Abstract
Purpose
DNA-PKcs is one of the DNA repair genes. It was recently found that hyperplasia and dysplasia of the intestinal mucosa and the production of aberrant crypt foci were developed in DNA-PKcs-null mice, and this suggests a suppressive role for DNA-PKcs in tumorigenesis.
Materials and Methods
To investigate the possible relationship between the clinico-pathologic characteristics and the survival of gastric cancer patients, the expression status of DNA-PKcs was determined in 279 consecutive gastric cancers. Immunohistochemical analysis was performed to evaluate the expression levels of DNA-PKcs protein by using the tissue array method.
Results
Out of 279 consecutive gastric cancers, 63 cases (22.6%) showed the loss of DNA-PKcs expression. The loss of DNA-PKcs expression was significantly associated with advanced cancer (p<0.001), lymphatic invasion (p=0.001), lymph node metastasis (p=0.009), and advanced pTNM stage (p=0.009). Univariate survival analysis revealed that patients with the loss of DNA-PKcs expression had significantly poorer survival than those patients with intact DNA-PKcs expression (p=0.004). Moreover, the loss of DNA-PKcs expression was identified to correlate with a lower survival in the subgroup of stage I gastric cancer patients (p=0.037).
Conclusion
The loss of DNA-PKcs expression was found in 23% of human gastric cancers and this was identified to significantly correlate with poor patient survival, especially for stage I gastric cancer patients.
Keywords: Immunohistochemistry, Stomach neoplasms, Survival analysis, DNA-PKcs
INTRODUCTION
DNA-PK is a serine/threonine kinase consisting of a 465-kDa catalytic subunit (DNA-PKcs) and a heterodimeric regulatory complex termed Ku, and Ku is composed of a 70-kDa polypeptide (Ku70) and an 86-kDa polypeptide (Ku86). DNA-PK is a critical regulator for the recombination of V, D and J immunoglobulin gene segments during the development of the immune system for immunoglobulin class switching, and it is also involved for non-homologous DNA double-strand break (DSB) repair (1). It is generally believed that Ku helps to recruit DNA-PKcs to DNA and Ku is required for the physiological activation of DNA-PK at the site of DNA damage (2,3). However, there is evidence that at least in vitro, DNA-PKcs can bind to linear DNA fragments and become activated for kinase activity in the absence of Ku (4,5). Recent studies have demonstrated that hyperplasia and dysplasia of the intestinal mucosa and the production of aberrant crypt foci were developed in DNA-PKcs-null mice, but not in Ku70-/- and Ku86-/- mice (6). This suggests that DNA-PKcs may play an important role as a tumor suppressor, at least in the intestinal mucosa. In contrast, Rigas et al. have reported that the expression of DNA-PKcs was not significantly altered in colon cancers compared to normal control, and this suggests a reduced role for DNA-PKcs in colonic carcinogenesis (7). In gastric cancers, the expression and functional status of DNA-PKcs has not been defined. In this study, we evaluated the expression status of DNA-PKcs in 279 consecutive gastric cancers and we evaluated the relationship of DNA-PKcs with the clinicopathologic characteristics, including patient survival.
MATERIALS AND METHODS
1) Tissue specimens
A total of 279 consecutive, surgically resected cases of gastric cancers were identified from the files of the Department of Pathology, Seoul National University College of Medicine from January 1, 1995, to June 30, 1995. The age, gender, tumor location, lymphatic invasion, vascular invasion and pTNM stage (8) were evaluated by reviewing the medical charts and the pathologic records. The tissue slides were reviewed for the histologic classification (according to the WHO classification and Lauren's classification) (9). The clinical outcome of the patients was followed from the date of surgery to the date of death or to December 1, 2000. The follow-up period was 1~72 months (mean period of follow-up: 52 months). The cases lost to follow-up and those deaths from any cause other than gastric cancer were regarded as censored data for the analysis of the survival rates.
2) Tissue array methods
Core tissue biopsies (2 mm in diameter) were taken from the individual paraffin-embedded gastric cancers (donor blocks), and they arranged in a new recipient paraffin block (a tissue array block), as previously described (10,11). Each tissue array block contained up to sixty samples, with a total of five blocks for the tissue array. Each block contained internal controls consisting of three non-neoplastic gastric mucosa samples from body, antrum and intestinal metaplasia. It has previously been proven that there is excellent agreement between the staining results obtained from different intra-tumoral areas of gastric carcinomas (10); therefore, a core was sampled from each case. An adequate case was defined as a tumor occupying more than 10% of the core area. Four µm were cut from each tissue array block, and it was deparaffinized and then dehydrated.
3) Immunohistochemistry
Immunohistochemical staining of DNA-PKcs was performed using DNA-PKcs stain (sc-1552, 1 : 100, Santa Cruz, Santa Cruz, CA) and a streptavidin peroxidase procedure (labeled streptavidin-biotin) after the antigen retrieval process. The immunolabeling level of each case was scored as positive (strong labeling), weak positive (faint staining), or negative (absence of staining), and the pattern of immunolabeling was scored as diffusely positive (≥ 50% of the tumor cells), focally positive (10~49% of the tumor cells), or negative (<10% of the tumor cells). For statistical analysis, the immunostainings were considered as having a loss of expression if less than 10% of the cells showed positive staining.
4) Statistical analyses
The chi-square test or Fisher's exact test (2-sided) was performed. Survival curves were plotted using the Kaplan-Meier product-limit method, and the differences between the survival curves were tested using the log-rank test. Multivariate survival analysis was performed using the Cox proportional hazards model. The results were considered to be statistically significant when the p values were less than 0.05. All statistical analyses were conducted using the SPSS 12.0 statistical software program (SPSS, Chicago, IL).
RESULTS
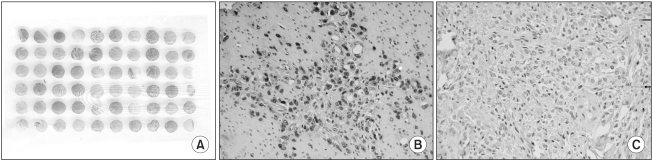
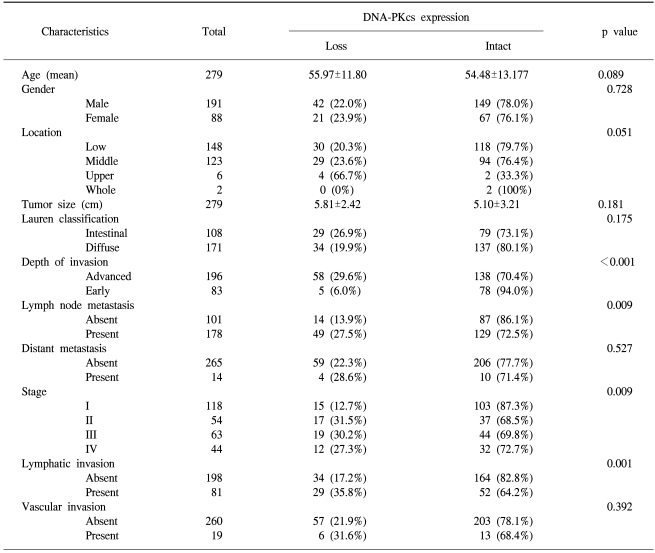
In the non-neoplastic gastric mucosa, DNA-PKcs was expressed in the nucleus of the epithelial cells, the stromal cells and the inflammatory cells. Out of the 279 consecutive gastric cancers, 63 cases (22.6%) showed the loss of DNA-PKcs expression (Fig. 1). Table 1 demonstrates the correlation between the DNA-PKcs expression status and the clinico-pathologic parameters. The loss of DNA-PKcs expression was more frequent in the cancers of the upper third of the stomach, although this showed borderline statistical significance (p=0.051). Regarding tumor progression, the patients with gastric cancer having the loss of DNA-PKcs expression were more likely to have advanced cancers (p<0.001), lymphatic invasion (p= 0.001), lymph node metastasis (p=0.009), and advanced pTNM stage (p=0.009). However, no significant correlation was found between the loss of DNA-PKcs expression and patients' age, gender, the tumor size or the histologic classification by Lauren (p>0.05).
Fig. 1.
Microscopic features of the immunohistochemical staining of DNA-PKcs in consecutive gastric cancers. (A) Tissue array slide containing 60 cores (×1.8), (B) DNA-PKcs-positive cancer (×400), and (C) DNA-PKcs-negative cancer (×400).
Table 1.
Correlation between DNA-PKcs expression and the clinico-pathologic parameters in consecutive gastric carcinomas
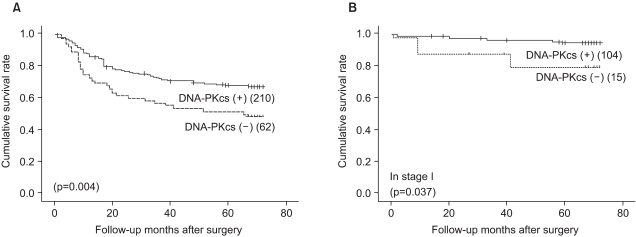
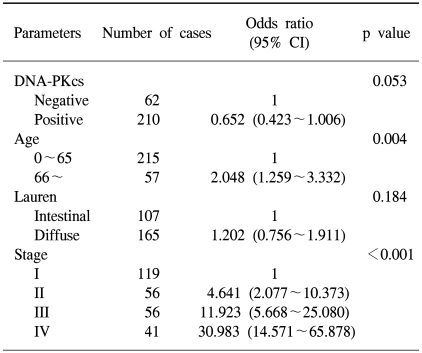
Univariate survival analysis by Kaplan-Meier survival curves showed that patients with the loss of DNA-PKcs expression had significantly poorer survival than those patients with intact DNA-PKcs expression (p=0.004) (Fig. 2A). The 5-year survival rate was 67.7±3.3% for patients with intact DNA-PKcs expression, while it was 51.3±6.4% for patients with the loss of DNA-PKcs expression. By multivariate analysis that included age, the Lauren histologic type, the pTNM stage and the expression status of DNA-PKcs, loss of DNA-PKcs expression was not an independent prognostic indicator, but it had borderline statistical significance (p=0.053) (Table 2). The Kaplan-Meier survival curves that were stratified according to the pTNM stage (stage I-IV) revealed that loss of DNA-PKcs expression correlated with a probability of lower survival for the patient subgroup having stage I cancers (p=0.037) (Fig. 2B).
Fig. 2.
Survival curves calculated using the Kaplan-Meier method. (A) The survival curves showed that DNA-PKcs-negative cancers have an unfavorable prognosis compared to DNA-PKcs positive cancers (p=0.004). (B) In the subgroup of stage I gastric cancer patients, loss of DNA-PKcs expression correlated with a lower survival probability (p=0.037).
Table 2.
Multivariate analysis using the Cox proportional hazard model
DISCUSSION
Cells are continually under assault by a plethora of DNA damaging agents, including reactive oxygen species, ultraviolet light, ionizing radiation and a variety of man-made and natural mutagenic chemicals. Non-repaired or incorrectly repaired DNA damage can result in gene mutation and alterations of the proteins that control cell growth or cell survival (12). Since DNA damage can arise in many different forms, various DNA repair systems have been evolved to protect cells from specific types of DNA damage. The DNA DSB repair apparatus is specific for DNA DSBs, which are the major lethal lesions induced by ionizing radiation (13). DNA-PKcs, one of the DNA DSB repair apparatus, has recently been suggested to be a tumor suppressor (6), but the implication for the loss of DNA-PKcs expression in gastric tumorigenesis has been poorly defined. At the present, DNA-PKcs was identified to be localized at the nucleus in the non-neoplastic gastric mucosa and the absence of DNA-PKcs expression was observed in 63 of 279 consecutive gastric cancers (22.6%).
Our study demonstrate that gastric cancers with the loss of DNA-PKcs expression are associated with advanced staging, a high prevalence of lymph node metastasis and poor patient survival. Several functions of DNA-PKcs are suggested to be associated with tumor progression and patient survival. A role for DNA-PKcs in DSB repair has been well documented. Genetic instability is an important factor in the rapid accumulation of genetic changes during carcinogenesis (14). Unrepaired DNA DSBs that are due to the functional loss of DNA-PKcs would result in the generation of acentric chromosome fragments during mitosis (12). Unstable chromosomes during cell proliferation may be associated with genetic changes, including changes in the genes that are involved with tumor progression, invasion and metastasis. In addition to DSB repair, DNA-PKcs has a regulatory function for p53. DNA-PKcs has been reported to destabilize the interaction between p53 and MDM2, and these events then lead to p53 protein stabilization (15). In various cancers, including gastric cancer, p53 alteration has been reported to be significantly associated with an advanced tumor stage or a poor prognosis (11).
Previous studies have shown that DNA-PKcs-deficient cells display a hypersensitivity to ionizing radiation (16). Defects of DNA DSB repair may be associated not only with tumor progression, but also with sensitizing tumor cells to DNA DSB producing agents, such as ionizing radiation and radiomimetic drugs like methyl methanesulfonate or bleomycin. However, careful interpretation and additional study is needed to determine the correlation between the DNA-PKcs expression status and effects of chemo or radiotherapy because of the complex interactions of the various genes and proteins in tumor cells.
The incidence of small and early gastric cancer is high in Asia and the rate has increased due to early diagnosis (17). The overall 5-year survival rate of patients with gastric cancers was 64.0±2.9% in this study: the 5 year survival rate was 92.2±2.5% for patients with stage I tumor and 42.5±4.0% for patients with stage II-IV tumor (data not shown). Although the pTNM stage is the most important prognostic factor for gastric cancer patients, a portion of the stage I gastric cancer patients show a poor outcome. We found that the loss of DNA-PKcs expression was associated with a poorer outcome for stage I gastric cancer patients, suggesting that DNA-PKcs expression could be a promising prognostic marker for early stage gastric cancers.
The expression of the tumor suppressor genes is controlled by some molecular mechanisms such as mutation, the loss of heterozygosity (LOH), and epigenetic mechanisms (DNA methylation and histone acetylation/deacetylation). Several possible mechanisms that regulate DNA-PKcs expression, other than mutations, have been studied because only extremely rare mutations of DNA-PKcs have been documented to date in human cancer (18). Cho et al. have demonstrated an allotypic imbalance (LOH and MSI) of DNA-PKcs with a concomitant decrease in DNA-PKcs expression in some low-grade, soft tissue sarcomas of the extremities in adults (19). In contrast to this result, DNA-PKcs down-regulation was noted to be a proteasome-dependent degradation that required tyrosine kinase activity in chronic myelogenous leukemia and it was associated with a marked DNA repair deficiency along with an increased sensitivity to ionizing radiation (20). Thus, one of the possible mechanisms involved in the down-regulation of DNA-PKcs could be its degradation through the ubiquitin-proteasome pathway, and this suggests that post-transcriptional regulation is involved. Because the promoter region of DNA-PKcs contains large CpG islands, epigenetic mechanisms could be considered as another possible mechanism for the loss of DNA-PKcs expression (21). To date, however, the mechanism for the loss of DNA-PKcs expression has not been clarified in gastric or colorectal cancers, and further study is needed.
CONCLUSIONS
Loss of DNA-PKcs expression was found in 63 out of 279 (22.5%) consecutive gastric cancers and this was significantly associated with tumor progression and poor patient survival. The loss of DNA-PKcs expression may be a potential prognostic marker, and especially for stage I gastric cancers.
Footnotes
This study was supported by grant no. 02-03-005 from the Seoul National University Bundang Hospital Research Fund.
References
- 1.Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan DW, Lees-Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 3.Suwa A, Hirakata M, Takeda Y, Jesch SA, Mimori T, Hardin JA. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci USA. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurimasa A, Ouyang H, Dong LJ, Wang S, Li X, Cordon-Cardo C, et al. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rigas B, Borgo S, Elhosseiny A, Balatsos V, Manika Z, Shinya H, et al. Decreased expression of DNA-dependent protein kinase, a DNA repair protein, during human colon carcinogenesis. Cancer Res. 2001;61:8381–8384. [PubMed] [Google Scholar]
- 8.American Joint Committee on Cancer. AJCC cancer staging manual. 6th ed. New York: Springer-Verlag; 2002. [Google Scholar]
- 9.International Agency for Research on Cancer (IARC) World Health Organization Classification of Tumors; pathology and genetics of tumors of the digestive system. Lyon: IARC Press; 2000. [Google Scholar]
- 10.Lee HS, Lee HK, Kim HS, Yang HK, Kim YI, Kim WH. MUC1, MUC2, MUC5AC, and MUC6 expressions in gastric carcinomas: their roles as prognostic indicators. Cancer. 2001;92:1427–1434. doi: 10.1002/1097-0142(20010915)92:6<1427::aid-cncr1466>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 11.Darwish NS, Kim MA, Chang MS, Lee HS, Lee BL, Kim YI, et al. Prognostic significance of CD24 expression in gastric carcinoma. Cancer Res Treat. 2004;36:298–302. doi: 10.4143/crt.2004.36.5.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindahl T. Genetic instability in cancer. In: Jackson SP, editor. DNA damage detection by DNA dependent protein kinase and related enzymes. New York: Cold Spring Harbor Laboratory Press; 1996. pp. 261–279. [PubMed] [Google Scholar]
- 13.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 15.Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 16.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, 3rd, Barron GM, et al. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi Y, Yoshikawa T, Tsuburaya A, Motohashi H, Karpeh MS, Brennan MF. Is gastric carcinoma different between Japan and the United States? Cancer. 2000;89:2237–2246. [PubMed] [Google Scholar]
- 18.Blunt T, Gell D, Fox M, Taccioli GE, Lehmann AR, Jackson SP, et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho NH, Cordon-Cardo C, Li GC, Kim SH. Allotype imbalance or microsatellite mutation in low-grade soft tissue sarcomas of the extremities in adults. J Pathol. 2002;198:21–29. doi: 10.1002/path.1177. [DOI] [PubMed] [Google Scholar]
- 20.Deutsch E, Dugray A, AbdulKarim B, Marangoni E, Maggiorella L, Vaganay S, et al. BCR-ABL down-regulates the DNA repair protein DNA-PKcs. Blood. 2001;97:2084–2090. doi: 10.1182/blood.v97.7.2084. [DOI] [PubMed] [Google Scholar]
- 21.Connelly MA, Zhang H, Kieleczawa J, Anderson CW. The promoters for human DNA-PKcs (PRKDC) and MCM4: divergently transcribed genes located at chromosome 8 band q11. Genomics. 1998;47:71–83. doi: 10.1006/geno.1997.5076. [DOI] [PubMed] [Google Scholar]