substrate through the formation of a hydrogen bond with the 3′-hydroxyl, or that Val is large enough to restrict movement of the substrate. In both cases, the affinity resembled that of the wild-type enzyme. Additionally, the kcat of L185V was increased by a factor of 42 compared with L185A, which represents half of the maximal velocity of the wild-type enzyme. Although the activity of L185S was increased almost 10-fold compared with L185A, the kcat was still only 10% that of the wild-type enzyme.
Moderate effects on substrate affinity were observed for V106A and K186V, as well as for S181A and T182A, with 4- and 2-fold decreases, respectively. However, the kcat values were substantially different. Whereas K186V exhibited a turnover number similar to that of the wild-type enzyme, the S181A and V106A mutations showed increases of 50 and 90%, respectively, and T182A showed a decrease to less than 10% of the wild-type value.
To determine whether kinetic parameters could be correlated with computational fitness scores, all mutated versions of the enzyme were modeled based on the predicted three-dimensional model of the wild-type enzyme, and salutaridine was docked into the substrate binding site. Substantially increased fitness scores were calculated for most of the mutations that showed strongly decreased substrate affinities, such as F104A, L185A, L266A, and N272A (Table 2). However, the scores of other mutations, such as I275A, or those with Km values similar to that of the wild-type enzyme did not always match the observed affinity resulting in a linear correlation with coefficient (r2) of 0.211. A similar outcome occurred when the fitness scores were compared with the corresponding catalytic efficiencies. However, it should be noted that Km values do not only reflect the affinity of the substrate with respect to corresponding fitness scores, but are also a kinetic constant of the substrate conversion.
TABLE 2.
Docking results for the catalytic and inhibitory conformation of salutaridine
Salutaridine was docked in both conformations to the three-dimensional model of each SalR variant. Most negative scores relate to highest expected affinity.
| Version | Fitness scorea |
|
|---|---|---|
| Catalytic | Inhibitory | |
| SalR-WT | −63.5 | −41.1 |
| F104A | −53.6 | −49.8 |
| V106A | −53.7 | −39.8 |
| D107A | −59.2 | −45.0 |
| S181A | −69.6 | −43.4 |
| T182A | −58.5 | −47.7 |
| L185A | −47.5 | −45.1 |
| Ser | −56.2 | −45.7 |
| Val | −54.6 | −46.4 |
| K186V | −68.2 | −40.7 |
| L266A | −54.6 | −43.3 |
| M271A | −63.4 | −43.7 |
| N272A | −49.1 | −44.9 |
| I275A | −59.6 | −43.2 |
| Val | −61.3 | −44.1 |
| F104A/I275A | −50.0 | −41.1 |
a The scores are crude indications for the affinity of the ligand to the enzyme.
In most cases, the simultaneous decrease in affinity for salutaridine and in catalytic efficiency was accompanied by an increase in the Km for the cofactor NADPH (r2 = 0.43) (Table 1). A strong decrease in affinity for NADPH by a factor of more than five was observed for the mutants with low affinity for salutaridine, such as D107A, L185A, M271A, N272A, and I275A, and for mutants exhibiting low catalytic efficiencies, including T182A, L185A, M271A, and N272A.
None of the mutations changed the substrate specificity of the enzyme. After incubation with a variety of potential substrates (for structures see supplemental Fig. S2), no new peaks could be detected in HPLC or GC chromatograms compared with control incubations lacking the enzyme (data not shown). Unfortunately, compounds with more similarity to salutaridine could not be tested due to their lack of availability. The compounds tested were chosen either because they possess a keto group as part of a six-member ring system, or because they are important BIAs that potentially undergo enzymatic reduction. Furthermore, the cyclohexanone and cyclohexenone derivatives are substrates for other SDRs of plant origin.
Circular Dichroism Spectra
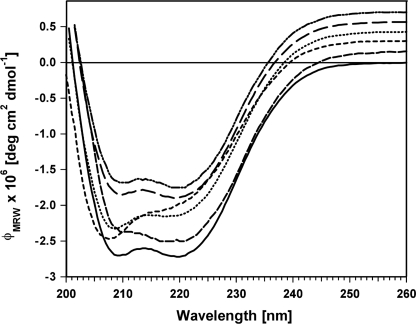
To evaluate the influence of each mutation on the secondary structure of SalR, far UV CD spectroscopy was performed at room temperature. The two minima at 209 and 220 nm in the spectrum of the wild-type enzyme account for the dominating proportion of α-helical elements (32) (Fig. 3). For most of the mutations, the CD spectra were essentially identical. Small changes in the minimum at 209 nm were detected, which were less pronounced for the M271A and N272A mutants and not detected with the L185A mutant. The minimum at 209 nm was similar for L185S and the wild-type enzyme, but the band at 220 nm was completely absent in L185S. K186V displayed a CD-spectrum similar to L185S with a smaller effect on the minimum at 220 nm.
FIGURE 3.
CD spectra of SalR and mutants. For clarity, the spectra have been shifted to more positive values. All curves normally end at 0.0 deg cm2 dmol−1 at 260 nm, as shown for the wild type. From bottom to top, at 260 nm the CD spectra are for wild-type SalR (solid line), L185A (medium dash line), L185S (small dash line), K186V (dotted line), M271A (large dash line), and N272A (double dot dash line).
Substrate Inhibition of Salutaridine Reductase
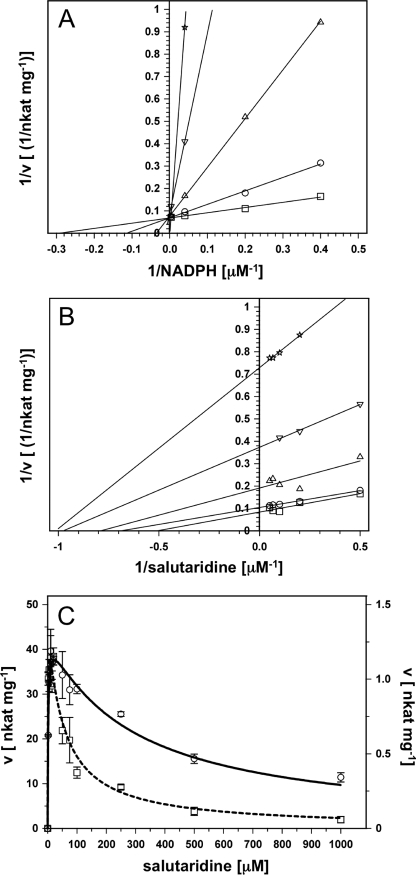
The wild-type enzyme exhibited strong substrate inhibition with a Ki of ∼180 μm and a decrease in velocity at salutaridine concentrations higher than 20 μm (see Table 1). Substrate inhibition is a common phenomenon in enzymes with a Ordered Bi Bi mechanism, as has been shown for a member of the SDR family, pteridine reductase (33, 34). To investigate the mechanism of substrate inhibition we performed product inhibition studies with NADP+, which inhibited SalR with a Ki of 1.2 μm (data not shown). This inhibition could be alleviated by increasing NADPH concentrations, indicating that inhibition was competitive (Fig. 4A). Using salutaridine as the variable substrate and at concentrations lower than those resulting in substrate inhibition, double reciprocal plots showed non-competitive/mixed inhibition (Fig. 4B). These results are consistent with a Ordered Bi Bi mechanism with NADPH as the leading substrate and the alkaloid product leaving before NADP+, as reported for alcohol dehydrogenase (35). For this type of reaction mechanism, substrate inhibition is often explained by the binding of surplus substrate to an enzyme-NADP+ complex (36–38). For SalR, this would result in a dead-end enzyme-salutaridine-NADP+ complex. If this were the case, substrate inhibition should be stronger in the presence of NADP+. An increase in NADP+ would shift the equilibrium toward the NADP+-enzyme complex, which provides a higher probability for salutaridine to form the dead-end complex with NADP+. When we performed the enzyme assays with variable salutaridine concentrations in the presence of 25 μm NADP+, the velocity/substrate concentration curve showed stronger substrate inhibition compared with assays performed in the absence of the inhibitor (Fig. 4C).
FIGURE 4.
NADP+ inhibition studies of wild-type SalR. A, double reciprocal plot of NADP+ inhibition at variable NADPH concentrations and at a constant salutaridine concentration of 20 μm. NADP+ concentrations (μm): 0 (squares), 2.5 (circles), 25 (triangles up), 100 (triangles down), and 250 μm (stars). B, double reciprocal plot of NADP+ inhibition at variable salutaridine concentrations at a constant NADPH concentration of 25 μm. NADP+ concentrations (μm): 0 (squares), 2.5 (circles), 25 (triangles up), 50 (triangles down), and 100 μm (stars). C, velocity/substrate concentration curve for SalR in the absence (solid line) or presence (dashed line) of 25 μm NADP+. The data were recorded in the presence of 250 μm NADPH. Note the difference in scale for the curve in the absence (left y axis) and presence (right y axis) of the inhibitor.
According to the size of the binding pocket of SalR, mutually exclusive binding in the productive and non-productive orientation must occur for salutaridine to act both as a substrate and an inhibitor. The docking experiments revealed one salutaridine orientation with an appreciable fitness score that would result in a dead-end enzyme-salutaridine-NADP+ complex (Table 2). The fitness score is substantially higher for the non-productive compared with the productive orientation, which corroborates the almost 80-fold difference between Ki and Km (see Table 1).
Considering the non-productive complex in the context of a three-dimensional coordinate system with three perpendicular axes, in which the piperidine ring is aligned with the vertical z axis, the salutaridine molecule is first rotated counterclockwise by 160° around the z axis, and then clockwise by 70° around the x axis (Fig. 2, B and C). Compared with the catalytic orientation, ring A rather than ring C is pointed toward the catalytic residues and the cofactor, resulting in a non-productive complex. In a coarse approximation, salutaridine can be regarded as a molecule possessing a 3-fold symmetry around an axis going through atoms C4a and C1 (Fig. 4C). Additionally, the benzyl and cyclohexadienone rings are similar in size and show a similar substitution pattern with one methoxy group and one oxygen function. Thus, it is plausible that the described rotation could occur and that the molecule would still fit into the binding pocket. The formation of hydrogen bonds between Ser181 and the 4′-methoxy and 3′-hydroxy groups, between Thr182 and the 3′-hydroxyl, and between Lys186 and the 6-methoxy support this conformation. Further contributions involve π-π and van der Waals interactions between Phe104 and ring C, and with C3 and C4 of the isoquinoline moiety, and between Leu266 and ring A, and with the 4′-methoxy group. Other hydrophobic interactions include Cγ2 of Val106 with C8, Cδ1 of Leu185 and with the 4′-methoxy group, and Cγ1 and Cδ1 of Ile275 with the N-methyl and Cα.
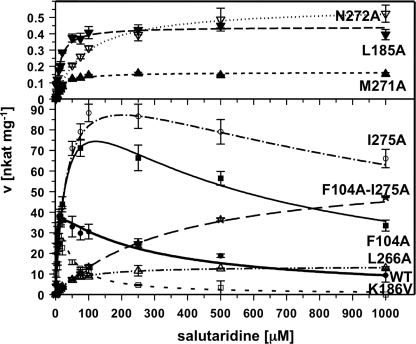
Several mutations had strong effects on substrate inhibition. Overall, a positive correlation (r2 = 0.43) between the Km and the Ki values was apparent, although several exceptions were found (see Table 1). For example, T182A and K186V showed weaker affinities for salutaridine in the catalytic binding, but stronger affinities for the inhibitory binding. In particular, K186V showed almost 10-fold stronger substrate inhibition compared with the wild-type enzyme (see Table 1 and Fig. 5). Conversely, the Km values for V106A and S181A were different compared with those of the wild-type enzyme, but substrate inhibition was similar (see Table 1). Enzyme variants exhibiting Km values close to that of the wild-type enzyme also had similar Ki values, as observed for L185V and I275V. The strongest reductions in substrate inhibition were measured for F104A, D107A, and I275A, with Ki values higher than 400 μm and 1 mm, respectively. Four mutations did not show any signs of substrate inhibition up to 1 mm salutaridine (Fig. 5). Whereas L266A and N272A also displayed the weakest affinities for catalytic salutaridine binding, the increase in Km values for L185A and M271A was comparable with other mutations that still exhibited substrate inhibition. Furthermore, the absence of substrate inhibition was accompanied by a strong decrease in kcat, especially for L185A, M271A, and N272A. Although L266A was devoid of substrate inhibition and showed slightly higher activity than the wild-type enzyme at higher salutaridine concentration, its activity was relatively low compared with the maximal velocity measured for I275A, F104A, or the wild-type enzyme (see Table 1 and Fig. 5). Although I275A and F104A mutants showed the highest catalytic rates, the reaction velocities were reduced by 30 and 45%, respectively, at 1 mm salutaridine (Fig. 5). Nevertheless, the kinetic parameters of these two substitutions prompted the creation of a F104A/I275A double mutant, with the expectation that a SalR enzyme variant could be produced with high reaction velocity and little, if any, substrate inhibition. Indeed, the resulting protein showed no substrate inhibition accompanied by a weak affinity for productive salutaridine binding (Table 1). In addition, the kcat of the F104A/I275A double mutant was almost 2-fold higher compared with the wild-type enzyme. Although still lower than that observed for the F104A and I275A single mutations, the velocity of the double mutation at 1 mm substrate was higher than that of F104A (Fig. 5). Based on the calculated Vmax, the velocity should also be higher than that of I275A when the concentration of salutaridine is increased further. As with other mutants, no change of substrate specificity was detected with the tested compounds.
FIGURE 5.
Velocity/substrate concentration curves for wild-type (WT) SalR and selected mutants exhibiting substantial substrate inhibition. The enzyme assays were performed as described under “Experimental Procedures” in the presence of 250 μm NADPH as cofactor. The activities were calculated based on signal intensities at 210 nm of salutaridine substrate and 7(S)-salutaridinol product after HPLC separation. The numerical parameters for all curves are shown in Table 1. The open symbols are: squares, K186V; triangles up, L266A; stars, F104A-I275A; circles, I275A; triangles down, N272A. The closed symbols are: circles, WT; triangles up, M271A; triangles down, L185A.
DISCUSSION
SalR belongs to a new class of SDRs exhibiting an extended length compared with other members of the family (11). Homology modeling and substrate docking suggest that some of the additional amino acids form a largely helical domain and are involved in substrate binding (19). The effects of site-directed mutagenesis on catalytic function support this assumption. Residues Phe104, Val106, and Asp107 all reside in this extended stretch of amino acids and their substitution with Ala resulted in pronounced effects on substrate affinity. In F104A, the strong π-π interactions with ring A of salutaridine in the wild-type enzyme were removed. The decrease in substrate affinity of D107A supports the assumption that Asp107 forms a hydrogen bond to the nitrogen in salutaridine (Fig. 2A). The opposite part of the substrate binding pocket consists of Leu266, Met271, Asn272, and Ile275. All Ala substitutions at this site caused a strong decrease in substrate affinity. According to the substrate docking, Ile275 and Leu266 interact with the isoquinoline moiety of salutaridine via hydrophobic interactions. In previous experiments, the effect on substrate affinity observed with the L266A mutant could be alleviated by replacement of Leu266 with Val instead of Ala (19). Similarly, enzyme performance could be restored to wild-type levels by replacing Ile275 with Val rather than Ala, demonstrating that the distance between Cγ1 (rather than Cδ1) and the isoquinoline moiety of the substrate is a crucial determinant of affinity. L266A also led to a decrease in the activity of the enzyme. The reactive ring C is sandwiched between the catalytic Tyr236 in addition to Thr182 and Leu266 (Fig. 2A). It is conceivable, that Leu266 guides ring C to the correct position at the catalytic center, which is disrupted by substitution with Ala. Substitutions of Met271 and Asn272 with Ala led to a substantial decrease in reductase activity. A direct interaction between Met271 and salutaridine is unlikely because of the large distance between this residue and the substrate. However, van der Waals interactions between Met271 and the nicotinamide ring of the cofactor could occur, and the M271A mutation might have resulted in improper cofactor alignment. Similarly, the side chain of Asn272 is in hydrogen-bonding distance to the amide of the nicotinamide ring. As for Met271, the effects of the N272A mutation are more likely related to an improper alignment of the cofactor than to a direct interaction with salutaridine. Furthermore, CD spectra for both mutations suggest subtle alterations in secondary structure (Fig. 3). It had been previously shown that replacing both residues by Thr leads to a complete loss of activity and to a shift toward more random coil elements (19).
The bottom of the substrate binding pocket consists of Ser181, Thr182, Leu185, and Lys186 (Fig. 2A). S181A exhibited only small differences in kinetic parameters compared with those of the wild-type enzyme. Ser181 presumably interacts with the oxygen of the 6-methoxy group by hydrogen bond formation. A substitution with Ala might compensate for the missing hydrogen bond by promoting hydrophobic interaction with the methyl group. The slightly lower affinity yet strongly reduced activity of T182A suggests that the hydrogen bond between Thr182 and the 3′-hydroxyl group is more important for proper alignment of the substrate than for initial substrate binding. Furthermore, the Thr182 side chain and Leu266 form the clamp that constrains ring C in the active site. When substituted, Ala might be too small to exert the same effect. The exceptionally low activity of L185A suggests that Leu185 is crucial for correct positioning of the substrate. Only Cδ1 might undergo hydrophobic interaction with the 6-methoxy group, whereas a specific interaction with the 3′-hydroxyl is unlikely. This suggests that Leu185 contributes to substrate binding via its voluminous side chain. Additionally, the CD spectrum of L185A indicates a role for Leu185 in the structural integrity of the enzyme (Fig. 3). The partial restoration of wild type activity suggests that Val can perform the same function as Leu to some extent. The L185S mutant showed increased substrate affinity, possibly by the formation of a hydrogen bond to the 3′-hydroxy group. However, the low activity showed that this interaction was not strong enough for proper substrate alignment. The K186V mutant exhibited an activity level similar to that of the wild-type enzyme, and a comparably moderate reduction in substrate affinity, suggesting that the contribution of the Nϵ amino group to substrate binding is of minor importance.
Taken together, the effects of the various amino acid substitutions corroborate the substrate docking shown in Fig. 2A, where substrate affinity is achieved through π-π and hydrophobic interactions involving Phe104, Leu266, and Ile275, and by hydrogen bonding with Asp107. The correct position of the substrate is determined through hydrophobic interactions with Leu185 and Leu266, and by hydrogen bonding with Thr182.
Several structural features underlying substrate inhibition in enzymes with Ordered Bi Bi mechanisms have been proposed. In d-3-phosphoglycerate deydrogenase, substrate inhibition involves additional allosteric binding of surplus hydroxypyruvic acid, the enzymatic substrate (39). For sulfotransferases, the crystal structure showed that the substrate adopts a different conformation and binds closer to the cofactor binding site after the cofactor has been desulfated (40, 41). In lactate dehydrogenase, substrate inhibition has been partially linked with positioning of the NAD+ cofactor rather than to substrate binding (42). For SalR, we have identified an alternative mode of salutaridine binding, which leads to an unproductive complex. All amino acids involved in substrate binding exert the same kind of interactions with salutaridine when it is bound as an inhibitor. However, these interactions occur with different parts of the salutaridine molecule depending on its function as either a substrate or an inhibitor (Fig. 2B). The concomitant increases in Km and Ki in almost all of the SalR mutants undermine this assumption (see Table 1). Only Asp107 is unable to form hydrogen bonds with salutaridine in the inhibitory mode. The lack of this interaction might explain the difference in the fitness scores and affinity constants between productive and non-productive salutaridine binding. The assumption of alternative salutaridine binding as the basis for substrate inhibition is supported by the low Ki of K186V. In the unproductive salutaridine docking, Lys186 might interact with the oxygen of the 6-methoxy group (Fig. 2B). Val at this position might allow salutaridine to slide deeper to the bottom of the substrate binding pocket by hydrophobic interaction with the 6-methoxy group. This in turn could lead to more efficient planar stacking between Phe104 and the cyclohexadienone ring, yielding stronger π-π interactions. Ile275 seems to push salutaridine into the substrate binding pocket by interaction with the N-methyl group, thereby fixing it to the enzyme in the unproductive binding mode. Substitution of Ile275 for Ala, an amino acid that is too small to exert the same effect, leads to weak substrate inhibition.
Several attempts to remove the substrate inhibition of enzymes have been reported. However, the removal of (or a decrease in) substrate inhibition is generally accompanied by reduced catalytic activity in many enzymes including lactate dehydrogenase, 17β-hydroxysteroid dehydrogenase, d-phosphoglycerate dehydrogenase, sinapyl alcohol dehydrogenase, and tetrahydroquinone dehalogenase (39, 42–45). Compared with wild-type SalR, the independent F104A and I275A mutants displayed weak substrate inhibition accompanied by similar or even increased reaction velocities. With the F104A/I275A double mutant created on the basis of these results, we have shown that it is possible to systematically engineer an enzyme devoid of substrate inhibition and to simultaneously maintain a high kcat value. The maximal velocity was even 2-fold higher compared with that of wild-type SalR. Furthermore, the mutant enzyme retained its substrate specificity with all compounds tested. Clearly, additional compounds especially those with structures more similar to salutaridine also require evaluation in this context.
Concerns about the supply of both licit and illicit opium poppy products have prompted several investigations on alternative production systems for morphinan alkaloids. Recently, the pathway to reticuline has been reconstituted in yeast (8), and the introduction of a human cytochrome P450 facilitated the production of salutaridine (7). Such studies have demonstrated a strong correlation between product yield and the use of specific enzyme variants. For example, coclaurine N-methyltransferase from opium poppy was more effective than the corresponding enzyme from Thalictrum flavum (7). The use of engineered enzymes might further enhance productivity in such systems. In this regard, the removal of substrate inhibition might prove invaluable as a means of achieving higher product yields. Whether the F104A/I275A double mutant of SalR might be superior to the wild-type enzyme in a yeast-based morphinan production system must still be investigated. Additionally, efficient in vitro enzymatic conversions of salutaridine to 7(S)-salutaridinol will likely be more efficient with the double mutant than with the wild-type SalR.
Supplementary Material
Acknowledgments
We thank Dr. Jessica Gifford and Dr. Dae-Kyun Ro for help with CD spectroscopy and GC analysis, respectively.
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (to P. J. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S2.
- BIA
- benzylisoquinoline alkaloid
- CD
- circular dichroism
- SalR
- salutaridine reductase
- SDR
- short chain dehydrogenase/reductase
- HPLC
- high pressure liquid chromatography.
REFERENCES
- 1.Ziegler J., Facchini P. J. (2008) Annu. Rev. Plant Biol. 59, 735–769 [DOI] [PubMed] [Google Scholar]
- 2.Allen R. S., Millgate A. G., Chitty J. A., Thisleton J., Miller J. A., Fist A. J., Gerlach W. L., Larkin P. J. (2004) Nat. Biotechnol. 22, 1559–1566 [DOI] [PubMed] [Google Scholar]
- 3.Allen R. S., Miller J. A., Chitty J. A., Fist A. J., Gerlach W. L., Larkin P. J. (2008) Plant Biotechnol. J. 6, 22–30 [DOI] [PubMed] [Google Scholar]
- 4.Frick S., Chitty J. A., Kramell R., Schmidt J., Allen R. S., Larkin P. J., Kutchan T. M. (2004) Transgenic Res. 13, 607–613 [DOI] [PubMed] [Google Scholar]
- 5.Frick S., Kramell R., Kutchan T. M. (2007) Metab. Eng. 9, 169–176 [DOI] [PubMed] [Google Scholar]
- 6.Larkin P. J., Miller J. A., Allen R. S., Chitty J. A., Gerlach W. L., Frick S., Kutchan T. M., Fist A. J. (2007) Plant Biotechnol. J. 5, 26–37 [DOI] [PubMed] [Google Scholar]
- 7.Hawkins K. M., Smolke C. D. (2008) Nat. Chem. Biol. 4, 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minami H., Kim J. S., Ikezawa N., Takemura T., Katayama T., Kumagai H., Sato F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grothe T., Lenz R., Kutchan T. M. (2001) J. Biol. Chem. 276, 30717–30723 [DOI] [PubMed] [Google Scholar]
- 10.Gerardy R., Zenk M. H. (1993) Phytochemistry 34, 125–132 [Google Scholar]
- 11.Ziegler J., Voigtländer S., Schmidt J., Kramell R., Miersch O., Ammer C., Gesell A., Kutchan T. M. (2006) Plant J. 48, 177–192 [DOI] [PubMed] [Google Scholar]
- 12.Oppermann U. C., Filling C., Berndt K. D., Persson B., Benach J., Ladenstein R., Jörnvall H. (1997) Biochemistry 36, 34–40 [DOI] [PubMed] [Google Scholar]
- 13.Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., Ghosh D. (1995) Biochemistry 34, 6003–6013 [DOI] [PubMed] [Google Scholar]
- 14.Tanaka N., Nonaka T., Nakanishi M., Deyashiki Y., Hara A., Mitsui Y. (1996) Structure 4, 33–45 [DOI] [PubMed] [Google Scholar]
- 15.Ghosh D., Wawrzak Z., Weeks C. M., Duax W. L., Erman M. (1994) Structure 2, 629–640 [DOI] [PubMed] [Google Scholar]
- 16.Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., Oppermann U. (2002) J. Biol. Chem. 277, 25677–25684 [DOI] [PubMed] [Google Scholar]
- 17.Persson B., Kallberg Y., Oppermann U., Jörnvall H. (2003) Chem. Biol. Interact. 143–144, 271–278 [DOI] [PubMed] [Google Scholar]
- 18.Ghosh D., Sawicki M., Pletnev V., Erman M., Ohno S., Nakajin S., Duax W. L. (2001) J. Biol. Chem. 276, 18457–18463 [DOI] [PubMed] [Google Scholar]
- 19.Geissler R., Brandt W., Ziegler J. (2007) Plant Physiol. 143, 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi H. W., Lee B. G., Kim N. H., Park Y., Lim C. W., Song H. K., Hwang B. K. (2008) Plant Physiol. 148, 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis E. M., Ringer K. L., McConkey M. E., Croteau R. (2005) Plant Physiol. 137, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ringer K. L., McConkey M. E., Davis E. M., Rushing G. W., Croteau R. (2003) Arch. Biochem. Biophys. 418, 80–92 [DOI] [PubMed] [Google Scholar]
- 23.Bjeldanes L. F., Rapoport H. (1972) J. Org. Chem. 37, 1453–1454 [DOI] [PubMed] [Google Scholar]
- 24.Rearick D. E., Gates M. (1970) Tetrahedron Lett. 7, 507–508 [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M., Ohno S., Adachi S., Nakajin S., Shinoda M., Nagahama Y. (1992) J. Biol. Chem. 267, 13451–13455 [PubMed] [Google Scholar]
- 27.MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 28.Pellegrini E., Field M. J. (2002) J. Phys. Chem. A 106, 1316–1326 [Google Scholar]
- 29.Korb O., Stützle T., Exner T. E. (2009) J. Chem. Inf. Model. 49, 84–96 [DOI] [PubMed] [Google Scholar]
- 30.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31.Sippl M. J. (1990) J. Mol. Biol. 213, 859–883 [DOI] [PubMed] [Google Scholar]
- 32.Hennessey J. P., Jr., Johnson W. C., Jr. (1981) Biochemistry 20, 1085–1094 [DOI] [PubMed] [Google Scholar]
- 33.Luba J., Nare B., Liang P. H., Anderson K. S., Beverley S. M., Hardy L. W. (1998) Biochemistry 37, 4093–4104 [DOI] [PubMed] [Google Scholar]
- 34.Nare B., Hardy L. W., Beverley S. M. (1997) J. Biol. Chem. 272, 13883–13891 [DOI] [PubMed] [Google Scholar]
- 35.Winberg J. O., McKinley-McKee J. S. (1994) Biochem. J. 301, 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duffel M. W., Jakoby W. B. (1981) J. Biol. Chem. 256, 11123–11127 [PubMed] [Google Scholar]
- 37.Gamage N. U., Duggleby R. G., Barnett A. C., Tresillian M., Latham C. F., Liyou N. E., McManus M. E., Martin J. L. (2003) J. Biol. Chem. 278, 7655–7662 [DOI] [PubMed] [Google Scholar]
- 38.Pocker Y., Raymond K. W. (1985) Alcohol 2, 3–8 [DOI] [PubMed] [Google Scholar]
- 39.Burton R. L., Chen S., Xu X. L., Grant G. A. (2007) J. Biol. Chem. 282, 31517–31524 [DOI] [PubMed] [Google Scholar]
- 40.Gamage N. U., Tsvetanov S., Duggleby R. G., McManus M. E., Martin J. L. (2005) J. Biol. Chem. 280, 41482–41486 [DOI] [PubMed] [Google Scholar]
- 41.Rehse P. H., Zhou M., Lin S. X. (2002) Biochem. J. 364, 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eszes C. M., Sessions R. B., Clarke A. R., Moreton K. M., Holbrook J. J. (1996) FEBS Lett. 399, 193–197 [DOI] [PubMed] [Google Scholar]
- 43.Gangloff A., Garneau A., Huang Y. W., Yang F., Lin S. X. (2001) Biochem. J. 356, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bomati E. K., Noel J. P. (2005) Plant Cell 17, 1598–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warner J. R., Behlen L. S., Copley S. D. (2008) Biochemistry 47, 3258–3265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.