Abstract
ECX21941 represents a very large family (over 600 members) of novel, ocean metagenome–specific proteins identified by clustering of the dataset from the Global Ocean Sampling expedition. The crystal structure of ECX21941 reveals unexpected similarity to Sm/LSm proteins, which are important RNA-binding proteins, despite no detectable sequence similarity. The ECX21941 protein assembles as a homopentamer in solution and in the crystal structure when expressed in Escherichia coli and represents the first pentameric structure for this Sm/LSm family of proteins, although the actual oligomeric form in vivo is currently not known. The genomic neighborhood analysis of ECX21941 and its homologs combined with sequence similarity searches suggest a cyanophage origin for this protein. The specific functions of members of this family are unknown, but our structure analysis of ECX21941 indicates nucleic acid-binding capabilities and suggests a role in RNA and/or DNA processing.
Keywords: Structural genomics, metagenomics, nucleic acid binding, Sm-like, viral protein
Introduction
The ECX21941 gene from the Global Ocean Sampling (GOS) metagenome dataset 1,2 encodes a protein with a molecular weight of 11.5 kDa (residues 1−104) and a calculated isoelectric point of 5.75. ECX21941 was selected for structure determination in a pilot project to explore structural diversity of proteins from the ocean metagenome using the semiautomated, high-throughput pipeline of the Joint Center for Structural Genomics (JCSG; http://www.jcsg.org) 3 as part of the National Institute of General Medical Sciences’ Protein Structure Initiative. ECX21941 is a representative of a very large, novel, ocean metagenome–specific family (over 600 members), and its function is unknown. Genomic neighborhood analysis of ECX21941 and homologous proteins suggests a cyanophage origin for this protein.
The structure of ECX21941 is similar that of Sm/LSm/Sm-like proteins despite lack of any detectable sequence similarity and further analysis confirmed that it is a very divergent member of this protein family. Sm and Sm-like (or Like Sm, LSm) proteins (PF01423 [PFAM], cd00600 [CDD]) form a very large (>1500 members) and evolutionary diverse 4 protein family with an open β-barrel fold with SH3-like topology and diverse functions that center around RNA processing. The Sm/LSm family is classified into 23 different groups by the NCBI Conserved Domains Database 5 and into seven structurally characterized families of proteins with Sm-like fold by the SCOP database (sunid: 50181) 6. In eukaryotes, they are essential for pre-mRNA splicing 7, telomere formation 8, trans splicing 9, and mRNA degradation 10,11 and are implicated in human autoimmune diseases 12. Sm-like proteins have also been reported and characterized in bacteria and archaea, and share similar RNA-binding features with their eukaryotic counterparts 13–15.
ECX21941 is the first structural representative of Sm-like proteins with a pentameric assembly of protomers, as observed in the crystal structure and in solution from protein expressed in Escherichia coli. Other known functional assemblies are homohexameric (bacteria and archaea) 16,17, homoheptameric (archaea) 18–21, or heteroheptameric/octameric (eukaryota) 22–25. It is still unclear what drives different oligomer arrangements in Sm and LSm proteins, particularly in vivo, and how these different potential oligomerization states affect molecular activity. The crystal structure of ECX21941 presented here should aid in biochemical analyses to determine whether it is involved in RNA-mediated regulation and/or post-transcriptional processing of RNAs 26–31.
Materials and Methods
Protein production and crystallization
The DNA encoding ECX21941 (GenBank: ECX21941.1, GI:142318367, GOS_2577746) was synthesized with codons optimized for Escherichia coli expression and cloned into plasmid pSpeedET (CodonDevices, Cambridge, MA). Since crystallization trials with the full-length construct were unsuccessful, the polymerase incomplete primer extension (PIPE) 32 method was used to delete part of the gene encoding the C-terminal residues 100−104. The final construct used encodes residues 1–99 of ECX21941 in addition to MGSDKIHHHHHHENLYFQG of an expression and purification tag followed by a tobacco etch virus (TEV) protease cleavage site at its N-terminus. The cloning junctions were confirmed by DNA sequencing. Protein expression was performed in a selenomethionine-containing medium using the Escherichia coli strain GeneHogs (Invitrogen). At the end of fermentation, lysozyme was added to the culture to a final concentration of 250 µg/mL, and the cells were harvested. After one freeze/thaw cycle, the cells were homogenized in Lysis Buffer [50 mM HEPES pH 8.0, 50 mM NaCl, 10 mM imidazole, 1 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP)] and passed through a Microfluidizer (Microfluidics). The lysate was clarified by centrifugation at 32,500 × g for 30 minutes and loaded onto nickel-chelating resin (GE Healthcare) pre-equilibrated with Lysis Buffer. The resin was washed with Wash Buffer [50 mM HEPES pH 8.0, 300 mM NaCl, 40 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP], and the protein was eluted with Elution Buffer [20 mM HEPES pH 8.0, 300 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP]. The eluate was buffer exchanged with HEPES Crystallization Buffer [20 mM HEPES pH 8.0, 200 mM NaCl, 40 mM imidazole, 1 mM TCEP] and treated with 1 mg of TEV protease per 15 mg of eluted protein. The digested protein was passed over nickel-chelating resin (GE Healthcare) pre-equilibrated with HEPES Crystallization Buffer, and the resin was washed with the same buffer. The flow-through and wash fractions were combined and concentrated for crystallization assays to 12.5 mg/mL by centrifugal ultrafiltration (Millipore). ECX21941 was crystallized using the nanodroplet vapor diffusion method 33 with standard JCSG crystallization protocols 3. Initial screening for diffraction was carried out using the Stanford Automated Mounting system (SAM) 34 at the Stanford Synchrotron Radiation Laboratory (SSRL, Menlo Park, CA). The crystallization reagent that produced the crystal used for structure solution contained 0.2 M calcium acetate and 20% (w/v) polyethylene glycol 3350 at pH 7.3. Ethylene glycol was added as a cryoprotectant to a final concentration of 10% (v/v). The crystal was indexed in the monoclinic space group C2 (Table I) 35,36. To determine its oligomeric state in solution, ECX21941 was analyzed using a 1 cm × 30 cm Superdex 200 column (GE Healthcare) coupled with miniDAWN static light scattering and Optilab differential refractive index detectors (Wyatt Technology). The mobile phase consisted of 20 mM Tris pH 8.0, 150 mM NaCl, and 0.02% (w/v) sodium azide. The molecular weight was calculated using ASTRA 5.1.5 software (Wyatt Technology).
Table I.
Summary of crystal parameters, data collection, and refinement statistics for ECX21941 (PDB: 3by7).
| Space group | C2 | ||
|---|---|---|---|
| Unit cell parameters | a = 108.25 Å, b = 77.18 Å, c = 71.47 Å, β = 113.82° | ||
| Data collection | λ1 MAD Se | λ2 MAD Se | λ3 MAD Se |
| Wavelength (Å) | 0.9537 | 0.9796 | 0.9794 |
| Resolution range (Å) | 27.1–2.60 | 27.0–2.60 | 28.0–2.60 |
| Number of observations | 20,192 | 19,957 | 70,849 |
| Number of unique reflections | 16,122 | 15,964 | 16,117 |
| Completeness (%) | 95.7 (92.4)a | 94.7 (84.1) | 95.9 (91.7) |
| Mean I/σ (I) | 9.5 (1.8)a | 10.1 (2.1) | 12.3 (2.6) |
| Rsym on I (%) | 4.7 (40.4)a | 4.1 (34.8) | 5.5 (29.6) |
| Highest resolution shell (Å) | 2.69–2.60 | 2.69–2.60 | 2.69–2.60 |
| Model and refinement statistics | |||
| Resolution range (Å) | 27.1–2.60 | Data set used in refinement | λ1 |
| Number of reflections (total) | 16,120b | Cutoff criteria | |F| > 0 |
| Number of reflections (test) | 825 | Rcryst | 0.235 |
| Completeness (% total) | 96.6 | Rfree | 0.285 |
| Stereochemical parameters | |||
| Restraints (RMS observed) | |||
| Bond angle (°) | 1.247 | ||
| Bond length (Å) | 0.013 | ||
| Average isotropic B-value (Å2) | 74.1c | ||
| ESU based on Rfree (Å) | 0.339 | ||
| Protein residues/atoms | 405/3024 | ||
| Water molecules | 7 | ||
Highest resolution shell.
Rsym = Σ|Ii-<Ii>| / Σ|Ii| where Ii is the scaled intensity of the ith measurement and <Ii> is the mean intensity for that reflection.
Rcryst = Σ| |Fobs|-|Fcalc| | / Σ|Fobs| where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5.1% of the total reflections chosen at random and omitted from refinement.
Typically, the number of unique reflections used in refinement is slightly less than the total number that were integrated and scaled. Reflections are excluded due to systematic absences, negative intensities, and rounding errors in the resolution limits and cell parameters.
This value represents the total B that includes TLS and residual B components.
Data collection, structure solution, and refinement
Multi-wavelength anomalous diffraction (MAD) data were collected at the Advanced Photon Source (APS; Chicago, IL) on beamline 23-ID-D at wavelengths corresponding to the high-energy remote (λ1), inflection (λ2), and peak (λ3) of a selenium MAD experiment. The datasets were collected at 100K using a MAR300 CCD detector. The MAD data were integrated and reduced using XDS and then scaled with the program XSCALE 37. Data statistics are summarized in Table I. Phasing was performed with SHELXD 38 and autoSHARP 39, and automated iterative model building was performed using ARP/wARP 40 and RESOLVE 41. The initial trace revealed five protein subunits in the asymmetric unit (ASU), with a main-chain completeness of ~75% (with ~60% side chains) and starting Rcryst/Rfree values of ~36%/40%. From this initial trace, one of the chains (chain A) was manually adjusted to correct sequence registry and side chain rotamers using Coot 42. Molecular replacement (PHASER 43) was then used to place the other four molecules in the ASU using this partially refined structure as the search molecule. Model adjustments and completion, were performed with Coot 42. Structure refinement was carried out using REFMAC5 applying tight main-chain and loose-side chain NCS restraints and one TLS group per protomer chain throughout the refinement, Residues 0–1 and 91–99 are omitted from all five chains due to weak electron density. The tip (residues 40–43) of the loop region spanning residues 37 to 46 was disordered to a varying extent in each protomer and, therefore, was omitted from the structure since it could not be reliably modeled into the relatively weak, discontinuous, electron density. In addition, some monomers have slightly larger omitted regions around residue 40 and at the C-terminus. A total of 57 residues have their side chains truncated due to lack of interpretable density. Refinement statistics are summarized in Table I.
Validation and deposition
Analysis of the stereochemical quality of the model was accomplished using AutoDepInputTool 44, MolProbity, SFcheck 4.0 35, and WHATIF 5.0 45. Protein quaternary structure analysis was performed using the PQS (Protein Quaternary Structure) server 46, the PISA (Protein Interfaces, Surfaces, and Assemblies) server 47, and PITA (Protein InTerfaces and Assemblies) software 48. Figure 1B was adapted from an analysis using PDBsum 49. Figure 1A, Figure 2B, and Figure 3 were prepared with PyMOL (DeLano Scientific 50). Electrostatics surface potentials (Figure 3B) were calculated using APBS 51 and rendered within PyMOL using the APBS plug-in. Missing side-chain atoms were added to the model in their favored rotamers position using Coot, prior to electrostatic calculations and rendering. Figure 2A was prepared using MUSTANG 52 for structure superposition, JOY 53 for structural features annotation, and PDBsum analysis or existing annotations from the CDD database to highlight functional residues 5.
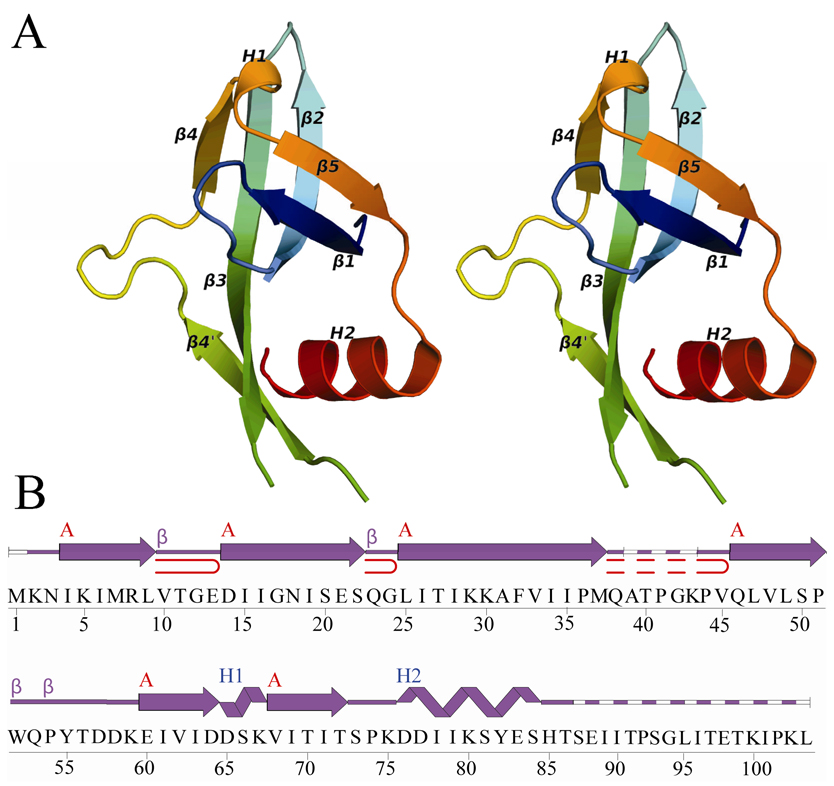
Figure 1.
Crystal structure of ECX21941 from the GOS ocean metagenome sequence dataset. (A) Stereo ribbon diagram of the ECX21941 monomer color-coded from N-terminus (blue) to C-terminus (red). Helices H1 and H2 and β-strands β1–β5 are indicated. (B) Diagram showing the secondary structural elements of ECX21941 superimposed on its primary sequence. The α-helices, β-strands, and β-turns are indicated. The β-sheet is indicated by a red A, and the β-hairpins are depicted as red loops. Disordered regions are depicted by a dashed line.
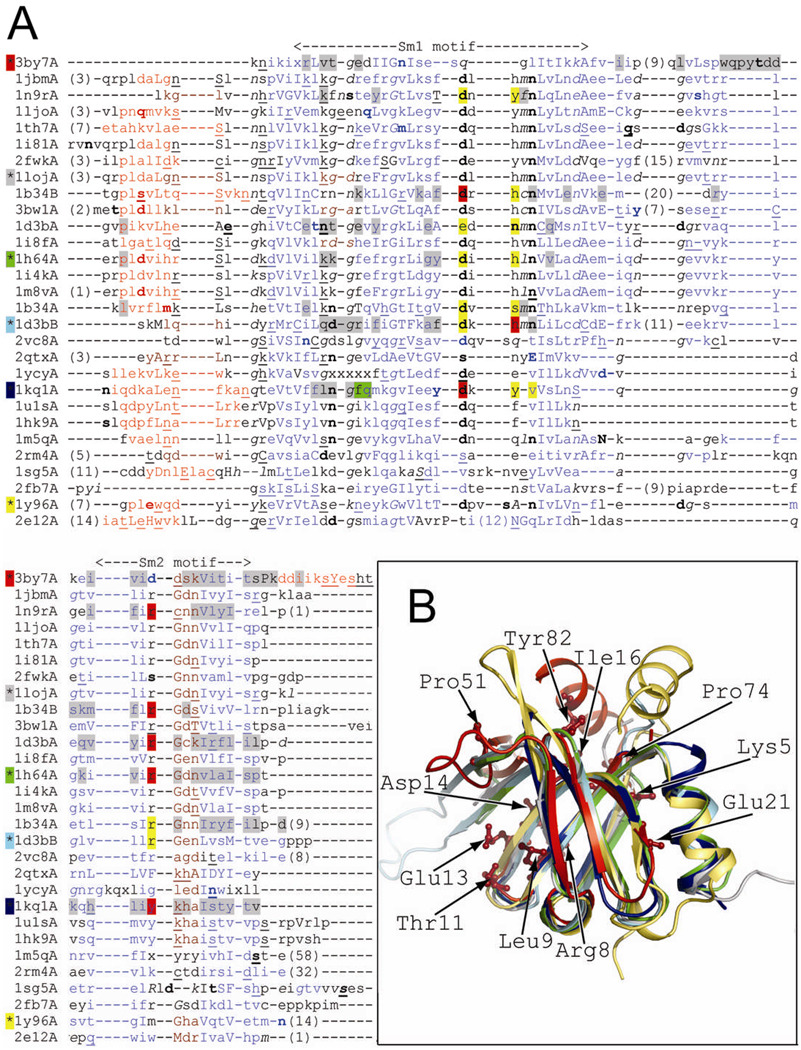
Figure 2.
(A) Structure-based sequence alignment. The structural features are annotated with JOY software 53 (solvent inaccessible, uppercase; solvent accessible, lowercase; positive φ, italic; hydrogen bond to main-chain amide, bold; hydrogen bond to main-chain carbonyl, underlined; α-helix, red; β-strand, blue; 310-helix, maroon; unknown, x). Residues involved in RNA or protein binding are highlighted (binding both RNA and adjacent subunit, red; RNA binding, yellow; adjacent subunit binding, gray; adjacent hexamer binding, green) according to PDBsum server (PDB code 3by7) or annotations from CDD database 5 (PDB codes: 1loj, 1h64, 1d3b, 1kq1, and 1y96). The background color of the asterisk preceding the PDB code corresponds to the color of the cartoon representation in Fig. 2B. The Sm1 and Sm2 motifs correspond to previously characterized structures, but not PDB code 3by7. (B) Structure superposition of ECX21941 (PDB code 3by7A, red) and structurally diverse representatives of proteins with Sm-like fold: archaeal protein SmAP1 from M. thermoautotrophicum (the most structurally similar protein; PDB code 1loj, gray), archaeal Sm1 from P. abyssi (PDB code 1h64A, green), human small nuclear ribonucleoprotein (snRNP)-associated protein B (SmB, PDB code 1d3bB, cyan), bacterial translational regulator Hfq from S. aureus (PDB code 1kq1A, navy blue), and gemin6 from the human SMN complex (PDB code 1y96A, yellow). Residues conserved among ECX21941 homologs (Lys5, Arg8, Leu9, Thr11, Glu13, Asp14, Ile16, Glu21, Pro51, Pro74, and Tyr82; PDB code 3by7) are shown by a stick representation.
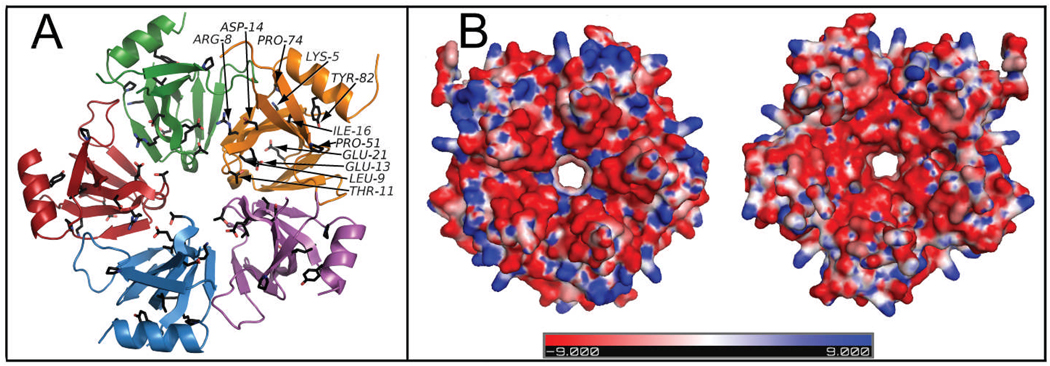
Figure 3.
(A) Residues conserved among the ECX21941 homologs are depicted in the schematic view of the pentamer by a black ball-and-stick representation. Each monomer is highlighted in a different color. For clarity, conserved residues are labeled on one monomer only. (B) The electrostatic surface potential of the pentamer formed by ECX21941 (top and bottom view) shows a ring of positively charged amino acids in blue (n.b. negatively charged amino acids are in red and neutral in white) on the outer periphery and a region of charged residues lining the entrance to the central pore. Lys67 constitutes the positively charged region at the entrance to the pore from the top side lining the central pore. Going from top to bottom, a negatively charged region is formed from Asp64, Asp65, and Ser66 prior to Lys67. The positively charged patch on the outer periphery of the top surface arises from Lys2, Lys5, Lys29, Lys30, Lys59, and Lys75.
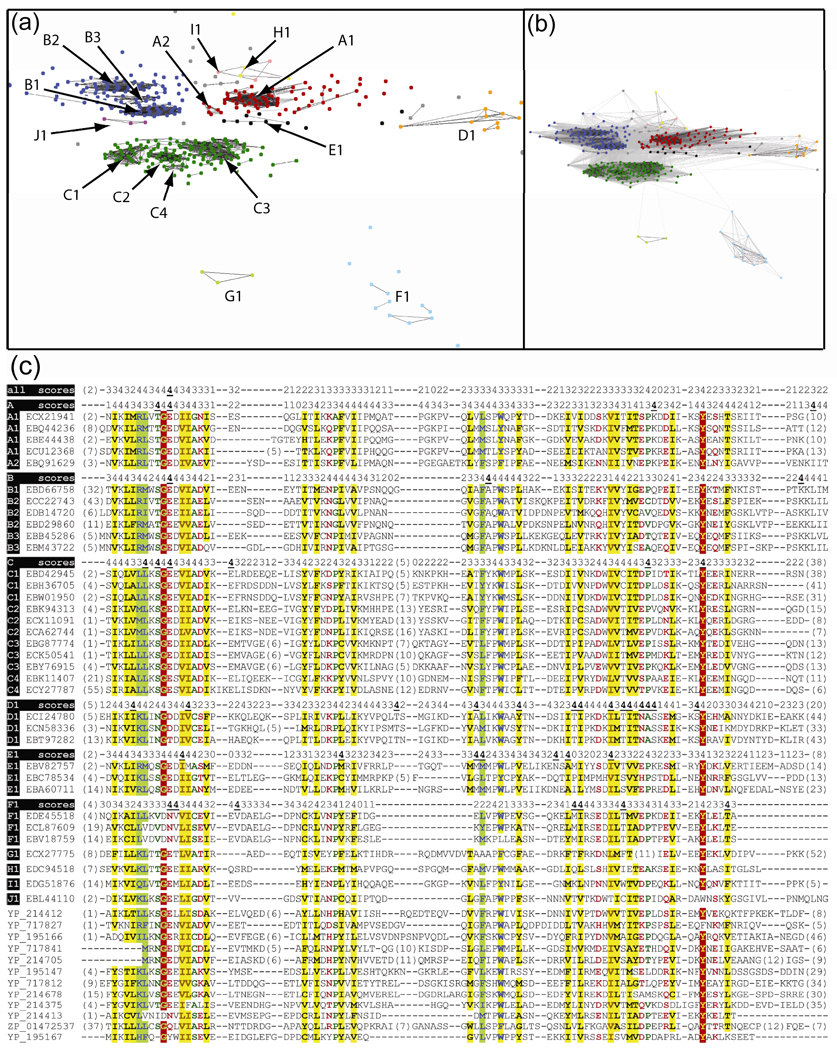
Putative homologs of ECX21941 were clustered using pairwise sequence similarities (CLANS software 54 with P-value = 5e-6) into groups of close homologs (Fig. 4). The genomic scaffolds encoding putative homologs with ≤ 85% sequence identity to ECX21941 were used in the genomic neighborhood analysis. CLANS software was used to identify groups of homologous proteins encoded by such scaffolds. Sequence conservation in Fig. 2B and 3A was calculated using Rate4Site 55. Selected scaffolds with different arrangements of the most frequently observed neighbors are shown in Fig. 5.
Figure 4.
(A) Clusters of ECX21941 homologs from marine metagenome–specific dataset of Sm-like proteins. Lines represent BLAST hits with P-value ≤ 5e-39. (B) The same as above, but the BLAST hits with P-value ≤ 5e-9 are shown. (C) Multiple sequence alignment of ECX21941 and its homologs from marine metagenome dataset. Prochlorococcus cyanophage P-SSM2 (YP_214412, YP_214375, and YP_214413), Synechococcus cyanophage syn9 (YP_717827, YP_717841, and YP_717812), Synechococcus cyanophage S-PM2 (YP_195166, YP_195147, and YP_195167), Prochlorococcus cyanophage P-SSM4 (YP_214705 and YP_214678), and Synechococcus sp. RS9916 (ZP_01472537). Numbers in parentheses indicate number of residues omitted for clarity. The conservation scores (0, not conserved; 4, highly conserved; 4 with bold and underlined font, the most conserved) for all homologs and for the most abundant clusters (A–F) were derived from an analysis using rate4site software using all available homologs of ECX21941 and not just those shown in the alignment. Residues are highlighted according to the amino acid properties. Red shading indicates conservation of single residue. Brown font with yellow shading indicates conservation of aliphatic residues (I, L, V). Green font indicates conservation of the smallest residues (A, G, and S). Blue font with yellow shading indicates conservation of aromatic residues (F, H, W, and Y). Dark green font indicates conservation of small residues (A, C, D, G, N, P, S, T, and V). Red font indicates conservation of polar residues (C, D, E, H, K, N, Q, R, S, and T). Blue font with green shading indicates conservation of big residues (E, F, H, I, K, L, M, Q, R, W, and Y). Black font with yellow shading indicates conservation of hydrophobic residues (A, C, F, G, H, I, L, M, T, V, W, and Y).
Figure 5.
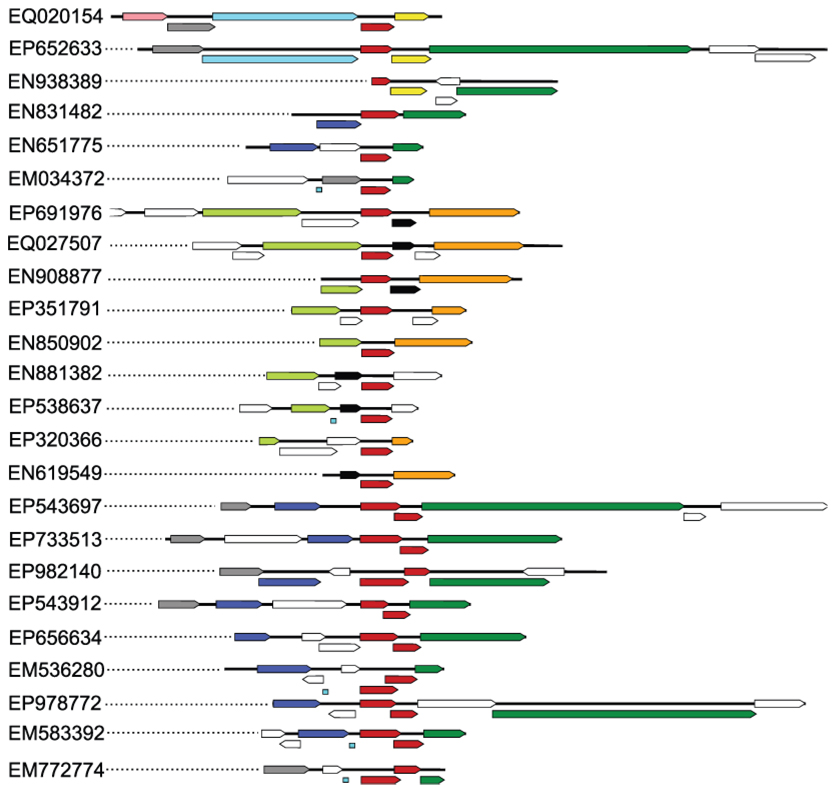
The conserved genomic neighborhood of ECX21941 (GenBank scaffold: EQ020154) and its homologs from an environmental metagenome dataset. The ECX21941 homologs with 85% or less protein sequence identity were found in 239 genomic scaffolds in about 24 different arrangements (shown above), where the ORF encoding ECX21941 or its homolog co-occur with at least two of the top ten most frequent neighbors (scaffolds containing just one conserved neighbor were used to calculate the neighbors’ frequency, but are not shown above). The first scaffold shown (GenBank: EQ020154) is for ECX21941. ECX21941 and its homologs (sometimes with partly overlapping, highly divergent sequences that were probably duplicated in some of the analyzed scaffolds) are highlighted in red, while the most frequently observed neighbors (frequency of observed neighbors per 239 genomic scaffolds is shown in parenthesis) are highlighted as follows: (99) dark green, protein of unknown function, similar to T4-like DNA polymerase from cyanophage P-SSM2 (GenBank: YP_214414); (49) dark blue, similar to heat shock protein IbpA (Hsp20 from cyanophage P-SSM2, GenBank: YP_214406) and proteins with conserved domains PRK10743 (NCBI), PRK11597 (NCBI), and COG0071 (NCBI); (33) light green, protein of unknown function, similar to T4-like clamp loader subunit from cyanophage P-SSM2 (YP_214393), containing conserved domain PRK00440 (NCBI) that is annotated as replication factor C small subunit; (26) yellow, similar to ferredoxin-containing conserved domains KOG3309 (NCBI) and COG0633 (NCBI); (25) cyan , similar to ribosomal protein S6 modification protein from marine gamma proteobacterium HTCC2080 (GenBank: ZP_01626003.1); (23) gray, similar to T4-like translational repressor (regA) from cyanophage P-SSM2; (ZP_01626003.1, pfam01818); (17) black, similar to hypothetical protein PSSM4_019 from cyanophage P-SSM4 (GenBank: YP_214580); (16) not shown, proteins similar to UvsW from cyanophage P-SSM4 (GenBank: YP_214677.1), with conserved domain of superfamily II DNA and RNA helicases (NCBI: COG1061); (15) orange, similar to putative cytosine-specific DNA methyltransferase from Synechococcus cyanophage syn9 (GenBank: YP_717828.1) and proteins with conserved domain COG0338 (NCBI) and pfam02086 (NCBI); (11) pink, similar to clamp loader subunit from Synechococcus cyanophage syn9 (GenBank: YP_717829.1), a protein without similarity to known conserved domains. Tiny blue squares indicate gaps in DNA sequences. Dotted lines connect graphical representations of each scaffold with their GenBank accession codes. Sporadic neighbors are shown as white arrows. Arrows representing overlapping ORFs are shifted below.
Atomic coordinates and experimental structure factors for ECX21941 from the GOS ocean metagenome dataset have been deposited in the PDB and are accessible under code 3by7.
Results and Discussion
The crystal structure of ECX21941 was determined to 2.6 Å resolution using the MAD method (Fig. 1). Data collection, model, and refinement statistics are summarized in Table I. The final model includes five protomers and seven water molecules in the ASU. The Matthews coefficient (Vm) 56 for ECX21941 is 2.5 Å3/Da, and the estimated solvent content is 49.8%. The Ramachandran plot produced by MolProbity 57 shows that 97.7% of the residues are in favored regions with no Ramachandran outliers.
The ECX21941 protomer is a single domain that, in general, adopts the characteristic twisted β-sheet seen in Sm and LSm proteins (Fig. 1; SCOP sunid 50181). This assignment is supported by a DALI 58 structure similarity search, which finds hits to numerous Sm and LSm proteins with Z-scores varying from 7.0 to 4.9, sequence identities ranging from 6% to 20%, and RMSDs ranging from 1.0 Å to 3.3 Å. Molecular weights of 50,360 Da and 50,020 Da were determined by two independent runs of analytical size exclusion chromatography in combination with static light scattering (SEC/SLS). Because ECX21941 has a calculated molecular weight of 11,137 Da (mass determined by LC/MS was 11,136 Da), SEC/SLS suggested that it forms a homo-pentamer in solution, consistent with quaternary structural analysis using the PQS, PISA, and PITA programs.
The structure and function of human and yeast hetero-heptameric/octameric Sm/LSm proteins are well characterized 24,25,59–61 (PDB codes: 2vc8, 1y96, 1n9r, 1d3b, 1b34, and 3bw1) . In bacteria (E. coli, Staphylococcus aureus, and Pseudomonas aeruginosa), the Sm-like Hfq protein forms a homo-hexamer (PDB codes: 1hk9 62, 1kq1 16, 1u1s 63, and 1ycy). Archaeal homo-heptameric Sm-like proteins have been characterized by crystallographic studies (PDB codes: 1i81 19, 1i4k 21, 1ljo 17, 1i8f 64, 1h64 65, 1loj, 1jbm 66, 1m5q 18, 1th7 20, and 2qtx 67) or biochemically 68,69.
From the numerous previously solved crystal structures of Sm and LSm proteins (five eukaryotic, 10 archaeal, and four bacterial), a brief comparative structural analysis is presented here using the following representative structures from the three kingdoms of life: human small nuclear ribonucleoprotein-associated protein B (PDB code: 1d3bB) and human gem-associated protein gemin6 (1y96); archaeal SmAP1 from Methanothermobacter thermautotrophicus (1loj) and archael Sm-related protein from Pyrococcus abyssi (1h64); and bacterial Hfq from S. aureus (1kq1). A superposition of the structure of ECX21941 with these representatives (Fig. 2B) reveals that, despite the lack of any discernible sequence similarity between ECX21941 and other Sm-like proteins (Fig. 2A), the overall structure of all of the monomers is very similar. All secondary structure elements are of similar length and have very similar orientations. However, the ECX21941 structure has some key distinguishing features: (a) the absence of an N-terminal helix; (b) the presence of a very pronounced C-terminal helix; (c) an insertion between strands β3 and β4 (also seen in SmB, PDB code 1d3b, chain B), which forms loop 4 in other Sm/LSm proteins (Fig. 1A); and (d) an insertion between β4' and β4, which forms loop 4' (flanked by Pro51 and Lys59 (Fig 2B and 4C) that is involved in interaction with the adjacent subunit and, hence, participates in oligomer formation (Fig. 1, 2, and 3A).
The presence of charged and aromatic amino acids (76–86) in the C-terminal α-helix (Lys80, Tyr82, His85, Lys100, and Lys103) and in loop 4 (Trp52, Tyr55, and Lys59) indicates they may be involved in nucleic acid interactions. The variation in size of loop 4 between the typical Sm1 and Sm2 motifs (motifs seen in previously characterized Sm/LSm proteins, but not in ECX21941) has also been observed in other Sm/LSm proteins (PDB codes 2fwkA, 1b34B, 1d3bB, and 2fb7A; Fig. 2A). Several proteins that are structurally similar to the Sm/LSm proteins, such as the Tudor domain (PDB codes 2e6n and 2o4×) and gemin6 (PDB code 1y96), have an α-helix at both termini.
The interaction interface between the protomers in the pentameric ring is formed by residues from β4 in one subunit with β5 in the adjacent monomer and by loop 4' (Fig. 2A and 3A). The length of loop 4 contributes to the overall thickness of the petameric ring by increasing its height. The absence of an N-terminal α-helix and the orientation of the C-terminal α-helix do not significantly impact the overall shape and diameter of the assembly. The ring formed by ECX21941 has a diameter of ~60 Å, a width of ~30 Å, and a central pore size of ~9.2 Å. In the hexameric E. coli Hfq (PDB code 1hk9 62), the ring has a diameter of ~65 Å, a width of ~28 Å, and a central pore size of ~11 Å at its most narrow region. The archaeal LSm protein 64 (PDB code 1i8f) has a heptameric ring structure of ~65 Å diameter and ~38 Å width, which is similar to the dimensions of the core of human Sm, as observed by electron microscopy 70.
In the absence of functional data, we cannot determine if the observed homopentameric assembly of ECX21941 represents its biologically relevant form. It could be a consequence of overexpressing the protein in E. coli. ECX21941 may form functional hetero-oligomers in vivo, as occur in the eukaryotic Sm protein complexes, either with other cyanophage Sm-like proteins, where multiple paralogs are commonly found in a particular phage (Fig. 4C) or with host cyanobacterial Sm-like proteins. Recently, a cyanobacterial Sm-like protein similar to the bacterial RNA chaperone Hfq 71 (ssr3341, NP_441518), was identified and characterized and a single homolog of this protein is found in various strains of Synechococcus sp. Interestingly, ssr3341 was found to regulate genes essential for motility of Synechocystis sp. PCC 6803. The loss of motility caused by insertional inactivation of ssr3341 was complemented by reintroduction of the wild-type gene, correlated with the re-establishment of type IV pili on the cell surface 72. Some of the type IV pili function as receptors for bacteriophages, including PO4 phage for P. aeruginosa and the cholera toxin phage (CTXΦ) for Vibrio cholerae 73,74. It is possible that the cyanophage-encoded ECX21941, or its homologs, could play a similar role in the regulation of type IV pili biogenesis that may affect the rate of transduction.
Analysis of the electrostatic surface of the ECX21941 assembly reveals the surface charge distributions that may be relevant for interaction with a ligand. The different views in Fig. 3B portray positively charged amino acids on the outer periphery of the ring and a region of charged residues at the entrance to the central pore (Fig. 3B). Lys67 constitutes the positively charged region at the entrance to the pore from the top side. A negatively charged region is composed of Asp64, Asp65 and Ser66 prior to Lys67 going from one side to the other. The positively charged patch on the outer periphery of the top surface is formed by Lys2, Lys5, Lys29, Lys30, Lys59, and Lys75, many of which are conserved in other Sm/LSm proteins (Fig. 2, 4C). Lys2 superimposes with Arg19 in 1hk9 and 1u1s (Fig. 2A). Lys29 is located in a similar position in 1b34A (Lys41), 1hk9A (Lys47), 1u1sA (Lys47), and 2qtxA (Lys53; Fig.2A). Lys75 corresponds to Arg66 in 1u1s and 1hk9, but its side chain faces the opposite direction, whereas Asp64, Asp65, and Ser66 correspond to residues (with different physicochemical properties) that in other Sm-like proteins are involved in RNA binding and/or oligomerization (Fig. 2A). In Sm-like proteins, the 310-helix (H1) typically contains Lys67 that faces the entrance to the central pore in a similar position to Lys67 in 1d3bA (SmD3 protein). Site-directed mutagenesis coupled with oligonucleotide binding assays, which are beyond the scope of this study, should reveal the functional importance of these residues.
ECX21941 has several hundred predicted homologs in the GOS metagenome dataset, some of which have homologs in cyanophage species (Fig. 4C). The sequence similarity scores between cyanophage proteins and the HMMER (http://hmmer.janelia.org/) profile (calculated using alignment of all Sm-like proteins from the GOS) are between 17.5 and 116.6 (E-values are between 8.6e-6 and 3.6e-34). The HMMER score for the only known similar cyanobacterial protein (GenBank: ZP_01472537) is 31.4 (E-value = 3.6e-9). A Sm-like protein (GenBank: ECL08690) from the GOS with identified similarity to this cyanobacterial protein (BLAST score = 101, E-value = 0.004) has a much higher sequence similarity to a cyanophage protein (GenBank: YP_214412; BLAST score = 191, E-value = 2e-13). One protein from Prochlorococcus cyanophage P-SSM2 has a detectable sequence similarity to ECX21941 (BLAST score 85, E-value = 0.31) and significant similarity scores to other Sm-like proteins from the GOS (HMMER score = 39.9 and E-value = 4.4e-11; BLAST hit to GenBank number ECV68329, with score = 189 and E-value = 3e-13).
The protein sequence clustering identified several groups of close homologs (Fig. 4), indicating similar diversity of marine metagenome-specific Sm-like proteins, as observed in previously known Sm-like proteins. Thus, it is unlikely that all marine metagenome-specific Sm-like proteins have the same function.
The analysis of the genomic neighborhood of ECX21941 and its homologs identified several frequently observed proteins (Fig. 5). Such an analysis is limited to the immediate neighborhood because genomic scaffolds in the metagenomic dataset are relatively short. A similar sequential arrangement of the conserved genomic neighbors was found in one case between a scaffold with an ECX21941 homolog (GenBank scaffold ID: EP543697; Fig. 5) and a genome of cyanophage P-SSM2 (GenBank: AJ630128), where the order is regA (GenBank: CAF34194.1), small heat shock protein (HSP20-like chaperone, GenBank: CAF34195.1), hypothetical protein (GenBank: CAF34196.1), hypothetical protein (GenBank: CAF34197.1), and DNA polymerase gp43 (GenBank: CAF34198.1).
The JCSG has developed The Open Protein Structure Annotation Network (TOPSAN), a wiki-based community project to collect, share, and distribute information about protein structures determined at PSI centers. TOPSAN offers a combination of automatically generated, as well as comprehensive, expert-curated annotations, provided by JCSG personnel and members from the research community. Additional information about ECX21941 is available at http://www.topsan.org/explore?pdbID=3by7.
Conclusions
The crystal structure of ECX21941 reveals, for the first time, a pentameric assembly of protomers for Sm-like proteins. The weak, but statistically significant, sequence similarity between ECX21941 and cyanophage proteins (Fig. 4C), a strong similarity between its homologs and cyanophage proteins, and a strong similarity between proteins from an ECX21941 conserved neighborhood to cyanophage proteins (Fig. 5), led to the conclusion that ECX21941 is likely to be the first known structural representative of a viral (cyanophage) Sm-like protein. The bacterial Sm-like protein Hfq has long been known as a host factor for phage Qbeta RNA replication 75. The RNA-binding residues in previously characterized Sm-like proteins 16 correspond to Gln23 and Asp64, which are not conserved among ECX21941 homologs (Fig. 4C), and are on the opposite side of the ring of highly conserved residues (Arg8, Thr11, Glu13, and Asp14; Fig. 2B, 3A). Thus, the function of ECX21941 is likely to be different and remains unknown. However, the genomic neighborhood of ECX21941 and its homologs is enriched in ORFs encoding DNA-processing proteins (Fig. 5) with annotations similar to several proteins known to be involved in a non-homologous DNA repair pathway, or to genes putatively regulated by attenuation (such as Lhr-like helicases).
Acknowledgments
Portions of this research were performed at the APS Beamline ID-23-D of the GM/CA-CAT and SSRL. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). The SSRL is a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health (National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences). The GOS sequence dataset was initially made available by the J. Craig Venter Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Grant Sponsor: National Institute of General Medical Sciences, Protein Structure Initiative; Grant Number: U54 GM074898.
References
- 1.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, Jaroszewski L, Cieplak P, Miller CS, Li H, Mashiyama ST, Joachimiak MP, van Belle C, Chandonia JM, Soergel DA, Zhai Y, Natarajan K, Lee S, Raphael BJ, Bafna V, Friedman R, Brenner SE, Godzik A, Eisenberg D, Dixon JE, Taylor SS, Strausberg RL, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci U S A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scofield DG, Lynch M. Evolutionary diversification of the Sm family of RNA-associated proteins. Mol Biol Evol. 2008;25:2255–2267. doi: 10.1093/molbev/msn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burge CB, Tuschl T, Sharp PA. Splicing of Precursors to mRNAs by the Spliceosomes. In: Gesteland TCaJA RF, editor. The RNA World. Volume 37. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 8.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 9.Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 10.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 12.Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- 14.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee T, Feig AL. The RNA binding protein Hfq interacts specifically with tRNAs. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toro I, Basquin J, Teo-Dreher H, Suck D. Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J Mol Biol. 2002;320:129–142. doi: 10.1016/S0022-2836(02)00406-0. [DOI] [PubMed] [Google Scholar]
- 18.Mura C, Phillips M, Kozhukhovsky A, Eisenberg D. Structure and assembly of an augmented Sm-like archaeal protein 14-mer. Proc Natl Acad Sci U S A. 2003;100:4539–4544. doi: 10.1073/pnas.0538042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins BM, Harrop SJ, Kornfeld GD, Dawes IW, Curmi PM, Mabbutt BC. Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J Mol Biol. 2001;309:915–923. doi: 10.1006/jmbi.2001.4693. [DOI] [PubMed] [Google Scholar]
- 20.Kilic T, Thore S, Suck D. Crystal structure of an archaeal Sm protein from Sulfolobus solfataricus. Proteins. 2005;61:689–693. doi: 10.1002/prot.20637. [DOI] [PubMed] [Google Scholar]
- 21.Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3'-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- 24.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 25.Naidoo N, Harrop SJ, Sobti M, Haynes PA, Szymczyna BR, Williamson JR, Curmi PM, Mabbutt BC. Crystal structure of Lsm3 octamer from Saccharomyces cerevisiae: implications for Lsm ring organisation and recruitment. J Mol Biol. 2008;377:1357–1371. doi: 10.1016/j.jmb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 27.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 28.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 31.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 33.Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, Scheibe D, Uber DC, Cornell EW, Nordmeyer RA, Kolbe WF, Jin J, Jones AL, Jaklevic JM, Schultz PG, Stevens RC. An approach to rapid protein crystallization using nanodroplets. J Appl Crystallogr. 2002;35:278–281. [Google Scholar]
- 34.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact sample cassettes and a small-scale robot. J Appl Crystallogr. 2002;2002:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Tickle IJ, Laskowski RA, Moss DS. Error estimates of protein structure coordinates and deviations from standard geometry by full-matrix refinement of gammaB- and betaB2-crystallin. Acta Crystallogr D Biol Crystallogr. 1998;54:243–252. doi: 10.1107/s090744499701041x. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 38.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 39.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. In: Doublié S, editor. Macromolecular Crystallography Protocols, Volume 2: Structure Determination. Volume 364, Methods in Molecular Biology. Totowa, NJ: Humana Press; 2006. pp. 215–230. [DOI] [PubMed] [Google Scholar]
- 40.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 41.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.McCoy AJ, Gross-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 45.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- 46.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Ponstingl H, Kabir T, Thornton JM. Automatic inference of protein quaternary structure from crystals. Journal of Applied Crystallography. 2003;36:1116–1122. [Google Scholar]
- 49.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeLano WL. The PyMOL Molecular Graphics System. 2002 [Google Scholar]
- 51.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 53.Mizuguchi K, Deane CM, Blundell TL, Johnson MS, Overington JP. JOY: protein sequence-structure representation and analysis. Bioinformatics. 1998;14:617–623. doi: 10.1093/bioinformatics/14.7.617. [DOI] [PubMed] [Google Scholar]
- 54.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 55.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18 Suppl 1:S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 56.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 57.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 59.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 60.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B"-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 61.Kambach C, Walke S, Nagai K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr Opin Struct Biol. 1999;9:222–230. doi: 10.1016/s0959-440x(99)80032-3. [DOI] [PubMed] [Google Scholar]
- 62.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Blaesi U, Moll I, Kachalova G, Yokoyama S, Vassylyev D, Garber M, Nikonov S. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr D Biol Crystallogr. 2005;61:141–146. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 64.Mura C, Cascio D, Sawaya MR, Eisenberg DS. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc Natl Acad Sci U S A. 2001;98:5532–5537. doi: 10.1073/pnas.091102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thore S, Mayer C, Sauter C, Weeks S, Suck D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J Biol Chem. 2003;278:1239–1247. doi: 10.1074/jbc.M207685200. [DOI] [PubMed] [Google Scholar]
- 66.Mura C, Kozhukhovsky A, Gingery M, Phillips M, Eisenberg D. The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs) Protein Sci. 2003;12:832–847. doi: 10.1110/ps.0224703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen JS, Boggild A, Andersen CB, Nielsen G, Boysen A, Brodersen DE, Valentin-Hansen P. An Hfq-like protein in archaea: crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA. 2007;13:2213–2223. doi: 10.1261/rna.689007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Urlaub H, Raker VA, Kostka S, Luhrmann R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001;20:187–196. doi: 10.1093/emboj/20.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arluison V, Mutyam SK, Mura C, Marco S, Sukhodolets MV. Sm-like protein Hfq: location of the ATP-binding site and the effect of ATP on Hfq-RNA complexes. Protein Sci. 2007;16:1830–1841. doi: 10.1110/ps.072883707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kastner B, Bach M, Luhrmann R. Electron microscopy of small nuclear ribonucleoprotein (snRNP) particles U2 and U5: evidence for a common structure-determining principle in the major U snRNP family. Proc Natl Acad Sci U S A. 1990;87:1710–1714. doi: 10.1073/pnas.87.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 72.Dienst D, Duhring U, Mollenkopf HJ, Vogel J, Golecki J, Hess WR, Wilde A. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology. 2008;154:3134–3143. doi: 10.1099/mic.0.2008/020222-0. [DOI] [PubMed] [Google Scholar]
- 73.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 74.Bradley DE. A pilus-dependent Pseudomonas aeruginosa bacteriophage with a long noncontractile tail. Virology. 1973;51:489–492. doi: 10.1016/0042-6822(73)90448-0. [DOI] [PubMed] [Google Scholar]
- 75.Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]