Abstract
Acting via the glucocorticoid receptor (GR), glucocorticoids exert potent anti-inflammatory effects partly by repressing inflammatory gene transcription occurring via factors such as NF-κB. In the present study, the synthetic glucocorticoid, dexamethasone, induces expression of MKP-1 (mitogen-activated protein kinase (MAPK) phosphatase-1) in human bronchial epithelial (BEAS-2B) and pulmonary (A549) cells. This correlates with reduced TNFα-stimulated p38 MAPK phosphorylation. Since NF-κB-dependent transcription and IL-8 protein, mRNA, and unspliced RNA (a surrogate of transcription rate) are sensitive to p38 MAPK inhibitors (SB203580 and SB239063), we explored the role of MKP-1 in repression of these outputs. Repression of TNFα-induced p38 MAPK phosphorylation, NF-κB-dependent transcription, and IL-8 expression by dexamethasone are sensitive to transcriptional or translational inhibitors. This indicates a role for de novo gene synthesis. Adenoviral expression of MKP-1 profoundly reduces p38 MAPK phosphorylation and IL-8 expression. Similarly, NF-κB-dependent transcription is significantly reduced to levels consistent with maximal p38 MAPK inhibition. Thus, MKP-1 attenuates TNFα-dependent activation of p38 MAPK, induction of IL-8 expression, and NF-κB-dependent transcription. Small interfering RNA knockdown of dexamethasone-induced MKP-1 expression partially reverses the repression of TNFα-activated p38 MAPK, demonstrating that MKP-1 participates in the dexamethasone-dependent repression of this pathway. In the presence of MKK6 (MAPK kinase 6), a p38 MAPK activator, dexamethasone dramatically represses TNFα-induced NF-κB-dependent transcription, and this is significantly reversed by MKP-1-targeting small interfering RNA. This reveals an important and novel role for transcriptional activation (transactivation) of MKP-1 in the repression of NF-κB-dependent transcription by glucocorticoids. We conclude that GR transactivation is essential to the anti-inflammatory properties of GR ligands.
Glucocorticoids are the most effective treatment for chronic inflammatory diseases, such as asthma (1). Their potent anti-inflammatory actions are primarily due to the ability to inhibit the expression of numerous proinflammatory mediators, including cytokines, chemotactic mediators, adhesion molecules, and other inflammatory proteins (1). These wide ranging effects on gene expression lead, in turn, to reduced inflammatory responses (e.g. by reducing the number of inflammatory cells that infiltrate the airways) (1).
At the molecular level, the suppressive effects of glucocorticoids are classically attributed to the repression of proinflammatory transcription factors, such as nuclear factor κB (NF-κB)2 and activator protein (AP)-1 (2). Under resting conditions, heterodimers of NF-κB, typically p50 (NFKB1) and p65 (RelA), are held in the cytoplasm by inhibitor of κB (IκB) proteins (3). Upon cell stimulation, for example by the inflammatory cytokines tumor necrosis factor α (TNFα) or interleukin (IL)-1β, signal transduction cascades lead to the phosphorylation and activation of the IκB kinase complex. This phosphorylates the IκB protein, typically IκBα at serines 32 and 36, to promote ubiquitination and subsequent degradation. NF-κB then translocates to the nucleus to bind κB response elements and activate the transcription of numerous inflammatory genes (4). One such gene is the neutrophil chemoattractant, IL-8 (CXCL8), which is strongly NF-κB-dependent in airway epithelial cells (5, 6), and may contribute to the (often neutrophilic) response that is observed in severe asthma (1).
The mechanism(s) by which glucocorticoids inhibit transcriptional activation of NF-κB is still the subject of considerable research activity and, indeed, debate (7, 8). Notwithstanding this, the effect is generally stated to occur via the binding of ligand-bound glucocorticoid receptor (GR) with a transcription factor, such as NF-κB, to directly inhibit transcriptional activity via a process that is referred to as “transrepression” (2, 9, 10). In this model, GR does not directly contact the DNA but binds indirectly, via the targeted transcription factor, to produce a tethering negative GRE (2). The recruitment of transcriptional repressors, such as histone deacetylases, exerts repression via the tethering negative GRE (11, 12). However, findings that implicate various inflammatory signal transduction cascades in the transcriptional activation of either NF-κB or AP-1 provide alternative, possibly parallel, mechanisms, to explain the repression that is exerted by glucocorticoids (7).
In addition to the classical IκB kinase-IκBα pathway, which allows NF-κB translocation and DNA binding, the transcriptional activation of NF-κB is also regulated by events that specifically impact on transactivation (13). Thus, small molecule inhibitors of protein kinase C and the p38 mitogen-activated protein kinase (MAPK) reduce NF-κB-dependent transcription yet do not affect NF-κB translocation or DNA binding (14, 15). In this context, inflammatory stimuli, including TNFα and IL-1β, initiate signaling via small GTPases to turn on MKK3 and -6 (MAPK kinases 3 and 6) and thereby activate p38 MAPK (16). Subsequently, p38 MAPK may contribute to p65-dependent transactivation (17), possibly by phosphorylating downstream substrates that contribute to transcriptional activation by NF-κB (18, 19). Thus, MSK1 (mitogen- and stress-activated kinase 1), a downstream target of p38 MAPK, phosphorylates p65 at serine 276, and this enhances transcriptional activation (20). Furthermore, the phosphorylation of p65 by MSK1 can control binding of the coactivator, CREB-binding protein, and transcription of the inflammatory gene, stem cell factor (21). Likewise, p38 MAPK is implicated in the phosphorylation of the co-activator, p300, leading to potentiation of its activity and an increased association with NF-κB (22, 23). In addition to the direct phosphorylation of NF-κB or co-activator molecules, the phosphorylation of histone H3 at serine 10 increases binding of p65 to specific promoters, and a p38-dependent mechanism is proposed (24). Similarly, the p38 MAPK has been implicated in the phosphorylation of the TATA-binding protein as a mechanism by which NF-κB-dependent transactivation may be enhanced (25).
Since MAPK pathways are potent activators of inflammatory responses, strict control mechanisms exist to prevent prolonged or inappropriate activation. In the case of p38 MAPK, negative feedback can occur via the dual specificity phosphatase, MKP-1 (MAPK phosphatase-1), which is also referred to as DUSP1 (dual specificity phosphatase 1) (26). However, in many cell types, MKP-1 is also profoundly up-regulated by glucocorticoids, and this contributes to the anti-inflammatory effects of these drugs by reducing the activation of MAPKs (26–28). Thus, cells that do not express (gene knock-out) or have reduced expression of MKP-1 (by siRNA silencing) have partially impaired responses to the glucocorticoid, dexamethasone (28–30).
Whereas many studies provide convincing evidence that the induction of MKP-1 by glucocorticoids contributes to repression via post-transcriptional mechanisms (28–30), the knowledge that p38 MAPK plays a role in transcriptional activation by NF-κB presents a possible mechanism of action occurring at the level of transcriptional control. As such, a number of studies show a role for MKP-1 in the regulation of proinflammatory genes (30–32), many of which are NF-κB-dependent. However, a possible role for MKP-1 in mediating transcriptional repression was not addressed in these studies. In pulmonary cells, the ability of dexamethasone to repress the expression of IL-8 mRNA involves both transcriptional and post-transcriptional processes, and this effect is prevented by transcriptional blockade (33). Since IL-8 expression is also NF-κB-dependent (5, 6) (see supplemental Fig. S1), we have selected pulmonary (A549) and bronchial airway epithelial (BEAS-2B) cells to examine whether glucocorticoid-induced MKP-1 expression can mediate the repression of NF-κB-dependent transcription and IL-8 expression.
EXPERIMENTAL PROCEDURES
Cell Culture
A549 and BEAS-2B cells were grown to confluence in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) or DMEM/Ham's F-12 medium (Invitrogen), respectively, both supplemented with 10% fetal calf serum (Invitrogen). Prior to experimentation, cells were incubated overnight in serum-free medium and then changed to fresh serum-free medium containing cytokine and drugs. TNFα (R&D Systems, Hornby, Canada) was dissolved in phosphate-buffered saline (Sigma) plus 0.1% bovine serum albumin (Sigma), dexamethasone (Sigma) was dissolved in Hanks' balanced salt solution (Sigma), cycloheximide (Sigma) was dissolved in sterile water, and actinomycin D, SB203580, SB239063, and SB202474 (EMD Biosciences, San Diego, CA) were dissolved in DMSO. Final concentrations of DMSO were <0.1%.
NF-κB-dependent Reporters and Luciferase Assay
The adenoviral NF-κB reporter, Ad5-NF-κB-luc, containing five copies of the classical NF-κB motif, as previously described (6), was introduced to preconfluent A549 and BEAS-2B cells at an MOI of 1 for 24 h in DMEM or DMEM/F-12 plus 10% fetal calf serum medium. BEAS-2B cells stably harboring the previously described and validated NF-κB-dependent luciferase reporter, pGL3.neo.TATA.3κBu (3κBu-luc) (34), were grown until confluent in DMEM plus 10% fetal calf serum medium containing 0.1 mg/ml G-418 (Promega, Madison, WI). All cells were incubated in serum-free medium overnight prior to changing to fresh serum-free medium containing experimental drugs and cytokine. At 6 h post-stimulation, cells were harvested in 1× reporter lysis buffer (Promega), and luminescence was measured using a BD Monolight Luminometer (BD Biosciences).
Western Blotting
Cells were lysed in 1× reporter lysis buffer (Promega) plus 1× complete protease inhibitor mixture (Roche Laval, Quebec, Canada) and phosphatase inhibitors (Sigma). Total cellular lysates were size-fractionated on 4–12% gradient NuPage® BisTris acrylamide gels (Invitrogen) and then electroblotted to Hybond-ECL membranes (GE Healthcare). Membranes were incubated with the following antisera according to the manufacturers' instructions: anti-MKP-1 (M-18) (catalog number sc-1102) and anti-IκB-α/MAD-3 (C-21) (catalog number sc-371) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); anti-phospho-p38 (catalog number 9211), anti-phospho-hsp27 (catalog number 2401), and anti-phospho-IκB-α (catalog number 9241) (Cell Signaling, Danvers, MA); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog number 4699-9555(ST)) (AbD Serotec, Raleigh, NC). After washing, membranes were incubated with anti-rabbit or anti-mouse horseradish peroxidase-linked immunoglobulins (Dako, Mississauga, Canada). Immune complexes were detected using ECL (GE Healthcare) and visualized by autoradiography.
RNA Isolation, cDNA Synthesis, and TaqMan or SYBR Green Real Time PCR
Total RNA was extracted using an RNeasy minikit (Qiagen, Mississauga, Canada), and reverse transcription reactions were prepared using 0.5 μg of RNA to obtain cDNA as previously described (33). The resulting cDNA was diluted 1:5 with RNase-free water and stored at 4 °C. An ABI 7900HT instrument (Applied Biosystems Inc., Foster City, CA) was utilized to carry out real time PCR analysis on 2.5 μl of cDNA using TaqMan Mastermix (Applied Biosystems) or SYBR GreenER Mastermix (Invitrogen) in a 20-μl reaction volume. Relative concentrations of cDNA were obtained using a cDNA standard curve of serial dilutions of a TNFα-stimulated sample. Amplification conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, and then 40 or 44 cycles of 95 °C for 15 s, 60 °C for 45 s. For TaqMan analysis, primer pairs and probe specific to the following genes were as follows: GAPDH (NM_002046) (4326317E; Applied Biosystems) and IL-8 (NM_000584) cytoplasmic (forward, 5′-CTG GCC GTG GCT CTC TTG-3′; reverse, 3′-TTA GCA CTC CTT GGC AAA ACT G-5′; probe, 5′-6FAM-CCT TCC TGA TTT CTG CAG CTC TGT GTG AA-TAMRA-3′). For SYBR green analysis, primer pairs specific to the following gene were as follows: Luciferase (M15077.1) (forward, 5′-CGC TGG AGA GCA ACT GCA TA-3′; reverse, 3′-CCA GGA ACC AGG GCG TAT CT-5′). Specificity of primers for SYBR green analysis was determined by analysis of the dissociation (melt) curves: 95 °C for 15 s, 60 °C for 20 s, 95 °C for 15 s with ramping to 95 °C over 20 min with primer specificity indicated by a single peak in the rate of change of fluorescence with temperature.
Real Time PCR Analysis of Unspliced Nuclear IL-8 RNAs
Since unspliced nuclear RNA accumulates transiently in the nucleus following transcriptional activation, this may be measured as a surrogate index of transcription rate (33, 35). To analyze unspliced nuclear RNA, probe and primer sets were designed that crossed an intron/exon junction (see supplemental Fig. S2 for a diagram). Unspliced IL-8 RNA was normalized to U6, an RNA polymerase III-dependent gene as GAPDH nuclear unspliced RNA was regulated in response to TNFα (data not shown). Since these probe and primer sets detect both unspliced RNA and genomic DNA, it was critical to assess the potential contribution by contaminating genomic DNA in each sample. Therefore, following total RNA extraction as described above, every RNA sample was subject to reverse transcription in the presence and absence of reverse transcriptase enzyme. Any amplification product in the reverse transcription-negative samples was attributed to genomic DNA contamination, and samples with greater than 10% genomic contamination were excluded from further analysis. RNA extraction, cDNA synthesis, and TaqMan real time PCR were carried out as described above. Probe and primer sequences were as follows: unspliced nuclear IL-8 (nIL-8A) (forward, 5′-CTC TTG GCA GCC TTC CTG AT-3′; reverse, 3′-CTG TTT CTG AAT AAA AAG GAT GTT TGT TAC-5′; probe, 5′-6FAM-TGA AGG TAA GCA CAT CTT TCT GAC CTA CAG CG-MGB-3′) and U6 (NR_004394) (forward, 5′-AAT TGG AAC GAT ACA GAG AAG ATT AGC-3′; reverse, 3′-GGA ACG CTT CAC GAA TTT GC-5′; probe, 5′-6FAM-TGG CCC CTG CGC AA-MGB-3′).
Enzyme-linked Immunosorbent Assay (ELISA)
Following stimulation of cells, supernatants were harvested, and ELISA for IL-8 was performed using a DuoSet ELISA kit (R&D Systems) according to the manufacturer's instructions.
Adenoviral Infection
A549 and BEAS-2B cells were grown to preconfluence (∼70%) and incubated for 24 h in DMEM or DMEM/F-12 plus 10% fetal calf serum containing the indicated MOI of the following adenoviral (Ad5) expression vectors: Ad5-MKP-1, containing a mouse MKP-1 cDNA expression cassette (36) (Seven Hills Bioreagents, Cincinnati, OH); Ad5-MKK6, containing a human cDNA expression cassette for activated MKK6 with the mutation S207E/S211E (37) (Seven Hills Bioreagents); Ad5-NF-κB-luc (see “NF-κB-dependent Reporters and Luciferase Assay”); or an empty Ad5 vector (null). In all cases, cells were then incubated overnight in serum-free medium before treatment with cytokine or drugs.
siRNA-mediated Gene Silencing
Cells were grown to ∼60–70% confluence in 12-well plates. Cells were washed with serum-free medium and then incubated with siRNA containing serum-free medium at 37 °C for 24 h prior to the addition of cytokine and drugs. Transfections were prepared as follows. siRNA (25 nm) was mixed with LipofectamineTM 2000 (1 μl of 1 μg/μl) (Invitrogen) in 100 μl of serum-free DMEM/F-12 and incubated at room temperature for 30 min before the addition to cells. The sequences for siRNA targeting were as follows: MKP-1 siRNA 1 (SI00374801; 5′-TAG CGT CAA GAC ATT TGC TGA-3′); MKP-1 siRNA 2 (SI00374808; 5′-CTG TAC TAT CCT GTA AAT ATA-3′) (all from Qiagen); green fluorescent protein siRNA (control siRNA) (P-002048-03-20; 5′-GGC AAG CTG ACC CTG AAG TTC-3′) (Dharmacon, Chicago, IL).
Data Presentation and Statistical Analysis
All graphical data are presented as mean ± S.E. Statistical analysis between groups was performed using one-way analysis of variance (ANOVA) with Bonferroni's or Dunnett's post-test or a paired t test as indicated. Significance between groups was assumed as follows. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
Effect of Dexamethasone and TNFα on MKP-1 Expression and Phosphorylation of p38 MAPK
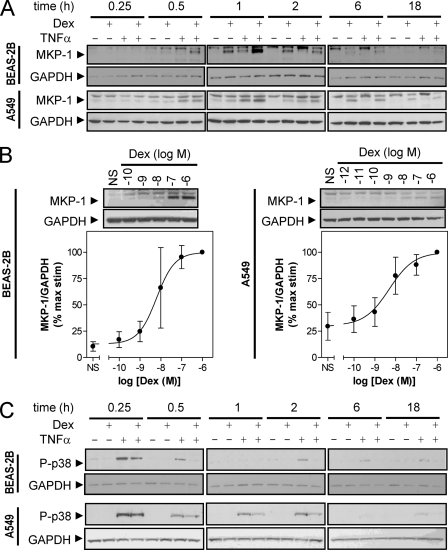
To examine the expression of MKP-1, BEAS-2B and A549 cells were treated with maximally effective concentrations of dexamethasone, TNFα, or dexamethasone plus TNFα for various times. In response to dexamethasone alone, expression of MKP-1 protein was first detected at 0.5 h (BEAS-2B) or 1 h (A549), and expression peaked at 1 h (BEAS-2B) or 6 h (A549). In each case, MKP-1 expression had returned to basal levels by 18 h (Fig. 1A). Treatment with TNFα also revealed induction of MKP-1 protein. This was detectable at 0.5 h for both BEAS-2B and A549 cells, reached a peak at 1 h, and had returned to basal levels by 2 h (A549) or 6 h (BEAS-2B) (Fig. 1A). In combination, dexamethasone plus TNFα potentiated the expression of MKP-1 protein at 0.5 h (A549) or 1 h (A549 and BEAS-2B) and 2 h (BEAS-2B) when compared with either treatment alone (Fig. 1A). In addition to MKP-1, our Western blot analyses revealed an immunoreactive band of slightly higher molecular weight. Since this was not induced by dexamethasone, and the manufacturer's data sheets (Santa Cruz Biotechnology) suggest this to be MKP-2, we do not comment further on this band (Fig. 1A).
FIGURE 1.
Expression of MKP-1 and phospho-p38 in BEAS-2B and A549 cells. A, BEAS-2B and A549 cells were either not stimulated or stimulated with dexamethasone (1 μm), TNFα (10 ng/ml), or a combination of both. Cells were harvested for protein after 0.25, 0.5, 1, 2, 6, or 18 h, and cell lysates were subject to Western blot analysis for MKP-1 and GAPDH. B, BEAS-2B and A549 cells were treated with various concentrations of dexamethasone as indicated. After 1 h (BEAS-2B) or 6 h (A549), cells were harvested for Western blot analysis of MKP-1 and GAPDH. Following densitometric analysis, data (n = 3–4), normalized to GAPDH were expressed as a percentage of maximum concentration of dexamethasone and plotted as means ± S.E. C, cells treated as in A were harvested for Western blot analysis of phospho-p38 MAPK (P-p38) and GAPDH. In all cases, blots representative of 3–4 such experiments are shown.
Subsequently, the times at which MKP-1 expression was maximal (1 h for BEAS-2B, 6 h for A549) were selected to examine the effect of dexamethasone concentration on MKP-1 expression. In each case, concentration-dependent increases in MKP-1 protein expression, with EC50 values of 6.2 × 10−9 and 5.0 × 10−9 m for BEAS-2B and A549 cells, respectively, were observed up to the maximally effective concentration of 1 μm (Fig. 1B).
Western blot analysis for the Thr180/Tyr182-phosphorylated (i.e. activated) form of p38 MAPK revealed maximal activation by TNFα at 15 min in both BEAS-2B and A549 cells (Fig. 1C). In BEAS-2B cells, this response generally returned to basal levels by 1 h before reappearing, to a lesser extent, at 2 h and then diminishing by 18 h (Fig. 1C). In A549 cells, TNFα-induced p38 MAPK phosphorylation decreased to a constant level at 0.5–2 h, before returning to basal levels by 6 h (Fig. 1C). In response to dexamethasone alone, there was no evidence of phosphorylated p38 MAPK in either cell line at any time point (Fig. 1C). However, in combination with TNFα, the presence of dexamethasone inhibited TNFα-induced phosphorylation of p38 MAPK. Thus, at 15 min, the inhibition of p38 MAPK phosphorylation by dexamethasone was partial but became more robust at later time points (Fig. 1C). This effect correlated with the induction of MKP-1 protein expression (Fig. 1A).
TNFα-induced NF-κB-dependent Transcription and IL-8 Expression Are p38 MAPK-dependent
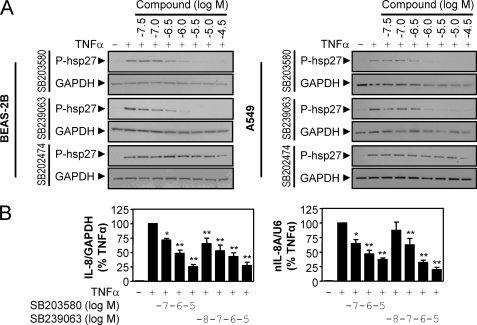
To examine whether NF-κB-dependent transcription and IL-8 expression were p38 MAPK-dependent in BEAS-2B and A549 cells, both cell lines were treated with various concentrations of SB203580, SB239063, and an inactive control compound, SB202474, prior to stimulation with TNFα. Western blot analysis of hsp27, a downstream target of p38 (16), revealed that SB203580 and SB239063 but not the inactive compound, SB202474, caused a concentration-dependent inhibition of hsp27 phosphorylation (Fig. 2A). This finding confirms the inhibition of p38 MAPK by SB203580 and SB239063 in BEAS-2B and A549 cells and thus validates the use of these compounds in the current study.
FIGURE 2.
Effect of the p38 MAPK inhibitors, SB203580 and SB239063, on NF-κB-dependent transcription and IL-8 expression. A, BEAS-2B and A549 cells were incubated with various concentrations of SB203580, SB239063, or SB202474 as indicated for 30 min prior to stimulation with TNFα (10 ng/ml) for 15 min. Cells were harvested for protein, and cell lysates were subjected to Western blot analysis for phospho-hsp27 (P-hsp27) and GAPDH. Blots representative of at least three such experiments are shown. B, BEAS-2B cells were incubated for 30 min with various concentrations of SB203580 or SB239063 before stimulation for 1 h with TNFα (10 ng/ml). Cells were harvested for RNA, and TaqMan real time PCR was carried out using probes and primers specific to either IL-8 and GAPDH mRNA or unspliced nuclear IL-8 transcript (IL-8A) and U6 nuclear RNA. Data (n = 7–8), normalized to GAPDH (IL-8) or U6 (IL-8A), were expressed as percentage of TNFα-stimulated cells and plotted as means ± S.E. Significance, relative to TNFα, using ANOVA with a Bonferroni post-test is indicated; *, p < 0.05; **, p < 0.01.
To examine NF-κB-dependent transcription, we utilized BEAS-2B cells stably harboring the NF-κB-dependent luciferase reporter, 3κBu-luc, that we had previously validated as being NF-κB-dependent (34). Likewise, the adenovirus-delivered reporter, Ad5-NF-κB-luc, is validated as NF-κB-dependent in both A549 and BEAS-2B cells (5, 38). In BEAS-2B and A549 cells infected with Ad5-NF-κB-luc, TNFα induced luciferase activity 211 ± 29- and 193 ± 37-fold respectively. In the BEAS-2B 3κBu-luc reporter cells, luciferase was induced 111 ± 31-fold by TNFα. In all three cases, the response was inhibited by increasing concentrations of SB203580 and SB239063 with maximal inhibition achieved between 1 and 30 μm (Table 1; see supplemental Fig. S3A). The potency of these effects (Table 1) is consistent with previously published values and agrees with the inhibition of phosphorylated hsp27 (Fig. 2A) (39, 40). However, in each case, the maximal inhibition by SB203580 and SB239063 is 28–65% (Table 1; see supplemental Fig. S3A). The control compound, SB202474, only affected luciferase activity at high concentrations, and this is attributed to off-target effects (supplemental Fig. S3A). Therefore, these data demonstrate that NF-κB-dependent transcription in these models is partly dependent on the p38 MAPK. Analysis of NF-κB DNA binding using EMSA revealed that inhibition of NF-κB-dependent transcription by SB203580 and SB239063 did not occur via inhibition of DNA binding (see supplemental Fig. S4A). In addition, the effect of SB203580 and SB239063 on phosphorylation of IκBα was examined, and this revealed that SB230580 and SB239063 did not affect this phosphorylation event (see supplemental Fig. S4B). These data suggest that p38 dependence of NF-κB occurs downstream of pathway activation and DNA binding.
TABLE 1.
Effect of p38 inhibitors, SB203580 and SB239063, on NF-κB-dependent transcription
BEAS-2B cells stably harboring the NF-κB-dependent reporter, 3κBu-luc, and BEAS-2B and A549 cells infected with the NF-κB-dependent reporter Ad5-NF-κB-luc were incubated with various concentrations of SB203580, SB239063, or SB202474 as in Fig. 2A. Cells were then stimulated for 6 h with TNF-α (10 ng/ml) before harvesting for luciferase activity determination. EC50 and maximal inhibition values were determined for each drug in each reporter. Maximal inhibition is expressed as means ± S.E. See the legend to Fig. 2 for further details.
| Reporter system | Treatment | EC50 | Maximal inhibition |
|---|---|---|---|
| m | % | ||
| BEAS-2B Ad5-NF-κB-luc | SB203580 | 2.5 × 10−7 | 35 ± 15 |
| SB239063 | 7.7 × 10−8 | 41 ± 7.2 | |
| BEAS-2B 3κBu.BG-luc | SB203580 | 3.5 × 10−8 | 65 ± 3.9 |
| SB239063 | 3.8 × 10−8 | 59 ± 3.1 | |
| A549 Ad5-NF-κB-luc | SB203580 | 7.2 × 10−7 | 59 ± 7.5 |
| SB239063 | 1.5 × 10−6 | 28 ± 33 |
To explore the role of the p38 MAPK pathway in IL-8 expression, the effects of SB203580 and SB239063 on the release of IL-8 protein that was induced by TNFα were tested. In untreated BEAS-2B cells, IL-8 release was 4.5 ± 1.5 pg/ml, and in the presence of TNFα, this was increased to 835 ± 161 pg/ml at 6 h. Prior incubation with SB203580 or SB239063 produced a concentration-dependent decrease in IL-8 release (EC50 = 4.8 × 10−8 and 6.6 × 10−8 m, respectively) with maximal inhibitions to 39 and 24% (respectively) of the TNFα-simulated response being achieved at 3–30 μm (supplemental Fig. S3B). Again, SB202474 revealed only modest effects at higher concentrations (supplemental Fig. S3B). Analysis of IL-8 mRNA and nIL-8A as a surrogate of transcription rate (see Ref. 33 and references therein) by real time PCR revealed significant dose-dependent inhibition of TNFα-induced IL-8 RNA expression by SB203580 and SB239063 (Fig. 2B). These data provide compelling evidence that the induction of IL-8 expression at the level of protein, mRNA, and transcription by TNFα is dependent on p38 MAPK in BEAS-2B cells. Analysis of the effect of SB203580 and SB239063 on IL-8 release in A549 cells revealed that this response was not unambiguously p38 MAPK-dependent, and therefore IL-8 expression in A549 cells was not examined further.
TNFα-induced p38 Phosphorylation, NF-κB-dependent Transcription, and IL-8 Expression Are Inhibited by Dexamethasone Pretreatment
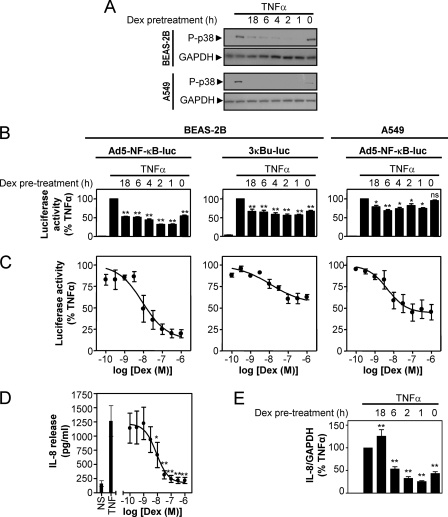
The finding that dexamethasone-dependent inhibition of p38 MAPK phosphorylation became more robust at later times prompted an examination of the effect of dexamethasone pretreatment on TNFα-stimulated p38 phosphorylation (Fig. 3A). In BEAS-2B cells, the inhibition of p38 MAPK phosphorylation by dexamethasone was maximal with 1- and 2-h pretreatments, and this declined with longer pretreatment times (Fig. 3A). In contrast, p38 MAPK phosphorylation in A549 cells was completely prevented by all of the dexamethasone pretreatments (Fig. 3A). In both cell types, the simultaneous addition of dexamethasone with TNFα was least effective at inhibiting p38 MAPK phosphorylation, and the magnitude of this effect was highly variable.
FIGURE 3.
Effect of dexamethasone pretreatment on p38 phosphorylation, NF-κB-dependent transcription, and IL-8 expression. A, BEAS-2B and A549 cells were pretreated with dexamethasone (1 μm) for 18, 6, 4, 2, 1, or 0 h before stimulation with TNFα (10 ng/ml). After 15 min, cells were harvested for protein and subjected to Western blot for phospho-p38 MAPK (P-p38) and GAPDH. Blots representative of at least three such experiments are shown. B, BEAS-2B and A549 cells infected with the Ad5-NF-κB-luc reporter and BEAS-2B cells stably harboring the reporter 3κB-luc were pretreated with dexamethasone and stimulated with TNFα as in A. After 6 h, cells were harvested for luciferase activity determination. Data (n = 6, left and middle; n = 10, right) expressed as a percentage of TNFα are plotted as means ± S.E. Significance, relative to TNFα-stimulated samples, using ANOVA with Dunnett's post-test is indicated; *, p < 0.05; **, p < 0.01; ns, not significant. C, BEAS-2B and A549 cells infected with the Ad5-NF-κB-luc reporter (left and right, respectively) and BEAS-2B cells stably harboring the reporter 3κBu-luc (middle) were incubated with various concentrations of dexamethasone as indicated for 1 (A549) or 2 h (BEAS-2B). Cells were then stimulated with TNFα (10 ng/ml) for 6 h prior to harvesting for luciferase activity determination. Data (n = 4 (left), n = 9 (middle), and n = 6 (right)) expressed as percentage of TNFα stimulated samples were plotted as means ± S.E. D, BEAS-2B cells were incubated with various concentrations of dexamethasone, as indicated, for 1 h prior to stimulation with TNFα (10 ng/ml) for 6 h. Supernatants were harvested, and IL-8 release was measured by ELISA. Data (n = 4) expressed as IL-8 release in pg/ml are plotted as means ± S.E. Significance, relative to TNFα-stimulated cells, using ANOVA with Dunnett's post-test is indicated; *, p < 0.05; **, p < 0.01. E, BEAS-2B cells were pretreated with dexamethasone (1 μm) for the indicated times before being stimulated with TNFα (10 ng/ml) for 1 h. Cells were then harvested for RNA, and real time PCR was carried out using IL-8 and GAPDH-specific probe and primer sets. Data (n = 5), normalized to GAPDH and expressed as a percentage of TNFα-stimulated samples, are plotted as means ± S.E. Significance, relative to TNFα-stimulated cells, using ANOVA with Dunnett's post-test is indicated; **, p < 0.01.
The effect of dexamethasone pretreatment on NF-κB-dependent transcription was also assessed. In BEAS-2B cells, luciferase activity from the Ad5-NF-κB-luc reporter was induced 230 ± 38-fold by TNFα (Fig. 3B, left). This response was significantly inhibited by all of the dexamethasone pretreatments, with maximum reductions to ∼32% of the response in TNFα-stimulated cells being achieved with 1–2-h pretreatment times (Fig. 3B, left). In BEAS-2B cells stably harboring the reporter 3κBu-luc, TNFα induced luciferase activity by 36 ± 2.9-fold (Fig. 3B, middle). Luciferase activity was significantly reduced by all of the dexamethasone pretreatments with maximal inhibition, to 56–57% of TNFα-stimulated cells, again occurring following 1–2-h pretreatments (Fig. 3B, middle). In A549 cells infected with the Ad5-NF-κB-luc reporter, luciferase activity was induced 269 ± 41-fold by TNFα, and dexamethasone maximally inhibited this to 69 ± 2.7% of the response in TNFα-stimulated cells. This occurred at 6 h (Fig. 3B, right), and a similar level of repression, to 74% of TNFα-stimulated cells, was observed at 1 h (Fig. 3B, right). Treatment of these reporters with increasing concentrations of dexamethasone revealed concentration-dependent inhibition of the TNFα-induced luciferase 13 ± 10% (BEAS-2B Ad5-NF-κB-luc), 55 ± 7.4% (BEAS-2B 3κBu-luc), and 44 ± 6.3% (A549 Ad5-NF- κB-luc) of the response in TNFα-stimulated cells (Fig. 3C). In each case, maximal inhibition occurred at ∼1 μm and EC50 values were 8.9 × 10−9 m (BEAS-2B and Ad5-NF-κB-luc), 1.2 × 10−8 m (BEAS-2B and 3κu-luc), and 4.6 × 10−9 m (A549 and Ad5-NF-κB-luc). As with the p38 inhibitors, the effects of dexamethasone on the NF-κB DNA binding and signaling pathway were examined. These data revealed that dexamethasone did not inhibit NF-κB DNA binding or phosphorylation of IκBα (see supplemental Fig. S4), suggesting that inhibition by dexamethasone occurs downstream of NF-κB pathway activation and DNA binding.
To examine the effect of dexamethasone treatment on IL-8 expression, BEAS-2B cells were treated with various concentrations of dexamethasone prior to stimulation with TNFα. Release of IL-8 protein was 160 ± 51 pg/ml in untreated cells, and this was induced by TNFα to 1300 ± 270 pg/ml. This induction was concentration-dependently inhibited by dexamethasone (EC50 = 7.2 × 10−9 m) with maximal inhibition to 15 ± 5.1% of the level produced by TNFα-stimulated cells (Fig. 3D). BEAS-2B cells were also pretreated with dexamethasone for the times indicated prior to stimulation with TNFα and subsequent analysis of IL-8 mRNA at 1 h. TNFα resulted in a 2200 ± 480-fold induction in IL-8 mRNA. Simultaneous treatment with dexamethasone or pretreatments for up to 6 h significantly inhibited this repression. Maximal inhibition, to 25 ± 1.7% of the response in TNFα-stimulated cells, was observed following a 1-h preincubation of dexamethasone (Fig. 3E).
Taken together, these data indicate that the maximal repression by dexamethasone of p38 phosphorylation, NF-κB-dependent transcription, and IL-8 expression by dexamethasone generally occurs with a pretreatment time of 1–2 h. Thus, inhibition by dexamethasone is time-dependent.
Inhibitors of Transcription and Translation Reverse the Inhibition of p38 Phosphorylation, NF-κB-dependent Transcription, and IL-8 mRNA Expression by Dexamethasone
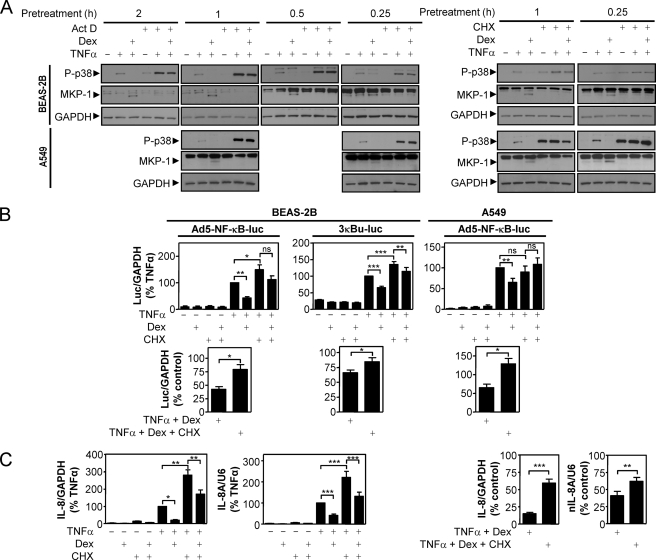
To investigate the potential requirement for new gene synthesis in the repression of the above responses by dexamethasone, the ability to exert repression was tested in the presence or absence of the transcriptional inhibitor, actinomycin D, or the translational inhibitor, cycloheximide. Initially, BEAS-2B and A549 cells were pretreated with dexamethasone for various times in the absence or presence of actinomycin D or cycloheximide. Cells were then stimulated with TNFα prior to the analysis of phosphorylated p38 MAPK. As described above, p38 MAPK phosphorylation was induced by TNFα, and this was repressed by all of the dexamethasone treatments (Fig. 4A). In the presence of either actinomycin D or cycloheximide, the dexamethasone-dependent repression of p38 MAPK phosphorylation was prevented. Importantly, this effect coincided with the loss of dexamethasone-induced MKP-1 expression, and this also confirms the efficacy of the actinomycin D and cycloheximide treatments (Fig. 4A). In the presence of actinomycin D or cycloheximide, TNFα showed enhanced phosphorylation of p38 MAPK. Likewise, cycloheximide also tended to increase basal levels of phosphorylated p38 MAPK. In each case, we attribute this effect to the inhibition of negative feedback processes (41).
FIGURE 4.
Effect of actinomycin D and cycloheximide on the dexamethasone-dependent repression of p38 MAPK phosphorylation, NF-κB activity, and IL-8 mRNA expression. A, BEAS-2B and A549 cells were incubated with dexamethasone (1 μm) for the indicated times in the absence or presence of actinomycin D (10 μg/ml) or cycloheximide (10 μg/ml) before being stimulated with TNFα (10 ng/ml) for 15 min. Cells were then harvested for protein and subjected to Western blot analysis for phospho-p38 MAPK, MKP-1, and GAPDH. Blots representative of at least three such experiments are shown. B, BEAS-2B and A549 cells infected with the Ad5-NF-κB-luc reporter and BEAS-2B cells stably harboring the reporter 3κBu-luc were pretreated with dexamethasone (1 μm) for 1 h in the absence or presence of cycloheximide (10 μg/ml) prior to stimulation with TNFα (10 ng/ml) for 1 h. RNA was harvested, and real time PCR was performed for luciferase and GAPDH. Top, data (n = 6 (left), n = 10 (middle), and n = 8 (right)), normalized to GAPDH and expressed as a percentage of TNFα-stimulated cells, are plotted as means ± S.E. Significance, using ANOVA with Bonferroni's post-test, is indicated; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. Lower panels, data, as above, were reanalyzed, and dexamethasone repression was expressed as a percentage of control (either TNFα or TNFα + cycloheximide) and plotted as means ± S.E. Significance, relative to TNFα + dexamethasone, using a paired t test is indicated; *, p < 0.05. C, using cDNA generated from the experiments in B, real time PCR was performed for IL-8, nIL-8A, and U6. Left two graphs, data (n = 12–14), normalized to GAPDH (IL-8) or U6 (nIL-8A) and expressed as a percentage of TNFα-stimulated cells, are plotted as means ± S.E. Significance, using ANOVA with Bonferroni's post-test, is indicated; *, p < 0.05; **, p < 0.01; ***, p < 0.001. Right two graphs, data, as above, were reanalyzed, and dexamethasone repression was expressed as a percentage of control (either TNFα or TNFα + cycloheximide) and plotted as means ± S.E. Significance, relative to TNFα + dexamethasone, using a paired t test is indicated; **, p < 0.01; ***, p < 0.001.
In order to test for the role of new gene synthesis in the dexamethasone-dependent repression of NF-κB-dependent transcription, the luciferase reporter cells were treated with dexamethasone (1 μm) in the absence or presence of the translational inhibitor, cycloheximide, to block the expression of any genes that might be induced by dexamethasone. Following treatment with TNFα for 1 h, the cells were harvested for RNA, and luciferase mRNA was analyzed using real time reverse transcription-PCR. Using this approach, TNFα induced luciferase mRNA expression from the Ad5-NF-κB-luc reporter 29 ± 11- and 250 ± 110-fold in BEAS-2B or A549 cells, respectively (Fig. 4B, top left and right). In the presence of dexamethasone, this induction was only 42 ± 12% (BEAS-2B) or 65 ± 9.8% (A549) of the response to TNFα alone. By contrast, in BEAS-2B cells, the presence of cycloheximide significantly reduced the magnitude of repression elicited by dexamethasone to 78.95 ± 22.79% of the response to TNFα (in the presence of cycloheximide) (Fig. 4B, top left and bottom left). Similarly, in A549 cells in the presence of cycloheximide, dexamethasone led to a response that was 128.89 ± 14.32% of the response to TNFα in the presence of cycloheximide (Fig. 4B, top right and bottom right). This indicates a complete block of the repressive ability of dexamethasone at the level of NF-κB-dependent transcription.
Likewise, in BEAS-2B cells stably harboring the reporter 3κBu-luc, TNFα induced luciferase mRNA 3.7 ± 0.18-fold (Fig. 4B, top middle), and this was repressed by dexamethasone to 66 ± 4.6% of control (Fig. 4B, top and bottom middle). However, as with the viral reporter, repression by dexamethasone was significantly reduced in the presence of cycloheximide to 85 ± 6.8% of control values (Fig. 4B, bottom middle).
Analysis of IL-8 mRNA and nIL-8A by real time PCR revealed a pattern of expression similar to that of the luciferase mRNA. IL-8 and nIL-8A RNA were induced 520 ± 110- and 170 ± 32-fold, respectively, by TNFα. In the presence of dexamethasone, this induction was inhibited to 15 ± 1.8% (IL-8) and 41 ± 6.6% (nIL-8A) of control (Fig. 4C). However, in the presence of cycloheximide, this was significantly reversed to 59 ± 5.7 and 62 ± 5.7% of control (Fig. 4C, left). Taken together, these data all suggest that new gene synthesis is required for the repression of p38 phosphorylation, NF-κB-dependent transcription, and IL-8 expression by dexamethasone.
Effect of MKP-1 Overexpression on p38 Phosphorylation, IL-8 Expression, and NF-κB-dependent Transcription
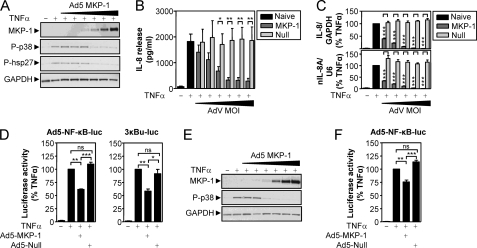
To investigate whether MKP-1 expression can elicit repression of the p38 MAPK pathway, BEAS-2B cells were infected with increasing MOIs of Ad5-MKP-1. Cells were then stimulated with TNFα for 15 min, and the expression of MKP-1, phospho-p38, and the downstream target, phospho-hsp-27, was analyzed by Western blot (Fig. 5A). Expression of MKP-1 protein was increased with increasing MOI of virus, and this correlated with the inhibition of TNFα-induced p38 and hsp-27 phosphorylation (Fig. 5A). These data validate the effectiveness of adenovirus-mediated overexpression at preventing activation of the p38 MAPK pathway.
FIGURE 5.
Effect of MKP-1 overexpression on p38 MAPK phosphorylation, NF-κB-dependent transcription, and IL-8 expression. A, BEAS-2B cells were infected with increasing MOIs (1, 3, 10, 30, 100, and 300) of Ad5-MKP-1 for 36 h before being stimulated with TNFα (10 ng/ml) for 15 min. Cells were then harvested for protein and subjected to Western blot analysis for MKP-1, phospho-p38 (P-p38), phospho-hsp27 (P-hsp27), and GAPDH. Blots representative of three such experiments are shown. B, BEAS-2B cells were infected with Ad5-MKP-1 as in A or with empty Ad5 vector (null) and then stimulated with TNFα (10 ng/ml) for 6 h prior to supernatants being harvested for analysis of IL-8 release by ELISA. Data (n = 6–9), expressed as IL-8 release in pg/ml, are plotted as means ± S.E. Significance, using a paired t test, relative to the appropriate concentrations of null virus is indicated; *, p < 0.05; **, p < 0.01. C, BEAS-2B cells were infected with Ad5-MKP-1 and null virus as in B (using only MOIs 3, 10, 30, 100, and 300) and then stimulated with TNFα (10 ng/ml). After 1 h, total RNA was extracted for real time PCR analysis of IL-8 and GAPDH mRNA or nuclear nIL-8A and U6 RNA. Data (n = 10), normalized to GAPDH (IL-8) or U6 (nIL-8A) and expressed as a percentage of TNFα-stimulated cells, are plotted as means ± S.E. Significance, relative to the appropriate concentration of null virus, using a paired t test is indicated; ***, p < 0.001. D, BEAS-2B cells infected with the Ad5-NF-κB-luc reporter or stably harboring the 3κBu-luc reporter were infected with MOI 30 of Ad5-MKP-1, null virus, or no virus for 36 h prior to stimulation with TNFα (10 ng/ml) for 6 h. Cells were then harvested for luciferase activity determination. Data (n = 4 (left) and n = 6 (right)), expressed as a percentage of TNFα-stimulated cells, are plotted as means ± S.E. Significance, using ANOVA with Bonferroni's post-test, is indicated; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant. E, A549 cells were treated as in A and then subjected to Western blot analysis for MKP-1, phospho-p38, and GAPDH. Blots are representative of three such experiments. F, A549 cells infected with the Ad5-NF-κB-luc reporter were treated as in D and harvested for luciferase activity determination. Data (n = 6), expressed as a percentage of TNFα-stimulated cells, are plotted as means ± S.E. Significance, using an ANOVA with Bonferroni's post-test is indicated; **, p < 0.01; ***, p < 0.001; ns, not significant.
Following this, BEAS-2B cells were infected with increasing MOIs of Ad5-MKP-1 or with an empty adenoviral vector (null) and stimulated with TNFα for 6 h prior to the analysis of IL-8 protein release by ELISA. IL-8 release was 71 ± 21 pg/ml in unstimulated cells, and this was induced to 1800 ± 280 pg/ml by TNFα. This induction was dose-dependently inhibited by increasing concentrations of Ad5-MKP-1 to a maximum of 6.2 ± 2.6% of TNFα-stimulated cells at MOI 300 (Fig. 5B). Importantly, the null virus had little effect on IL-8 release. In parallel experiments, IL-8 mRNA and nuclear RNA were induced 790 ± 190- and 270 ± 88-fold, respectively, by TNFα. Again, this induction was dose-dependently inhibited by increasing MOI of Ad5-MKP-1, to a maximum of 1.1 ± 0.22% (IL-8) and 0.85 ± 0.11% (nIL-8A) of TNFα-stimulated cells, whereas the null virus was without effect.
Since maximal inhibition of p38 and hsp-27 phosphorylation by Ad5-MKP-1 occurred at MOI 30, this MOI was used to treat BEAS-2B cells previously infected with the Ad5-NF-κB-luc reporter or stably harboring the 3κBu-luc reporter. Luciferase activity was induced 43 ± 6.9-fold (Ad5-NF-κB-luc) and 88 ± 23-fold (3κBu-luc) by TNFα, and this induction was significantly inhibited to 62 ± 0.85% (Ad5-NF-κB-luc) and 58 ± 4.1% (3κBu-luc) of TNFα-stimulated cells (Fig. 5D). Again, the empty virus had no effect on luciferase activity (Fig. 5D).
In addition, A549 cells were infected as in Fig. 5A, and Western blot analysis revealed increasing expression of MKP-1 with increasing MOI of Ad5-MKP-1 (Fig. 5E). This correlated with the inhibition of p38 phosphorylation, and maximal effect was observed at MOI 30 (Fig. 5E). As in Fig. 5D, A549 cells infected with the Ad5-NF-κB-luc reporter were infected with MOI 30 of Ad5-MKP-1. Luciferase activity was induced 140 ± 12-fold by TNFα, and this was repressed by Ad5-MKP-1 to 76 ± 3.5% of TNFα-stimulated cells (Fig. 5F). The empty virus had no significant effect on luciferase activity (Fig. 5F). These data clearly show that MKP-1 can inhibit the p38 phosphorylation, NF-κB-dependent transcription, and both IL-8 protein and RNA expression that is induced by TNFα.
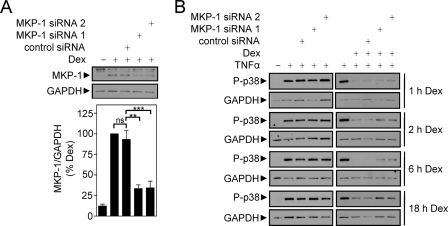
Effect of siRNA-mediated Knockdown of MKP-1 on Dexamethasone-dependent Repression of p38 Phosphorylation, NF-κB-dependent Transcription, and IL-8 Expression
To explore the relationship between MKP-1 and dexamethasone-mediated repression of the above outputs, BEAS-2B cells were initially incubated with MKP-1-specific or control siRNA for 24 h prior to stimulation with dexamethasone for 1 h. MKP-1 expression was induced by dexamethasone, and this was reduced to 33 ± 4.5 and 34 ± 8.1% by MKP-1 siRNA 1 and MKP-1 siRNA 2, respectively (Fig. 6A). These data confirmed that the upper, nonspecific immunoreactive band believed to be MKP-2 is not MKP-1. The control siRNA had no significant effect on dexamethasone-induced MKP-1 expression (Fig. 6A). Following this analysis, BEAS-2B cells were incubated with siRNA prior to treatment with dexamethasone for 1, 2, 6, or 18 h and then stimulated with TNFα for 15 min. Analysis of p38 phosphorylation by Western blot revealed that TNFα stimulated phosphorylation of p38, and this was repressed by dexamethasone at each pretreatment time (Fig. 6B). However, in the presence of MKP-1-specific siRNAs, the ability of dexamethasone to repress phospho-p38 was reduced, thereby confirming a role for MKP-1 in this response (Fig. 6B).
FIGURE 6.
Effect of MKP-1 targeting siRNA on dexamethasone-dependent repression of p38 MAPK phosphorylation. A, BEAS-2B cells were incubated with control or MKP-1-specific siRNAs for 24 h prior to stimulation with dexamethasone for 1 h. Cells were then harvested for protein and subjected to Western blot analysis for MKP-1 and GAPDH. Blots representative of 10 such experiments are shown. Following densitometric analysis, data (n = 10), normalized to GAPDH, were expressed as a percentage of dexamethasone-stimulated cells and plotted as means ± S.E. B, BEAS-2B cells were incubated with siRNAs as in A and then pretreated with dexamethasone for the times indicated prior to stimulation with TNFα (10 ng/ml) for 15 min. Cells were then harvested and subjected to Western blot analysis for phospho-p38 (P-p38) and GAPDH. Blots representative of six such experiments are shown.
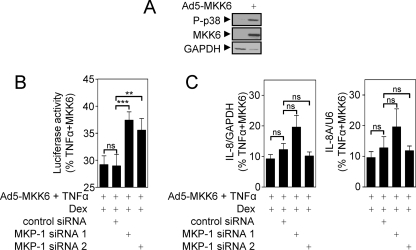
To examine the effect of dexamethasone-induced MKP-1 on NF-κB-dependent transcription, BEAS-2B cells infected with the Ad5-NF-κB-luc reporter were incubated with control or MKP-1 specific siRNAs for 24 h. The cells were then pretreated with dexamethasone for 1 h before stimulation with TNFα for 6 h. Analysis of luciferase activity revealed that the presence of MKP-1-specific siRNAs did not prevent the ability of dexamethasone to repress NF-κB-dependent transcription (data not shown). Two major possibilities may account for these data: 1) there may be no role for MKP-1 in this repression, or 2) there is redundancy in the pathways by which glucocorticoids down-regulate NF-κB-dependent transcription. Since NF-κB-dependent transcription is both p38-dependent (Fig. 2) and inhibited by MKP-1 (Fig. 5), we believe the lack of effect by the siRNA is likely to be due to redundancy. To address this, BEAS-2B cells were infected with both the Ad5-NF-κB-luc reporter and Ad5-MKK6 as a mechanism to maximally activate the p38 MAPK and potentially enhance the sensitivity of the system to inhibition by MKP-1. As expected, MKK6 overexpression induced the phosphorylation of p38 MAPK (Fig. 7A) and also enhanced NF-κB-dependent transcription from the BEAS-2B NF-κB-luc reporter by 25 ± 5.1% (data not shown). BEAS-2B cells infected with the Ad5-NF-κB-luc reporter, in the presence of Ad5-MKK6, were pretreated with dexamethasone and stimulated with TNFα (Fig. 7B). As before, TNFα + Ad5-MKK6 robustly induced luciferase activity, and dexamethasone significantly repressed (p < 0.01) this induction to 29 ± 1.6% of the response achieved in TNFα + Ad5-MKK6-stimulated cells (Fig. 7B). This level of inhibition was unaffected in the presence of control siRNA (Fig. 7B). However, both the MKP-1-specific siRNAs, siRNA 1 and 2, significantly reversed the ability of dexamethasone to inhibit NF-κB-dependent transcription to 37 ± 1.5% (siRNA 1) and 36 ± 2.2% (siRNA 2) of TNFα + Ad5-MKK6-stimulated cells (Fig. 7B). This shows that the endogenously expressed MKP-1 that is induced by dexamethasone does inhibit NF-κB-dependent transcription.
FIGURE 7.
Effect of MKP-1 targeting siRNA on dexamethasone-dependent repression of NF-κB-dependent transcription and IL-8 expression. A, BEAS-2B cells were infected with Ad5-MKK6 for 36 h prior to harvesting for Western blot analysis of phospho-p38, MKK6, and GAPDH. Blots representative of four such experiments are shown. B, BEAS-2B cells were infected with the Ad5-NF-κB-luc reporter and Ad5-MKK6 for 24 h prior to incubation with control or MKP-1-specific siRNAs for 24 h. Cells were then pretreated with dexamethasone for 1 h prior to stimulation with TNFα for 6 h and harvesting for luciferase activity determination. Data (n = 7) are expressed as percentage of TNFα + MKK6-stimulated cells and plotted as means ± S.E. Significance, using an ANOVA with Bonferroni's post-test, is indicated; **, p < 0.01; ***, p < 0.001; ns, not significant. C, BEAS-2B cells were infected with Ad5-MKK6 and incubated with siRNAs as in B. Cells were then pretreated with dexamethasone for 1 h and stimulated with TNFα for 1 h. Total RNA was extracted and converted to cDNA, and real time PCR was carried out for IL-8, GAPDH, nIL-8A, and U6. Data (n = 7) normalized to GAPDH (IL-8) or U6 (nIL-8A) were expressed as percentage of TNFα + MKK6-stimulated cells and plotted as means ± S.E. Significance, using ANOVA with Bonferroni's post-test is indicated; ns, not significant.
In parallel experiments, BEAS-2B cells were incubated with control or MKP-1-specific siRNAs for 24 h prior to pretreatment with dexamethasone for 1 h and stimulation with TNFα for 1 h. Analysis of IL-8 and nIL-8A expression revealed that the presence of MKP-1-specific siRNAs had no significant effect on the ability of dexamethasone to repress IL-8 or nIL-8A expression (data not shown). Cells were then infected with Ad5-MKK6 prior to incubation with siRNAs and stimulated as above. IL-8 mRNA expression was induced 220 ± 31-fold by TNFα + Ad5-MKK6, and nIL-8A was induced 210 ± 1.4-fold. This induction of IL-8 and nIL-8A was repressed by dexamethasone to 9.6 ± 1.9 and 9.2 ± 1.4% of TNFα + Ad5-MKK6-stimulated cells, respectively. However, in the presence of MKP-1-specific siRNAs, there was again no significant effect on the dexamethasone-mediated repression (Fig. 7C). This effect is likely to be due to a greater degree of redundancy as the number of pathways leading to a response becomes greater and more complex.
DISCUSSION
An increasing body of data now indicates that the anti-inflammatory properties of glucocorticoids are due not only to direct transrepression of inflammatory transcription factors but also to the up-regulation of multiple anti-inflammatory genes (7, 8, 26, 35). Induction of one such gene, MKP-1, by glucocorticoids has previously been characterized in a number of cell types (26). In the current study, MKP-1 protein expression was strongly induced by dexamethasone in both BEAS-2B and A549 cells, and this effect correlated with the repression of p38 MAPK phosphorylation, an established substrate for MKP-1 (26). Importantly, EC50 values for the dexamethasone-dependent induction of MKP-1 were in the midnanomolar range, and this is consistent with the potencies described for the repression of inflammatory genes, such as COX-2 and IL-8, as well as for NF-κB-dependent transcription, in both A549 and BEAS-2B cells (33, 42, 43) (and the current study). Consequently, MKP-1 is a pharmacologically plausible, but previously untested, candidate for mediating these repressive effects of dexamethasone. This view is strengthened by our demonstration that NF-κB-dependent transcription in BEAS-2B and A549 cells and IL-8 expression in BEAS-2B cells are at least partially dependent on the p38 MAPK pathway.
Previous studies have shown that the inhibition of COX-2 and IL-8 mRNA expression by dexamethasone is both time-dependent and sensitive to inhibition by transcriptional and translational blockade (33, 43). Since these genes are highly NF-κB-dependent in these cells and the repression by dexamethasone is partly mediated at a transcriptional level, we sought to investigate the role of p38 MAPK inhibition by MKP-1 in these responses. Examination of the p38 MAPK phosphorylation that is induced by TNFα revealed that the repression exerted by dexamethasone became increasingly robust with pretreatment with dexamethasone. This is consistent with observations made at the level of both COX-2 and IL-8 transcription and mRNA expression and suggests that time-dependent events (e.g. the induction of new gene expression) are necessary for full repression by dexamethasone (33, 43). The fact that actinomycin D and cycloheximide blocked dexamethasone-dependent repression of p38 MAPK phosphorylation supports this concept and is in concordance with previously published data (28). Likewise, the observation that cycloheximide reduced, but did not totally prevent, the ability of dexamethasone to repress NF-κB-dependent transcription and IL-8 RNA synthesis is also consistent with a role for dexamethasone-induced gene expression. Conversely, since the repression exerted by dexamethasone was not completely reversed by cycloheximide, these data also support the existence of repressive mechanisms that do not require new gene synthesis. Thus, a component of the overall repression is clearly consistent with existing models of transrepression in which inflammatory transcription is directly targeted by GR and the recruitment of histone deacetylases (8, 11).
In the current study, overexpression of MKP-1 profoundly repressed the p38 MAPK phosphorylation and IL-8 expression that was induced by TNFα. By contrast, maximally effective concentrations of MKP-1 adenovirus vector affected NF-κB-dependent transcription to a lesser degree, and this is consistent with the reduced efficacy of both the pharmacological inhibitors of p38 MAPK and indeed dexamethasone on this response. In both these cases, the repression of NF-κB is primarily exerted at a point downstream of IκBα phosphorylation and DNA binding (44–46) (the current study). Thus, a common point (or points) of control appears to be likely. In terms of regulation by p38 MAPK kinase, a number of potential targets, including TATA-binding protein (25), CREB-binding protein (21), histones (24), MSK1 (47), and many others are suggested, and it will be of considerable interest to examine the effect of dexamethasone on these regulatory steps.
The above data clearly show that MKP-1 can inhibit p38 phosphorylation, NF-κB-dependent transcription, and IL-8 expression. To investigate the role of dexamethasone-induced MKP-1, we used siRNA to selectively silence the MKP-1 expression that was induced by dexamethasone. In these studies, we were able to achieve 66–67% knockdown of dexamethasone-induced MKP-1, and as expected, this resulted in a partial reversal of dexamethasone-mediated repression of the p38 MAPK phosphorylation that was induced by TNFα. These data agree with previous studies indicating p38 to be a major target of MKP-1 (26). However, when examining the effect of MKP-1 knockdown on the dexamethasone-dependent repression of NF-κB-dependent transcription, we initially saw no effect of MKP-1 silencing. Reasons for this result may include insufficiency of knockdown. Certainly, this is a potential issue, since the very high level of inducibility of MKP-1 expression by dexamethasone appears to result in lower levels of knockdown than we can routinely achieve with constantly expressed genes. In addition, the knockdown of MKP-1 may result in a compensatory up-regulation of another regulatory protein; however, evidence either for or against this mechanism is not currently available. An alternative explanation is that MKP-1 is not involved in the repressive effect exerted by dexamethasone on NF-κB-dependent transcription. However, given that 1) MKP-1 is induced by dexamethasone over a concentration range and time that is consistent with the inhibition of NF-κB, 2) MKP-1 overexpression inhibits p38 phosphorylation and NF-κB-dependent transcription, 3) MKP-1 actually does play a role in the repression of p38 MAPK by dexamethasone, and finally 4) NF-κB-dependent transcription is partly p38 MAPK dependent, we believe that dexamethasone-induced MKP-1 does play a role in the repression of NF-κB-dependent transcription. Since there are numerous pathways, including the IκB kinase, p38 MAPK, protein kinase C, and many other signaling cascades, that are required for activation of NF-κB-dependent transcription (13–15, 17), a likely explanation for our observation is that there is redundancy in the repression of NF-κB-dependent transcription by dexamethasone. This point is amply illustrated by the finding that the simple activation of the p38 MAPK pathway, by MKK6, is not sufficient to induce NF-κB-dependent transcription (data not shown). Thus in addition to the effects due to MKP-1, repression of NF-κB-dependent transcription by dexamethasone may occur via classical transrepression as well as via the induction of a variety of other glucocorticoid-inducible genes, including IκBα (7). For example, glucocorticoid-inducible leucine zipper is profoundly induced by dexamethasone in both A549 and BEAS-2B cells (33, 48). This protein has previously been shown to profoundly repress NF-κB-dependent transcription (49), including in BEAS-2B cells, in response to dexamethasone (50).
In order to address this issue, we overexpressed MKK6 to chronically activate p38 MAPK and increase the sensitivity of NF-κB-dependent transcription to inhibition by MKP-1 (as opposed to other glucocorticoid effector processes). As expected, this manipulation produced an up-regulation of p38 MAPK phosphorylation and also enhanced the NF-κB-dependent transcription that was induced by TNFα. In this system, siRNA-mediated knockdown of MKP-1 partially reversed the repressive effect of dexamethasone on NF-κB-dependent transcription. These data confirm, for the first time, that the induction of MPK-1 expression by dexamethasone exerts a repressive effect on NF-κB-dependent transcription. In the current study, parallel analysis of IL-8 expression showed that knockdown of dexamethasone-induced MKP-1 did not affect the ability of dexamethasone to repress IL-8 mRNA or nuclear RNA, even in the presence of MKK6 overexpression. This result was despite IL-8 protein, mRNA, and unspliced nuclear RNA all being highly dependent on the p38 MAPK pathway and being profoundly inhibited by overexpression of MKP-1. However, activation of IL-8 expression obviously involves a greater number of signal transduction pathways than the activation of either p38 MAPK- or NF-κB-dependent transcription. Thus, with each step downstream from, in this case, the TNFα receptor toward the final release of IL-8, there is an increased likelihood of redundancy between the various mechanisms of repression that may be exerted by dexamethasone (7). Therefore, although MKP-1 can inhibit p38 MAPK phosphorylation, NF-κB-dependent transcription, and IL-8 expression, the knockdown of dexamethasone-induced MKP-1 expression is only sufficient to partially reverse the repressive effects of dexamethasone on p38 phosphorylation and NF-κB-dependent transcription but not IL-8 expression.
The finding that induction of MKP-1 expression contributes to the repression that is exerted by glucocorticoids has important consequences with respect to the screening strategies that are now routinely used to identify new anti-inflammatory ligands of GR that may show reduced side effect profiles (51). Many of the side effects of glucocorticoids have been widely attributed to the induction of metabolic genes, including tyrosine aminotransferase or phosphoenolpyruvate carboxykinase, whereas repression of factors such as NF-κB or AP-1 was attributed to classical transrepression. This has led to considerable interest in developing ligands of GR that show reduced transactivation yet maintain transrepression activity (51). However, a complete loss of GR transactivation would impair the ability to induce MKP-1, and this would impact not only on the repression of NF-κB (current study) but also on AP-1 (52) and on the expression of various inflammatory genes, including cyclooxygenase-2, GROα (growth-regulated oncogene α), and IL-6 (31, 32, 53). Somewhat paradoxically, most screens for transrepression make use of standard reporter gene assays for NF-κB, AP-1, or parts of the promoter regions of relevant inflammatory genes, such as E-selectin (7). In this situation, our data predict that repression exerted by glucocorticoids or, indeed, novel GR ligands will represent a composite response involving a component that is attributed to MKP-1 and other components that are due to further glucocorticoid-inducible genes as well as classical transrepressive mechanisms. Thus, it is not surprising that the prototypical dissociated glucocorticoid, RU24858 (54), which shows dissociated characteristics in terms of inability to transactivate simple GRE-dependent transcription and ability to transrepress both AP-1 and NF-κB, also induces MKP-1 expression (33). This idea finds support from data in which the dimerization-defective GR mutant (A458T) (GRdim) was defective at inducing simple GRE-dependent transcription (55) yet caused transrepression of inflammatory genes and also induced MKP-1 (29, 56). Thus, prior models have not previously excluded the contribution that MKP-1 may exert toward the “apparent transrepression” of inflammatory gene expression by glucocorticoids. Since MKP-1 has recently been implicated in the repressive activity of glucocorticoids acting on AP-1-dependent transcription (52), we strongly suggest the need to critically reevaluate previous data with respect to transrepression by GR in the light of a possible role for proteins such as MKP-1. In this context, we specifically urge against the use of the term “transrepression,” unless a role for transactivation has been specifically excluded. In this context, it is commonly noted that “transrepression” by glucocorticoids acting via GR occurs at a lower concentration of glucocorticoid than that needed to achieve transactivation from simple GRE-dependent reporters (57). Thus, in A549 cells, simple GRE-dependent transcription showed an EC50 of 54.5 nm for dexamethasone, whereas both repression of NF-κB and inflammatory gene expression produced EC50 values in the low nanomolar range (33, 42) (and the current study). Likewise, in BEAS-2B cells, the induction of simple GRE-dependent transcription by dexamethasone occurred with an EC50 of 19 nm (48), yet repression of NF-κB-dependent transcription and IL-8 expression occurred at EC50 values in the 5–10 nm range (the current study), and this is consistent with the induction of MKP-1 by dexamethasone. Thus, the induction of MKP-1 occurs at a lower concentration of glucocorticoid than for the induction of simple GRE-dependent transcription, and this effect may help explain this longstanding discrepancy. Importantly, these data mean that a given glucocorticoid may show differential potencies with respect to each different effector response. In the context of G-protein-coupled receptors, the idea that the receptor shows differential coupling efficiency is not new; however, this idea is still relatively underdeveloped in the field of nuclear hormone receptors (58).
In summary, we show in human BEAS-2B and A549 epithelial cells that dexamethasone profoundly induces MKP-1 expression to exert repression of the p38 MAPK pathway. This leads to repression of the NF-κB-dependent transcription that is induced by TNFα. Therefore, we describe MKP-1 as an effector protein that is induced by glucocorticoids and plays a novel role in the repression of NF-κB-dependent inflammatory gene transcription. This effect represents transcriptional repression occurring via transactivation and must be carefully distinguished from mechanisms of transrepression. We conclude that in generating new anti-inflammatory ligands of GR, a complete loss of transactivation ability would lead to a loss of MKP-1 expression, and this may result in reduced anti-inflammatory potential. We therefore advocate the selection of improved GR ligands on the basis of differential gene expression profiles rather than on transrepression and transactivation.
Supplementary Material
This work was supported by a start-up grant from the Alberta Heritage Foundation for Medical Research (AHFMR) (to R. N.) and operating grants from the Canadian Institutes of Health Research (CIHR) (to R. N.), a Lung Association of Alberta and the North West Territories Studentship (to E. M. K.), and an Izaak Walton Killam Postdoctoral Fellowship (to N. S. H.). Real time PCR was performed by virtue of an equipment and infrastructure grant from the Canadian Fund of Innovation and the Alberta Science and Research Authority.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- NF-κB
- nuclear factor κB
- AP
- activator protein
- Ad5
- adenovirus serotype 5
- GR
- glucocorticoid receptor
- IκB
- inhibitor of κB
- IL
- interleukin
- MAPK
- mitogen-activated protein kinase
- MOI
- multiplicity of infection
- TNF
- tumor necrosis factor
- GRE
- glucocorticoid receptor element
- CREB
- cAMP-response element-binding protein
- DMEM
- Dulbecco's modified Eagle's medium
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ELISA
- enzyme-linked immunosorbent assay
- ANOVA
- analysis of variance
- nIL-8A
- unspliced nuclear IL-8
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- 6FAM
- 6-carboxyfluorescein
- TAMRA
- 5-carboxytetramethylrhodamine
- MGB
- minor groove binder.
REFERENCES
- 1.Barnes P. J. (2008) Nat. Rev. Immunol. 8, 183–192 [DOI] [PubMed] [Google Scholar]
- 2.De Bosscher K., Vanden Berghe W., Haegeman G. (2003) Endocr. Rev. 24, 488–522 [DOI] [PubMed] [Google Scholar]
- 3.Karin M., Delhase M. (2000) Semin. Immunol. 12, 85–98 [DOI] [PubMed] [Google Scholar]
- 4.Karin M., Ben-Neriah Y. (2000) Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 5.Catley M. C., Chivers J. E., Holden N. S., Barnes P. J., Newton R. (2005) Br. J. Pharmacol. 145, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton R., Holden N. S., Catley M. C., Oyelusi W., Leigh R., Proud D., Barnes P. J. (2007) J. Pharmacol. Exp. Ther. 321, 734–742 [DOI] [PubMed] [Google Scholar]
- 7.Newton R., Holden N. S. (2007) Mol. Pharmacol. 72, 799–809 [DOI] [PubMed] [Google Scholar]
- 8.De Bosscher K., Haegeman G. (2009) Mol. Endocrinol. 23, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Bosscher K., Schmitz M. L., Vanden Berghe W., Plaisance S., Fiers W., Haegeman G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13504–13509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheinman R. I., Gualberto A., Jewell C. M., Cidlowski J. A., Baldwin A. S., Jr. (1995) Mol. Cell Biol. 15, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito K., Barnes P. J., Adcock I. M. (2000) Mol. Cell Biol. 20, 6891–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito K., Jazrawi E., Cosio B., Barnes P. J., Adcock I. M. (2001) J. Biol. Chem. 276, 30208–30215 [DOI] [PubMed] [Google Scholar]
- 13.Schmitz M. L., Mattioli I., Buss H., Kracht M. (2004) ChemBioChem 5, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 14.Beyaert R., Cuenda A., Vanden Berghe W., Plaisance S., Lee J. C., Haegeman G., Cohen P., Fiers W. (1996) EMBO J. 15, 1914–1923 [PMC free article] [PubMed] [Google Scholar]
- 15.Catley M. C., Cambridge L. M., Nasuhara Y., Ito K., Chivers J. E., Beaton A., Holden N. S., Bergmann M. W., Barnes P. J., Newton R. (2004) J. Biol. Chem. 279, 18457–18466 [DOI] [PubMed] [Google Scholar]
- 16.Kyriakis J. M., Avruch J. (2001) Physiol. Rev. 81, 807–869 [DOI] [PubMed] [Google Scholar]
- 17.Vanden Berghe W., Plaisance S., Boone E., De Bosscher K., Schmitz M. L., Fiers W., Haegeman G. (1998) J. Biol. Chem. 273, 3285–3290 [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S., Hayden M. S. (2008) Nat. Rev. Immunol. 8, 837–848 [DOI] [PubMed] [Google Scholar]
- 19.Newton R., Holden N. S. (2006) Drug Discov. Today Dis. Mech. 3, 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., Haegeman G. (2003) EMBO J. 22, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reber L., Vermeulen L., Haegeman G., Frossard N. (2009) PLoS ONE 4, e4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bratton M. R., Frigo D. E., Vigh-Conrad K. A., Fan D., Wadsworth S., McLachlan J. A., Burow M. E. (2009) Carcinogenesis 30, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha R. N., Jana M., Pahan K. (2007) J. Immunol. 179, 7101–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saccani S., Pantano S., Natoli G. (2002) Nat. Immunol. 3, 69–75 [DOI] [PubMed] [Google Scholar]
- 25.Carter A. B., Knudtson K. L., Monick M. M., Hunninghake G. W. (1999) J. Biol. Chem. 274, 30858–30863 [DOI] [PubMed] [Google Scholar]
- 26.Clark A. R., Martins J. R., Tchen C. R. (2008) J. Biol. Chem. 283, 25765–25769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kassel O., Sancono A., Krätzschmar J., Kreft B., Stassen M., Cato A. C. (2001) EMBO J. 20, 7108–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasa M., Abraham S. M., Boucheron C., Saklatvala J., Clark A. R. (2002) Mol. Cell Biol. 22, 7802–7811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham S. M., Lawrence T., Kleiman A., Warden P., Medghalchi M., Tuckermann J., Saklatvala J., Clark A. R. (2006) J. Exp. Med. 203, 1883–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang B. N., Jude J. A., Panettieri R. A., Jr., Walseth T. F., Kannan M. S. (2008) Am. J. Physiol. Lung Cell Mol. Physiol 295, L186–L193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Issa R., Xie S., Khorasani N., Sukkar M., Adcock I. M., Lee K. Y., Chung K. F. (2007) J. Immunol. 178, 7366–7375 [DOI] [PubMed] [Google Scholar]
- 32.Quante T., Ng Y. C., Ramsay E. E., Henness S., Allen J. C., Parmentier J., Ge Q., Ammit A. J. (2008) Am. J. Respir. Cell Mol. Biol. 39, 208–217 [DOI] [PubMed] [Google Scholar]
- 33.Chivers J. E., Gong W., King E. M., Seybold J., Mak J. C., Donnelly L. E., Holden N. S., Newton R. (2006) Mol. Pharmacol. 70, 2084–2095 [DOI] [PubMed] [Google Scholar]
- 34.Holden N. S., Gong W., King E. M., Kaur M., Giembycz M. A., Newton R. (2007) Br. J. Pharmacol. 152, 891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipson K. E., Baserga R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9774–9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bueno O. F., De Windt L. J., Lim H. W., Tymitz K. M., Witt S. A., Kimball T. R., Molkentin J. D. (2001) Circ. Res. 88, 88–96 [DOI] [PubMed] [Google Scholar]
- 37.Kaiser R. A., Bueno O. F., Lips D. J., Doevendans P. A., Jones F., Kimball T. F., Molkentin J. D. (2004) J. Biol. Chem. 279, 15524–15530 [DOI] [PubMed] [Google Scholar]
- 38.Tudhope S. J., Catley M. C., Fenwick P. S., Russell R. E., Rumsey W. L., Newton R., Barnes P. J., Donnelly L. E. (2007) J. Immunol. 179, 6237–6245 [DOI] [PubMed] [Google Scholar]
- 39.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. (1995) FEBS Lett. 364, 229–233 [DOI] [PubMed] [Google Scholar]
- 40.Underwood D. C., Osborn R. R., Bochnowicz S., Webb E. F., Rieman D. J., Lee J. C., Romanic A. M., Adams J. L., Hay D. W., Griswold D. E. (2000) Am. J. Physiol. Lung Cell Mol. Physiol 279, L895–L902 [DOI] [PubMed] [Google Scholar]
- 41.Newton R., Stevens D. A., Hart L. A., Lindsay M., Adcock I. M., Barnes P. J. (1997) FEBS Lett. 418, 135–138 [DOI] [PubMed] [Google Scholar]
- 42.Chivers J. E., Cambridge L. M., Catley M. C., Mak J. C., Donnelly L. E., Barnes P. J., Newton R. (2004) Eur. J. Biochem. 271, 4042–4052 [DOI] [PubMed] [Google Scholar]
- 43.Newton R., Seybold J., Kuitert L. M., Bergmann M., Barnes P. J. (1998) J. Biol. Chem. 273, 32312–32321 [DOI] [PubMed] [Google Scholar]
- 44.Bergmann M., Hart L., Lindsay M., Barnes P. J., Newton R. (1998) J. Biol. Chem. 273, 6607–6610 [DOI] [PubMed] [Google Scholar]
- 45.Newton R., Hart L. A., Stevens D. A., Bergmann M., Donnelly L. E., Adcock I. M., Barnes P. J. (1998) Eur. J. Biochem. 254, 81–89 [DOI] [PubMed] [Google Scholar]
- 46.Nissen R. M., Yamamoto K. R. (2000) Genes Dev. 14, 2314–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beck I. M., Vanden Berghe W., Vermeulen L., Bougarne N., Vander Cruyssen B., Haegeman G., De Bosscher K. (2008) EMBO J. 27, 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaur M., Chivers J. E., Giembycz M. A., Newton R. (2008) Mol. Pharmacol. 73, 203–214 [DOI] [PubMed] [Google Scholar]
- 49.Ayroldi E., Migliorati G., Bruscoli S., Marchetti C., Zollo O., Cannarile L., D'Adamio F., Riccardi C. (2001) Blood 98, 743–753 [DOI] [PubMed] [Google Scholar]
- 50.Eddleston J., Herschbach J., Wagelie-Steffen A. L., Christiansen S. C., Zuraw B. L. (2007) J. Allergy Clin. Immunol. 119, 115–122 [DOI] [PubMed] [Google Scholar]
- 51.Uings I. J., Farrow S. N. (2005) Curr. Opin. Pharmacol. 5, 221–226 [DOI] [PubMed] [Google Scholar]
- 52.Diefenbacher M., Sekula S., Heilbock C., Maier J. V., Litfin M., van Dam H., Castellazzi M., Herrlich P., Kassel O. (2008) Mol. Endocrinol. 22, 1767–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho I. J., Kim S. G. (2009) Mol. Endocrinol. 23, 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vayssière B. M., Dupont S., Choquart A., Petit F., Garcia T., Marchandeau C., Gronemeyer H., Resche-Rigon M. (1997) Mol. Endocrinol. 11, 1245–1255 [DOI] [PubMed] [Google Scholar]
- 55.Reichardt H. M., Kaestner K. H., Tuckermann J., Kretz O., Wessely O., Bock R., Gass P., Schmid W., Herrlich P., Angel P., Schütz G. (1998) Cell 93, 531–541 [DOI] [PubMed] [Google Scholar]
- 56.Reichardt H. M., Tuckermann J. P., Göttlicher M., Vujic M., Weih F., Angel P., Herrlich P., Schütz G. (2001) EMBO J. 20, 7168–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonat C., Rahmsdorf H. J., Park K. K., Cato A. C., Gebel S., Ponta H., Herrlich P. (1990) Cell 62, 1189–1204 [DOI] [PubMed] [Google Scholar]
- 58.Simons S. S., Jr. (2006) Curr. Top. Med. Chem. 6, 271–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.